Significance
A way to treat genetically defined diseases is to target the mRNAs that emerge from mutated genes with complementary oligonucleotides that recruit enzymes to destroy the mRNA. There are several limitations to this approach. Highly structured RNA repeat expansions cannot be directly targeted with oligonucleotides and require direct injection into the central nervous system to achieve therapeutic tissue distribution. In this study, we report the design of a brain penetrant small molecule that targets the toxic RNA structure that causes the most common form of inherited ALS. In mouse models, the compound enters the brain and binds the target to eliminate only the disease-causing mRNA by facilitating its degradation by natural decay processes.
Keywords: drug design, RNA, repeat expansion disorders, RNA-targeted degradation, induced proximity
Abstract
A hexanucleotide repeat expansion in intron 1 of the C9orf72 gene is the most common genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia, or c9ALS/FTD. The RNA transcribed from the expansion, r(G4C2)exp, causes various pathologies, including intron retention, aberrant translation that produces toxic dipeptide repeat proteins (DPRs), and sequestration of RNA-binding proteins (RBPs) in RNA foci. Here, we describe a small molecule that potently and selectively interacts with r(G4C2)exp and mitigates disease pathologies in spinal neurons differentiated from c9ALS patient-derived induced pluripotent stem cells (iPSCs) and in two c9ALS/FTD mouse models. These studies reveal a mode of action whereby a small molecule diminishes intron retention caused by the r(G4C2)exp and allows the liberated intron to be eliminated by the nuclear RNA exosome, a multi-subunit degradation complex. Our findings highlight the complexity of mechanisms available to RNA-binding small molecules to alleviate disease pathologies and establishes a pipeline for the design of brain penetrant small molecules targeting RNA with novel modes of action in vivo.
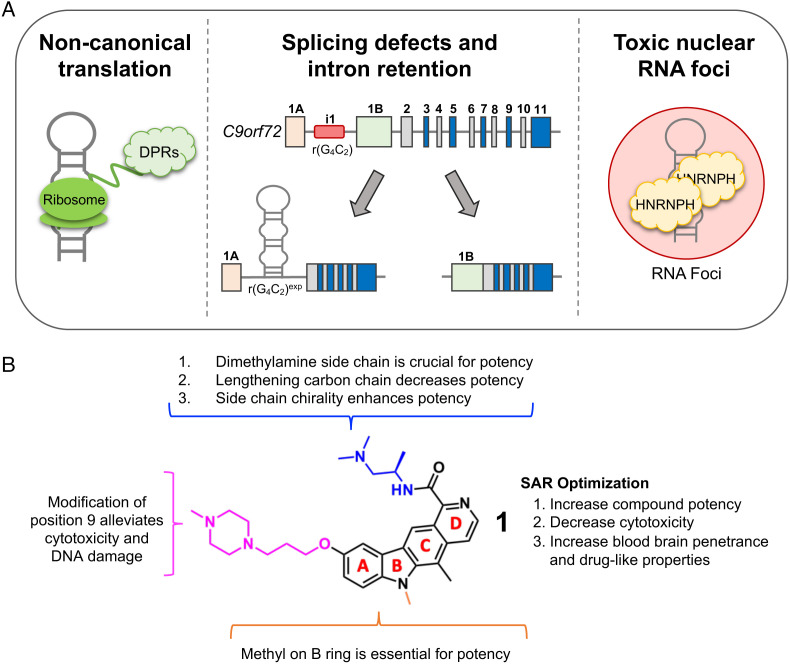
A hexanucleotide repeat expansion of GGGGCC (G4C2exp) located in the first intron of chromosome 9 open-reading frame 72 (C9orf72) is the most common genetic cause of familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), collectively referred to as c9ALS/FTD (1, 2). These diseases are characterized by devastating deficits in cognitive, behavioral, language, and/or motor functions (3, 4). Healthy individuals generally retain fewer than 30 G4C2 repeats in the C9orf72 gene, while affected individuals have greater than 65 repeats with repeat lengths commonly in the thousands (5, 6). The toxic RNA derived from the repeat expansion, r(G4C2)exp, induces disease pathology by at least two mechanisms: (i) noncanonical translation of the expanded repeat [repeat associated non-AUG (RAN) translation] that generates toxic dipeptide repeat proteins (DPRs) (7–12) and (ii) formation of toxic foci composed of the repeat expansion (12, 13) and various RNA-binding proteins (RBPs), which the RNA sequesters and inactivates (9, 14). A driving force of these pathomechanisms is the aberrant retention of the repeat-containing intron 1, which in healthy cells is spliced out and eliminated by native RNA degradation pathways (Fig. 1A) (15).
Fig. 1.
Hallmarks of c9ALS/FTD disease pathology caused by r(G4C2)exp and identification of a lead small molecule that alleviates disease-associated pathologies. (A) Schematic of disease pathologies caused by r(G4C2)exp in c9ALS/FTD and the secondary structure of the hairpin formed by the RNA repeat expansion. (B) Structure of lead compound 1 with SAR indicated.
Intriguingly, r(G4C2)exp, but not short repeats of non-pathological length, folds into stable structures (16, 17), suggesting that structure-specific small molecules may reduce its pathology. Further, some microsatellite diseases are caused by the same repeat expansion harbored in different genes, and different repeating sequences can share similar RNA structures, such as r(CGG)exp and r(G4C2)exp. Therefore, targeting pathological mRNAs by recognizing disease-driving RNA structures with small molecules could lead to the development of a single compound that could target all gene products that harbor a disease-driving microsatellite expansion (18). Herein, we report the identification of a lead small molecule 1 that binds r(G4C2)exp in cells. The molecule was optimized to be potent, selective, and brain penetrant, while mechanistic studies demonstrate that it facilitates the decay of the toxic repeat by native RNA quality control mechanisms. Eliminating the repeat expansion mitigates the downstream pathology in various c9ALS/FTD patient-derived cell lines, including spinal neurons differentiated from induced pluripotent stem cell (iPSC), and in two mouse models of c9ALS/FTD.
Results
Identifying Lead Chemical Matter to Inhibit r(G4C2)exp-Associated Pathologies.
Using a previously validated cellular assay to identify selective inhibitors of r(G4C2)exp RAN translation (SI Appendix, Fig. S1A) (16), we studied the Repurposing, Focused Rescue, and Accelerated Medchem (ReFRAME) small molecule library (19) to identify new scaffolds that alleviate c9ALS/FTD pathologies. The library is a collection of small molecules that have undergone preclinical evaluation or clinical development (n = 10,928). From this screen, we identified 69 small molecules (0.6% hit rate) that inhibited RAN translation, but not canonical translation (SI Appendix, Fig. S1A).
Since c9ALS/FTD is a disease of the central nervous system (CNS), we refined the pool of hit compounds by their potential for blood–brain barrier (BBB) penetrance. This potential can be quantified by assessing the physicochemical properties of a compound and comparing them to known small molecule medicines that cross the BBB, or a Central Nervous System Multiparameter Optimization Desirability (CNS-MPO) score (20, 21). Although originally developed for protein-targeted medicines, a CNS-MPO score is a composite of the calculated partition coefficient (cLogP), molecular weight, pKa of the most basic center, topological polar surface area, and number of hydrogen-bond donors, assigning a score to each feature from 0 to 1 relative to CNS penetrant drugs, and thus can also be used for RNA-targeted medicines. CNS-MPO scores >3.5 indicate that a small molecule has high potential for BBB penetrance.
Applying a CNS-MPO score cutoff of >4.0 to the hits from the ReFRAME screen afforded 16 lead compounds, the IC50s of which were measured in our RAN translation assay as both a secondary validation and to rank potency (SI Appendix, Fig. S1 A and B). Compounds 2–10 inhibited RAN translation dose dependently with IC50s ranging from ~300 nM to ~1,000 nM (SI Appendix, Fig. S1C). Interestingly, these nine compounds are structurally similar to each other and to a previously designed compound that targets r(G4C2)exp (16, 17). All hits share a flat aromatic core-scaffold capable of forming π–π stacking interactions with the RNA, and five have a dimethylamine substituted side chain (highlighted in red SI Appendix, Fig. S1B; 2 (MPO = 4.45), 3 (MPO = 5.23), 4 (MPO = 5.16), 9 (MPO = 4.58), and 10 (MPO = 4.67)).
SAR of Lead Molecule 2.
Of the molecules emanating from the ReFRAME library screen, we selected 2 for lead optimization due to its structural simplicity and potency (IC50 = ~300 nM; SI Appendix, Fig. S1B). Derivatives were designed and synthesized to elucidate structure–activity relationships (SAR; Fig. 1B and SI Appendix, Fig. S2A) and improve compound potency and drug-like properties. After a battery of SAR validations including assessing inhibition of RAN translation, toxicity in cellular models and patient-derived cells, and abundance of poly(GP) (one of six DPRs) in patient-derived lymphoblastoid cell lines (LCLs) (SI Appendix, Fig. S2 B–E), compound 1 was identified as our lead small molecule (Fig. 1B and SI Appendix, Fig. S2). Further evaluation of 1 was carried out through comprehensive characterization in vitro, in patient-derived cells including iPSCs and spinal neurons derived thereof (iPSNs), and two bacterial artificial chromosome (BAC) transgenic mouse models.
Target Engagement of r(G4C2) Repeats by 1 In Vitro Using Chem-CLIP.
A series of studies were completed to understand the RNA targets engaged by 1. We previously developed a method named Chemical Cross-Linking and Isolation by Pull Down (Chem-CLIP) to assess direct target engagement of RNA-binding compounds in vitro and in cells (SI Appendix, Fig. S3A) (22). Chem-CLIP equips an RNA-binding small molecule with cross-linking and purification modules. Here, 1 was functionalized with a diazirine cross-linking module and a biotin purification handle to generate Chem-CLIP probe 35 (SI Appendix, Fig. S3 B, Top). A control Chem-CLIP probe lacking the RNA-binding module, 36, was also synthesized (SI Appendix, Fig. S3 B, Bottom) to assess non-specific cross-linking of the diazirine.
Previous work has demonstrated that the toxic RNA repeat preferentially folds into a hairpin structure with an array of 1×1 GG nucleotide internal loops, over a G-quadruplex (16, 17). Further, this hairpin structure, and not the G-quadruplex, is RAN translated (16). To test which structure is targeted by 1, we folded r(G4C2)8 in the absence or presence of K+, with K+ favoring formation of the G-quadruplex. Also studied were a fully base-paired RNA [r(GGCC)10], the corresponding DNA repeats d(G4C2)8 folded in the absence or presence of K+, and the fully base-paired DNA duplex d(GGCC)10. In vitro, 35 dose-dependently pulled down radioactively labeled r(G4C2)8 folded into the hairpin structure, with ~40% of the RNA pulled down at a concentration of 500 nM (SI Appendix, Fig. S3C). In contrast, control probe 36 failed to pull down the RNA over the same 500-fold concentration range studied for 35 (SI Appendix, Fig. S3C). To confirm that 1 and 35 bind the same site within r(G4C2)8, we performed competitive (C-)Chem-CLIP studies with a constant concentration of Chem-CLIP probe 35 (500 nM) and increasing concentrations of 1. Indeed, 1 dose dependently reduced the percentage of RNA pulled down by 35 (SI Appendix, Fig. S3D), confirming the compounds bind to the same site. As expected, all other nucleic acids studied were pulled down to a significantly lower extent (P < 0.0001) by the Chem-CLIP probe, demonstrating the compound’s selectivity for the hairpin structure (SI Appendix, Fig. S3E).
Binding Affinity and Selectivity of 1.
As in vitro Chem-CLIP studies showed selective, direct target engagement of r(G4C2) repeats by 1 versus other nucleic acids, the affinity and selectivity of 1 for r(G4C2)8, both in the presence and absence of K+, were measured using a direct, in-solution binding assay carried out by microscale thermophoresis. Also studied were d(G4C2)8, fully base-paired r(GGCC)8, as well as r(GGC)16 and r(CGG)16 (causative of fragile X-associated tremor ataxia syndrome), both of which display 1×1 GG internal loops similar to those found in the hairpin form of r(G4C2)exp (SI Appendix, Fig. S4A). In summary, 1 most avidly bound r(G4C2)8 in the hairpin form with a Kd of 560 ± 160 nM (SI Appendix, Fig. S4). Binding of 1 to r(G4C2)8 in the presence of K+, which shifts the equilibrium towards the G-quadruplex form, as well as to r(GGC)16 and r(CGG)16 was ~two-fold weaker (SI Appendix, Fig. S4). No saturable binding was observed to the fully base-paired RNA or d(G4C2)8 (Kds > 20 μM) (SI Appendix, Fig. S4).
Compound 1 Directly Engages r(G4C2)exp in Patient-Derived Cells.
Our preliminary studies both in vitro and in patient-derived LCLs indicate that 1 directly engages r(G4C2)exp to reduce poly(GP) abundance (a known DPR and pharmacodynamic marker for ALS; SI Appendix, Figs. S2–S4). We therefore performed further target validation studies in c9ALS patient-derived iPSCs. Compound 1 was neither toxic to (SI Appendix, Fig. S5A) nor affected cellular proliferation of the iPSCs at concentrations up to 1 µM (SI Appendix, Fig. S5B). Cellular target validation was carried out using two methods, ASO-Bind-Map (22, 23) and Chem-CLIP. ASO-Bind-Map evaluates target engagement through competition between an antisense oligonucleotide (ASO) and a small molecule. A small molecule that binds and thermodynamically stabilizes an RNA can impede molecular recognition by an ASO that shares the same binding site, protecting the RNA from subsequent degradation by ribonuclease H (RNase H; SI Appendix, Fig. S6A). ASO-Bind-Map allows a simplified way to assess target engagement in cells at a site specified by the base-pairing complementarity of the ASO. Patient-derived iPSCs were treated with varying concentrations of 1, followed by transfection with an ASO complementary to r(G4C2)exp. Treatment with the r(G4C2)exp-targeting ASO alone caused a significant (P = 0.0002) decrease in C9orf72 mRNA abundance (measured by qRT-PCR using primers complementary to the exon 2–exon 3 junction present in all C9orf72 splicing isoforms) (SI Appendix, Fig. S6C). Compound 1 protected the RNA from RNase H-mediated degradation in a dose-dependent manner, validating 1’s binding site in cells (SI Appendix, Fig. S6B). Notably, a scrambled ASO had no effect on C9orf72 mRNA abundance in the presence or absence of 1 (SI Appendix, Fig. S6B).
Direct cellular target engagement in patient-derived iPSCs was further evaluated with Chem-CLIP probe 35. To quantify the abundance of RNA enriched by 35, qRT-PCR was performed with primer sets specific to various regions of C9orf72 transcripts. Repeat-containing C9orf72 is alternatively processed into variants that: (i) contain exon 1a and intron 1, which harbors r(G4C2)exp and (ii) that lack exon 1a and intron 1 but contain exon 1b (SI Appendix, Fig. S6C). Thus, the abundance of C9orf72 splicing isoforms with or without the repeat expansion can be measured using the two different sets of primers. Chem-CLIP pulldown fractions of RNA harvested from iPSCs were analyzed using both primer sets. Compound 35 selectively pulled down r(G4C2)exp-containing intron 1 in c9ALS patient-derived iPSCs, enriching the RNA abundance by ~two-fold in the pulled down fraction as compared to the starting lysate (normalized to β-actin; SI Appendix, Fig. S6D). This enrichment was not observed for transcripts lacking the repeat expansion (SI Appendix, Fig. S6E). Further, no enrichment of either transcript (with exon 1a and intron 1 or with exon 1b) was observed in iPSCs from healthy donors (SI Appendix, Fig. S6 D and E).
Compound 1 Alleviates r(G4C2)exp-Associated Pathways in Patient-Derived Cells.
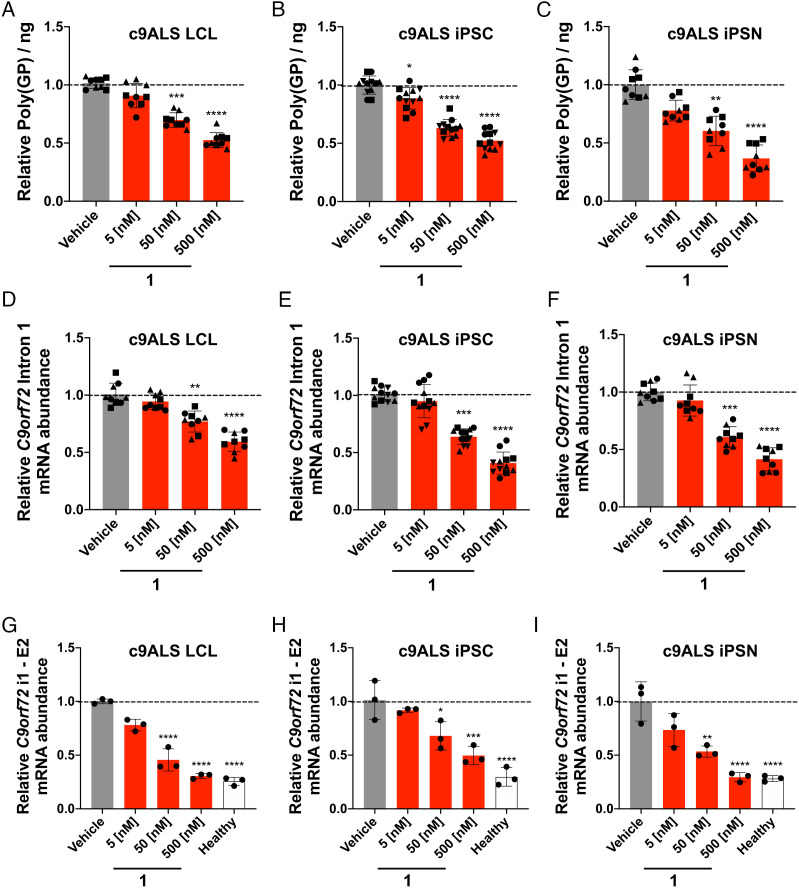
We next expanded our studies to evaluate compound 1 comprehensively across several patient-derived cell lines including LCLs, iPSCs and iPSNs. Across four discrete patient-derived LCLs, 1 dose dependently decreased poly(GP) abundance as determined using an electrochemiluminescent immunoassay (Fig. 2A). This reduction was statistically significant at doses of 50 and 500 nM, which yielded decreases in poly(GP) abundance of 30 ± 6% (P < 0.0001) and 48 ± 6% (P < 0.0001), respectively. In agreement with studies in LCLs, 1 dose dependently reduced poly(GP) in patient-derived iPSCs (n = 4 C9orf72 iPSC lines) with a 48 ± 8% (P < 0.0001) decrease in poly(GP) observed upon treatment with 500 nM compound (Fig. 2B). Importantly, despite the reduction of poly(GP), wild-type (WT) C9ORF72 protein levels were unaffected, as determined by Western blotting (SI Appendix, Fig. S7). These results indicate that 1 is selectively inhibiting the aberrant translation of DPRs while having no effect on the canonical translation of C9ORF72 protein. Moreover, in iPSNs differentiated from three different iPSC lines, 500 nM of 1 reduced poly(GP) by 64 ± 12% (P < 0.0001) (Fig. 2C).
Fig. 2.
Compound 1 selectively alleviates disease-associated defects in patient-derived cells. (A) Effect of 1 on poly(GP) abundance in protein lysate extracted from c9ALS patient-derived LCLs (n = 3 C9orf72 LCLs, three replicates per concentration in each line). (B) Effect of 1 on poly(GP) abundance in protein lysate extracted from c9ALS patient-derived iPSCs (n = 4 C9orf72 iPSC lines, three replicates per concentration in each line). (C) Effect of 1 on poly(GP) abundance in protein lysate extracted from patient-derived iPSNs (n = 3 C9orf72 iPSN lines, three replicates per concentration in each line). (D) Effect of 1 on C9orf72 intron 1 abundance, which harbors r(G4C2)exp, in c9ALS patient-derived LCLs, as determined by qRT-PCR with intron 1-specific primers (n = 3 C9orf72 LCLs, three replicates per concentration in each line). (E) Effect of 1 on C9orf72 intron 1 abundance in c9ALS patient-derived iPSCs, as determined by qRT-PCR with intron 1-specific primers (n = 4 C9orf72 iPSC lines, three replicates per concentration in each line). (F) Effect of 1 on C9orf72 intron 1 abundance in c9ALS patient-derived iPSNs, as determined by qRT-PCR with intron 1-specific primers primers (n = 3 C9orf72 iPSN lines, three replicates per concentration in each line). (G) Effect of 1 on C9orf72 intron 1 3′ splice in a c9ALS patient-derived LCL, as determined by qRT-PCR with primers spanning the intron 1–exon 2 junction (n = 1 C9orf72 LCL, three replicates per concentration). (H) Effect of 1 on C9orf72 intron 1 3′ splice in a c9ALS patient-derived iPSC line, as determined by qRT-PCR with primers spanning the intron 1–exon 2 junction (n = 1 C9orf72 iPSC line, three replicates per concentration). (I) Effect of 1 on C9orf72 intron 1 3′ splice in a c9ALS patient-derived iPSN, as determined by RT-qPCR with primers spanning the intron 1–exon 2 junction (n = 1 C9orf72 iPSN line, three replicates per concentration). RNA quantification was measured relative to GAPDH. Vehicle indicates 0.1% (v/v) dimethyl sulfoxide (DMSO). *P < 0.05, **P < 0.01, ***P < 0.001 , ****P < 0.0001, as determined by a One Way ANOVA with multiple comparisons. Error bars are reported as SD.
Compound 1 Selectively Eliminates C9orf72 Intron 1, Aberrantly Retained in c9ALS/FTD Patient-Derived Cells.
As r(G4C2)exp is harbored in an aberrantly retained intron, it is possible that the small molecule facilitates liberation of the intron, which could then be subsequently decayed by endogenous cellular pathways (24). To examine whether the abundance of intron 1 was indeed reduced upon compound treatment, we employed primers: (i) specific for intron 1 that harbors r(G4C2)exp; (ii) that span the exon 2–exon 3 junction common to all splicing isoforms with or without r(G4C2)exp; and (iii) specific for exon 1b transcripts, which lack r(G4C2)exp (SI Appendix, Fig. S6C). Whether in c9ALS patient-derived LCLs, iPSCs, or iPSNs, the following trends were observed upon treatment with 1: intron 1 abundance was dose-dependently reduced (n = 3 patient-derived LCLs; n = 4 patient-derived iPSC lines; n = 3 patient-derived iPSN lines), while the abundance of transcripts amplified with primers spanning the exon 2–exon 3 junction or specific to exon 1b was unchanged (Fig. 2 D–F and SI Appendix, Fig. S8). Importantly, no changes were observed across all transcripts in lymphoblastoid, iPSC or iPSN lines from healthy donors (n = 1 LCL; n = 4 iPSC lines; n = 3 iPSN cell lines), (SI Appendix, Fig. S9). As the abundance of the downstream exon 2–exon 3 junction was unaffected by 1, it suggests selective decay of the repeat-containing intron 1, rather than degradation of an entire repeat-containing transcript. This is interesting as a previously reported dimeric small molecule that also binds the 1×1 GG internal loops of the r(G4C2)exp does not exhibit this intron-specific decay (25).
Previously, the Swanson laboratory reported that r(G4C2)exp causes retention of C9orf72 intron 1 (15). Compared to control BAC transgenic mice expressing r(G4C2)37, mice expressing r(G4C2)500 have a ~five-fold higher intron retention ratio of transcripts with the intron 1 5′ splice site, and a ~1.5-fold higher intron retention ratio of transcripts with the intron 1 3’ splice site (15). In c9ALS patient-derived LCLs, iPSCs and iPSNs, we observed an intron retention ratio of transcripts with the 3′ splice site of intron 1 of 2.0 ± 0.4 (P < 0.01), 3.7 ± 0.4 (P < 0.0001) and 3.7 ± 0.7 (P < 0.0001), respectively (SI Appendix, Fig. S10 A and B, and Table S2). These ratios were approximately ~three-fold greater than those observed in corresponding cells from healthy donors. Similar results were observed when using primers located within intron 1 (SI Appendix, Fig. S10 A and C, and Table S2). Upon treatment with 1 (50 nM), a decrease in intron 1 abundance was observed in c9ALS patient-derived LCLs, iPSCs and iPSNs using primers for the 3′ splice site of intron 1 (Fig. 2 G–I), with treatment at the 500 nM dose showing C9orf72 intron retention (i1–E2 primers) abundance similar to those in cells from healthy donors. Collectively, these results suggest that 1 facilitates intron 1 splicing and promotes its subsequent decay.
To assess the selectivity of this decay, we evaluated changes caused by 1 in the transcriptome of c9ALS patient-derived iPSCs by RNA sequencing (RNA-seq). Treatment with 100 nM of 1 elicited a significant (P = 0.02) decrease in intron 1 abundance, as assessed by the ratio of intron 1 to exon 2 reads (SI Appendix, Fig. S11 A and B), while total read counts of C9orf72 showed no significant decrease, further supporting the selective processing of the retained intron rather than degradation of the entire transcript (SI Appendix, Fig. S11C). Among 26,957 genes detected, in patient-derived iPSCs, only four genes (0.015%) were significantly affected by compound treatment [false discovery rate (FDR) <0.01; two upregulated and two downregulated] demonstrating selectivity of 1 for r(G4C2)exp (SI Appendix, Fig. S11D). Three of these four genes are also dysregulated in c9ALS iPSCs not treated with 1, meaning their dysregulation is a function of disease pathology, not an off-target of 1. In fact, the dysregulation of these three genes is shifted towards that of healthy cells’ expression levels upon treatment with 1. Furthermore, 289 of the 391 genes (~74%) found to be dysregulated in c9ALS iPSCs were rescued back to that of healthy iPSC expression levels upon treatment with 1 (SI Appendix, Fig. S11). Analysis of total RNA from iPSCs derived from healthy donors showed no significant differences in intron 1 splicing or total C9orf72 read counts for cells treated with 1 (100 nM) compared to vehicle (SI Appendix, Fig. S11 E–G). Of the 27,465 genes detected in healthy iPSCs, 0.098% were significantly affected (P = 0.05), with 13 genes upregulated and 14 genes downregulated (FDR<0.01) transcriptome-wide (SI Appendix, Fig. S11H), demonstrating good overall selectivity. Importantly, none of the genes affected in c9ALS iPSCs and iPSCs derived from healthy donors overlapped, suggesting these are not off-targets modulated by 1.
Sequestration of hnRNP H Causes Retention of Intron 1, Which Is Alleviated by 1.
Sequestration of RBPs by r(G4C2)exp, particularly heterogeneous nuclear ribonucleoprotein H (hnRNP H) (9, 14), causes alternative pre-mRNA splicing defects (8, 9, 26), including intron retention (27) that can be alleviated by small molecules (24). We therefore hypothesized that hnRNP H binding to the repeat-containing intron reduces spliceosome access, leading to intron retention.
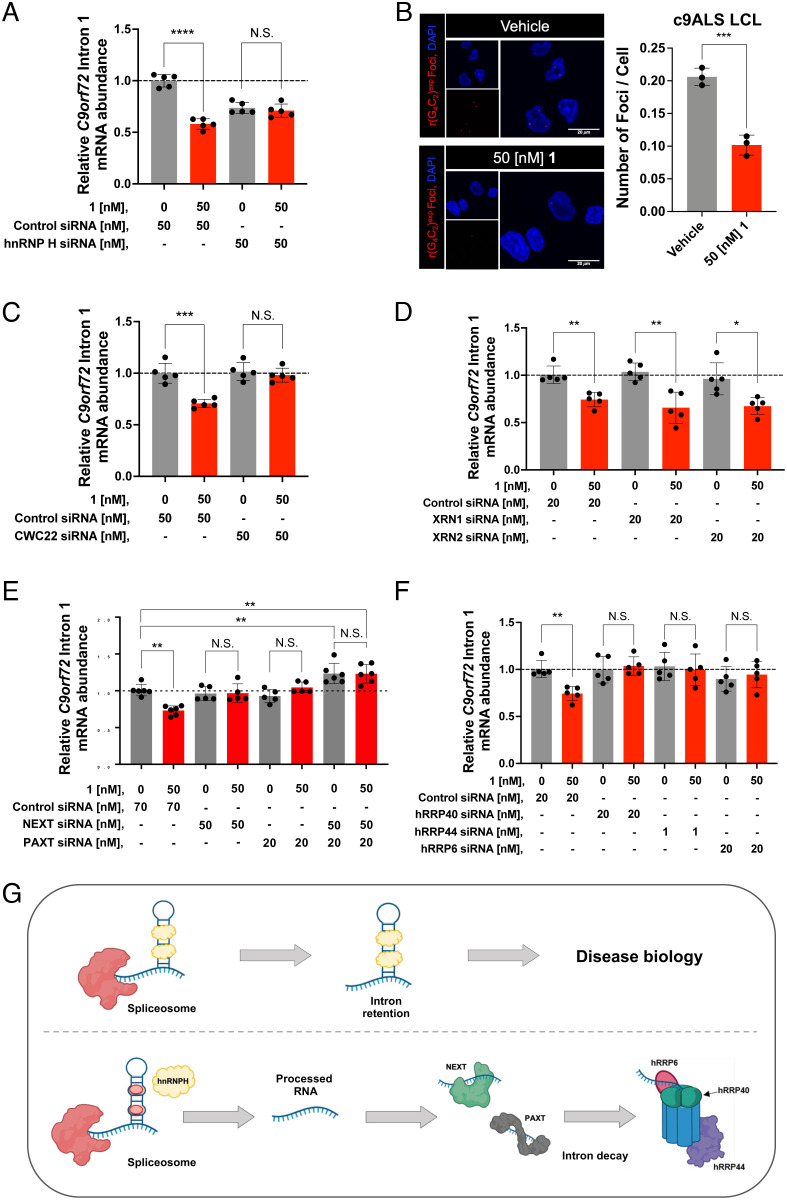
To test this hypothesis, we treated c9ALS iPSCs with an siRNA pool targeting hnRNP H or a randomized siRNA as a control. We first confirmed knockdown of hnRNP H by Western blotting and qRT-PCR, both of which were reduced by ~75% upon treatment of 50 nM siRNA pool (SI Appendix, Fig. S12 A–C). Further, this knockdown exacerbated deregulation of the alternative splicing of Trans-Activation-Responsive RNA-Binding Protein 2 (TARBP2) exon 7 (SI Appendix, Fig. S12D), a known substrate processed by hnRNP H (9, 14). Additionally, the siRNAs targeting hnRNP H reduced C9orf72 intron 1 abundance by 26 ± 5% (P < 0.001), supporting the hypothesis that hnRNP H binding prevents splicing and subsequent decay of the intron (Fig. 3A). Thus, we sought to answer if 1’s mode of action to reduce intron 1 abundance is displacement of hnRNP H.
Fig. 3.
Compound 1 selectively degrades C9orf72 intron 1 via the nuclear exosome and RNA carrier proteins. (A) Effect of co-treating a c9ALS patient-derived iPSC line with 1 and an siRNA targeting hnRNP H, on the abundance of C9orf72 intron 1, as determined by qRT-PCR using intron 1-specific primers (n = 1 c9ALS iPSC line, five replicates per concentration). (B) Left: Representative images of r(G4C2)exp foci in a c9ALS patient-derived LCL imaged by confocal microscopy treated with vehicle or 1. Right: Quantification of relative number of r(G4C2)exp foci per cell (n = 1 c9ALS LCL, three replicates; 200 nuclei counted per biological sample). (C) Effect of co-treating c9ALS patient-derived iPSCs with 1 and an siRNA targeting CWC22, on the abundance of C9orf72 intron 1, as determined by qRT-PCR using intron 1-specific primers (n = 1 c9ALS iPSC line, five replicates per condition). (D) Effect of co-treating c9ALS patient-derived iPSCs with 1, and siRNAs targeting either XRN1 or XRN2 on the abundance of C9orf72 intron 1, as determined by qRT-PCR using intron 1-specific primers (n = 1 c9ALS iPSC line, five replicates per condition). (E) Effect of treating c9ALS patient-derived iPSCs with 1, siRNAs targeting NEXTPAXT, or both, on the abundance of C9orf72 intron 1, as determined by qRT-PCR using intron 1-specific primers (n = 1 c9ALS iPSC line, five replicates per condition). (F) Effect of co-treating c9ALS patient-derived iPSCs with 1 and siRNAs targeting various components of the exosome (hRRP6, hRRP40, or hRRP44), on the abundance of C9orf72 intron 1, as determined by qRT-PCR using intron 1-specific primers (n = 1 c9ALS iPSC line, five replicates per condition). (G) Schematic representation of the RNA decay mechanism of action upon treatment with 1. RNA abundance was measured and quantified relative to GAPDH. Vehicle indicates 0.1% (v/v) DMSO. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, as determined by an Unpaired t test with Welch’s correction. Error bars are repoted as SD.
In vitro, 1 dose-dependently displaced hnRNP H protein from r(G4C2)8 (SI Appendix, Fig. S12E), as assayed by a previously described time-resolved fluorescence resonance energy transfer assay (16). To demonstrate that 1 displaces hnRNP H from r(G4C2)exp in patient-derived cells, we assessed the alternative splicing of TARBP2 exon 7 as well as the formation of toxic RNA foci, which occurs in a subset of CNS cells in c9ALS patients (12, 13). Indeed, 1 (50 nM; co-treated with a randomized siRNA): (i) shifted the TARBP2 exon 7 splicing pattern (increased inclusion) in patient-derived iPSCs toward WT (SI Appendix, Fig. S12D); and (ii) significantly (P = 0.0009) reduced the average number of r(G4C2)exp foci per nucleus in patient-derived LCLs (from 0.21 ± 0.01 to 0.10 ± 0.2; Fig. 3B).
As these data demonstrate displacement of hnRNP H from r(G4C2)exp in patient-derived cells, we investigated the effects of co-treating iPSCs with 1 and either the siRNA pool targeting hnRNP H or a randomized siRNA control, as a complement to our earlier investigations of intron 1 abundance upon treatment with 1. As expected, co-treatment of 1 (50 nM) and a control siRNA resulted in a 42 ± 5% (P < 0.0001) reduction in C9orf72 intron 1 abundance compared to vehicle treatment, as assessed by qRT-PCR using primers specific for C9orf72 intron 1 (Fig. 3A), similar to treatment with 50 nM of 1 alone (Fig. 2D). Interestingly, treatment with the hnRNP H-targeting siRNA alone or combined with 50 nM of 1 reduced intron 1 abundance to a similar extent – by 26 ± 5% and 29 ± 6%, respectively (Fig. 3A). Collectively, these data suggest a mechanism of action in which 1 binds r(G4C2)exp, displaces sequestered hnRNP H, and facilitates subsequent intron processing.
Compound 1 Selectively Interfaces the Toxic r(G4C2)exp Intron with Endogenous Decay Pathways.
To establish that the liberation of the C9orf72 r(G4C2)exp-containing intron is necessary for 1’s mechanism of action, we knocked down CWC22 spliceosome-associated protein homolog (CWC22), a component of the spliceosome required for pre-mRNA splicing, with an siRNA pool (SI Appendix, Fig. S12 F–H). Co-treatment of 1 (50 nM) and the anti-CWC22 siRNAs ablated decay of C9orf72 intron 1 by 1, as assessed by qRT-PCR using primers specific for C9orf72 intron 1 (Fig. 3C), suggesting that the intron must be liberated to be a substrate for native decay pathways.
To explore the fate of the liberated intron, targeted siRNAs were used to elucidate the endogenous decay pathways that contribute to its degradation. Interestingly, the RAN translation of r(G4C2)exp suggests that the intron in which it is harbored may be both nuclear and cytoplasmic. Candidates for its degradation therefore include both cytoplasmic and nuclear decay pathways such as the nuclear or cytoplasmic exosome (3′–5′ exoribonuclease complexes) or the 5′–3′ exoribonucleases XRN2 (nuclear) or XRN1 (cytoplasmic) (28–31).
To assess whether endogenous 5′–3′ RNA decay mechanisms play a role in C9orf72 intron 1 decay, we co-treated c9ALS iPSCs with 1 (50 nM) and siRNAs targeting XRN1 (cytoplasmic) or XRN2 (nuclear) (SI Appendix, Fig. S13). However, upon analysis by qRT-PCR using primers specific for C9orf72 intron 1, no change in the abundance of C9orf72 intron 1 transcripts was observed compared to the control siRNA-treated samples (Fig. 3D). Therefore, these data exclude native 5′–3′ RNA decay pathways from the mechanism of 1-mediated C9orf72 intron 1 decay.
We therefore evaluated the role of the exosome in intron 1 decay. The nuclear exosome comprises nine core proteins and two catalytic subunits (32, 33). Substrates are delivered to the first catalytic subunit of the protein complex, exosome component 10 (hRRP6/EXOSC10) (34), by nuclear exosome targeting complex [NEXT; unadenylated substrates (pA−)] and poly(A) tail exosome targeting connection [PAXT; adenylated substrates (pA+)] (35, 36). The transcript is then funneled through the exosome core (contains hRRP40) and passed to the second catalytic subunit, exosome complex exonuclease RRP44 (hRRP44/DIS3), where the transcript is fully degraded (33). A 3′–5′ exosome-mediated RNA decay mechanism also functions in the cytoplasm, but with the exclusion of NEXT and PAXT, and the inclusion of the cytoplasmic exonuclease hDIS3L (37).
To investigate whether C9orf72 intron 1 is subject to exosomal decay, we co-treated c9ALS iPSCs with 1 and targeted siRNAs that individually knocked down NEXT, PAXT, hRRP6, hRRP40, or hRRP44 (SI Appendix, Figs. S14 and S15). The siRNAs targeting NEXT and PAXT ablated the activity of 1, as assessed by measuring C9orf72 intron 1 abundance by qRT-PCR (Fig. 3E), suggesting the nuclear exosome plays a role in 1-mediated C9orf72 intron 1 decay. Additionally, co-knockdown of NEXT and PAXT resulted in an observed increase in C9orf72 intron 1 abundance, suggesting that both pA− and pA+ intron 1 isoforms exist and are being transported to the nuclear exosome (Fig. 3E and SI Appendix, Fig. S14 G and H). Previous work has shown unadenylated nuclear RNAs can become adenylated and thereby converted into PAXT substrates, supporting this hypothesis (37). Additionally, C9orf72 intron 1 contains several canonical polyadenylation signals, one of which is near the 3′ end of the intron, which may also contribute to the production of (pA+) isoforms.
To further investigate the proposed nuclear exosome-mediated decay pathway, we co-treated c9ALS iPSCs with 1 (50 nM) and siRNAs targeting the hRRP44, hRRP40, or hRRP6 components of the exosome. Indeed, all three siRNAs rendered 1 inactive, confirming that the small molecule interfaces the repeat expansion with the exosome, facilitating its decay through native RNA quality control pathways (Fig. 3 F and G). As hRRP6 and hRRP44 are primarily nuclear (38), and both NEXT and PAXT are involved in the mechanism of C9orf72 intron 1 decay, our data suggest that the observed decay is driven by the nuclear, not the cytoplasmic exosome.
Compound 1 Ameliorates c9ALS/FTD Pathology In Vivo.
As 1 successfully rescued c9ALS/FTD pathologies in various types of patient-derived cells, we next assessed the compound’s therapeutic potential in a C9orf72 BAC transgenic mouse model (C9BAC) that expresses 500 G4C2 repeats, referred to as +/+PWR500 mice (39). These mice have been reported to present several pathological hallmarks of c9ALS/FTD including the presence of RNA foci and DPRs (39, 40). Mice were treated daily by intraperitoneal (i.p.) injection with 10 mg/kg of 1 (or vehicle) for 2 wk. During the treatment period, no significant changes in bodyweight were observed in compound- or vehicle-treated mice (SI Appendix, Fig. S16A).
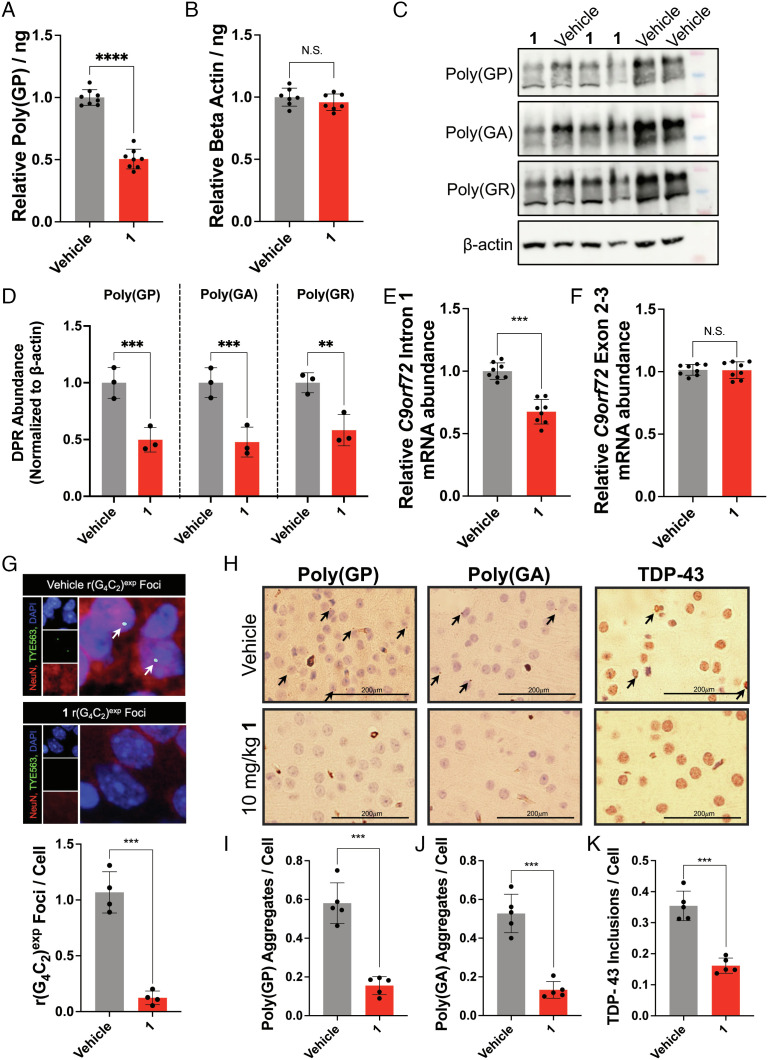
After the treatment period, the abundance of the DPR protein poly(GP) was measured in total brain tissue, affording a 52 ± 7% (P < 0.0001) decrease in poly(GP) abundance in 1-treated mice (Fig. 4A). Importantly, no change in β-actin protein abundance, produced by canonical translation, was observed in either +/+PWR500 or in WT mice (Fig. 4B and SI Appendix, S16B). These findings were additionally corroborated by Western blot analysis of total protein harvested from brain tissue to visualize the abundance of several DPRs upon compound treatment. Indeed, both poly(GP) and poly(GA) were reduced by 50 ± 11% and 53 ± 13%, respectively, with no observed effects on β-actin protein abundance (Fig. 4 C and D). As anticipated from studies in patient-derived cells, the abundance of intron 1-containing transcripts was also reduced by 27 ± 6% (P < 0.001) in 1-treated mice as compared to vehicle-treated mice (Fig. 4E). No effect was observed on total C9orf72 transcript abundance (Fig. 4F) or transcripts lacking the repeat expansion (exon 1b isoform; SI Appendix, Fig. S16 C and D). As a further confirmation of these data, we measured the intron retention ratio as previously published by the Swanson laboratory (15). Upon treatment with 1, the intron retention ratio of transcripts with the 3′ splice site decreased by 0.4 ± 0.1 (P < 0.05) (SI Appendix, Fig. S16E). RNA-seq analysis of total RNA harvested from brain tissue of treated and non-treated mice found only four of the 25,668 genes detected (0.015%, FDR = 0.01) to be significantly affected by treatment, with all four genes being upregulated by compound 1 (SI Appendix, Fig. S16F), demonstrating compound selectivity in vivo. Notably, the four genes upregulated by 1 were unaffected in c9ALS iPSCs and in iPSCs derived from healthy donors (SI Appendix, Fig. S11 D and H). Furthermore, the average number of r(G4C2)exp-containing foci per nucleus was decreased from 1.07 ± 0.18 foci/nucleus in vehicle-treated mice to 0.12 ± 0.06 (P < 0.001) foci/nucleus in 1-treated mice (Fig. 4G and SI Appendix, S16G).
Fig. 4.
Compound 1 alleviates c9ALS-FTD-associated pathologies in a preclinical mouse model. (A) Effect of 1 on poly(GP) abundance in brain tissue harvested from +/+PWR500 mice (39). Relative poly(GP) abundance was normalized to total protein abundance in vehicle-treated mice (n = 8 mice per treatment group). (B) Effect of 1 on the abundance of β-actin abundance (control protein) in brain tissue harvested from +/+PWR500 mice. Relative β-actin abundance was normalized to total protein abundance in vehicle-treated mice (n = 8 mice per treatment group). (C) Representative Western blot showing poly(GP), poly(GA), poly(GR), and β-actin protein levels upon treatment of vehicle or 10 mg/kg of 1 treated in +/+PWR500 mice. (D) Quantification of poly(GP), poly(GA), and poly(GR), abundance (normalized to β-actin) upon treatment with 10 mg/kg of 1 (n = 3). (E) Effect of 1 on r(G4C2)exp-containing intron 1 abundance in the C9orf72 BAC mouse model +/+PWR500 as determined by qRT-PCR analysis of total RNA isolated from treated and untreated mice with intron 1-specific primers (n = 8 mice per treatment group). (F) Effect of 1 on C9orf72 exon 2–3 abundance, common to transcripts both those containing and lacking r(G4C2)exp, in +/+PWR500 mice, as determined by qRT-PCR analysis of total RNA isolated from treated and untreated mice with exon 2–3-specific primers (n = 8 mice per treatment group). (G) Top: Representative images of 1’s effect on r(G4C2)exp-containing foci in the cortex of +/+PWR500 mice, as determined by RNA-fluorescence in situ hybridization (FISH); Bottom: Quantification of average number of foci per nucleus in the cortex of brain tissue (n = 4 mice per treatment group). (H) Representative histological images of cortex from +/+PWR500 mice treated with 1 or vehicle, visualizing poly(GP), poly(GA), or TDP-43. Poly(GP) and poly(GA) aggregates appear as punctate spots, as indicated with arrows. TDP-43 appears as aggregates located around the cell are indicated with arrows. (I) Quantification of poly(GP) aggregates from histological analysis, including images in H (n = 5 mice per treatment group; all cells (~200) counted per section; three sections). (J) Quantification of poly(GA) aggregates from histological analysis, including images in H (n = 5 mice per treatment group; all cells (~200) counted per section; three sections). (K) Quantification of TDP-43 inclusions from histological analysis, including images in H (n = 5 mice per treatment group; all cells (~200) counted per section; three sections). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, as determined by an Unpaired t test with Welch’s correction (A–E and G–I). Error bars are reported as SD.
We next assessed the presence of poly(GP) aggregates, poly(GA) aggregates, and TAR DNA-binding protein 43 (TDP-43) inclusions, a pathological hallmark of ALS/FTD (15), in sagittal brain sections of treated and untreated mice by immunohistochemical analysis. The number of poly(GP) aggregates was reduced from 0.58 ± 0.11 to 0.16 ± 0.05 (P = 0.0003) aggregates per cell in the cortex of mice treated with 1 (P = 0.0003), consistent with reduction of poly(GP) abundance measured in brain lysates (Fig. 4 H and I). Likewise, poly(GA) aggregates, also produced by RAN translation, were diminished from 0.53 ± 0.10 aggregates per cell in untreated mice to 0.13 ± 0.04 (P = 0.0003) in 1-treated mice (P = 0.0003; Fig. 4 H and J), while inclusions containing TDP-43 decreased from 0.35 ± 0.05 to 0.16 ± 0.02 (P = 0.0002) per cell throughout the cortex (P = 0.0002; Fig. 4 H and K). Notably, global and choline acetyltransferase (ChAT) staining in the cortex of +/+PWR500 mice was unaffected by compound treatment (Fig. 4H).
We confirmed the therapeutic potential of 1 in a second C9orf72 BAC mouse model that expresses 100–1,000 G4C2 repeats, dubbed Lutzy (39, 40). Following the same treatment regimen used for +/+PWR500 mice, the following findings were observed in 1-treated mice: (i) no observable toxicity, as assessed by changes in bodyweight and ChAT staining of brain tissue (SI Appendix, Fig. S17A); (ii) a significant (P = 0.0005) decrease in intron 1 abundance in total RNA harvested from whole-brain tissues (SI Appendix, Fig. S17B); (iii) reduced poly(GP) abundance in lysates harvested from whole-brain tissues (SI Appendix, Fig. S17C); (iv) reduction of poly(GP) and poly(GA) aggregates in sagittal brain sections of the cortex (SI Appendix, Fig. S17 D and E); and (v) no effect on the abundance of the exon 1b isoform, which does not contain the repeat expansion, (SI Appendix, Fig. S18 A and B) or on the abundance of β-actin (SI Appendix, Fig. S18C). Additionally, in WT mice, treatment with 1 had no effect on poly(GP) abundance (expressed ~10-fold less than in diseased mice) or β-actin abundance (SI Appendix, Fig. S18 D and E). Taken together, these data confirm activity of 1 in mouse brain tissue when administered via i.p. injection.
Discussion
Herein, we report the identification and optimization of a drug-like, BBB penetrant molecule, 1, that alleviates pathologies associated with c9ALS/FTD in various patient-derived cells, including spinal neurons differentiated from iPSCs, and in two c9ALS/FTD mouse models. These studies suggest that by using physicochemical properties such as CNS-MPO scores derived from data collected from protein-focused small molecules, a brain penetrant RNA-specific targeting small molecule can be obtained. Indeed, when compared to previously developed compounds, both a binding small molecule and a ribonuclease targeting chimera (RiboTAC) (25), 1 has drastically improved CNS-MPO (4.29 vs. 1.00 and 0.62, respectively) and Quantitative Estimate of Drug-likeness (0.321 vs. 0.061 and 0.009, respectively) scores, without sacrificing activity (SI Appendix, Table S9). Future directions for the development of 1 include studying the alleviation of nuclear pore defects (cellular phenotype) (41) in patient-derived iPSNs as well as alleviation of behavioral phenotypes in an adeno-associated virus (AAV) rodent model of c9ALS. (13)
Mechanistic studies revealed that the compound directly engages the r(G4C2)exp RNA target, alleviating C9orf72 intron 1 retention and accumulation. That is, the small molecule interfaces the repeat expansion with natural splicing and decay processes, in this case the spliceosome and the nuclear RNA exosome, by displacing sequestered RBPs. While it has been previously observed that displacement of RBPs sequestered by RNA repeat expansions can rescue alternative pre-mRNA splicing (24), this work provides a comprehensive mechanistic evaluation, identifying a mechanism in which a repeat-containing intron is subjected to nuclear exosomal decay in a disease-selective manner. It is likely that other compounds that bind to RNA targets can affect an RNA-mediated process to facilitate elimination of an RNA target.
Previously, other small molecules have been identified that bind the G-quadruplex or hairpin folds formed by r(G4C2)exp. These include small molecules that bind and stabilize the G-quadruplex structure over the hairpin structure, driving reduction of RNA foci and DPRs (42), and compounds like TMPyP4 porphyrin that distorts RNA G-quadruplexes, thus ablating interactions of binding partners such as heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) or serine-/arginine-rich splicing factor 1 (ASF/SF2) which drive downstream phenotypic changes (43). Alternatively, molecules have been identified that selectively bind the hairpin structure and prevent sequestration of RBPs and inhibit RAN translation (16) or that stabilize a novel conformational state of the r(G4C2)exp hairpin (44). We recently described a dimeric small molecule that rescued c9ALS/FTD pathologies in patient-derived cells but did not affect intron 1 or exon 2–3 abundance (25). Notably, facilitating intron splicing and downstream decay is not common to all small molecules that target r(G4C2)exp. This observation demonstrates that small molecules can have various modes of action from simple binding that prevents interactions with proteins (45, 46), be they processing factors (42, 43, 47) or the ribosome (16), to interfacing with endogenous RNA degradation pathways (25, 48). However, through selective liberation and decay of the repeat-containing intron by 1, rescue of disease pathways can be elicited, the effects of which can be retained for longer than simple, transiently binding small molecules (25).
Furthermore, in the field of targeted protein degradation, chimeric compounds have facilitated protein decay (49, 50); however, monomeric degraders or glues with low molecular weights have also been found to facilitate noncanonical interactions of proteins with quality control components (51). This study represents an example of a mechanistically defined way in which small molecules targeting RNA can produce a similar output. Unique from ASOs which degrade through RNase H, our compound acts by affecting the processing of a pre-mRNA, thereby liberating the toxic intron and allowing for it to be decayed by native quality control (51, 52).
Importantly, the observation that small molecules can be designed to target the repeats themselves and provide a selective functional output in vivo suggests that a single compound could target all mRNAs that emanated from genes that harbor the same disease-driving repeat expansion, provided the small molecule penetrates the disease-affected tissues. In support of this notion, it has been reported that a small molecule targeting the r(CUG)exp, which was initially found to improve disease in myotonic dystrophy type 1 (repeat located in the 3′ untranslated region (UTR)), also improves disease pathology in Fuch’s endothelial corneal dystrophy, where r(CUG)exp is in an intron (48). Notably, the core structure of 1 is a derivative of a previously elucidated small molecule (16), identified by Inforna, thus highlighting that compounds identified by rational design and phenotypic screening can agnostically reveal structures unique to the r(G4C2)exp hairpin that can then be elaborated to a drug-like lead molecule.
Collectively, this work represents a significant advancement in the field of RNA-targeting small molecules, demonstrating that small molecule binders can be designed to selectively degrade toxic RNA introns in a mechanistically defined manner. This understanding of RNA-targeting small molecule mechanism of action opens many possibilities. For example, small molecules could be used to selectively target toxic regions of RNA without affecting abundance of the full transcript, thereby mitigating the potential for harmful loss-of-function consequences. This also expands the possibilities for the design of small molecules with a degradation-driven mechanism of action and oligonucleotide-like functional outputs but with compounds that have the added benefit of brain penetrance.
Materials and Methods
General Methods.
General experimental procedures are provided in SI Appendix. All studies with patient-derived cell lines were approved by and performed in accordance with the Scripps Institutional Review Board (IRB) Protocol Number IRB-20-7686 and the University of California San Diego’s Embryonic Stem Cell Research Oversight Committee Study #202000. All cell lines were de-identified prior to their acquisition and use.
Cell Culture.
HEK293T cells (CRL-3216) were acquired from American Tissue Culture Collection (SI Appendix, Table S5) and were maintained in Dulbecco's Modified Eagle Medium (Corning) supplemented with 10% (v/v) fetal bovine serum (FBS; Sigma Aldrich), 1% (v/v) Penicillin–Streptomycin Solution (Corning) and 1% glutaGRO supplement (Corning) at 37°C and 5% CO2. LCLs (cultured at passage number ≤20) were acquired from the Coriell Institute (SI Appendix, Table S5) and were maintained in Roswell Park Memorial Institute 1640 medium supplemented with 10% (v/v) FBS and 1% (v/v) Penicillin–Streptomycin Solution. LCLs in 6-well plates were treated with compound for 4 d in growth medium (2 mL medium total volume; final concentration of 0.1% (v/v) DMSO). ALS patient-derived iPSCs (cultured at passage number ≤14) were provided by the Laboratory for Neurodegenerative Research, Johns Hopkins University School of Medicine (SI Appendix, Table S5) or purchased from the Cedars Sinai Induced Pluripotent Stem Cell Core. All iPSCs were maintained in Matrigel-coated (356234, Corning) plates with mTeSR™1 feeder-free medium (Basal medium; STEMCELL Technologies), according to the manufacturer’s instructions. iPSCs were treated with compound in Basal medium (final concentration of 0.1% (v/v) DMSO) for 4 d in Matrigel-coated 6-well plates. Fresh medium containing compound was added to the cells on days 1 and 3. Cells were harvested on day 4 for analysis. For experiments with siRNAs, siRNAs were transfected on days 1 and 3 using Lipofectamine RNAiMax (Life Technologies), following the manufacturer’s recommendations. After transfection, cells were incubated for 1 h at 37°C, followed by compound addition to a final concentration of 50 nM (final concentration of 0.1% (v/v) DMSO). Cells were harvested on day 4 for analysis. Differentiated motor neurons (iPSNs) were derived from the aforementioned iPSCs (SI Appendix, Table S5). The differentiation protocol can be found in SI Appendix and was carried out as previously described (25). Beginning on day 15 of differentiation, cells were treated with compound diluted in Stage 3 Differentiation Medium (final concentration of <0.1% (v/v) DMSO). Fresh medium containing compound was added to the iPSNs every 3–4 d until reaching full maturity at day 32, after which cells were harvested for analysis.
Small Molecule Screening for Inhibitors of RAN Translation.
Compounds in the ReFRAME library (Calibr) were evaluated for inhibiting RAN translation of r(G4C2)exp in HEK293T cells, cultured as described above. HEK293T cells were grown in 100 mm diameter dishes to ~80% confluency and then co-transfected in growth medium with a plasmid encoding (G4C2)66-no ATG-GFP (RAN translation) and a control plasmid encoding mCherry (to assess canonical translation) using JetPrime transfection reagent (Polyplus), according to the manufacturer’s instructions. The cells were then seeded into 384-well plates and incubated for 2 h before addition of compound using a Biomek NXP Laboratory Automation Workstation equipped with a 10 nL 384-pin head. After treatment with compound for 24 h, the compound-containing medium was removed, and the cells were lysed with a solution of 100 mM potassium phosphate, pH 7.8 containing 0.2% Triton X-100. Fluorescence was measured using a BioTek FLx800 plate reader using an excitation wavelength of 530/25 nm and an emission wavelength of 590/35 nm for mCherry; the excitation and emission wavelengths for GFP were 485/20 nm and 528/20 nm, respectively. Background fluorescence was determined by measuring fluorescence intensities in untransfected cells. The background-corrected ratio of GFP to mCherry was used to determine the effect of compound on RAN and canonical translation, respectively.
Measuring Abundance of C9orf72 Variants by qRT-PCR.
After the indicated treatment period, total RNA was extracted using a Quick-RNA Miniprep Kit (Zymo Research) per the manufacturer’s protocol. Reverse transcription was performed using 1 µg of total RNA, as determined by Nanodrop UV spectrophotometer (ThermoFisher), and a qScript™ cDNA Synthesis Kit (Quantabio) per the manufacturer’s protocol. qRT-PCR was performed on a QuantStudioTM Real-Time PCR Instrument (Applied Biosystems) using Power SYBR Green Master Mix (Applied Biosystems). Expression levels of mRNAs were normalized to GAPDH, β-actin, or 18S rRNA as indicated. Please refer to SI Appendix, Table S2 for a list of primers.
RNA-Seq.
All sequencing data described in this paper were deposited in Mendeley Data (https://doi.org/10.17632/k3ph59xrtz.1) (53). A detailed protocol for RNA processing and sequencing analysis can be found in SI Appendix.
Measuring Poly(GP) Levels Using an Electrochemiluminescence Immunoassay.
After treatment, total protein was extracted into CoIP2 buffer [50 mM Tris-HCl, pH 7.4, 300 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA), 1% (v/v) Triton-X 100, 2% (w/v) sodium dodecyl sulfate, 0.01% Protease and Phosphatase inhibitors (Fisher Scientific)] by incubating on ice for 5 min followed by sonication (3-s intervals at 35% power for approximately 20 s). Detergent was removed using a Pierce™ Detergent Removal Spin Column 0.5 mL (Thermo Scientific), following the manufacturer’s protocol. Protein concentration was measured by Pierce™ Micro BCA Protein Assay Kit (Thermo Scientific). Poly(GP) levels were measured using the electrochemiluminescence immunoassay described in SI Appendix.
Mouse Studies.
All animal studies were completed as approved by the Scripps Florida Institutional Animal Care and Use Committee. Mice were treated by daily i.p. injection of 10 mg/kg of compound prepared in 10% (v/v) DMSO / 90% (v/v) saline for a period of 2 wk. Mouse weights were recorded over the dosing period. Mice were euthanized 24 h after the last dose, and tissue was harvested for study. Please refer to SI Appendix for a detailed description of all mouse studies.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This study was funded by the NIH (P01 NS099114 to M.D.D. and L.P.; DP1 NS096898 and R35 NS116846 to M.D.D.; and R35 NS097273 to L.P.), Target ALS (to M.D.D.), the Nelson Family Fund (to M.D.D.), the Fuirst Family Fund (to M.D.D.), and the Scheller Graduate Student Fellowship (to S.M.M). Fig. 3G was created with BioRender.com.
Author contributions
J.A.B. and M.D.D. designed research; J.A.B., S.M.M., R.F., Y.L., R.I.B., H.A., P.R.A.Z., Q.M.R.G., and A.J.A. performed research; T.F.G., Y.-J.Z., L.P., and T.H.J. contributed new reagents/analytic tools; J.A.B. and Y.T. analyzed data; J.A.B., S.M.M., J.L.C.-D., and M.D.D. wrote the paper; and T.F.G., Y.-J.Z., L.P., and T.H.J. contributed expertise and experimental recommendations.
Competing interests
M.D.D. is a founder of Expansion Therapeutics. Some of the compounds in this paper are described in WO2022/055922.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
RNA-seq data have been deposited in Mendeley Data (https://doi.org/10.17632/k3ph59xrtz.1) (53). All other data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.DeJesus-Hernandez M., et al. , Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton A. E., et al. , A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleveland D. W., Rothstein J. D., From Charcot to Lou Gehric-deciphering selective sotor neuron death in ALS. Nat. Neurosci. 2, 806–819 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Sasha Bozeat C. A. G., Lambon Ralph Matthew A., Hodges John R., Which neuropsychiatric and behavioural features distinguish frontal and temporal variants of frontotemporal dementia from Alzheimer’s disease? Neurol. Neurosurg. Psychiatry 69, 178–186 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Blitterswijk M., et al. , Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): A cross-sectional cohort study. Lancet Neurol. 12, 978–988 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck J., et al. , Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am. J. Hum. Genet. 92, 345–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zu T., et al. , RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 110, 4968–4977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper-Knock J., et al. , Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137, 2040–2051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y.-B., et al. , Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 5, 1178–1186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ash P. E., et al. , Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori K., et al. , Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Gendron T. F., et al. , Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chew J., et al. , Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener. 14, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar V., Hasan G. M., Hassan M. I., Unraveling the role of RNA mediated toxicity of C9orf72 repeats in C9-FTD/ALS. Front. Neurosci. 11, 711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sznajder Ł. J., et al. , Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. U.S.A. 115, 4234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z. F., et al. , The hairpin form of r(G4C2)exp in c9ALS/FTD is repeat-associated non-ATG translated and a target for bioactive small molecules. Cell Chem. Biol. 26, 179–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Z., et al. , Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 84, 239 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Falese J. P., Donlic A., Hargrove A. E., Targeting RNA with small molecules: From fundamental principles towards the clinic. Chem. Soc. Rev. 50, 2224–2243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janes J., et al. , The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc. Natl. Acad. Sci. U.S.A. 115, 10750–10755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wager T. T., et al. , Defining desirable central nervous system drug space through the alignment of molecular properties, in vitro ADME, and safety attributes. ACS Chem. Neurosci. 1, 420–434 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wager T. T., Hou X., Verhoest P. R., Villalobos A., Moving beyond rules: The development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem. Neurosci. 1, 435–449 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velagapudi S. P., et al. , Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 113, 5898–5903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Childs-Disney J. L., et al. , A massively parallel selection of small molecule-RNA motif binding partners informs design of an antiviral from sequence. Chem 4, 2384–2404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benhamou R. I., Angelbello A. J., Wang E. T., Disney M. D., A toxic RNA catalyzes the cellular synthesis of its own inhibitor, shunting it to endogenous decay pathways. Cell Chem. Biol. 27, 223–231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bush J. A., et al. , Ribonuclease recruitment using a small molecule reduced c9ALS:FTD r(G4C2) repeat expansion in vitro and in vivo ALS models. Sci. Transl. Med. 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conlon E. G., et al. , The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q., Conlon E. G., Manley J. L., Rio D. C., Widespread intron retention impairs protein homeostasis in C9orf72 ALS brains. Genome Res. 30, 1705–1715 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson L., et al. , Rapid depletion of DIS3, EXOSC10, or XRN2 reveals the immediate impact of exoribonucleolysis on nuclear RNA metabolism and transcriptional control. Cell Rep. 26, 2779–2791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garneau N. L., Wilusz J., Wilusz C. J., The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113–126 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Nagarajan V. K., Jones C. I., Newbury S. F., Green P. J., XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta Gene. Regul. Mech. 1829, 590–603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilleren P. J., Parker R., Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell 12, 1453–1465 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Liu Q., Greimann J. C., Lima C. D., Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–1237 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Schneider C., Tollervey D., Threading the barrel of the RNA exosome. Trends Biochem. Sci. 38, 485–493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meola N., et al. , Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520–533 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Schmid M., Jensen T. H., Controlling nuclear RNA levels. Nat. Rev. Genet. 19, 518–529 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Wu G., et al. , A two-layered targeting mechanism underlies nuclear RNA sorting by the human exosome. Cell Rep. 30, 2387–2401 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Kowalinski E., et al. , Structure of a cytoplasmic 11-subunit RNA exosome complex. Mol. Cell 63, 125–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lubas M., et al. , Exonuclease hDIS3L2 specifies an exosome-independent 3’-5’ degradation pathway of human cytoplasmic mRNA. EMBO J. 32, 1855–1868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., et al. , C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90, 521–534 (2016). [DOI] [PubMed] [Google Scholar]
- 40.O’Rourke J. G., et al. , C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 88, 892–901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K., et al. , Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simone R., et al. , G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 10, 22–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamiri B., Reddy K., Macgregor R. B. Jr., Pearson C. E., TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 289, 4653–4659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ursu A., et al. , A small molecule exploits hidden structural features within the RNA repeat expansion that causes c9ALS/FTD and rescues pathological hallmarks. ACS Chem. Neurosci. 12, 4076–4089 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abulwerdi F. A., et al. , Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol. 14, 223–235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donlic A., et al. , Discovery of small molecule ligands for MALAT1 by tuning an RNA-binding scaffold. Angew Chem. Int. Ed. 57, 13242–13247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen L., et al. , Rationally designed small molecules that target both the DNA and RNA causing myotonic dystrophy type 1. J. Am. Chem. Soc. 137, 14180–14189 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Angelbello A. J., et al. , A small molecule that binds an RNA repeat expansion stimulates its decay via the exosome complex. Cell Chem. Biol. 28, 34–45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakamoto K. M., et al. , Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U.S.A. 98, 8554 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spradlin J. N., Zhang E., Nomura D. K., Reimagining druggability using chemoproteomic platforms. Acc. Chem. Res. 54, 1801–1813 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Mayor-Ruiz C., et al. , Rational discovery of molecular glue degraders via scalable chemical profiling. Nat. Chem. Biol. 16, 1199–1207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanan E. J., et al. , Monomeric targeted protein degraders. J. Med. Chem. 63, 11330–11361 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Bush Jessica, Tong Yuquan, Global RNA-seq for a RNA targeted small molecule that triggers elimination of r(G4C2)exp in c9ALS/FTD Mendeley Data
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
RNA-seq data have been deposited in Mendeley Data (https://doi.org/10.17632/k3ph59xrtz.1) (53). All other data are included in the article and/or SI Appendix.