Significance
Consumption of fruits and vegetables has been associated with protection from cardiometabolic disease (CMD)-related mortality. While these beneficial effects have traditionally been attributed to flavonoids, these molecules are poorly available for small intestinal absorption and subsequent systemic distribution. Here, we demonstrate that host health effects of flavonoid consumption actually hinge on their metabolism by dedicated members of the gut microbiota. We next show that a single gut microbial flavonoid catabolite is sufficient to reduce diet-induced CMD burden in mice. Metagenomics analysis revealed the individuality of flavonoid metabolism among the human gut microbiota. Our study highlights the importance of microbial metabolism and its resulting bioactive catabolites in the design of dietary and probiotic interventions to complement traditional CMD treatment strategies.
Keywords: gut microbiome, flavonoid metabolism, cardiometabolic disease, obesity, AMP-activated protein kinase
Abstract
The molecular mechanisms by which dietary fruits and vegetables confer cardiometabolic benefits remain poorly understood. Historically, these beneficial properties have been attributed to the antioxidant activity of flavonoids. Here, we reveal that the host metabolic benefits associated with flavonoid consumption hinge, in part, on gut microbial metabolism. Specifically, we show that a single gut microbial flavonoid catabolite, 4-hydroxyphenylacetic acid (4-HPAA), is sufficient to reduce diet-induced cardiometabolic disease (CMD) burden in mice. The addition of flavonoids to a high fat diet heightened the levels of 4-HPAA within the portal plasma and attenuated obesity, and continuous delivery of 4-HPAA was sufficient to reverse hepatic steatosis. The antisteatotic effect was shown to be associated with the activation of AMP-activated protein kinase α (AMPKα). In a large survey of healthy human gut metagenomes, just over one percent contained homologs of all four characterized bacterial genes required to catabolize flavonols into 4-HPAA. Our results demonstrate the gut microbial contribution to the metabolic benefits associated with flavonoid consumption and underscore the rarity of this process in human gut microbial communities.
A balanced diet is one of the most influential drivers of human health (1). This notion has become increasingly important in our industrialized era, characterized by pervasive human metabolic syndrome (2, 3). More recently, diet has become appreciated as a focal determinant of gut microbial community structure, function, and resilience where dietary choices are recognized to rapidly alter the human gut microbiome (4). Moreover, diet-derived metabolites from the human gut microbiota have been causally linked to cardiovascular and metabolic disease pathogenesis (5–8). Consequently, microbial metabolites arising from specific dietary components, such as trimethylamine (TMA), imidazole propionate, and short-chain fatty acids (SCFAs), have gained recognition as the central mediators of human health and disease (5–9).
Flavonoids represent a key molecular component of plant-based diets. They have been attributed antioxidant, antiobesogenic, and chemoprotective properties through the scavenging of free radicals and activation of molecular effectors implicated in human disease (10). Dietary flavonoids are largely glycosylated, limiting their absorption in the small intestine and thereby their systemic distribution (10). Consequently, upon passing into the colon, flavonoids become a substrate for gut microbial catabolism. Notable prior studies have highlighted that dietary flavonoids attenuate diet-induced obesity in a microbe-dependent manner (11, 12). Several human gut bacteria that are capable of breaking down flavonoid substrates through reduction and subsequent cleavage of the central nonaromatic ring, followed by hydrolysis to yield monophenolic acid degradation products, have been isolated (13). The four types of bacterial enzymes required in the flavone/flavonol catabolic pathway, namely flavone reductase (FLR), chalcone isomerase (CHI), enoate reductase (EnoR), and phloretin hydrolase (PHY), have been identified and biochemically characterized (14–17). Together, these enzymes represent just one mechanism by which commensal gut bacteria metabolize host dietary inputs; additional homologous or nonhomologous pathways may remain undiscovered.
Results
Flavonoids Reduce the Obesogenic Effects of a High Fat Diet.
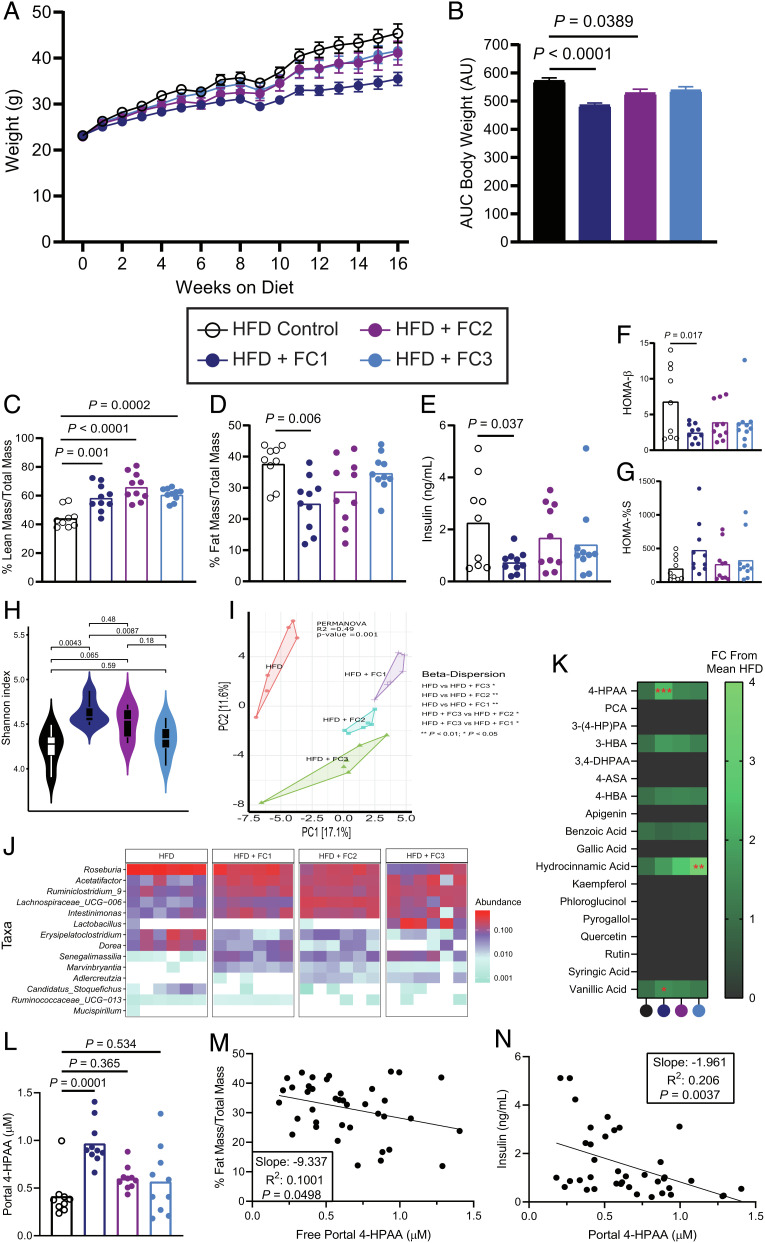
Since ingested flavonoids themselves have poor bioavailability, we hypothesized that their microbial monophenolic acid catabolites are responsible for prior ascribed antiobesogenic properties (11, 12). To identify candidate catabolites, we used a comparative targeted metabolomics analysis of mice that were supplemented three different flavonoid composites (FC1, FC2, and FC3) on a high fat diet (HFD) background. For 16 wk, we provided mice with ad libitum access to a high fat control diet, or the same HFD base, supplemented with 1% w/w of FC1, FC2, or FC3. Metabolic parameters including body weight, lean mass, fat mass, and glucose homeostasis were tracked for the duration of the experiment. We observed that FC1 mice were markedly protected from HFD-induced obesity (Fig. 1 A–D and SI Appendix, Fig. S1 A–D). Moreover, FC1 mice had more lean mass and less fat mass and were less hyperinsulinemic than HFD control mice 16 wk after initiation of differential diet treatments and comparable body weight to a low fat diet control (Fig. 1 E–G and SI Appendix, Fig. S1 E and F). Analysis of the cecal microbial composition by 16S rRNA sequencing revealed that FC1 mice had significantly more diverse cecal microbial communities that clustered distinctly from the HFD control mice using NMDS, and from the other FC mice (Fig. 1 H–J and SI Appendix, Figs. S2 A–C and S3 A–I). Of note, Lachnospiraceae UCG-006 was highly abundant in the cecal communities of FC mice compared to the HFD controls (Fig. 1J and SI Appendix, Figs. S2 B and C and S3 F). Moreover, other Lachnospiraceae clades (NK4A136 and UCG-009) were significantly more abundant in FC1 mice when compared to the HFD control group (SI Appendix, Figs. S2 B and C and S3 G).
Fig. 1.
Flavonoid composites reduce HFD-induced obesity and alter the cecal microbiome. (A) Body weights of 6-wk-old male C57BL/6 mice fed a high fat control diet, or the same diet supplemented with 1% w/w FC1, FC2, or FC3 for 16 wk; n = 9–10 per group, error bars represent SEM. (B) Mean cumulative area under the curve (AUC) for body weights after 16 wk, error bars represent SEM. (C) and (D) Lean and fat mass after 16 wk as measured by EchoMRI and normalized to total body mass, n = 9–10 per group. (E) Plasma insulin after 16 wk on either control or experimental diets following a 4-h fast, n = 9–10 per group. (F) Homeostatic model assessment of β-cell function (HOMA-β), n = 9–10 per group. (G) Percent insulin sensitivity after 16 wk, n = 9–10 per group. (H) Shannon alpha diversity estimates for cecal microbiomes based on 16S rRNA profiles of all the four groups. Statistical analysis was performed via ANOVA. (I) Principal component analysis (PCA) plot based on the Bray–Curtis dissimilarity index between the cecal 16S rRNA profile of all the four groups. Statistical analysis was performed with PERMANOVA where R2 values are noted for comparisons with significant P-values and stand for percentage variance explained by the variable of interest. (J) Heatmap of the significantly differentially abundant bacterial taxa across the cecal 16S rRNA profiles of all the four groups. n = 6 for all 16S rRNA sequencing analyses. (K) Heatmap of portal plasma flavonoids and microbial flavonoid catabolites measured by LC-MS/MS, n = 9–10 per group. Statistical analysis was performed with one-way ANOVA with Tukey’s multiple comparison test. (L) Portal plasma concentration of the microbial flavonol catabolite 4-HPAA measured by LC-MS/MS. (M) Simple linear regression analysis of fat mass proportion and portal plasma 4-HPAA. (N) Simple linear regression analysis of terminal plasma insulin and portal plasma 4-HPAA. P values shown were calculated using one-way ANOVA with Dunnett’s multiple comparisons test; n = 9–10 per group. Individual points represent individual mice, and bars represent group means.
Provided that mice are likely to contain commensal gut microbes capable of catabolizing flavonoids into monophenolic acids, we expected that these microbe-derived monophenolic acids would be enriched in the portal plasma of FC mice compared to the HFD control. A targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) approach revealed six known microbial flavonoid catabolites at detectable levels in the portal plasma (18) (Fig. 1K and SI Appendix, Fig. S3 J–N). Of the six metabolites, only 4-hydroxyphenylacetic acid (4-HPAA) and 4-hydroxy-3-methoxybenzoic acid were significantly enriched in the portal plasma of FC1 mice compared to the HFD control (Fig. 1 K and L and SI Appendix, Fig. S3J). Between these two metabolites, 4-HPAA had the largest fold difference in portal plasma concentration relative to the HFD control (2.33 ± 0.35, mean fold change ±95% confidence interval, n = 9–10). In addition, 4-HPAA correlated negatively with %fat mass (Fig. 1M; R2 = 0.1001, P = 0.0498) and plasma insulin levels (Fig. 1N; R2= 0.206, P = 0.004). Finally, plasma levels of 4-HPAA have been previously reported to correlate negatively with indices of obesity in a cohort of nondiabetic obese human subjects (19). Hence, we selected 4-HPAA as our candidate molecule to test the hypothesis that a single microbial flavonoid catabolite would be sufficient to abrogate key parameters of HFD-induced metabolic disease.
A Single Microbial Flavonoid Catabolite Reverses Hepatic Steatosis.
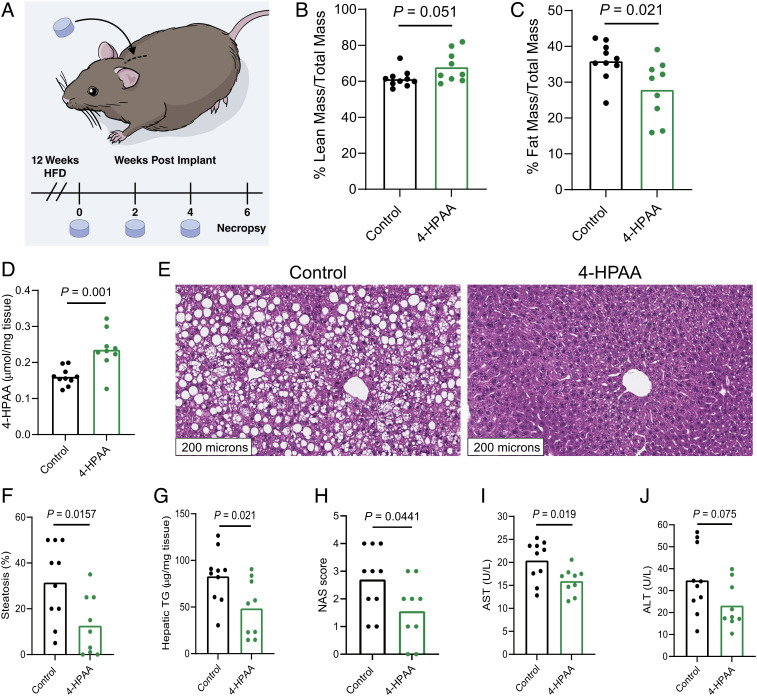
To test whether the metabolically beneficial effects of FC1 could at least in part be attributed to the microbial flavonoid catabolite 4-HPAA, we implanted subcutaneous slow-release pellets delivering 4-HPAA (350 μg/d) into mice prefed a HFD to establish obesity and compared them to obese control mice that received an implanted sham scaffold for 6 wk (Fig. 2A). This subcutaneous delivery method has been used previously to study different microbial metabolites at a single-metabolite resolution (6). Its main advantage is that the delivered molecule is independent of further modification by gut microbial metabolism, a limitation of providing 4-HPAA in the diet or drinking water (6). After 25 d, we surveyed global metabolism and energy substrate utilization using indirect calorimetry data to compare the 4-HPAA and the scaffold-only control mice at isothermal 30°C, room temperature 23°C, and under cold challenge at 4°C. We observed that 4-HPAA-treated mice were more prone to utilize carbohydrates as an energy source as measured by the respiratory exchange ratio (RER) during the light cycle under cold exposure (SI Appendix, Fig. S4A). At the time of sacrifice, the mRNA expression of uncoupling protein 1 (Ucp1), a key regulator of nonshivering thermogenesis that mediates oxygen consumption in cold conditions (20), was increased in brown adipose tissue of 4-HPAA-treated mice (SI Appendix, Fig. S4B). This suggests that 4-HPAA may modulate metabolic flexibility during cold exposure.
Fig. 2.
4-HPAA is sufficient to reverse HFD-induced hepatic steatosis and liver injury. (A) 4-wk-old male C57BL/6 mice were fed a HFD for 12 wk to induce obesity. After the induction of diet-induced obesity, mice were randomly assigned to receive a subcutaneously implanted control scaffold pellet, or a pellet releasing 350 µg 4-HPAA per day for 2 wk. New pellets were implanted every 2 wk for a total of 6 wk. Lean mass (B) and fat mass (C) were measured by Echo MRI, n = 9–10 per group. (D) 4-HPAA accumulates in the liver as measured by LC-MS/MS, n = 9–10 per group. (E–G) After a total of 18 wk of HFD feeding, H&E-stained sections of the liver revealed profound hepatic steatosis in scaffold control mice, while 4-HPAA-treated mice had marked reversal of steatosis and triglyceride deposition, n = 6–10 per group. (H) NAFLD activity score (NAS) incorporating histologic assessments of steatosis, hepatocellular ballooning, and inflammation. (I and J) Quantification of liver injury biomarkers in peripheral plasma, n = 9–10 per group. Statistical analysis was performed using unpaired two-tailed Student’s t test. Individual points represent individual mice, and bars represent group means. (Fig. 2A reprinted with permission, Cleveland Clinic Foundation ©2022. All Rights Reserved.)
After 6 wk of continuous 4-HPAA exposure, modest changes in gross anthropometrics (Fig. 2 B and C) were overshadowed by striking liver-specific effects of 4-HPAA. Consistent with our previous findings (21), 4-HPAA accumulates in the liver as it undergoes rapid first-pass hepatic metabolism (Fig. 2D). Remarkably, after just 6 wk, subcutaneous 4-HPAA administration led to a marked reversal of hepatic steatosis when compared to control mice (Fig. 2 E–G). We observed moderate differences in the NAS which incorporates histological assessments of steatosis, hepatocellular ballooning, and lobular inflammation (Fig. 2H), yet 4-HPAA-treated mice had lower plasma concentrations of the liver injury marker aspartate aminotransferase but not alanine aminotransferase (Fig. 2 I and J).
Next, we measured the mRNA expression of several genes implicated in lipid metabolism and inflammation in the liver (SI Appendix, Fig. S4C) and observed a reduction in the fatty acid transporter Cd36, the fatty acid desaturase Scd1, and the pro-inflammatory cytokine Tnfa in 4-HPAA-treated mice with a concomitant increase in Pgc1a, a master regulator of mitochondrial biogenesis, and Lcn13, a regulator of glucose and lipid metabolism, respectively (22–24). In addition, we measured the mRNA expression of lipid metabolism and inflammation-related genes in the gonadal white adipose tissue as it plays a central role in maintaining metabolic homeostasis (SI Appendix, Fig. S4D). 4-HPAA-treated mice also had increased mRNA levels of lipoprotein lipase (Lpl), the adipocyte-specific isoform of peroxisome proliferator-activated receptor gamma (Pparg2, a master regulator of glucose homeostasis and lipid metabolism), as well as a reduction of the expression of macrophage-associated glycoprotein Cd68. Taken together, our data suggest that 4-HPAA confers a distinct metabolic benefit that is regulated in part by tissue-specific transcriptional reprogramming.
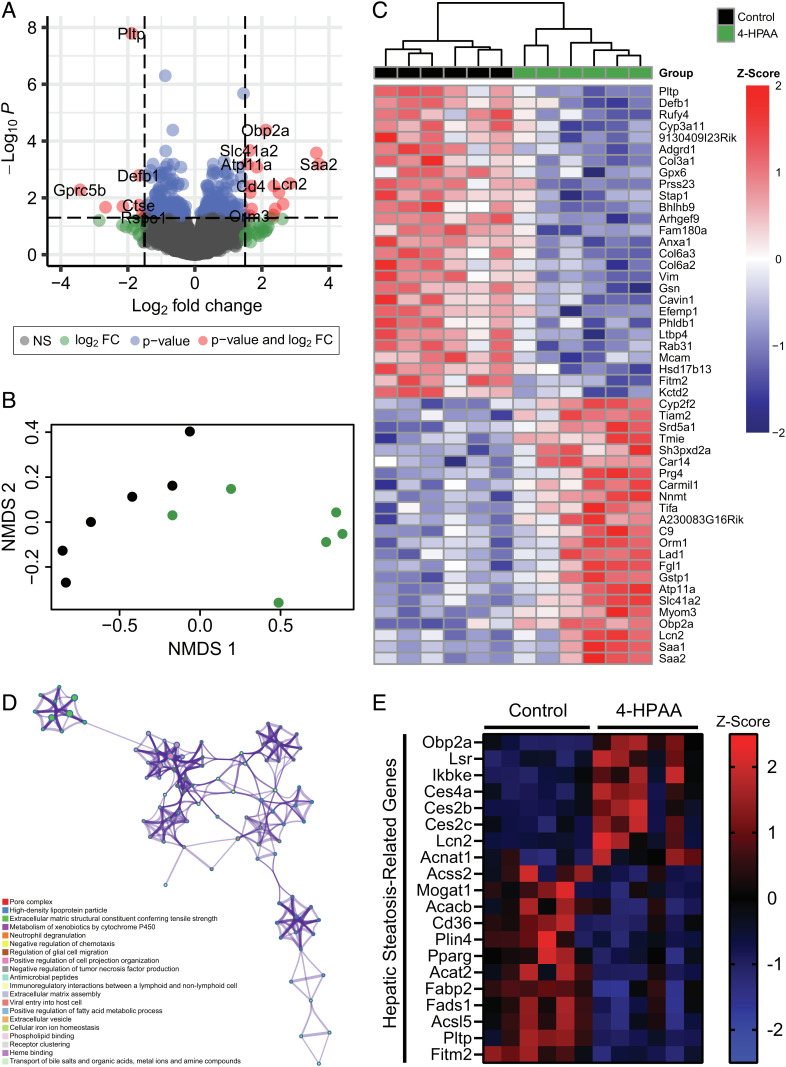
To obtain a more comprehensive mechanistic overview that expands upon these targeted transcriptional data, we performed unbiased bulk RNA sequencing on the livers of mice treated with either control or 4-HPAA subcutaneous pellets (Fig. 3). In total, 304 genes were significantly differentially expressed between control and 4-HPAA-treated mice (Fig. 3A). Moreover, the control and 4-HPAA treatments were distinguishable in both nonmetric multidimensional scaling (NMDS) analysis and unsupervised hierarchical clustering analysis (Fig. 3 B and C). Ontology enrichment analysis revealed the potential interaction of various molecular pathways including the negative regulation of tumor necrosis factor production and positive regulation of fatty acid metabolic processes (Fig. 3D). Furthermore, when examining differentially expressed genes implicated in hepatic steatosis, there is a clear pattern suggesting that 4-HPAA treatment may be implicated in the improvement of hepatic steatosis (Fig. 3E). Together, these data suggest that 4-HPAA treatment may reduce hepatic steatosis at least in part through transcriptional remodeling within the liver.
Fig. 3.
4-HPAA influences the hepatic transcriptome. (A) Volcano plot of RNA-Seq hepatic transcriptome from control and 4-HPAA-treated mice after 6 wk. (B) NMDS of hepatic RNA-Seq data representing the distribution of samples in space based on the top 500 differentially expressed genes by adjusted P value and log2 fold change between control (black) and 4-HPAA-treated (green) mice. (C) Heatmap of differentially expressed genes with hierarchical clustering arranged by adjusted P value with Z-score values scaled by row. (D) Metascape-enriched ontology clusters based on the top 100 most significant differentially expressed genes between control and 4-HPAA-treated mice. Each ontology term is represented by a circle node where the size of the node is proportional to the number of genes that fall into that ontology assignment. (E) Heatmap representing the Z-score of genes implicated in hepatic lipid metabolism and subsequent related disorders in control and 4-HPAA-treated mice. A sample size of n = 6 per group was used for all RNA-Seq-related analyses.
4-HPAA Activates Hepatic AMPKα Signaling.
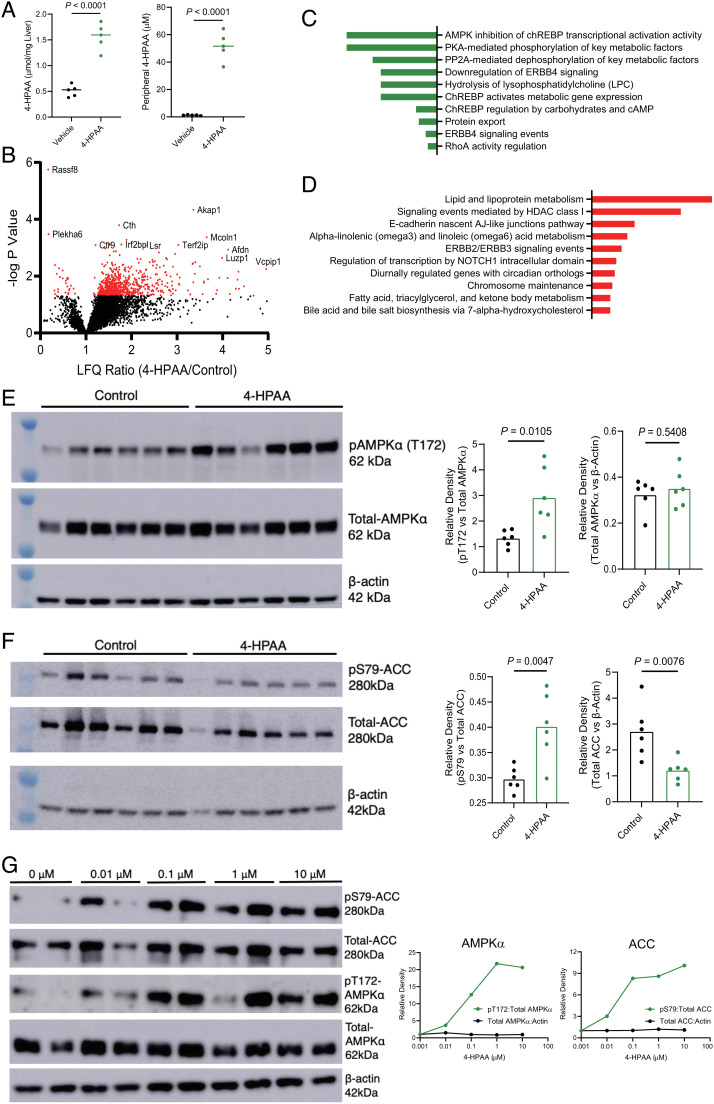
We next set out to uncover the signaling cascade(s) that could be linked to 4-HPAA-induced hepatic transcriptional changes. Since the portal vein drains gut microbe-derived metabolites from the mesentery into the liver (21), we utilized a portal vein injection model to recapitulate this physiological route and followed this by an unbiased phosphoproteomic analysis. Here, we injected either a saline vehicle control, 75 μg, or 150 μg 4-HPAA directly into the portal vein of anesthetized mice and harvested hepatic tissue 10 min later for the enrichment of phosphorylated peptides and subsequent LC-MS analysis. At end point, injection of the 150 µg dose had resulted in significantly increased concentrations of 4-HPAA, achieving 50 µM in peripheral plasma and 1.5 µmol/mg tissue in the liver (Fig. 4A). The subsequent unbiased proteomics approach revealed numerous phosphopeptides that were differentially abundant in 4-HPAA-treated and saline vehicle control groups (Fig. 4B). Pathway analysis of all parent genes mapping to phosphopeptides down-regulated in 4-HPAA-treated mice implicated the AMP-activated protein kinase (AMPK) pathway and AMPK-related metabolic effectors (Fig. 4 C and D). Moreover, up-regulated phosphopeptides were implicated in key metabolic pathways such as lipid and lipoprotein metabolism, which fall within the domain of AMPK-related molecular functions.
Fig. 4.
Untargeted phosphoproteomic analysis reveals that 4-HPAA activates the AMPK pathway and downstream effectors to reduce de novo hepatic lipogenesis. (A) 4-HPAA concentration in the liver and peripheral plasma, 10 min after animals received an intraportal injection of 150 µg 4-HPAA or saline control. (B) Volcano plot of label-free quantitation (LFQ) ratio vs. −log P value for 4-HPAA/control hepatic phosphoproteomic analysis following portal vein injection of 4-HPAA or a saline vehicle control. (C) Pathway enrichment analysis of all hepatic phosphopeptides down-regulated following 4-HPAA/control portal vein injections. (D) Pathway enrichment analysis of top 125 up-regulated hepatic phosphopeptides following portal vein injection of 4-HPAA or a saline vehicle control, n = 5 per group for A–D. (E) Western blot analysis of pAMPKα (T172), total AMPKα, and β-actin with densitometric quantification, n = 6 per group. (F) Western blot analysis of pACC (S79), total ACC, and β-actin with densitometric quantification, n = 6 per group. (G) Primary murine hepatocytes were treated with either DMSO vehicle control (0 μM) or 0.01, 0.1, 1, and 10 μM 4-HPAA for 30 min at which point the protein expression of pACC (S79), total ACC, pAMPKα (T172), total AMPKα, and β-actin was measured via western blot analysis with densitometric quantification, with dots representing the mean of two biological replicates. Statistical analysis for panels A and B was performed using unpaired two-tailed Student’s t test. Individual points represent individual mice, and bars represent group means.
With the data from this unbiased approach in hand, we next sought to determine whether AMPK signaling is up-regulated in the livers of mice treated with subcutaneous 4-HPAA. Together with insight provided by pathway analysis of the phosphoproteomic data, we hypothesized that 4-HPAA treatment would lead to the activation of AMPK based on four additional premises: i) literature reports on the activation of AMPK by the structurally distinct monophenolic acids such as gallic acid, vanillic acid, and caffeic acid (25–27); ii) the observation that exogenously administered 4-HPAA accumulates in the liver; iii) our observed transcriptional data that are consistent with AMPK activation (Fig. 3)(28); and iv) physiologic hormone-driven activation of AMPK by adiponectin and leptin is potently antisteatotic(29–31). We next tested our hypothesis experimentally by immunoblot analysis of the liver tissues from our subcutaneous delivery experiment. This analysis revealed increased phosphorylation of AMPKα (Fig. 4E) and its downstream effector acetyl-CoA carboxylase (ACC, a central mediator of de novo fatty acid synthesis; Fig. 4E) in 4-HPAA-treated mice when compared to the scaffold control group. This observation was specific to the α subunit of AMPK, as subcutaneous 4-HPAA administration did not lead to the phosphorylation of the AMPKα subunit (SI Appendix, Fig. S5). Since subcutaneous 4-HPAA delivery could bypass first-pass metabolism, we confirmed dose-dependent AMPK and ACC activation in livers from animals that received direct administration of 4-HPAA via the portal vein as an alternative delivery method (SI Appendix, Fig. S6) (21).
To determine whether 4-HPAA can activate AMPKα in a cell autonomous manner, we treated primary mouse hepatocytes with physiologically relevant concentrations of 4-HPAA (0.01–10 μM) for 30 min. Mirroring our in vivo observations, 4-HPAA treatment of primary mouse hepatocytes led to the phosphorylation of AMPKα and ACC in a dose-dependent manner in vitro (Fig. 4G). Surprisingly, the peak phosphorylation of AMPKα was observed after treatment with 1 μM 4-HPAA, the same concentration as observed in the portal blood of our FC1 mice (Figs. 1 L and 4 G). These results indicate that 4-HPAA acts locally in the liver—the primary site of accumulation—to activate AMPKα and downstream signaling events that in turn activates fatty acid oxidation and blunts de novo lipogenesis.
Human Flavonoid Catabolism is Rare, Underscoring its Therapeutic Potential.
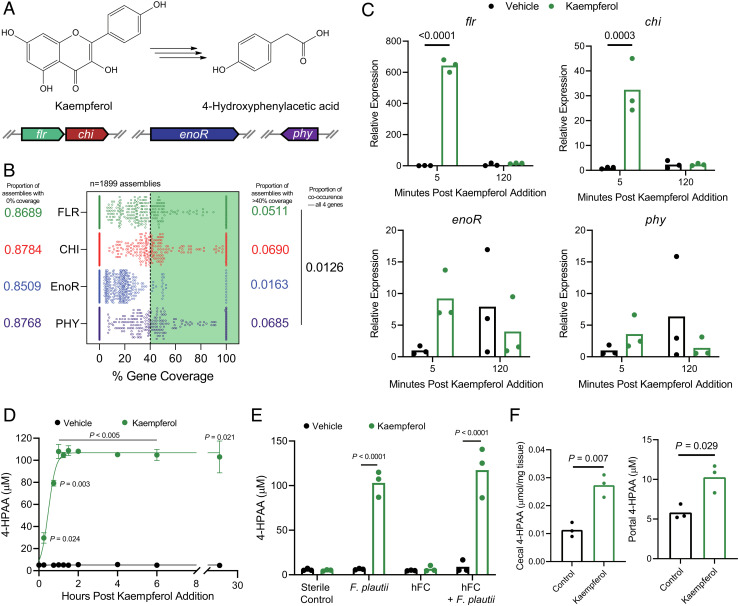
While our FC mice harbored flavonoid-catabolizing gut microbiota members capable of producing 4-HPAA, clinical studies have shown that flavonoid catabolism is less prominent among human gut microbiota and displays marked interindividual variation (32, 33). Several human gut commensals capable of catabolizing dietary flavonols into monophenolic acids have been identified, the most studied example being the eponymous Flavonifractor plautii (formerly Clostridium oribscindens) (SI Appendix, Fig. S7) (18, 34). Notably, F. plautii belongs to the Lachnospiraceae family, which accounted for one of the major changes in the microbiota of our FC mice (Fig. 1J and SI Appendix, Fig. S2 B and C). Recently, the complete set of genes required for flavonol catabolic activity was characterized. In total, four genes are required for the stepwise degradation of flavones and flavonols into monophenolic acids such as 4-HPAA. These four genes encode a FLR, CHI, EnoR, and PHY (14–17) (Fig. 5A). To predict the flavonol catabolic capacity of human metagenomes, we calculated the incidence of co-occurrence for this complete catabolic gene set in the metagenomic sequencing data (n = 1,899 assemblies) from two publicly available repositories (35, 36). Strikingly, although F. plautii was present in 28% of human microbiomes in these datasets (Datasets S1 and S2), the incidence of any one catabolic gene (>40% sequence similarity to the reference gene set) does not exceed seven percent. Moreover, the co-occurrence of all the four genes—required for the complete flavonol degradation pathway—is exceptionally rare, with an incidence of roughly one in a hundred (Fig. 5B). These data suggest that not all F. plautii strains can catabolize flavonols and support the recent report that F. plautii isolates are subject to interstrain competition in the presence of flavonoids (37). Overall, this highlights the rarity of a complete flavonol catabolic gene set in human microbiomes, although the possibility exists that other catabolic pathways remain undiscovered.
Fig. 5.
Flavonol- and flavone-catabolizing genes are rarely found in human microbiomes. (A) Graphical depiction of the complete bacterial gene set required to catabolize the flavonol kaempferol into 4-HPAA. (B) Computational screening of 1,899 assemblies of metagenomic data derived from human fecal samples representing over 1,300 human subjects for each of the bacterial genes required to catabolize flavonols/flavones. Assemblies with >40% coverage to the parent Flavonifractor plautii YL31 gene of interest were considered present. (C) Relative expression of F. plautii flavonol-catabolizing genes as measured by RT-qPCR following the addition of a vehicle control or kaempferol over time, n = 2–3 per group. (D) F. plautii rapidly converts kaempferol into 4-HPAA as measured by LC-MS/MS, n = 3 per group. (E) F. plautii converts kaempferol into 4-HPAA after 24 h of anaerobic incubation in vitro and retains its function when added to a nonconverting human fecal community (hFC). (F) Portal and cecal 4-HPAA concentrations in mice monocolonized with F. plautii after oral gavage with kaempferol or saline control. Individual points represent technical replicates in panels C, D, and E. Individual points in panel F represent biological replicates, with bars representing group means. Statistical analysis for panels C and E was performed using two-way ANOVA with Tukey’s correction for multiple comparisons (n = 3 per group). Statistical analysis for panel D was performed using a two-tailed unpaired t test with Holm–Šídák correction for multiple comparisons (n = 3 per group), and for panel F with a two-tailed Student’s t test (n = 3 per group).
When presented with a flavonol substrate, F. plautii rapidly induces the transcription of the genes implicated in the flavonol degradation pathway (Fig. 5C). The transcription of these genes is down-regulated upon the depletion of kaempferol, and subsequent production of 4-HPAA (Fig. 5 C and D). These data underscore the dynamic capacity of F. plautii to rapidly respond to the environmental availability of kaempferol.
Having deduced the predicted rarity of flavonol-catabolizing genes in human metagenomes together with their confirmed transcription in vitro, we next set out to test in vitro conversion of the flavonol kaempferol into 4-HPAA by stable human-derived polymicrobial communities. In addition, we tested whether community converter capacity could be induced by supplementation with an isolate of F. plautii with known flavonol-catabolizing capacity (Fig. 5E). In this experiment, we used a human-derived fecal microbiome that lacked catabolic activity toward kaempferol as a negative control and scaffold community into which we spiked in F. plautii. Following a 24-h growth period in anaerobic conditions at 37°C, we provided either a vehicle control or kaempferol substrate and incubated the communities for an additional 24 h. Using a targeted LC-MS/MS approach to measure 4-HPAA, the complete conversion of kaempferol substrate to 4-HPAA was observed in the F. plautii-containing community but not the scaffold negative control community. Together, these data support our hypothesis that a single microbe with catabolic activity toward flavonoids can be introduced into complex human fecal microbial communities to produce 4-HPAA in vitro. The utility of this approach is emphasized by the rarity of complete flavonoid catabolic gene sets in human metagenomes and represents the potential to confer flavonoid catabolism to humans.
Finally, we aimed to test the hypothesis that F. plautii could stably monocolonize the gastrointestinal tract of a germ-free mouse and that 4-HPAA levels could be subsequently modulated via the administration of a kaempferol substrate. Germ-free mice were colonized with F. plautii and after 2 wk, two doses of kaempferol were administered via oral gavage (1 d and 2 h prior to end point, respectively). Control animals received equivalent volumes of sterile saline. As expected, increased levels of 4-HPAA were observed in the portal plasma and cecum of kaempferol-treated mice when compared to the saline controls (Fig. 5F). These data suggest that 4-HPAA production can be modulated in vivo with single-microbe resolution.
Discussion
Dietary intervention is a common approach to combat cardiometabolic disease (CMD), often complementing traditional pharmaceutical therapies. The human gut microbiota extensively metabolizes dietary input, yielding a plethora of microbe-derived metabolites with poorly characterized influences on host physiology. Pertinent to this study, dietary flavonoids have been reported to abrogate diet-induced obesity in a microbe-dependent manner (11, 12). Moreover, several commensal gut bacteria are known to catabolize flavonoids into monophenolic acids (18). However, the contribution of these diet-derived gut microbial metabolites to CMD remains unclear. In an effort to understand the role of microbial flavonoid catabolites in obesity-related CMD, we used a multifaceted approach to study the interface of diet, the gut microbiota, and host physiology with single-molecule resolution.
Our study uncovers the functional importance of a single flavonoid-derived microbial catabolite, 4-HPAA, in abrogating HFD-induced hepatic steatosis. In addition, we establish the ability of 4-HPAA to activate AMPKα and modulate its downstream effectors. Remarkably, our investigation of the presence of F. plautii flavone/flavonol-catabolizing (4-HPAA producing) genes present in human metagenomic data revealed that although 28% of the 1,899 assemblies contained any one particular strain of F. plautii, just over one percent contained all the four genes required to produce 4-HPAA from a flavonoid precursor. Paired with the notable intraindividual variation of flavonoid catabolic activity in humans (32, 33), these data underscore the need to consider the microbial contribution to dietary intervention as a complementary strategy to combat CMD. Stratification by “responder” status based on the co-occurrence of the complete flavonoid-catabolizing gene set may inform, in part, the efficacy of intervention. While theoretically possible, the ability of a probiotic flavonoid-catabolizing F. plautii strain to stably colonize the gut remains to be determined and may prove challenging due to strain-level competition, particularly in the presence of a flavonoid substrate (37), and the frequency of nonsynonymous mutations over time (38). As an alternative, additional flavonoid-catabolizing microbiota members can be isolated and characterized or a next-generation synbiotic could be engineered by inserting the flavonoid-catabolizing gene set into a stably colonizing commensal host.
Our study has several limitations. First, studying human-relevant dietary substrates in a mouse model is reductive in that the human and mouse microbiota are not entirely comparable entities (39). Consequently, the development of humanized gnotobiotic mouse models has served as a welcomed advance in the study of diet–gut microbiota interactions (40). However, even this state-of-the-art approach has its own limitations as the colonization of the complete donor community is often not possible due to specific microbe–host dependencies (40). Second, since germ-free mice are resistant to diet-induced obesity (41), we were unable to test the antiobesogenic properties of FC1-HFD or 4-HPAA in the absence of the gut microbiota. Third, the possibility exists that undiscovered bacterial catabolic pathways may catabolize flavones/flavonols into 4-HPAA. To this point, we simply used the knowledge at our disposal, focusing on the known catabolic pathway. Similarly, aromatic amino acids (AAA) such as tyrosine may be metabolized by the gut microbiota to form 4-HPAA as an intermediate in p-cresol production (42–44). The contribution of AAA-derived 4-HPAA to the resolution of hepatic steatosis remains unclear and warrants further investigation. Last, although the subcutaneous administration provides a high level of resolution, this approach discounts the physiological circulation of gut microbial metabolites, first draining through the mesentery into the portal vein before delivery to the liver. Future studies can overcome this challenge by continuously infusing metabolites of interest directly into the portal vein, mirroring the natural circulation of microbial metabolites in vivo (21).
In conclusion, we used an array of in vitro, in vivo, and in silico analyses to reveal the gut microbial contribution of flavonoid catabolism in the context of overnutrition-induced metabolic disease and identified 4-HPAA, a single microbe-derived metabolite sufficient to abrogate obesity-driven hepatic steatosis. We see this as an example of how a single gut microbial metabolite stemming from the diet can profoundly impact host physiology and is a step toward combined diet–probiotic interventional therapies for CMDs.
Methods
Animal Studies.
Male C57BL/6 mice were purchased from Jackson Laboratory and housed under specific pathogen-free conditions within the Biological Resources Unit of Cleveland Clinic Lerner Research Institute. For the flavonoid composite feeding study, 6-wk-old mice were randomly assigned ad libitum access to a standard HFD (Diet D12492, Research Diets, Inc.), or the same diet supplemented with 1% w/w Flavonoid Composite 1 (Elderberry extract powder, Product Code 70120034, Artemis International), Flavonoid Composite 2 (Blackcurrant extract powder, Product Code 24140500, Artemis International), or Flavonoid Composite 3 (Aronia berry extract powder, Product Code 24198000, Artemis International) for 16 wk (Dataset S4). For the subcutaneous implant experiment, 4-wk-old mice were fed a standard HFD with 60 kcal% lard-derived fat (Diet D12492, Research Diets, Inc.) ad libitum for 12 wk to induce obesity-related metabolic syndrome. Next, scaffold control pellets or pellets releasing 350 μg 4-HPAA per day for 2 wk were subcutaneously implanted (Innovative Research of America). New pellets were implanted every 14 d for a total of 6 wk, while the animals remained on ad libitum HFD. For the portal vein injection with subsequent hepatic phosphoproteomic analysis, 9.5-wk-old male C57BL/6 mice were fasted for 12 h after which they were anesthetized with isoflurane at 3% for induction and 2% for maintenance. Next, a midline laparotomy with dissection was performed and the duodenum was moved to expose the portal vein. The GI tract was kept moist with occasional saline wetting. Using a 31-gauge insulin syringe, 100 μL of either 0.9% sterile saline vehicle or 150 ug 4-HPAA was injected over the course of 1 min. The needle remained in place and was supported by a gauze for 10 min to allow for hepatic distribution of 4-HPAA. After 10 min, a small volume of portal blood was aspirated in the syringe (~10 μL) and then flushed to ensure any remaining 4-HPAA is ejected from the syringe. A small aliquot of portal blood was collected using the same syringe used to inject the treatment. The needle was removed from the portal vein and pressure was immediately applied using a cotton swab to control bleeding followed by peripheral blood collection from the inferior vena cava. Finally, the left lobe of the liver was excised for flash freezing and subsequent downstream analysis. For the gnotobiotic mouse experiment, 24-wk-old male germ-free B6N mice from Charles River maintained in a sterile environment using the Allentown cage system were placed on a sterile phytochemically depleted diet (Research Diets OSD D11112201 – 1.5Vii). 1 wk later, all mice were given two oral gavages (200 μL) of F. plautii ATCC 49531 3 d apart. After a 2-wk colonization period, the mice were given oral gavages of either a sterile saline vehicle control or 500 μg kaempferol 24 h and 2 h prior to sacrifice. Tissues were harvested as described above. All experiments and procedures were approved by an Institutional Animal Care and Use Committee.
Phenotyping of Mouse Models.
Body weight and food consumption were measured at baseline and weekly thereafter for the duration of the flavonoid composite feeding experiment and subcutaneous pellet experiment. Lean and fat mass values were assessed at baseline, 25 d after the first pellet, and at the end of both mouse experiments using EchoMRI body composition analysis (EchoMRI, LLC). Plasma insulin levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit (Product No. 90080, Crystal Chem). Enzymatic analysis of hepatic triglycerides was performed as previously described (45). AST and ALT were measured as previously described (21). To assess pathologic changes in liver morphology, H&E-stained sections were assessed in a blinded manner by a board-certified pathologist for steatosis, inflammation, and ballooning.
Oxymax/CLAMS Metabolic Cage Studies.
For indirect calorimetry studies and activity monitoring, mice (n = 6 per group) were single-housed in the Oxymax/Comprehensive Animal Monitoring System (CLAMS) metabolic cage monitoring system (Columbus Instruments) 25 d after the initial pellet implant. The animals were allowed to acclimate for 24 h under murine isothermal conditions(46) (30°C) before 24 h of continuous data monitoring at 30°C, 24 h at 23°C, and 24 h at 4°C. Data analysis was performed as previously described(21) using CalR(47).
Cecal Microbiome Sequencing and Analysis.
DNA was extracted from mouse cecal contents and food samples using the QIAGEN PowerSoil Pro kit using the manufacturer’s protocol. 16S rRNA amplicon sequencing was done for the V3 and V4 region using Illumina iSeq 100 system from mouse cecal contents. Raw 16S amplicon sequences and metadata were demultiplexed using the split_libraries_fastq.py script implemented in QIIME1.9.1 (48). The demultiplexed fastq file was split into sample specific fastq files using the split_sequence_file_on_sample_ids.py script from QIIME1.9.1 (48). Individual fastq files without nonbiological nucleotides were processed using the Divisive Amplicon Denoising Algorithm (DADA) pipeline (49). The output of the DADA2 pipeline (feature table of amplicon sequence variants) was processed for alpha and beta diversity analysis using the phyloseq (50) and microbiomeSeq (http://www.github.com/umerijaz/microbiomeSeq) packages in R. Alpha diversity estimates were measured within group categories using the estimate_richness function of the phyloseq package (50). NMDS was performed using the Bray–Curtis dissimilarity matrix (51) between groups and visualized by using the ggplot2 package (52). We assessed the statistical significance (P < 0.05) throughout, and whenever necessary we adjusted P-values for multiple comparisons according to the Benjamini–Hochberg method to control false discovery rate (FDR) (53) while performing multiple testing on taxa abundance according to sample categories. We performed an ANOVA among sample categories while measuring the alpha diversity measures using the plot_anova_diversity function in the microbiomeSeq package (http://www.github.com/umerijaz/microbiomeSeq). Permutational multivariate analysis of variance (PERMANOVA) with 999 permutations was performed on all principal coordinates obtained during principal coordinate analysis with the ordination function of the microbiomeSeq package. Linear regression (parametric) and Wilcoxon (nonparametric) tests were performed on amplicon sequence variant abundances against metadata variable levels using their base functions in R (54).
Human Microbiome Metagenomic Analysis.
Longitudinal shotgun microbiome data from Poyet et al. (35) and Zou et al. (36) were downloaded using Entrezpy (44, 55). The shotgun microbiome data were quality trimmed using nesoni pipeline (https://github.com/Victorian-Bioinformatics-Consortium/nesoni). MetaPhlAn2 (METAgenomic PHyLogenetic ANalysis) was used for species-level metagenomic profiling of the quality-trimmed reads (56). Using megahit assembler, the shotgun microbiome data were assembled into contigs (57). The contigs were screened for FLR (EC 1.1.1.234), CHI (EC 5.5.1.6), EnoR (EC 1.3.1.3), and phloretin hydrolase (EC 3.7.1.4) using abricate (https://github.com/tseemann/abricate). The quality-trimmed reads were also mapped on the nucleotide sequences of FLR (EC 1.1.1.234), CHI (EC 5.5.1.6), EnoR (EC 1.3.1.3), and phloretin hydrolase (EC 3.7.1.4) using BBMap (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbmap-guide/). A count matrix was generated using featurecount function implemented in Subread package (58, 59). To independently validate these results, we mapped an equal number of reads from each sample on the protein coding genes of the F. plautii genome using BBMAP software. Next, we parsed the output (bam files) to estimate the number of reads mapped to each gene and the overall genome. Finally, we evaluated the prevalence of the marker genes (i.e., FMN reductase, acetyl-CoA hydrolase and pyridine nucleotide-disulfide oxidoreductase, phloretin hydrolase) at different relative abundance cutoffs (SI Appendix, Fig. S8). Using 0.03% as the cutoff, F. plautii was detected in the 134 samples (out of 465 samples). In addition, we analyzed the 652 F. plautii genomes currently present in the UHGG database. We created a local database and performed BLASTP analysis using marker genes (FMN reductase, acetyl-CoA hydrolase and pyridine nucleotide-disulfide oxidoreductase, phloretin hydrolase) as query. We then performed best blast hit (BBH) analysis on the blast output using biopython. BLAST parsing revealed that 197 F. plautii MAGs had all the four marker genes. However, it is important to note that the percent completion of these MAGs varies from 65 to 99%; therefore, it is not possible for us to determine whether some of these marker genes were actually absent in these MAGs or whether they are absent due to the lack of sequencing depth.
LC-MS/MS Analysis of Monophenolic Acids.
Plasma, liver, and bacterial supernatant were prepared for LC-MS/MS analysis and quantified as previously described (21). Briefly, stable-isotope dilution high-performance liquid chromatography tandem mass spectrometry (LC–MS/MS) was used for quantification of levels of 4-HPAA using an AB SCIEX Q-Trap 4,000 triple quadrupole mass spectrometer equipped with an electrospray ionization source operating in negative ion mode. Bacterial supernatants were prepared by first pelleting 24-h cultures by spinning at 15,000 × g for 10 min. Next, up to 500 μL was transferred to an Amicon Ultra-0.5 mL 3K filter (Millipore Catalog No. UFC500324) and centrifuged at 15,000 × g for 30 min. 20 μL of the filtered supernatant was prepared as previously described for analysis (21). The d6(methyl)- isotopolog of 4-HPAA was used as an internal standard (CDN Isotopes Product No. D-7842) at 10 μM for plasma and liver samples and 50 μM for bacterial supernatant. 4-HPAA was monitored using multiple reaction monitoring of precursor and characteristic product ions in negative mode as follows: m/z 151.2 → 107.0 for 4-HPAA; m/z 157.2 → 113.0 for d6-4-HPAA; m/z 120.7 → 77.0 for benzoic acid; m/z 136.8 → 92.9 for 3-hydroxybenzoic acid (3-HBA); m/z 136.8 → 92.9 for 4-hydroxybenzoic acid (4-HBA); m/z 148.9 → 104.9 for 3-PPA 3-phenylpropanoic acid (3-HPPA); and m/z 166.6 → 151.9 for 4-hydroxy-3-methoxybenzoic acid (4-H-3-MBA). For LC-MS/MS analysis of samples from gnotobiotic studies, a Shimadzu 8,050 triple quadrupole mass spectrometer equipped with an electrospray ionization source operating in negative ion mode was used. The column used was a Kinetex 2.6 µm C18 100 Å, LC Column 50 × 2.1 mm (Part No. 00B-4462-AN) with H2O + 5 mM ammonium acetate + 0.1% acetic acid as mobile phase A and methanol + 10% acetonitrile + 0.1% acetic acid as mobile phase B. The d6(methyl)- isotopolog of 4-HPAA was used as an internal standard (CDN Isotopes Product No. D-7842) at 10 µM. 4-HPAA was monitored using multiple reaction monitoring of precursor and characteristic product ions in negative mode as m/z 151.1 → 107.1 for 4-HPAA and m/z 157.0 → 113.1 for d6-4-HPAA.
Cell Culture and Immunoblotting.
Primary hepatocytes were isolated from two male C57BL/6 mice using methods described elsewhere (60) and plated in 6-well collagen I-coated plates at a density of 3 × 105 cells/well in Williams’ E medium containing 10% fetal bovine serum. Primary hepatocytes were serum-starved in plain William's E medium for 2 h prior to treatment with a DMSO vehicle control or 4-HPAA (TCI America Product No. H0290) for 30 min. Cell pellets or whole liver homogenates were prepared from primary hepatocytes or murine liver, respectively, in a modified RIPA buffer with protease and phosphatase inhibitors. Western blotting was performed as previously described (61). Images were captured using a GE Amhersham Imager 6000, and the accompanying software (version 1,1.1) was used for densitometric protein expression analysis. Primary antibodies (pS79-ACC, Cell Signaling, #11818; ACC, Cell Signaling, #3676; pT172-AMPKα, Cell Signaling, #2535; AMPKα, Cell Signaling, #5831; pS182-AMPKβ1, Cell Signaling, #4186; AMPKβ1, Cell Signaling, #4178) were prepared 1:1,000 in TBST buffer with 5% (w/v) BSA. Rabbit secondary (Cell Signaling, #7074) and β-actin (Cell Signaling, #12620) antibodies were prepared 1:5,000 in TBST with 5% (w/v) nonfat dried milk.
RNA Sequencing and Analysis.
RNA was isolated via the RNeasy Plus Mini Kit (Qiagen, Hong Kong) from mouse liver following the manufacturer’s protocol. RNA samples were checked for quality and quantity using the Bioanalyzer (Agilent, Santa Clara, CA, USA). RNA-Seq libraries were generated using the Illumina mRNA TruSEQ Directional library kit and sequenced using an Illumina NovaSeq 6000 (both according to the manufacturer’s instructions). RNA-Seq was performed by the University of Chicago Genomics Facility. Raw sequence files were deposited in NCBI’s Sequence Read Archive (accession number GSE188967). Paired-end 100 bp reads were controlled for quality with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) before trimming Illumina adapters from the 3′ ends using cutadapt (62). STAR was used to align the trimmed reads to the Mus musculus genome (NCBI GRCm38.93) (63). Read counts for each gene were calculated using HTseq (64) and loaded into R (http://www.R-project.org/) (R Development Core Team, 2015) where DESeq2 (65) (v.1.28.1, https://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to perform differential expression analysis on genes with at least one count per sample with alpha set to 0.05. P-values were adjusted using the Benjamini–Hochberg correction procedure (53), and genes with P < 0.05 were considered statistically significant. Heatmaps were generated of the top 50 differentially expressed transcripts using pheatmap (66) and RColorBrewer (67). NMDS analysis was performed using the plotMDS function of edgeR (68) using the top 500 differentially expressed genes as sorted by adjusted P value and log2 fold change. Pathway analysis on the top 100 differentially expressed genes was performed using Metascape (69). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (70) and are accessible through GEO Series accession number GSE188967.
Hepatic Phosphoproteomic Analysis.
Flash-frozen liver samples were inhomogenized in urea lysis buffer (20 mM HEPES, 9M urea, 1X HALT protease/phosphatase inhibitor, pH 8.0) using Tissue Tearor (BIOSPEC PRODUCTS, INC, model # 985370). Protein concentrations were measured using Pierce BCA kit, and a 1-mg aliquot of protein was used for in-solution digestion. The samples were reduced by dithiothreitol and alkylated by iodoacetamide. 25 µg of sequencing-grade trypsin was added to each sample and incubated at room temperature overnight. The digested samples were desalted using C18 cartridges. The desalted samples were lyophilized and spiked with phosphopeptides standard (Protea Biosciences Group, Inc. #PS-180-1) prior to phosphopeptide enrichment using Hi-Select Fe-NTA Phosphopeptide Enrichment Kit (ThermoScientific A32992), and the eluted peptides were dried immediately in a SpeedVac (Thermo Fisher Scientific, Waltham, MA, USA). Each sample was reconstituted in 30 µL 0.1% formic acid for LC-MS analysis. The LC-MS system was a Thermo Scientific Fusion Lumos mass spectrometry system (ThermoScientific, San Jose, CA, USA). The HPLC column was a Dionex 15 cm × 75 μm id Acclaim Pepmap C18, 2 μm, 100 Å reversed-phase capillary chromatography column (ThermoScientific, San Jose, CA, USA). A 5-µL aliquot of the extracts was injected, and the peptides were eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 µL/min and were introduced into the source of the mass spectrometer online. The microelectrospray ion source was operated at 1.9 kV. The digest was analyzed using the data-dependent multitask capability of the instrument acquiring full-scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans. The data were analyzed by using Proteome Discoverer 2.4 (Thermo Fisher Scientific, Waltham, MA, USA) to search all CID spectra collected in the experiment against the mouse SwissProtKB databases (17,017 entries) with an automatically generated decoy database (reversed sequences). The search was performed looking for fully tryptic peptides with a maximum of two missed cleavages. Oxidation of methionine and phosphorylation (S, T, Y) were set as dynamic modifications. The precursor mass tolerance for these searches was set to 10 ppm, and the fragment ion mass tolerance was set to 0.5 Da. A 1% FDR was applied to identify high-confidence peptides. The site of modification within each peptide was identified using the PhosphoRS mode in Proteome Discoverer 2.4. Phosphopeptide quantitation was performed by aligning chromatograms, normalizing to total peptide amount, and calculating the normalized LFQ intensities for each peptide. These LFQ values were used to compare phosphopeptides identified in the control and 4-HPAA samples, and three filters were used to identify differentially expressed phosphopeptides including: (1) LFQ values in at least three samples, (2) a twofold difference in abundance, and (3) a P-value < 0.05. Pathway analysis was performed using Enrichr (71).
Microbial Culturing.
Microbial culture experiments were performed in an anaerobic chamber (Coy Laboratory Products, Inc.) under the following conditions: 85% N2, 5% H2, 10% CO2, and <25 PPM O2. To examine the conversion of kaempferol substrate into 4-HPAA, a roughly 1-cm section of human fecal material prepared using the FAST technique (72) was suspended in 10 mL of a 50:50 (w:w) mixture of Wilkins–Chalgren (73) and Gifu Anaerobic Medium (74), vortexed for 2 min, and incubated at 37°C for 24 h. Next, the resulting microbial culture was mixed 1:1 (v:v) with fresh media containing either DMSO vehicle or kaempferol hydrate (TCI America Product No. K0018) at a final concentration of 100 μM and cultured at 37°C in a deep-well 96-well plate for 24 h. The reaction was stopped by freezing the 96-well plate at −80°C. For kinetic RT-qPCR studies, F. plautii was grown to mid-log phase at which point either a DMSO vehicle control or kaempferol was added. The supernatants and pellets were harvested at 5, 15, and 120 min postkaempferol addition for subsequent LC-MS/MS and RT-qPCR, respectively.
Quantitative Real-Time Polymerase Chain Reaction.
The bacterial pellet used for RNA extraction was obtained by centrifuging 10 mL of culture at 4,400 × g for 30 min. The supernatant was removed, and RNA was isolated using TRIzol reagent according to the manufacturer’s recommended protocol (Invitrogen Product No. 15596026). RNA was further treated to remove DNA contamination using the DNA-free kit using the provided protocol (Invitrogen Product No. AM1906). cDNA synthesis was performed using qScript cDNA Supermix (Quanta Bio Product No. 95048). RT-qPCR analysis was performed using the Applied Biosystems 7,500 Real-Time PCR System. Genes of interest were normalized to the F. plautii groEL gene, and relative mRNA expression levels were calculated based on the ΔΔ-CT method.
Whole-Genome Sequencing and Analysis.
DNA was extracted from Flavonifractor plautii ATCC 49531 using the QIAGEN DNeasy PowerSoil Pro kit. Illumina library preparation preceded 2 × 151 bp sequencing using the NextSeq 2000 platform. The reads were trimmed and quality controlled using Trimmomatic (75) and then assembled using SPAdes (76) and annotated via Prokka (77). Genome visualization, annotation, and phylogenetic comparisons were performed using PATRIC with RASTk (78, 79). The closest reference and representative genomes were identified by Mash/MinHash (80). PATRIC global protein families (PGFams)(81) were selected from these genomes to determine the phylogenetic placement of this genome. The protein sequences from these families were aligned with MUSCLE (82), and the nucleotides for each of those sequences were mapped to the protein alignment. The joint set of amino acid and nucleotide alignments were concatenated into a data matrix, and RaxML (83) was used to analyze this matrix, with fast bootstrapping being used to generate the support values in the tree (84).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
We would like to thank Artemis International for providing the flavonoid composites. This work was supported in part by the National Institutes of Health grants T32 GM088088 (L.J.O.), R01 AI153173 (J.C.), R01 HL120679 (J.M.B.), R01 DK130227 (J.M.B.), P01 HL147823 (A.M.H., Z.W., J.M.B., and S.L.H.), P50 AA024333 (L.E.N., D.S.A., and J.M.B.), and U01 AA026938 (L.E.N. and J.M.B.). J.C. is additionally supported by seed funding from the Cleveland Clinic Foundation, a Research Grant from the Prevent Cancer Foundation (PCF2019-JC), an American Cancer Society Institutional Research Grant (IRG-16-186-21), and a Jump Start Award (CA043703) from the Case Comprehensive Cancer Center. The phosphoproteomic analysis was performed using a Thermo Scientific Fusion Lumos LC-MS purchased with an NIH-shared instrument grant 1S10OD023436-01 (B.B.W.). We would also like to thank Brandon Stelter from the Cleveland Clinic Center for Medical Art and Photography for creating Fig. 2A.
Author contributions
L.J.O., J.M.B., and J.C. designed research; L.J.O., K.S., W.M., B.D., I.C., V.V., R.B., K.F., A.J.H., D.O., I.N., Z.W., D.S.A., B.B.W., N.S., and A.M.H. performed research; L.E.N. and S.L.H. contributed new reagents/analytic tools; L.J.O., W.M., I.C., I.N., Z.W., D.S.A., B.B.W., N.S., C.M., P.P.A., S.L.H., J.M.B., and J.C. analyzed data; and L.J.O., J.M.B., and J.C. wrote the paper.
Competing interests
The authors declare a competing interest. J.C. is a Scientific Advisor for Seed Health, Inc. Z.W. and S.L.H. report being named as co-inventor on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics. S.L.H. reports being a paid consultant for Procter & Gamble, and Zehna Therapeutics, having received research funds from Procter & Gamble, Roche Diagnostics and Zehna therapeutics, and being eligible to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland Heart Lab, a fully owned subsidiary of Quest Diagnostics, Zehna Therapeutics and Procter & Gamble. The other authors declare they have no competing interests.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
J. Mark Brown, Email: brownm5@ccf.org.
Jan Claesen, Email: claesej@ccf.org.
Data, Materials, and Software Availability
All data are available in the main text or the supplementary materials. Raw sequence files from the liver RNA-Seq were deposited in NCBI’s Sequence Read Archive (accession number GSE188967).
Supporting Information
References
- 1.Snetselaar L. G., Jesus J. M. D., DeSilva D. M., Stoody E. V., Dietary guidelines for Americans, 2020-2025, Nutr. Today 56, 287–295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manus M. B., Evolutionary mismatch. Evol. Med. Public Health 2018, 190–191, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pontzer H., Wood B. M., Raichlen D. A., Hunter-gatherers as models in public health. Obesity Rev. 19, 24–35 (2018). [DOI] [PubMed] [Google Scholar]
- 4.David L. A., et al. , Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z., et al. , Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh A., et al. , Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 175, 947–961.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Koeth R. A., et al. , Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang W. H. W., et al. , Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan J., et al. , The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Williamson G., Kay C. D., Crozier A., The bioavailability, transport, and bioactivity of dietary flavonoids: A review from a historical perspective. Compr. Rev. Food Sci. Food Saf. 17, 1054–1112 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Burke A. C., et al. , Intervention with citrus flavonoids reverses obesity, and improves metabolic syndrome and atherosclerosis in obese Ldlr-/- mice. J. Lipid Res. 59, 1714–1728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito D., et al. , Black currant anthocyanins attenuate weight gain and improve glucose metabolism in diet-induced obese mice with intact, but not disrupted, gut microbiome. J. Agric. Food Chem. 63, 6172–6180 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Winter J., Moore L. H., Dowell V. R., Bokkenheuser V. D., C-ring cleavage of flavonoids by human intestinal bacteria. Appl. Environ. Microbiol. 55, 6 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herles C., Braune A., Blaut M., First bacterial chalcone isomerase isolated from Eubacterium ramulus. Arch. Microbiol. 181, 428–434 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Braune A., Gütschow M., Blaut M., An NADH-dependent reductase from eubacterium ramulus catalyzes the stereospecific heteroring cleavage of flavanones and flavanonols. Appl. Environ. Microbiol. 85, e01233-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoefer L., Braune A., Blaut M., Cloning and expression of a phloretin hydrolase gene from eubacterium ramulus and characterization of the recombinant enzyme. Appl. Environ. Microbiol. 70, 6131–6137 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang G., et al. , Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria. Nat. Commun. 12, 790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborn L. J., Claesen J., Brown J. M., Microbial flavonoid metabolism: A cardiometabolic disease perspective. Annu. Rev. Nutr. 41, 433–454 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Hoyles L., et al. , Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ukropec J., Anunciado R. P., Ravussin Y., Hulver M. W., Kozak L. P., UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1-/- mice. J. Biol. Chem. 281, 31894–31908 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Orabi D., et al. , A surgical method for continuous intraportal infusion of gut microbial metabolites in mice. JCI Insight 6, e145607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho K. W., Zhou Y., Sheng L., Rui L., Lipocalin-13 regulates glucose metabolism by both insulin-dependent and insulin-independent mechanisms. Mol. Cell. Biol. 31, 450–457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng L., Cho K. W., Zhou Y., Shen H., Rui L., Lipocalin 13 protein protects against hepatic steatosis by both inhibiting lipogenesis and stimulating fatty acid β-oxidation. J. Biol. Chem. 286, 38128–38135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y., Rui L., Lipocalin 13 regulation of glucose and lipid metabolism in obesity. Vitam. Horm. 91, 369–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doan K. V., et al. , Gallic acid regulates body weight and glucose homeostasis through AMPK activation. Endocrinology 156, 157–168 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Jung Y., et al. , Vanillic acid attenuates obesity via activation of the AMPK pathway and thermogenic factors in vivo and in vitro. FASEB J. 32, 1388–1402 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Liao C.-C., Ou T.-T., Wu C.-H., Wang C.-J., Prevention of diet-induced hyperlipidemia and obesity by caffeic acid in C57BL/6 mice through regulation of hepatic lipogenesis gene expression. J. Agric. Food. Chem. 61, 11082–11088 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Mihaylova M. M., Shaw R. J., The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long Y. C., Zierath J. R., AMP-activated protein kinase signaling in metabolic regulation. J. Clin. Invest. 116, 1776–1783 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao P., Saltiel A. R., From overnutrition to liver injury: AMP-activated protein kinase in nonalcoholic fatty liver diseases. J. Biol. Chem. 295, 12279–12289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith B. K., et al. , Treatment of nonalcoholic fatty liver disease: Role of AMPK. Am. J. Physiol. Endocrinol. Metab. 311, E730–E740 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Rechner A. R., et al. , Colonic metabolism of dietary polyphenols: Influence of structure on microbial fermentation products. Free Radical Biol. Med. 36, 212–225 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Cassidy A., Minihane A.-M., The role of metabolism (and the microbiome) in defining the clinical efficacy of dietary flavonoids. Am. J. Clin. Nutr. 105, 10–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter J., Popoff M. R., Grimont P., Bokkenheuser V. D., Clostridium orbiscindens sp. nov., a human intestinal bacterium capable of cleaving the flavonoid C-ring. Int. J. Syst. Bacteriol. 41, 355–357 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Poyet M., et al. , A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 25, 1442–1452 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Zou Y., et al. , 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 37, 179–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Castaño G. P., Rey F. E., Caro-Quintero A., Acosta-González A., Gut-derived Flavonifractor species variants are differentially enriched during in vitro incubation with quercetin. PLoS One 15, e0227724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yilmaz B., et al. , Long-term evolution and short-term adaptation of microbiota strains and sub-strains in mice. Cell Host Microbe 29, 650–663.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Nguyen T. L. A., Vieira-Silva S., Liston A., Raes J., How informative is the mouse for human gut microbiota research? Dis Model Mech. 8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J. C., Im S.-H., Of men in mice: The development and application of a humanized gnotobiotic mouse model for microbiome therapeutics. Exp. Mol. Med. 52, 1383–1396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bäckhed F., Manchester J. K., Semenkovich C. F., Gordon J. I., Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U.S.A. 104, 979–984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Hou Y., Wang G., Zheng X., Hao H., Gut microbial metabolites of aromatic amino acids as signals in host-microbe interplay. Trends Endocrinol. Metab. 31, 818–834 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Lu J., et al. , Fructooligosaccharide decreases the production of uremic toxin precursor through modulating gut microbes mediated tyrosine metabolism pathway. Future Foods 4, 100069 (2021). [Google Scholar]
- 44.Dodd D., et al. , A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helsley R. N., et al. , Obesity-linked suppression of membrane-bound O-Acyltransferase 7 (MBOAT7) drives non-alcoholic fatty liver disease. eLife 8, e49882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon C. J., The mouse thermoregulatory system: Its impact on translating biomedical data to humans. Physiol. Behav. 179, 55–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mina A. I., et al. , CalR: A web-based analysis tool for indirect calorimetry experiments. Cell Metab. 28, 656–666.e1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso J. G., et al. , QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Callahan B. J., et al. , DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMurdie P. J., Holmes S., phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McMurdie P. J., Holmes S., Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10, e1003531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Springer Publishing Company, Incorporated, 2009). [Google Scholar]
- 53.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995). [Google Scholar]
- 54.Tiit E.-M., Nonparametric statistical methods. Myles Hollander and Douglas A. Wolfe, Wiley, Chichester, 1999. No. of pages: xiii+779. Price: £ 39.95. ISBN 0-471-19045-4. Statist. Med. 19, 1386–1388 (2000) [Google Scholar]
- 55.Buchmann J. P., Holmes E. C., Entrezpy: A python library to dynamically interact with the NCBI Entrez databases. Bioinformatics 35, 4511–4514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Truong D. T., et al. , MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W., MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Liao Y., Smyth G. K., Shi W., The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCullough R. L., et al. , Differential contribution of complement receptor C5aR in myeloid and non-myeloid cells in chronic ethanol-induced liver injury in mice. Mol. Immunol. 75, 122–132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown J. M., et al. , Conjugated linoleic acid induces human adipocyte delipidation: Autocrine/paracrine regulation of MEK/ERK signaling by adipocytokines. J. Biol. Chem. 279, 26735–26747 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 63.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S., Pyl P. T., Huber W., HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolde R., pheatmap Pretty Heatmaps. R Package Version 1.0.12. https://CRAN.R-project.org/package=pheatmap (2019). [Google Scholar]
- 67.Neuwirth E., RColorBrewer ColorBrewer Palettes. R Package Version 1.1-3. https://CRAN.R-project.org/package=RColorBrewer (2014). [Google Scholar]
- 68.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar R., Domrachev M., Lash A. E., Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic. Acids. Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen E. Y., et al. , Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romano K. A., et al. , Fecal aliquot straw technique (FAST) allows for easy and reproducible subsampling: Assessing interpersonal variation in trimethylamine-N-oxide (TMAO) accumulation. Microbiome 6, 91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilkins T. D., Chalgren S., Medium for use in antibiotic susceptibility testing of anaerobic bacteria. Antimicrob. Agents Chemother. 10, 926–928 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gotoh A., et al. , Use of gifu anaerobic medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Biosci. Biotechnol. Biochem. 81, 2009–2017 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A., Using SPAdes De Novo assembler. Curr. Prot. Bioinformatics. 70, e102 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Davis J. J., et al. , The PATRIC bioinformatics resource center: Expanding data and analysis capabilities. Nucleic. Acids. Res. 48, D606–D612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brettin T., et al. , RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ondov B. D., et al. , Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis J. J., et al. , PATtyFams: Protein families for the microbial genomes in the PATRIC database. Front. Microbiol. 7, 118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids. Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stamatakis A., Hoover P., Rougemont J., A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
All data are available in the main text or the supplementary materials. Raw sequence files from the liver RNA-Seq were deposited in NCBI’s Sequence Read Archive (accession number GSE188967).