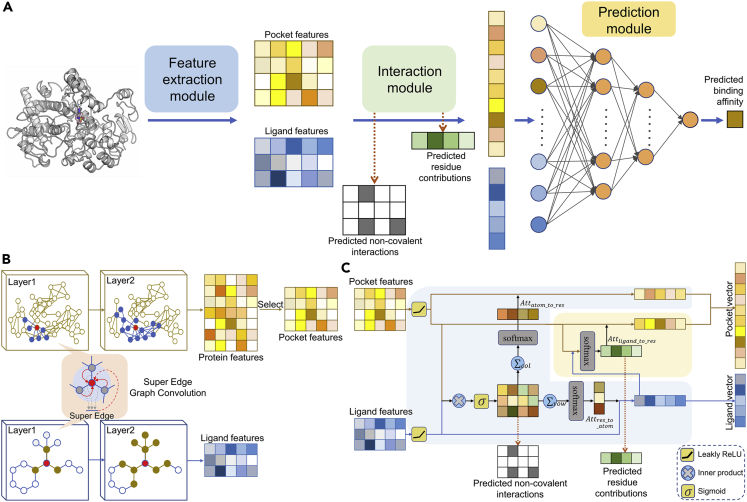
Figure 1.
Architecture of SEGSA_DTA
(A) Architecture overview of SEGSA_DTA. The SEGSA_DTA is shown to predict the binding affinity of protein–ligand pairs. In this model, a feature extraction module is first used to extract pocket and ligand features from each protein–ligand pair. Each row of the pocket or ligand features represents a residue or an atom. Then, the pocket and ligand features are processed by the interaction module to produce the pocket and ligand vectors, with the predicted non-covalent interactions and the predicted residue contributions. Finally, the pocket and ligand vectors are concatenated and fed into the prediction module, a fully connected deep neural network with two hidden layers, to predict the binding affinity.
(B) Feature extraction module. The input protein graph and ligand graph are subjected to super edge graph convolution network to extract protein and ligand features, respectively. The pocket features are selected from the protein features according to protein pocket residues.
(C) Interaction module. The interaction module is structurally a multi-supervised attention module, including a supervised bidirectional attention block (pale blue background color) and a supervised unidirectional attention block (pale yellow background color).