Abstract
Pseudogenes are important resources for investigation of genome evolution and genomic diversity because they are nonfunctional but have regulatory effects that influence plant adaptation and diversification. However, few systematic comparative analyses of pseudogenes in closely related species have been conducted. Here, we present a turnip (Brassica rapa ssp. rapa) genome sequence and characterize pseudogenes among diploid Brassica species/subspecies. The results revealed that the number of pseudogenes was greatest in Brassica oleracea (CC genome), followed by B. rapa (AA genome) and then Brassica nigra (BB genome), implying that pseudogene differences emerged after species differentiation. In Brassica AA genomes, pseudogenes were distributed asymmetrically on chromosomes because of numerous chromosomal insertions/rearrangements, which contributed to the diversity among subspecies. Pseudogene differences among subspecies were reflected in the flavor-related glucosinolate (GSL) pathway. Specifically, turnip had the highest content of pungent substances, probably because of expansion of the methylthioalkylmalate synthase-encoding gene family in turnips; these genes were converted into pseudogenes in B. rapa ssp. pekinensis (Chiifu). RNA interference-based silencing of the gene encoding 2-oxoglutarate-dependent dioxygenase 2, which is also associated with flavor and anticancer substances in the GSL pathway, resulted in increased abundance of anticancer compounds and decreased pungency of turnip and Chiifu. These findings revealed that pseudogene differences between turnip and Chiifu influenced the evolution of flavor-associated GSL metabolism-related genes, ultimately resulting in the different flavors of turnip and Chiifu.
Key words: turnip genome, comparative genomics, pseudogene evolution, GSL biosynthesis, flavor
Pseudogenes are distributed asymmetrically among diploid Brassica species/subspecies. Differences in MAM pseudogenes have contributed to the contrasting flavors of two Brassica rapa subspecies, turnip and Chiifu.
Introduction
The “triangle of U model” represents the genetic relationships among Brassica species (Nagaharu, 1935), including three diploid species, Brassica rapa (AA, 2n = 20) (Cheng et al., 2016; Zhang et al., 2018), Brassica nigra (BB, 2n = 16), and Brassica oleracea (CC, 2n = 18) (Liu et al., 2014). Hybridizations involving these species resulted in three amphidiploid species: Brassica juncea (AABB, 2n = 36) (Yang et al., 2016), Brassica napus (AACC, 2n = 38) (Chalhoub, 2014), and Brassica carinata (BBCC, 2n = 34) (Johnston et al., 2005; Song et al., 2021). These analyses of Brassica reference genome sequences structurally characterized Brassica ancestral genes and revealed candidate genes controlling target traits. Because Brassica is closely related to Arabidopsis thaliana, both underwent paleopolyploidization events (γ, ∼300 million years ago [mya]; β, ∼112–235 mya; and α, ∼20–100 mya) (Bowers et al., 2003). Brassicaceae lineage-specific polyploidization events (whole-genome triplication [WGT], ∼12.4–22.5 mya) (Beilstein et al., 2010; Wang et al., 2011b) occurred after their divergence from the Arabidopsis lineage. These events were followed by a diploidization that involved substantial genomic recombination, pseudogenization, and eventual gene loss (Bowers et al., 2003; Lysak et al., 2005; Town et al., 2006; Mun et al., 2009; Wang et al., 2011b; Cheng et al., 2013; Chalhoub, 2014; Guo et al., 2021; Yang et al., 2022). Gene loss is an important process in the two-step evolutionary model of Brassica diploid plants (least fractionated subgenome [LF], medium fractionated subgenome [MF1], and most fractionated subgenome [MF2]) (Wang et al., 2011b; Cheng et al., 2012). Ancient polyploidization events generated Brassica vegetable and oilseed crops with various shapes and tastes that developed under natural conditions or through human activities (Wang et al., 2011b; Graham and May, 2011). Brassica vegetable crops include B. rapa (Chinese cabbage, pak choi, and turnip) and B. oleracea (broccoli, cabbage, and cauliflower), and oilseed crops include B. napus, B. juncea, and B. carinata. The same Brassica species may exhibit morphological diversity, and different Brassica species can form morphologically similar organs.
Pseudogenes are genetic elements related to functional genes, but they are nonfunctional because of disabling mutations that have occurred during long-term evolution (Xie et al., 2019). Pseudogenes may include in-frame stop codons, frameshifts, and truncated gene sequences (Zhang et al., 2003). Although pseudogenes are nonfunctional, they can affect plant development and adaptation (Gujas et al., 2012; Wu et al., 2017; Xie et al., 2019; Xu et al., 2019). They have also been associated with intellectual disabilities in humans (Green et al., 2017) and incipient balancing selection in bacteria (Will et al., 2010). Polyploidization events have produced thousands of pseudogenes in plant genomes (Wolfe, 2001; Xie et al., 2019). For example, analyses of B. napus and B. oleracea indicated that some genes in Brassica ancestors were lost or underwent pseudogenization, mainly affecting flowering time (Schiessl et al., 2014). Pseudogenes related to production of anticancer phytochemicals and morphological variations represent the consequences of genome duplications and genetic divergence via polyploidization, resulting in biochemical and morphological changes in B. oleracea (Liu et al., 2014). Brassica pseudogenes contributed to genome evolution after ancient polyploidization events, but there have been relatively few genome-wide, multispecies analyses of their rates of evolution and surrounding chromatin environment.
Glucosinolates (GSLs) and the products of their hydrolysis, especially isothiocyanates, are important secondary metabolites related to the typical flavors (bitterness and pungency) of Brassicaceae plants (Bell et al., 2018). Degradation of glucoraphanin generates products (e.g., sulforaphane) that reportedly have anticancer activities (Fimognari et al., 2002; Tortorella et al., 2015). GSLs and their hydrolysates that affect plant flavors vary greatly between species. Aliphatic GSLs (e.g., sinigrin, gluconapin, and progoitrin) and indolic GSLs (e.g., glucobrassicin and neoglucobrassicin) are responsible for the bitter taste of broccoli and some cauliflower varieties (Engel et al., 2002; Jones et al., 2006; Stotz et al., 2011). Isothiocyanate compounds derived from sinigrin, gluconapin, gluconasturtiin, glucoputranjivin, glucosinalbin, glucobrassicanapin, and glucoraphasatin are associated with the pungency of several Brassicaceae crops, including cabbage, broccoli, kale, wasabi, caper, maca, and radish (Bell et al., 2018). In other species, such as papaya, glucotropaeolin production leads to increased pungency (Bell et al., 2018). Because pungency is an undesirable trait, decreasing the abundance of these compounds may be critical for satisfying consumer taste preferences (Drewnowski and Gomez-Carneros, 2000; Suzuki et al., 2006; Bell et al., 2018). However, the effects of ancient polyploidization events on the flavor-related traits of Brassica crops have not been resolved at the genomic level.
Turnip (Brassica rapa ssp. rapa; AA, 2n = 20), an important B. rapa crop, is one of the oldest known taproot vegetables (Liang et al., 2006; Zhang et al., 2014). It was initially cultivated in Europe in 2500–2000 BC, but it subsequently spread to other parts of the world (Song et al., 1990; Wu et al., 2019). The turnip taproot is often used in French and Japanese cooking (Sasaki and Takahashi, 2002). In China, turnip has traditionally been cultivated on the Qinghai–Tibet Plateau as an edible crop used as animal feed; it is also valued for its pharmaceutical properties (Zheng et al., 2018). Thus, turnips were domesticated and cultivated by our ancestors (Ignatov et al., 2008; Cheng et al., 2016; Qi et al., 2017). Many studies have confirmed that turnips are a good source of vitamin C, dietary fiber, folate, niacin, and calcium (Parveen et al., 2015; Ma et al., 2016). However, the turnip taproot has a pungent taste, possibly because of the considerable abundance of aliphatic GSLs, which are the most abundant GSLs in turnip (Bell et al., 2018). Accordingly, the taste of turnip varies substantially from that of other B. rapa crops with the AA genome, and this has influenced the acceptance of turnips by consumers. A draft genome assembly at the scaffold level and a genome assembly using PacBio and chromosome conformation capture (Hi-C) technologies at the chromosome level have recently been constructed on the basis of an analysis of a European turnip (ECD04) (Park et al., 2021; Yang et al., 2022). Large-scale resequencing of B. rapa and a pan-genome revealed the diverse morphotypes and structural variations that arose during intraspecific diversification of B. rapa, providing researchers with useful genomic information on Chinese and European turnips (Cheng et al., 2016; Cai et al., 2021). However, studies of differences in the evolution of flavors between turnip and other B. rapa crops have been lacking. For the benefit of people living in need of turnips on the Tibetan plateau or other environmentally hostile areas, the effect of the evolution of the GSL biosynthesis pathway on the pungency of turnips should be elucidated.
In this study, we compared pseudogene evolution among Brassica diploid species. We analyzed a chromosome-level turnip genome sequence that was obtained using Illumina and PacBio data and was assembled according to information generated by Hi-C technologies. The results of this investigation highlight the differences in the pseudogenes of Brassica species. The objective of this study was to clarify the dynamics of Brassica pseudogene evolution and the effects of GSL metabolism on turnip pungency. The data presented here provide useful insights into turnip genome evolution and will serve as an important resource for breeding programs interested in optimizing the content of beneficial GSLs in turnips and other Brassica crops.
Results
Genome sequencing, assembly, and annotation
For the turnip genome sequencing analysis, a single plant was collected in Nangqen, Qinghai province, China (96°29′24″, 32°12′36″) (Figure 1A). Genomic DNA was extracted and sequenced using PacBio and Illumina sequencing strategies. We obtained 44.93 Gb of PacBio reads, which corresponded to about 110× coverage of the 446.09-Mb genome; the genome size was estimated on the basis of k-mer statistics (Supplemental Tables 1 and 2). The assembled genome was 409.69 Mb, which included 2437 contigs with an N50 value of 1.21 Mb (Table 1), making it similar in size to the assembled Chiifu A03 genome (403.20 Mb with a contig N50 value of 4.29 Mb) (Sun et al., 2022). To evaluate the genome assembly quality, Illumina short reads were mapped to the assembly, which resulted in a mapping efficiency of 96.69% (Supplemental Table 3). The genome integrity, determined on the basis of benchmarking universal single-copy orthologs (BUSCO), was 97.20% (Supplemental Table 4).
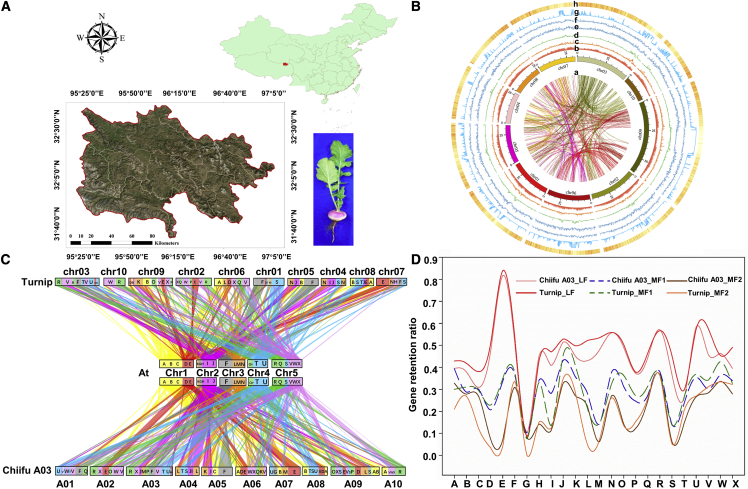
Figure 1.
Genome sequencing of turnip.
(A) Turnip sample collection sites in Tibet.
(B) Overview of the turnip draft genome assembly. chr1–chr10, circular representation of the pseudomolecules. a, gene duplications; b, total number of repetitive elements; c, copia elements; d, gypsy elements; e, terminal inverted repeat elements; f, large retrotransposon derivative elements; g, GC density; h, gene density. The presented data are for a 100-kb window.
(C) Structure and segmental collinearity of the genomes of turnip, Chiifu A03, and A. thaliana. Syntenic blocks are labeled according to the A. thaliana genome (A–X).
(D) Gene retention ratio of the three subgenomes (LF, MF1, and MF2) of turnip and Chiifu A03 on the basis of a comparison with A. thaliana (A–X blocks). The x axis presents the physical position of each A. thaliana block (A–X). The y axis presents the percentage of the retained orthologous genes corresponding to a gene in the A. thaliana A–X blocks.
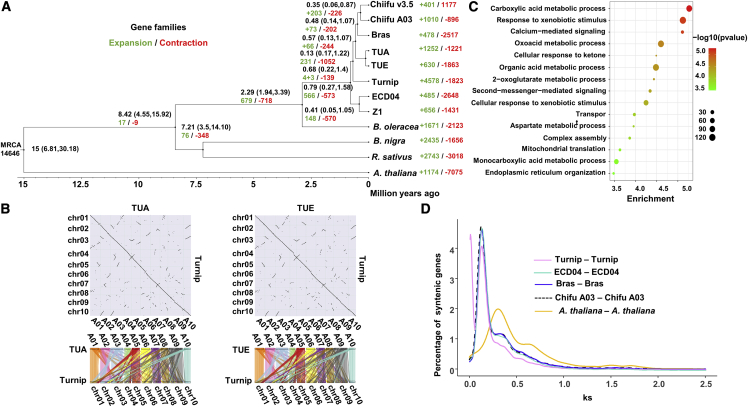
Figure 2.
Evolution of the turnip genome.
(A) Phylogenetic tree, with the number of gene family expansion and contraction events indicated by green and red numbers, respectively, below each species name. The estimated divergence times (million years ago) are indicated at each node (95% credibility intervals).
(B) Dot plot for the segmental collinearity between the turnip and TUA genomes and between the turnip and TUE genomes. The TUA and TUE chromosomes are indicated by different colors, and the orthologous chromosomal segments in turnip are indicated by the same color. Conserved collinear blocks of gene models are presented for the 10 turnip chromosomes and the TUA and TUE genomes.
(C) GO enrichment analysis of the most significantly expanded gene families in turnip. The enrichment factor indicates −Log10 (P value). The 15 most significantly enriched pathways are shown.
(D) Distribution of the synonymous substitution rate (Ks).
Table 1.
Summary of the turnip genome assembly and annotation.
| Primary genome assembly | |
|---|---|
| Sequenced genome size (Mb) | 409.69 |
| Number of contigs | 2437 |
| Contig N50 (bp) | 1 214 203 |
| Contig N90 (bp) | 54 084 |
| Maximum contig size (bp) | 8 713 068 |
| Total size (bp) | 409 691 509 |
| Chromosome-level genome assembly | |
| Number of chromosomes/scaffolds | 10/1501 |
| Scaffold N50 | 37 215 573 |
| Scaffold N90 | 26 283 954 |
| GC content (%) | 35.14 |
| Maximum scaffold size | 57 405 788 |
| Total size | 355 929 172 |
To anchor the scaffolds to chromosomes, 26.79 Gb of clean reads (65.50×) were obtained via Hi-C library sequencing (Supplemental Table 5). Read pairs were mapped to the draft assembly using BWA (version 0.7.10-r789) (Li and Durbin, 2009). We determined that 85.28% of the reads were correctly mapped to the genome (Supplemental Table 6), including 22.01% uniquely mapped read pairs. LACHESIS (Burton et al., 2013) was used to group, sort, and orient all contigs, and 1501 scaffolds were successfully anchored to 10 pseudochromosomes (chr01–chr10) (Figure 1B; Supplemental Table 7). The scaffold N50 value for the final assembly was 37.22 Mb. Thus, the chromosome-level genome assembly for turnip comprised pseudochromosomes ranging in length from 23.02 to 57.40 Mb (Supplemental Figure 1; Supplemental Table 7). The number of pseudochromosomes was consistent with the previously reported number of chromosomes in the Brassica AA genome (i.e., n = 10 and 2n = 20) (Zhang et al., 2018).
The de novo prediction of repetitive sequences in the turnip genome indicated that repetitive sequences represented 42.60% of the assembled genome; this proportion was lower than that of Chiifu A03 (50.31%) (Sun et al., 2022) and ECD04 (46.9%) (Yang et al., 2022) (Supplemental Table 8). The long terminal repeat (LTR) retrotransposon was the most common repetitive sequence, accounting for 17.13% of the genome, and this proportion was between that of Chiifu A03 (19.62%) and ECD04 (20.77%) (Supplemental Table 8; Sun et al., 2022; Yang et al., 2022). Protein-coding genes were predicted via ab initio gene predictions, homology-based predictions, and RNA sequencing (RNA-seq) and then integrated using EVidenceModeler (version 1.1.1) (Haas et al., 2008). In total, 56 832 genes were obtained. In addition, 98.57% of the genes were annotated by screening databases (e.g., NR) (Supplemental Table 9), and microRNA target genes were predicted (Supplemental Table 10).
We next assessed the quality of the turnip genome. Specifically, the LTR assembly index score for the turnip genome was 13.8, indicative of good assembly continuity. Genome annotation completeness was estimated to be 95.30% on the basis of the BUSCO assessment. Illumina paired-end reads for TUA, TUE, A03, and ECD04 and reads derived from 48A resequencing data for a turnip population (Yang et al., 2019) were mapped to turnip chromosomes (Supplemental Figure 2; Supplemental Table 11). Approximately 96.47%–97.42% of these reads were mapped to the turnip chromosomes, indicating that the turnip genome assembly contained almost all of the information provided by the Illumina reads and 48A resequencing data. Similarly, 96.34%–98.56% of the Illumina paired-end reads and reads obtained from48A resequencing data were mapped to the TUA, TUE, A03, and ECD04 chromosomes (Supplemental Figure 3; Supplemental Tables 12, 13, 14, and 15). These results confirmed that the turnip genome used in this study was assembled and annotated appropriately.
The whole-genome triplication (WGT) in Brassica species was followed by extensive gene loss and frequent reshuffling of triplicated genomic blocks (Liu et al., 2014). Triplicated regions, which were determined on the basis of homologous gene pairing between A. thaliana and turnip, as well as Chiifu A03, were constructed; they were related to the 24 ancestral crucifer karyotype blocks (A–X) in A. thaliana (Schranz et al., 2006). Most of the regions in the turnip genome shared a conserved syntenic block with the A. thaliana genome (Figure 1C). The WGT-derived triplicated blocks in the turnip and Chiifu A03 genomes were partitioned into the LF, MF1, and MF2 subgenomes (Figure 1D; Wang et al., 2011b). These syntenic blocks occupied most of the genome assemblies of A. thaliana (25 054 genes, 90.59% of 27 655 genes), turnip (26 973 genes, 47.46%), and Chiifu A03 (26 738 genes, 55.96%), providing the foundation for comparative analyses (Supplemental Table 16).
Comparative genomics analysis
To analyze turnip evolution, 3593 single-copy orthologs from 12 species, including Brassica species with an AA genome (Chiifu v3.5, Chiifu A03, B. rapa L. ssp. chinensis [Bras], TUA, TUE, ECD04, B. rapa Z1 [Z1], and turnip) (Cai et al., 2021; Istace et al., 2021; Li et al., 2021; Sun et al., 2022; Yang et al., 2022; Zhang et al., 2022), a BB genome (B. nigra) (Perumal et al., 2020), and a CC genome (B. oleracea) (Cai et al., 2020), as well as Raphanus sativus (Kitashiba et al., 2014), were used for protein sequence alignments, with the A. thaliana genome (Cheng et al., 2017) serving as an outgroup (Figure 2A and Supplemental Figure 4). Turnips and other B. rapa subspecies, which were derived from a common ancestral Brassica species (AA genome), were clustered together on a branch. The divergence time between B. nigra and B. oleracea was approximately 8.42 mya, whereas the divergence time between B. oleracea and B. rapa subspecies was about 2.29 mya. The timing of this evolutionary process was consistent with the findings of an earlier study (Guo et al., 2021). The divergence times between turnip and ECD04 and between turnip and other B. rapa subspecies were approximately 0.79 and 0.68 mya, respectively. LTR retrotransposons were actively inserted into the turnip, ECD04, and Chiifu A03 genomes approximately 1.34, 1.88, and 1.65 mya, respectively (i.e., before the divergence between turnip and ECD04 and between turnip and B. rapa subspecies). A comparative analysis of the timing of LTR retrotransposon insertion into the genomes revealed that turnip and B. rapa subspecies had similar evolutionary histories (Supplemental Figure 5).
The MCScanX package (Wang et al., 2012) was used to analyze the collinearity between turnip and Chiifu A03, ECD04, TUA, and TUE (Figure 2B and Supplemental Figure 6). A total of 643 and 631 large syntenic blocks were detected between the turnip genome and the TUA and TUE genomes, respectively. Moreover, 335.62 Mb (97.00%) and 336.31 Mb (97.20%) of the sequences on the 10 turnip chromosomes were revealed to be collinear, covering almost all of the TUA and TUE chromosomes, respectively (Figure 2B). To verify the accuracy and continuity of the assembled sequences, sequence collinearity between the turnip genome and the Chiifu A03, ECD04, TUA, and TUE genomes was determined using the nucmer program of the MUMmer package (v4.0rc1) (Marcais et al., 2018), after which NGenomeSyn was used to detect highly similar sequence segments (Supplemental Figure 7). This analysis indicated that turnip is closely related to TUA, TUE, ECD04, and Chiifu A03. However, there were numerous chromosomal insertions/rearrangements that differentiated turnip from TUA, TUE, ECD04, and Chiifu A03. This is in accordance with previous studies that detected many genomic insertions/rearrangements after the WGT event in Brassica (Liu et al., 2014). An earlier analysis of retained or lost genes after the WGT in Brassica revealed over-retention of genes involved in metabolic pathways (Liu et al., 2014; Lou et al., 2012). To further resolve the diversity between turnip and B. rapa subspecies, we detected 4578 and 1010 gene families that had expanded in the turnip and Chiifu A03 genomes, respectively (Figure 2A). Gene Ontology (GO) enrichment analysis indicated that the expanded gene families in turnip were mainly related to metabolic processes and responses to environmental stimuli (Figure 2C), whereas those in Chiifu A03 were mainly associated with S-glycoside catabolism (Supplemental Figure 8). This implies that there may be differences in the survival strategies of turnip growing on the Tibetan plateau and domesticated Chiifu A03 (Cheng et al., 2016; Zhao et al., 2005). On the basis of duplicated gene pairs, we calculated the age distribution of the synonymous substitution rate (Ks) (Figure 2D). The results indicated that, other than the common polyploidization events among Brassica species, there were no additional species-specific WGD events in turnip, consistent with the results of the phylogenetic analysis of the Brassica AA genome. This may also explain the similarity in the genomic structures of turnip and B. rapa subspecies.
Comparative analysis of pseudogenes in diploid Brassica genomes
To systematically identify candidate pseudogenes in the genomes of eight diploid Brassica species, including the AA genomes of turnip, ECD04, Chiifu A03, Chiifu v3.5, Bras, and Z1, the BB genome of B. nigra (Bni), and the CC genome of B. oleracea (Bol), we used prediction software and performed homology searches with stringent filters to minimize noise and enhance positive signals (Figure 3A and Supplemental Figure 9A). Among the examined species, Bol (CC genome) and Bni (BB genome) had the most and fewest pseudogenes, respectively. The AA genomes included 3933–5847 pseudogenes. Turnip, ECD04, Chiifu A03, Chiifu v3.5, and Bras had similar numbers of pseudogenes (Figure 3A). We subsequently determined the pseudogene evolution rate by estimating the Ks values for the pseudogenes and their functional paralogs (Figure 3B). The Chiifu A03 pseudogenes evolved faster than the turnip pseudogenes, possibly because of the high artificial selection pressure to which Chiifu A03 was subjected.
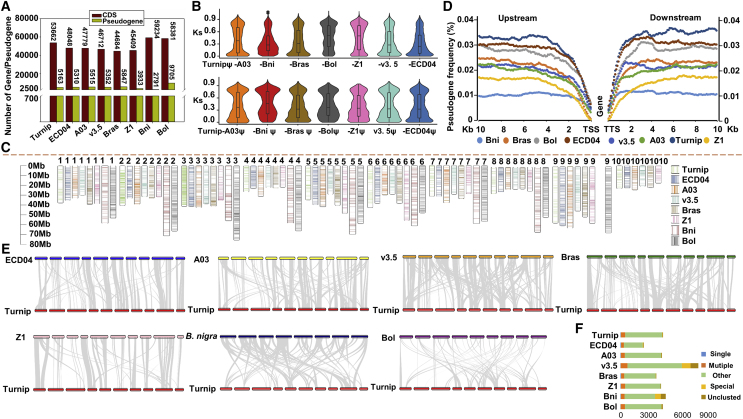
Figure 3.
Identification and comparison of pseudogenes in diploid Brassica species.
(A) Number of genes/pseudogenes in eight diploid Brassica genomes, including the AA genomes of turnip, ECD04, Chiifu A03, Chiifu v3.5, B. rapa L. ssp. chinensis (Bras), and B. rapa Z1, as well as the BB genome of B. nigra (Bni) and the CC genome of B. oleracea (Bol).
(B) Comparison of pseudogene evolution rates (Ks) in eight diploid Brassica species/subspecies. Turnipψ–others indicates the pseudogenes of turnip and functional genes of other species/subspecies. Turnip–othersψ indicates the pseudogenes of other species/subspecies and functional genes of turnip.
(C) Comparison of pseudogene distribution between the turnip genome and the genomes of other diploid Brassica species/subspecies. Numbers represent the corresponding chromosome numbers.
(D) Distribution of pseudogenes 10 kb upstream and downstream of coding sequences (CDSs) in eight diploid Brassica species/subspecies. The presence of a pseudogene downstream of the transcription termination site or upstream of the transcription start site of each gene is indicated by 1, whereas the absence of a pseudogene is indicated by 0, with a total window size of 100 kb. The data presented on the y axis were calculated by dividing the number of genes with an upstream or downstream pseudogene (1) by the total number of genes.
(E) Comparative analysis of pseudogenes between turnip and other diploid Brassica species/subspecies. Syntenic blocks were determined according to alignment of the turnip chromosomes with the chromosomes of other diploid Brassica species/subspecies.
(F) Comparison of the number of unique and shared pseudogene families between turnip and other diploid Brassica species/subspecies.
Anchoring of the Brassica pseudogenes to each chromosome revealed an asymmetrical distribution, with the greatest variation in the distribution of pseudogenes on chr02 of turnip and on A02 of the other species. Considering the rearrangement results for chromosome A02, we hypothesized that genomic rearrangements may be responsible for the differences in pseudogene distribution (Figure 3C and Supplemental Figure 9B). We analyzed the distribution of pseudogenes 10 kb upstream and downstream of coding sequences (CDSs) in eight Brassica diploid species. Turnip had the most pseudogenes in the upstream/downstream regions, with similar pseudogene distribution trends detected in the AA genomes of Bras, Chiifu v3.5, and Chiifu A03. By contrast, the BB genome (Bni) had the fewest pseudogenes in the upstream/downstream regions. This result provides further evidence of the asymmetrical distribution of pseudogenes in eight Brassica diploid species (Figure 3D). In addition, we identified syntenic blocks of pseudogenes in Brassica. More specifically, the synteny between turnip and other species/subspecies (i.e., the percentage of syntenic regions) was as follows: 38.40% (Chiifu A03), 43.10% (Bras), 40.20% (Chiifu v3.5), 41.00% (ECD04), 29.20% (Z1), 21.20% (Bni), and 19.90% (Bol) (Figure 3E and Supplemental Figure 9C). The lowest synteny between turnip and Bol may be related to the relatively extensive chromosomal rearrangements and asymmetrical gene loss in duplicated genomic blocks that occurred in the Bol genome (Liu et al., 2014). To further characterize the pseudogenes specific to turnip, syntenic orthologs of pseudogenes in Brassica were identified (Figure 3F).
We then performed a GO enrichment analysis of the pseudogenes in eight diploid Brassica species/subspecies by annotating their closest functional paralogs (Supplemental Figure 10). The turnip pseudogenes were mainly annotated with the GO terms “multicellular organismal process,” “anatomical structure development,” and “macromolecule biosynthetic process,” probably because some nonfunctional or dispensable genes became pseudogenes. The Chiifu A03 pseudogenes are related to “cellular localization,” “signal transduction,” and “signaling,” suggesting that Chiifu A03 had more pseudogenes related to stress responses. These findings may reflect functional differences in pseudogenes between the two subspecies, which may be the result of selection pressure during domestication. We assessed the functions associated with the Pfam domains encoded by the pseudogenes on the basis of the annotations of their functional paralogs (Supplemental Figure 11). A relatively small proportion of the pseudogenes were related to core genes, such as transcription factor genes (0.20%–1.41%) and kinase genes (0.86%–2.59%), but most of the pseudogenes were functionally unknown and unclassified. Accordingly, there were relatively few regulatory genes among the pseudogenes.
Effect of pseudogenes on the GSL biosynthesis pathway
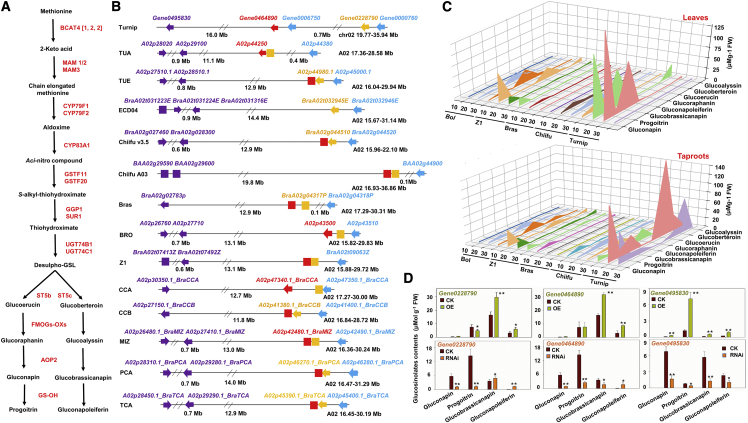
GSLs and the products of their hydrolysis are determinants of the unique taste of Brassica crops (Bell et al., 2018). Compared with other Brassica species, turnips contain more aliphatic GSLs (Yang et al., 2020). On the basis of the pseudogene functional annotations, we analyzed the genes involved in the aliphatic GSL metabolic pathway (Figure 4A and Supplemental Figure 12); these genes are the predominant GSL-related genes in the Brassica AA genome. Genes encoding methylthioalkylmalate (MAM) synthase were identified in turnip, ECD04 (Yang et al., 2022), Chiifu A03 (Sun et al., 2022), Bras (Li et al., 2021), and 10 other representative Brassica species/subspecies (Cai et al., 2021), including TUA, TUE, Chiifu v3.5, BRO, Z1, CCA, CCB, MIZ, PCA, and TCA (Figure 4B, Supplemental Figure 13; Supplemental Table 17). Turnip had the most MAM functional genes, which were distributed mainly on chr02 but also on chr03 and chr04. In the other species/subspecies, the MAM genes were distributed on chromosomes A02, A03, and A04 (Supplemental Figure 14). Syntenic relationships were detected between the turnip MAM genes on chr03 and chr04 and the MAM genes on chromosomes A03 and A04 in the other species/subspecies. However, all five MAM genes on chr02 in turnip were functional, whereas the syntenic regions on chromosome A02 in the other species/subspecies contained 1–3 MAM pseudogenes. There were two main pseudogene types, those on blocks homologous to a turnip MAM gene (Gene0495830), which had a premature termination codon (Supplemental Figure 15), and those on blocks homologous to turnip MAM genes (Gene0228790 or Gene0464890), which became pseudogenes. Because of the decrease in MAM gene family size in the other species/subspecies as a result of the development of pseudogenes, we hypothesized that MAM genes (Gene0495830, Gene0228790, and Gene0464890) may be critical for explaining the differences in GSL synthesis between turnip and the other species/subspecies.
Figure 4.
Comparison of pseudogenes and determination of flavor-related metabolite content in the aliphatic GSL metabolic pathways of diploid Brassica species.
(A) Overview of the aliphatic GSL biosynthesis pathway.
(B) Analysis of synteny among MAM genes in 14 diploid Brassica species/subspecies. Turnip chromosome chr02 with MAM genes and the collinear chromosome A02 in other Brassica species/subspecies are presented. Identical colors represent the same homologous region. Arrows indicate the gene orientation on the chromosome. Boxes represent pseudogenes into which those on blocks homologous to turnip MAM (Gene0495830, Gene0228790, or Gene0464890) were converted in other species/subspecies. Specifically, MAM (Gene0495830) in turnip was converted into pseudogenes because of codon termination in other species/subspecies. Syntenic regions in both genomes, with one turnip genome containing a functional gene (Gene0228790 or Gene0464890) and the other containing a homologous sequence with clear markers indicative of a pseudogene, are presented.
(C) Differences in aliphatic GSL content in the leaves and taproots of Brassica species/subspecies 10, 20, and 30 days after germination (n = 4) as determined by HPLC-MS/MS. AA genome: turnip, Chiifu, Bras, and B. rapa Z1; CC genome: Bol.
(D) Pungency-related compound content in Chiifu hairy roots overexpressing MAM genes (Gene0228790, Gene0464890, and Gene0495830) (top graphs) and in turnip hairy roots in which these three genes were silenced via RNAi (bottom graphs). The compound content was determined by HPLC-MS/MS. Error bars indicate the standard deviation (n = 4). Asterisks indicate significant differences between the transgenic turnip/Chiifu hairy roots and the control (Student’s t-test; ∗P < 0.05, ∗∗P < 0.01).
To test this hypothesis, we first compared the aliphatic GSL content in Brassica species with the AA genome (turnip, Chiifu, Bras, and Z1) and Bol (CC genome) (Figure 4C). The aliphatic GSL content varied among organs (leaves/taproots) and developmental stages (10, 20, and 30 days). Compared with their abundance in other Brassica species/subspecies, several aliphatic GSLs were more abundant in turnip taproots (gluconapin, progoitrin, glucobrassicanapin, glucoraphanin, glucoerucin, and glucoberteroin) or turnip leaves (gluconapin and glucobrassicanapin). These compounds are the main sources of the pungency of Brassica plants (Bell et al., 2018; Kusznierewicz et al., 2013; Depree et al., 1998). We also obtained hairy roots from Chiifu and turnip plants that had been transformed using Agrobacterium rhizogenes for overexpression or silencing (via RNA interference [RNAi]) of the MAM genes (Gene0228790, Gene0464890, and Gene0495830) (Figure 4D and Supplemental Figure 16). The glucobrassicanapin and gluconapoleiferin contents determined by liquid chromatography-mass spectrometry (MS) were significantly higher in Chiifu hairy roots overexpressing Gene0228790, Gene0464890, and Gene0495830 than in the control. As expected, the RNAi-mediated silencing of Gene0228790, Gene0464890, and Gene0495830 in turnip hairy roots decreased the gluconapin, progoitrin, and glucobrassicanapin content. These results imply that MAM is important for development of the pungent flavor of turnips.
Effect of 2-oxoglutarate-dependent dioxygenase (AOP2)-encoding gene on the GSL biosynthesis pathway
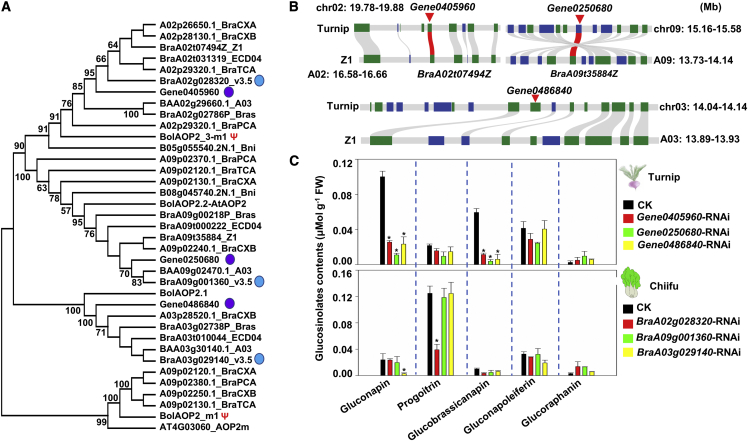
Degradation of glucoraphanin produces sulforaphane, one of the best anticancer compounds identified to date (Fimognari, 2002; Tortorella et al., 2015). Other genes that influence the production of specific anticancer- and flavor-related aliphatic GSLs are AOP genes, which encode enzymes that convert sulforaphane-related GSLs into GSLs that lack anticancer properties (Liu et al., 2014). We identified AOP2 genes in turnip, Bol (CC genome), Bni (BB genome), and nine other representative Brassica AA genome species/subspecies (ECD04, Chiifu A03, Bras, Chiifu v3.5, Z1, CXA, CXB, PCA, and TCA) (Cai et al., 2021; Figure 5A). Most species/subspecies had three AOP2 genes, but among the three Bol AOP2 genes, one was functional, whereas the other two were pseudogenes. This may help to explain the high glucoraphanin levels in Bol (inactive AOP2 genes) but not in Chiifu (three active AOP2 genes) (Wang et al., 2011a; Liu et al., 2014). Two AOP2 genes in Z1 had syntenic relationships with two AOP2 genes in turnip, but there was no synteny between Z1 AOP2 genes and one of the turnip AOP2 genes (Gene0486840) (Figure 5B).
Figure 5.
Effect of AOP2 genes on the GSL biosynthesis pathway.
(A) Neighbor-joining trees of AOP gene families in genomes of 13 species, including the AA genome of CXA, CXB, Z1, ECD04, TCA, Chiifu v3.5, turnip, Chiifu A03, Bras, and PCA, the BB genome of Bni, and the CC genome of Bol, with A. thaliana serving as an outgroup, were constructed by aligning the CDSs with 1000 bootstrap replicates. Three AOP2 genes were present in the turnip (purple circles; Gene0405960, Gene0250680, and Gene0486840) and Chiifu (blue circles; BraA02g028320, BraA09g001360, and BraA03g029140) genomes. Red asterisks and Ψ represent pseudogenes in Bol.
(B) Analysis of the synteny between the AOP2 genes in B. rapa Z1 and turnip. Of the three AOP2 genes in turnip, B. rapa Z1 lacked a collinear gene for Gene0486840.
(C) Pungency-related compound and glucoraphanin content in AOP2-RNAi turnip and Chiifu hairy roots. Error bars indicate the standard deviation (n = 4). Asterisks indicate significant differences between the transgenic turnip/Chiifu hairy roots and the control (Student’s t-test; ∗P < 0.05, ∗∗P < 0.01).
The three AOP2 genes in turnip were expressed in taproots and leaves at different developmental stages, implying that the sulforaphane-related GSLs in turnip are converted by AOP2 to GSLs that lack anticancer activities (Figure 5A, Supplemental Table 18, and Supplemental Figure 17). Thus, we attempted to inactivate all AOP2 genes to decrease pungency and increase sulforaphane formation. We used RNAi technology to silence the expression of three AOP2 genes in turnip (Gene0405960, Gene0250680, and Gene0486840) and Chiifu (BraA02g028320, BraA09g001360, and BraA03g029140) and then analyzed the pungency and accumulation of glucoraphanin in the resulting samples (Figure 5C and Supplemental Figure 18). As expected, inhibition of AOP2 expression significantly decreased the content of pungency-related compounds (gluconapin and glucobrassicanapin) in turnip and enhanced glucoraphanin accumulation in turnip and Chiifu. These findings are relevant for future attempts to modulate turnip pungency (Supplemental Figure 19).
Discussion
Pseudogenes are important for research on evolution and comparative genomics because they represent the molecular remnants of ancient genes that existed in the genome millions of years ago (Zou et al., 2009; Moghe et al., 2014; Xie et al., 2019; Xu et al., 2019). The only cross-species comparisons of pseudogenes in plants have focused on the evolution and expression signatures of pseudogenes in the Arabidopsis and rice genomes (Zou et al., 2009), the pseudogenization of duplicated genes in wild radish (Raphanus raphanistrum) and three other Brassicaceae species (Moghe et al., 2014), and the evolutionary origins of pseudogenes and their associations with regulatory sequences among seven angiosperms (Xie et al., 2019). The diversity in pseudogene evolution among Brassica species remains unclear. The comparison of Brassica pseudogenes revealed in this study suggests that the CC genome had more pseudogenes than the AA genome and fewer pseudogenes derived from a common ancestor, with relatively few syntenic blocks, indicating that the pseudogenes in the AA genome appeared after the divergence from Bol. The pseudogenes were asymmetrically distributed on the chromosomes among the subspecies. The Chiifu A03 pseudogenes evolved faster than the turnip pseudogenes. Hence, although Chiifu A03 and turnip have the AA genome, the genetic diversification of these two subspecies may be related to selection pressure imposed during domestication.
Pseudogenes usually result from gene duplications or retrotranspositions related to WGD events (Wolfe, 2001; Xie et al., 2019). The small difference in the number of pseudogenes among Brassica species with the AA genome is consistent with the lack of additional WGD and LTR events after divergence in these species. The asymmetrical distribution of pseudogenes on the chromosomes was due to numerous chromosomal insertions/rearrangements in the Brassica AA genomes (Lou et al., 2012; Liu et al., 2014). These pseudogene differences were revealed by gene functional annotations, which indicated that core genes, including those encoding transcription factors, were generally not converted to pseudogenes. Previous studies have determined that Brassica crops exhibit extreme morphological characteristics and diverse environmental adaptability because of artificial selection during domestication and breeding (Cheng t al., 2016; Qi et al., 2017). This phenomenon demonstrates that plant survival is the first priority and that the diversity in specific characteristics increased after domestication. Accordingly, the metabolic differences may have been the result of selection for agriculturally desirable traits by humans, especially the flavor-related characteristics of domesticated/semi-domesticated Chiifu and turnip.
The production of four aliphatic GSLs (gluconapin, progoitrin, glucobrassicanapin, and gluconapoleiferin) influences formation of the distinct flavors of Brassica crops (Bell et al., 2018). The variability in aliphatic GSL structures is due mainly to two major genetic loci (MAM and AOP) (Keurentjes et al., 2006; Wentzell et al., 2007; Liu et al., 2014). Specifically, MAM controls the variability in aliphatic GSL carbon chain length, whereas AOP is responsible for modification of side chain structure (Benderoth et al., 2009; Liu et al., 2014). Detailed analysis of five Brassica species in this study revealed considerable variation in the relative content of aliphatic GSLs among species and subspecies, consistent with the findings of earlier studies (Chen et al., 2008; Yang et al., 2020). Previous research confirmed that there were significant differences in the GSL content of Chinese cabbage (Chiifu) germplasm, due largely to genetic changes because of selection pressure influenced by consumer preference (Kang et al., 2006). Thus, the high concentrations of pungency-related substances in turnip plants grown on the Tibetan plateau may be related to their minimal domestication by humans. In Bol, two nonfunctional AOP2 genes are associated with decreased accumulation of these four metabolites, which, in turn, is related to an increase in anticancer GSL content (Liu et al., 2014); this may also be the result of flavor-related selection pressure during domestication. We determined that the Chiifu flavor preferred by humans is influenced mainly by nonfunctional MAM genes, whereas the pungency of turnip is mainly associated with expansion of the MAM gene family. This is in accordance with the results of GSL content surveys and explains why gluconapin and glucobrassicanapin are abundant in turnip (Padilla et al., 2007; Lee et al., 2013) but not in Chiifu. This inspired us to attempt to convert AOP2 in turnip into a nonfunctional gene via RNAi. Doing so will enhance accumulation of anticancer substances and optimize turnip flavor. Therefore, agriculturally important Brassica crop traits may be improved by focusing on GSL pathway-related genes. The data generated in this study will help researchers and breeders develop crops with more desirable flavors and a greater abundance of anticancer compounds, satisfying worldwide consumer demands.
Methods
Plant materials
Turnip seeds collected from Nangqen county, Qinghai province (N 32°12′11″, E 96°28′50″) were sown in a seedling raising plate. Seedlings were cultivated under controlled greenhouse conditions (12 h light [28°C]/12 h dark [25°C] cycle, 200 mmol photons m−2 s−1 light intensity, and 75%–80% relative humidity) and were watered appropriately. Genomic DNA was extracted from the leaves and used for subsequent genomic DNA sequencing analysis and construction of Hi-C libraries.
Illumina and PacBio sequencing
Genomic DNA was extracted from leaves according to a standard cetyltrimethylammonium bromide (CTAB) method. DNA quality was assessed using a 2100 Bioanalyzer (Agilent Technologies, USA). The Illumina sequencing library was constructed and then sequenced (150-bp paired-end mode) using the Illumina X Ten platform as described by the manufacturer. For PacBio sequencing, 10 μg genomic DNA was sheared, and then approximately 20-kb fractions were selected using the BluePippin Selection system (Sage Science, USA). The library was sequenced using the Pacific Biosciences Sequel platform.
Estimation of genome size
The turnip genome size was estimated on the basis of a k-mer frequency analysis of the Illumina short reads using the Jellyfish program (version 2.2.6) (http://www.genome.umd.edu/jellyfish.html) with a k-mer frequency of 21. The heterozygosity ratio was estimated using the GenomeScope online tool (http://qb.cshl.edu/genomescope/). Finally, genome size was calculated using the following formula: genome size = k-mer coverage/mean k-mer depth.
De novo assembly and genome refinement
The PacBio SMRT (Single Molecule Real Time) analysis package (https://www.pacb.com) was used for quality control screening of the raw reads with the following parameters: readScore, 0.75; minSubReadLength, 500, including removal of sequencing adapters and low-quality short reads. Errors in the PacBio long reads were corrected using the error correction module embedded in Canu (version 1.3) (Koren et al., 2017) with correctedErrorRate set to 0.045. De novo sequence assembly was performed using the default parameters of Canu to produce contigs. The clean PacBio reads were then aligned with the assembled contigs using Basic Local Alignment with Successive Refinement (version 1.3.1) (Chaisson and Tesler, 2012). The contigs were further corrected using Quiver from the SMRT Analysis package. Illumina paired-end reads from the same turnip species were aligned to the optimized contigs using BWA (version 0.7.10-r789) (Li, 2014). The assembled sequences were polished using Pilon (version 1.22) (Walker et al., 2014) with the following parameters: -mindepth 10 -changes -fix bases. The completeness of the genome assembly was evaluated using BUSCO (version 4.0.6) and the embryophyta_odb10 single-copy gene dataset (Simao et al., 2015).
Hi-C library construction
Leaves from individual turnip plants were harvested and immersed in a formaldehyde solution to crosslink and fix chromatin. The leaf cells were lysed, and HindIII endonuclease was used to digest the fixed chromatin. The DNA ends were marked with biotin-14-dCTP (2′-deoxycytidine 5′-triphosphate), and the blunt ends were ligated to each other using DNA ligase. Next, the nuclear complexes were reverse crosslinked during incubation with proteinase K at 65°C. The DNA was purified and sheared (100–500 bp) by sonication. The biotin-labeled fragments were enriched using streptavidin magnetic beads. Poly(A) tails were added to the fragment ends using the Klenow fragment (exo-) before adding the Illumina paired-end sequencing adapter in a ligation mixture. PCR amplification of fractions was performed using 12 cycles, and the PCR products were sequenced using the Illumina HiSeq platform (150-bp paired-end reads).
Pseudomolecule construction by Hi-C
Clean Hi-C reads were used with HiC-Pro (Servant et al., 2015) to map the Hi-C sequencing reads to the assembled contigs using the BWA-aln algorithm without any mismatches and with detection of valid contacts (Li and Durbin, 2009). The preassembled contigs split into 50-kb segments (on average) combined with uniquely matched Hi-C data were clustered, ordered, and directed onto the pseudochromosomes using LACHESIS software (Burton et al., 2013). Orientation errors with obvious discrete chromatin interaction patterns were manually adjusted to improve the chromosome-scale assembly quality. The final chromosome assemblies were divided into 100-kb bins with equal lengths. The interaction signals generated by the valid mapped read pairs between each bin were visualized in a heatmap.
Genome annotation
To annotate repetitive sequences, the assembled turnip genome was screened using LTR_FINDER (version 1.05) (Xu and Wang, 2007), MITE-Hunter (Han and Wessler, 2010), RepeatScout (version 1.0.5) (Price et al., 2005), and PILER-DF (version 2.4) (Edgar and Myers, 2005). All isolated sequences were then classified using PASTEClassifier (Hoede et al., 2014) and mapped using the Repbase database and RepeatMasker software (version 4.0.6) (Tarailo-Graovac and Chen, 2009). Using a substitution rate (r) of 7.3 × 10−9 substitutions per site per year (Exposito-Alonso et al., 2018), the insertion date (T) was calculated for each LTR retrotransposon (T = K/2r; K, genetic distance). Next, ab initio, homology-based, and RNA-seq-based prediction methods were combined to annotate gene models. The ab initio predictions were obtained using Genscan (Haas et al., 2008), Augustus (version 2.4) (Stanke and Morgenstern, 2005), GlimmerHMM (version 3.0.4) (Majoros et al., 2004), GeneID (version 1.4) (Blanco et al., 2007), and SNAP (version 2006-07-28) (Korf, 2004). GeMoMa software (version 1.3.1) was used to predict homologous species (mainly A. thaliana, B. juncea, B. napus, and Chiifu v3.0). Protein sequences were downloaded from the Brassica database (http://brassicadb.cn/#/). The RNA-seq reads were mapped to the genome assembly using HISAT and StringTie (Kim et al., 2015). TransDecoder (http://transdecoder.github.io) and GeneMarkS-T (Tang et al., 2015) were used to identify transcripts according to the mapping results. Finally, all prediction results were integrated using EVidenceModeler (version 1.1.1) (Haas et al., 2008). Turnip genes were functionally annotated using the eggNOG, GO, KEGG_ko, and Pfam databases and the eggNOG online service (http://eggnog-mapper.embl.de/; Huerta-Cepas et al., 2019; Cantalapiedra et al., 2021). DIAMOND BLASTP (default alignment parameter) (Buchfink et al., 2021) was used to align turnip proteins to sequences in the NR and Swiss-Prot databases, with the best hit (-k 1) used for annotations.
Phylogenetic tree construction
Orthologous groups were identified using OrthoFinder (version 2.3.12) and all-versus-all BLASTP alignments (E < 1e−5) with protein sequences encoded by the genomes of the following 12 species: turnip, B. rapa ssp. pekinensis (Chiifu-401-41, v3.5) (Zhang et al., 2022), B. rapa L. ssp. pekinensis cv. A03 (Chiifu A03) (Sun et al., 2022), European turnip ECD04 (Yang et al., 2022), Bras (Li et al., 2021), B. rapa ssp. rapa (TUA and TUE) (Cai et al., 2021), and B. rapa (Z1) (Chaisson and Tesler, 2012) with the AA genome; Bol (To1000, v2.0) (Cai et al., 2020) with the CC genome; Bni (Ni100_V2) (Perumal et al., 2020) with the BB genome; Raphanus sativus (Kitashiba et al., 2014; http://radish.kazusa.or.jp); and A. thaliana (Athaliana_447_Araport11) (Cheng et al., 2017) (E < 1e−5, inflation factor = 1.5). Protein sequences encoded by single-copy genes were used to generate a multiple sequence alignment concatenated to a super alignment matrix. A maximum-likelihood phylogenetic tree was constructed according to the PROTCATJTT model in RAxML software (version 8.2.12) (Stamatakis, 2014). Species divergence times were estimated using MCMCtree in PAML (Yang, 2007) with an independent substitution rate (clock = 2) and GTR substitution model l. A Markov chain Monte Carlo analysis was run for 10 000 generations using a burn-in of 1000 iterations. Calibration points were applied according to the core Brassicaceae origin time of 21.3–29.8 mya (Guo et al., 2017). Homozygous gene pairs were identified for turnip, Chiifu A03, ECD04, and A. thaliana, and Ks values were calculated using WGDI (https://github.com/SunPengChuan/wgdi).
Determination of syntenic relationships between turnip and its relatives
Homologous genes were analyzed using MCScanX (Wang et al., 2012) with the following parameters: E <1e−10; Gap_penalty, −3. Syntenic blocks were defined as those with at least five syntenic genes. The sequence collinearity between turnip and other genomes was assessed using the nucmer program of the MUMmer package (v4.0rc1) (Marcais et al., 2018), and the syntenic relationships were visualized using NGenomeSyn (https://github.com/Hewm2008/NGenomeSyn). We assigned and partitioned multiple turnip or Chiifu A03 chromosomal segments that matched the same A. thaliana (Athaliana_447_Araport11) segment (24 ancestral crucifer blocks A–X) into the LF, MF1, and MF2 subgenomes (Schranz et al., 2006; Wang et al., 2011b).
Gene family expansion analysis
The expansion and contraction of gene families were determined using CAFE5 (default parameters) (https://github.com/hahnlab/CAFE5). Functional annotations were performed using eggNOG-mapper (http://eggnog-mapper.embl.de/). The GO annotation analysis was performed using TBtools_windows-x64_1_098685 (eggNOG-mapper Helper), and the results were visualized using online tools (http://www.bioinformatics.com.cn/).
Identification of pseudogenes in the Brassica diploid genomes
Pseudogenes were identified using two methods. Pseudogenes were first identified by examining the assembled genomes of turnip and seven other Brassica species (Chiifu v3.5, Chiifu A03, Z1, ECD04, Bol, Bni, and Bras) as described previously (Xie et al., 2019). In brief, the analysis consisted of five major steps. First, we identified intergenic regions (masked genic and transposon regions) with sequences similar to known proteins using Exonerate (https://github.com/nathanweeks/exonerate). The following steps focused on intergenic non-TE (transposable element) regions. We preliminarily screened the candidate pseudogene regions by comparing the genomic regions with known proteins; we accepted alignments with an E value of less than 1e−5, identity of 20% or greater, match length of 30 amino acids or more, and match length of 5% or greater of the query sequence. In chromosomal segments with multiple hits, the alignment hit with the best match was retained. Next, homologous segments were linked into contigs according to the distance between the hits on the chromosome (Gc) and the distance on the query protein (Gq); the distance was set to 50 bp. The candidate contigs were then realigned using a more accurate alignment program, tfasty, with the following parameters: -A -m 3 ‘q’. Accurate sequences and the positions of frameshifts and stop codons as well as insertions and deletions were determined in this step. In the final step, Exonerate was used to identify pseudogene–functional paralog pairs.
The second method was as follows. The CDSs and protein sequences were extracted from each genome using the gffread tool in Cufflinks (version 2.2.1). The annotated genes on the genome were masked to obtain the new genome mask_gene_genome.fa; the above pep was done after the query sequence using GenBlastA (version 1.0.4) (Gough and Chothia, 2002) to the new genome for homologous gene prediction using the following parameters: genblasta -P wublast -pg tblastn -q query.pep.fa -t mask_gene_genome.fa -p T -e 1e-5 -g T -f F -a 0.5 -d 100000 -r 10 -c 0.5 -s 0. Pseudogene prediction was performed using GeneWise v0.2 (Lees et al., 2012) to obtain the final results with the following parameters: -Identity 0.95 -cover 0.95.
Finally, the predicted pseudogenes were combined with the pseudogenes identified in the abovementioned search to obtain the final number of pseudogenes for each examined species. Numerical computations were performed at the Heifei Advanced Computing Center.
Pseudogene annotation and evolutionary analysis
Pseudogenes were annotated according to their functional paralogs in the non-redundant protein sequence (NR), Swiss-Prot, kyoto encyclopedia of genes and genomes (KEGG), gene ontology (GO), clusters of orthologous genes (COG), nucleotide sequence (NT), and Pfam databases. To evaluate the level of the selective constraint on the pseudogenes, we calculated the Ks and Ka values for each pseudogene and its closest functional paralog (Xie et al., 2019). First, collinear blocks in the genomes of turnip and the other species/subspecies were compared using MCScanX (Wang et al., 2012). We then extracted the pseudogene–functional paralog pairs, the pseudogenes in the other species/subspecies and the closest functional paralogs in turnip, and the turnip pseudogenes and the closest functional paralogs in the other species/subspecies. We subsequently extracted the paired nucleotide sequences separately and translated them into protein sequences for a comparison using multiple alignment using fast fourier transform (MAFFT, version 7.487) (Katoh and Standley, 2013). The protein sequence comparison results were converted to CDS comparison results using ParaAT. Finally, selection pressure was calculated using the KaKs Calculator (version 2.0) (Wang et al., 2010). A Fisher’s test with KaKs < 3 was also performed.
Pseudogene Pfam domain analysis
We annotated all pseudogenes according to their functional paralogs in the Pfam database (Pfam-A.hmm) using HMMER 3.1b2 (February 2015) (http://hmmer.org/) with ≤1e−5 set as the threshold.
GSL extraction and analysis
GSL content was measured as described previously (Yang et al., 2020). In brief, 200 mg plant tissue was added to 80% (v/v) methanol solution containing 50 μL 1 mM sinalbin as an internal standard. The solution was mixed and then centrifuged. The collected supernatant was added to DEAE-Sephadex A-25 ion-exchange columns. The columns were washed with 80% methanol, double distilled H2O (ddH2O), and 20 mM [2(N-morpholino)ethanesulfonic acid] MES buffer (pH 5.2) before 30 μL sulfatase solution was applied. After overnight incubation at room temperature, the eluted desulfo-GSLs were separated using a high-performance liquid chromatography system (HPLC, Agilent 1100) and a ultra-performance liquid chromatography/mass spectrometry/mass spectrometry (UPLC-MS/MS) system (LCMS-8040 system, Shimadzu) with a reverse-phase C18 column and a water–acetonitrile gradient. GSL content was calculated on the basis of the peak areas at 229 nm relative to the peak area of the internal standard using the recommended relative response factors reported in DIN EN ISO 9167. The results were calculated in terms of μmol/g fresh weight.
Generation of transgenic turnip and Chiifu hairy roots
The full-length CDSs of turnip BrrMAM genes (Gene0495830, Gene0464890, and Gene0228790) were cloned into the binary vector pRI101-AN to generate 35S::BrrMAM-GFP constructs. For the RNAi constructs, the reverse complementary sequences of BrrMAM genes (Gene0495830, Gene0464890, Gene0228790) and AOP2 genes (Gene0405960, Gene0250680, and Gene0486840 in turnip; BraA02g08320, BraA09g001360, and BraA03g029140 in Chiifu) were cloned into pRI101-AN-FLAG vectors. The resulting recombinant plasmids and the negative control vectors (35S::GFP and 35S::FLAG) were inserted separately into A. rhizogenes strain LBA9402 cells.
Turnip and Chiifu hairy root cultures were established as described previously (Chung et al., 2016; Yin et al., 2020). In brief, a cotyledon infection method was used to insert the abovementioned genes into the turnip and Chiifu roots. The 35S::GFP and 35S::FLAG vectors were used as controls. The GSL content in the hairy roots was quantified using the UPLC-MS/MS system as described above. All primers are listed in Supplemental Table 19.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, Pan-Third Pole Environment Study for a Green Silk Road (Pan-TPE) (XDA2004010306); the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502); the National Natural Science Foundation of China (32070362, 41771123, and 32100315); the “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences; the Natural Science Foundation of Yunnan Province (202101AT070190); Digitalization, Development, and Application of Biotic Resource (202002AA100007); the Poverty Alleviation through Science and Technology Projects of the Chinese Academy of Sciences (KFJ-FP-201905); the Technology Transfer into Yunnan Project (202003AD150005); and the Postdoctoral Research Funding Projects of Yunnan Province (to X. Yin).
Author contributions
Conceptualization and Supervision, Yongping Yang and Yunqiang Yang; Methodology and Software, Y.Z., D.Y., X. Yang, and Z.Z.; Validation, X. Yin, X.S., X.K., and X.L.; Formal Analysis, X. Yin, G.W., Y.D., and Yunqiang Yang; Writing–Original Draft, X. Yin; Writing–Review & Editing, X. Yin and Yunqiang Yang; Funding Acquisition, Yongping Yang, Yunqiang Yang, and Y.D.
Acknowledgments
No conflict of interest is declared.
Published: September 2, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Yunqiang Yang, Email: yangyunqiang@mail.kib.ac.cn.
Yongping Yang, Email: yangyp@mail.kib.ac.cn.
Supplemental information
Data availability
Raw Illumina and PacBio sequencing data and genome assembly data have been deposited in the Genome Sequence Archive in the China National Genomics Data Center (accession numbers CRA005412 and GWHBFXQ00000000).
References
- Beilstein M.A., Nagalingum N.S., Clements M.D., Manchester S.R., Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2010;107:18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L., Oloyede O.O., Lignou S., Wagstaff C., Methven L. Taste and flavor perceptions of glucosinolates, isothiocyanates, and related compounds. Mol. Nutr. Food Res. 2018;62:1700990. doi: 10.1002/mnfr.201700990. [DOI] [PubMed] [Google Scholar]
- Benderoth M., Pfalz M., Kroymann J. Methylthioalkylmalate synthases: genetics, ecology and evolution. Phytochemistry Rev. 2009;8:255–268. doi: 10.1007/s11101-008-9097-1. [DOI] [Google Scholar]
- Blanco E., Parra G., Guigó R. Using geneid to identify genes. Curr. Protoc. Bioinformatics. 2007;Chapter 4 doi: 10.1002/0471250953.bi0403s18. Unit 4.3. [DOI] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Buchfink B., Reuter K., Drost H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021;18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Wu J., Liang J., Lin R., Zhang K., Cheng F., Wang X. Improved Brassica oleracea JZS assembly reveals significant changing of LTR-RT dynamics in different morphotypes. Theor. Appl. Genet. 2020;133:3187–3199. doi: 10.1007/s00122-020-03664-3. [DOI] [PubMed] [Google Scholar]
- Cai X., Chang L., Zhang T., Chen H., Zhang L., Lin R., Liang J., Wu J., Freeling M., Wang X. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021;22:166. doi: 10.1186/s13059-021-02383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021;38:5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J., Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinf. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F., Liu S., Parkin I.A.P., Tang H., Wang X., Chiquet J., Belcram H., Tong C., Samans B., et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhu Z., Gerendás J., Zimmermann N. Glucosinolates in Chinese Brassica campestris vegetables: Chinese cabbage, purple cai-tai, choysum, pakehoi, and turnip. Hortscience. 2008;43:571–574. doi: 10.21273/hortsci.43.2.571. [DOI] [Google Scholar]
- Cheng C.-Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- Cheng F., Mandáková T., Wu J., Xie Q., Lysak M.A., Wang X. Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell. 2013;25:1541–1554. doi: 10.1105/tpc.113.110486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Wu J., Fang L., Sun S., Liu B., Lin K., Bonnema G., Wang X. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One. 2012;7:e36442. doi: 10.1371/journal.pone.0036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Sun R., Hou X., Zheng H., Zhang F., Zhang Y., Liu B., Liang J., Zhuang M., Liu Y., et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016;48:1218–1224. doi: 10.1038/ng.3634. [DOI] [PubMed] [Google Scholar]
- Chung I.-M., Rekha K., Rajakumar G., Thiruvengadam M. Production of glucosinolates, phenolic compounds and associated gene expression profiles of hairy root cultures in turnip (Brassica rapa ssp rapa) Biotech. 2016;3:6. doi: 10.1007/s13205-016-0492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depree J.A., M. Howard T., P. Savage G. Flavour and pharmaceutical properties of the volatile sulphur compounds of Wasabi (Wasabia japonica) Food Res. Int. 1998;31:329–337. doi: 10.1016/S0963-9969(98)00105-7. [DOI] [Google Scholar]
- Drewnowski A., Gomez-Carneros C. Bitter taste, phytonutrients, and the consumer: a review. Am. J. Clin. Nutr. 2000;72:1424–1435. doi: 10.1093/ajcn/72.6.1424. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Myers E.W. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21:i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- Engel E., Baty C., le Corre D., Souchon I., Martin N. Flavor-active compounds potentially implicated in cooked cauliflower acceptance. J. Agric. Food Chem. 2002;50:6459–6467. doi: 10.1021/jf025579u. [DOI] [PubMed] [Google Scholar]
- Exposito-Alonso M., Becker C., Schuenemann V.J., Reiter E., Setzer C., Slovak R., Brachi B., Hagmann J., Grimm D.G., Chen J., et al. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 2018;14:e1007155. doi: 10.1371/journal.pgen.1007155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J., Chothia C. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 2002;30:268–272. doi: 10.1093/nar/30.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham N., May S. Springer; New York: 2011. Genetics and Genomics of the Brassicaceae. [Google Scholar]
- Green C., Willoughby J., DDD Study, Study D.D.D. De novo SETD5 loss-of-function variant as a cause for intellectual disability in a 10-year old boy with an aberrant blind ending bronchus. Am. J. Med. Genet. 2017;173:3165–3171. doi: 10.1002/ajmg.a.38461. [DOI] [PubMed] [Google Scholar]
- Gujas B., Alonso-Blanco C., Hardtke C.S. Natural Arabidopsis brx Loss-of-function alleles confer root adaptation to acidic soil. Curr. Biol. 2012;22:1962–1968. doi: 10.1016/j.cub.2012.08.026. [DOI] [PubMed] [Google Scholar]
- Guo N., Wang S., Gao L., Liu Y., Wang X., Lai E., Duan M., Wang G., Li J., Yang M., et al. Genome sequencing sheds light on the contribution of structural variants to Brassica oleracea diversification. BMC Biol. 2021;19:93. doi: 10.1186/s12915-021-01031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Liu J., Hao G., Zhang L., Mao K., Wang X., Zhang D., Ma T., Hu Q., Al-Shehbaz I.A., et al. Plastome phylogeny and early diversification of Brassicaceae. BMC Genom. 2017;18:176. doi: 10.1186/s12864-017-3555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Wessler S.R. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoede C., Arnoux S., Moisset M., Chaumier T., Inizan O., Jamilloux V., Quesneville H. PASTEC: an automatic transposable element classification tool. PLoS One. 2014;9:e91929. doi: 10.1371/journal.pone.0091929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov A.N., Artemyeva A.M., Hida K. Origin and expansion of cultivated Brassica rapa in Eurasia: linguistic facts. 5th International Symposium on Brassicas/16th International Crucifer Genetics Workshop (Brassica) Acta Hortic. 2008;867:81–88. doi: 10.17660/ActaHortic.2010.867.9. [DOI] [Google Scholar]
- Fimognari C., Nüsse M., Cesari R., Iori R., Cantelli-Forti G., Hrelia P. Growth inhibition, cell-cycle arrest and apoptosis in human T-cell leukemia by the isothiocyanate sulforaphane. Carcinogenesis. 2002;23:581–586. doi: 10.1093/carcin/23.4.581. [DOI] [PubMed] [Google Scholar]
- Istace B., Belser C., Falentin C., Labadie K., Boideau F., Deniot G., Maillet L., Cruaud C., Bertrand L., Chèvre A.M., et al. Sequencing and chromosome-scale Assembly of plant genomes, Brassica rapa as a use case. Biology. 2021;10:732. doi: 10.3390/biology10080732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.S., Pepper A.E., Hall A.E., Chen Z.J., Hodnett G., Drabek J., Lopez R., Price H.J. Evolution of genome size in Brassicaceae. Ann. Bot. 2005;95:229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.B., Faragher J.D., Winkler S. A review of the influence of postharvest treatments on quality and glucosinolate content in broccoli (Brassica oleracea var. italica) heads. Postharvest Biol. Technol. 2006;41:1–8. doi: 10.1016/j.postharvbio.2006.03.003. [DOI] [Google Scholar]
- Kang J.Y., Ibrahim K.E., Juvik J.A., Kim D.H., Kang W.J. Genetic and environmental variation of glucosinolate content in Chinese cabbage. Hortscience. 2006;41:1382–1385. doi: 10.21273/hortsci.41.6.1382. [DOI] [Google Scholar]
- Katoh K., Standley D.M. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J.J.B., Fu J., de Vos C.H.R., Lommen A., Hall R.D., Bino R.J., van der Plas L.H.W., Jansen R.C., Vreugdenhil D., Koornneef M. The genetics of plant metabolism. Nat. Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba H., Li F., Hirakawa H., Kawanabe T., Zou Z., Hasegawa Y., Tonosaki K., Shirasawa S., Fukushima A., Yokoi S., et al. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 2014;21:481–490. doi: 10.1093/dnares/dsu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusznierewicz B., Iori R., Piekarska A., Namieśnik J., Bartoszek A. Convenient identification of desulfoglucosinolates on the basis of mass spectra obtained during liquid chromatography-diode array-electrospray ionisation mass spectrometry analysis: method verification for sprouts of different Brassicaceae species extracts. J. Chromatogr. A. 2013;1278:108–115. doi: 10.1016/j.chroma.2012.12.075. [DOI] [PubMed] [Google Scholar]
- Lee J.G., Bonnema G., Zhang N., Kwak J.H., de Vos R.C.H., Beekwilder J. Evaluation of glucosinolate variation in a collection of turnip (Brassica rapa) germplasm by the analysis of intact and desulfo glucosinolates. J. Agric. Food Chem. 2013;61:3984–3993. doi: 10.1021/jf400890p. [DOI] [PubMed] [Google Scholar]
- Lees J., Yeats C., Perkins J., Sillitoe I., Rentzsch R., Dessailly B.H., Orengo C. Gene3D: a domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Res. 2012;40:D465–D471. doi: 10.1093/nar/gkr1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Su T., Zhao X., Wang W., Zhang D., Yu Y., Bayer P.E., Edwards D., Yu S., Zhang F. Assembly of the non-heading pak choi genome and comparison with the genomes of heading Chinese cabbage and the oilseed yellow sarson. Plant Biotechnol. J. 2021;19:966–976. doi: 10.1111/pbi.13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y.S., Kim H.K., Lefeber A.W.M., Erkelens C., Choi Y.H., Verpoorte R. Identification of phenylpropanoids in methyl jasmonate treated Brassica rapa leaves using two-dimensional nuclear magnetic resonance spectroscopy. J. Chromatogr. A. 2006;1112:148–155. doi: 10.1016/j.chroma.2005.11.114. [DOI] [PubMed] [Google Scholar]
- Liu S., Liu Y., Yang X., Tong C., Edwards D., Parkin I.A.P., Zhao M., Ma J., Yu J., Huang S., et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou P., Wu J., Cheng F., Cressman L.G., Wang X., McClung C.R. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell. 2012;24:2415–2426. doi: 10.1105/tpc.112.099499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A., Pecinka A., Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Wang Y., Xuan Z. Analysis and comparison of nutritional compositions in Xinjiang turnip (Brassica rapa L.) Science & Technology of Food Industry. 2016;37:360–364. doi: 10.13386/j.issn1002-0306.2016.04.064. [DOI] [Google Scholar]
- Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghe G.D., Hufnagel D.E., Tang H., Xiao Y., Dworkin I., Town C.D., Conner J.K., Shiu S.-H. Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. Plant Cell. 2014;26:1925–1937. doi: 10.1105/tpc.114.124297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J.H., Kwon S.J., Yang T.J., Seol Y.J., Jin M., Kim J.A., Lim M.H., Kim J.S., Baek S., Choi B.S., et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935;7:389–452. [Google Scholar]
- Padilla G., Cartea M.E., Velasco P., de Haro A., Ordás A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry. 2007;68:536–545. doi: 10.1016/j.phytochem.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Park S.G., Noh E., Choi S., Choi B., Shin I.G., Yoo S.I., Lee D.J., Ji S., Kim H.S., Hwang Y.J., et al. Draft genome assembly and transcriptome dataset for European turnip (Brassica rapa L. ssp. rapifera), ECD4 carrying clubroot resistance. Front. Genet. 2021;12:651298. doi: 10.3389/fgene.2021.651298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen T., Hussain A., Someshwar Rao M. Growth and accumulation of heavy metals in turnip (Brassica rapa) irrigated with different concentrations of treated municipal wastewater. Nord. Hydrol. 2015;46:60–71. doi: 10.2166/nh.2014.140. [DOI] [Google Scholar]
- Perumal S., Koh C.S., Jin L., Buchwaldt M., Higgins E.E., Zheng C., Sankoff D., Robinson S.J., Kagale S., Navabi Z.-K., et al. A high-contiguity Brassica nigra genome localizes active centromeres and defines the ancestral Brassica genome. Native Plants. 2020;6:929–941. doi: 10.1038/s41477-020-0735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Jones N.C., Pevzner P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Qi X., An H., Ragsdale A.P., Hall T.E., Gutenkunst R.N., Chris Pires J., Barker M.S. Genomic inferences of domestication events are corroborated by written records in Brassica rapa. Mol. Ecol. 2017;26:3373–3388. doi: 10.1111/mec.14131. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Takahashi T. A flavonoid from Brassica rapa flower as the UV-absorbing nectar guide. Phytochemistry. 2002;61:339–343. doi: 10.1016/s0031-9422(02)00237-6. [DOI] [PubMed] [Google Scholar]
- Schiessl S., Samans B., Huettel B., Reinhard R., Snowdon R.J. Capturing sequence variation among flowering-time regulatory gene homologs in the allopolyploid crop species Brassica napus. Front. Plant Sci. 2014;5:3389. doi: 10.3389/fpls.2014.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Lysak M.A., Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.-J., Vert J.-P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Song K., Osborn T.C., Williams P.H. Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPS).3. genome relationships in Brassica and related genera and the origin of Brassica-oleracea and B-rapa (SYN campestris) Theor. Appl. Genet. 1990;79:497–506. doi: 10.1007/bf00226159. [DOI] [PubMed] [Google Scholar]
- Song X., Wei Y., Xiao D., Gong K., Sun P., Ren Y., Yuan J., Wu T., Yang Q., Li X., et al. Brassica carinata genome characterization clarifies U's triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021;186:388–406. doi: 10.1093/plphys/kiab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz H.U., Sawada Y., Shimada Y., Hirai M.Y., Sasaki E., Krischke M., Brown P.D., Saito K., Kamiya Y. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 2011;67:81–93. doi: 10.1111/j.1365-313X.2011.04578.x. [DOI] [PubMed] [Google Scholar]
- Sun X., Li X., Lu Y., Wang S., Zhang X., Zhang K., Su X., Liu M., Feng D., Luo S., et al. Construction of a high-density mutant population of Chinese cabbage facilitates the genetic dissection of agronomic traits. Mol. Plant. 2022;15:913–924. doi: 10.1016/j.molp.2022.02.006. [DOI] [PubMed] [Google Scholar]
- Suzuki C., Ohnishi-Kameyama M., Sasaki K., Murata T., Yoshida M. Behavior of glucosinolates in pickling cruciferous vegetables. J. Agric. Food Chem. 2006;54:9430–9436. doi: 10.1021/jf061789l. [DOI] [PubMed] [Google Scholar]
- Tang S., Lomsadze A., Borodovsky M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015;43:e78. doi: 10.1093/nar/gkv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Current Protocols in Bioinformatics. 2009;25:4. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Tortorella S.M., Royce S.G., Licciardi P.V., Karagiannis T.C. Dietary sulforaphane in cancer chemoprevention: the role of epigenetic regulation and HDAC inhibition. Antioxidants Redox Signal. 2015;22:1382–1424. doi: 10.1089/ars.2014.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town C.D., Cheung F., Maiti R., Crabtree J., Haas B.J., Wortman J.R., Hine E.E., Althoff R., Arbogast T.S., Tallon L.J., et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Dev. Reprod. Biol. 2010;8:77–80. doi: 10.1016/s1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu J., Sun S., Liu B., Cheng F., Sun R., Wang X. Glucosinolate biosynthetic genes in Brassica rapa. Gene. 2011;487:135–142. doi: 10.1016/j.gene.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., Bai Y., Mun J.H., Bancroft I., Cheng F., et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.-h., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell A.M., Rowe H.C., Hansen B.G., Ticconi C., Halkier B.A., Kliebenstein D.J. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 2007;3:1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will J.L., Kim H.S., Clarke J., Painter J.C., Fay J.C., Gasch A.P. Incipient balancing selection through adaptive loss of aquaporins in natural saccharomyces cerevisiae populations. PLoS Genet. 2010;6:e1000893. doi: 10.1371/journal.pgen.1000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- Wu D., Liang Z., Yan T., Xu Y., Xuan L., Tang J., Zhou G., Lohwasser U., Hua S., Wang H., et al. Whole-genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol. Plant. 2019;12:30–43. doi: 10.1016/j.molp.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Wu W., Liu X., Wang M., Meyer R.S., Luo X., Ndjiondjop M.N., Tan L., Zhang J., Wu J., Cai H., et al. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Native Plants. 2017;3:17064. doi: 10.1038/nplants.2017.64. [DOI] [PubMed] [Google Scholar]
- Xie J., Li Y., Liu X., Zhao Y., Li B., Ingvarsson P.K., Zhang D. Evolutionary origins of pseudogenes and their association with regulatory sequences in plants. Plant Cell. 2019;31:563–578. doi: 10.1105/tpc.18.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.-C., Niu X.-M., Li X.-X., He W., Chen J.-F., Zou Y.-P., Wu Q., Zhang Y.E., Busch W., Guo Y.-L. Adaptation and phenotypic diversification in Arabidopsis through loss-of-function mutations in protein-coding genes. Plant Cell. 2019;31:1012–1025. doi: 10.1105/tpc.18.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Liu D., Wang X., Ji C., Cheng F., Liu B., Hu Z., Chen S., Pental D., Ju Y., et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016;48:1225–1232. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- Yang Y., Pu Y., Yin X., Du J., Zhou Z., Yang D., Sun X., Sun H., Yang Y. A splice variant of BrrWSD1 in turnip (Brassica rapa var. rapa) and its possible role in wax ester synthesis under drought stress. J. Agric. Food Chem. 2019;67:11077–11088. doi: 10.1021/acs.jafc.9b04069. [DOI] [PubMed] [Google Scholar]
- Yang Y., Hu Y., Yue Y., Pu Y., Yin X., Duan Y., Huang A., Yang Y., Yang Y. Expression profiles of glucosinolate biosynthetic genes in turnip (Brassica rapa var. rapa) at different developmental stages and effect of transformed flavin-containing monooxygenase genes on hairy root glucosinolate content. J. Sci. Food Agric. 2020;100:1064–1071. doi: 10.1002/jsfa.10111. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z., Jiang Y., Gong J., Li Q., Dun B., Liu D., Yin F., Yuan L., Zhou X., Wang H., et al. R gene triplication confers European fodder turnip with improved clubroot resistance. Plant Biotechnol. J. 2022;20:1502–1517. doi: 10.1111/pbi.13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Yang Y., Lv Y., Li Y., Yang D., Yue Y., Yang Y. BrrICE1.1 is associated with putrescine synthesis through regulation of the arginine decarboxylase gene in freezing tolerance of turnip (Brassica rapa var. rapa) BMC Plant Biol. 2020;20:504. doi: 10.1186/s12870-020-02697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Cai X., Wu J., Liu M., Grob S., Cheng F., Liang J., Cai C., Liu Z., Liu B., et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018;5:50. doi: 10.1038/s41438-018-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Zhao J., Lens F., de Visser J., Menamo T., Fang W., Xiao D., Bucher J., Basnet R.K., Lin K., et al. Morphology, carbohydrate composition and vernalization response in a genetically diverse collection of Asian and European turnips (Brassica rapa subsp. rapa) PLoS One. 2014;9:e114241. doi: 10.1371/journal.pone.0114241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Guo J., Cai X., Li Y., Xi X., Lin R., Liang J., Wang X., Wu J. Improved reference genome annotation of Brassica rapa by pacific biosciences RNA sequencing. Front. Plant Sci. 2022;13:841618. doi: 10.3389/fpls.2022.841618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Harrison P.M., Liu Y., Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang X., Deng B., Lou P., Wu J., Sun R., Xu Z., Vromans J., Koornneef M., Bonnema G. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 2005;110:1301–1314. doi: 10.1007/s00122-005-1967-y. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Luo L., Liu Y., Yang Y., Wang C., Kong X., Yang Y. Effect of vernalization on tuberization and flowering in the Tibetan turnip is associated with changes in the expression of FLC homologues. Plant Divers. 2018;40:50–56. doi: 10.1016/j.pld.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Lehti-Shiu M.D., Thibaud-Nissen F., Prakash T., Buell C.R., Shiu S.-H. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 2009;151:3–15. doi: 10.1104/pp.109.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina and PacBio sequencing data and genome assembly data have been deposited in the Genome Sequence Archive in the China National Genomics Data Center (accession numbers CRA005412 and GWHBFXQ00000000).