Significance
Some proteins can refold into their native structures from a denatured state entirely on their own, whereas others require the assistance of molecular chaperones. Over three decades, biochemists have performed refolding assays on purified proteins in which denaturant-unfolded enzymes have been reactivated in a chaperone-dependent manner, but a systematic assessment of which proteins need chaperones to refold—and which do not—has been missing. To and coauthors use a limited proteolysis–mass spectrometry approach to globally interrogate refolding on a whole E. coli extract. Their results provide a map to understand what types of proteins are more reliant on chaperones to refold, and also highlight a cohort of proteins that are unable to fully refold even when chaperones are supplied.
Keywords: chaperones, proteomics, protein folding, GroEL, refoldability
Abstract
The journey by which proteins navigate their energy landscapes to their native structures is complex, involving (and sometimes requiring) many cellular factors and processes operating in partnership with a given polypeptide chain’s intrinsic energy landscape. The cytosolic environment and its complement of chaperones play critical roles in granting many proteins safe passage to their native states; however, it is challenging to interrogate the folding process for large numbers of proteins in a complex background with most biophysical techniques. Hence, most chaperone-assisted protein refolding studies are conducted in defined buffers on single purified clients. Here, we develop a limited proteolysis–mass spectrometry approach paired with an isotope-labeling strategy to globally monitor the structures of refolding Escherichia coli proteins in the cytosolic medium and with the chaperones, GroEL/ES (Hsp60) and DnaK/DnaJ/GrpE (Hsp70/40). GroEL can refold the majority (85%) of the E. coli proteins for which we have data and is particularly important for restoring acidic proteins and proteins with high molecular weight, trends that come to light because our assay measures the structural outcome of the refolding process itself, rather than binding or aggregation. For the most part, DnaK and GroEL refold a similar set of proteins, supporting the view that despite their vastly different structures, these two chaperones unfold misfolded states, as one mechanism in common. Finally, we identify a cohort of proteins that are intransigent to being refolded with either chaperone. We suggest that these proteins may fold most efficiently cotranslationally, and then remain kinetically trapped in their native conformations.
Protein folding represents the culmination of the central dogma of molecular biology—enabling the primary information encoded in nucleic acids and translated into polypeptides, to take shape into functional macromolecules. The striking accuracy of AI-based structure predictors has given new credence to Anfinsen’s dogma that protein three-dimensional structures is encoded at the amino acid sequence level (1, 2); nevertheless, the journey by which proteins navigate their energy landscapes to locate their native structures is complex, involving (and sometimes requiring) many cellular processes and factors (3, 4). While it is well understood that molecular chaperones are required for specific proteins to refold from their denatured forms (5–8), how these findings generalize to the proteome-scale is less clear; moreover, the potential influence of the cellular milieu is typically not captured in most in vitro chaperone refolding experiments.
Traditional protein refolding assays monitor structure or activity recovered by a denatured protein molecule following dilution from denaturant (9); however, activity-based readouts are challenging to generalize to whole proteomes. Pioneering work by Kerner et al. introduced a high-throughput method to survey the clients of GroEL/GroES (Escherichia coli’s group I chaperonin) by identifying proteins that are enriched in a fraction coprecipitating with chaperonin (6), an approach that has since been extended to survey several other chaperone systems, such as DnaK (10, 11). High-throughput measurements of protein precipitation, conducted on individually over-expressed proteins with and without chaperones (12, 13), or on whole extracts following heat treatment (14, 15) have also been reported.
Nevertheless, a systematic dissection of which proteins require chaperone assistance to refold from the denatured state remains lacking, even for the relatively simple E. coli proteome. This is because pull-down approaches cannot unambiguously assess a protein’s dependency (obligatory use) on a chaperone to refold, since they cannot discriminate the possibility that a putative client interacts with a chaperone without requiring it. Indeed, many proteins that were presumed to be obligate chaperonin clients based on their enrichment in chaperonin coprecipitation studies were later found to remain soluble in vivo during GroE knockdown (16). Furthermore, a recent study (17) estimated that a third of soluble E. coli proteins are intrinsically nonrefoldable, meaning they cannot fully reassume their native forms following complete denaturation, even under conditions without appreciable precipitation. However, how many (and what kinds) of intrinsically nonrefoldable proteins can be rescued by chaperones—as opposed to requiring cotranslational folding (18–20)—is not known. A particularly underexplored question is when chaperones are required for refolding in the presence of the full complement of metabolites, ions, and small molecules in the cytosol, which can potentially supply additional “chemical” chaperones (21–23).
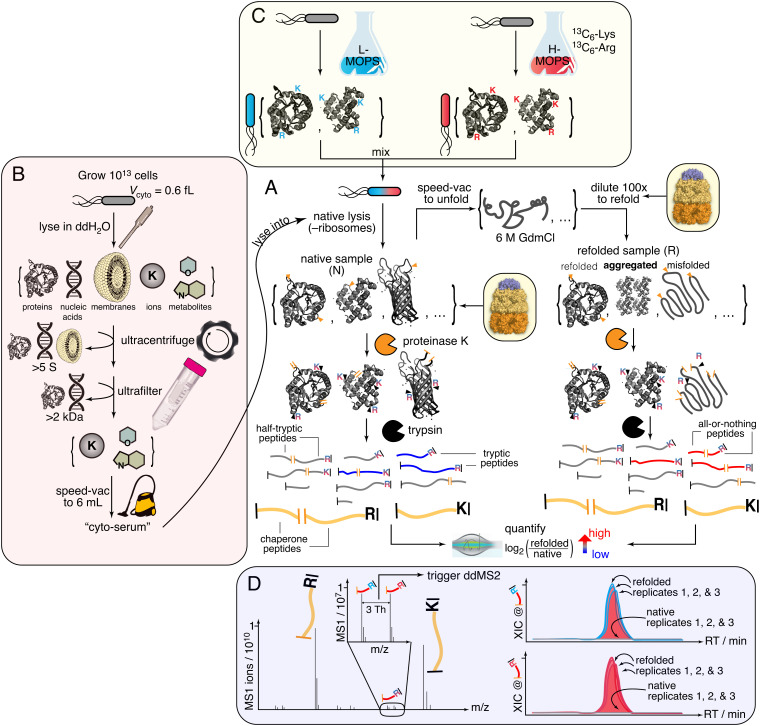
To address these questions, we sought to generalize the traditional biochemical experiment of refolding unfolded proteins by dilution from denaturant—with or without chaperones (6–8, 23–25)—to the E. coli proteome. To do so, we developed a limited proteolysis–mass spectrometry (LiP–MS) approach to probe protein structures globally during refolding (Fig. 1 and SI Appendix, Fig. S1A) (26–29). In this experiment, E. coli lysates are fully unfolded by overnight incubation in 6 M guanidinium chloride (GdmCl), returned to native conditions by rapid dilution, and the conformational ensembles of the proteins in the mixture probed by pulse proteolysis with proteinase K (PK), which cleaves only in regions that are solvent-exposed or flexible. Using liquid chromatography–tandem mass spectrometry (LC–MS/MS), we sequence and quantify tens of thousands of peptide fragments from the refolding reactions to assess regions of proteolytic susceptibility and compare their abundances to “native” samples that are identical except were never unfolded. Hence, to observe structural differences among proteins that cannot refold to their native forms, the limited proteolysis profiles of the PK-treated native and refolded samples are compared with each other.
Fig. 1.
Limited proteolysis–mass spectrometry (LiP–MS) to interrogate the refoldability of the E. coli proteome in a cytosol-like milieu. (A) The core portion of the experiment. (B) Preparation of cyto-serum. (C) A pseudo-SILAC method is used in which replicate E. coli cultures are grown with either light (L) or heavy (H) lysine (Lys) and arginine (Arg). L/H pairs of cultures are mixed together and colysed. Consequentially, peptides derived from the proteome will exist as isotopomeric pairs. (D) Coeluting isotopomer pairs are preferentially isolated for data-dependent MS2 (ddMS2) scans, enabling high coverage of the E. coli proteome during MS analysis. Example of all-or-nothing peptide, in which feature is absent from native samples at masses corresponding to both the L- and H-peptides.
Using this approach, we interrogate protein refolding in the cytosolic milieu with the molecular chaperones, GroEL/ES (Hsp60/Hsp10) and DnaK/DnaJ/GrpE (Hsp70/40). We discover that protein isoelectric point (pI) emerges unexpectedly as a key explanatory variable for refoldability: basic proteins are generally efficient refolders, particularly in the cytosolic milieu, while acidic proteins are more frequently reliant on GroEL to refold. GroEL can restore many intrinsically nonrefoldable proteins, especially acidic proteins, proteins with high molecular weight (MW), proteins with many domains, and domains with α/β architectures. The cohort of proteins that GroEL refolds overlaps extensively with those which DnaK can restore, suggesting a mechanism in common for these two distinct molecular machines. Finally, our study sheds light on a small group of proteins that are recalcitrant to refolding with either chaperone, a group that we hypothesize is adapted to fold cotranslationally and unfold slowly, which would obviate the need for chaperone assistance after biosynthesis. This group heavily represents proteins involved in core and ancient metabolic processes, namely glycolysis and translation.
Results
A Method to Interrogate Refolding the E. coli Proteome in Cytosol with Chaperonin.
The E. coli cytosol is an idiosyncratic medium predominantly buffered by glutamate and replete with a wide array of cofactors, metabolites, and ions with concentrations spanning over six orders of magnitude (30, 31). To probe the effect this medium exerts on protein folding, we isolate the cytosolic medium by culturing cells to the end of log phase and lysing them into pure water (Fig. 1B). Macromolecules larger than 2 kDa are depleted by ultracentrifugation and subsequent ultrafiltration of the supernatant (see Methods and SI Appendix, Fig. S2A). The filtrate is then reduced under vacuum until its volume equals that of the combined internal volume of the original cellular population, given the estimated E. coli cytoplasm volume of 0.6 fL/cell (32). The resulting liquid, which we refer to as ‘cyto-serum,’ consists of all the stable and free ions, metabolites, and cofactors present in the E. coli cytosol near their physiological concentrations. Cyto-serum is a nonviscous off-yellow (λmax 258 nm) liquid with a pH of ~7 (SI Appendix, Fig. S2 B–E).
We use cyto-serum as a lysis buffer to resuspend separate E. coli cell pellets (grown to the end of log phase in MOPS media (33)), which are natively lysed by cryogenic pulverization, a mechanical lysis method chosen because it keeps large and weakly bound protein assemblies intact (34, 35) (Fig. 1). Use of cyto-serum as a lysis buffer enables us to maintain proteins at suitably low concentrations for refolding (0.116 mg/ml, ca. 4 µM), while keeping the small molecule constituents of the cytosol near their physiological concentrations.
In preliminary experiments, we tested whether cyto-serum would be suitable for global refolding experiments by measuring the levels of aggregation that accrue after 2 h. Pelleting assays detected low levels of precipitation (6 ± 2% of protein), slightly higher than our previously optimized condition that used a Tris buffer at pH 8.2 (3 ± 1%, SI Appendix, Fig. S2D). Nevertheless, this 3% increase in precipitation is close to what we previously observed for refolding in a defined buffer at neutral pH (17, 36), thereby confirming that alkaline pH helps suppress aggregation, and that the cytosolic components do not increase aggregation levels beyond an expected effect from pH. To further investigate aggregate formation (including smaller soluble nonprecipitating aggregates), we performed sedimentation velocity analytical ultracentrifugation and mass photometry on these refolding reactions (SI Appendix, Fig. S3). Both techniques showed that the molecular size distributions of the refolded samples were similar to native extracts, confirming the absence of soluble aggregates. These studies show that complex mixtures of proteins are less aggregation-prone than most of these individual proteins are when they are overexpressed (12), and allow us to focus on interrogating soluble misfolded states without the confounding effect of aggregation. Moreover, we confirmed through reactivation studies on two metabolic enzymes that similar levels of refolding occur in lysates as do on purified enzymes at early times (≤5 min) before aggregation could start (SI Appendix, Fig. S4 A and B).
Following these tests, we proceeded to perform global refolding experiments by diluting unfolded E. coli extracts with cyto-serum supplemented with 4 µM GroEL and 8 µM GroES (Fig. 1; ca. 100-fold higher concentration than their natural abundances in diluted lysate). Note that all chaperone concentrations are given in protomers (and not in terms of complexes), and these are similar to those used in GroEL refolding assays on single purified clients (7, 13, 37–39). A superstoichiometric amount of GroES was chosen to suppress GroEL’s futile ATPase activity (38, 40) stimulated by the high K+ concentrations of the cytosol. Because it is important to compare compositionally identical native and refolded samples, GroEL-assisted refolding reactions were referenced against compositionally identical native samples that were also supplemented with chaperones and cyto-serum (cf. Fig. 1A and SI Appendix, Fig. S1). This step is essential because even though native proteins should not “need” GroEL, if a correctly refolded protein has a propensity to associate transiently with GroEL (as a “triage complex” (41, 42)), such an interaction would still affect its proteolysis profile and therefore needs to be present in the reference sample.
In preliminary LC–MS/MS experiments, we detected low coverage of the proteome because >80% of the total protein content in these refolding reactions are the added chaperone and cyto-serum adds many nonprotein contaminants (SI Appendix, Fig. S5A). To address this challenge, we developed an isotope-labeling strategy to distinguish peptides belonging to refolding clients from those belonging to chaperonin proteins or from other cellular contaminants (Fig. 1C). Three replicate E. coli cultures are grown in two different MOPS media: one with natural abundance (light) isotopes of Arg and Lys, and a second with [13C6]Arg and [13C6]Lys (heavy). Pairs of light and heavy media are mixed together (for each biological replicate) prior to lysis and initiating the unfolding/refolding/LiP–MS workflow. In this way, peptides from client proteins will be present in the sample as a pair of isotopomers that coelute during liquid chromatography and generate a signature twin-peak feature (Fig. 1D) that distinguish them from chaperone-derived peptides despite being several orders of magnitude lower in intensity (SI Appendix, Fig. S5C). The mass spectrometer is then instructed to preferentially select peaks with the correct spacing for data-dependent isolation and MS2 acquisition. We confirmed that coeluting isotopomers generate fragmentation spectra with expected mass-shifts in the y-ions (SI Appendix, Fig. S5D).
We refer to this strategy as ‘pseudo-SILAC’ because it uses stable isotope labeling to direct the mass spectrometer to select the correct features, as opposed to performing quantifications. Instead, we calculate refolded/native abundance ratios by comparing the areas under the curve between runs (known as label-free quantification), because of its superior dynamic range (43–45) and ability to confidently identify when a feature is absent from a particular sample. We note that even though pseudo-SILAC is not as necessary for experiments without chaperones, we applied it to all conditions in this study uniformly to remove any potential source of bias when comparing chaperone to nonchaperone conditions.
GroEL/GroES Rescues Many Nonrefoldable Proteins.
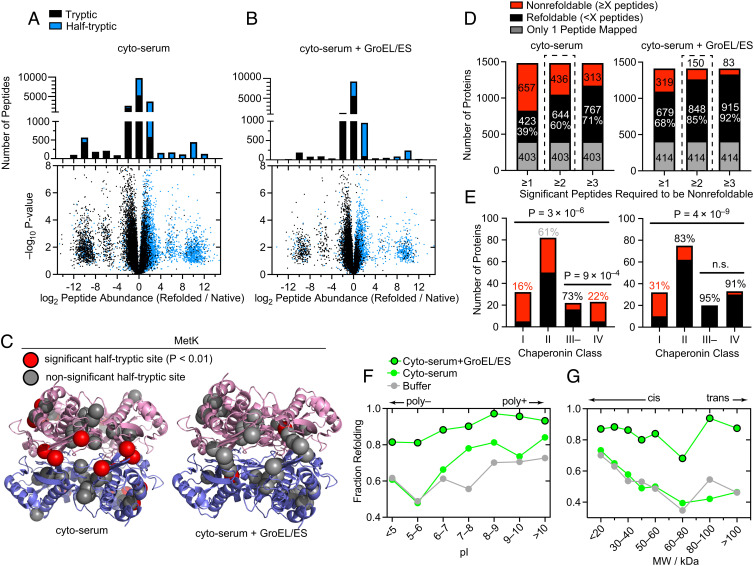
GroEL/GroES significantly remodels the refolding profile of the E. coli proteome (Fig. 2). To summarize these data, we present peptide-level volcano plots and abundance ratio histograms (Fig. 2 A and B) for refolding in cyto-serum without and with chaperonin after 1 min, where the differences are the most apparent. Half-tryptic peptides are shown in blue, and demarcate locations where PK cleaved (cf. Fig. 1); full-tryptic peptides are shown in black and represent the absence of a PK cut. The observation that most peptides that are more abundant in the refolded samples (right-hand side) are half-tryptic (86% without GroEL, 90% with GroEL), and that most peptides that are more abundant in the native samples (left-hand side) are full-tryptic (80% without GroEL, 81% with GroEL; P < 10–15 by the Mann–Whitney U test for both) imply that the refolded proteome is globally more susceptible to proteolysis than the native proteome—further evidence that refolding occurred with minimal aggregation (see also SI Appendix, Fig. S3). To further test this interpretation, for each half-tryptic peptide we used the AlphaFold database to calculate the relative solvent accessible surface area (rSASA) of each PK cut site in the context of its native protein structure (SI Appendix, Fig. S6). We found that sites which became much more accessible in the refolded form were typically very buried in their native structural contexts (median rSASA 15% without GroEL, 13% with GroEL), as expected.
Fig. 2.
GroEL/ES is a versatile chaperone that assists the refolding of many E. coli proteins. (A) Volcano plots and associated peptide histogram comparing peptide abundances from three native and three refolded E. coli lysates after 1 min of refolding. Effect sizes reported as ratio of averages, and P-values are calculated using the t test with Welch’s correction for unequal variance (n = 3). Data correspond to #1 in SI Appendix, Fig. S1B. (B) Similar to panel A, except where 4 µM GroEL and 8 µM GroES were present in the native samples and added to the refolding reaction. Data correspond to #4 in SI Appendix, Fig. S1B. (C) Structure of MetK (PDB: 1P7L), indicating sites where proteolytic susceptibility is the same (gray spheres) or significantly different (red spheres) in the refolded samples compared to native. Left, locations of 9 PK cut-sites with significantly different susceptibility in the refolded sample, after refolding in cyto-serum (red spheres). Right, location of one PK cut-site with significantly different susceptibility in the refolded sample, after refolding in cyto-serum and GroEL/ES. (D) Bar charts showing the total number of refoldable or nonrefoldable proteins after 5 min, without and with GroEL/ES. Bars correspond to alternative cutoff schemes. ≥2 is used for the rest of the study. In gray are proteins with only 1 peptide quantified, which are not used. Data correspond to #2, 5 in SI Appendix, Fig. S1B. (E) Bar charts indicating the number of refolding and nonrefolding proteins associated with one of four chaperonin classes (as defined by (6, 16)), in experiments without and with chaperonin. Percentages indicate percentage refolding within that category. P-values for the all-way comparison are from chi-square test; for the two-way III– v. IV comparison are from Fisher’s exact test. (F) Fraction of proteins that refold in either Tris buffer (gray (47)), cyto-serum (green), or cyto-serum with GroEL/ES (green, black border), separated on the basis of individual proteins’ isoelectric point (pI). Data from the 5-min refolding time. (G) Fraction of proteins that refold in either Tris buffer (gray (47)), cyto-serum (green), or cyto-serum with GroEL/ES (green, black border), separated on the basis of individual proteins’ molecular weight (MW).
These experiments showed strong technical reproducibility. For instance, ~90% of peptides had a refolded/native abundance ratio within a factor of 1.4 on separate performances of the experiment (SI Appendix, Fig. S7 A and B), and when these ratios were plotted against each other, R2 was between 0.74 and 0.81. When only peptides that were deemed significant in their respective experiments were considered, R2 rose to 0.87–0.91 (SI Appendix, Fig. S7 C and D).
Points on the flanking lobes correspond to peptides that were detected only in the refolded or native samples. We refer to these as ‘all-or-nothing’ peptides and assign a limit-of-detection abundance to them in samples where they are not detected. All-or-nothing peptides represent nonrefoldable regions within proteins that were completely inaccessible to PK in the native conformation but became proteolytically susceptible when that region failed to refold. After refolding with GroEL, many fewer all-or-nothing peptides were detected (1,736 (9.5%) without GroEL, 691 (5.6%) with GroEL), signifying fewer proteins that were structurally distinct from their native forms. Utmost caution is warranted in calling all-or-nothing peptides since they are based on missing data; however, a stringent filtering process we have adopted (see SI Appendix, Methods) also makes them reproducible over technical replicates of the experiment (SI Appendix, Fig. S7 C–F).
We mapped peptides back to their parent proteins and labeled an individual protein nonrefoldable if we could identify two or more peptides with a significant abundance difference in the refolded samples relative to the native samples (>twofold effect-size, P < 0.01 by t test with Welch’s correction for unequal population variances). Applying these cutoffs, 90–93% of peptides are given the same call (significant or not) between replicates of the experiment, and 87–89% of proteins are assigned the same status (refoldable or not) (SI Appendix, Fig. S7 E–H). The majority of these significant peptides are not all-or-nothing (for which a 64-fold effect-size is used as a cutoff), and represent cases where a site is more susceptible to PK in the refolded samples but not completely inaccessible in the native. For MetK, there is only one such significant peptide after refolding with GroEL (Fig. 2C)—many fewer than after refolding on its own—consistent with its known status as an obligate GroEL client (46). Phosphoglucose isomerase (Pgi) was identified by our experiment as a GroEL-dependent refolder, which we independently confirmed by an enzyme reactivation assay on purified Pgi (SI Appendix, Fig. S4C). By this metric, the proteome was the most refoldable at the 5-min time point both with and without chaperonin (SI Appendix, Fig. S8A), hence we chose to focus on it for further analysis. After 5 min, in cyto-serum 60% (of 1,080 proteins) are refoldable intrinsically (SI Appendix, Data S1), and with the addition of GroEL/GroES, this rises to 85% (of 998 proteins) (Fig. 2D and SI Appendix, Data S2), using a ≥2 peptide cutoff to call a protein nonrefoldable (as used previously (17)). The overall refoldability rates do depend on this admittedly arbitrary cutoff employed to call a protein nonrefoldable; however, the ≥2 peptide cutoff can be viewed as a compromise between not allowing too much weight to be assigned to a single significant peptide, and not making it too difficult to call a protein nonrefoldable with lower coverage. Importantly, none of the key trends we describe in the following depend sensitively on this choice (Fig. 2D and SI Appendix, Fig. S10 G–I).
To contextualize this experiment, we first sought to compare these results to two landmark studies interrogating E. coli chaperonin usage across the proteome. Kerner et al. formalized a classification system based on the enrichment level of various proteins in the fraction that coprecipitates with a tagged GroEL/ES complex (6). Class I proteins are those that are depleted in the GroEL fraction relative to their level in the cytoplasm, while class III proteins are those that are highly enriched in the GroEL fraction. Complementing this study, Fujiwara et al. used an E. coli strain in which GroEL expression is arabinose dependent and measured which proteins precipitate in the E. coli cytoplasm after GroEL expression is cut off by shifting cells from arabinose to glucose (16). Many (40%) of the class III proteins were still soluble in the cytoplasm without chaperonin and were renamed class III–. On the other hand, those whose solubility in the cytoplasm is expressly chaperonin-dependent were renamed class IV.
Our refolding assay is strikingly consistent with Fujiwara’s subclassification (Fig. 2E) (16). In the chaperonin-null condition (Fig. 2E), the majority (73%) of class III– proteins are refoldable, whereas only a minority (22%) of class IV proteins are. The observation concerning class III– proteins implies, intriguingly, that there are many proteins that associate strongly with GroEL in vivo that do not actually require it. The strong alignment between class IV and nonrefoldability implies that without GroEL, most class IV proteins populate misfolded states which aggregate at the high concentrations of the cellular environment, but in our assay instead persist as soluble misfolded states that do not aggregate but also cannot correct themselves. The observation that a few class IV proteins are refoldable in our assay suggests that in these situations, GroEL’s function is to serve as an obligatory holdase, a function that is no longer necessary when aggregation is suppressed. With chaperonin added to the refolding reactions, both class III– and class IV proteins are nearly completely refoldable (95% and 91% respectively, Fig. 2E). This finding implies that the majority of GroEL’s obligate clients (class IV) require it actively (e.g., either as a foldase or unfoldase), not merely as an infinite-dilution chamber (e.g., holdase) (5). We note that class IV proteins are actually more refoldable in Tris buffer pH 8.2 (which further suppresses aggregation) than in cyto-serum (47), hence in the cytosol, GroEL’s assistance is even more needed than it is in an alkaline refolding buffer.
Proteins with higher isoelectric points (pI > 8) tend to be intrinsically refoldable and especially so in the cytosol, whereas proteins with lower isoelectric points (pI < 7) are less intrinsically refoldable, a difference that is largely mitigated by GroEL (Fig. 2F). Proteins with a high MW tend to be less intrinsically refoldable, but GroEL smooths over this difference as well (with an important exception for proteins sized 60–80 kDa), exerting its most prominent rescuing power on proteins of greatest MW (Fig. 2G). The discontinuity for proteins sized 60–80 kDa has previously been attributed to the dimensions of the GroEL cavity, which is known not to accommodate proteins larger than 60 kDa (6). However, we find that GroEL is extremely effective at assisting the largest E. coli proteins. These observations support the view that the unsealed trans cavity of GroEL is also an active chaperone, that out-of-cage refolding occurs, and are consistent with previous works that have found activity of GroEL on large substrates (48–52).
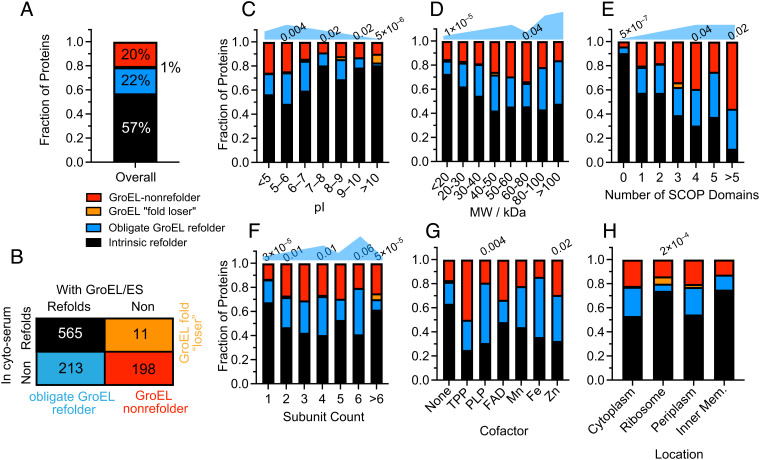
Our data further elucidate the types of proteins that tend to be obligate GroEL refolders (Fig. 3). To make this assessment, we pooled together the data from the “no GroEL” condition (native and refolded) and from the “GroEL” condition (native and refolded), selected the subset of proteins that were confidently assessed in both conditions, and assigned them statuses based on their refolding outcomes in the two conditions (Fig. 3 A and B and SI Appendix, Fig. S1 and Data SA). Inspection of the distribution of obligate GroEL refolders, broken down by pI range (Fig. 3C), shows that obligate GroEL refoldability peaks for mildly acidic proteins (5 < pI < 6; 26%), is lower for proteins that are neutrally charged in the cytosol (7 < pI < 8; 11%), and is lowest for basic proteins (pI > 10; 2%). Indeed, among polybasic proteins (pI > 10) there are three examples (7.3%) of proteins that lose their intrinsic capacity to refold in the presence of chaperonin (note only 1% of all proteins overall are in this category). This may be because some basic proteins could get stuck in the GroEL cavity, whose lumen is negatively charged (53, 54). Such a tendency might explain why polybasic proteins generally have been optimized to refold on their own (Figs. 2F and 3C), as they might otherwise unproductively bind too tightly within GroEL. Low-MW proteins are the least likely to require GroEL, and high-MW proteins are the most (Fig. 3D). The large (>80 kDa) obligate GroEL refolders are all (100%) multidomain proteins, wherein potentially one nonnative domain could fit in the unsealed trans cavity. Indeed, we find a robust trend that proteins with more domains up to 5 become progressively more reliant on GroEL (Fig. 3E), though proteins with >5 domains appear to be poor refolders even with GroEL (P = 0.02 by chi-square test). Together, these findings provide support for the view that the trans mechanism or out-of-cage refolding is effective at resolving misfolded domains in the context of large multidomain proteins.
Fig. 3.
Defining the scope of obligate GroEL refolders across the proteome. (A) Frequency of proteins that refolded in both conditions (intrinsic refolder; black), only with GroEL/ES (obligate GroEL refolder; blue), only without GroEL/ES (GroEL “fold loser”; orange) or did not refold in either (GroEL-nonrefolder; red). Data used for this figure correspond to #b in SI Appendix, Fig. S1C (SI Appendix, Data SA). Numbers listed above bars indicate P-values by the chi-square test that the category has a different GroEL usage profile than the proteome overall. Blue shapes qualitatively denote the need-level for GroEL. (B) Truth table showing the number of proteins in each of the categories described in A. Analysis covers 987 proteins for which at least two peptides could be confidently quantified in both conditions. (C) As A, except proteins are separated based on isoelectric point (pI). (D) As A, except proteins are separated based on molecular weight (MW). (E) As A, except proteins are separated based on the number of domains in the protein, as defined by the SCOP database. (F) As A, except proteins are separated based on the number of subunits in the complex to which they are part. (G) As A, except proteins are separated based on their bound cofactor. (H) As A, except proteins are separated by their cellular location.
We also found a few correlations between GroEL usage patterns and subunit composition, cellular location, and cofactors (Fig. 3 F–H). Monomers and assemblies of all sizes benefit from GroEL’s assistance. Tetramers and hexamers are most likely to be obligate GroEL refolders (32% and 39%, respectively), consistent with several model GroEL clients being tetramers like MetF (7) and DapA (55, 56). Proteins in large complexes with >6 subunits are the least reliant on GroEL (Fig. 3F). We find that GroEL benefits cofactor-harboring proteins, particularly proteins that host TPP, PLP, Fe2+, and Zn2+, which are generally less refoldable on their own (17), and have high propensities to be obligate GroEL refolders (between 38% and 50%, Fig. 3G). Finally, we find that GroEL is effective at recovering proteins in all E. coli locations (Fig. 3H), including the periplasm. The observation is unusual because GroEL is strictly a cytosolic chaperone, and when extracted from cells does not coprecipitate periplasmic proteins (6). Hence, even though periplasmic proteins use a distinct suite of chaperones in vivo (57), GroEL can act as an effective substitute during in vitro refolding.
To control for the possibility that the trends described here arose from a coverage bias (e.g., a certain class of proteins are “easier” to label as nonrefoldable because they have more quantified peptides per protein), we assessed the frequency of significant peptides for each class without respect to which protein they arose. All of the key trends remain statistically significant at the peptide level as well (P-values range from 10–5 to 10–31 by the chi-square test), and the peptide significance frequencies overlay well the refoldability frequencies at the protein level (SI Appendix, Fig. S10 A–F and Datas S1–S3). Furthermore, the protein-level trends are not sensitive to the peptide cutoff to call a protein nonrefoldable (SI Appendix, Fig. S10 G–I), and hence can be considered robust.
Effect of Chaperonin on Refolding Kinetics.
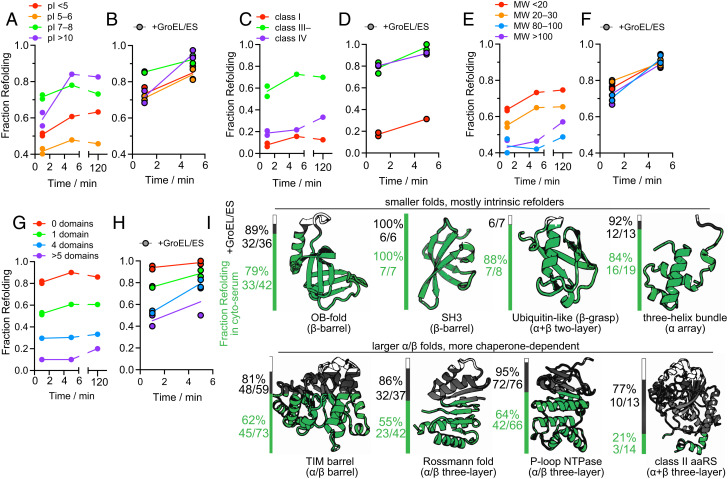
Classic protein folding kinetic studies, typically carried out on small single-domain proteins, record folding times on the ms–s timescales (58). Because of the duration of the PK incubation time (1 min), our experiments do not afford the same level of temporal resolution; however, comparisons between refoldability levels at the 1-min and 5-min time points can provide insight into the types of proteins that refold slowly (i.e., require more than 1 min)—both with and without chaperonin (Fig. 4). In cyto-serum, overall refoldability increases from 52 to 60% from 1 to 5 min, a similar uptick as to what we observe in the chaperonin refolding experiment (77 to 85%). However, from 5 min to 2 h the overall refoldability in cyto-serum slightly decreases, which we attribute to a mix of degradation and aggregation (SI Appendix, Fig. S8 A, B, and E). Specifically, the inefficiently refolding proteins that slowly aggregate could cause low-stability refoldable proteins that transiently populate unfolded conformations to join the aggregate. Hence, the ‘optimal’ refolding time is one that gives most proteins sufficient time to refold but before inefficient refolders have time to aggregate. With chaperonin, refolding decreases precipitously at 2 h (down to 74%). Though we initially thought this was due to depletion of ATP, measurements of ATP concentration in global refolding reactions revealed that ATP hydrolysis occurs at a modest rate (SI Appendix, Fig. S9), starting at 600 µM, and plateauing at 280 µM at longer refolding times. Hence, the more likely explanation for the downturn in apparent foldability at later times is GroEL-dependent reactivation of proteases (such as Lon and ClpP) that subsequently degrade the sample.
Fig. 4.
Kinetic of protein refolding in cyto-serum, with or without GroEL/ES. (A, B) Fraction of proteins that refold after 1 min, 5 min, or 120 min in either cyto-serum (A), or in cyto-serum with GroEL/ES (B, black borders), separated on the basis of individual proteins’ pI. Data correspond to #1–5 in SI Appendix, Fig. S1B. Proteins with high pI are more likely to refold slowly both with and without GroEL. (C, D) As A and B, except proteins are separated based on chaperonin class (6, 16). (E, F) As A and B, except proteins are separated based on molecular weight (MW). (G, H) As A and B, except proteins are separated based on number of domains, as defined by the SCOP database. (I) Fraction of domains that refold in either cyto-serum (green) or in cyto-serum with GroEL/ES (black), separated based on which fold the domain is assigned to in the SCOP hierarchy. Raw counts (number of domains refolding/total number of domains) are provided as well. Data correspond to #2, 5 in SI Appendix, Fig. S1B.
Despite the similar increase in refoldability percentages from 1 to 5 min, the types of proteins that benefit from additional time were distinct without and with chaperonin. In the GroEL-null condition (Fig. 4 A, C, E, and G and SI Appendix, Data S1K), slow refolders tend to have high pI (>10; Fig. 4A) or be class III– (Fig. 4C). These features are readily explainable: highly polycationic proteins would have significantly more intra-chain repulsion that would slow down compaction, and class III– proteins are those which populate kinetically trapped intermediates that, given time, can self-correct. Such proteins employ GroEL in vivo as a nonobligatory holdase. Conspicuously absent from this set are proteins with low pI (<6, polyanions), class IV proteins, and proteins with high MW or many domains (Fig. 4 A, C, E, and G). In all cases, it is because rather than fold slowly, proteins in these categories tend to be intrinsically nonrefoldable. On the other hand, it is interesting to notice an enrichment for very slow refolding (i.e., requiring more than 5 min) for proteins with higher MW (Fig. 4E).
With chaperonin, proteins with low pI (<5 or 5–6) are still not particularly slow refolding, but now for the opposite reason: because GroEL is generally expeditious at refolding them, so they have mostly refolded within 1 min (Fig. 4B and SI Appendix, Data S2K). Proteins with high pI (>10) show similar kinetics with chaperonin as they do without. This may be because such proteins could bind too tightly to GroEL’s negatively charged lumen, which would render it a less efficient chaperone for these clients (and in a few rare cases, preclude folding). Both class III– and class IV proteins are refolded rapidly by GroEL (Fig. 4D), consistent with kinetic models that suggest these proteins form intermediates that rapidly sort to GroEL (42, 60). Finally, we find few differences in the rate for folding high-MW or low-MW proteins, a contrast with chaperone-null conditions in which high-MW proteins that fail to refold quickly generally do not recover within 5 min (Fig. 4 E and F).
GroEL/ES is Crucial for Folding α/β Folds.
Because our PK susceptibility measurements can be resolved down to individual residue locations, it is possible to assign nonrefolding sites to specific structural domains within proteins. Using the SCOP database (structural classification of proteins (61, 72)), such domains can be grouped into fold types, reflecting deep evolutionary relationships between polypeptides that share a common topology despite having very different sequences and functions. The intrinsic refoldability levels of different folds in cyto-serum largely preserve trends previously observed (Fig. 4I) (17). Small domains with ‘simple’ topologies (low contact order (62)) tend to be the most refoldable, such as OB-folds (79%), 3-helical bundles (84%), ubiquitin-like folds (88%), and SH3 barrels (100%). The specialized folds that are unique to aminoacyl-tRNA synthetases (aaRSs) are generally the least intrinsically refoldable, namely the adenine nucleotide-hydrolase-like fold (46%, the core of class I synthetases), and the class II aaRS core fold (21%). TIM barrels display slightly lower-than-average levels of refoldability in cyto-serum (62%, average is 64%).
GroEL has a profoundly restorative effective on these fold types (Fig. 4I), elevating the refolding frequencies of the class I and class II aaRS folds to 83% and 77%, respectively. In our experiment, GroEL rescued many TIM barrels (raising their refolding frequency to 81%) which is consistent with the previous observation that GroEL has a strong preference to coprecipitate TIM barrel-containing proteins (6, 55). However, we found additionally that GroEL had very pronounced effects on assisting Rossmann-folds (of both the NADH-binding (55 to 87%) and SAM-binding (73 to 100%) sublineages), P-loop NTPases (64 to 95%), and PRTase-like domains (29 to 100%). All the fold types that disproportionately benefit from GroEL have α/β architectures (63, 64) (except for the class II aaRS fold, which is α+β). In the presence of GroEL, we find that all fold types are highly refoldable, implying that GroEL smooths over the intrinsic differences in refoldability associated with different protein topologies.
DnaK is Also a Versatile Chaperone That Complements GroEL.
A second key chaperone in E. coli is DnaK (Hsp70), which operates with its cochaperone DnaJ (Hsp40) and a nucleotide exchange factor, GrpE (3, 24, 65, 66, 69). In experiments conceptually similar to those described in the previous sections (Fig. 4), we performed global refolding assays in which 5 µM DnaK, 1 µM DnaJ, and 1 µM GrpE were supplemented into the cyto-serum refolding dilution buffer (as well as to the native samples, as in Fig. 1A). We chose these concentrations and cochaperone ratios drawing from work showing their utility to facilitate folding for a wide variety of clients (13, 69). Initial analysis provided poor coverage (759 proteins total; SI Appendix, Fig. S8 A and B), because DnaK, DnaJ, and GrpE (abbreviated as DnaKJE) are cleaved by PK at many locations and accounted for 1,038 (11%) of all peptides quantified. To rectify this matter, we matched between runs from the GroE refolding samples (see Methods and SI Appendix, Fig. S1). With this change, the DnaK experiment’s coverage improved: we could quantify 11445 peptides (SI Appendix, Fig. S8D), making refoldability assessments on 901 proteins (SI Appendix, Fig. S8C), comparable to that of the GroEL experiment (998 proteins, 12,562 peptides).
DnaK results in 79% of the E. coli proteome refolding after 5 min (Fig. 5 A and B and SI Appendix, Data S3), comparable but slightly less to that of GroEL (85%). Indeed, virtually all the refoldability trends we found for GroEL were echoed with DnaK. This includes: a flattened pI-dependence (SI Appendix, Fig. S11A), a flattened MW-dependence with a less pronounced dip at 60–80 kDa (SI Appendix, Fig. S11B), and very little dependence on subunit stoichiometry (SI Appendix, Fig. S11C). The most salient difference is DnaK is somewhat worse at refolding large >80 kDa proteins (77%) compared to GroEL (91%). Class I proteins remain challenging candidates for DnaK, though class III– proteins demonstrated a noticeable preference (an effect that is not statistically significant however, on account of low counts) for DnaK over GroEL at both the 1-min and 5-min time points (SI Appendix, Fig. S11D). Mannose-6-phosphate isomerase (ManA) was identified by this experiment as a DnaK-dependent refolder, which we independently confirmed by an enzyme reactivation assay on purified ManA (SI Appendix, Fig. S4D).
Fig. 5.
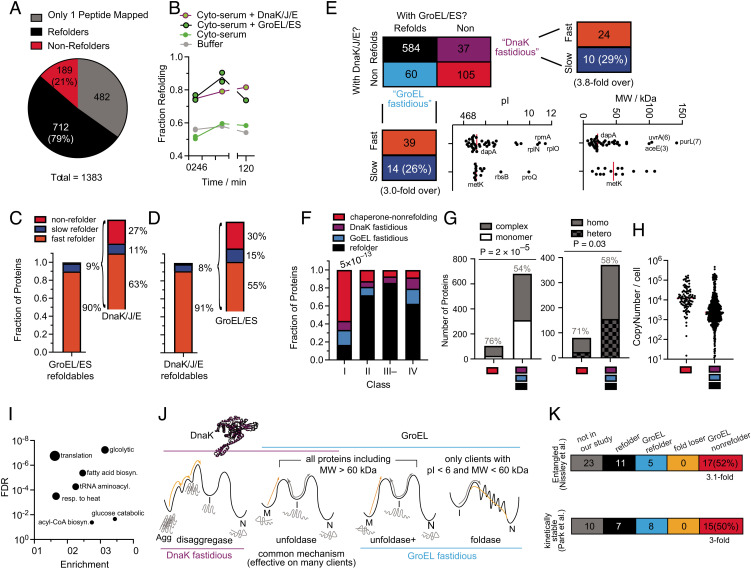
DnaK, DnaJ, GrpE refold many E. coli proteins, with only a few that are fastidious for one chaperone over the other. (A) Pie charts showing the number of (non)refoldable proteins for refolding experiments in cyto-serum with DnaKJE. Data correspond to #e in SI Appendix, Fig. S1C (SI Appendix, Data S3). (B) Fraction of proteins that refold after 1 min, 5 min, or 120 min in buffer (gray), cyto-serum (green), cyto-serum with GroEL/ES (green, black borders), or cyto-serum with DnaK/J/E (green, purple borders). (C) Frequency of slow refolding with GroEL. Of the 66 proteins that refold slowly with GroEL, bar to the right shows the frequency of proteins that refolded fast (within 1 min), slow (not within 1 min but within 5 min), or not at all in the cyto-serum/DnaKJE experiment. Uses #4, 5 in SI Appendix, Fig. S1B. (D) Frequency of slow refolding with DnaKJE. Of the 49 proteins that refold slowly with DnaK, bar to the right shows the frequency of proteins that refolded fast, slow, or not at all in the cyto-serum/GroE experiment. Uses #d, e in SI Appendix, Fig. S1C. (E) Truth table summarizing the results comparing refolding with GroE or DnaKJE (uses #e in SI Appendix, Fig. S1C). Analysis covers 786 proteins for which at least two peptides could be confidently quantified in both conditions. Proteins that refold only with GroE are called “GroEL fastidious” (light blue) and those only with DnaKJE are called “DnaK fastidious” (purple). pI and MW distributions for the GroEL fastidious proteins are given, broken down by whether they are fast GroEL refolders or slow GroEL refolders. (F) Frequency of proteins that refolded in both conditions (black), only with GroEL (light blue), only with DnaK (purple), or did not refold in either (chaperone-nonrefolder; red), separated on the basis of chaperonin class (6, 16). Numbers listed above bars indicate P-value by the chi-square test. (G) Left, Number of chaperone-nonrefolding proteins that are monomeric or in constitutive complexes. Gray percentages represent fraction in complexes. P-value according to Fisher’s exact test. Right, number of chaperone-nonrefolding proteins in complexes that are homomeric or heteromeric. Gray percentages represent fraction homomeric. P-value according to Fisher’s exact test. (H) Abundance of the 105 chaperone-nonrefolding proteins, compared to the other 681 in this analysis, according to Li et al. (68). (I) Gene ontology enrichment analysis of the 105 chaperone-nonrefolding proteins, compared to the E. coli genome, using PantherDB (91). (J) A model for the overlapping, but distinct, activities of DnaK and GroEL. (K) Further analyses on GroEL-nonrefolding proteins, correlating with separate studies which identified proteins that were found in a computational model to form entangled near-native states that would bypass recognition from chaperones (Top, 47); or that were found to be kinetically stable by remaining undigested by proteases for days (Bottom; 29).
Our results suggest that a refolding problem that is ‘challenging’ for one chaperone is not necessarily challenging for another. For instance, when we look at the minority of GroEL refolders that required more than 1 min to refold (slow refolders, 66 proteins in total), the majority are refolded quickly by DnaK (Fig. 5C). Ipso facto, for the minority of DnaK refolders that required more than 1 min to refold (51 proteins in total), the majority are refolded quickly by GroEL (Fig. 5D and SI Appendix, Data S3K). Hence, the strengths of these chaperones are complementary for certain clients.
In a comparative analysis that pooled together both the GroEL and DnaK refolding conditions at the 5-min time point, we identify 786 proteins for which two or more peptides were detected in each condition (Fig. 5E), thereby permitting an independent assessment of refoldability under both conditions (SI Appendix, Data SB, see SI Appendix, Fig. S8F for other time points). We find that most proteins that refold under GroEL also refold under DnaK, with only a small subset of proteins that appear to be specialized for GroEL (60 total) or DnaK (37 total). We will refer to the clients that can only refold with one chaperone or the other as ‘fastidious’ clients.
While the GroEL-fastidious clients mostly refold rapidly with GroEL (74%), we do find a surprisingly large number that refold slowly with GroEL (26%), threefold more frequent than slow GroEL-refolding in general (cf. Fig. 5E). It is instructive to divide the GroEL-fastidious clients into subgroups that refold quickly with GroEL and slowly with GroEL. The fast-refolding GroEL-fastidious clients are disproportionately acidic (the median pI of this group is 5.13 with 3 ribosomal proteins discounted) and low-MW (with three exceptions, though these high-MW proteins have many smaller domains). These proteins therefore most likely utilize GroEL’s foldase activity (folding inside the cage (5, 59), Fig. 5J). On the other hand, the GroEL-fastidious clients that refold slowly are perhaps those with highly entrenched misfolded states that require higher energy inputs to unfold and many iterative annealing cycles to fully correct. These proteins therefore likely employ GroEL’s stronger unfoldase activity (Fig. 5J). This hypothesis is supported by the fact that this group includes the well-known obligate GroEL client, MetK.
DnaK-fastidious clients also have a surprisingly large number of cases that refold slowly with DnaK (29%), 3.8-fold more frequent than slow DnaK refolding in general. Though we could not detect any obvious feature shared by the DnaK-fastidious refolders, it is likely that the misfolded forms these proteins populate are more easily recognized by the DnaJ/DnaK system. We reached this assessment by cross-comparing our data to a study from Calloni et al. (10), which measured enrichment factors for DnaK clients that coisolate with a DnaK pull-down (SI Appendix, Fig. S11D). Proteins which were not detected by Calloni (which are presumed to not be DnaK clients), were indeed significantly over-represented with refolders and had few DnaK-fastidious refolders. On the other hand, we found significant enrichments (upto 2.2-fold) for DnaK-fastidious refolders among proteins with modest DnaK-enrichment factors (< fivefold) at the 1-min timepoint—but not those with >fivefold enrichment or at the later timepoint. This result makes sense, because proteins which DnaK can refold rapidly should be detected in pull-downs but would not accumulate to large portions of steady-state fractional DnaK occupancy. On the other hand, at the 5-min timepoint, the category with the greatest proportion of DnaK-fastidious refolders is the one with the highest enrichment factor (>10-fold). As expected, we found that class IV proteins were enriched to be GroEL-fastidious (2.2-fold), but the effect is not statistically significant on account of fewer proteins being simultaneously detected in the GroEL and DnaK refolding experiments (Fig. 5F).
Discussion
Revising the Scope of Obligate GroEL Refolders.
Our study is consistent with aspects of, but also necessitates revision to, the consensus model of which E. coli proteins require GroEL for efficient refolding. The consensus model is strongly influenced by the classic work by Kerner et al. in which rapid depletion of ATP was used to entrap GroEL clients within the cis cavity of the GroEL/ES complex (6). Pull-down on a His-tagged GroES then resulted in coprecipitation of GroEL interactors, which were identified with mass spectrometry. Proteins that were highly enriched in the GroEL fraction, which were termed class III proteins, were found to be generally low-abundance, between 30 and 60 kDa, and over-represent TIM barrel folds. By analyzing protein refoldability levels in cyto-serum vs. those in cyto-serum supplemented with GroEL and GroES, we can assess which of the proteins that get entrapped with in GroEL also depend on it to refold. Our results concur with the finding that TIM barrels tend to be more GroEL-dependent (Fig. 4I). On the other hand, the findings that GroEL is particularly important for refolding high-MW and low-pI proteins (Fig. 3) in E. coli have not been described. Why were these patterns not previously observed? Thoughtful reflection on what pull-down assays can and cannot show is instructive in this matter. High-MW proteins cannot be entrapped within the sealed GroEL/ES cis cavity, and therefore would be systematically excluded from pull-down assays. Indeed, previous work has highlighted several examples in which GroEL restored the activity of high-MW proteins that cannot fit inside the cavity, particularly aconitase (AcnB, 93 kDa) (48), which our study confirms can refold to a native structure in the presence of GroEL. Our experiment also confirms DNA gyrase (GyrA (97 kDa) and GyrB (90 kDa)) and MetE (85 kDa) can refold in the presence of GroEL. Previous work showing that GroEL can refold high-MW proteins has been explained by positing that the trans cavity can also bind misfolded clients (49, 50), and that out-of-cage refolding occurs (53). Our results suggest that these two activities represent critical functions of GroEL. While E. coli does not have many proteins with MW greater than 80 kDa, these observations suggest that GroEL plays a significant role in their biogenesis, echoing the observation that eukaryotic TriC/CCT has been shown to principally operate on large proteins (71).
A second key feature that emerges from our set of obligate GroEL-refolders is the outsize role GroEL plays in refolding acidic proteins (pI < 6). The negatively charged cavity walls of GroEL (53, 54) would be expected to create a ‘repulsive field’ for acidic proteins that could facilitate their compaction, overcoming the inter-residue electrostatic repulsion within a protein chain that would counter its tendency to collapse. Supporting this view is the further observation that the group of slow GroEL refolders has few proteins with low pI (Figs. 4B and 5E). Indeed, the primary work which established the potential foldase activity of the GroEL cavity (5) found that inside the cage, GroEL/ES accelerates productive folding (foldase) of R. rubrum RuBisCo but merely prevents aggregation of B. taurus rhodanese (holdase). Consistent with our model, RrRuBisCo has a low pI (of 5.6) while BtRhodanese does not (6.9). PepQ, whose folding is also catalyzed by the GroEL cavity (25), also is acidic (pI of 5.7). There is a plausible reason why this key relationship with pI was not detected previously: because GroEL refolds acidic protein expeditiously, they would not accumulate within it to become a large steady-state fraction of GroEL occupancy. Such assertions raise the obvious question: What about cationic proteins? Our study shows that E. coli protein with high pI are generally efficient intrinsic refolders, and particularly so in the cytosolic medium (Fig. 2F and (17, 47)), thereby bypassing GroEL.
DnaK’s Activities in Relation to GroEL’s.
Hsp70s and the menagerie of cochaperone J-domain proteins have attracted interest in recent years, due to their importance in several diseases and the discovery that they can disperse amyloid fibrils (65, 79). Our approach provides a means to compare DnaK’s activity to GroEL’s proteome-wide under the same conditions. Overall, DnaK and GroEL refold a similar clientele with only a small number that are specialized (fastidious) for one or the other. These observations are consistent with prevailing ideas that the proteostasis network is integrated (60) and that the DnaK and GroEL systems are complementary (42), with a large amount of redundancy built in. This finding is consistent with an emerging view that most chaperones share a common mechanism that can be effective on many clients, namely, unfoldase activity on misfolded states (Fig. 5A) (52, 59, 69, 73–75), thereby providing those molecules with further opportunities to refold properly (the iterative annealing mechanism (76)).
However, DnaK and GroEL also have aspects that make them unique (Fig. 5J). In addition to acting as an unfoldase, DnaK is part of the E. coli disaggregase system, while GroEL’s cavity can also act as a foldase (54, 60). GroEL’s unfoldase activity may be its more general function, with foldase activity reserved for smaller (<60 kDa) acidic (pI < 6) clients. It is possible that GroEL is a stronger unfoldase because its apical domain movements (which couple to unfolding) are driven by cooperative binding/hydrolysis of 7 ATPs. In a few cases, GroEL’s “strong unfoldase” activity may be required for a handful of clients that populate misfolded states that are deeply energetically entrenched (with MetK and DapA as important examples). MetK in particular forms true topological knots (47), expected to result in entrenched misfolded states.
DnaK is known to play a key role in promoting disaggregase activity, a critical function that was probably rendered less important in our assay because of the low aggregation occurring in our dilute refolding reactions. Our results show that under such permissive conditions, DnaK and GroEL can act relatively interchangeably (Fig. 5E), consistent with there being very few biophysical profiles that benefit synergistically from these client systems according to the FoldEco model (42). Under conditions with greater aggregation, cooperation between multiple chaperone systems would likely be more significant (70).
Chaperonins Potentiated an Expansion of α/β Folds.
Are certain types of protein topologies better at folding themselves than others? Our study suggests that under cellular-like conditions, small all-β domains refold the best, specifically, ubiquitin-like folds, SH3 barrels, and OB-folds. These findings support the theory that all-β domains were the earliest globular proteins, the immediate descendants of amyloids (77, 78). On the other hand, the most expansive and versatile folds are all α/β, and include TIM barrels, Rossmanns, and P-loop NTP hydrolases, though these folds all display stronger dependence on GroEL (63). The current view is that Hsp60s (relatives of GroEL) are very ancient, and possibly the only chaperone system the last universal common ancestor (LUCA) possessed (80). In light of this, we theorize that these fold-types coemerged with chaperonin, and that the emergence of chaperonin led to a great expansion of protein functional space attendant with them (81). Once these larger, more topologically complex domains could be efficiently folded, their functional versatility became accessible, and they became the most dominant architectures of the protein world.
Possible Models for Chaperone-Nonrefolders.
One important feature of the DnaK/GroEL cross-correlation dataset (Fig. 5E) is that there are some proteins that do not refold with either GroEL or DnaK, and in fact the most predictive descriptor for whether a protein cannot refold with GroEL is whether it cannot refold with DnaK and ipso facto (odd’s ratio = 51.4; P-value < 10–66 by Fisher’s exact test). We will refer to these 105 proteins in the following as chaperone-nonrefolders for brevity’s sake, though what is implied is ‘proteins that do not refold with either GroEL or DnaK.’ How do these proteins locate their native states in the first place?
Notwithstanding the important caveat that misfolded states encountered during refolding from denaturant are possibly distinct from those populated in vivo, we enumerate four potential explanations: (i) these proteins require a combination of chaperone systems acting synergistically to refold (e.g., the DnaK and GroEL systems together); (ii) these proteins require longer incubation times and/or different concentrations of GroEL, DnaK, and their cochaperones to refold; (iii) these proteins require the service of chaperones not considered in this study to refold, such as HtpG (E. coli’s Hsp90), trigger factor (TF), ClpB, or small heat shock proteins (IbpA, IbpB); and (iv) these proteins have a strong preference to fold cotranslationally on the ribosome and would be challenging to refold from the denatured state with any set of chaperones.
The first two explanations can be mostly ruled out. Of the 33 class IV proteins assessed in our GroEL refolding assay (which are bona fide obligate GroEL clients), 30 (91%) refolded in vitro with GroEL alone (Fig. 2E). This result could not have been obtained if GroEL-requiring proteins generally required DnaK simultaneously. Similarly, the high refolding proportion of the challenging class IV substrates in our assay would seem to dismiss the possibility that the conditions and timescales used in these experiments were inadequate to elicit the native function of GroEL. While there are several well-known examples of multiple chaperones operating in a cascade (such as in preventing aggregation of BtRhodanese (82) and in refolding heat-denatured porcine malate dehydrogenase (70)), we note that neither of these are native E. coli proteins.
The latter two explanations both deserve consideration. Our study did not cover the other E. coli chaperone systems such as TF (3, 83), small heat shock proteins (70, 84), the ClpB disaggregase (85–87), or HtpG—a foldase that operates with the DnaK system (88, 89)—all of which might play important roles in refolding certain clients. Nevertheless, several additional lines of evidence support the view that the 105 chaperone-nonrefolders fold cotranslationally. We find a striking overrepresentation of class I proteins in this group (Fig. 5F). Class I proteins bypass the (predominantly) post-translational GroEL chaperone system, which is consistent with them completing most of their folding on the ribosome. We note that this amounts to a revision to the typical view of class I proteins: Whereas it had been assumed that class I proteins do not strongly engage GroEL because they are efficient intrinsic folders, our data suggest that this may be because more of their folding is completed cotranslationally. Moreover, the majority of these proteins are in complexes (80 out of 105, 76%), of which the majority (57 out of 80, 71%) are in homocomplexes (Fig. 5G). Homomers have been shown to be the most likely to assemble during translation in a “co-co” fashion (wherein nascent chains assemble while both are in translation) (90). Our study suggests that this mode of assembly may be obligatory in some situations. We find that chaperone-nonrefolders are also generally highly abundant proteins (Fig. 5H) and overrepresent core metabolic processes, including tRNA aminoacylation (Fig. 5I). Moreover, the high representation of synthetases is also notable given that previous refolding assays on purified ThrS showed that no combination of GroEL and DnaK can reactivate it beyond ~50% (6). We also find that proteins which form noncovalent lasso entanglements (47) are threefold more likely to not refold with GroEL (Fig. 5K), a subtle form of misfolding that is hard for chaperones to detect but which can be avoided through properly scheduled cotranslational folding (67). As further evidence that chaperone-nonrefolding proteins ultimately populate native-like conformations that evade chaperone detection is our finding that in the long-term, ATP levels stabilized in our GroEL-refolding assays rather than run out (cf. SI Appendix, Fig. S9).
Finally, we find that kinetically stable proteins (29) are threefold more likely to not refold with GroEL (Fig. 5K). One would imagine that proteins which fold well cotranslationally, but inefficiently from a denatured form, should ideally have very slow unfolding rates such that the denatured form would not appreciably populate during biological timescales. Our observed correlation supports this notion. On the other hand, we did not identify significant correlation between nonrefolding proteins and those whose aggregation level is mitigated by chaperones during over-expressed in vitro translation (12, 13; SI Appendix, Fig. S12): this discrepancy highlights the inherent difference between cotranslational folding and refolding from denaturant.
While our study cannot unambiguously determine which proteins have a heightened preference to fold cotranslationally, the evidence presented here does build a case that such a category of proteins exist in the E. coli proteome, and that chaperone-nonrefolders (as defined here) are enriched with them.
Materials and Methods
Detailed methods are provided in the SI Appendix, as are tables describing reagents and resources, data availability, and mass spec parameters. In brief: cyto-serum was prepared by growing E. coli cells (strain K12) to OD600 2.0, lysing cells in Millipore Water by sonication, removing macromolecules by ultracentrifugation (16,000 g for 15 min, then 40,000 rpm in SW55 Ti rotor for 20 h) and ultrafiltration (2k MWCO), and reducing the volume in a vacuum centrifuge to the original combined cellular volume. To perform global refolding experiments, E. coli cells were grown in MOPS in media in pairs to OD600 0.8, with one set of cultures containing 0.5 mM [13C6]L-arginine and 0.4 mM [13C6]L-lysine, and the other with 0.5 mM L-arginine and 0.4 mM L-lysine. Pairs of cell pellets were mixed and lysed into cyto-serum by cryogenic pulverization, clarified (16,000 g for 15 min), ribosome-depleted (33,300 rpm in SW55 Ti rotor for 90 min), and normalized to 3.3 mg/ml by BCA assay. Global unfolding was conducted by adding solid GdmCl and reduction in vacuo to 11.6 mg/ml (protein) and 6 M (GdmCl) final concentration and overnight incubation; refolding was initiated by 100-fold dilution with cyto-serum. Either GroEL/ES (4 µM, 8 µM, respectively) DnaK/DnaJ/GrpE (5 µM, 1 µM, 1 µM), or no chaperone was supplemented. After desired refolding time, limited proteolysis was conducted with PK (1:100 w/w ratio, 1 min) before quenching by immersion in an oil bath (110˚C), and addition of urea to 8 M. Standard proteomics mass spec sample preparation followed, and data were analyzed with custom scripts built in Python.
Supplementary Material
SI Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Acknowledgments
We thank Susan Marqusee, Ed O’Brien, and Dan Nissley for thoughtful discussion. We thank Philip Mortimer for maintaining the Mass Spectrometer Facility at JHU Department of Chemistry. We thank Di Wu and Grzegorz Piszczek at the National Institutes of Health (Bethesda, MD) for expertise on and assistance with mass photometry experiments; Funding: S.D.F. acknowledges support from the NIH Director’s New Innovator Award (DP2GM140926) and from the NSF Division of Molecular and Cellular Biology (MCB2045844). K.G.F. acknowledges support from NIGMS (R01GM079440). T.D. was supported by an NIH training grant (T32GM008403).
Author contributions
P.T., K.G.F., and S.D.F. designed research; P.T., Y.X., S.O.L., T.D., and S.D.F. performed research; S.D.F. contributed new reagents/analytic tools; P.T. and S.D.F. analyzed data; and P.T. and S.D.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Raw proteomic data are available via the ProteomeXchange under accession codes PXD030869. Processed (quantified) peptide data are available on Dryad at DOI: https://doi.org/10.5061/dryad.bnzs7h4dg. Summary data used to construct figures are provided online as SI Appendix, Data SA–SB, S1–S3, and S1K–S3K. Python programs are available on GitHub at https://github.com/FriedLabJHU/Refoldability-Tools/.
Supporting Information
References
- 1.Anfinsen C. B., Principles that govern the folding of protein chains. Science 181, 223–230 (1973). [DOI] [PubMed] [Google Scholar]
- 2.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science. 353, aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Tyedmers J., Mogk A., Bukau B., Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Bio. 11, 777–788 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Brinker A., et al. , Dual Function of protein confinement in chaperonin-assisted protein folding. Cell 107, 223–233 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Kerner M. J., et al. , Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Singh A. K., Balchin D., Imamoglu R., Hayer-Hartl M., Hartl F. U., Efficient catalysis of protein folding by GroEL/ES of the obligate chaperonin substrate MetF. J. Mol. Biol. 432, 2304–2318 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Viitanen P. V., et al. , Chaperonin-facilitated refolding of ribulose bisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are potassium dependent. Biochemistry 29, 5665–5671 (1990). [DOI] [PubMed] [Google Scholar]
- 9.Anfinsen C. B., Haber E., Sela M., White F. H., The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc. Natl. Acad. Sci. U.S.A. 47, 1309–1314 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calloni G., et al. , DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 1, 251–264 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Willmund F., et al. , The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niwa T., et al. , Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 4201–4206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niwa T., Kanamori T., Ueda T., Taguchi H., Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc. Natl. Acad. Sci. U.S.A. 109, 8937–8942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mateus A., et al. , Thermal proteome profiling in bacteria: Probing protein state in vivo. Mol. Sys. Biol. 14, e8242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarzab A., et al. , Meltome atlas—thermal proteome stability across the tree of life. Nat. Methods 17, 495–503 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara K., Ishihama Y., Nakahigashi K., Soga T., Taguchi H., A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29, 1552–1564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.To P., Whitehead B., Tarbox H. E., Fried S. D., Nonrefoldability is pervasive across the E. coli Proteome. J. Am. Chem. Soc. 143, 11435–11448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorov A. N., Baldwin T. O., Cotranslational protein folding. J. Biol. Chem. 272, 32715–32718 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Frydman J., Erdjument-Bromage H., Tempst P., Hartl F. U., Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat. Struct. Biol. 6, 697–705 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Liu K., Maciuba K., Kaiser C. M., The ribosome cooperates with a chaperone to guide multi-domain protein folding. Mol. Cell 74, 310–319.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamant S., Eliahu N., Rosenthal D., Goloubinoff P., Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J. Biol. Chem. 276, 39586–39591 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Bandyopadhyay A., et al. , Chemical chaperones assist intracellular folding to buffer mutational variations. Nat. Chem. Biol. 8, 238–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daugherty D. L., Rozema D., Hanson P. E., Gellman S. H., Artificial chaperone-assisted refolding of citrate synthase. J. Biol. Chem. 273, 33961–33971 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Sekhar A., Santiago M., Lam H. N., Lee J. H., Cavagnero S., Transient interactions of a slow-folding protein with the Hsp70 chaperone machinery. Protein Sci. 21, 1042–1055 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weaver J., et al. , GroEL actively stimulates folding of the endogenous substrate protein PepQ. Nat. Commun. 8, 15934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Souza N., Picotti P., Mass spectrometry analysis of the structural proteome. Curr. Opin. Struct. Biol. 60, 57–65 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Feng Y., et al. , Global analysis of protein structural changes in complex proteomes. Nat. Biotech. 32, 1036–1044 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Park C., Marqusee S., Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat. Methods 2, 207–212 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Park C., Zhou S., Gilmore J., Marqusee S., Energetics-based protein profiling on a proteomic scale: Identification of proteins resistant to proteolysis. J. Mol. Biol. 368, 1426–1437 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett B. D., et al. , Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szatmári D., et al. , Intracellular ion concentrations and cation-dependent remodelling of bacterial MreB assemblies. Sci. Rep. 10, 12002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips R., Kondev J., Theriot J., Garcia H. G., Orme N., Physical Biology of the Cell. Taylor & Francis Group, 10.1201/9781134111589 (2012). [DOI] [Google Scholar]
- 33.Neidhardt F. C., Bloch P. L., Smith D. F., Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris C. L., An aminoacyl-tRNA synthetase complex in Escherichia coli. J. Bacteriol. 169, 2718–2723 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace E. W. J., et al. , Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., et al. , A systematic protein refolding screen method using the DGR approach reveals that time and secondary TSA are essential variables. Sci. Rep. 7, 9355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weissman J. S., Rye H. S., Fenton W. A., Beechem J. M., Horwich A. L., Characterization of the active intermediate of a GroEL–GroES-mediated protein folding reaction. Cell 84, 481–490 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Rye H. S., et al. , Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature 388, 792–798 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Sadat A., et al. , GROEL/ES buffers entropic traps in folding pathway during evolution of a model substrate. J. Mol. Biol. 432, 5649–5664 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Todd M. J., Viitanen P. V., Lorimer G. H., Hydrolysis of adenosine 5’-triphosphate by Escherichia coli GroEL: Effects of GroES and potassium ion. Biochemistry 32, 8560–8567 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Gottesman S., Wickner S., Maurizi M. R., Protein quality control: triage by chaperones and proteases. Genes Dev. 11, 815–823 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Powers E. T., Powers D. L., Gierasch L. M., FoldEco: A model for proteostasis in E. coli. Cell Rep. 1, 265–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong A. T., Leprevost F. V., Avtonomov D. M., Mellacheruvu D., Nesvizhskii A. I., MSFragger: Ultrafast and comprehensive peptide identification in shotgun proteomics. Nat. Methods 14, 513–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palomba A., et al. , Comparative evaluation of maxquant and proteome discoverer MS1-based protein quantification tools. J. Proteome Res. 20, 3497–3507 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nahnsen S., Bielow C., Reinert K., Kohlbacher O., Tools for label-free peptide quantification. Mol. Cell Proteomics. 12, 549–556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ying B., Taguchi H., Kondo M., Ueda T., Co-translational Involvement of the Chaperonin GroEL in the folding of newly translated polypeptides. J. Biol. Chem. 280, 12035–12040 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Nissley D. A., et al. , Universal protein misfolding intermediates can bypass the proteostasis network and remain soluble and less functional. Nat. Commun. 13, 3081 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaudhuri T. K., Farr G. W., Fenton W. A., Rospert S., Horwich A. L., GroEL/GroES-mediated folding of a protein too large to be encapsulated. Cell 107, 235–246 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Chaudhuri T. K., Verma V. K., Maheshwari A., GroEL assisted folding of large polypeptide substrates in Escherichia coli: Present scenario and assignments for the future. Prog. Biophys. Mol. Biol. 99, 42–50 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Farr G. W., et al. , Folding with and without encapsulation by cis- and trans-only GroEL–GroES complexes. EMBO J. 22, 3220–3230 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paul S., Singh C., Mishra S., Chaudhuri T. K., The 69 kDa Escherichia coli maltodextrin glucosidase does not get encapsulated underneath GroES and folds through trans mechanism during GroEL/ GroES-assisted folding. FASEB J. 21, 2874–2885 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Priya S., et al. , GroEL and CCT are catalytic unfoldases mediating out-of-cage polypeptide refolding without ATP. Proc. Natl. Acad. Sci.U.S.A. 110, 7199–7204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Y. C., et al. , Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Tang Y., Chang H., Chakraborty K., Hartl F. U., Hayer-Hartl M., Essential role of the chaperonin folding compartment in vivo. EMBO J. 27, 1458–1468 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Georgescauld F., et al. , GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding. Cell 157, 922–934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ambrose A. J., Fenton W., Mason D. J., Chapman E., Horwich A. L., Unfolded DapA forms aggregates when diluted into free solution, confounding comparison with folding by the GroEL/GroES chaperonin system. FEBS Lett. 589, 497–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mas G., Thoma J., Hiller S., Bacterial Cell Walls and Membranes. Subcell Biochem. 92, 169–186 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Bartlett A. I., Radford S. E., An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat. Struct. Mol. Biol. 16, 582–588 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Balchin D., Hayer-Hartl M., Hartl F. U., Recent advances in understanding catalysis of protein folding by molecular chaperones. FEBS Lett. 594, 2770–2781 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Santra M., Farrell D. W., Dill K. A., Bacterial proteostasis balances energy and chaperone utilization efficiently. Proc. Natl. Acad. Sci. U.S.A. 114, E2654–E2661 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pandurangan A. P., Stahlhacke J., Oates M. E., Smithers B., Gough J., The SUPERFAMILY 2.0 database: A significant proteome update and a new webserver. Nucleic Acids Res. 47, gky1130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plaxco K. W., Simons K. T., Baker D., Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 277, 985–994 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Cheng H., et al. , ECOD: An evolutionary classification of protein domains. PLoS Comput. Biol. 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schaeffer R. D., Liao Y., Cheng H., Grishin N. V., ECOD: New developments in the evolutionary classification of domains. Nucleic Acids Res. 45, D296–D302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenzweig R., Nillegoda N. B., Mayer M. P., Bukau B., The Hsp70 chaperone network. Nat. Rev. Mol. Cell. Biol. 20, 665–680 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Mayer M. P., Gierasch L. M., Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 294, 2085–2097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang Y., et al. , How synonymous mutations alter enzyme structure and function over long time scales. Nat. Chem., in press. [DOI] [PMC free article] [PubMed]
- 68.Li G. W., Burkhardt D., Gross C., Weissman J. S., Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Imamoglu R., Balchin D., Hayer-Hartl M., Hartl F. U., Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat. Commun. 11, 365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veinger L., Diamant S., Buchner J., Goloubinoff P., The small heat-shock protein ibpb from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J. Biol. Chem. 273, 11032–11037 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Yam A. Y., et al. , Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly-made proteins with complex topologies. Nat. Struct. Mol. Biol. 15, 1255–1262 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gough J., Karplus K., Hughey R., Chothia C., Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Sharma S. K., Rios P. D. L., Christen P., Lustig A., Goloubinoff P., The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat. Chem. Biol. 6, 914–920 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Lin Z., Madan D., Rye H. S., GroEL stimulates protein folding through forced unfolding. Nat. Struct. Mol. Biol. 15, 303–311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macošek J., Mas G., Hiller S., Redefining molecular chaperones as chaotropes. Front. Mol. Biosci. 8, 683132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thirumalai D., Lorimer G. H., Chaperonin-mediated protein folding. Annu. Rev. Bioph. Biom. 30, 245–269 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Petrov A. S., et al. , History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. U.S.A. 112, 15396–15401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bowman J. C., Petrov A. S., Frenkel-Pinter M., Penev P. I., Williams L. D., Root of the tree: The significance, evolution, and origins of the ribosome. Chem. Rev. 120, 4848–4878 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Gao X., et al. , Human Hsp70 disaggregase reverses Parkinson’s-linked α-synuclein amyloid fibrils. Mol. Cell 59, 781–793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rebeaud M. E., Mallik S., Goloubinoff P., Tawfik D. S., On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life. Proc Natl. Acad. Sci. U.S.A. 118, e2020885118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lindquist S., Protein folding sculpting evolutionary change. Cold Spring Harb. Symp. Quant. Biol. 74, 103–108 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Langer T., et al. , Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356, 683–689 (1992). [DOI] [PubMed] [Google Scholar]
- 83.Kaiser C. M., et al. , Real-time observation of trigger factor function on translating ribosomes. Nature 444, 455–460 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Eyles S. J., Gierasch L. M., Nature’s molecular sponges: Small heat shock proteins grow into their chaperone roles. Proc. Natl. Acad. Sci. U.S.A. 107, 2727–2728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maurizi M. R., Xia D., Protein binding and disruption by Clp/Hsp100 chaperones. Structure. 12, 175–183 (2004). [DOI] [PubMed] [Google Scholar]
- 86.Deville C., Franke K., Mogk A., Bukau B., Saibil H. R., Two-step activation mechanism of the ClpB disaggregase for sequential substrate threading by the main ATPase motor. Cell Rep. 27, 3433–3446.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Avellaneda M. J., et al. , Processive extrusion of polypeptide loops by a Hsp100 disaggregase. Nature 578, 317–320 (2020). [DOI] [PubMed] [Google Scholar]
- 88.Genest O., Hoskins J. R., Camberg J. L., Doyle S. M., Wickner S., Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc. Natl. Acad. Sci. U.S.A. 108, 8206–8211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luengo T. M., Kityk R., Mayer M. P., Rüdiger S. G. D., Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Bertolini M., et al. , Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. Science 371, 57–64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mi H., et al. , Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Data Availability Statement
Raw proteomic data are available via the ProteomeXchange under accession codes PXD030869. Processed (quantified) peptide data are available on Dryad at DOI: https://doi.org/10.5061/dryad.bnzs7h4dg. Summary data used to construct figures are provided online as SI Appendix, Data SA–SB, S1–S3, and S1K–S3K. Python programs are available on GitHub at https://github.com/FriedLabJHU/Refoldability-Tools/.