Significance
Spiral Ganglion neurons (SGNs) are responsible for relaying complex sound information from the cochlea to the auditory brainstem, and their loss contributes to age-related and acquired hearing loss. Physiological, anatomical, and molecular approaches have identified four different subtypes of SGNs in the adult cochlea. However, when and how these subtypes become specified developmentally is unknown. Using single-cell RNA-Seq, we characterized the timing and order of SGN subtype specification. We further identified transcription factors that potentially regulate downstream genes and are differentially expressed by SGNs as they split into specific subtypes. This work contributes a comprehensive data set and analysis of gene expression across SGN development that will be an outstanding resource for studies of development, regeneration, and neuronal function.
Keywords: cochlea, hearing, neuronal subgroups, single-cell RNASeq, auditory
Abstract
The afferent innervation of the cochlea is comprised of spiral ganglion neurons (SGNs), which are characterized into four subtypes (Type 1A, B, and C and Type 2). However, little is known about the factors and/or processes that determine each subtype. Here, we present a transcriptional analysis of approximately 5,500 single murine SGNs collected across four developmental time points. All four subtypes are transcriptionally identifiable prior to the onset of coordinated spontaneous activity, indicating that the initial specification process is under genetic control. Trajectory analysis indicates that SGNs initially split into two precursor types (Type 1A/2 and Type 1B/C), followed by subsequent splits to give rise to four transcriptionally distinct subtypes. Differential gene expression, pseudotime, and regulon analyses were used to identify candidate transcription factors which may regulate the subtypes specification process. These results provide insights into SGN development and comprise a transcriptional atlas of SGN maturation across the prenatal period.
Neural encoding of the dynamic aspects of complex sounds, such as frequency and intensity, requires complex and heterogeneous neuronal populations. In mammals, the afferent innervation to the cochlea is composed of the spiral ganglion neurons (SGNs; Fig. 1A). Broadly, the spiral ganglion (SG) contains two types of neurons. Type 1 SGNs constitute 90 to 95% of the population and form unitary synapses with inner hair cells (IHCs), while Type 2 SGNs, which comprise the remaining 5 to 10%, form en passant synapses with multiple outer hair cells (OHCs). Type 1 SGNs can be further divided into three subtypes based on different physiological and morphological characteristics including threshold and spontaneous firing rate (1–3). One subtype is characterized by low thresholds and high spontaneous firing rates (high SR), and they are likely more adapted to responding to quiet, low-intensity sounds (1). A second subtype has high thresholds and low spontaneous rates (low SR) and they are thought to play a role in discerning auditory stimuli in high-background noise environments (4). A third sub-type is characterized by SR between low and high fibers (med SR) (1–3). Recent studies have demonstrated that low SR SGNs are lost at a more rapid rate during aging, suggesting a crucial role in age-related hearing loss and expected outcomes for cochlear implant patients (5–8). The function of Type 2s is less clear, although it seems likely that they play a role in the perception of pain and possibly cochlear damage (9, 10).
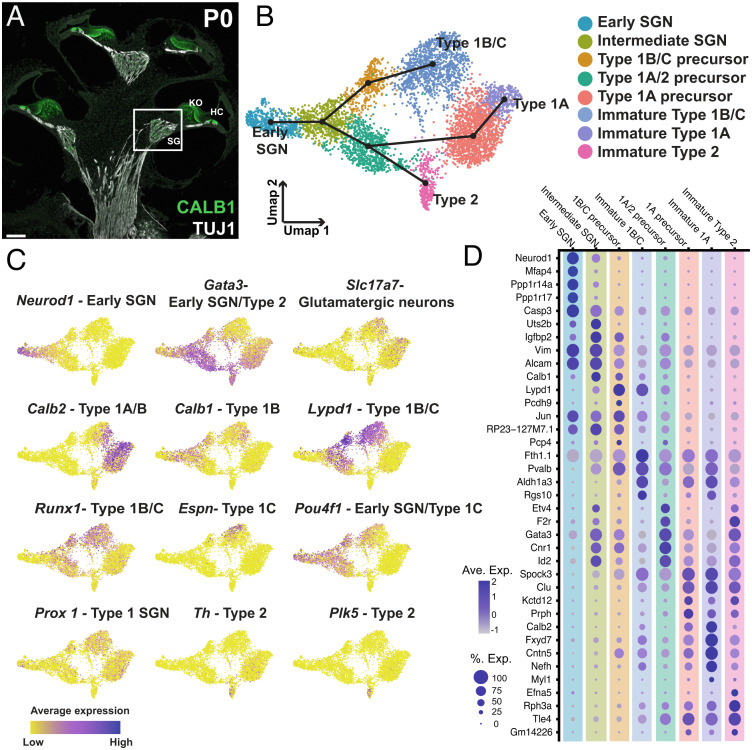
Fig. 1.
Characterization of SGN phenotypes in the combined dataset. (A) Midmodiolar cross section of a P1 cochlea with TUJ1- labeling of SGNs including their central and peripheral processes and CALB1-labeling of SGN cell bodies (SG; box), hair cells (HC), and the medial portion of Kolliker’s organ (KO). (B) UMAP of 5,441 SGNs collected at E14, E16, E18, or P1 with cluster identities indicated. Slingshot trajectory plot overlayed on the UMAP indicates order in which SGN subtypes are specified beginning in the "Early SGN" cluster. (C) Feature plots of the same data as in B, displaying gene expression levels of known markers for SGN subtypes and developmental phases across the dataset (see text for details). (D) Dot plot showing the top five differentially expressed genes for each of the clusters identified in B. (Scale bar in A, 100 µm.)
Recently, single-cell RNASeq was used to characterize SGNs in the adult mouse cochlea. Results identified three transcriptionally unique Type 1 subtypes. While definitive proof of the identity of these subtypes is not yet available, correlative data suggest that these three subtypes correspond to high (Type 1A), low (Type 1C), and medium (Type 1B) spontaneous rate SGNs (5, 11, 12). While the results presented above provided significant insights regarding the composition of the SG, the question of how/when these subtypes arise during development was not fully addressed.
In the mouse, SGNs are born between embryonic day (E) 9 and E12 and subsequently go through a prenatal period of development, presumed to be largely under genetic control, followed by a period of development in the early postnatal weeks characterized by spontaneous and subsequently evoked activity after hearing onset. Two recent single-cell RNASeq studies demonstrated that disruption of both spontaneous and evoked activity leads to changes in the proportion of SGNs which are specified into each of the Type 1 subtypes, suggesting key roles for postnatal activity in subtype specification (5, 11). However, the third study found evidence for the presence of all three Type 1 subtypes at the time of birth (12). This suggests that the initial process of subtype specification occurs during the embryonic period, a result that is consistent with other studies suggesting that Type 2 SGNs are specified as early as E14 (13–15). Considering that different Type 1 subtypes appear to be disproportionately affected during age-related hearing loss, an understanding of the transcriptional pathways that specify each subtype could be useful in the development of potential therapies to preserve/restore hearing.
To characterize the transcriptional regulation of subtype formation in the SG, we utilized single-cell RNASeq to profile SGNs across prenatal development. Our results demonstrate that all four SGN subtypes (1A, B, C, and 2) are present by E18 and moreover, that the different subtypes develop through two transient precursor cell types, one of which expresses characteristics of Type 1A and Type 2 SGNs while the other expresses characteristics of Type 1B and Type 1C. Subsequently, both Type 1A/2 and Type 1B/C precursors split to generate all four SGN subtypes. These results suggest that events occurring during the prenatal period, likely under transcriptional control, are responsible for the initial development of all four adult SGN subtypes. This initial transcriptional process is followed by activity-dependent events during the early postnatal period which are required to further refine and mature SGNs into their fully differentiated adult states.
Results
Initial Dataset Overview.
To build a transcriptional map of mouse SG development, we collected SGs at four different time points—E14, E16, E18, and P1. Briefly, ganglia and the sensory epithelia were grossly dissected from all other associated tissues (see Methods). Ganglia were then dissociated and single cells from each sample were captured using the 10X Chromium platform. Three independent biological replicates were collected at E14, E16, and E18 with two biological replicates collected at P1. In addition, for E16, each replicate was separated into apical and basal halves prior to collection. After sequencing, initial quality control and doublet removal was conducted (see Methods), and then unbiased clustering analysis was used to identify different cell types including SGNs, mesenchymal cells, and glia (SI Appendix, Fig. S1). The SGNs represented a small proportion of the total population of cells captured at each time point but were easily identifiable based on expression of a range of SG neuronal markers such as Tubb3, Nefl, and Pou4f1 and neuronal developmental genes, such as Neurod1 (SI Appendix, Fig. S1B). SGNs were then removed from the overall dataset and analyzed separately—5,441 neurons in total, representing 8.5% of all cells collected. The other cell types collected in this dataset will be analyzed and published later. To ensure that the SGN data set included only neurons, we compared the expression of the neuronal marker Tubb3 with the glial marker Sox10 in the SGN-selected cells for each independent collection from each time point (SI Appendix, Fig. S1D). Results indicate uniform expression of Tubb3 with no expression of Sox10 in the SGN cells from each sample and time point.
Next, the SGNs from all 14 biological replicates were combined and reanalyzed (16). When visualized on a dimensionally reduced Uniform Manifold Approximation and Projection (UMAP) plot, the datasets produced a gradient of time points along the UMAP1 axis from E14 at one end, representing the most immature cells, to E18 and P1 at the opposite end, representing the most mature (SI Appendix, Fig. S2). Consistent with the basal to apical development of SGNs, cells collected from the base at E16 were located closer to cells collected at E18 or P1, while cells from the E16 apex were located closer to the E14 cells. The distribution of cells in the combined UMAP had a striking pattern in which cells appeared to separate along two distinct paths with increasing maturity. To explore this pattern in more depth, unbiased clustering was applied to identify unique transcriptional cell types. Results indicated eight distinct clusters (Fig. 1B). To identify specific cell types, expression of previously determined markers of SGN subtypes, such as Neurod1 and Gata3 in immature SGNs, Calb2 in Type 1A, Lypd1, Calb1, Espn, and Pou4f1 in type 1B/C, and Th and Plk5 in Type 2s were localized to specific clusters (Fig. 1C). In addition, several additional clusters, which featured expression of both early and late markers and, therefore, appeared to represent intermediate stages of development, were also present (Fig. 1 B and C). To confirm that the observed distribution of cells represents a developmental process, we sought to model this computationally using Slingshot (17), a trajectory inference method, to fit a developmental trajectory to the data. As a first step, Slingshot generates a minimum spanning tree to identify lineages of cells starting at the same origin and then diverging to unique endpoints. Consistent with our hypothesis, Slingshot generated a trajectory with an initial bifurcation into two branches and then a subsequent split between the Type 1A and Type 2 clusters within one of those branches (Fig. 1B, overlay on UMAP), suggesting that Type 1A SGNs are initially more closely related transcriptionally to Type 2 SGNs than they are to the other two Type 1 subtypes. Finally, we identified additional markers for each cluster in the trajectory by comparing gene expression for each cluster versus all other clusters (Fig. 1D and Dataset S1).
Specification of SGN Type 1B/C.
While Type 1A and Type 2 SGNs formed two distinct clusters in the merged data set, Type 1Bs and Type 1Cs were located in a single cluster even when clustering parameters were varied, suggesting high transcriptional similarity between the two groups. However, analysis of feature plots for Type 1B or 1C markers such as Calb1 (1B), Calb2 (1A/B), Lypd1, Pou4f1, or Espn (all 1C) indicated regional specificity within the cluster (Fig. 1C). This suggested that 1B and 1C SGNs may be specified by P1 but that their transcriptional similarity relative to all other neurons in this larger dataset, the majority of which were from more immature time points, was insufficient to render unique clusters. To visualize transcriptional differences between these clusters at this particular point in development, the E18 and P1 SGNs were isolated and reclustered separately. This dataset of 1,303 neurons grouped into eight clusters and Slingshot analysis recapitulated the bifurcation from Intermediate SGNs to Type1A/2 and Type 1B/C precursors (Fig. 2 A and B). However, in contrast with the analysis of the complete dataset, both initial branches of the trajectory now split a second time to give rise to all four SGN subtypes (Fig. 2B).
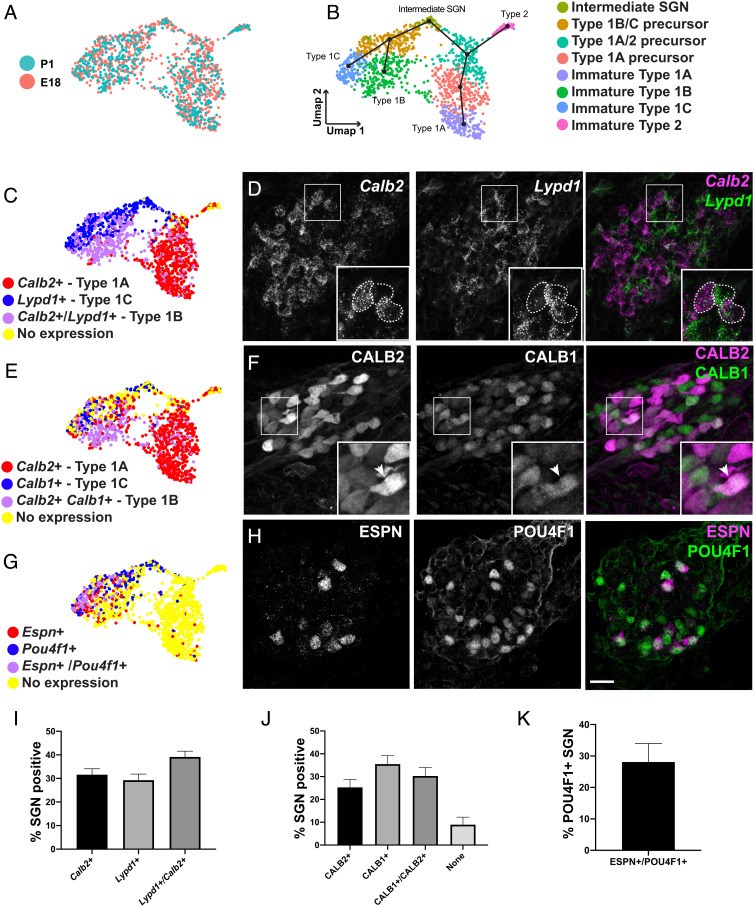
Fig. 2.
All SGN subtypes are specified by E18. (A, B) UMAP of 1,303 SGNs collected at E18 or P1 colored by time point (A) or cluster identity (B). Slingshot trajectory plot overlayed on the UMAP in B indicates order in which all four SGN subtypes are specified beginning in the "Intermediate SGN" cluster. (C, E, and G) Feature plots of the same data as in B displaying binarized gene expression of markers of Type 1 subtypes along with visualization of expression of the same markers in E18 SGN sections. (D) Fluorescent in situ hybridization confirms expression of Lypd1 and Calb2 at E18. Inset: example of three SGNs expressing either Calb2 (Type 1A) or Lypd1 (Type 1C) or both (Type 1B). (F) Immunohistochemistry confirms expression of CALB2, CALB1, or both in different SGNs at E18. Inset: arrowhead indicates a neuron expressing both markers (Type 1B). (H) Immunohistochemistry confirms expression of Type 1C markers ESPN, POU4F1, or both in a subset of SGNs at E18. (I–K) Quantification of proportion of SGNs expressing each marker above at E18. About 1/3 of SGNs express each of the three Type 1 subtypemarker combinations (I–K). Approximately 30% of POU4F1+ SGNs also express ESPN at E18. Data presented as mean ± SEM. (Scale bar in H (same in D and F), 20 µm.)
To validate these results, expression of subtype specific markers was examined in feature plots and by single-molecule fluorescent in situ hybridization (smFISH) or immunohistochemistry in E18 tissue sections. Comparison of expression of Calb2+ (Type 1A), Lypd1+ (Type 1C) and Calb2+/Lypd1+ (Type 1B) was consistent with the presence of all three Type 1 cell types in the UMAP analysis (Fig. 2C). Quantification in tissue sections found that roughly 1/3 of SGNs were either Calb2+ (31.6%), Lypd1+ (29.3%), or Calb2+/Lypd1+ (39.2%), also consistent with the presence of the three subtypes (Fig. 2 D and I). Similarly, immunohistochemistry for Calbindin 1 (CALB1) and Calretinin (CALB2) distinguished between Type 1A (CALB2+, 25.3% of all SGNs) and Type 1B (CALB1+/CALB2+, 30.3% of all SGNs)(Fig. 2 E, F, and J). Finally, we used two Type 1C markers, POU class 4 homeobox 1 (POU4F1) and Espin (ESPN), to localize this subtype (Fig. 2 G and H). ESPN, a late marker for Type 1Cs, always colocalized with POU4F1 (Fig. 2H). In contrast, only 28% of POU4F1 SGNs also expressed ESPN (Fig. 2K). However, this is consistent with the demonstrated broad expression of Pou4f1 in immature SGNs, which is only maintained in Type ICs as the neuron mature.
Markers for Type 2 SGNs.
The development and functional role(s) of Type 2 SGNs has been difficult to determine, in part because of their low abundance and limited definitive markers, especially at early postnatal time points. While the number of Type 2 SGNs that were captured was limited (47 out of 1,303 total SGNs in our E18/P1 combined dataset; Fig. 3A), the population was sufficient to identify new marker genes. As a first step, the expression of several adult markers of Type 2 SGNs was examined in our E18/P1 dataset. Th and Calca (adult markers of apical and basal Type 2 SGNs, respectively (10, 18)) were strongly expressed in the Type 2 cluster, although some Th expression was present in a subset of Type 1A neurons (Fig. 3B). In contrast, two other adult Type 2 markers, Prph and Gata3, were also widely expressed in Type 1 SGNs, and, in particular, in Type 1A cells, a result that is consistent with their apparent shared origin but suggests that these are not unique markers for Type 2s at early developmental stages (Fig. 3C). Finally, Epha4 has been proposed to have higher expression levels in Type 1 compared with Type 2 SGNs at P0 (19); however, in our dataset, this gene appeared to be equally expressed throughout the Type 1 and Type 2 populations (Fig. 3D).
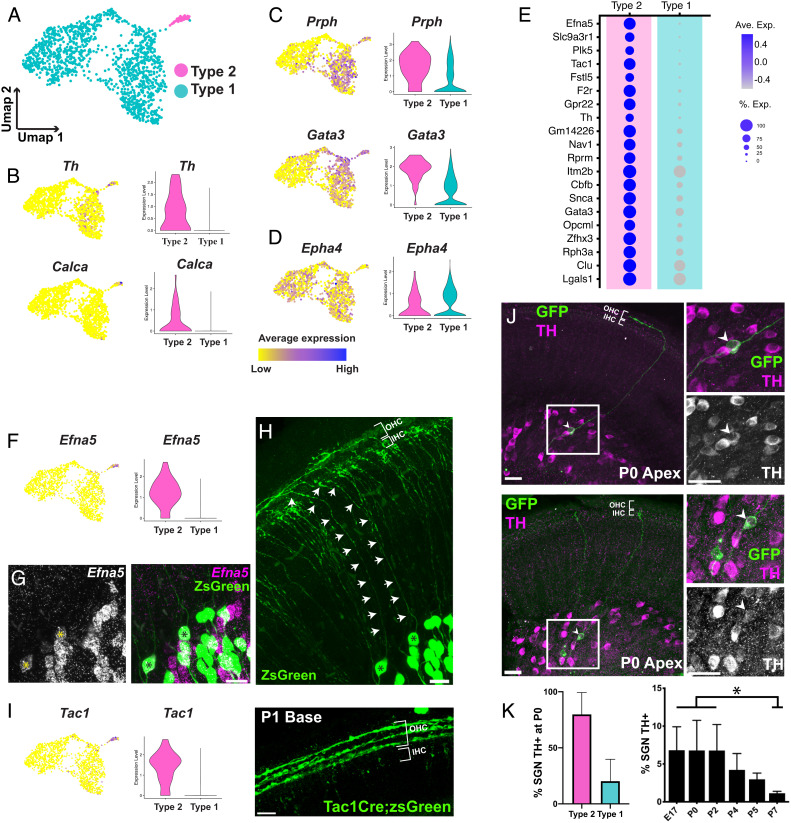
Fig. 3.
Markers of Type 2 SGNs at P1. (A) UMAP of E18/P1 dataset from Fig. 2 colored based on Type 1 and Type 2 identity. (B–D) Feature and violin plots indicating the expression of several established markers of Type 2 (B and C) and Type 1 (D) SGNs in the E18/P1 dataset (see text for details). (E) Dot plot displaying the top 20 genes differentially expressed in the Type 2 cluster in comparison to all the Type 1 clusters. (F) Feature and violin plots showing that Efna5 expression is specific to the Type 2 cluster. (G) Fluorescent in situ hybridization for Efna5 at P1 (Left Panel) combined with genetic sparse labeling of SGNs using Neurog1CreERT2R26RZSGreen (Right Panel). Two double-positive cells are indicated with asterisks. (H) Tracing of the peripheral processes for the two cells indicated in G confirms that their peripheral processes terminate in the OHC region and are therefore Type 2 SGNs. (I) Feature and violin plots showing that Tac1 expression is specific to the Type 2 cluster. Right hand panel: Genetic labeling using Tac1CreR26RZSGreen at P1 shows strong labeling in Type 2 peripheral processes innervating the three rows of OHCs. (J) At P0, TH immunohistochemistry combined with genetic sparse labeling using Neurog1CreERT2R26RmT/mG shows TH is expressed by Type 2 SGNs innervating the OHCs (Upper Panel). Right hand panels: High magnification images of the boxed region illustrating TH expression in a labeled Type 2 SGN (arrows). Lower Panel: similar view as above but illustrating TH expression in a Type I SGN. (K) Quantification of Type 1 and Type 2 SGNs that are positive for TH at P0. Approximately 80% of Type 2s and 20% Type 1s are TH+. Right hand panel: The proportion of SGNs that are TH+ significantly decreases across the 1st postnatal week, suggesting ongoing refinement of gene expression in Type 2 SGNs during this period. Data presented as mean ± SEM *P ≤ 0.05. (Scale bar in G–I, 20 µm, J, 40 µm.)
To identify new markers which would be useful for the identification of Type 2 SGNs at birth, we examined differentially expressed genes in Type 2 SGNs versus all Type 1 SGNs in the E18/P1 dataset. Among the top 20 markers generated by this analysis were Efna5, Tac1, and Th (Fig. 3E and Dataset S1). To validate the specificity of these three markers, smFISH for Efna5 was combined with genetic sparse labeling in Neurog1CreERT2;R26RZSGreen cochlea to trace the peripheral processes of Efna5+ cell bodies to their termination in the organ of Corti at either the IHCs (Type 1) or OHCs (Type 2). Results indicated that all Efna5+ SGNs were Type 2s (Fig. 3 F–H). Next cochleae from Tac1Cre;R26RZSGreen mice were analyzed at P1. Although some ZSGreen+ peripheral processes terminated in the IHC region, an overwhelming majority of labeled SGN processes extended into the OHC region (Fig. 3I).
Lastly, we used antibody labeling to examine the expression pattern of Tyrosine Hydroxylase (TH) in the early postnatal period. Colocalization of TH expression with Neurog1CreERT2;R26RmTmG sparse-labeling indicated that 20.2% (+/− 8.7%) of TH-labeled SGNs were Type 1s (Fig. 3 J and K). This finding was largely consistent with results from the single-cell analysis which indicated expression of Th in some Type 1A SGNs (Fig. 3B). In addition, TH immuno-labeling in SG sections indicated a significant decrease in the percentage of TH+ neurons across the perinatal period (Fig. 3K and SI Appendix, Fig. S3), indicating possible further postnatal refinement of Type 1 and Type 2 gene expression.
Separation of Developing SGNs into Two Precursors.
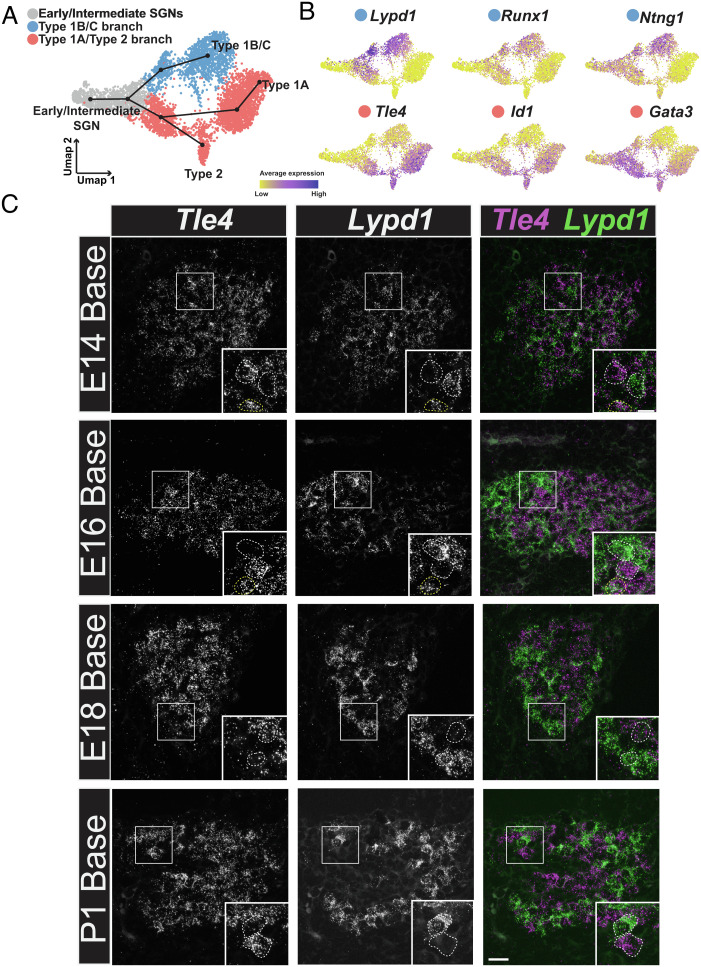
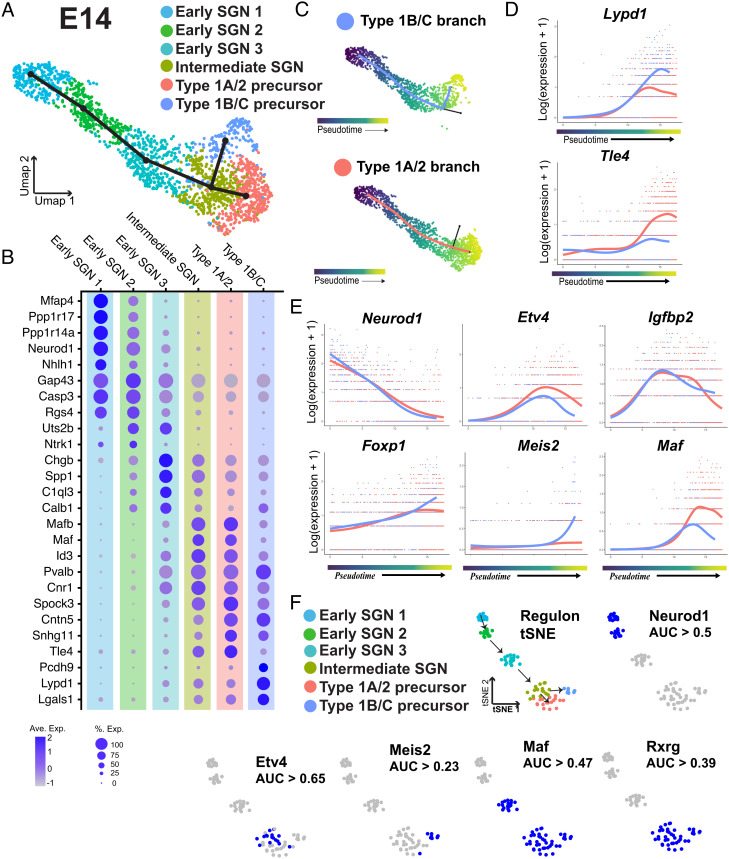
To identify markers of the initial split of SGNs into Type 1A/2 and Type 1B/C precursor paths, gene expression along each branch was analyzed (Fig. 4A). Lypd1, Runx1, and Ntng1 are all initially expressed by cells along both branches but are only maintained in the Type 1B/C path. Tle4, Id1, and Gata3 are all largely expressed only by cells along the Type 1A/2 path (Fig. 4B). Interestingly, these genes show two different patterns of expression with Lypd1/Runx1/Ntng1 initially showing broad expression in both branches followed by downregulation in the 1A/2 path. In contrast, Tle4/Id1/Gata3 are only upregulated in the 1A/2 path. This suggests that the Type 1A/2 precursor may require specific, positive induction while the 1B/C precursor may represent a default phenotype. To validate the presence of two initial developmental branches and to determine the timing of this split, smFISH was performed for Lypd1 and Tle4 in cochlear tissue sections collected at E14, E16, E18, or P1. Neither marker was observed in the less mature apical cochlear region at E14, however, in the more mature base, three subsets of SGNs, some expressing high levels of Lypd1 with limited Tle4, some expressing high levels of Tle4 with limited Lypd1, and some expressing both, were observed (Fig. 4C). These results are consistent with a trajectory in which some SGN precursors are positive for both markers before committing to one branch or the other. It is important to consider that some Type 1B/C SGNs collected at E18 or P1 were weakly positive for Tle4 and strongly positive for Lypd1 (Fig. 4B). The presence of these cells could suggest that the cells that were identified as intermediate at E14 are actually Type 1B/C SGNs. However, in contrast with Type 1B/C SGNs, Tle4/Lypd1 double-positive cells collected at E14 show roughly equivalent levels of expression of Tle4 and Lypd1 (Fig. 5B). In addition, since these cells were collected at a very immature stage of SGN development, Tle4/Lypd1 positive cells from this time point are unlikely to represent Type 1B/C cells. Moreover, if E14 Tle4/Lypd1 positive cells did represent an early population of Type 1B/C cells, then those cells would be expected to cluster with Type 1B/C cells collected at older ages. Instead, all E14 cells cluster with other cells collected at E14 (Fig. 1B and SI Appendix, Fig. S2), suggesting that these cells are not Type 1B/C cells and instead are a transcriptionally distinct, less mature cell type. Finally, as the SGN matures, the amount of overlap between the markers appears to decrease such that by E18 only roughly 30% of SGNs are double positive for Lypd1 and Tle4 (Fig. 4C and SI Appendix, Fig. S4).
Fig. 4.
SGN progenitors initially split into two precursors. (A) UMAP from Fig. 2 showing the combined dataset across all ages and the Slingshot trajectory for the predicted order of specification into four subtypes. UMAP is colored based on the initial split into two precursor branches. (B) Feature plots displaying genes which are expressed along the Type 1B/C or Type 1A/2 branches. (C) Fluorescent in situ hybridization for Tle4 (Type 1A/2 branch) and Lypd1 (Type 1B/C branch) in tissue sections of basal SG from E14, E16, E18, and P1. Insets: examples of individual SGNs expressing either Tle4 or Lypd1 (white dashed line). At E14 and E16, examples of SGNs in a transitional state expressing both Tle4 and Lypd1 are also shown (yellow dashed line). (Scale bar in C, 20 µm.)
Fig. 5.
Characterization of the SGN progenitor split into Type 1B/C and Type 1A/2 branches. (A) UMAP of 1,765 SGNs collected at E14 with cluster identities indicated. Slingshot trajectory plot overlayed on the UMAP identifies a split into two branches. (B) Dot plot showing the top five differentially expressed markers for each of the clusters identified in A. (C) Pseudotime plots of the cells in each branch. (D) Expression plots of Lypd1 and Tle4 across pseudotime for each branch. Tle4 is specifically upregulated in the 1A/2 branch while Lypd1 expression is only maintained in the 1B/C branch. (E) Expression plots for transcription factors identified using tradeSeq. Top row: Etv4 and Igfbp2 are examples of transcription factors with gene expression that peaks as the two branches are splitting. Bottom row: examples of transcription factors identified as differentially expressed at the endpoints of the two branches. (F) Top row: tSNE plot based on identification of transcriptional regulons using SCENIC. Clusters are colored based on the clusters identified in A. Arrows indicate order of specification based on Slingshot analysis. Regulon tSNE plot of Neurod1 shows expected activity in early SGN clusters, blue dots indicate the regulon is active. Bottom row: examples of regulons which were both significantly restricted to specific clusters and were predicted to be active based on AUC thresholding (see Methods).
Since the single-cell and smFISH analyses indicated that the initial specification into the 1A/2 and 1B/C branches begins as early as E14, we examined the E14 dataset in more detail. Reclustering indicated six clusters, three of which, labeled as Intermediate SGN, Type 1A/2, and Type 1B/C, were present in the full dataset (Fig. 5A). However, with reclustering, the Early SGN cluster resolved into three clusters, designated as Early SGN 1–3, that aligned linearly along the UMAP1 axis (Fig. 5A). Differential expression identified marker genes for each phase (Fig. 5B and Dataset S1). Neurod1 was most highly expressed in Early SGN1, while the 1A/2 and 1B/C marker genes Tle1 and Lypd1 were expressed in two spatially separated clusters on the other side of the projection (SI Appendix, Fig. S7). The presence of these additional early SGN states was validated by analyzing the dataset with an additional single-cell analysis tool, Monocle (20, 21), which generated a spatially equivalent UMAP and set of transcriptional clusters (SI Appendix, Fig. S5). To confirm that the Early SGN1-3 clusters were comparable between the Seurat and Monocle analyses, we examined the top 10 marker genes for Early SGN1-3 using Monocle and found that 87% of these genes were also present in the top 20 marker genes for the same clusters as calculated by Seurat. However, whether these clusters represent distinct states, which every developing SGN moves through, or is reflective of the gradient of maturation along the cochlear axis at this time point, cannot be determined from these results. Slingshot trajectory analysis using Early SGN1 as a starting point but without specifying any particular endpoint identified a bifurcation between two developmental branches in the Intermediate SGN cell cluster (Fig. 5A and SI Appendix, Fig. S6). This result suggests that at E14, as SGNs mature, they begin to separate along either the 1A/2 or 1B/C branches. However, while Slingshot places the location of this split in the Intermediate SGN cluster, it is entirely possible that cells in slightly earlier and later clusters are also part of this bifurcation. Therefore, to identify genes involved in this split, we used Slingshot to perform a pseudotime analysis which was less dependent on discrete clusters. The resulting pseudotime analysis divided the cells into two trajectories, one which branched toward the Type 1A/2 fate and the other which branched toward the Type 1B/C fate (Fig. 5C). The analysis also assigned pseudotime values to each SGN as a measure of their progression through development. Once the SGNs were ordered in pseudotime, we were able to analyze the changes in expression of genes across SGN maturation. Next, we applied tradeSeq (22) to discover genes which are expressed in patterns significantly associated with pseuodtime and which are differentially expressed between the two developmental branches. As noted previously, Lypd1 increases along both branches before expression drops in the 1A/2 branch toward the end of the projection (Fig. 5D). In contrast, expression of Tle4 is low in both branches before rapidly increasing only in the 1A/2 branch.
Among other genes that were identified as differentially expressed by tradeSeq between the 1A/2 and 1B/C branches were several transcription factors including Etv4, Igfbp2, Foxp1, Meis2, and Maf (Fig. 5E and Dataset S2). The tradeSeq analysis indicates different patterns of expression that could be indicative of unique roles for each transcription factor (Fig. 5E). For instance, Meis2 shows a rapid upregulation in the 1B/C branch at a pseudotime point that correlates with the split between the two branches.
To examine if any candidate transcription factors might be controlling gene expression along the branches, we used Single-Cell rEgulatory Network Inference and Clustering (SCENIC) to identify transcriptional regulons (23). SCENIC analysis aims to infer the regulatory activity of transcription factors in a dataset by identifying sets of coregulated genes, or regulons, with common transcription factor binding sites. For this analysis, cells were subsetted as described previously (24). The resulting analysis generates a t-distributed stochastic neighbor embedding (tSNE) graph of unique cell clusters based on expression of transcriptional regulons. The SCENIC analysis generated cellular clusters that were comparable to those generated through analysis of the full transcriptional data set (Fig. 5F). We then examined the most significantly correlated regulons for different clusters (Fig. 5F and Dataset S5). The Neurod1 regulon was active in the two most immature clusters, as would be expected based on its role in SGN development (25–27) (Fig. 5F). Activity of the Etv4 and Meis2 regulons mirrored the results from the tradeSeq analysis, with Etv4 activity concentrated in the final common cluster while Meis2 activity is restricted to the endpoint of the Type 1B/C branch (Fig. 5F). Similarly, the Maf regulon was active in the entire latter half of the pseudotime projection even though Maf gene expression appears lower in the Type 1B/C branch by comparison with the 1A/2 branch. Finally, the Rxrg regulon was also active in the final common cluster as well as both the 1A/2 and 1B/C clusters.
Development of the Four SGN Subtypes.
Based on the preceding analysis, separation of the 1B/C and 1A/2 precursors into their subsequent final phenotypes seemed likely to begin around E16. While the E16 data set was separated into apical and basal halves prior to collection, for the initial analysis, these data sets were combined to produce a dataset which grouped into six clusters (SI Appendix, Fig. S8 A and B). As was the case for the cells collected at E14, the Intermediate SGN, Type 1B/C precursor, and Type 1A/2 precursor clusters were present at E16. To identify the remaining clusters, differential expression analysis was conducted to identify markers (SI Appendix, Fig. S8C and Dataset S1). Analysis of these markers plus known markers of the different SGN subtypes (SI Appendix, Fig. S8 C and D) allowed us to identify one cluster as containing Type 1As and two clusters containing SGNs expressing markers consistent with 1B and 1C. In order to tease apart the 1B and 1C clusters, we compared the expression of 1C markers in less mature apical SGNs with more mature neurons from the base (SI Appendix, Fig. S9). In the 1B cluster, the proportion of SGNs expressing Pou4f1 was lower in the base than in the apex, indicating that this gene is downregulated in this population with maturation (SI Appendix, Fig. S9A). In the 1C cluster, expression of Pou4f1 stayed the same in basal and apical SGNs. Espn was not significantly expressed in apical SGNs but was detected in the basal 1C cluster (SI Appendix, Fig. S9A). This expression pattern was confirmed by immuno-labeling for ESPN in the SG at E16. We found little to no labeling in the apex; however, ESPN-positive SGNs were identifiable in the base (SI Appendix, Fig. S9C). The expression patterns of both Pou4f1 and Espn suggest that Type 1B and Type 1C SGNs are first becoming specified at E16. While a distinct Type 2 cluster was not observed at this time point, when we compared expression of Efna5 and Tac1, two Type 2 markers from our E18/P1 dataset, we found both were expressed specifically in the Type 1A/Type 2 precursor cluster (SI Appendix, Fig. S9B). This indicates that Type 2 SGNs are beginning to become specified at this time point; however, the small number of these cells in the data set may be insufficient to generate an independent cluster.
Next, Slingshot analysis was used to identify potential regulators of the bifurcation between the Type 1B/C and Type 1A/2 branches. However, Slingshot failed to render a clean trajectory, most likely because of the presence of some 1B neurons in the 1A cluster (SI Appendix, Fig. S8A). While we cannot rule out the possibility that some Type 1B SGNs are derived from the 1A branch, our results are more consistent with mixing of these clusters due to an upregulation of genes, such as Calb2, that are expressed in both Type 1B and Type 1A neurons as they mature.
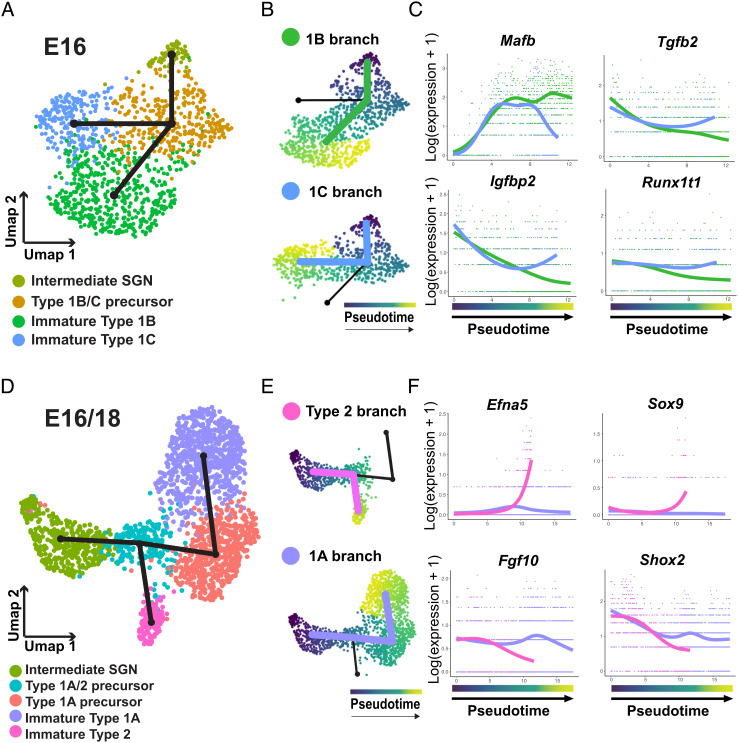
In order to identify genes that might be driving the specification of 1B and 1C subtypes, we removed 1A/2 precursors and immature 1A cells from the dataset and reanalyzed only cells along the 1B/C branch. (Fig. 6A). Slingshot analysis generated a single bifurcation trajectory going from the Intermediate SGN cell cluster to a split in the Type 1B/C precursor cluster leading to immature 1B and immature 1C clusters as the end points (Fig. 6A). tradeSeq was then used to identify differentially expressed genes along the 1B or 1C branches (Fig. 6B and Dataset S3). Among the top results were several transcription factors, including Mafb, which is known to be expressed in SGNs, and showed prolonged expression only in the 1B branch, and Tgfb2, Igfbp2, and Runx1t1, all of which showed general decreased expression in the 1B versus 1C branch (Fig. 6C).
Fig. 6.
Characterization of Type 1A/2 and Type 1B/C precursor splits. (A) UMAP of 1,127 SGNs from the Type 1B/C branch collected at E16. Slingshot trajectory plot overlayed on the UMAP indicates split between Type 1B and C branches. (B) UMAP of the cells in each branch ordered in pseudotime. (C) Plots of the expression of transcription factors identified by tradeSeq analysis as differentially expressed at the endpoints of the two branches. (D) UMAP plot of 1,977 SGNs combined from the Type 1A/2 branch collected at E16 or E18. Slingshot trajectory plot overlayed on the UMAP indicates split between Type 1A and Type 2 branches. (E) UMAPs of the cells in each branch ordered in pseudotime. (F) Plots of the expression of transcription factors identified by tradeSeq analysis as differentially expressed as the two branches split and at the endpoints of the two branches.
To identify additional candidate transcription factors for these branches, SCENIC analysis was performed (SI Appendix, Fig. S10 and Dataset S6). Consistent with previous results, the Pou4f1 regulon was identified in all clusters except immature 1B while the Etv4 regulon was observed only in precursor and Intermediate SGN clusters (SI Appendix, Fig. S10A). To validate these results, the expression of both POU4F1 and Etv4 was localized in sections from E16 cochlea. As has been previously reported (28), POU4F1 protein expression was present in a subset of SGNs, with a wider expression pattern in the apex than the base (SI Appendix, Fig. S10B). This is consistent with the progressive downregulation of the Pou4f1 regulon in non-1C neurons. Etv4 mRNA expression was stronger in the apical cochlear turn, consistent with the finding that the Etv4 regulon is active in the “Type 1B/C precursor” cluster (SI Appendix, Fig. S10C).
Finally, to identify genes that may drive the development of Type 1A and Type 2 SGNs, we combined the E16 and E18 datasets. This was necessary because the small number of Type 2 neurons in the E16 dataset was insufficient to form a unique cluster. We then created a subset of cells containing the Intermediate SGN, Type 1A/2 precursor, Type 1A precursor, immature 1A, and immature Type 2 clusters. Slingshot analysis created a single bifurcating trajectory that originated in the Intermediate SGN cluster and then split along Type 1A and Type 2 branches (Fig. 6 D and E). tradeSeq and SCENIC analysis identified candidate genes and transcriptional regulons for both branches (Fig. 6F and SI Appendix, Fig. S11 and Datasets S4 and S7). In particular, Efna5 and Sox9 were found to be specifically upregulated in developing Type 2s while Type 1As were found to persistently express Fgf10 and Shox2 (Fig. 6F).
Discussion
Neurons within the SG can be subdivided into four subtypes based on physiological, anatomical, and transcriptional criteria. While the specific roles of each subtype are not fully understood, all are believed to be required for normal auditory function. Moreover, the loss of particular subtypes over time likely contributes to age-related hearing loss (6, 7, 29). Despite the importance of SGN neurons for auditory function, how the various subtypes are specified during development is not well understood. Here, we used single-cell RNASeq to better understand the initial genetic regulation of SGN development during the prenatal period. Using markers established in the adult SG, we were able to identify all three Type 1 subtypes and Type 2 SGNs, as well as a range of phases that developing SGNs may move through as they mature. While our focus was on understanding the process of subtype development, this dataset also contains a wealth of information regarding other processes that occur in developing SGNs at the same time, such as peripheral and central axon outgrowth and synapse formation. The full dataset will be available for others to access through the gEAR portal (https://umgear.org/).
Previous single-cell RNASeq studies have shown that all four subtypes of SGNs appear to be transcriptionally distinct in the early postnatal period (12), with mature expression profiles by the third postnatal week (5, 11, 12). However, the timing and manner in which SGNs separate into those subtypes were unknown. Our results indicate that all SGN subtypes can be identified transcriptionally as early as E16, with reliable expression of subtype markers present at E18. This places the timing of initial SGN subtype formation in the prenatal period, prior to the onset of spontaneous activity (30, 31). Although a recent study has shown that some spontaneous activity is present in the SGNs at E16, these events were largely restricted to the basal neurons, were few in number, and were uncorrelated with support cell and inner hair cell activity (32). Therefore, it is likely that initial SGN subtype development is under genetic regulation rather than dependent on activity. Evidence from knockout models indicates that spontaneous activity plays an important role in shaping the transcriptional phenotypes of the SGN subtypes (5, 11). However, our data demonstrate that all four SGN subtypes are already present to some degree by E16, suggesting that the role of activity may be to maintain, refine, or more fully specify subtype that have already initiated their developmental process.
Our analysis identified three branch points along the SGN developmental trajectory. The first of these occurs at E14 when immature SGNs split along two branches giving rise to Type 1B/C and Type 1A/Type 2 precursors. Two potential regulatory genes were noted at this split, Etv4 and Meis2. Although both of these genes have been reported to be expressed in the developing SGN previously (12, 33–35), their functions have not been studied. However, in other sensory systems, both genes play roles in neuronal specification and development. For example, in the dorsal root ganglia, Etv4 expression is restricted to a specific subtype of neurons and appears to interact with (Nerve Growth Factor) (NGF) signaling to mediate developmental processes such as axon outgrowth and guidance (36). Similarly, Meis2 is involved in both neurogenesis and the specification of specific neuron subtypes in the olfactory bulb (37). Based on these results and the timing of their expression in the SG, both genes seem likely to play roles in SGN subtype formation.
Another interesting aspect of the initial split between the Type 1B/C and Type 1A/2 branches is that many of the genes which mark the Type 1B/C branch, such as Pou4f1, appear to be initially expressed by all developing SGNs and then are downregulated in the Type 1A/2 branch (28, 38). By contrast, markers of the 1A/2 branch generally only turn on as 1A/2 SGN precursors appear. This suggests that the Type 1B/C branch represents a default phenotype and that suppression of this choice is required to move a neuron onto the Type 1A/Type 2 branch. Interestingly, Tle4, an early marker of Type 1A/2 SGN precursors, is a known transcriptional repressor (39, 40) and has been shown to compete with Meis2 and associate with Runx family genes, to repress downstream specification pathways (41–44), suggesting a similar role in the SG.
Recently, a similar analysis of SGN development was published by Petitpré and colleagues (45). While the collection and sequencing methods used in that study differed from ours, the results were remarkably similar, including the time point at which SGNs begin to specify into subtypes and the identification of predicted regulatory genes. The major difference between the two studies is the question of whether the Type 1B subtype originates from a shared lineage with the 1A precursors, as Petitpré et al. conclude, or with the Type 1C precursors, as we conclude. These two competing hypotheses will need to be tested using in vivo techniques such as lineage tracing in future studies.
The development and function of Type 2 SGNs are poorly understood due, in part, to the lack of reliable markers for this cell type, particularly during the prenatal and early postnatal periods. The analysis of the combined E18/P1 SGN dataset identified a number of genes, including Efna5, Th, and Tac1, that are restricted to Type 2 SGNs and can, potentially, be used to enhance studies of these neurons. In fact, one of the genes, Tac1, has recently been used as a cre-driver to label Type 2 SGNs (46), highlighting the usefulness of this approach. Interestingly, Efna5 had previously been proposed to be restricted to a subpopulation of Type 1 SGNs at P0 (19); however, the evidence we present here, along with findings from Petitpré et al. (12) in older animals, indicates that it is a very specific marker for neonatal Type 2 SGNs.
The trajectory analyses presented here also provide insights regarding the timing of Type 2 development. While Type 2 SGNs could be identified in cells collected at E16, those cells and Type 1A cells continue to share a high degree of transcriptional similarity into the early postnatal period. This result was surprising considering the significant differences between Type 1 and Type 2 SGNs in terms of morphology and function. It seemed more likely that Type 2s would separate along their own branch at an early developmental time point. In fact, previous studies suggested that Type 2 SGNs first begin to extend peripheral processes toward the HCs at E14 (13, 15). However, our results suggest that these cells are extending processes prior to commitment to their final subtype. Whether this means that some aspects of subtype specification may be determined through environmental interactions remains to be determined. Finally, our analysis identified several transcription factors, in particular Sox9, which show higher levels of expression and regulon activity during Type 2 development and, therefore, may play a role in Type 2 SGN specification.
It is important to note that there are inherent limitations in our study regarding the timing of the Type 2 branch point. Type 2s represent only 5 to 10% of the total adult SGN population and so make up a small proportion of our overall dataset. As a result, it is possible that the Type 2 split may occur earlier than E16 but the combination of a low number of these cells in our data set along with a high degree of transcriptional similarity between neurons at this time point prevented the identification of these cells. Therefore, while our current data support a split between the Type 1A and Type 2 subtypes at E16, verification of this event will require further studies with larger populations of SGNs.
Overall, the dataset presented here provides a wealth of information regarding the development of neuronal diversity in the SG. Future work will focus on understanding the exact roles of the candidate genes highlighted by this study.
Materials and Methods
All mice were maintained within the Porter Neuroscience Research Center Shared Animal Facility. All animal care and housing were conducted in accordance with the NIH guidelines for animal use (Protocols 1254 and 1262).
Single-Cell RNASeq Collections.
Tissue collections, SG dissection, dissociation, and single-cell capture using the 10X Genomics Chromium Controller were carried out using standard techniques (see SI Appendix, Methods). Libraries were sequenced on an Illumina NextSeq and were aligned to the Ensembl mouse MM10 assembly using Cell Ranger 3.0.1 analysis software (10X Genomics).
Data Analysis.
Data was processed and analyzed using the following R-based packages: Seurat (v3.2) (47), DoubletFinder (v2.0.3) (48), Harmony (v1.0) (49), Slingshot (v1.8) (17), tradeSeq (v1.4)(20), Monocle 3 (21, 50), and SCENIC (23). See SI Appendix, Methods for detailed methods of analysis.
Immunohistochemistry and smFISH.
Immunohistochemistry and RNAScope in situ hybridization were carried out using standard techniques (see SI Appendix, Methods). Probes and antibodies are listed in SI Appendix, Tables S1 and S2.
Subtype Marker Quantification.
The proportion of SGNs expressing a particular subtype marker were assessed by counting the number of positive SGNs for each marker against the total number of SGNs labeled with a pan neuronal marker—either Green Fluorescent Protein (GFP); using MAPTEGFP tissue TUJ1, or Neurofilament (using wildtype CD1 tissue). See SI Appendix, Methods for comprehensive methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Acknowledgments
We would like to thank the staff of the Shared Animal Facility, Porter Neuroscience Research Center for their excellent animal care and Dr. Lisa Goodrich for providing Neurog1CreERT2 transgenic mice. We would also like to thank the NIDCD/NIDCR Genomics and Computational Biology Core (GCBG) for outstanding support with sequencing and data analysis, further technical assistance from Hui Cheng (NIDCD Bioinformatics and Biostatistics Core), Likhitha Kolla for data analysis, and YeonSoo Kim for RNAScope experiments. We thank Dr. Michael Hoa and Dr. Francois Lallemend for providing comments on an earlier version of this manuscript. This research was supported by funds from the NIDCD Division of Intramural Research to M.W.K. (DC000059) and the GCBC (DC000086). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Author contributions
T.R.S. and M.W.K. designed research; T.R.S. performed research; T.R.S. analyzed data; and T.R.S. and M.W.K. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. L.V.G. is a guest editor invited by the Editorial Board.
Contributor Information
Tessa R. Sanders, Email: tessa.sanders@nih.gov.
Matthew W. Kelley, Email: kelleymt@nidcd.nih.gov.
Data, Materials, and Software Availability
Single-cell gene expression data have been deposited in the Gene Expression Omnibus data repository under accession code: GSE195500. Gene by cell expression matrix and data visualizations presented in this paper will be available through the gEAR Portal.
Supporting Information
References
- 1.Taberner A. M., Liberman M. C., Response properties of single auditory nerve fibers in the mouse. J. Neurophys. 93, 557–569 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Liberman M., Single-neuron labeling in the cat auditory nerve. Science 216, 1239–1241 (1982). [DOI] [PubMed] [Google Scholar]
- 3.Liberman M. C., Auditory-nerve response from cats raised in a low-noise chamber. J. Acoust. Soc. Am. 63, 442–455 (1978). [DOI] [PubMed] [Google Scholar]
- 4.Costalupes J. A., Young E. D., Gibson D. J., Effects of continuous noise backgrounds on rate response of auditory nerve fibers in cat. J. Neurophys. 51, 1326–1344 (1984). [DOI] [PubMed] [Google Scholar]
- 5.Shrestha B. R. et al. , Sensory neuron diversity in the inner ear is shaped by activity. Cell 174, 1229–1246.e1217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmiedt R., Mills J., Boettcher F., Age-related loss of activity of auditory-nerve fibers. J. Neurophys. 76, 2799–2803 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Sergeyenko Y., Lall K., Liberman M. C., Kujawa S. G., Age-related cochlear synaptopathy: An early-onset contributor to auditory functional decline. J. Neurosci. 33, 13686–13694 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatelin V., et al. , Cochlear implant outcomes in the elderly. Otol. Neurotol. 25, 298–301 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Flores E. N., et al. , A non-canonical pathway from cochlea to brain signals tissue-damaging noise. Curr. Biol. 25, 606–612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J. S., Vyas P., Glowatzki E., Fuchs P. A., Opposing expression gradients of calcitonin-related polypeptide alpha (Calca/Cgrpα) and tyrosine hydroxylase (Th) in type II afferent neurons of the mouse cochlea. J. Comp. Neurol. 526, 425–438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun S. et al. , Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell 174, 1247–1263.e1215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petitpré C., et al. , Neuronal heterogeneity and stereotyped connectivity in the auditory afferent system. Nat. Commun. 9, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koundakjian E. J., Appler J. L., Goodrich L. V., Auditory neurons make stereotyped wiring decisions before maturation of their targets. J. Neurosci. 27, 14078–14088 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Druckenbrod N. R., Goodrich L. V., Sequential retraction segregates SGN processes during target selection in the cochlea. J. Neurosci. 35, 16221–16235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coate T. M., Spita N. A., Zhang K. D., Isgrig K. T., Kelley M. W., Neuropilin-2/semaphorin-3F-mediated repulsion promotes inner hair cell innervation by spiral ganglion neurons. eLife 4, e07830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tessa R., Kelley Matthew W. Spiral ganglion subtype specification occurs prior to birth in the mouse https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE195500
- 17.Street K., et al. , Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vyas P., Wu J. S., Zimmerman A., Fuchs P., Glowatzki E., Tyrosine hydroxylase expression in type II cochlear afferents in mice. J. Assoc. Res. Otolaryngol. 18, 139–151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Defourny J., et al. , Ephrin-A5/EphA4 signalling controls specific afferent targeting to cochlear hair cells. Nat. Commun. 4, 1–12 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Trapnell C., et al. , The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao J., et al. , The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Berge K., et al. , Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aibar S., et al. , SCENIC: Single-cell regulatory network inference and clustering. Nat. Met. 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolla L., et al. , Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 11, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim W.-Y., et al. , NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development 128, 417–426 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evsen L., Sugahara S., Uchikawa M., Kondoh H., Wu D. K., Progression of neurogenesis in the inner ear requires inhibition of transcription by neurogenin1 and neurod1. J. Neurosci. 33, 3879–3890 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu M., et al. , Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 14, 2839–2854 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherrill H. E., et al. , Pou4f1 defines a subgroup of Type I spiral ganglion neurons and is necessary for normal inner hair cell presynaptic Ca2+ signaling. J. Neurosci. 39, 5284–5298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu P., et al. , Primary neural degeneration in the human cochlea: Evidence for hidden hearing loss in the aging ear. Neuroscience 407, 8–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tritsch N. X., Bergles D. E., Developmental regulation of spontaneous activity in the Mammalian cochlea. J. Neurosci. 30, 1539–1550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babola T. A., et al. , Homeostatic control of spontaneous activity in the developing auditory system. Neuron 99, 511–524.e515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babola T. A., et al. , Purinergic signaling controls spontaneous activity in the auditory system throughout early development. J. Neurosci. 41, 594–612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso M. B. D., et al. , Meis2 is required for inner ear formation and proper morphogenesis of the cochlea. Front. Cell Dev. Biol. 9, 679325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C., et al. , Comprehensive transcriptome analysis of cochlear spiral ganglion neurons at multiple ages. Elife 9, e50491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orvis J., et al. , gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat. Met. 18, 843–844 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontanet P., Irala D., Alsina F. C., Paratcha G., Ledda F., Pea3 transcription factor family members Etv4 and Etv5 mediate retrograde signaling and axonal growth of DRG sensory neurons in response to NGF. J. Neurosci. 33, 15940–15951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agoston Z., et al. , Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141, 28–38 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Deng M., Yang H., Xie X., Liang G., Gan L., Comparative expression analysis of POU4F1, POU4F2 and ISL1 in developing mouse cochleovestibular ganglion neurons. Gene Expr. Patterns 15, 31–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Li X., Ning F., Shang Y., Hu X., TLE4 acts as a corepressor of Hes1 to inhibit inflammatory responses in macrophages. Protein Cell 10, 300–305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milili M., Gauthier L., Veran J., Mattei M. G., Schiff C., A new Groucho TLE4 protein may regulate the repressive activity of Pax5 in human B lymphocytes. Immunology 106, 447–455 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agoston Z., Schulte D., Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development 136, 3311–3322 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Brady G., Whiteman H. J., Spender L. C., Farrell P. J., Downregulation of RUNX1 by RUNX3 requires the RUNX3 VWRPY sequence and is essential for Epstein-Barr virus-driven B-cell proliferation. J. Virol. 83, 6909–6916 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Javed A., et al. , Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF (alpha)/AML/PEBP2 (alpha)) dependent activation of tissue-specific gene transcription. J. Cell Sci. 113, 2221–2231 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Shin T. H., et al. , TLE4 is a critical mediator of osteoblast and Runx2-dependent bone development. Front. Cell Dev. Biol. 9, 671029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petitpré C., et al. , Single-cell RNA-sequencing analysis of the developing mouse inner ear identifies molecular logic of auditory neuron diversification. Nat. Commun. 13, 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nowak N., Wood M. B., Glowatzki E., Fuchs P. A., Prior acoustic trauma alters type II afferent activity in the mouse cochlea. eNeuro, 8, ENEURO.0383-21.2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuart T., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korsunsky I., et al. , Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Met. 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu X., et al. , Reversed graph embedding resolves complex single-cell trajectories. Nat. Met. 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Data Availability Statement
Single-cell gene expression data have been deposited in the Gene Expression Omnibus data repository under accession code: GSE195500. Gene by cell expression matrix and data visualizations presented in this paper will be available through the gEAR Portal.