Abstract
Objectives
To determine the role of MYL4 regulation of lysosomal function and its disturbance in fibrotic atrial cardiomyopathy.
Background
We have previously demonstrated that the atrial-specific essential light chain protein MYL4 is required for atrial contractile, electrical, and structural integrity. MYL4 mutation/dysfunction leads to atrial fibrosis, standstill, and dysrhythmia. However, the underlying pathogenic mechanisms remain unclear.
Methods and results
Rats subjected to knock-in of a pathogenic MYL4 mutant (p.E11K) developed fibrotic atrial cardiomyopathy. Proteome analysis and single-cell RNA sequencing indicate enrichment of autophagy pathways in mutant-MYL4 atrial dysfunction. Immunofluorescence and electron microscopy revealed undegraded autophagic vesicles accumulated in MYL4p.E11K rat atrium. Next, we identified that dysfunctional MYL4 protein impairs autophagy flux in vitro and in vivo. Cardiac lysosome positioning and mobility were regulated by MYL4 in cardiomyocytes, which affected lysosomal acidification and maturation of lysosomal cathepsins. We then examined the effects of MYL4 overexpression via adenoviral gene-transfer on atrial cardiomyopathy induced by MYL4 mutation: MYL4 protein overexpression attenuated atrial structural remodeling and autophagy dysfunction.
Conclusions
MYL4 regulates autophagic flux in atrial cardiomyocytes via lysosomal mobility. MYL4 overexpression attenuates MYL4 p.E11K induced fibrotic atrial cardiomyopathy, while correcting autophagy and lysosomal function. These results provide a molecular basis for MYL4-mutant induced fibrotic atrial cardiomyopathy and identify a potential biological-therapy approach for the treatment of atrial fibrosis.
Keywords: Myosin light chain 4, Atrial cardiomyopathy, Autophagy, Lysosome
List of highlights
-
•
Mutation of Myosin light chain 4 (MYL4) leads to atrial fibrosis, standstill, and dysrhythmia.
-
•
The loss-of-function MYL4 blocks autophagy flux in atrial cardiomyocytes, leading to accumulation of undegraded vesicles.
-
•
MYL4 regulates lysosome positioning and mobility in atrial cardiomyocytes.
-
•
MYL4 overexpression attenuates atrial fibrosis caused by MYL4 p.E11K, while correcting atrial autophagy function.
Abbreviations
- ATG10
Autophagy Related 10
- ATG8A
Autophagy-related 8a
- BAX
B-Cell Lymphoma 2–Associated X Protein
- BCL-2
B-Cell Lymphoma 2
- HO
Homozygote
- HSPA8
Heat Shock Protein Family A Member 8
- LAMP-2A
Lysosomal-Associated Membrane Protein 2
- LC3B
Microtubule Associated Protein 1 Light Chain 3 Beta
- MAP1B
Microtubule Associated Protein 1B
- MU
Mutation
- MYH6
Myosin Heavy Chain 6
- MYL3
Myosin Light Chain 3
- MYL4
Myosin Light Chain 4
- NRAMs
Neonatal Rat Atrial Cardiomyocytes
- NRCM
Neonatal Rat Cardiomyocytes
- NRVMs
Neonatal Rat Ventricular Cardiomyocytes
- p62
Sequestosome 1
- PBS
Phosphate Buffered Saline
- TUNEL
Terminal Deoxynucleotidyl Transferase Dutp Nick End Labeling
- WT
Wild Type
1. Background
Atrial cardiomyopathy is a common condition involving alterations in atrial structure, architecture, contraction, and/or electrophysiology [1]. Only a limited number of studies have investigated the mechanisms underlying atrial cardiomyopathy. Myl4 encodes atrial essential light chain protein, which is expressed almost exclusively in the atria of adults [2]. We previously identified a myosin light chain 4 (MYL4) loss-of-function mutation in an atrial standstill/arrhythmia family and established a MYL4 p.E11K knock-in rat model (MYL4p.E11K rats) that reproduces the clinical phenotype with atrial-specific cardiomyocyte apoptosis and fibrotic lesions [3].
Myosins are essential cytoskeletal motor proteins that transport intracellular particles along actin filaments to various cellular locations [4], which was involved in many physiologic processes. Myosin I regulate cellular autophagy function by trafficking lysosome in its maturation or fusion with autophagosomes [5,6]. All myosins contain several light chains. There are two types of light chains in the myosin chaperone, the essential light chain (ELC) and the regulatory light chain. MYL4 is an atrial-specific ELC that is necessary for ATPase activity of the myosin head and also for actin binding [7], a process that directly determines motor function.
Here, we aimed to evaluate the potential role of dysregulation of autophagy and lysosomal function in the atrial cardiomyopathy caused by MYL4 mutation. We also aimed to determine whether MYL4 gene-transfer can correct the autophagy/lysosomal dysfunction caused by MYL4 mutation, and whether this intervention can prevent the associated atrial fibrosis.
2. Methods
2.1. Animal models
All animal procedures were performed in accordance with NIH guidelines (Guide for the Care and Use of Laboratory Animals) and approved by the Animal Care and Use Committees of Shanghai Tenth People's Hospital (NO.SHDSYY-2020-3334). MYL4p.E11K rat strains were generated by CRISPR/Cas-mediated genome editing as reported previously [3].
2.2. Primary cultures of neonatal rat cardiomyocytes
Isolation and culture of primary neonatal rat cardiomyocytes (NRCMs) were prepared from heart of 1- to 3-days old WT and MYL4p.E11K rats. In brief, the separated atriums and ventricles were digested in 1% collagenase II. The supernatant was stopped by culture medium (20% FBS, 1% penicillin and streptomycin in complete high glucose DMEM medium) and filtered by 100-μm cell strainer. Then collected supernatant supernatants was pre-plated for 60 min to remove fibroblasts and endothelial cells. The residual supernatant with cardiomyocytes was centrifuged at 300×g for 8 min and replanted in collagen-coated dishes at 5% CO2 and 37 °C for 24 h. Cells were treated with 30 mM ammonium chloride (NH4Cl) (Sigma, USA) for 12 h to inhibit endosome-lysosome system acidification.
2.3. Plasmid transfection and adenovirus infection
The plasmid targeting MYL4 (hU6-MCS-CMV-GFP-SV40-Neomycin) and control plasmid (CON036) were purchased from GENECHEM Incorporation (China). NRCMs were seeded in 12-well plate at the density of 5 × 105 cells/ml and transfected with 1.6 μg DNA using lipofectamine 2000 (Thermo Fisher, USA) in OPTI-MEN (Thermo Fisher, USA) for 24 h. The adenovirus for rat mutated MYL4 (Ad-MU-MYL4), WT MYL4 (Ad-WT-MYL4) and empty vector (Ad-Vector) were purchased from GENECHEM Incorporation (China) and the construction details and infection of NRCMs were described previously [3]. For localized virus delivery, 10 μl 1E+10 PFU/ml Ad-WT-MYL4 or Ad-Vector was delivered by intramyocardial injection in MYL4p.E11K rats’ left atrium.
Adenoviruses encoding mRFP-GFP-LC3 was purchased from Hanbio Technology Corporation (China). Cardiomyocytes were infected with the virus at MOI 600 according to the manufacturer's instruction. 10 μl Ad-mRFP-GFP-LC3 was injected in 1-week MYL4 rat left ventricle for investigation of autophagy flux in vivo. After 3 weeks, survived animals were sacrificed by isoflurane.
2.4. Histological analysis
For histological analysis, tissues were sliced at 5 μm thickness sections. Modified Masson's Trichrome Stain Kit (Scy Teklaboratories, USA) was used to determine interstitial fibrosis, which can distinguish blue fibrosis and red muscle. All sections were scanned with digital microscopy (Olympus, Japan) equipped with a 20X objective lens for 15 to 20 images per sample. All images were acquired under same conditions. The automatic exposure and white balance were turned off. Images were analyzed with Image J software (version 1.48v; National Institutes of Health). The area of fibers in deconvoluted color images was measured with ‘Threshold’ tool. Blood vessels, perivascular tissue and epicardium were excluded when measuring fibrotic area for fibrosis quantification. For transmission electron microscopy observation, rat hearts were fixed in 2% glutaraldehyde and immersed in 2% osmium tetroxide and 1% aqueous uranyl acetate for 1 h. The sample was washed by a series of ethanol solution and incubated into propylene oxide and EMbed 812 mixtures for 1 h, followed by polymerization at 70 °C. After sliced at 80 nm sections, 5% uranyl acetate and Reynold's lead citrate were used to stain. Sections were observed by a 40–120 kV transmission electron microscope (Hitachi H600 Electron Microscope, Hitachi, Japan). At least 10 fields of each sample were analyzed.
2.5. Western blot analysis
Rat atrium tissue lysate and whole cells were prepared by 1x cell lysis (Cell Signaling Technologies, USA) and 1x protease inhibitors (Cat. 04693159001; Roche Molecular Biochemicals, USA). The crude extracts were centrifuged at 13000×g, 4 °C for 10 min and the supernatant was collected. The insoluble sediment was resuspended in 8 mM guanidine hydrochloride in complete lysis. The protein concentration was measured by bicinchoninic acid protein assay and equal amounts of protein samples were separated on 12% polyacrylamide gels, transferred onto PVDF membrane. Membranes were blocked in 5% non-fat milk or 3% BSA at room temperature and incubated with primary antibodies overnight at 4 °C. The primary antibodies targeted against Caspase-3 (Cell Signaling Technology, 9662s), BCL-2 (Abcam, ab32124), BAX (Abcam, ab32503), Vinculin (Santa Cruz Biotechnology, sc-73614), LAMP-2A (Abcam, ab125068), LC3B (Abcam, ab192890), p62 (Abcam, ab109012), MYL4 (Abnova, HOOOO4635-M01), Cathepsin B (Abcam, ab214428) or Cathepsin L (Santa Cruz Biotechnology, sc-390367) at a 1:1000 dilution. After 1-h HRP-conjugated secondary antibodies incubation, bends were visualized using chemiluminescence (ECL, TANON, China) and viewed under Amersham Imager 600 system (GE Healthcare, USA). The relative intensity of each band was normalized to Vinculin.
2.6. Quantitative real-time PCR
RNA was extracted by Trizol reagent (Invitrogen, USA) and purified RNA (100 ng) was reverse-transcribed using HiScript III RT SuperMix (Vazyme, China). The quantitative RT-PCR was performed on 2 μg cDNA product using FastStart Universal SYBR Green Master (Roche, USA) on a Roche Lightcycler. Information of primers is presented in Supplementary Table 1.
2.7. Immunofluorescence and confocal microscopy
Rat heart tissue section and cells were fixed in 4% (w/v) paraformaldehyde (PFA)/PBS for 15 min, followed by permeabilization with 0.2% Triton X‐100 for 10 min and block with 5% BSA for 30 min at room temperature. First antibodies targeted against LAMP2 (Santa Cruz Biotechnology, sc-20004), LC3B (Abcam, ab192890) or MYL4 (Invitrogen, PA5-84091) at a dilution in 1:100 were incubated at 4 °C overnight. Second antibodies were incubated for 1 h and cell nucleus was stained by DAPI for 15 min at room temperature. Fluorescence images were obtained using Nikon fluorescent microscope (Nikon, Japan).
2.8. Time-lapse live-cell imaging
NRCMs were incubated with Hoechst 33342 (Beyotime, China) and Lyso-Tracker Green (Beyotime, China) at the concentrations of 1:2000 and 1:20000 respectively for 30 min and washed by HSBB solution for 3 min. Then NRCMs were incubated with completed culture medium and imaged using Nikon confocal microscope. The moving distance of lysosome was measured by Image J software.
2.9. Live-cell lysosomal acidification
NRCM lysosome acidification was detected by fluorescence intensity using Lyso- Sensor Green (Yeasen, China) with 30 min incubation. This dye is concentrated in acid cell component, of which fluorescence intensity is higher in lower pH component. Each group was captured for 10 to 15 fields using Nikon confocal microscope. All the images were analyzed by Image J software to measure lysosome acidification. The average pH value in knock-down group was normalized to the average value of control group.
2.10. Single-cell RNA sequencing database analysis
The scRNA-seq data were downloaded from the Gene Expression Omnibus database (GEO; GSE128908). Seurat's (version 3.1.1 in R[8]) standard process was conducted to analyze the sequencing results. In brief, genes expressed in less than 10 cells and cells with fewer than 200 unique molecular identifiers (UMIs) or mitochondrial gene expression exceeding 60% were excluded. Clustering of filtered cell data set was identified by highly variable genes on their mean expression from 0.05 to 10 and dispersion between 1.5 and 20. The clusters were presented by t-distributed stochastic neighbor embedding (t-SNE) using dimensionality reduction. The Shared Nearest Neighbor (SNN) graph was constructed with 50 nearest neighbors and 20 dimensions of PCs as input. Clusters were identified using the above graph with resolution parameter of 0.6. Differential gene expression among clusters used MAST test in Seurat and gene expression over 25% cells was considered as significant difference. P value < 0.05 and |log2foldchange| > 1 was set as the threshold for significantly differential expression. GO enrichment and KEGG pathway enrichment analysis of DEGs were respectively performed using R based on the hypergeometric distribution.
2.11. 4D label-free quantitative proteomics
NRAMs (1X10 [6]) obtained from neonatal WT and MYL4 rat atrium were thawed in SDT (4%SDS,100 mM Tris-HCl, 1 mM DTT, pH7.6) buffer on ice. The amount of protein was quantified with the BCA Protein Assay Kit (Bio-Rad, USA). 20 μg of protein for each sample were mixed with 5X loading buffer respectively and boiled for 5 min. The proteins were separated on 12.5% SDS-PAGE gel (constant current 14 mA, 90 min). Protein bands were visualized by Coomassie Blue R-250 staining. The lysate was digested by trypsin according to filter-aided sample preparation (FASP) procedure and then desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 ml, Sigma). Digested peptides were concentrated by vacuum centrifugation and reconstituted in 40 μl of 0.1% (v/v) formic acid. The LC-MS/MS analysis was operated on the timsTOF Pro mass spectrometer (Bruker) in positive ion mode with 100–1700 m/z and 0.6 to 1.6 1/k0, coupled to Nanoelute (Bruker Daltonics) for 60 min. The peptides were loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm*2 cm, nanoViper C18) connected to the C18-reversed phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% Formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% Formic acid) at a flow rate of 300 nl/min controlled by IntelliFlow technology. The mass spectrometer was operated in positive ion mode. The mass spectrometer collected ion mobility MS spectra over a mass range of m/z 100–1700 and 1/k0 of 0.6–1.6, and then performed 10 cycles of PASEF MS/MS with a target intensity of 1.5 k and a threshold of 2500. Active exclusion was enabled with a release time of 0.4 min. MaxQuant 1.6.14.0 software is used for identification and quantitation analysis of MS raw data. The original data files were available on the iProX database (https://www.iprox.cn/page/home.html; IPX0005144000). The studied proteins were blasted against the online Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://geneontology.org/) to retrieve their KEGG orthology identifications and were subsequently mapped to pathways in KEGG. Enrichment analysis were performed by the Fisher’ exact test and p-value was adjusted by Benjamini- Hochberg correction for multiple testing. Both functional categories and pathways with p-values below 0.05 was considered statistically significant.
2.12. Statistical analysis
Data were analyzed using the package for social sciences (SPSS) for Windows 10. Numerical variables with a normal distribution are presented as the mean ± SEM. For intergroup comparisons of numerical variables, an independent sample t-test and Mann-Whitney U test were used. Bonferroni corrections were used to adjust the p-value to reduce type I errors in multiple comparisons. A two-tailed p < 0.05 was considered to be statistically significant.
3. Results
3.1. Autophagy-associated pathways are identified by proteome analysis of the atria of MYL4p.E11K rats
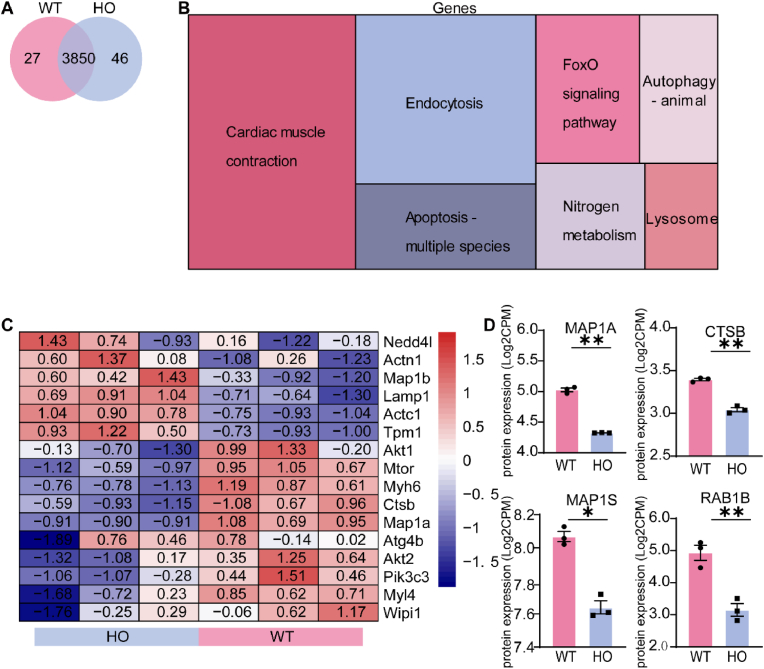
We first separated atrial cardiomyocytes and confirmed the presence of atrial cardiomyocyte markers (Supplemental Figs. 1A–D). To identify proteins enriched in the atria of MYL4p.E11K rats, we performed 4D label-free quantification proteome analysis on WT and MYL4p.E11K rat atrial cardiomyocytes N = 3). On average, 3896 proteins per MYL4 and 3877 proteins per WT sample were detected (Fig. 1A). A total of 203 proteins were upregulated and 158 proteins were downregulated in the MYL4p.E11K group. These differentially expressed proteins were significantly enriched in cardiac muscle contraction-, metabolism-, and protein degradation-related pathways (Fig. 1B). Interestingly, the MYL4 rat samples demonstrated downregulation of autophagy, a canonical protein degradation pathway (Fig. 1C). The levels of certain autophagy-related proteins MAP1A, CTSB, MAP1S and RAB1B were significantly decreased (Fig. 1D).
Fig. 1.
Autophagy-associated pathways are identified via proteome analysis of the atria of MYL4p.E11Krats. (A) Venn diagram showing the protein expression overlap in WT and MYL4 rat atrial cardiomyocytes. (B) Relative expression differences between WT and MYL4 groups as illustrated by a Treemap of condensed gene ontology (GO) terms. The sizes of boxes corresponded to the number of significant terms associated with the GO category. (C) Heat map analysis of expression pattern of cardiomyopathy and autophagy associated genes. (D) Differently expressed genes between WT and MYL4p.E11K rats in the autophagy pathway. CTSB = cathepsin B; DAPI = 40,6-diamidino-2-phenylindole; FoxO = forkhead box class O; HO = homozygote; RAB1B = member RAS oncogene family; MAP1A = microtubule associated protein 1A; MAP1S = microtubule associated protein 1S; WT = wild type. Two-tailed Student's t-test is used to compare two groups; Data are presented as mean ± SEM. *p < 0.05, **p < 0.001; n = 3.
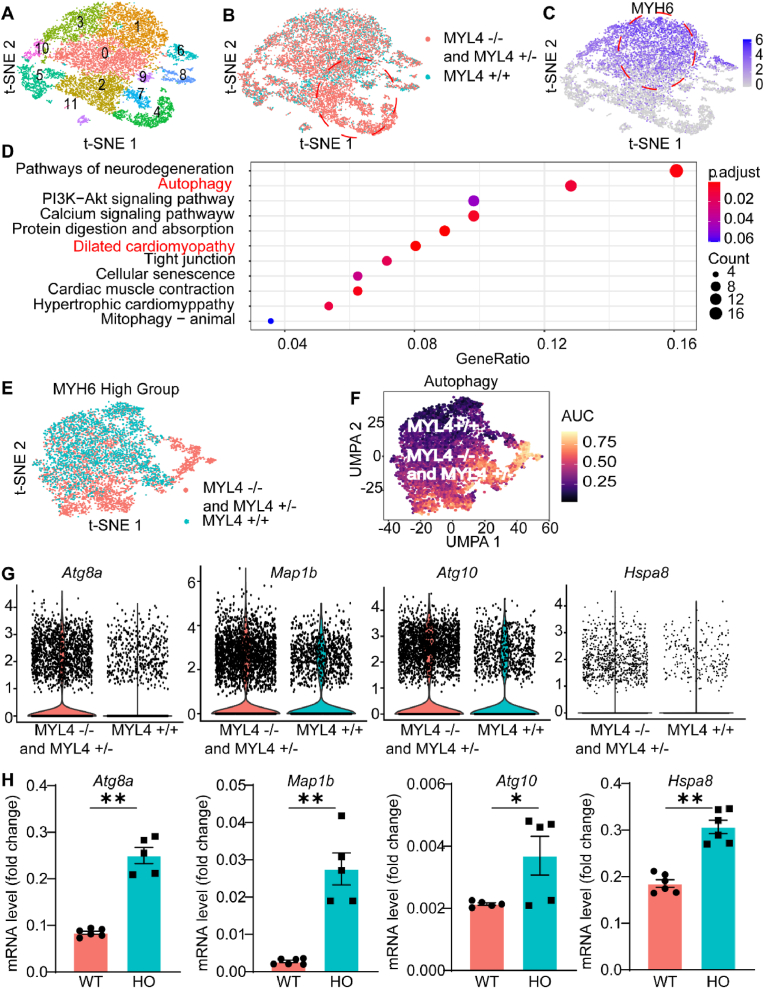
We re-analyzed the single-cell RNA sequencing (scRNA-seq) data from the Gene Expression Omnibus (GEO; GSE128908) that recorded scRNA-seq results of MYL4 ± and MYL4−/− hESC-atrial cells as well as MYL4 +/+ controls [9]. After filtration and quality control, 14,861 digested single cells were subjected to subsequent analysis. We then defined 11 cell clusters by cluster analysis and evaluated gene expression differences among MYH6+ hESC-atrial cells (Fig. 2A–C). The mutant cell lines were significantly enriched for the dilated cardiomyopathy pathway and the autophagy-related pathway according to scoring of individual cells for pathway activities [10] (Fig. 2D–F). Fig. 2G indicates that MYL4−/− cells highly expressed the autophagy-related genes Atg8a, Map1b, Atg10, and Hspa8. The altered gene expression was further confirmed by RT-qPCR (Fig. 2H) in samples collected from MYL4p.E11K rat atrium. Taken together, these findings suggest that MYL4 dysfunction strongly affects atrial cardiomyocytes autophagy related gene expression.
Fig. 2.
Upregulation of autophagy and dilated cardiomyopathy pathway in MYL4 mutants. (A, B) t-SNE plot of single-cell clusters in combined WT and mutant populations. The mutant unique cluster (#2,7,4) is circled in red. (C) t-SNE overlay of atrial lineage maker MYH6 expressed in cardiomyocyte fraction. (D) Dotplot heatmap showed GO biological process terms enriched in cluster marker genes. (E, F) t-SNE plot of MYH6 high group in combined WT and mutant populations and selected transcription factor network activity AUC score cluster distributions. (G) Violin plots of selected top differentially expressed genes between WT and mutant lines in autophagy pathway. (H) Quantitative RT-PCR analysis of top different genes expression in MYL4 p.E11K rat atrium tissue. Atg8a = autophagy-related protein 8a; Atg10 = autophagy related 10; Hspa8 = heat shock protein family A member 8; Map1b = microtubule associated protein 1B. Other abbreviations are the same as those in Fig. 1. Data are presented as mean ± SEM. Two-tailed Student's t-test is used to compare two groups; *p < 0.05, **p < 0.001; for MYL4+/+ group, n = 4, for MYL4 ± and MYL4−/−group, n = 8. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Autophagy flux is impaired in the atria of MYL4p.E11K rats
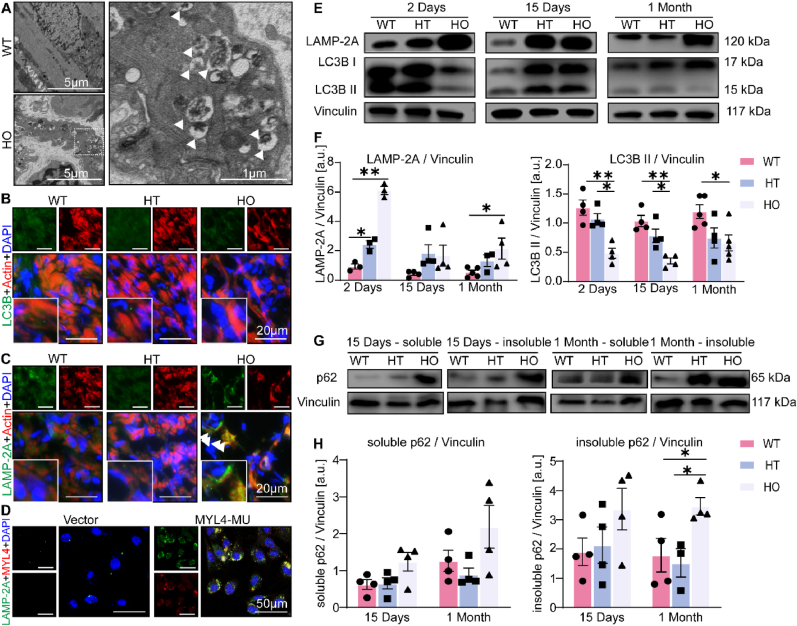
Autophagy and the ubiquitin-proteasome system are essential for controlling protein quality and quantity by degrading misfolded proteins [11]. We overexpressed WT and mutant MYL4 protein, as well as empty vector as negative control, in primary NRAMs and measured the ubiquitination levels by Western blot. The ubiquitination levels remained similar in all groups (Supplemental Fig. 2A), suggesting mutant MYL4 does not affect the level of cellular ubiquitination. Therefore, we next determined whether MYL4 mutation triggers autophagy in atrial cardiomyocytes. The rat atria were observed using a transmission electron microscope. Numerous multivesicular bodies were observed in the pericellular area, and these are highlighted by the white arrowheads in Fig. 3A. We then examined multivesicular bodies using immunofluorescence and observed that they were LAMP-2A-positive accumulations in the atrium of MYL4p.E11K rats, but not LC3B positive autophagosomes (Fig. 3B and C). We overexpressed the mutant MYL4 p.E11K protein in NRAMs and confirmed the presence of increased LAMP-2A positive puncta in vitro (Fig. 3D).
Fig. 3.
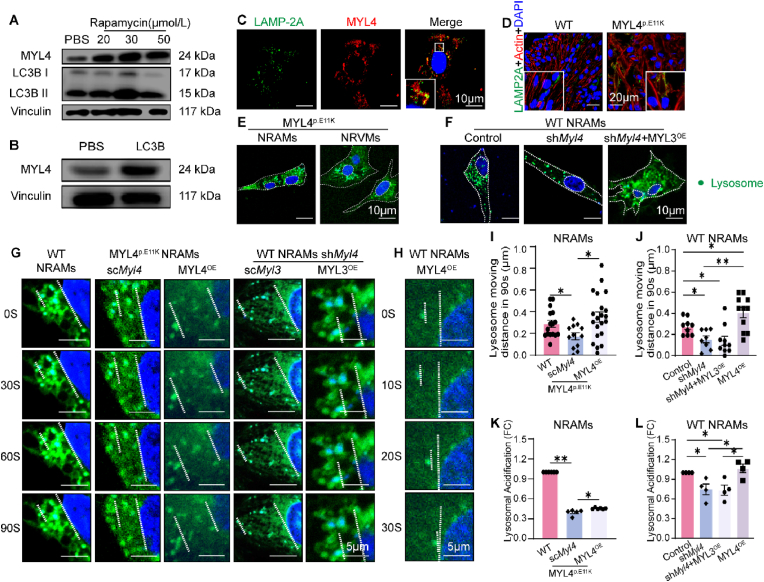
MYL4 mutation impairs autophagy function in the MYL4p.E11Krat atrium with accumulation of autophagy-lysosomal vesicles. (A) Representative transmission electron micrograph (15,000X) acquired from the atrium tissue of an 11-week MYL4p.E11K rat with normal ECG revealed the presence of double-membrane autophagic vesicles containing cellular materials (arrows). (B, C) Confocal microscopic appearance of autophagosomes (LC3B) and lysosomes (LAMP-2A) in atrial sections of 11-week-old heterozygous and homozygous MYL4p.E11K and WT rats. (D) Confocal immunofluorescence revealed the accumulated LAMP-2A was co-localized with the overexpressed MYL4 p.E11K protein in neonatal atrial cardiomyocytes. (E, F) Representative Western blot and statistical results of atrium tissues from heterozygous and homozygous MYL4p.E11K and WT rats. n = 3–5 (G, H) Representative protein bands and statistical analysis of soluble and insoluble p62 protein from atrium tissue of heterozygous and homozygous MYL4p.E11K and WT rats; n = 4. HT = heterozygote; LAMP-2A = lysosomal-associated membrane protein 2; LC3B = microtubule associated protein 1 light chain 3 beta; MU = mutation.; p62 = sequestosome 1. Other abbreviations are the same as those in Fig. 1. Data are presented as mean ± SEM. Two-tailed Student's t-test was used to compare the two groups; *p < 0.05, **p < 0.001.
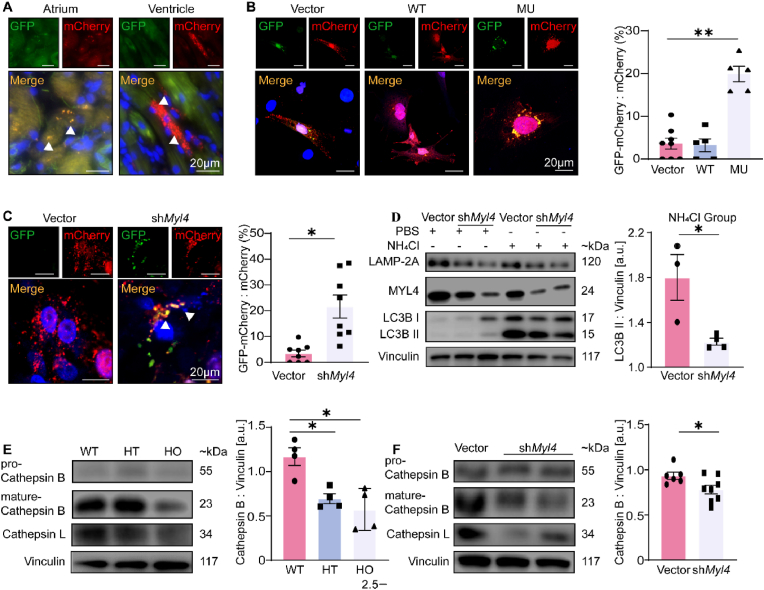
Next, we targeted the classical nodal points of autophagy, measuring the amounts of lysosome protein (LAMP-2A) and autophagosome formation (LC3B-II) in MYL4p.E11K rats of different ages by western blotting (Fig. 3E and F, Supplemental Fig. 2B). Compared with WT atrium, we observed a marked increase in LAMP-2A, an index of lysosome quantity and a decrease in autophagosome formation (LC3B II/Vinculin ratio). Furthermore, we observed that the MYL4 rat atrium exhibited significantly increased levels of both soluble and insoluble p62 proteins (Fig. 3G and H), suggesting autophagy was inhibited in MYL4 rat atrium. The level of LAMP-2A and LC3B in MYL4 rat ventricular tissue changed at an early age (2 days), but then remained steady (Supplemental Fig. 3). Then we further confirmed an atrial-specific suppression of autophagic flux in MYL4 rat by overexpressing mCherry-GFP-LC3B in situ (Fig. 4A). The mCherry-GFP-LC3B protein first exhibits yellow fluorescence (autophagosome) that becomes red fluorescence (autolysosome) when autophagosomes fuse with acidified lysosomes. In vitro, we also observed accumulation of mCherry-GFP-LC3B dots (yellow, autophagosome) in NRAMs exposed to mCherry-GFP-LC3B and MYL4 p.E11K overexpression adenovirus (Fig. 4B and 20.93 ± 19.90% vs 3.86 ± 1.503% from WT group, p = 0.001). To exclude the possibility of protein metastability caused by protein overexpression, we knocked down MYL4 expression in NRAMs. The autophagic flux of atrial myocytes was also significantly suppressed in MYL4 knock-down NRAMs (Fig. 4C), with reduced lysosome and autophagosome formation (Fig. 4D, Supplemental Fig. 2C). Cathepsin maturation was also disturbed: although the pro-cathepsins were present at the same level in both groups, mature cathepsin B was decreased in both MYL4 rat atrial tissue and MYL4 knockdown NRAMs (Fig. 4E and F). These results suggest that mutant MYL4 increased lysosome formation and that loss of functional MYL4 inhibited the autophagy process by disrupting lysosome function.
Fig. 4.
Loss of functional MYL4 inhibits autophagy flux and lysosome maturation in the atrium. (A) Atrial and ventricular tissues obtained from MYL4p.E11K rats were assessed using a fluorescence microscope. Representative images of mCherry-GFP-LC3B transfection are displayed. (B, C) Confocal microscopy and statistical analysis revealed inhibited autophagy in NRAMs with overexpressed mutant MYL4 proteins or knocked down MYL4 protein; n = 5–8. Representative images of mCherry-GFP-LC3B transfection are presented. (D) MYL4 knock-down reduced autophagosome (LC3B) and lysosome (LAMP-2A) formation in NRAMs; n = 3–4. (E, F) MYL4 dysfunction interfered with lysosome maturation in vivo and in vitro; n = 4–8. Abbreviations are the same as those in Fig. 1, Fig. 2. Data are presented as mean ± SEM. Two-tailed Student's t-test is used to compare two groups; *p < 0.05, **p < 0.001.
3.3. MYL4 depletion interferes with lysosome function by regulating lysosome positioning and motility in atrial cardiomyocytes
Lysosomes are highly dynamic cellular organelles, and interactions with the actin cytoskeleton contribute to their spatial distribution [12]. To better understand MYL4 affect lysosome function, we induced autophagy in neonatal atrial cardiomyocytes (NRAMs) using rapamycin. MYL4 protein was increased in a concentration-dependent manner in response to rapamycin treatment (Fig. 5A), and overexpression of LC3B protein in NRAMs also increased MYL4 protein levels (Fig. 5B). Lysosome marker LAMP-2A was also found to co-localize with MYL4 in atrial cardiomyocytes (Fig. 5C). In addition, we found a significantly different expression of myosins in MYL4p.E11K rat atrium based on quantitative proteomics data (Supplementary Table 2). These results imply that MYL4 might play a role in cardiomyocyte autophagy by regulating lysosome. Indeed, numerous lysosomes were predominantly concentrated along the plasma membrane in the MYL4p.E11K rat atrium tissue, while the WT rat atrium exhibited lysosomes mostly surrounding the cell nucleus (Fig. 5D). In isolated MYL4p.E11K or MYL4 knock-down WT atrial cardiomyocytes, lysosomes had the similar change of distribution from perinuclear to peripheral area, which MYL3, the ventricular ELC isoforms, merely had effect on restoration (Fig. 5E and F).
Fig. 5.
MYL4 depletion interfered with lysosome positioning and the motility of neonatal atrial cardiomyocytes. (A, B) Representative Western blot indicating that MYL4 was associated with autophagy function. (C) Co-localization of lysosomes (green) and MYL4 (red) in cardiomyocytes. (D, E) The distribution of lysosomes (green dots) was altered in MYL4 rat atrium and atrial cardiomyocytes. (F) Depletion of MYL4 caused dispersal of the perinuclear lysosome (green dots) population in atrial cardiomyocytes. (G–J) Lysosome (green dots) movement was regulated by MYL4 depletion or overexpression. Representative images used nucleus as location reference. (K, L) MYL4 regulated acidification of lysosome in atrial cardiomyocytes; n = 4–21. MYL4OE = MYL4 overexpression; PBS = phosphate buffered saline. NRAMs = neonatal rat atrial cardiomyocytes; NRVMs = neonatal rat ventricular cardiomyocytes. Other abbreviations are the same as those in Fig. 1. Data are presented as mean ± SEM. Two-tailed Student's t-test is used to compare two groups; *p < 0.05, **p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, we measured lysosomal mobility in live NRAMs using confocal microscopy (Fig. 5G). Compared to WT cells, the moving distance was significantly decreased in atrial cardiomyocytes from neonatal MYL4p.E11K rats (174.4 ± 31.2 nm vs. 288.6 ± 32.3 nm from WT NRAMs, p = 0.02, Fig. 5I), which can be reversed by overexpressing MYL4 protein (351.4 ± 46.5 nm vs. 174.4 ± 31.2 nm from MYL4 p.E11K scMyl4, p = 0.0118, Fig. 5I). Similarly, the WT atrial cardiomyocytes exhibited a significantly decreased moving distance after MYL4 knock-down (154.3 ± 32.5 nm vs. 274.4 ± 26.0 nm from NRAMs control, p = 0.0107, Fig. 5J), which MYL3 overexpression exerted a slight influence on. We also observed that MYL4 overexpression significantly promoted lysosome motility in WT atrial cardiomyocytes (406.8 ± 48.3 nm vs. 274.4 ± 26.0 nm from NRAMs control, p = 0.0364, Fig. 5J).
Parallelly, we showed lysosome position and mobility in neonatal ventricular cardiomyocytes (NRVMs) in Supplemental Fig. 4. MYL4 and MYL3 are atrial- and ventricular-specific essential light chains respectively [7]. It's notable that MYL4 mutation (Fig. 5E) or MYL4 knock-down merely disturbed lysosome transportation, but MYL4 overexpression can rescue MYL3 knock-down induced lysosome mis-location. In summary, although MYL4 can regulate lysosome distribution and movement in both atrial and ventricular cardiomyocytes, its dysfunction mainly affects atrial cardiomyocytes, which is consistent with the MYL4p.E11K rat phenotype.
3.4. MYL4 overexpression rescues lysosome acidification and attenuates fibrotic atrial cardiomyopathy while correcting autophagy function
Lysosome acidification is central to its degradation function. Juxtanuclear lysosomes are more acidic and exhibit higher hydrolase activity [13]. Accordingly, we evaluated lysosomal acidification in cardiomyocytes using a lysosome sensor consisting of a fluorescent dye accumulated in acid cell components, where its fluorescence becomes stronger at lower pH levels. In NRAMs of MYL4p.E11K rat and MYL4 knockdown NRAMs, lysosomal acidification was significantly reduced (0.40-fold change, p = 0.001; 0.75-fold change, p = 0.02, Fig. 5K and L, respectively). In contrast, MYL4 overexpression rescued lysosome maturation in MYL4 p.E11K atrial cardiomyocytes (0.455 vs. 0.40-fold change from MYL4 p.E11K scMyl4, p = 0.0091, Fig. 5K).
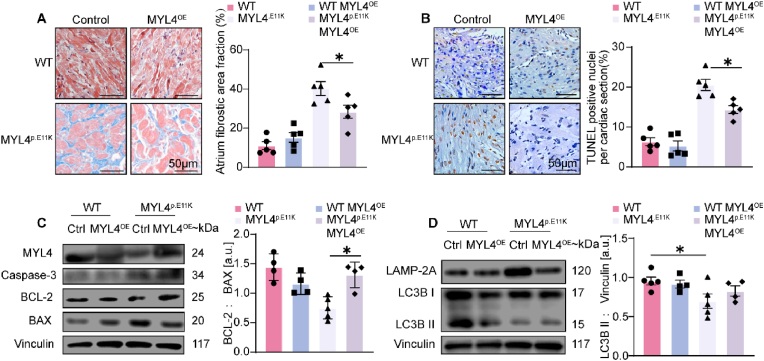
In a previous study, we demonstrated marked fibrosis and apoptosis in MYL4 p.E11K rat atrium from an early age (2 days after birth) [3]. To verify whether MYL4 is necessary and sufficient to rescue atrial autophagy function and atrial cardiomyopathy, we overexpressed WT MYL4 protein in 1-week old MYL4p.E11K rat's heart then sacrificed after 1 month. As shown in Fig. 6 A and B, there were significant reductions in fibrosis and apoptosis in MYL4p.E11K rats treated with MYL4 overexpression. The level of apoptotic protein caspase-3 and Bcl-2/Bax ratio were markedly decreased in MYL4p.E11K rat atrium overexpressing exogenous WT MYL4 (Fig. 6C). Moreover, lysosome accumulation as well as conversion of LC3BI to LC3BII (Fig. 6D, Supplemental Fig. 5) was restored after MYL4 overexpression. Taken together, these findings demonstrate a novel role for MYL4 as a critical regulator of autophagy and survival of cardiomyocytes in fibrotic atrial cardiomyopathy.
Fig. 6.
In suit MYL4 overexpression attenuates MYL4 p.E11K caused fibrotic atrial cardiomyopathy. (A, B) Representative images of Masson staining and TUNEL staining of atrial sections. Quantification of fibrotic area and apoptotic ratio were on right panel; n = 5. (C) Apoptotic protein Caspase-3 and BCL-2/BAX were measured by Western blot analysis; n = 4–5. (D) Western blot analysis for autophagy function in atrium; n = 4–5. Ctrl = control; BAX = B-cell lymphoma 2–associated X protein; BCL-2 = B-cell lymphoma 2. Other abbreviations are the same as those in Fig. 1. Data were presented as mean ± SEM. Two-tailed Student's t-test is used to compare two groups; *p < 0.05, **p < 0.001.
4. Discussion
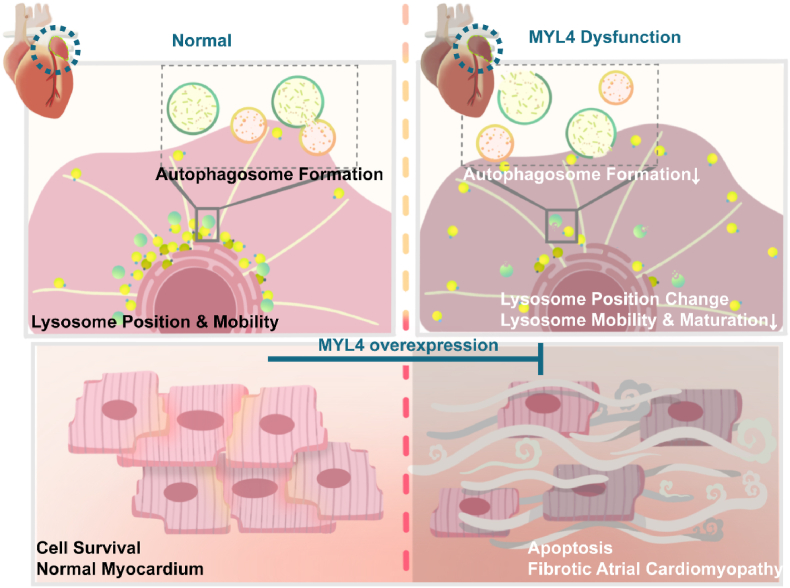
In the present study, we have made novel observations about the potential role of autophagy flux as a mediator of the consequences of MYL4 dysfunction, as well as of the potential therapeutic value of enhancing MYL4 expression. First, we demonstrated that the mutant MYL4 protein activates cell autophagy. Second, autophagy progression in cardiomyocytes is accompanied by MYL4 expression. Loss of MYL4 protein impairs lysosome maturity as well as the conversion of LC3B II. Third, we demonstrated that MYL4 dysfunction disturbed lysosomal position and movement, ultimately resulting in autophagy flux deficiency. Finally, we alleviated fibrotic atrial cardiomyopathy by MYL4 overexpression in vivo. The mechanisms underlying MYL4 related autophagy in the context of atrial cardiomyopathy are summarized in Fig. 7.
Fig. 7.
Proposed mechanisms underlying MYL4-mediated regulation of cellular autophagy function via lysosome positioning and mobility in atrial cardiomyocytes. Left panel, MYL4 promoted lysosome mobility, autophagy flux, and cell survival in the atrium. Right panel, loss of functional MYL4 caused spontaneous development of lysosome mis-distribution with atrial enlargement, increased apoptosis, and fibrotic lesions. MYL4 overexpression attenuated atrial apoptosis and fibrosis in MYL4p.E11K rats. ER = endoplasmic reticulum; MYL4 = myosin light chain 4.
The term cardiomyopathy was first introduced by Brigden in 1957 [14], where it was referred to as “isolated non-coronary myocardial disease”. Several studies have established a clear definition, classification, and treatment approach for ventricular cardiomyopathies. However, atrial cardiomyopathy was not systemically defined and analyzed until 2016. A working group with representation from the European Heart Rhythm Association (EHRA), the Heart Rhythm Society (HRS), the Asian Pacific Heart Rhythm Society (APHRS), and Sociedad Latino Americana de Estimulacion Cardiacay Electrofisiologia (SOLAECE) proposed the definition of atrial cardiomyopathy as “structural, architectural, contractile or electrophysiological changes” [15]. Myl4 encodes the atrial-specific essential light chain (ELC). MYL4 is expressed in the whole heart of human embryos [16], whereas it is substituted by MYL3 in the ventricle after birth and remains only in the atrium for the entire life [2,16,17]. Notably, MYL4 protein is re-expressed in hypertrophic or dilative ventricles [18] and results in an accelerated shortening velocity and isometric tension production [19]. These studies suggest that MYL4 plays an important role in cardiac development and heart function. Recently, several studies based on genome sequencing and bioinformation analysis have confirmed that mutation of MYL4 directly induces a series of atrial cardiomyopathies, including early onset AF [20,21], atrial standstill [3], fibrotic atrial cardiomyopathy [22] and metastable atrial state [9]. SNPs of MYL4 are also associated with atrial fibrillation one-set and outcomes after ablation [23]. Our group previously identified a rare missense variant of MYL4 (c.31G > A [p.E11K]) from an atrial standstill family and established a knock-in rat model [3]. MYL4p.E11K rats have extensive fibrosis deposition in the atrium and present decreased P-wave amplitude and prolonged PR intervals at 15 days after birth. Although atrial fibrosis can be caused by various facts such as genetics, aging, hypertension, diabetes mellitus, chronic heart failure and atrial fibrillation [[24], [25], [26], [27]]. These data suggested loss-of-functional MYL4, instead of aging and other facts, accounts for severe inherited atrial cardiomyopathy without affection on ventricle in MYL4p.E11K rats [3]. In this study, we revealed that in situ overexpression of WT Myl4 reversed MYL4 p.E11K caused fibrotic atrial cardiomyopathy, while correcting autophagy function of atrium. Based on our MYL4p.E11k rat model, we found MYL4 dysfunction impairs atrium autophagy function after early birth, which is consistent with the timing of the disease phenotype. In addition, the loss of functional MYL4 affected autophagy associated protein expression and lysosome, the autophagy central component, in atrium specifically. In our previous study, MYL4 p.E11K also showed specifical atrial cardiomyopathy in both clinical manifestation and animal model. Above all, our study provides a novel insight of idiopathic fibrotic atrial cardiomyopathy from autophagy aspect.
Autophagy is an evolutionarily conserved cellular mechanism that maintains intracellular homeostasis [28]. Through the degradation of aging and damaged or superfluous cell components, autophagy is essential for cells in both the physiological environment and in the context of stress/nutrient limitation to promote cell survival [29,30]. Approximately 40% of the cells in the heart are myocytes that exhibit limited capacity for proliferation and are long-lived into adulthood. Hence, appropriate autophagy function as an inherent repair process is important. Deletion of autophagy-relevant genes spontaneously develops a series of cardiomyopathy conditions. For example, cardiomyocyte-specific deletion of Atg5 leads to a dilated ventricle, cardiac hypertrophy, and impaired contractility [31,32], and LAMP2−/− mice present with vacuolar cardioskeletal myopathy [33,34]. Studies have also reported that after coronary artery bypass surgery, patients are more likely to experience postoperative atrial fibrillation if their cardiac autophagy is inefficient [35]. Moreover, pharmacological or genetic enhancement of autophagy represents a potential therapeutic strategy for the treatment of cardiovascular diseases [36] such as atherosclerosis [37] and cardiomyopathy [38]. However, current studies examining cardiovascular disease primarily focus on the upstream regulators of the autophagy pathway, and the role of autophagy components such as autophagosomes and lysosomes remains unclear. Lysosomes are the center of autophagy and are necessary for the completion of degradation [39]. Further studies demonstrate lysosome interfaces with other organelles, such as mitochondria [40,41], regulating metabolic process as well as ROS production [[42], [43], [44]]. Under physiological conditions, lysosomes are scattered throughout the cytoplasm and concentrated around the nucleus where they form a ‘perinuclear cloud’ in non-polarized cells [45]. Some lysosomes are relatively static; however, many lysosomes move between the cell center and periphery in a bidirectional manner. Impaired motility of lysosomes underlies a number of diseases, including neurodegeneration, cancer, and immune deficiency [12]. Intracellular lysosomes are transported by active directional motor proteins [46] such as kinesins, dyneins, and myosins. Our study unveils the link between lysosomal dysfunction and fibrotic atrial cardiomyopathy in a mutant myosin light chain model. We observed that the position and mobility of lysosomes in cardiomyocytes are both dependent upon the myosin essential light chain. MYL4 function is strongly associated with lysosomal distribution, mobility, acidification, and hydrolase maturation in atrial cardiomyocytes. However, the loss-of-functional MYL4 only has interference on lysosome in atrial cardiomyocytes. Despite the potential limitation of the MYL4 knock-down efficiency, we also didn't find change of lysosome function in isolated ventricular cardiomyocytes from global MYL4p.E11K rats. This explained why gene mutation of MYL4 leads to atrial-specific cardiomyopathy and provides a novel understanding of lysosomal activity in the context of fibrotic atrial cardiomyopathy.
Although our study has revealed the important roles of MYL4 and cellular autophagy in atrial cardiomyopathy, some aspects deserve further study. First, the role of MYL4 in lysosomal trafficking remains unclear. Lysosomes are highly dynamic and move by motor proteins along cytoskeletal components [47]. There are approximately 31 active myosins, and of these, the non-cortical myosin plays a critical role in lysosomal trafficking. Unfortunately, we failed to identify the myosins that were affected by MYL4 dysfunction. Second, in most murine experiments, we only used male rats to avoid individual differences induced by gender. Indeed, Wang et al. found a higher MYL4 proteins expression in cardiac tissue obtained from male subjects. Thus, further studies should replicate our results in female rats for assessment of its reproducibility. Finally, although MYL4 overexpression by adenovirus injection is efficient, a precise approach is required. Autophagy exerts a bidirectional effect on cell survival, and previous studies have demonstrated that the intensity, duration, and contingent activation of autophagy are key determinants of cardiovascular diseases outcomes [48].
In summary, we determined that MYL4 plays a key role in the pathogenesis of fibrotic atrial cardiomyopathy by regulating the autophagic flux of atrial cardiomyocytes through lysosomal functions, including lysosomal mobility and maturation. We also developed a therapeutic approach for fibrotic atrial cardiomyopathy via MYL4 overexpression that attenuated atrial structural remodeling. Our study provides new insights into the pathogenesis of atrial fibrosis as well as a novel treatment for it.
Authors’ contribution
(I) Conception and design: Yuan Zhong, Wenhui Peng, Hailing Li, Jun Wang and Stanley Nattel; (II) Administrative support: Yawei Xu, Wenhui Peng, and Hailing Li; (III) Funding acquisition: Kai Tang, Wenhui Peng, and Hailing Li (IV)Provision of study materials or patients: Kai Tang, Wenhui Peng, and Hailing Li; (V) Collection and assembly of data: Yuan Zhong, Shiyu Gong, Qing Yu, Guangxi E and Nuerbiyemu Maimaitiaili; (VI) Data analysis and interpretation: Yuan Zhong, Ming Zhai, and Hailing Li; (VII) Manuscript writing: All authors; (VIII) Final approval of manuscript: All authors.
Funding
This work was supported by Chinese National Natural Science Foundation No. 81700291, 82070230 and 91939101 and Clinical Research Plan of Shanghai Hospital Development Center No. SHDC2020CR1040B and SHDC2020CR4019.
Availability of data and materials
All data and materials are available on reasonable request to the corresponding author.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
We also thank Pro. Zhiping Xie for his advice on autophagy related experiment.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102606.
Contributor Information
Yawei Xu, Email: xuyawei@tongji.edu.cn.
Wenhui Peng, Email: pwenhui@tongji.edu.cn.
Hailing Li, Email: 1986lihailing@tongji.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Goette A., et al. EHRA/HRS/APHRS/SOLAECE expert consensus on atrial cardiomyopathies: definition, characterization, and clinical implication. Heart Rhythm. 2017;14:e3–e40. doi: 10.1016/j.hrthm.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummins P., Lambert S.J. Myosin transitions in the bovine and human heart. A developmental and anatomical study of heavy and light chain subunits in the atrium and ventricle. Circ. Res. 1986;58:846–858. doi: 10.1161/01.res.58.6.846. [DOI] [PubMed] [Google Scholar]
- 3.Peng W., et al. Dysfunction of myosin light-chain 4 (MYL4) leads to heritable atrial cardiomyopathy with electrical, contractile, and structural components: evidence from genetically-engineered rats. J. Am. Heart Assoc. 2017;6 doi: 10.1161/jaha.117.007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neefjes J., Jongsma M.M.L., Berlin I. Stop or go? Endosome positioning in the establishment of compartment architecture, dynamics, and function. Trends Cell Biol. 2017;27:580–594. doi: 10.1016/j.tcb.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Brandstaetter H., Kishi-Itakura C., Tumbarello D.A., Manstein D.J., Buss F. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome-lysosome fusion. Autophagy. 2014;10:2310–2323. doi: 10.4161/15548627.2014.984272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordonnier M.N., Dauzonne D., Louvard D., Coudrier E. Actin filaments and myosin I alpha cooperate with microtubules for the movement of lysosomes. Mol. Biol. Cell. 2001;12:4013–4029. doi: 10.1091/mbc.12.12.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logvinova D.S., Levitsky D.I. Essential light chains of myosin and their role in functioning of the myosin motor. Biochemistry. Biokhimiia. 2018;83:944–960. doi: 10.1134/s0006297918080060. [DOI] [PubMed] [Google Scholar]
- 8.Rahul, et al. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33(35) doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghazizadeh Z., et al. Metastable atrial state underlies the primary genetic substrate for MYL4 mutation-associated atrial fibrillation. Circulation. 2020;141:301–312. doi: 10.1161/circulationaha.119.044268. [DOI] [PubMed] [Google Scholar]
- 10.Corridoni D., et al. Single-cell atlas of colonic CD8(+) T cells in ulcerative colitis. Nat. Med. 2020;26:1480–1490. doi: 10.1038/s41591-020-1003-4. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Robbins J. Proteasomal and lysosomal protein degradation and heart disease. J. Mol. Cell. Cardiol. 2014;71:16–24. doi: 10.1016/j.yjmcc.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pu J., Guardia C.M., Keren-Kaplan T., Bonifacino J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016;129:4329–4339. doi: 10.1242/jcs.196287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson D.E., Ostrowski P., Jaumouillé V., Grinstein S. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 2016;212:677–692. doi: 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brigden W. Uncommon myocardial diseases: the non-coronary cardiomyopathies. Lancet (London, England) 1957;273:1243–1249. doi: 10.1016/s0140-6736(57)91537-4. [DOI] [PubMed] [Google Scholar]
- 15.Goette A., et al. EHRA/HRS/APHRS/SOLAECE expert consensus on atrial cardiomyopathies: definition, characterization, and clinical implication. Europace : European pacing, arrhythmias, and cardiac electrophysiology : journal of the working groups on cardiac pacing, arrhythmias, and cardiac cellular electrophysiology of the European Society of Cardiology. 2016;18:1455–1490. doi: 10.1093/europace/euw161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price K.M., Littler W.A., Cummins P. Human atrial and ventricular myosin light-chains subunits in the adult and during development. Biochem. J. 1980;191:571–580. doi: 10.1042/bj1910571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auckland L.M., Lambert S.J., Cummins P. Cardiac myosin light and heavy chain isotypes in tetralogy of Fallot. Cardiovasc. Res. 1986;20:828–836. doi: 10.1093/cvr/20.11.828. [DOI] [PubMed] [Google Scholar]
- 18.Schaub M.C., Tuchschmid C.R., Srihari T., Hirzel H.O. Myosin isoenzymes in human hypertrophic hearts. Shift in atrial myosin heavy chains and in ventricular myosin light chains. Eur. Heart J. 1984;5(Suppl F):85–93. doi: 10.1093/eurheartj/5.suppl_f.85. [DOI] [PubMed] [Google Scholar]
- 19.Morano M., et al. Regulation of human heart contractility by essential myosin light chain isoforms. J. Clin. Invest. 1996;98:467–473. doi: 10.1172/jci118813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudbjartsson D.F., et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 21.Gudbjartsson D.F., et al. A frameshift deletion in the sarcomere gene MYL4 causes early-onset familial atrial fibrillation. Eur. Heart J. 2017;38:27–34. doi: 10.1093/eurheartj/ehw379. [DOI] [PubMed] [Google Scholar]
- 22.Orr N., et al. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat. Commun. 2016;7 doi: 10.1038/ncomms11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong Y., et al. Rs4968309 in myosin light chain 4 (MYL4) associated with atrial fibrillation onset and predicts clinical outcomes after catheter ablation in atrial fibrillation patients without structural heart disease. Circ. J. : official journal of the Japanese Circulation Society. 2019;83:1994–2001. doi: 10.1253/circj.CJ-19-0415. [DOI] [PubMed] [Google Scholar]
- 24.Meyer K., Hodwin B., Ramanujam D., Engelhardt S., Sarikas A. Essential role for premature senescence of myofibroblasts in myocardial fibrosis. J. Am. Coll. Cardiol. 2016;67:2018–2028. doi: 10.1016/j.jacc.2016.02.047. [DOI] [PubMed] [Google Scholar]
- 25.Mehdizadeh M., Aguilar M., Thorin E., Ferbeyre G., Nattel S. The role of cellular senescence in cardiac disease: basic biology and clinical relevance. Nat. Rev. Cardiol. 2022;19:250–264. doi: 10.1038/s41569-021-00624-2. [DOI] [PubMed] [Google Scholar]
- 26.Hinderer S., Schenke-Layland K. Cardiac fibrosis - a short review of causes and therapeutic strategies. Adv. Drug Deliv. Rev. 2019;146:77–82. doi: 10.1016/j.addr.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Everett T.H.t., Olgin J.E. Atrial fibrosis and the mechanisms of atrial fibrillation. Heart Rhythm. 2007;4:S24–S27. doi: 10.1016/j.hrthm.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galluzzi L., et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroemer G., Mariño G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lum J.J., DeBerardinis R.J., Thompson C.B. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 31.Taneike M., et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6:600–606. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- 32.Nakai A., et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka Y., et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 34.Nishino I., et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 35.Garcia L., et al. Impaired cardiac autophagy in patients developing postoperative atrial fibrillation. J. Thorac. Cardiovasc. Surg. 2012;143:451–459. doi: 10.1016/j.jtcvs.2011.07.056. [DOI] [PubMed] [Google Scholar]
- 36.Bravo-San Pedro J.M., Kroemer G., Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ. Res. 2017;120:1812–1824. doi: 10.1161/circresaha.117.311082. [DOI] [PubMed] [Google Scholar]
- 37.Qiao L., et al. Deficient chaperone-mediated autophagy promotes inflammation and atherosclerosis. Circ. Res. 2021;129:1141–1157. doi: 10.1161/circresaha.121.318908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma X., et al. Transcription factor EB activation rescues advanced αB-crystallin mutation-induced cardiomyopathy by normalizing desmin localization. J. Am. Heart Assoc. 2019;8 doi: 10.1161/jaha.118.010866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klionsky D.J., et al. fourth ed. 2021. Guidelines for the use and interpretation of assays for monitoring autophagy; pp. 1–382. (Autophagy). 17. [DOI] [Google Scholar]
- 40.Yu W., et al. TBC1D15/RAB7-regulated mitochondria-lysosome interaction confers cardioprotection against acute myocardial infarction-induced cardiac injury. Theranostics. 2020;10:11244–11263. doi: 10.7150/thno.46883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trivedi P.C., Bartlett J.J., Pulinilkunnil T. Lysosomal biology and function: modern view of cellular debris Bin. Cells. 2020;9 doi: 10.3390/cells9051131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence R.E., Zoncu R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019;21:133–142. doi: 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- 43.Lim C.Y., Zoncu R. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 2016;214:653–664. doi: 10.1083/jcb.201607005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubota C., et al. Constitutive reactive oxygen species generation from autophagosome/lysosome in neuronal oxidative toxicity. J. Biol. Chem. 2010;285:667–674. doi: 10.1074/jbc.M109.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jongsma M.L., et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell. 2016;166:152–166. doi: 10.1016/j.cell.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reits E.A., Neefjes J.J. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 2001;3:E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- 47.Cabukusta B., Neefjes J. Mechanisms of lysosomal positioning and movement. Traffic (Copenhagen, Denmark) 2018;19:761–769. doi: 10.1111/tra.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Q.Q., et al. vol. 131. Clinical science; London, England: 1979. pp. 2319–2345. (Mechanisms Contributing to Cardiac Remodelling). 2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials are available on reasonable request to the corresponding author.
Data will be made available on request.