Abstract
Cheese microbiota contributes to various biochemical processes that lead to the formation of volatile compounds and the development of flavour during ripening. Nonetheless, the role of these microorganisms in volatile aroma compounds production is little understood. This work reports for the first time the dynamics and odour impact of volatile compounds, and their relationship to microbial shifts during the ripening of a raw ewe milk-derived cheese (Idiazabal). By means of SPME-GC-MS, 81 volatile compounds were identified, among which acids predominated, followed by esters, ketones and alcohols. The ripening time influenced the abundance of most volatile compounds, thus the moments of greatest abundance were determined (such as 30–60 days for acids). Through Odour Impact Ratio (OIR) values, esters and acids were reported as the predominant odour-active chemical families, while individually, ethyl hexanoate, ethyl 3-methyl butanoate, ethyl butanoate, butanoic acid or 3-methyl butanal were notable odorants, which would provide fruity, rancid, cheesy or malt odour notes. Using a bidirectional orthogonal partial least squares (O2PLS) approach with Spearman's correlations, 12 bacterial genera were reported as key bacteria for the volatile and aromatic composition of Idiazabal cheese, namely Psychrobacter, Enterococcus, Brevibacterium, Streptococcus, Leuconostoc, Chromohalobacter, Chryseobacterium, Carnobacterium, Lactococcus, Obesumbacterium, Stenotrophomonas and Flavobacterium. Non-starter lactic acid bacteria (NSLAB) were highly related to the formation of certain acids, esters and alcohols, such as 3-hexenoic acid, ethyl butanoate or 1-butanol. On the other hand, the starter LAB (SLAB) was related to particular ketones production, specifically 3-hydroxy-2-butanone; and environmental and/or non-desirable bacteria to certain ketones, hydrocarbons and sulphur compounds formation, such as 2-propanone, t-3-octene and dimethyl sulphone. Additionally, the SLAB Lactococcus and Psychrobacter, Brevibacterium and Chromohalobacter were described as having a negative effect on aroma development caused by NSLAB and vice versa. These results provide novel knowledge to help understand the aroma formation in a raw ewe milk-derived cheese.
Keywords: Volatile composition, Odour-active compounds, Microbiota, Correlation analysis, O2PLS, CCorA
Graphical abstract
Highlights
-
•
Volatile profile evolution during ripening of Idiazabal cheese was studied.
-
•
81 volatile compounds were identified in Idiazabal cheese, 18 not described so far.
-
•
Esters and acids were predominant odour-active compounds.
-
•
Bacterial genera were highly correlated to volatile and odour-active compounds.
-
•
5 LAB and 7 environmental/non-desirable bacteria were key bacteria for cheese aroma.
1. Introduction
Idiazabal cheese is a traditional semi-hard or hard cheese from the Basque Country (southwestern Europe), which is manufactured from raw milk of Latxa and/or Carranzana autochthonous breed sheep. Its production is regulated by its Protected Designation of Origin (PDO) since 1996 (Official Journal of the European Communities, 1996). The Idiazabal cheese making process is strictly regulated and establishes a minimum ripening time of 60 days (Boletín Oficial del Estado, 1993). Nonetheless, producers may employ different flock management and cheese making practices that, in turn, affect the quality of the milk and final cheese. The most notable differences are related to the flock management and grazing practices, such as valley or mountain grazing (Abilleira et al., 2010a; Valdivielso et al., 2016); the use of artisanal or commercial rennet (Virto et al., 2003) or the technological conditions used for cheese making and ripening (Aldalur et al., 2021).
During cheese ripening, various biochemical processes take place that are responsible for the synthesis of volatile compounds and, consequently, flavour development (Fox et al., 2017; Thierry et al., 2017). Volatile compounds originate primarily from three groups of metabolic pathways: the metabolism of residual lactose, lactate and citrate; the lipolysis and subsequent metabolism of the released free fatty acids (FFAs); and the proteolysis and metabolism of the resulting peptides and free amino acids (FAAs) (Le Quéré and Buchin, 2022). It has been reported that ripening time affects the volatile composition of Idiazabal cheese (Barron et al., 2005a, 2007). However, these studies only focused on certain time points of ripening and did not comprehensively characterize how volatile chemical families and individual compounds evolve throughout this process. This aspect is of special interest since the presence or absence, abundance and proportions of each compound characterize the type of cheese and its aroma (Le Quéré and Buchin, 2022). In fact, the imbalance or excessive concentrations of several compounds have been related to off-flavours (Zabaleta et al., 2016). Moreover, it is generally recognized that not all volatile compounds contribute to cheese aroma (Starowicz, 2021), that is to say, not all are odour-active compounds (Fox et al., 2017; Natrella et al., 2020). Despite the large work done to elucidate the flavour and key aromatic compounds of Idiazabal cheese (Barron et al., 2005a, 2007; Abilleira et al., 2010b; Valdivielso et al., 2016), it is unknown how the odour-impact of volatile compounds evolve during ripening.
Cheese microbiota contributes to numerous biochemical reactions involved in the formation of flavour compounds (Bertuzzi et al., 2018; Le Quéré and Buchin, 2022) and, indeed, has been described as responsible for the particular sensory properties of raw milk cheeses, such as Manchego or Erronkari/Roncal (Ballesteros et al., 2006). The microbiota of Idiazabal cheese has recently been characterized by means of high-throughput sequencing (HTS) (Santamarina-García et al., 2022a), allowing a better understanding of microbial communities than culture-based methods (Yeluri Jonnala et al., 2018). Overall, it has been observed that the ripening time modulates the bacterial composition. Specifically, the starter LAB (SLAB) (Lactococcus) predominates up to 30 or 60 days and then, non-starter LAB (NSLAB) (Lactobacillus, Leuconostoc, Enterococcus, Streptococcus and Carnobacterium) proliferate, while the relative abundance of non-desirable and/or environmental bacteria is reduced (such as Pseudomonas, Staphylococcus or Chromohalobacter). Moreover, bacterial composition differs largely among producers and several bacterial genera not reported previously in any raw ewe milk and derived cheeses have been identified (such as Buttiauxella or Obesumbacterium) (Santamarina-García et al., 2022a).
In recent times, several studies that attempt to elucidate the relationship between microbial communities and volatile compounds formation in fermented products have been published (Zhong et al., 2021; Xia et al., 2022). However, few studies have focused on cheese (Zheng et al., 2018; Chen et al., 2021a) and information on raw ewe milk-derived cheeses is scarce (Cardinali et al., 2021). In addition, although the aim of these studies was to understand the association between microbial communities and the aroma formation in fermented products, only one work has analysed the correlation to odour-active volatile compounds in fermented milk (Xia et al., 2021). In order to elucidate such correlations, complex chemometric approaches are needed and the multivariate bidirectional orthogonal partial least squares (O2PLS) (Trygg and Wold, 2003) is one of the most useful approaches (Bouhaddani et al., 2016). Nonetheless, its combination with other parameters, such as correlation coefficients, is the most appropriate approach (Galindo-Prieto et al., 2014).
Therefore, this study aimed to (1) characterize how volatile compounds and their odour impact evolve during the ripening of raw ewe milk-derived Idiazabal cheese, in order to (2) investigate how they relate to shifts during ripening of the unique microbiota found in this PDO cheese and (3) highlight the relevance of appropriate advanced statistical approaches to obtain insights and improve cheese quality. Moreover, the potential differences among producers producing the same type of cheese were also analysed. To the best of our knowledge, no such a comprehensive study has been conducted to date on any type of cheese.
2. Materials and methods
2.1. Cheese sampling
Four artisanal Idiazabal PDO cheese producers (identified as A, B, C and D) were selected for the sampling. Idiazabal cheeses were produced from raw milk of Latxa sheep, each producer employing the milk of its own flock and following the specifications issued by Idiazabal Designation of Origin Regulatory Board (Boletín Oficial del Estado, 1993). Briefly, the milk was tempered to 25 °C and the mesophilic lyophilized starter culture Choozit MM 100 LYO 50 DCU (mixture of Lactococcus lactis subsp. lactis, Lactococcus lactis subsp. cremoris and Lactococcus lactis subsp. lactis biovar. diacetylactis) (DuPont NHIB Ibérica S.L., Barcelona, Spain) was added. Milk was coagulated using artisanal rennet or commercial rennet NATUREN® 195 Premium (Chr. Hansen Holding A/S, Hørsholm, Denmark). Cheese ripening was carried out in chambers maintained at 80–95% relative humidity and 8–14 °C temperature. Cheese samples for analysis were collected in duplicate at six time points during ripening (1, 7, 14, 30, 60 and 120 days) (n = 48). Samples were collected and transported to the laboratory under refrigerated conditions (3 °C) and then stored in a freezer (−80 °C). Before analysis, cheese samples were defrosted for 24 h at 5 °C and kept at room temperature for 1 h.
2.2. Solid-phase microextraction (SPME) methodology
The preparation of Idiazabal cheese samples and the SPME procedure was carried out as previously described (Valdivielso et al., 2016). Briefly, removing the rind, 15 g of cheese were ground together with 20 g of anhydrous Na2SO4 (reagent grade, Scharlab, Barcelona, Spain). Afterwards, 5 μL of cyclohexanone (0.5 μg/L) (≥99.5%, Sigma-Aldrich, Madrid, Spain) were added as internal standard (IS) solution and it was re-homogenized. The extraction of the volatile compounds was carried out by adding 2.5 g of the aforementioned mixture to a 10 mL amber vial (Agilent Technologies). The vials were sealed with a PTFE septa and a steel magnetic cap (18 mm PTFE/SIL, Agilent Technologies) prior to placing them in the sample tray at room temperature for analysis. SPME procedure was carried out employing a PAL RSI 85 autosampler (CTC CombiPAL, Zwingen, Switzerland) equipped with a temperature-controlled air incubator. After 15 min of pre-equilibration time at 60 °C, volatile compounds were trapped onto a 50/30 μm DVB/Carboxen/PDMS fibre (57298-U, Supelco, Madrid, Spain) at 60 °C for 30 min.
2.3. Gas chromatography–mass spectrometry (GC-MS) analysis
Volatile compounds were analysed in a 7820A Gas Chromatograph system, equipped with a split/splitless injector and coupled to a 5975 series MS detector (Agilent Technologies), essentially as previously described (Barron et al., 2005b). Volatile compounds were desorbed from the fibre in the front injection port for 10 min at 240 °C in splitless mode (split valve was opened at 200 mL/min after 10 min of the injection). Then, volatile compounds were separated in a Supelcowax-10 fused silica capillary column (59.5 m length, 0.25 mm i.d.; 0.25 μm film thickness) (Supelco, Madrid, Spain) and helium (99.999% purity, Air liquid, Madrid, Spain) was employed as the carrier gas at a constant pressure of 16 psi. Oven was held at 40 °C for 10 min, then raised at a rate of 5 °C/min until 110 °C, increased again at 10 °C/min until 240 °C, and finally held at 240 °C for 15 min. Volatile compounds were then transferred to the MS detector through a transfer line at 280 °C and MS detector operated at 150 °C in full scan mode with 70 eV as total ion current.
The chromatographic data obtained were analysed with MSD ChemStation Data Analysis version 5.52 (Agilent Technologies). Volatile compounds were tentatively identified by comparing their mass spectra (match factor >800) with those of the National Institute of Standards and Technology spectra library (NIST version 2.0, Gaithersburg, USA). Mean linear retention index (LRI) value of each chromatographic peak was calculated from the analysis of each Idiazabal cheese sample (4 replicates) and the saturated alkanes standard mixture (3 replicates × 2 times through the experiment) (certified reference material 49452-u, C7-C40, Sigma-Aldrich). Then, positive identification of volatile compounds was carried out by comparing the LRI and mass spectra with those of commercially available high purity standards (≥90%, supplied by Sigma-Aldrich and Honeywell Fluka, Madrid, Spain). The limit of detection (LOD) was established at twice the noise (arbitrary units) of the chromatogram. Peak area quantification was carried out by the total ion current (TIC) and the content of volatile compounds was expressed as relative abundance (peak area in arbitrary units relative to the internal standard (IS)), as expressed by the following equation:
The mean abundance of the volatile compounds of each cheese sample was obtained from the peak areas (>LOD) of each compound, as long as they were detected in at least two of the four sample replicates. The relative abundance of volatile compounds in cheese samples was expressed as mean ± standard deviation.
2.4. Statistical analysis of volatile composition
Different packages and software were employed for data analysis. The IBM SPSS statistical package version 26.0 (IBM SPSS Inc., Chicago, 2019) was used for data preparation and analysis. Kruskal-Wallis analysis of variance with Bonferroni correction was performed in IBM SPSS package to determine the effect of ripening time and producer factors on volatile chemical families and individual compounds. Hierarchical Clustering Analysis (HCA) of volatile compounds was performed using log transformed, when necessary, and Unit Variance (UV) scaled data and plotted into a heat-map with RStudio version 1.3.959 and R version 3.6.3 (R Core Team, 2020) with “gplots” package (Warnes et al., 2020). The aim was to analyse volatile compounds groupings during ripening time. Then, trends along ripening were analysed through a Principal Component Analysis (PCA) using the SIMCA software version 15.0.0.4783 (Umetrics AB, Umeå, Sweden). The number of principal components (PC) was determined by eigenvalues (greater than 2.5) and cross validation. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) was also performed in SIMCA, in order to analyse whether samples differed according to the producer. Variable Influence on Projection (VIP) values and loadings weights were used to analyse the importance of each volatile compound in the model.
2.5. Calculation of Odour Impact Ratio
To obtain a measure of the odour impact of each volatile compound detected in Idiazabal cheese samples, Odour Impact Ratio (OIR) was calculated as Abilleira et al. (2010b) described, with minor modifications, and as the following equation expresses:
Compounds with OIR values greater than 1 indicate that the abundance of the volatile compound was higher than the odour threshold (OT) and thus, were odour-active compounds. OTs are usually calculated in water, however, values could change depending on the matrix (van Gemert, 2011). Therefore, available OT values measured in cheese were taken to avoid the matrix effect (listed in Table 2).
Table 2.
Estimated OIR values throughout ripening time (1, 7, 14, 30, 60 and 120 days) and sensory description of each volatile compound detected in Idiazabal cheese samples (n = 48).
| Volatile compounds | OTa |
OIRbvalues during ripening (days) |
Described odour notes | Referencesc | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 14 | 30 | 60 | 120 | ||||
| Acids | |||||||||
| Acetic acid | 22000 | <1 | Sour, vinegar, pungent, acid | 1, 2, 3 | |||||
| n- Butanoic acid | 50 | 33.6 | 138 | 186 | 213 | 241 | 132 | Rancid, cheesy, putrid, sharp, sour, sweat | 1, 2, 3 |
| n- Pentanoic acid | 137 | <1 | <1 | 8.28 | 30.4 | <1 | 3.13 | Sweat, putrid, sharp, sour, cheesy, burned | 1, 3 |
| n- Hexanoic acid | 290 | 10.9 | 43.5 | 57.9 | 84.6 | 88.4 | 53.7 | Sweat, sour, pungent, goat, rancid, cheesy, foot, faecal | 1, 2, 3 |
| (E)-3-Hexenoic acid | – | – | Pungent, sweat, vinegar, cheesy, green | 4, 5 | |||||
| n- Heptanoic acid | 3000 | <1 | Sweat, rancid, faecal | 2, 3 | |||||
| n- Octanoic acid | 450 | 2.61 | 8.28 | 11.5 | 17.7 | 21.4 | 14.1 | Sweat, goat, soapy, waxy, musty, faecal, dust, cleaner | 1, 2, 3 |
| n- Nonanoic acid | 4000 | <1 | Faecal, burned, fruity | 3 | |||||
| n- Decanoic acid | 10000 | <1 | Fatty, soapy, dust, waxy, burned | 2, 3 | |||||
| Alcohols | |||||||||
| Ethanol | 8 | <1 | Alcohol, winey, sweet, ethereal | 1 | |||||
| 1-Propanol | 5700 | <1 | Alcohol, winey, sweet | 1 | |||||
| 1-Butanol | 800 | <1 | Winey, sweet, fruity, fusel oil | 1 | |||||
| 3-Methyl-1-butanol | 250 | <1 | Alcohol, winey, fruity, burned, herbal | 1, 3 | |||||
| 1-Pentanol | 4000 | <1 | Alcohol, sharp, harsh | 1 | |||||
| 1-Hexanol | 50 | <1 | Winey, oily, flower, fruity, | 1, 2 | |||||
| 2-Ethyl-1-hexanol | 830 | <1 | Green, citrus, floral, oily, sweet | 5, 6 | |||||
| 1-Octanol | 42 | <1 | Fatty, waxy, citrus, oily, walnut, moss, chemical, metal, burned | 7, 8 | |||||
| 2-Butanol | 59 | <1 | <1 | <1 | 1.50 | 13.8 | 7.19 | Winey, alcohol, sweet, fruity, fusel oil | 1, 2 |
| 2-Pentanol | 41 | <1 | Alcohol, slightly green, winey, fruity | 1, 2 | |||||
| 3-Methyl-2-butanol | 420 | <1 | Fruity | 5 | |||||
| 2-Hexanol | 82 | <1 | Herbal, green, chemical, winey, fruity, fatty, terpenic, cauliflower | 5, 9 | |||||
| 2-Heptanol | 70 | <1 | Earthy, sweet, fruity, oily, green, herbal | 1, 2, 3 | |||||
| 2-Nonanol | 75 | <1 | Fatty, mild, green, melon, coconut | 7, 9, 10 | |||||
| 2,3-Butanediol | 11000 | <1 | Fruity | 3 | |||||
| Menthol | 920 | <1 | Minty, cooling | 11 | |||||
| 2-Methyl-3-pentanol | 420 | <1 | Grilled, bread | 3 | |||||
| 2-Propen-1-ol | 5000 | <1 | Pungent, mustard | 5 | |||||
| 2-Nonen-1-ol | 130 | <1 | Green, fatty, melon | 5 | |||||
| (E)-4-Hexen-1-ol | 100 | <1 | Green, herbal, musty, tomato | 5 | |||||
| 6-Heptene-2,4-diol | – | – | – | ||||||
| Aldehydes | |||||||||
| Hexanal | 5 | <1 | Green, herbal, sharp | 1, 3 | |||||
| Heptanal | 3 | 1.21 | <1 | <1 | <1 | <1 | <1 | Fatty, fruity, soapy, green, waxy, herbal | 10, 12, 13, 14, 15 |
| Octanal | 1.5 | <1 | Fruity, green, citrus, fatty, fatty-fruity, lemon | 10, 12, 13, 14, 15, 16 | |||||
| Nonanal | 10 | <1 | 1.42 | 1.52 | 2.42 | 1.44 | <1 | Sweet, fatty-floral, floral-waxy, rosy, citrus, peas, plastic | 1, 2, 3 |
| 3-Methyl-butanal | 0.200 | 15.0 | 19.8 | 6.46 | 12.2 | 42.0 | 217 | Malt, chocolate, toffee, green | 1 |
| Benzaldehyde | 325 | <1 | Almond, cherry stone, burned sugar | 7, 14, 17 | |||||
| Benzeneacetaldehyde | 5.50 | <1 | <1 | 1.45 | <1 | <1 | <1 | Floral, honey, daisy, green, violet-like, hyacinth, styrene, rosy, dry fruit, sweet | 6, 10, 14, 16, 18, 19 |
| Ketones | |||||||||
| 2-Propanone | 840 | <1 | Sweet, fruity, ethereal, nauseating | 1 | |||||
| 2-Butanone | 30 | <1 | <1 | <1 | 65.4 | 48.8 | 8.09 | Sweet, ethereal, slightly nauseating | 1 |
| 2-Pentanone | 70000 | <1 | Sweet, fruity, ethereal | 1, 2 | |||||
| 2-Heptanone | 5 | 3.51 | 6.92 | 7.93 | 32.5 | 16.4 | 34.0 | Musty, blue cheese, pungent, soapy, flower | 1, 2, 3 |
| 2-Octanone | 41 | <1 | Fruity, musty, floral, green, herbal, mouldy, humidity, soapy, musty, blue-cheese | 6, 8, 9, 10, 18 | |||||
| 2-Nonanone | 5 | 2.48 | 5.75 | 6.91 | 18.2 | 12.9 | 19.8 | Musty, floral, fruity, soapy | 1, 2 |
| 8-Nonen-2-one | – | – | Blue cheese, fruity baked | 6, 8, 10 | |||||
| 2-Undecanone | 6.2 | <1 | <1 | <1 | 1.14 | 1.06 | <1 | Fruity, herbal | 2 |
| (E,E)-6,10-Dimethyl-5,9-dodecadien-2-one | – | – | – | ||||||
| 2,3-Butanedione | 3 | 20.1 | 25.6 | 28.7 | 8.29 | 7.56 | <1 | Buttery, sweet, cream, caramel | 7, 10, 12, 13, 14, 15, 16, 20 |
| 3-Hydroxy-2-butanone | 850 | <1 | Buttery, flower | 1, 3 | |||||
| 2-Hydroxy-3-pentanone | 2500 | <1 | Fatty, truffle, earthy, nutty | 5, 6, 10 | |||||
| Esters | |||||||||
| Methyl hexanoate | 390 | <1 | Citrus, pineapple, ethereal | 9, 16 | |||||
| Methyl 4-methyl pentanoate | – | – | Strawberry, roasted cocoa | 21, 22 | |||||
| Ethyl butanoate | 1 | 4.32 | 27.0 | 53.8 | 78.3 | 62.8 | 288 | Fruity, apple, pineapple, banana, sweet, flower | 1, 2, 3 |
| Ethyl 2-methyl butanoate | – | – | Sweet, fruity | 10, 13 | |||||
| Ethyl 3-methyl butanoate | 0.1 | <1 | <1 | <1 | <1 | 442 | 339 | Fruity, olive, sweet | 2, 3 |
| Ethyl pentanoate | 8.7 | <1 | Fruity, sweet, acid, apple, pineapple, green, berry, tropical | 5, 15, 18 | |||||
| Ethyl hexanoate | 1 | 18.7 | 183 | 343 | 453 | 480 | 216 | Fruity, apple, pineapple, banana, mouldy, flower | 1, 2, 3 |
| Ethyl hex-4-enoate | – | – | – | ||||||
| Ethyl heptanoate | 2.2 | <1 | <1 | 2.17 | 2.62 | 1.75 | 15.4 | Fruity, pineapple, sweet, banana, berry, cognac and slightly green with a seedy nuance | 5, 9, 18 |
| Ethyl octanoate | 65 | <1 | <1 | 1.10 | 1.85 | 1.55 | <1 | Fruity, winey, pineapple, apricot, burned, earthy, flower | 1, 2, 3 |
| Ethyl nonanoate | 377 | <1 | Cheesy, fruity | 3 | |||||
| Ethyl decanoate | 23 | <1 | <1 | 19.9 | 43.6 | 3.82 | 13.9 | Fruity, winey, fatty, flower, humidity | 2, 3 |
| Ethyl dodecanoate | 400 | <1 | <1 | <1 | <1 | <1 | 3.23 | Flower, vanilla | 3 |
| Propyl butanoate | 124 | <1 | Fruity, sweet, pineapple, banana | 1, 2 | |||||
| Propyl hexanoate | – | – | Fruity, pineapple, blackberry, fatty | 1, 2 | |||||
| Propyl octanoate | – | – | Coconut | 5 | |||||
| Butyl butanoate | 100 | <1 | Fruity, pineapple, banana, sweet, fatty | 1, 2 | |||||
| Butyl hexanoate | 700 | <1 | Flower, fruity, pineapple, mouldy | 2, 3 | |||||
| Pentyl butanoate | 210 | <1 | Sweet, fruity, banana, pineapple, cherry, tropical | 5 | |||||
| Hexyl hexanoate | 6400 | <1 | Green, sweet, waxy, fruity with tropical and berry notes | 5 | |||||
| 1-Methylpropyl butanoate | – | – | Sweet, fruity, pineapple, rum, cherry, apple, overripe fruit | 5 | |||||
| 2-Methylpropyl hexanoate | – | – | Apple | 1 | |||||
| 2-Propenyl hexanoate | 200 | <1 | Pineapple, fatty-fruity | 1 | |||||
| 3-Methylbutyl hexanoate | 320 | <1 | Fruity, sweet, pineapple with a slightly pungent sour cheesy note | 5 | |||||
| Hydrocarbons | |||||||||
| Heptane | 950 | <1 | Solvent, sweet-ethereal, diffusive | 1 | |||||
| 1,3-Pentadiene | 2500 | <1 | Plastic, paint, kerosene | 23 | |||||
| t-3-Octene | – | – | Sharp, herbal, leather-like | 2 | |||||
| 1,3-Octadiene | 5600 | <1 | Woody-moss | 13 | |||||
| Toluene | 1 | 20.2 | 42.9 | 24.9 | 112 | 19.9 | 68.0 | Fruity, sweet-gassy, hydrocarbon | 1 |
| Sulphur compounds | |||||||||
| Dimethyl sulphide | 1.2 | 1.29 | <1 | <1 | 14.3 | <1 | <1 | Unpleasant wild radish, cabbage, sulphurous, pomegranate, corn, earthy, rancid | 10, 12, 15, 20 |
| Dimethyl sulphone | 2.5 | 10.0 | 3.22 | 2.96 | 24.5 | 3.10 | 2.71 | Sweet, flower, sulphurous, hot milk, burned | 9, 10, 14 |
| Terpenes | |||||||||
| D-Limonene | 70 | <1 | Grass | 3 | |||||
OT expressed as μg/L or μg/kg. Data are taken from the following: Abilleira et al. (2010b); Wang et al. (2021); Natrella et al. (2020); Sarhir et al. (2021); Majcher and Jeleń (2011); Kubícková and Grosch (1998); Attaie (2009); van Gemert (2011).
OIR values calculated as mean relative abundance from 4 producers (A, B, C and D) at each ripening time (1, 7, 14, 30, 60, 120 days)/odour threshold. OIR with values higher than 1 are bold coloured.
Described odour notes taken from: (1) Barron et al. (2005a); (2) Abilleira et al. (2010b); (3) Zabaleta et al. (2016); (4) Câmara et al. (2020); (5) The Good Scents Company Information System (2021); (6) Poveda et al. (2008); (7) Juric et al. (2003); (8) Jung et al. (2013); (9) Moio et al. (2000); (10) Curioni and Bosset (2002); (11) Zhang et al. (2022); (12) Natrella et al. (2020); (13) Sympoura et al. (2009); (14) Wang et al. (2020); (15) Karagul Yuceer et al. (2009); (16) Whetstine et al. (2005); (17) Chen et al. (2021b); (18) Qian and Reineccius (2002); (19) Fox et al. (2017); (20) Boscaini et al. (2003); (21) Campo et al. (2006); (22) Takeoka et al. (1995); (23) Horwood et al. (1981).
2.6. Correlation between bacterial communities and volatile compounds
An HTS analysis was performed to characterize the bacterial communities and the shifts that occur during ripening of the collected Idiazabal cheese samples, as described by Santamarina-García et al. (2022a). Briefly, 10 g of cheese were suspended in 90 mL of 2% (w/v) sterile sodium citrate (pH 8.0), and homogenized six times (each for 20 s ON and 10 s OFF) in a stomacher (Masticator Basic 400; IUL Instruments, Königswinter, Germany). Then, 1.5 mL of the resulting suspension was centrifuged (8000×g for 10 min at 4 °C) and the fat-containing supernatant was discarded. The obtained pellet was resuspended in 600 μL of sodium citrate, and centrifuged three times (8000×g for 10 min at 4 °C). DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA) and the 16S rRNA gene library was prepared using Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA, USA). The V3–V4 regions of the 16S rRNA gene were amplified by PCR (forward primer: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′; reverse primer: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′), as described by Klindworth et al. (2013). Then, 16S rRNA gene sequencing was performed on the Illumina MiSeq platform using the MiSeq Reagent Kit v3 (2 × 300 bp) (Illumina Inc.). MiSeq Reporter software was used for quality filtering and trimming of raw reads and taxonomic classification was performed using the MG-RAST web data analysis tool (Meyer et al., 2008), based on the Silva SSU database (Pruesse et al., 2007). Bacterial abundance was presented as relative abundance (%) based on the identified sequences.
To study the relationship between the identified bacterial genera and the dynamics of volatile aroma compounds, an O2PLS approach was applied to log transformed, when necessary, and UV scaled data in SIMCA. Main bacterial genera were selected as X-variables and volatile compounds as Y-variables. The model was validated, among others, by R2 and Q2 values, Permutation test or Inner Relation plot. The key bacterial genera for the volatile composition of Idiazabal cheese were identified based on VIP values and loading weights, together with Spearman's Rank Correlations calculated in SPSS and interpreted in a heat map with an HCA analysis performed in R with “pheatmap” package (Kolde, 2019). The resulted correlations were verified by a Canonical Correlation Analysis (CCorA) multivariate statistical approach performed in R with “vegan” package (Oksanen et al., 2020).
3. Results and discussion
3.1. Changes in volatile composition
Table 1 summarizes the average relative abundance of the individual volatile compounds identified during the ripening time of the collected Idiazabal cheese samples from the four producers (A, B, C and D). The average relative abundance percentage of the identified volatile chemical families is reported in Supplementary Table 1. A total of 81 volatile compounds were identified, which could be grouped into seven families according to their chemical structure, namely acids (9 individual acids), alcohols (21), aldehydes (7), ketones (12), esters (24), hydrocarbons (5), sulphur compounds (2) and terpenes (1). Esters and alcohols constituted the highest number of individual compounds, as previously reported for Idiazabal cheese (Barron et al., 2005a, 2007; Abilleira et al., 2010b; Valdivielso et al., 2016), although a higher number of alcohols were identified in the present study. Overall, 18 individual compounds not reported previously in Idiazabal cheese were detected. Compared to the latest works on raw ewe milk-derived cheeses, a greater number of individual compounds have been identified (Gaglio et al., 2019a; Cardinali et al., 2021).
Table 1.
Average relative abundance ± standard deviation of individual volatile compounds identified throughout ripening (1, 7, 14, 30, 60 and 120 days) of Idiazabal cheese samples from 4 producers (n = 48).
| ID | LRIa | Volatile compounds |
Ripening time (days)b |
P-valuec |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 14 | 30 | 60 | 120 | RT | P | |||
| Acids | ||||||||||
| C1 | 1647 | Acetic acidd | 8.06 ± 22.8 | 353 ± 542 | ND | 24.3 ± 36.4 | 61.6 ± 101 | 120 ± 243 | NS | NS |
| C2 | 1698 | n- Butanoic acidd | 1681 ± 1594 | 6884 ± 5928 | 9289 ± 7950 | 10674 ± 6878 | 12053 ± 7633 | 6624 ± 8670 | * | ** |
| C3 | 1880 | n- Pentanoic acidd | 10.5 ± 14.4 | 55.1 ± 50.4 | 1135 ± 3050 | 4166 ± 11566 | 48.1 ± 57.5 | 428 ± 942 | NS | * |
| C4 | 1921 | n- Hexanoic acidd | 3168 ± 2911 | 12617 ± 9968 | 16782 ± 13507 | 24546 ± 15744 | 25632 ± 19485 | 15567 ± 22362 | NS | *** |
| C5 | 2067 | (E)-3-Hexenoic acidd,f | ND | ND | ND | 682 ± 1692 | 196 ± 365 | 2682 ± 5030 | NS | *** |
| C6 | 2105 | n- Heptanoic acidd | ND | 102 ± 113 | 435 ± 817 | 183 ± 199 | 192 ± 210 | 151 ± 237 | NS | ** |
| C7 | 2192 | n- Octanoic acidd | 1174 ± 1231 | 3727 ± 2899 | 5186 ± 4099 | 7982 ± 5158 | 9623 ± 8291 | 6362 ± 7524 | * | *** |
| C8 | 2395 | n- Nonanoic acidd | ND | ND | ND | ND | ND | 307 ± 617 | ||
| C9 | 2396 | n- Decanoic acidd | 490 ± 502 | 1439 ± 1119 | 2185 ± 1695 | 4073 ± 2525 | 4441 ± 3872 | 2959 ± 3862 | * | ** |
| Total straight-chain acids | 6532 ± 6275 | 25177 ± 20619 | 35012 ± 31119 | 52331 ± 43801 | 52247 ± 40014 | 35200 ± 49486 | * | *** | ||
| Alcohols | ||||||||||
| C10 | 936 | Ethanold | 171 ± 58.5 | 696 ± 931 | 743 ± 1065 | 1288 ± 2102 | 443 ± 505 | 145 ± 154 | NS | NS |
| C11 | 1040 | 1-Propanold | ND | ND | ND | ND | ND | 287 ± 723 | ||
| C12 | 1145 | 1-Butanole | ND | 4.76 ± 8.93 | 10.6 ± 19.8 | 10.6 ± 19.6 | 20.3 ± 31.4 | 22.4 ± 24.9 | * | *** |
| C13 | 1205 | 3-Methyl-1-butanole | 7.46 ± 15.1 | 34.1 ± 29.8 | 58.2 ± 87.8 | 52.3 ± 55.9 | 29.5 ± 16.7 | 6.51 ± 7.76 | * | *** |
| C14 | 1249 | 1-Pentanole | 7.76 ± 6.20 | 4.60 ± 8.55 | ND | 19.4 ± 52.6 | ND | ND | ** | NS |
| C15 | 1351 | 1-Hexanole | ND | ND | 8.12 ± 15.5 | 8.69 ± 16.1 | 17.4 ± 20.8 | 26.0 ± 20.6 | *** | NS |
| C16 | 1482 | 2-Ethyl-1-hexanold | 9.66 ± 7.22 | 18.2 ± 22.2 | 5.41 ± 5.99 | 62.9 ± 147 | 15.8 ± 8.09 | 6.71 ± 9.78 | NS | * |
| C17 | 1552 | 1-Octanole | ND | ND | ND | ND | ND | 3.15 ± 4.24 | ||
| Total primary alcohols | 196 ± 87.1 | 757 ± 1001 | 825 ± 1194 | 1442 ± 2393 | 526 ± 582 | 497 ± 945 | NS | NS | ||
| C18 | 1025 | 2-Butanole | ND | ND | ND | 88.5 ± 124 | 812 ± 1102 | 424 ± 334 | *** | NS |
| C19 | 1120 | 2-Pentanole | ND | ND | ND | 1.60 ± 2.97 | 3.41 ± 8.25 | 27.0 ± 18.2 | *** | NS |
| C20 | 1122 | 3-Methyl-2-butanold,f | ND | ND | ND | ND | 4.41 ± 8.43 | 7.14 ± 13.3 | NS | NS |
| C21 | 1218 | 2-Hexanole | ND | ND | ND | ND | ND | 1.44 ± 2.75 | ||
| C22 | 1316 | 2-Heptanold | ND | ND | ND | 30.7 ± 82.2 | 3.73 ± 6.99 | 12.2 ± 19.4 | * | NS |
| C23 | 1509 | 2-Nonanole | ND | ND | ND | ND | ND | 7.29 ± 11.4 | ||
| C24 | 1640 | 2,3-Butanediold | ND | 27.1 ± 76.7 | ND | 32.1 ± 59.6 | ND | ND | NS | * |
| C25 | 1646 | Menthold,f | 6.65 ± 6.63 | 13.0 ± 10.4 | 27.5 ± 41.2 | 11.2 ± 12.6 | ND | ND | ** | ** |
| Total secondary alcohols | 6.65 ± 6.63 | 40.1 ± 87.1 | 27.5 ± 41.2 | 164 ± 281 | 823 ± 1126 | 479 ± 399 | *** | NS | ||
| C26 | 1352 | 2-Methyl-3-pentanold | 9.41 ± 17.4 | ND | ND | ND | ND | ND | ||
| Total tertiary alcohols | 9.41 ± 17.4 | ND | ND | ND | ND | ND | ||||
| C27 | 1115 | 2-Propen-1-old,f | ND | ND | 10.7 ± 20.9 | 26.8 ± 49.7 | 31.8 ± 81.4 | 1.79 ± 5.06 | NS | *** |
| C28 | 1396 | 2-Nonen-1-old,f | 0.497 ± 0.921 | ND | ND | ND | ND | ND | ||
| C29 | 1407 | (E)-4-Hexen-1-old,f | ND | ND | ND | ND | ND | 2.09 ± 3.94 | ||
| C30 | 1320 | 6-Heptene-2,4-diold,f | 8.00 ± 10.0 | 8.23 ± 18.7 | ND | ND | ND | ND | ** | * |
| Total allyl alcohols | 8.50 ± 11.0 | 805 ± 18.7 | 10.7 ± 20.9 | 26.8 ± 49.7 | 31.8 ± 81.4 | 3.88 ± 9.01 | NS | *** | ||
| Total alcohols | 221 ± 122 | 1602 ± 1107 | 863 ± 1256 | 1633 ± 2724 | 1381 ± 1789 | 980 ± 1353 | * | NS | ||
| Aldehydes | ||||||||||
| C31 | 1083 | Hexanale | ND | ND | 1.55 ± 2.88 | ND | ND | ND | ||
| C32 | 1187 | Heptanald | 3.62 ± 6.94 | ND | ND | ND | ND | ND | ||
| C33 | 1287 | Octanale | 0.0911 ± 0.258 | ND | ND | ND | ND | ND | ||
| C34 | 1399 | Nonanale | 7.93 ± 5.03 | 14.2 ± 4.22 | 15.2 ± 4.82 | 24.2 ± 35.2 | 14.4 ± 8.11 | 9.73 ± 12.1 | NS | NS |
| Total straight-chain aldehydes | 11.6 ± 12.2 | 14.2 ± 4.22 | 16.7 ± 7.70 | 24.2 ± 35.2 | 14.4 ± 8.11 | 9.73 ± 12.1 | NS | NS | ||
| C35 | 919 | 3-Methyl-butanald | 2.99 ± 5.54 | 3.96 ± 4.83 | 1.29 ± 2.44 | 2.44 ± 4.63 | 8.40 ± 10.8 | 43.5 ± 31.5 | *** | NS |
| Total branched-chain aldehydes | 2.99 ± 5.54 | 3.96 ± 4.83 | 1.29 ± 2.44 | 2.44 ± 4.63 | 8.40 ± 10.8 | 43.5 ± 31.5 | *** | NS | ||
| C36 | 1541 | Benzaldehydee | ND | ND | ND | 3.90 ± 11.0 | 12.4 ± 23.0 | 1.80 ± 2.24 | * | ** |
| C37 | 1667 | Benzeneacetaldehyded,f | ND | ND | 7.98 ± 15.6 | ND | ND | ND | ||
| Total aromatic aldehydes | ND | ND | 7.98 ± 15.6 | 3.90 ± 11.0 | 12.4 ± 23.0 | 1.80 ± 2.24 | NS | NS | ||
| Total aldehydes | 14.6 ± 17.7 | 18.2 ± 9.05 | 26.0 ± 25.7 | 30.5 ± 50.8 | 35.2 ± 41.9 | 55.0 ± 45.8 | * | NS | ||
| Ketones | ||||||||||
| C38 | 817 | 2-Propanoned | 13.8 ± 16.0 | 13.1 ± 16.9 | 10.6 ± 12.6 | 56.9 ± 142 | ND | ND | * | *** |
| C39 | 904 | 2-Butanonee | 2.84 ± 5.25 | ND | 25.2 ± 21.3 | 1962 ± 2305 | 1465 ± 1572 | 243 ± 335 | *** | NS |
| C40 | 978 | 2-Pentanonee | ND | ND | ND | 107 ± 129 | 57.2 ± 77.8 | 98.9 ± 73.9 | *** | NS |
| C41 | 1184 | 2-Heptanonee | 17.5 ± 9.45 | 34.6 ± 12.7 | 39.7 ± 17.2 | 163 ± 226 | 82.1 ± 60.3 | 170 ± 211 | ** | * |
| C42 | 1288 | 2-Octanonee | 1.82 ± 2.83 | 28.4 ± 74.6 | 1.57 ± 2.90 | 22.5 ± 57.0 | 1.92 ± 3.82 | 3.28 ± 6.14 | NS | *** |
| C43 | 1392 | 2-Nonanonee | 12.4 ± 8.40 | 28.7 ± 13.1 | 34.5 ± 20.8 | 91.0 ± 69.7 | 64.7 ± 56.1 | 99.2 ± 111 | * | * |
| C44 | 1447 | 8-Nonen-2-oned | ND | ND | ND | ND | ND | 8.91 ± 11.2 | ||
| C45 | 1604 | 2-Undecanonee | ND | ND | 2.72 ± 5.07 | 7.09 ± 10.8 | 6.59 ± 7.77 | 0.716 ± 1.35 | * | NS |
| Total methyl ketones | 48.4 ± 42.0 | 105 ± 117 | 114 ± 79.8 | 2409 ± 2940 | 1678 ± 1778 | 624 ± 749 | *** | NS | ||
| C46 | 918 | (E,E)-6,10-Dimethyl-5,9- dodecadien-2-oned,f | ND | ND | ND | ND | 29.2 ± 64.9 | ND | ||
| C47 | 981 | 2,3-Butanedionee | 60.3 ± 72.1 | 76.9 ± 79.1 | 86.2 ± 76.7 | 24.9 ± 46.4 | 22.7 ± 46.6 | ND | ** | ** |
| C48 | 1292 | 3-Hydroxy-2-butanoned | 349 ± 564 | 477 ± 401 | 533 ± 451 | 380 ± 377 | 179 ± 245 | 66.8 ± 110 | * | *** |
| C49 | 1367 | 2-Hydroxy-3-pentanoned,f | ND | ND | 4.69 ± 8.77 | 1.45 ± 2.85 | ND | ND | NS | ** |
| Total diketones and other ketones | 409 ± 636 | 554 ± 480 | 624 ± 536 | 406 ± 427 | 230 ± 357 | 66.8 ± 110 | * | *** | ||
| Total ketones | 457 ± 678 | 659 ± 597 | 738 ± 616 | 2815 ± 3367 | 1908 ± 2135 | 691 ± 859 | ** | * | ||
| Esters | ||||||||||
| C50 | 1186 | Methyl hexanoatee | ND | 3.81 ± 7.83 | 3.33 ± 6.17 | ND | ND | ND | NS | ** |
| C51 | 1187 | Methyl 4-methyl pentanoated,f | 2.87 ± 5.34 | 7.40 ± 8.64 | 3.33 ± 6.17 | ND | 3.84 ± 7.20 | 4.53 ± 8.66 | NS | ** |
| Total methyl esters | 2.87 ± 5.34 | 11.2 ± 16.5 | 6.66 ± 12.3 | ND | 3.84 ± 7.20 | 4.53 ± 8.66 | NS | ** | ||
| C52 | 1038 | Ethyl butanoated | 4.32 ± 3.12 | 27.0 ± 45.3 | 53.8 ± 97.8 | 78.3 ± 116 | 62.8 ± 81.3 | 288 ± 362 | ** | ** |
| C53 | 1055 | Ethyl 2-methyl butanoated | ND | ND | ND | ND | 1.77 ± 3.28 | 3.11 ± 5.75 | NS | ** |
| C54 | 1069 | Ethyl 3-methyl butanoated | ND | ND | ND | ND | 44.2 ± 112 | 33.9 ± 65.4 | NS | ** |
| C55 | 1134 | Ethyl pentanoatee | ND | ND | ND | ND | 2.12 ± 4.92 | 2.42 ± 4.54 | NS | ** |
| C56 | 1234 | Ethyl hexanoatee | 18.7 ± 12.0 | 183 ± 267 | 343 ± 554 | 453 ± 584 | 480 ± 647 | 216 ± 270 | * | * |
| C57 | 1292 | Ethyl hex-4-enoated,f | ND | ND | ND | 7.24 ± 14.1 | 7.70 ± 15.1 | 0.292 ± 0.559 | NS | *** |
| C58 | 1335 | Ethyl heptanoatee | ND | ND | 4.78 ± 8.89 | 5.77 ± 10.7 | 3.86 ± 9.01 | 33.8 ± 53.2 | NS | *** |
| C59 | 1434 | Ethyl octanoatee | 5.27 ± 3.72 | 31.7 ± 45.9 | 71.5 ± 117 | 120 ± 139 | 101 ± 171 | 58.3 ± 95.9 | * | ** |
| C60 | 1537 | Ethyl nonanoated,f | ND | ND | ND | ND | ND | 1.18 ± 2.20 | ||
| C61 | 1641 | Ethyl decanoated | 4.47 ± 3.82 | 20.3 ± 24.1 | 458 ± 1275 | 1002 ± 2560 | 87.9 ± 140 | 320 ± 559 | ** | NS |
| C62 | 1855 | Ethyl dodecanoated | ND | ND | ND | ND | 0.640 ± 1.80 | 1291 ± 2894 | NS | * |
| Total ethyl esters | 32.7 ± 22.7 | 262 ± 383 | 930 ± 2051 | 1667 ± 3424 | 791 ± 1185 | 2248 ± 4312 | *** | ** | ||
| C63 | 1122 | Propyl butanoatee | ND | ND | ND | ND | 3.35 ± 8.57 | 6.74 ± 7.88 | ** | *** |
| C64 | 1320 | Propyl hexanoatee | ND | ND | ND | ND | 14.6 ± 19.6 | 27.1 ± 28.0 | *** | NS |
| C65 | 1521 | Propyl octanoatee | ND | ND | ND | ND | ND | 5.68 ± 8.53 | ||
| Total propyl esters | ND | ND | ND | ND | 18.0 ± 28.2 | 39.5 ± 44.4 | *** | NS | ||
| C66 | 1218 | Butyl butanoated | ND | ND | ND | ND | 2.20 ± 5.05 | 2.98 ± 3.31 | ** | * |
| C67 | 1412 | Butyl hexanoated | ND | ND | ND | ND | 3.34 ± 9.45 | 7.81 ± 7.31 | *** | NS |
| Total butyl esters | ND | ND | ND | ND | 5.54 ± 14.5 | 10.8 ± 10.6 | *** | NS | ||
| C68 | 1267 | Pentyl butanoated,f | ND | ND | 2.11 ± 3.90 | 3.62 ± 6.81 | 2.45 ± 5.45 | 127 ± 237 | NS | *** |
| Total pentyl esters | ND | ND | 2.11 ± 3.90 | 3.62 ± 6.81 | 2.45 ± 5.45 | 127 ± 237 | NS | *** | ||
| C69 | 1414 | Hexyl hexanoated | ND | ND | ND | ND | ND | 3.08 ± 6.05 | ||
| Total hexyl esters | ND | ND | ND | ND | ND | 3.08 ± 6.05 | ||||
| C70 | 1131 | 1-Methylpropyl butanoated,f | ND | ND | ND | ND | ND | 5.56 ± 7.07 | ||
| C71 | 1323 | 2-Methylpropyl hexanoated | ND | ND | ND | ND | 11.4 ± 22.1 | 35.5 ± 32.4 | *** | NS |
| C72 | 1372 | 2-Propenyl hexanoated | ND | ND | ND | ND | 2.54 ± 5.47 | 0.485 ± 0.900 | NS | ** |
| C73 | 1461 | 3-Methylbutyl hexanoated,f | ND | ND | ND | ND | 1.55 ± 3.49 | 40.2 ± 74.8 | NS | ** |
| Total branched-alkyl esters | ND | ND | ND | ND | 15.5 ± 31.0 | 81.7 ± 115 | *** | NS | ||
| Total esters | 35.6 ± 28.0 | 273 ± 400 | 939 ± 2067 | 1671 ± 3431 | 836 ± 1271 | 2515 ± 4734 | *** | ** | ||
| Hydrocarbons | ||||||||||
| C74 | 700 | Heptanee | 7.46 ± 17.2 | 13.0 ± 17.6 | 7.10 ± 17.2 | 11.9 ± 14.2 | 5.45 ± 15.4 | ND | NS | NS |
| Total saturated hydrocarbons | 7.46 ± 17.2 | 13.0 ± 17.6 | 7.10 ± 17.2 | 11.9 ± 14.2 | 5.45 ± 15.4 | ND | NS | NS | ||
| C75 | 688 | 1,3-Pentadiened | ND | ND | ND | ND | ND | 2.42 ± 4.55 | ||
| C76 | 837 | t-3-Octened | 35.6 ± 54.4 | 35.7 ± 44.8 | 36.9 ± 47.0 | 239 ± 621 | 16.3 ± 33.0 | 17.5 ± 33.5 | NS | *** |
| C77 | 956 | 1,3-Octadiened,f | 5.93 ± 11.3 | 7.08 ± 13.9 | 7.50 ± 14.7 | 98.5 ± 268 | 6.71 ± 15.3 | 6.22 ± 11.7 | NS | *** |
| C78 | 1041 | Toluenee | 20.2 ± 12.2 | 42.9 ± 28.4 | 24.9 ± 19.4 | 112 ± 261 | 19.9 ± 23.5 | 68.0 ± 112 | NS | NS |
| Total unsaturated hydrocarbons | 61.7 ± 77.9 | 85.7 ± 87.1 | 69.3 ± 81.2 | 449 ± 1150 | 42.9 ± 71.8 | 94.1 ± 162 | NS | *** | ||
| Total hydrocarbons | 69.2 ± 95.1 | 98.7 ± 105 | 76.4 ± 98.4 | 461 ± 1164 | 48.4 ± 87.2 | 94.1 ± 162 | NS | *** | ||
| Sulphur compounds | ||||||||||
| C79 | 746 | Dimethyl sulphidee | 1.55 ± 2.86 | ND | ND | 17.2 ± 46.3 | ND | ND | NS | ** |
| C80 | 1934 | Dimethyl sulphoned | 25.0 ± 34.0 | 8.05 ± 15.7 | 7.41 ± 14.0 | 61.4 ± 163 | 7.76 ± 14.5 | 6.77 ± 12.5 | NS | *** |
| Total sulphur compounds | 26.6 ± 36.8 | 8.05 ± 15.7 | 7.41 ± 14.0 | 78.6 ± 209 | 7.76 ± 14.5 | 6.77 ± 12.5 | NS | *** | ||
| Terpenes | ||||||||||
| C81 | 1195 | D-Limonenee | 0.151 ± 0.430 | 1.35 ± 3.83 | ND | 1.02 ± 2.89 | ND | ND | NS | NS |
| Total terpenes | 0.151 ± 0.430 | 1.35 ± 3.83 | ND | 1.02 ± 2.89 | ND | ND | NS | NS | ||
LRI: linear retention index.
ND: not detected.
RT: ripening time factor effect; P: producer factor effect; NS: P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Tentatively identified volatile compounds.
Positively identified volatile compounds.
Volatile compounds not previously described in Idiazabal cheese.
Acids were the predominant chemical family (79.5–88.9% of the total abundance) (Supplementary Table 1), as observed previously (Barron et al., 2005a, 2007; Abilleira et al., 2010b; Valdivielso et al., 2016), and the highest abundances were found between 30 and 60 days of ripening (P ≤ 0.05) (Table 1). All the identified acids were straight-chain fatty acids, predominating hexanoic (44.2–51.1% of total acids), butanoic (18.8–27.3%) and octanoic acids (14.8–18.4%). These results agree with previous studies (Barron et al., 2005b, 2007; Abilleira et al., 2010b; Valdivielso et al., 2016). However, 3-hexenoic acid was identified for the first time and no branched-chain acids were detected in this study. Volatile acids also predominate in other raw ewe milk-derived cheeses, such as Vastedda della valle del Belìce cheese, and similar changes during ripening have been reported (Delgado-Martínez et al., 2019; Gaglio et al., 2019a). Straight-chain fatty acids are also abundant in other raw ewe milk-derived cheeses (Gaglio et al., 2019a; Cardinali et al., 2021), although, in some cases, such as in Torta del Casar, acetic acid, branched-chain or long-chain FFAs predominate (Delgado-Martínez et al., 2019). Moreover, clear differences were observed among Idiazabal cheese producers (P ≤ 0.001). Samples from producer B, who used commercial rennet, showed lower abundance of acids compared to producers A, C and D that used artisanal rennet. This was expected due to the higher pregastric lipase activity of artisanal rennet compared to commercial rennet (Virto et al., 2003) and its sn-3 stereospecificity (Thierry et al., 2017; Amores et al., 2021). Lipoprotein lipase (LPL) from raw milk and microbial lipases and esterases are also important lipolytic agents in raw milk cheeses (Thierry et al., 2017; Le Quéré and Buchin, 2022). However, LPL activity is very low in Idiazabal cheese (Chávarri et al., 1998).
Esters were the second family in order of importance throughout ripening (0.859–11.3%) (Supplementary Table 1), whose abundance increased until 120 days (P ≤ 0.001) (Table 1). Ethyl esters predominated (89.4–99.8% of total esters throughout ripening), specifically, decanoic, hexanoic, dodecanoic and butanoic ethyl esters. Moreover, several esters were identified for the first time in Idiazabal cheese, such as methyl 4-methyl pentanoate or pentyl butanoate. Among minor esters, a significant increase was also observed for propyl, butyl and branched-alkyl esters during ripening (P ≤ 0.01). These results partially agree with previous works in Idiazabal cheese, as far as identified compounds and their abundance are concerned (Barron et al., 2007; Abilleira et al., 2010b; Valdivielso et al., 2016). Compared to other raw ewe milk-derived cheeses, there are notable differences, although ethyl esters have also been described as predominant (Delgado-Martínez et al., 2019; Gaglio et al., 2019a; Cardinali et al., 2021). Significant differences among producers were only observed for methyl esters (P ≤ 0.01), with producers B and C showing the highest abundance; and for ethyl and pentyl esters (P ≤ 0.01 and P ≤ 0.001, respectively), for which producer A presented the highest abundance. In general, the presence of esters has been associated to LAB esterase activity (Liu et al., 2004; Bertuzzi et al., 2018) and consequently, the observed differentiation among producers could be related to the different LAB composition.
Ketones abundance was remarkable (3.68–7.16%) (Supplementary Table 1), which also changed during ripening time (P ≤ 0.01) (Table 1). Methyl ketones abundance increased until 30 days and decreased afterwards but remaining predominant, while diketones and other ketones showed the greatest abundance at the beginning of the ripening (Table 1). These results would explain why methyl ketones have been described as predominant in Idiazabal cheese before (Barron et al., 2005a, 2007; Valdivielso et al., 2016). Individually 2-butanone predominated, as observed in previous studies (Barron et al., 2007; Abilleira et al., 2010b; Valdivielso et al., 2016), together with 3-hydroxy-2-butanone that has previously been related to mountain grazing (Valdivielso et al., 2016). Some ketones, such as 2-hydroxy-3-pentanone or 6,10-dimethyl-5,9-dodecadien-2-one, have not been identified in Idiazabal cheese to date (Table 1). Differences among producers were only observed for diketones and other ketones (P ≤ 0.001), with producers C and D showing the highest abundance. Compared to other raw ewe milk-derived cheeses, there are great differences in the identified ketones and their abundance during ripening (Delgado-Martínez et al., 2019; Gaglio et al., 2019a; Cardinali et al., 2021). Gezginc et al. (2021) have also reported 2-butanone as the most common ketone in Tulum cheese, whereas 2-heptanone, 2-nonanone and hydroxyacetone predominate in Gran Ovino cheese (Gaglio et al., 2019b). Ketones are formed by catabolism of FFAs, involving the oxidation to β-ketoacids and decarboxylation (Fox et al., 2017; Bertuzzi et al., 2018), mainly by mould and fungi (Fox et al., 2017; Le Quéré and Buchin, 2022).
Alcohols were another abundant chemical family (3.40–5.46%) (Supplementary Table 1), which showed the highest abundance at 30 days of ripening (P ≤ 0.05) (Table 1). Primary alcohols were remarkable during ripening, although a decrease was observed from 30 to 120 days, when the abundance of secondary alcohols notably increased (Table 1). However, the ripening time effect was only significant for secondary alcohols (P ≤ 0.001). These results would explain the great abundance of secondary alcohols reported before (Barron et al., 2005a, 2007; Valdivielso et al., 2016). Ethanol and 2-butanol predominated, which partially agrees with previous studies (Barron et al., 2007; Abilleira et al., 2010b) and several alcohols were identified for the first time in Idiazabal cheese, such as 3-methyl-2-butanol or 2-propen-1-ol. Differences among producers were only observed for allyl alcohols (P ≤ 0.001), with producer A presenting the highest abundance. Compared to other raw ewe milk-derived cheeses, the evolution of alcohols and the predominant compounds differ (Fernández-García et al., 2004; Delgado-Martínez et al., 2019; Gaglio et al., 2019a; Cardinali et al., 2021). Gezginc et al. (2021) have also observed ethanol and 2-butanol as predominant in Tulum cheese, while Gaglio et al. (2019b) have reported 1-hexanol in Gran Ovino cheese. Alcohols biosynthesis has been related to lactose metabolism, reduction of carbonyl compounds (Le Quéré and Buchin, 2022) or several enzymatic and non-enzymatic reactions from amino acids or methyl ketones (Yvon and Rijnen, 2001; Bertuzzi et al., 2018; Le Quéré and Buchin, 2022).
Hydrocarbons, sulphur compounds, aldehydes and terpenes were minor compounds (<4%) (Supplementary Table 1), as observed before in Idiazabal cheese (Barron et al., 2005a, 2007; Abilleira et al., 2010b; Valdivielso et al., 2016). However, there are differences compared to other raw ewe milk-derived cheeses (Delgado-Martínez et al., 2019; Gaglio et al., 2019a; Cardinali et al., 2021), such as the high abundance of aromatic hydrocarbons reported for Vastedda della valle del Belìce cheese (Gaglio et al., 2019a). The ripening time only had a significant effect on aldehydes (P ≤ 0.05) (Table 1), with a predominance of straight-chain aldehydes at 30 days of ripening, aromatic aldehydes at 60 days and branched-chain aldehydes at 120 days. Nevertheless, the ripening time effect was only significant for the relative abundance of branched-chain aldehydes (P ≤ 0.001) (Table 1). Individually, nonanal and 3-methyl-butanal were abundant, unlike in previous studies in Idiazabal cheese (Barron et al., 2007; Abilleira et al., 2010b; Valdivielso et al., 2016), and benzeneacetaldehyde was identified for the first time. The evolution of aldehydes and the predominant compounds differ for other raw ewe milk-derived cheeses (Fernández-García et al., 2004; Delgado-Martínez et al., 2019; Gaglio et al., 2019a; Cardinali et al., 2021). The presence of straight-chain aldehydes in cheese is related to the autoxidation of unsaturated fatty acids, both free and esterified (Bertuzzi et al., 2018). The presence of branched-chain and aromatic aldehydes, on the other hand, is related to the degradation of amino acids by non-enzymatic or enzymatic reactions by yeast or LAB (Yvon and Rijnen, 2001; Bertuzzi et al., 2018; Le Quéré and Buchin, 2022). Anyway, aldehydes are transitory compounds because of their rapid reduction to alcohols or oxidation to acids (Le Quéré and Buchin, 2022), which explains the low abundance observed. In addition, it should be noted that among minor compounds, significant differences among producers were observed only for unsaturated hydrocarbons and sulphur compounds (P ≤ 0.001), with producer B showing the highest abundance in both cases. Overall, the formation of sulphur compounds is related to enzymatic reactions of sulphur-containing amino acids, mainly methionine, by microorganisms (Fox et al., 2017; Le Quéré and Buchin, 2022), while hydrocarbons mainly originate from degradation of carotene (Povolo et al., 2007).
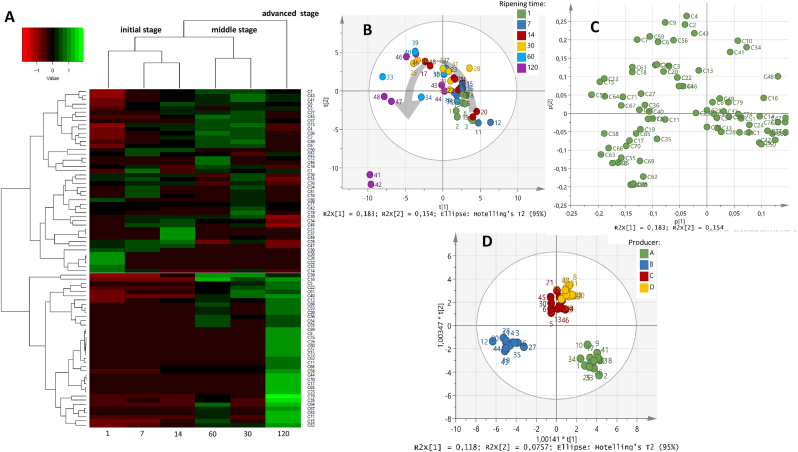
Subsequently, the dynamics of volatile compounds during ripening were analysed by multivariate analysis. Through an HCA, the volatile composition was divided into three stages: initial stage (1–14 days), middle stage (30–60 days) and advanced stage (120 days) (Fig. 1A). Acetic and heptanoic acids, methyl esters, most diketones, most straight-chain aldehydes and benzeneacetaldehyde, some alcohols (such as 2-methyl-3-pentanol or menthol), saturated hydrocarbons and terpenes characterized the initial stage. Most abundant acids, few ethyl esters, most methyl ketones and the rest of diketones, some primary and secondary alcohols, nonanal and benzaldehyde, unsaturated hydrocarbons and sulphur compounds characterized the middle stage. Finally, 3-hexenoic and nonanoic acids, most esters, the rest of primary and secondary alcohols, 3-methyl butanal and 8-nonen-2-one characterized the advanced stage. The characteristic presence of some volatile compounds at the initial and middle stage of ripening would explain why they have either not been identified or have been detected in smaller abundance in previous studies (Barron et al., 2005a, 2007; Abilleira et al., 2010b; Valdivielso et al., 2016). PCA approach confirmed the HCA results (Fig. 1B–C).
Fig. 1.
“Analysis of volatile composition evolution during ripening (1, 7, 14, 30, 60 and 120 days) of Idiazabal cheese by means of HCA (A) and PCA (scores and loadings plots, B and C respectively) and scores plot of the OPLS-DA model based on the producer (D). Volatile compounds are labeled according to the ID (Table 1). The scale values of the HCA correspond to log transformed and UV scaled data.”
To study differences in the volatile composition among producers of the Idiazabal cheeses analysed, an OPLS-DA approach was applied. Despite its limitations, the model reported a clear distinction between producers, specifically, between A, B and C-D (Fig. 1D). Octanoic acid, ethyl octanoate or 2-octanone were some of the most important volatile compounds for such differentiation. The observed differentiation could be due to several reasons, such as the type of rennet used (Virto et al., 2003), as mentioned above; or the level of rennet lipase employed, since the higher the level, the greater FFAs release and sensory scores in Idiazabal cheese (Amores et al., 2021). Moreover, the sheep grazing system could also have affected the volatile composition of Idiazabal cheese, since clear differences have been reported in cheeses made from milk from extensive mountain grazing, compared to indoor feeding and part-time grazing sheep (Valdivielso et al., 2016). Finally, cheese making and ripening conditions (De Filippis et al., 2016) or microbial composition (Fox et al., 2017), have also been reported to affect the volatile composition of cheese, although it has not been studied in Idiazabal cheese.
3.2. OIR values
It is well known that not all volatile compounds contribute to cheese aroma, mainly due to their OTs (Starowicz, 2021). Therefore, OIR values were calculated to elucidate the key aromatic compounds of Idiazabal cheese (Table 2). Overall, esters and acids showed the highest OIR values during ripening and, consequently, were the most odour-active compounds. Esters OIR values increased as ripening progressed and all the identified odour-active esters were ethyl esters, predominating ethyl 3-methyl butanoate, ethyl butanoate and ethyl hexanoate (Table 2). Therefore, odour-active esters corresponded to predominant esters (Table 1). The detected odour-active esters differ compared to other cheeses, although ethyl butanoate, ethyl hexanoate and ethyl octanoate are mainly reported (Qian and Reineccius, 2003; Taylor et al., 2013; Wang et al., 2020). Low OTs are reported for esters (Curioni and Bosset, 2002; Liu et al., 2004; Wang et al., 2021), hence they are described as potent odorants in several cheeses such as Grana Padano, providing sweet, floral and fruity odour notes and reducing the sharpness of FFAs (Curioni and Bosset, 2002; Liu et al., 2004; Bertuzzi et al., 2018). Fruity odour notes are related to ethyl esters (Moio et al., 2000; Curioni and Bosset, 2002; Liu et al., 2004) and specifically, apple, pineapple, olive or banana notes to the predominant odour-active esters (Table 2).
Volatile acids showed the highest OIR values at middle stage of ripening (Table 2), which is of special interest since excessive concentrations have been related to rancid or putrid odour notes in Idiazabal cheese (Barron et al., 2005b; Zabaleta et al., 2016). The predominant butanoic, hexanoic and octanoic acids (Table 1), together with pentanoic acid, were odour-active acids (Table 2). Therefore, a greater number of odour-active acids was observed compared to a previous study (Abilleira et al., 2010b). Volatile acids are notable aromatic compounds of various cheeses (Curioni and Bosset, 2002; Le Quéré and Buchin, 2022) and have been reported as key odorants in several cheeses, such as Cheddar, although there are differences among types of cheeses (Attaie, 2009; Taylor et al., 2013; Wang et al., 2020; Wang et al., 2021). It is noteworthy that there is no information on the OT of 3-hexenoic acid, which has been related to pungent or sweat odour notes (Table 2).
Some of the detected ketones presented high OIR values (Table 2), confirming their contribution to cheese aroma due to their low perception thresholds (Curioni and Bosset, 2002). The only odour-active diketone was 2,3-butanedione, which showed the highest OIR values at the initial stage of ripening and remained up to middle stage, providing buttery odour notes (Table 2). It has been described as an important odorant of many cheeses and fermented dairy products, such as Parmigiano Reggiano cheese (Qian and Reineccius, 2003; Attaie, 2009; Taylor et al., 2013). Within methyl ketones, the predominant 2-butanone (Table 1), together with 2-undecanone, showed high OIR values at middle stage of ripening and, therefore, less ripened Idiazabal cheeses would present sweet or fruity odour notes (Table 2). The 2-heptanone and 2-nonanone also showed high OIR values that increased until the end of the ripening, providing musty, blue cheese or floral odour notes. Methyl ketones are remarkable odorants of different cheeses, such as Cheddar, surface-ripened and blue-veined cheeses (Curioni and Bosset, 2002; Natrella et al., 2020; Wang et al., 2020; Wang et al., 2021). However, it is noteworthy that information on the odour impact of 2-butanone is scarce.
Aldehydes OIR values were in general low (<1) (Table 2). The 3-methyl butanal, which was the predominant aldehyde in the analysed Idiazabal cheeses (Table 1), was by far the most odour-active aldehyde, whose OIR values increased during ripening providing malt or chocolate notes (Table 2). Nonanal, which was also an abundant aldehyde (Table 1), presented lower OIR values, providing sweet or fatty-floral odour notes at initial and middle stages of ripening (Table 2). Within minor aldehydes, heptanal and benzeneacetaldehyde were also odour-active compounds and, although presenting low OIR values, they would contribute with fatty, fruity or floral odour notes at the initial stage of ripening (Table 2). These aldehydes have been previously reported as odour-active compounds in different cheeses (Natrella et al., 2020; Wang et al., 2020; Wang et al., 2021). Branched-chain aldehydes and, specifically 3-methyl butanal, are the most notorious odour-active aldehydes for cheese aroma, such as for Parmigiano Reggiano (Curioni and Bosset, 2002; Qian and Reineccius, 2003; Natrella et al., 2020). Straight-chain aldehydes are related to unpleasant green and herbaceous notes when exceed certain thresholds and nonanal is the most common odorant, remarkably for the aroma of Grana Padano cheese (Curioni and Bosset, 2002; Natrella et al., 2020; Wang et al., 2021). It is noteworthy that benzeneacetaldehyde has been reported as a remarkable odorant of Gruyère or Bovine Mozzarella cheeses (Curioni and Bosset, 2002; Wang et al., 2021).
Alcohols presented low OIR values (<1) and 2-butanol, which was one of the predominant alcohols (Table 1), was the only odour-active alcohol. It presented the highest OIR values between middle and advanced stages of ripening, providing winey or sweet odour notes (Table 2). 2-butanol is formed by reduction of 2-butanone (main ketone identified, as mentioned in section 3.1.), which, in turn, comes from the catabolism of butanoic acid (one of the predominant acids) and degradation of 2,3-butanediol (Urbach, 1993; Le Quéré and Buchin, 2022). Microbial degradation of 2,3-butanedione also leads to the production of 2-butanol (Urbach, 1993). However, information about its odour impact is scarce (Juric et al., 2003; Abilleira et al., 2010b). These results would confirm the small contribution of alcohols to cheese aroma, mainly due to their high OTs (Barlow et al., 1989).
Within the minor compounds identified (Table 1), hydrocarbons were minor odorants in cheese due to their high OTs, although they are precursors for other flavour compounds (Arora et al., 1995). The results obtained indicated toluene as the only odour-active hydrocarbon, which presented high OIR values throughout the entire ripening process, providing fruity, sweet-gassy or hydrocarbon odour notes (Table 2). Toluene originates from the degradation of carotene in milk (Villeneuve et al., 2013), although no information has been found in relation to its odour impact in cheese.
Both identified sulphur compounds presented high OIR values (Table 2). Dimethyl sulphide was an odour-active compound between initial and middle stage of ripening and dimethyl sulphone throughout the entire process. Therefore, sweet, flower or sulphurous odour notes would characterize the aroma of ripened Idiazabal cheese, but not unpleasant wild radish or cabbage (Table 2). Sulphur compounds are key odorants of many cheeses because of their low OTs (Curioni and Bosset, 2002; Le Quéré and Buchin, 2022), although they have not been reported for Idiazabal cheese. In other cheeses, such as Swiss, only dimethyl sulphide has been detected (Taylor et al., 2013; Natrella et al., 2020). Dimethyl sulphide is produced from sulphur-containing amino acids in the rumen (Villeneuve et al., 2013; Le Quéré and Buchin, 2022) and its oxidation results in the formation of dimethyl sulphone (Kilcawley et al., 2018).
The identified terpene (D-limonene) was not an odour-active compound (Table 2), as observed for Cheddar cheese (Wang et al., 2021). Information about the contribution of terpenes to the aroma of cheese is scarce (Curioni and Bosset, 2002). Terpenes are mainly attributed to the secondary metabolism of plants (Kilcawley et al., 2018), although, to a lesser extent, LAB can also synthetize them (Belviso et al., 2011). Consequently, they are mainly transferred from herbs or forages to milk fat (Viallon et al., 2000; Bertuzzi et al., 2018) and are potential pasture biomarkers (Abilleira et al., 2011; Kilcawley et al., 2018).
3.3. Correlation between bacterial communities and volatile compounds
According to a previous publication (Santamarina-García et al., 2022a), overall, the SLAB Lactococcus was the predominant genus during ripening of the analysed Idiazabal cheeses, although after 30 or 60 days of ripening its abundance decreased and the NSLAB began to proliferate. Specifically, the proliferation of Lactobacillus was promoted in cheeses collected from all producers (from a mean 0.0949% at 1 day of ripening to 8.96% at 120 days), while for the rest it depended on the producer. Leuconostoc proliferation was promoted in cheeses from producer A (from 4.48% at 1 day to 31.0% at 120 days), while in those from producers A and D, Streptococcus and Enterococcus proliferated (from 0.750% at 1 day to 4.52% at 120 days and from 0.675% at 1 day to 2.12% at 120 days, respectively). Additionally, the abundance of environmental or non-desirable bacteria decreased during ripening, although some genera remained abundant (>1%), namely Obesumbacterium, Hafnia, Staphylococcus, Buttiauxella, Psychrobacter, Raoultella, Serratia, Brevibacterium and Erwinia. Specifically, the abundance of Buttiauxella decreased during ripening, while that of Staphylococcus increased and the rest showed an increase at intermediate points of ripening (at 7, 14 or 30 days). Detailed data can be found in Santamarina-García et al. (2022a).
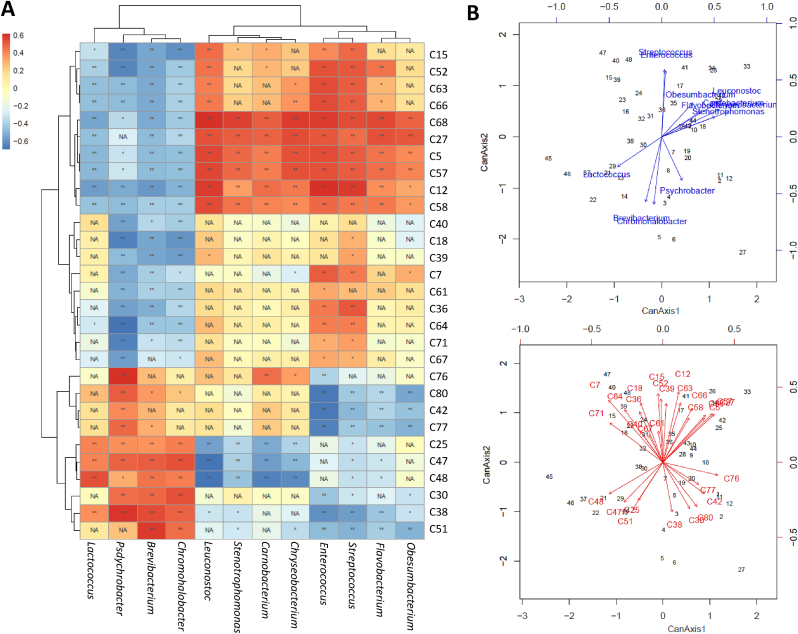
To elucidate the contribution of each bacterial genera to the volatile composition of Idiazabal cheese, an O2PLS approach with Spearman's correlations was applied (Fig. 2A). The obtained correlations were further confirmed through a CCorA (Fig. 2B). In this sense, 12 bacterial genera were reported as key bacteria related to volatile compounds production (Fig. 2A), namely Psychrobacter, Enterococcus, Brevibacterium, Streptococcus, Leuconostoc, Chromohalobacter, Chryseobacterium, Carnobacterium, Lactococcus, Obesumbacterium, Stenotrophomonas and Flavobacterium. The fact that LAB are reported as key bacteria is not surprising, since their genetic and metabolic diversity is considered responsible for flavour diversification in fermented products (Singh et al., 2006). Associations between these LAB and volatile compounds have previously been reported, such as for fermented yak milk (Jiang et al., 2020; Li et al., 2022). Nonetheless, information on Carnobacterium is scarce. Recent studies have also highlighted the notable contribution of environmental and/or non-desirable bacteria to the volatile composition of fermented products, such as Kazak cheese (Zheng et al., 2018; Meng et al., 2021).
Fig. 2.
“Correlation heatmap between key bacterial genera and volatile compounds (only those volatile compounds with at least one significant correlation greater than 0.500 are shown) (A) and verification of the resulted correlations through CCorA analysis (B). Volatile compounds are labeled according to the ID (Table 1). Significant correlations are represented by **P ≤ 0.01 and *P ≤ 0.05 and non-significant correlations (P > 0.05) by NA.”
Among LAB, Enterococcus and Streptococcus, followed by Leuconostoc, showed the largest number of high positive correlations with volatile compounds. These LAB showed positive correlations with volatile acids, mainly with 3-hexenoic and octanoic acids. These results could confirm that lipases and esterases from LAB are important lipolytic agents in cheese (Thierry et al., 2017), as reported in our previous publication (Santamarina-García et al., 2022b), and would affect the aroma of Idiazabal cheese (Table 2). Association between these LAB and acids production have also been observed for other fermented products such as sour meat (Meng et al., 2021; Zhong et al., 2021), although there is no information on 3-hexenoic acid. It is noteworthy the low correlation of these LAB to other acids, like butanoic or hexanoic acids, indicating a greater impact of the pregastric lipase of the rennet used (Virto et al., 2003). Enterococcus, Streptococcus and Leuconostoc were also highly correlated to alcohols, mainly 1-butanol and 2-propen-1-ol. This would indicate the implication of LAB metabolism in alcohols biosynthesis in cheese (Zuljan et al., 2016), as have been observed for other fermented food, such as Kazak fermented cheese (Zheng et al., 2018; Meng et al., 2021; Zhong et al., 2021). High correlations were also observed between these LAB and esters, such as pentyl butanoate, ethyl 4-hexenoate or ethyl butanoate, some of which were odour-active compounds (Table 2). This could be indicative of LAB esterase activity (Santamarina-García et al., 2022b) and its impact on the aroma of Idiazabal cheese (Liu et al., 2004). Correlations between these LAB and ethyl butanoate have already been reported (Meng et al., 2021; Zhong et al., 2021), but there is no information on the rest of the esters. Streptococcus also showed high correlation to benzaldehyde, whose biosynthesis pathway is not very well known (Le Quéré and Buchin, 2022). Nonetheless, benzaldehyde production from phenylalanine by Lactobacillus has been demonstrated (Groot and de Bont, 1998). Finally, Carnobacterium and Lactococcus showed fewer high positive correlations to volatile aroma compounds. Lactococcus was the only LAB positively correlated to ketones, specifically to 3-hydroxy-2-butanone, whose production by the starter Lactococcus lactis has been proven (Le Quéré and Buchin, 2022) and suggested for Idiazabal cheese (Barron et al., 2007).
On the other hand, environmental and/or non-desirable bacteria also presented positive correlations with volatile compounds. Psychrobacter showed the largest number of correlations, mainly with 2-propanone, t-3-octene and dimethyl sulphone; followed by Brevibacterium that was correlated to 2-propanone, 2,3-butanedione and methyl 4-methyl pentanoate; and Chromohalobacter that was correlated to 6-heptene-2,4-diol and 2,3-butanedione. Chryseobacterium, Obesumbacterium, Flavobacterium and Stenotrophomonas showed fewer correlations, mainly with 2-propen-1-ol and pentyl butanoate. Associations between these 7 genera and other volatile compounds production have been observed in fermented products, such as Beitang shrimp paste (Yao et al., 2021; Zhao et al., 2022). However, to the best of our knowledge, the production of the aforementioned compounds by these bacteria has not been reported so far. Lipase and/or protease activities have been described for the aforementioned genera, for example Chryseobacterium and Stenotrophomonas (Mukhia et al., 2021; Rathakrishnan and Gopalan, 2022), except for Obesumbacterium. These activities could lead to the release of FFAs or FAAs that are substrates for the production of several volatile compounds (Le Quéré and Buchin, 2022). Taking into account the OIR values (Table 2), an interesting contribution of these environmental and/or non-desirable bacteria to the aromatic composition of Idiazabal cheese could be deduced. The metabolic potential of other environmental and/or non-desirable bacteria has also been reported (Zhao et al., 2021; Xia et al., 2022). For instance, Li et al. (2022) have observed a strong correlation between Pseudomonas and linalool in fermented red sufu. Similarly, Yang et al. (2021) have reported a correlation between Staphylococcus and 3-methyl butanal in fermented sausages.
In addition, several negative correlations were observed between key bacterial genera and volatile compounds (Fig. 2A). NSLAB were mainly negatively correlated to ketones (such as 2-propanone and 2,3-butanedione) and Enterococcus also to hydrocarbons (such as 1,3-octadiene) and sulphur compounds (such as dimethyl sulphone). Within environmental and/or non-desirable bacteria, Psychrobacter, Brevibacterium and Chromohalobacter showed the greatest number of negative correlations, mostly with alcohols and esters (such as 1-butanol or ethyl butanoate). These correlations could be related to volatile compounds degradation abilities by the aforementioned bacteria, which have been little described so far (Fonseca et al., 2021). Therefore, taking into account the bacterial dynamics during ripening (Santamarina-García et al., 2022a), the volatile composition evolution and the aroma development could be partially understood. Moreover, it is noteworthy that Psychrobacter, Brevibacterium and Chromohalobacter were negatively related to those volatile compounds that NSLAB could have synthetized and vice versa. Considering the obtained OIR values (Table 2), Psychrobacter, Brevibacterium and Chromohalobacter could have a negative effect on the aroma development caused by NSLAB and conversely. Negative correlations between bacterial genera and volatile compounds of fermented products have already been reported (Zhong et al., 2021; Li et al., 2022) and Zheng et al. (2018) have classified Lactobacillus as a negative factor for flavour development in Kazak cheese. Moreover, the fact that Lactococcus was positively related to ketones production while the rest of LAB were negatively related, could corroborate the previously observed competitive inhibition between autochthonous LAB and those that made up the starter (Santamarina-García et al., 2022a).
In summary, the results obtained would indicate that bacterial communities would have a notable role in the evolution of the volatile and aromatic composition of Idiazabal cheese during ripening. Thus, the different microbial composition observed between producers (Santamarina-García et al., 2022a) and compared to other raw ewe milk-derived cheeses (Gaglio et al., 2019a; Dimov et al., 2021), would partially explain the observed differences in the volatile composition.
4. Conclusion
The results showed that mainly ripening time modulated the volatile composition of raw ewe milk-derived Idiazabal cheese, with a predominance of acids, followed by esters, ketones and alcohols. In terms of odour impact, esters and acids were the predominant odour-active families during ripening and individually, ethyl hexanoate, ethyl 3-methyl butanoate, ethyl butanoate, butanoic acid or 3-methyl butanal were some of the most important aromatic compounds, providing fruity, rancid, cheesy or malt odour notes. In terms of functional relationship, Psychrobacter, Enterococcus, Brevibacterium, Streptococcus, Leuconostoc, Chromohalobacter, Chryseobacterium, Carnobacterium, Lactococcus, Obesumbacterium, Stenotrophomonas and Flavobacterium were key bacteria for the volatile and aromatic composition of Idiazabal cheese. Overall, non-starter lactic acid bacteria (NSLAB) showed high positive correlations to certain acids, esters and alcohols, whereas the starter LAB (SLAB) was related to particular ketones formation and environmental and/or non-desirable bacteria to certain ketones, hydrocarbons and sulphur compounds production. It is noteworthy that the SLAB Lactococcus and Psychrobacter, Brevibacterium and Chromohalobacter were described as having a negative impact on aroma development caused by NSLAB and vice versa.
CRediT authorship contribution statement
Gorka Santamarina-García: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, and, Visualization. Gustavo Amores: Conceptualization, Methodology, Validation, Formal analysis, Resources, Data curation, Writing – review & editing, and, Supervision. Igor Hernández: Conceptualization, Methodology, Validation, Investigation, Resources, Data curation, Writing – review & editing. Lara Morán: Methodology, Validation, and, Resources. Luis Javier R. Barrón: Resources, Writing – review & editing, and, Funding acquisition. Mailo Virto: Conceptualization, Methodology, Validation, Resources, Writing – review & editing, Supervision, and, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the Idiazabal PDO cheesemakers for collaborating with this study and the technical and human support provided by SGIker (UPV/EHU/ERDF, EU), especially the chemometric support of L. Bartolomé. Financial support was provided by the Basque Government (IT944-16 and IT1568-22). G. Santamarina-García thanks the University of the Basque Country (UPV/EHU) for the predoctoral fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2022.100425.
Contributor Information
Gorka Santamarina-García, Email: gorka.santamarina@ehu.eus.
Gustavo Amores, Email: gustavo.amores@ehu.eus.
Igor Hernández, Email: igor.hernandezo@ehu.eus.
Lara Morán, Email: laramoran84@hotmail.com.
Luis Javier R. Barrón, Email: luisjavier.rbarron@ehu.eus.
Mailo Virto, Email: mailo.virto@ehu.eus.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abilleira E., Virto M., Nájera A.I., Salmerón J., Albisu M., Pérez-Elortondo F.J., Ruiz de Gordoa J.C., de Renobales M., Barron L.J.R. Effects of seasonal changes in feeding management under part-time grazing on the evolution of the composition and coagulation properties of raw milk from ewes. J. Dairy Sci. 2010;93:3902–3909. doi: 10.3168/jds.2009-2983. [DOI] [PubMed] [Google Scholar]
- Abilleira E., Schlichtherle-Cerny H., Virto M., de Renobales M., Barron L.J.R. Volatile composition and aroma-active compounds of farmhouse Idiazabal cheese made in winter and spring. Int. Dairy J. 2010;20:537–544. doi: 10.1016/j.idairyj.2010.02.012. [DOI] [Google Scholar]
- Abilleira E., Virto M., Nájera A.I., Albisu M., Pérez-Elortondo F.J., Ruiz de Gordoa J.C., de Renobales M., Barron L.J.R. Effects of seasonal changes in feeding management under part-time grazing on terpene concentrations of ewes' milk. J. Dairy Res. 2011;78:129–135. doi: 10.1017/S0022029910000750. [DOI] [PubMed] [Google Scholar]
- Aldalur A., Bustamante M.Á., Salmerón J., Barron L.J.R. Relationships between cheese-processing conditions and curd and cheese properties to improve the yield of Idiazabal cheese made in small artisan dairies: a multivariate approach. J. Dairy Sci. 2021;104:253–269. doi: 10.3168/JDS.2020-18926. [DOI] [PubMed] [Google Scholar]
- Amores G., Pérez-Elortondo F.J., Albisu M., Barron L.J.R. Short communication: to what extent do environmental or technological conditions affect the sensory differentiation of raw Ewe milk cheeses produced in valley or mountain farms? J. Dairy Sci. 2021;104:301–307. doi: 10.3168/JDS.2020-18358. [DOI] [PubMed] [Google Scholar]
- Arora G., Cormier F., Lee B. Analysis of odor-active volatiles in cheddar cheese headspace by multidimensional GC/MS/sniffing. J. Agric. Food Chem. 1995;43:748–752. doi: 10.1021/jf00051a035. [DOI] [Google Scholar]
- Attaie R. Quantification of volatile compounds in goat milk Jack cheese using static headspace gas chromatography. J. Dairy Sci. 2009;92:2435–2443. doi: 10.3168/JDS.2008-1732. [DOI] [PubMed] [Google Scholar]
- Ballesteros C., Poveda J.M., González-Viñas M.A., Cabezas L. Microbiological, biochemical and sensory characteristics of artisanal and industrial Manchego cheeses. Food Control. 2006;17:249–255. doi: 10.1016/j.foodcont.2004.10.008. [DOI] [Google Scholar]
- Barlow I., Lloyd G., Ramshaw E., Miller A., McCabe G., McCabe L. Correlations and changes in flavours and chemical parameters of Cheddar cheeses during maturation. Aust. J. Dairy Technol. 1989;5:7–18. [Google Scholar]
- Barron L.J.R., Redondo Y., Aramburu M., Pérez-Elortondo F.J., Albisu M., Nájera A.I., de Renobales M. Variations in volatile compounds and flavour in Idiazabal cheese manufactured from Ewe's milk in farmhouse and factory. J. Sci. Food Agric. 2005;85:1660–1671. doi: 10.1002/jsfa.2175. [DOI] [Google Scholar]
- Barron L.J.R., Redondo Y., Flanagan C.E., Pérez-Elortondo F.J., Albisu M., Nájera A.I., de Renobales M., Fernández-García E. Comparison of the volatile composition and sensory characteristics of Spanish PDO cheeses manufactured from ewes' raw milk and animal rennet. Int. Dairy J. 2005;15:371–382. doi: 10.1016/j.idairyj.2004.08.005. [DOI] [Google Scholar]
- Barron L.J.R., Redondo Y., Aramburu M., Gil P., Pérez-Elortondo F.J., Albisu M., Nájera A.I., de Renobales M., Fernández-García E. Volatile composition and sensory properties of industrially produced Idiazabal cheese. Int. Dairy J. 2007;17:1401–1414. doi: 10.1016/j.idairyj.2007.04.001. [DOI] [Google Scholar]
- Belviso S., Giordano M., Dolci P., Zeppa G. Degradation and biosynthesis of terpenoids by lactic acid bacteria isolated from cheese: first evidence. Dairy Sci. Technol. 2011;91:227–236. doi: 10.1007/s13594-011-0003-z. [DOI] [Google Scholar]
- Bertuzzi A.S., McSweeney P.L.H., Rea M.C., Kilcawley K.N. Detection of volatile compounds of cheese and their contribution to the flavor profile of surface-ripened cheese. Compr. Rev. Food Sci. Food Saf. 2018;17:371–390. doi: 10.1111/1541-4337.12332. [DOI] [PubMed] [Google Scholar]
- Boletín Oficial del Estado Orden del 30 de noviembre por la que se aprueba el Reglamento de la Denominación de Origen “Idiazabal” y su Consejo Regulador. BOE. 1993;289:34591–34596. https://www.mapa.gob.es/images/es/boe_289_031293_tcm30-523120.pdf [Google Scholar]
- Boscaini E., Van Ruth S., Biasioli F., Gasperi F., Märk T.D. Gas Chromatography−Olfactometry (GC-O) and proton transfer Reaction−Mass spectrometry (PTR-MS) analysis of the flavor profile of Grana Padano, Parmigiano Reggiano, and Grana trentino cheeses. J. Agric. Food Chem. 2003;51:1782–1790. doi: 10.1021/JF020922G. [DOI] [PubMed] [Google Scholar]
- Bouhaddani S. el, Houwing-Duistermaat J., Salo P., Perola M., Jongbloed G., Uh H.-W. Evaluation of O2PLS in Omics data integration. BMC Bioinf. 2016;17:S11. doi: 10.1186/S12859-015-0854-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Câmara J.S., Lourenço S., Silva C., Lopes A., Andrade C., Perestrelo R. Exploring the potential of wine industry by-products as source of additives to improve the quality of aquafeed. Microchem. J. 2020;155 doi: 10.1016/J.MICROC.2020.104758. [DOI] [Google Scholar]
- Campo E., Ferreira V., López R., Escudero A., Cacho J. Identification of three novel compounds in wine by means of a laboratory-constructed multidimensional gas chromatographic system. J. Chromatogr. A. 2006;1122:202–208. doi: 10.1016/J.CHROMA.2006.04.048. [DOI] [PubMed] [Google Scholar]
- Cardinali F., Ferrocino I., Milanović V., Belleggia L., Corvaglia M.R., Garofalo C., Foligni R., Mannozzi C., Mozzon M., Cocolin L., Osimani A., Aquilanti L. Microbial communities and volatile profile of Queijo de Azeitão PDO cheese, a traditional Mediterranean thistle-curdled cheese from Portugal. Food Res. Int. 2021;147 doi: 10.1016/J.FOODRES.2021.110537. [DOI] [PubMed] [Google Scholar]
- Chávarri F., Santisteban A., Virto M., de Renobales M. Alkaline phosphatase, acid phosphatase, lactoperoxidase, and lipoprotein lipase activities in industrial Ewe's milk and cheese. J. Agric. Food Chem. 1998;46:2926–2932. doi: 10.1021/JF970968N. [DOI] [Google Scholar]
- Chen C., Huang K., Yu H., Tian H. The diversity of microbial communities in Chinese milk fan and their effects on volatile organic compound profiles. J. Dairy Sci. 2021;104:2581–2593. doi: 10.3168/JDS.2020-19053. [DOI] [PubMed] [Google Scholar]
- Chen C., Zhou W., Yu H., Yuan J., Tian H. Characterization of major odor-active compounds responsible for nutty flavor in Cheddar cheese according to Chinese taste. Flavour Fragrance J. 2021;36:171–181. doi: 10.1002/FFJ.3627. [DOI] [Google Scholar]
- Curioni P.M.G., Bosset J.O. Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int. Dairy J. 2002;12:959–984. doi: 10.1016/S0958-6946(02)00124-3. [DOI] [Google Scholar]
- De Filippis F., Genovese A., Ferranti P., Gilbert J.A., Ercolini D. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Martínez F.J., Carrapiso A.I., Contador R., Ramírez M.R. Volatile compounds and sensory changes after high pressure processing of mature “Torta del Casar” (raw Ewe's milk cheese) during refrigerated storage. Innovat. Food Sci. Emerg. Technol. 2019;52:34–41. doi: 10.1016/J.IFSET.2018.11.004. [DOI] [Google Scholar]
- Dimov S.G., Gyurova A., Zagorchev L., Dimitrov T., Georgieva-Miteva D., Peykov S. NGS-based metagenomic study of four traditional Bulgarian green cheeses from tcherni vit. LWT--Food Sci. Technol. 2021;152 doi: 10.1016/j.lwt.2021.112278. [DOI] [Google Scholar]
- Fernández-García E., Gaya P., Medina M., Nuñez M. Evolution of the volatile components of raw ewes' milk Castellano cheese: seasonal variation. Int. Dairy J. 2004;14:39–46. doi: 10.1016/S0958-6946(03)00148-1. [DOI] [Google Scholar]
- Fonseca H.C., Melo D. de S., Ramos C.L., Menezes A.G.T., Dias D.R., Schwan R.F. Sensory and flavor-aroma profiles of passion fruit juice fermented by potentially probiotic Lactiplantibacillus plantarum CCMA 0743 strain. Food Res. Int. 2021;152 doi: 10.1016/J.FOODRES.2021.110710. [DOI] [PubMed] [Google Scholar]
- Fox P.F., Guinee T.P., Cogan T.M., McSweeney P.L.H. Fundamentals of Cheese Science. second. Springer US; New York: 2017. [DOI] [Google Scholar]
- Gaglio R., Cruciata M., Scatassa M.L., Tolone M., Mancuso I., Cardamone C., Corona O., Todaro M., Settanni L. Influence of the early bacterial biofilms developed on vats made with seven wood types on PDO Vastedda della valle del Belìce cheese characteristics. Int. J. Food Microbiol. 2019;291:91–103. doi: 10.1016/J.IJFOODMICRO.2018.11.017. [DOI] [PubMed] [Google Scholar]
- Gaglio R., Todaro M., Scatassa M.L., Franciosi E., Corona O., Mancuso I., Di Gerlando R., Cardamone C., Settanni L. Transformation of raw ewes' milk applying “Grana” type pressed cheese technology: development of extra-hard “Gran Ovino” cheese. Int. J. Food Microbiol. 2019;307 doi: 10.1016/J.IJFOODMICRO.2019.108277. [DOI] [PubMed] [Google Scholar]
- Galindo-Prieto B., Eriksson L., Trygg J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS) J. Chemometr. 2014;28:623–632. doi: 10.1002/CEM.2627. [DOI] [Google Scholar]
- Gezginc Y., Karabekmez-Erdem T., Tatar H.D., Dağgeçen E.C., Ayman S., Akyol İ. Metagenomics and volatile profile of Turkish artisanal Tulum cheese microbiota. Food Biosci. 2021;45 doi: 10.1016/j.fbio.2021.101497. [DOI] [Google Scholar]
- Groot M.N.N., de Bont J.A.M. Conversion of phenylalanine to benzaldehyde initiated by an aminotransferase in Lactobacillus plantarum. Appl. Environ. Microbiol. 1998;64:3009. doi: 10.1128/aem.64.8.3009-3013.1998. /pmc/articles/PMC106807/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwood J.F., Lloyd G.T., Ramshaw H., Stark W. An off-flavour associated with the use of sorbic acid during Feta cheese maturation. Aust. J. Dairy Technol. 1981;36:38–40. [Google Scholar]
- Jiang Y., Li N., Wang Q., Liu Z., Lee Y.K., Liu X., Zhao J., Zhang H., Chen W. Microbial diversity and volatile profile of traditional fermented yak milk. J. Dairy Sci. 2020;103:87–97. doi: 10.3168/jds.2019-16753. [DOI] [PubMed] [Google Scholar]
- Jung H.J., Ganesan P., Lee S.J., Kwak H.S. Comparative study of flavor in cholesterol-removed Gouda cheese and Gouda cheese during ripening. J. Dairy Sci. 2013;96:1972–1983. doi: 10.3168/JDS.2012-5644. [DOI] [PubMed] [Google Scholar]
- Juric M., Bertelsen G., Mortensen G., Petersen M.A. Light-induced colour and aroma changes in sliced, modified atmosphere packaged semi-hard cheeses. Int. Dairy J. 2003;13:239–249. doi: 10.1016/S0958-6946(02)00156-5. [DOI] [Google Scholar]
- Karagul Yuceer Y., Tuncel B., Guneser O., Engin B., Isleten M., Yasar K., Mendes M. Characterization of aroma-active compounds, sensory properties, and proteolysis in Ezine cheese. J. Dairy Sci. 2009;92:4146–4157. doi: 10.3168/JDS.2009-2124. [DOI] [PubMed] [Google Scholar]
- Kilcawley K.N., Faulkner H., Clarke H.J., O'Sullivan M.G., Kerry J.P. Factors influencing the flavour of bovine milk and cheese from grass based versus non-grass based milk production systems. Foods. 2018;7:37. doi: 10.3390/FOODS7030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:1–11. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. Package “pheatmap”. 2019. https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf Retrieved from.
- Kubícková J., Grosch W. Quantification of potent odorants in camembert cheese and calculation of their odour activity values. Int. Dairy J. 1998;8:17–23. doi: 10.1016/S0958-6946(98)00014-4. [DOI] [Google Scholar]
- Le Quéré J.-L., Buchin S. In: Encyclopedia of Dairy Sciences. McSweeney P.L.H., McNamara J.P., editors. Academic Press; Amsterdam: 2022. Cheese flavor; pp. 79–90. [DOI] [Google Scholar]
- Li K., Tang J., Zhang Z., Wu Z., Zhong A., Li Z., Wang Y. Correlation between flavor compounds and microorganisms of Chaling natural fermented red sufu. LWT--Food Sci. Technol. 2022;154 doi: 10.1016/J.LWT.2021.112873. [DOI] [Google Scholar]
- Liu S.Q., Holland R., Crow V.L. Esters and their biosynthesis in fermented dairy products: a review. Int. Dairy J. 2004;14:923–945. doi: 10.1016/J.IDAIRYJ.2004.02.010. [DOI] [Google Scholar]
- Majcher M.A., Jeleń H.H. Key odorants of Oscypek, a traditional Polish Ewe's milk cheese. J. Agric. Food Chem. 2011;59:4932–4937. doi: 10.1021/jf2002602. [DOI] [PubMed] [Google Scholar]
- Meng Y., Chen X., Sun Z., Li Y., Chen D., Fang S., Chen J. Exploring core microbiota responsible for the production of volatile flavor compounds during the traditional fermentation of Koumiss. LWT--Food Sci. Technol. 2021;135 doi: 10.1016/j.lwt.2020.110049. [DOI] [Google Scholar]
- Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moio L., Piombino P., Addeo F. Odour-impact compounds of Gorgonzola cheese. J. Dairy Res. 2000;67:273–285. doi: 10.1017/S0022029900004106. [DOI] [PubMed] [Google Scholar]
- Mukhia S., Kumar A., Kumar R. Generation of antioxidant peptides from soy protein isolate through psychrotrophic Chryseobacterium sp. derived alkaline broad temperature active protease. LWT--Food Sci. Technol. 2021;143 doi: 10.1016/j.lwt.2021.111152. [DOI] [Google Scholar]
- Natrella G., Faccia M., Lorenzo J.M., De Palo P., Gambacorta G. Short communication: sensory characteristics and volatile organic compound profile of high-moisture mozzarella made by traditional and direct acidification technology. J. Dairy Sci. 2020;103:2089–2097. doi: 10.3168/JDS.2019-17059. [DOI] [PubMed] [Google Scholar]
- Official Journal of the European Communities Commission Regulation (EC) No 1107/96 of 12 June 1996 on the registration of geographical indications and designations of origin under the procedure laid down in Article 17 of Council Regulation (EEC) No 2081/92. Q. J. Electron. Commer. QJEC. 1996;148:1–10. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A31996R1107 [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., Mcglinn D., Minchin P.R., O’hara R.B., Simpson G.L., Solymos P., Henry M., Stevens H., Szoecs E., Wagner H. Package “vegan. 2020. https://cran.r-project.org/web/packages/vegan/vegan.pdf Retrieved from.
- Poveda J.M., Sánchez-Palomo E., Pérez-Coello M.S., Cabezas L. Volatile composition, olfactometry profile and sensory evaluation of semi-hard Spanish goat cheeses. Dairy Sci. Technol. 2008;88:355–367. doi: 10.1051/dst:2007021. [DOI] [Google Scholar]
- Povolo M., Contarini G., Mele M., Secchiari P. Study on the influence of pasture on volatile fraction of ewes' dairy products by solid-phase microextraction and gas chromatography-mass spectrometry. J. Dairy Sci. 2007;90:556–569. doi: 10.3168/JDS.S0022-0302(07)71539-4. [DOI] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M.C., Reineccius G. Identification of aroma compounds in Parmigiano-Reggiano cheese by gas chromatography/olfactometry. J. Dairy Sci. 2002;85:1362–1369. doi: 10.3168/jds.S0022-0302(02)74202-1. [DOI] [PubMed] [Google Scholar]
- Qian M.C., Reineccius G. Quantification of aroma compounds in Parmigiano Reggiano cheese by a dynamic headspace gas chromatography-mass spectrometry technique and calculation of odor activity value. J. Dairy Sci. 2003;86:770–776. doi: 10.3168/JDS.S0022-0302(03)73658-3. [DOI] [PubMed] [Google Scholar]
- R Core Team The R Project for statistical computing. 2020. https://www.r-project.org
- Rathakrishnan D., Gopalan A.K. Isolation and characterization of halophilic isolates from Indian salterns and their screening for production of hydrolytic enzymes. Environmental Challenges. 2022;6 doi: 10.1016/J.ENVC.2021.100426. [DOI] [Google Scholar]
- Santamarina-García G., Hernández I., Amores G., Virto M. Characterization of microbial shifts during the production and ripening of raw Ewe milk-derived idiazabal cheese by high-throughput sequencing. Biology-Basel. 2022;11:769. doi: 10.3390/BIOLOGY11050769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamarina-García G., Amores G., López de Armentia E., Hernández I., Virto M. Relationship between the dynamics of gross composition, free fatty acids and biogenic amines, and microbial shifts during the ripening of raw Ewe milk-derived idiazabal cheese. Animals. 2022;12:3224. doi: 10.3390/ani12223224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhir S.T., Amanpour A., Bouseta A., Selli S. Fingerprint of aroma-active compounds and odor activity values in a traditional Moroccan fermented butter “Smen” using GC–MS–Olfactometry. J. Food Compos. Anal. 2021;96 doi: 10.1016/j.jfca.2020.103761. [DOI] [Google Scholar]
- Singh T.K., Cadwallader K.R., Drake M. In: Handbook of Food Products Manufacturing. Hui Y.H., editor. John Wiley & Sons, Inc.; New Jersey: 2006. Biochemical processes in the production of flavor in milk and milk products; pp. 715–748. [DOI] [Google Scholar]
- Starowicz M. Analysis of volatiles in food products. Separations. 2021;8:157. doi: 10.3390/SEPARATIONS8090157. [DOI] [Google Scholar]
- Sympoura F., Cornu A., Tournayre P., Massouras T., Berdagué J.L., Martin B. Odor compounds in cheese made from the milk of cows supplemented with extruded linseed and α-tocopherol. J. Dairy Sci. 2009;92:3040–3048. doi: 10.3168/JDS.2008-1802. [DOI] [PubMed] [Google Scholar]
- Takeoka G.R., Buttery R.G., Turnbaugh J.G., Benson M. Odor thresholds of various branched esters. LWT--Food Sci. Technol. 1995;28:153–156. doi: 10.1016/S0023-6438(95)80028-X. [DOI] [Google Scholar]
- Taylor K., Wick C., Castada H., Kent K., Harper W.J. Discrimination of Swiss cheese from 5 different factories by high impact volatile organic compound profiles determined by odor activity value using selected ion flow tube mass spectrometry and odor threshold. J. Food Sci. 2013;78:C1509–C1515. doi: 10.1111/1750-3841.12249. [DOI] [PubMed] [Google Scholar]
- The Good Scents Company Information System Providing information for the flavor, fragrance, food and cosmetic industries. 2021. https://www.thegoodscentscompany.com
- Thierry A., Collins Y.F., Mukdsi M.C.A., McSweeney P.L.H., Wilkinson M.G., Spinnler H.E. In: Cheese: Chemistry, Physics and Microbiology. fourth ed. McSweeney P.L.H., Fox P.J., Cotter P.D., Everett D.W., editors. Academic Press; London: 2017. Lipolysis and metabolism of fatty acids in cheese; pp. 423–444. [DOI] [Google Scholar]
- Trygg J., Wold S. O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integral OSC filter. J. Chemometr. 2003;17:53–64. doi: 10.1002/cem.775. [DOI] [Google Scholar]
- Urbach G. Relations between cheese flavour and chemical composition. Int. Dairy J. 1993;3:389–422. doi: 10.1016/0958-6946(93)90025-U. [DOI] [Google Scholar]
- Valdivielso I., Albisu M., de Renobales M., Barron L.J.R. Changes in the volatile composition and sensory properties of cheeses made with milk from commercial sheep flocks managed indoors, part-time grazing in valley, and extensive mountain grazing. Int. Dairy J. 2016;53:29–36. doi: 10.1016/j.idairyj.2015.09.007. [DOI] [Google Scholar]
- van Gemert L.J. Odour Thresholds: Compilations of Odour Threshold Values in Air, Water and Other Media. Second. Oliemans Punter & Partners BV; Zeist: 2011. [Google Scholar]
- Viallon C., Martin B., Verdier-Metz I., Pradel P., Garel J.-P., Coulon J.-B., Berdagué J.-L. Transfer of monoterpenes and sesquiterpenes from forages into milk fat. Lait. 2000;80:635–641. doi: 10.1051/LAIT:2000150. [DOI] [Google Scholar]
- Villeneuve M.P., Lebeuf Y., Gervais R., Tremblay G.F., Vuillemard J.C., Fortin J., Chouinard P.Y. Milk volatile organic compounds and fatty acid profile in cows fed timothy as hay, pasture, or silage. J. Dairy Sci. 2013;96:7181–7194. doi: 10.3168/JDS.2013-6785. [DOI] [PubMed] [Google Scholar]
- Virto M., Chávarri F., Bustamante M.A., Barron L.J.R., Aramburu M., Vicente M.S., Pérez-Elortondo F.J., Albisu M., de Renobales M. Lamb rennet paste in ovine cheese manufacture. Lipolysis and flavour. Int. Dairy J. 2003;13:391–399. doi: 10.1016/S0958-6946(03)00007-4. [DOI] [Google Scholar]
- Wang B., Wang J., Xu L.Y., Zhang J.H., Ai N.S., Cao Y.P. Characterization of the key odorants in kurut with aroma recombination and omission studies. J. Dairy Sci. 2020;103:4164–4173. doi: 10.3168/JDS.2019-17521. [DOI] [PubMed] [Google Scholar]
- Wang J., Yang Z.J., Xu L.Y., Wang B., Zhang J.H., Li B.Z., Cao Y.P., Tan L. Key aroma compounds identified in Cheddar cheese with different ripening times by aroma extract dilution analysis, odor activity value, aroma recombination, and omission. J. Dairy Sci. 2021;104:1576–1590. doi: 10.3168/JDS.2020-18757. [DOI] [PubMed] [Google Scholar]
- Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M., Venables B., Galili T. Package “gplots”. 2020. https://cran.r-project.org/web/packages/gplots/gplots.pdf Retrieved from.
- Whetstine M.E.C., Cadwallader K.R., Drake M.A. Characterization of aroma compounds responsible for the rosy/floral flavor in Cheddar cheese. J. Agric. Food Chem. 2005;53:3126–3132. doi: 10.1021/JF048278O. [DOI] [PubMed] [Google Scholar]
- Xia Y., Yu J., Liu H., Feng C., Shuang Q. Novel insight into physicochemical and flavor formation in koumiss based on microbial metabolic network. Food Res. Int. 2021;149 doi: 10.1016/J.FOODRES.2021.110659. [DOI] [PubMed] [Google Scholar]
- Xia Y., Oyunsuren E., Yang Y., Shuang Q. Comparative metabolomics and microbial communities associated network analysis of black and white horse- sourced koumiss. Food Chem. 2022;370 doi: 10.1016/J.FOODCHEM.2021.130996. [DOI] [PubMed] [Google Scholar]
- Yang P., Zhong G., Yang J., Zhao L., Sun D., Tian Y., Li R., Rong L. Metagenomic and metabolomic profiling reveals the correlation between the microbiota and flavor compounds and nutrients in fermented sausages. Food Chem. 2021;375 doi: 10.1016/J.FOODCHEM.2021.131645. [DOI] [PubMed] [Google Scholar]
- Yao Y., Zhou X., Hadiatullah H., Zhang J., Zhao G. Determination of microbial diversities and aroma characteristics of Beitang shrimp paste. Food Chem. 2021;344 doi: 10.1016/J.FOODCHEM.2020.128695. [DOI] [PubMed] [Google Scholar]
- Yeluri Jonnala B.R., McSweeney P.L.H., Sheehan J.J., Cotter P.D. Sequencing of the cheese microbiome and its relevance to industry. Front. Microbiol. 2018;9:1020. doi: 10.3389/fmicb.2018.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon M., Rijnen L. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 2001;11:185–201. doi: 10.1016/S0958-6946(01)00049-8. [DOI] [Google Scholar]
- Zabaleta L., Gourrat K., Barron L.J.R., Albisu M., Guichard E. Identification of odour-active compounds in ewes' raw milk commercial cheeses with sensory defects. Int. Dairy J. 2016;58:23–30. doi: 10.1016/j.idairyj.2016.01.018. [DOI] [Google Scholar]
- Zhang J., Li M., Zhang H., Pang X. Comparative investigation on aroma profiles of five different mint (Mentha) species using a combined sensory, spectroscopic and chemometric study. Food Chem. 2022;371 doi: 10.1016/J.FOODCHEM.2021.131104. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang Y., Li C., Li L., Yang X., Wu Y., Chen S., Zhao Y. Novel insight into physicochemical and flavor formation in naturally fermented tilapia sausage based on microbial metabolic network. Food Res. Int. 2021;141 doi: 10.1016/J.FOODRES.2021.110122. [DOI] [PubMed] [Google Scholar]
- Zhao D., Hu J., Chen W. Analysis of the relationship between microorganisms and flavour development in dry-cured grass carp by high-throughput sequencing, volatile flavour analysis and metabolomics. Food Chem. 2022;368 doi: 10.1016/j.foodchem.2021.130889. [DOI] [PubMed] [Google Scholar]
- Zheng X., Liu F., Shi X., Wang B., Li K., Li B., Zhuge B. Dynamic correlations between microbiota succession and flavor development involved in the ripening of Kazak artisanal cheese. Food Res. Int. 2018;105:733–742. doi: 10.1016/j.foodres.2017.12.007. [DOI] [PubMed] [Google Scholar]
- Zhong A., Chen W., Duan Y., Li K., Tang X., Tian X., Wu Z., Li Z., Wang Y., Wang C. The potential correlation between microbial communities and flavors in traditional fermented sour meat. LWT--Food Sci. Technol. 2021;149 doi: 10.1016/j.lwt.2021.111873. [DOI] [Google Scholar]
- Zuljan F.A., Mortera P., Alarcón S.H., Blancato V.S., Espariz M., Magni C. Lactic acid bacteria decarboxylation reactions in cheese. Int. Dairy J. 2016;62:53–62. doi: 10.1016/j.idairyj.2016.07.007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.