Abstract
Chemoresistance remains a major challenge in the current treatment of acute myeloid leukemia (AML). The bone marrow microenvironment (BMM) plays a complex role in protecting leukemia cells from chemotherapeutics, and the mechanisms involved are not fully understood. Antileukemia drugs kill AML cells directly but also damage the BMM. Here, we determined antileukemia drugs induce DNA damage in bone marrow stromal cells (BMSCs), resulting in resistance of AML cell lines to adriamycin and idarubicin killing. Damaged BMSCs induced an inflammatory microenvironment through NF-κB; suppressing NF-κB with small molecule inhibitor Bay11-7082 attenuated the prosurvival effects of BMSCs on AML cell lines. Furthermore, we used an ex vivo functional screen of 507 chemokines and cytokines to identify 44 proteins secreted from damaged BMSCs. Fibroblast growth factor-10 (FGF10) was most strongly associated with chemoresistance in AML cell lines. Additionally, expression of FGF10 and its receptors, FGFR1 and FGFR2, was increased in AML patients after chemotherapy. FGFR1 and FGFR2 were also widely expressed by AML cell lines. FGF10-induced FGFR2 activation in AML cell lines operates by increasing P38 MAPK, AKT, ERK1/2, and STAT3 phosphorylation. FGFR2 inhibition with small molecules or gene silencing of FGFR2 inhibited proliferation and reverses drug resistance of AML cells by inhibiting P38 MAPK, AKT, and ERK1/2 signaling pathways. Finally, release of FGF10 was mediated by β-catenin signaling in damaged BMSCs. Our data indicate FGF10-FGFR2 signaling acts as an effector of damaged BMSC-mediated chemoresistance in AML cells, and FGFR2 inhibition can reverse stromal protection and AML cell chemoresistance in the BMM.
Keywords: acute myeloid leukemia, bone marrow stromal cells, DNA damage, FGF10, chemoresistance
Abbreviations: ADR, adriamycin; AML, acute myeloid leukemia; Ara-C, cytarabine; BM, bone marrow; BMM, bone marrow microenvironment; BMSC, bone marrow stromal cell; BTC, betacellulin; CM, conditioned medium; CYR61, cysteine-rich 61; DDR, DNA damage response; EG-VEGF, endocrine gland–derived vascular endothelial growth factor; ERK1/2, extracellular signal–regulated kinase 1/2; FBS, fetal bovine serum; FGF10, fibroblast growth factor-10; FGFR, fibroblast growth factor receptor; GO, Gene Ontology; GEPIA, Gene Expression Profiling Interactive Analysis; GF, growth factor; GH, growth hormone; HUVEC, human umbilical vein endothelial cell; IDA, idarubicin; IL-6, interleukin 6; IL-8, interleukin 8; LiCl, lithium chloride; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; ND, newly diagnosed; NS, normal saline; PARP1, poly(ADP-ribose) polymerase 1; PTEN, phosphatase and tensin homolog; qRT–PCR, quantitative RT–PCR; STAT, signal transducer and activator of transcription; TCGA, The Cancer Genome Atlas
Acute myeloid leukemia (AML) is a highly aggressive and heterogeneous hematologic malignancy characterized by clonal expansion, arrested differentiation of myeloid precursors in the bone marrow (BM), along with the inhibition of normal hematopoiesis (1). The prognosis of AML patients depends on cytogenetic alterations, molecular variants, and immunophenotypic factors (2, 3). Despite a diverse range of treatment options for AML have been introduced over the past several decades, a substantial proportion of patients with AML relapse or are refractory to the primary therapy, leading to higher mortality and shorter overall survival. The inherent and/or acquired drug resistance and recurrence of leukemia are still more difficult issues for current treatment. Therefore, it is imperative to identify additional therapeutic targets for AML treatment improvement.
The bone marrow microenvironment (BMM), which is comprised of immune and stromal cell types and extracellular components (e.g., cytokines, growth factors (GFs), hormones, and extracellular matrix), provides a permissive niche for leukemia cell survival and plays critical roles in the pathogenesis and progression of AML (4, 5, 6). The chemoresistance of AML is closely linked to the crosstalk between leukemic cells and BMM (7, 8). In particular, the interactions of BM stromal cells (BMSC) and leukemia cells activate secretion of soluble factors, such as interleukin 6 (IL-6), cysteine-rich 61 (CYR61), and C-X-C motif chemokine ligand 8, and protective signaling pathways including the PI3K–protein kinase B (Akt), which thus promote survival and chemoresistance of AML cells (9, 10, 11). However, the molecular mechanisms of stroma-induced chemoresistance in AML cells remain poorly elucidated.
Recent data indicate that anticancer drugs induce genotoxic stress in microenvironmental cells initiating a DNA damage response (DDR) and secretion of downstream cytokines important in mediating acquired drug resistance (12). Sun et al. (13) reported that mitoxantrone and docetaxel damage DNA in fibroblasts from patients with prostate cancer resulting in the release of diverse proteins, which promote proliferation of prostate cancer cells. These proteins also mediate drug resistance through secretion of Wnt family member wingless-type MMTV integration site family member 16B (WNT16B) and activation of Wnt signaling. Gilbert et al. (14) demonstrated that adriamycin (ADR) causes DNA damage in mice thymus endothelial cells initiating a stress response and release of IL-6 and TIMP metallopeptidase inhibitor 1 (TIMP-1), molecules that promote survival of lymphoma cells. Another study showed that drug-induced damage to endothelial and stromal cells reduces sensitivity of breast cancer cells to anticancer drugs via secretion of tumor necrosis factor alpha, activation of NF-κB signaling, and increasing C-X-C motif chemokine ligand 1/2 expression (15). Recent studies report that antileukemia drugs damage BMM-type cells and promote drug resistance of leukemic cells (16, 17), the mechanism of which is unclear.
In the present study, we tested that the research hypothesis drug-induced DDRs in BMSCs promotes resistance of AML cell lines to antileukemia drugs. Our results support the notion that antileukemia drug–induced alterations in BMSCs including changes in the NF-κB P65 and P38 mitogen-activated protein kinase (MAPK) signaling pathways and release of DNA damage proteins, which affect survival of AML cell lines. We also determined release of fibroblast growth factor-10 (FGF10) is mediated by β-catenin signaling in damaged BMSCs and promotes AML drug resistance in a paracrine manner by activating fibroblast growth factor receptor 2 (FGFR2), P38 MAPK, AKT, extracellular signal–regulated kinase 1/2 (ERK1/2), and signal transducer and activator of transcription 3 (STAT3) signaling pathways. FGFR2 inhibition or gene silencing reverses drug resistance by inhibiting P38 MAPK, AKT, and ERK1/2 signaling pathways. Targeting FGF10–FGFR2 signaling could be a strategy to overcome drug resistance in patients with AML.
Results
Antileukemia drugs induce DNA damage in BMSCs
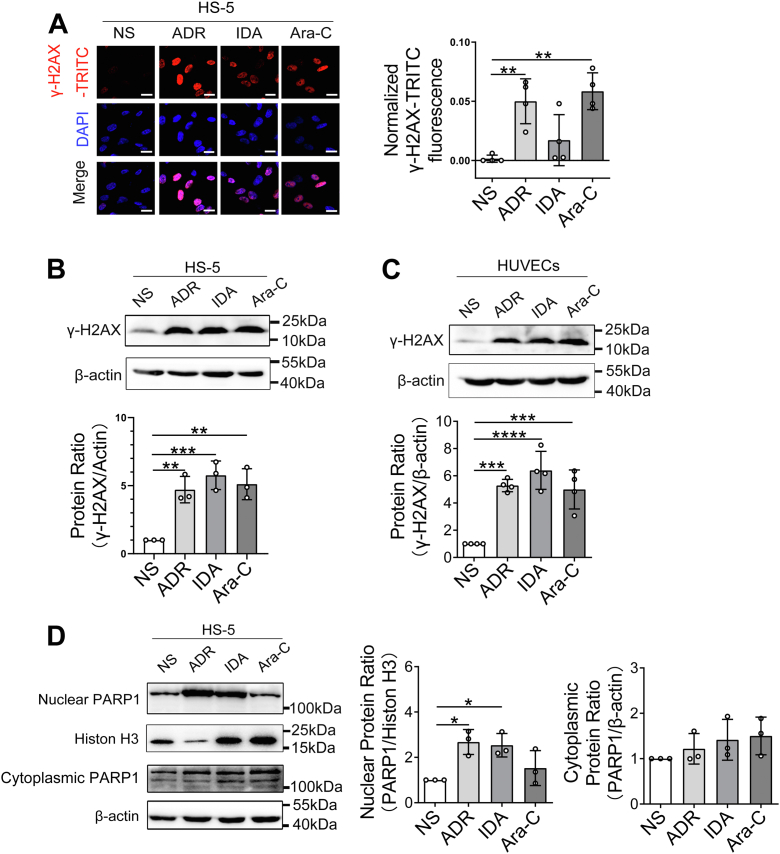
To assess for chemotherapy-induced damage responses in BMM, we first treated the human BMSC line HS-5 with ADR, idarubicin (IDA), cytarabine (Ara-C), or normal saline (NS) and determined phosphorylation of histone H2AX on Ser139 (γ-H2AX) by immune fluorescence and Western blotting. We found that each chemotherapy drug substantially increased numbers of γ-H2AX foci in HS-5 cells compared with controls (Fig. 1, A and B). HS-5 cells also had higher levels of the DNA damage marker poly(ADP-ribose) polymerase 1 (PARP1; Fig. 1D). Similar to our observations in HS-5 cells described previously, we also detected increase of γ-H2AX in human umbilical vein endothelial cells (HUVECs) (Fig. 1C). In order to further analyze the regulatory mechanisms of DNA damage in BMSCs, we performed high-throughput RNA-Seq using HS-5 cells after IDA exposure. The results showed a total of 864 genes associated with DDR, of which 716 genes were upregulated and 148 genes were downregulated (Fig. S1, A and B). The hierarchical biclustering analysis indicated significant DNA damage–associated genes in IDA-treated HS-5 cells (Fig. S1C). Gene Ontology (GO) enrichment analysis of the transcriptome from damaged HS-5 cells indicated that many differential genes fell into DNA replication, DNA repair, and cellular response to DNA damage stimulus (Fig. S1D). In addition, by conducting Kyoto Encyclopedia of Genes and Genomes analysis, we demonstrated that the changed genes were related to several pathways related to DNA damage, such as P53 and FOXO signaling pathway (Fig. S1E). Thus, our results suggest that BMSCs undergo considerable transcriptional rewiring of DNA damage after drug exposure.
Figure 1.
Antileukemia drugs induce DNA damage in bone marrow stromal cells in vitro. HS-5 and HUVECs cells were exposed to 200 μg/l ADR, 20 μg/l IDA, 400 μg/l Ara-C or NS as a vehicle control for 24 h. A, cells were probed with antibodies recognizing γ-H2AX foci (red and pink signals) by immune fluorescence, and nuclei were counterstained with DAPI (blue). The scale bar represents 20 μm. Quantitative analysis of γ-H2AX-TRITC fluorescence intensity was performed, and data were shown normalized to cells treated with NS as a control. Western blotting analysis of γ-H2AX protein levels in HS-5 cells (B) and HUVECs (C) after drug exposures. β-actin was the loading control. D, DNA damage marker PARP1 was analyzed in HS-5 cell lysates after drug exposures by Western blotting. Western blots were quantified using β-actin or histone H3 as a loading control, and data were normalized to 1.0 using samples treated with NS as a negative control. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p< 0.001, and ∗∗∗∗p< 0.0001. ADR, adriamycin; Ara-C, cytarabine; NS, normal saline; DAPI, 4′,6-diamidino-2-phenylindole; HUVEC, human umbilical vein endothelial cell; IDA, idarubicin; PARP1, poly(ADP-ribose) polymerase 1; TRITC, tetramethylrhodamine-isothiocyanate.
DNA damage of BMSCs confers chemoresistance and inhibits apoptosis in AML cell lines
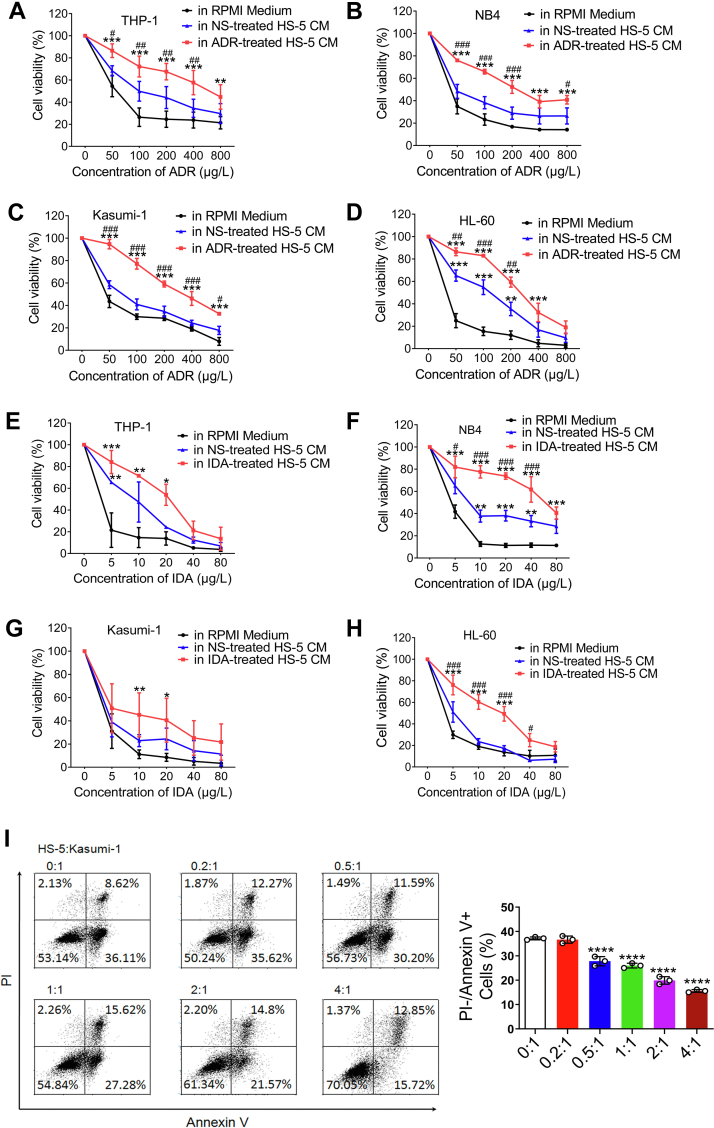
The effects of damaged BMSCs on the survival of AML cell lines THP-1, NB4, Kasumi-1, and HL-60 were studied after 72 h of culture in conditioned medium (CM) from HS-5 cells treated with ADR or IDA. Controls were CM from HS-5 cells treated with NS or RPMI1640 with 0.5% fetal bovine serum (FBS). CM from HS-5 cells treated with ADR significantly increased proportions of surviving AML cells exposed to ADR compared with HS-5 CM or RPMI1640 medium (Fig. 2, A–D). Similar results were found in CM from HS-5 cells treated with IDA (Fig. 2, E–H). However, CM from HS-5 cells had little impact on the survival of THP-1 and Kasumi-1 cells compared with RPMI1640 medium except for HL-60 and NB4 cells (Fig. 2, D and F). Besides, CM from IDA- or ADR-treated HS-5 cells had little effect on proliferation of the lymphoid cell lines, THP1 and NB4, compared with controls (Fig. S2). Next, we developed an in vitro model in which Kasumi-1 cells were directly cocultured with HS-5 cells to determine if there was protection from apoptosis induced by ADR. As shown in Figure 2I, HS-5 contact significantly decreased the proportion of apoptotic cells comparatively to culture alone. Altogether, these results support a prosurvival and antiapoptosis effect of damaged BMSCs on AML cells related to some secreted factors and cell–cell interactions.
Figure 2.
Damaged bone marrow stromal cells confer resistance to antileukemia drugs and inhibit apoptosis in AML cell lines. THP-1, NB4, Kasumi-1, and HL-60 cells were cultured with CM from 200 μg/l ADR-treated, 20 μg/l IDA-treated, and NS-treated HS-5 cells. RPMI1640 with 0.5% FBS was chosen as control. Each group was treated with ADR or IDA at the indicated concentrations for 72 h. CCK-8 assay was performed to determine percent viable cells. A–D, CM from ADR-treated HS-5 cells. E–H, CM from IDA-treated HS-5 cells. The difference between CM from ADR-treated HS-5 cells and RPMI medium was marked with asterisk, and pound sign was represented for the comparison between ADR-treated HS-5 CM and HS-5 CM. I, Kasumi-1 cells were directly cocultured with HS-5 cells in the indicated proportions and treated with 100 μg/l ADR for 48 h. Cells were stained with annexin V/PI and viability assayed by flow cytometry. Percent cells are shown in each quadrant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p< 0.001, and ∗∗∗∗p< 0.0001. #p < 0.05, ##p < 0.01, and ###p< 0.001. ADR, adriamycin; AML, acute myeloid leukemia; CCK-8, Cell Counting Kit-8; CM, conditioned medium; FBS, fetal bovine serum; IDA, idarubicin; NS, normal saline; PI, propidium iodide.
Cytotoxic damage induced a spectrum of secretory proteins in BMSCs
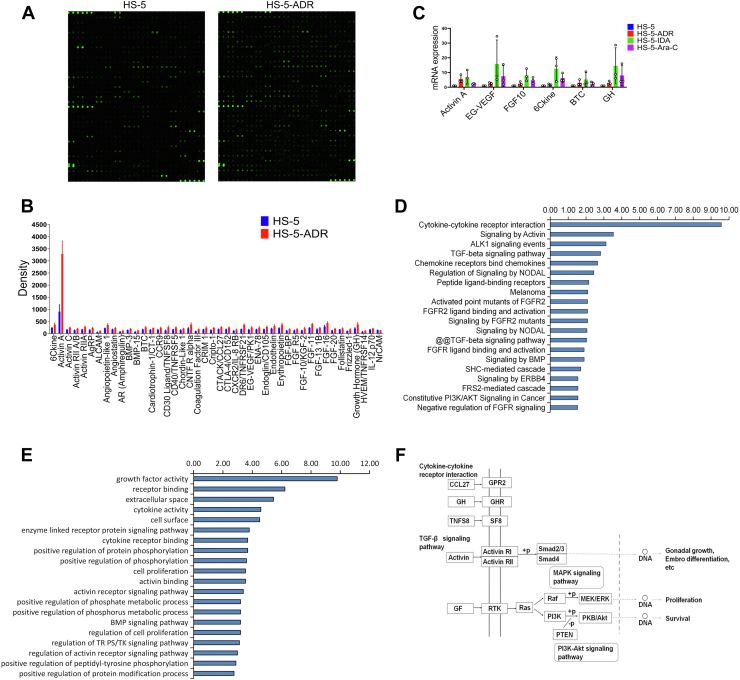
Recent work has shown that DNA damage induces a secretory phenotype in cultured cells (18). Genotoxic cancer therapy induces DNA damage in benign cells and initiate DDR, resulting in the secretion of damage-associated proteins, including IL-6 and interleukin 8 (IL-8) (19). In addition, CYR61 has been recently considered as a secreted matricellular protein and is associated with cell-intrinsic chemoresistance in other malignancies (10). In the present study, we first examined whether antileukemia drug exposure increased the expression of IL-6, IL-8, and CYR61 in damaged HS-5 cells. Consistent with the previous reports, the mRNA expressions of IL-6 and IL-8 were significantly increased in HS-5 cells after treated with ADR or IDA compared with controls, but no significant change of CYR61 was found (Fig. S3). Furthermore, we used a human chemokine and cytokine antibody array including 507 specific antibodies (Datas S1 and S2; Fig. 3A) to determine through which factors CM from damaged HS-5 cells conferred drug resistance to AML cell lines. As shown in Figure 3B and Table 1, levels of 44 proteins were significantly increased in CM from ADR-treated HS-5 cells compared with control. Among 44 proteins, 23 proteins were induced >1.5-fold change. Next, we analyzed transcript levels in HS-5 cells after treatment with ADR, IDA, or Ara-C by real-time quantitative RT–PCR (qRT–PCR). The mRNA levels of activin A, endocrine gland–derived vascular endothelial growth factor (EG-VEGF), FGF10, 6Ckine, betacellulin (BTC), and growth hormone (GH) were increased consistent with data from the protein microarrays (Fig. 3C).
Figure 3.
Drug-induced damage to bone marrow stromal cells produces diverse proteins.A, a human chemokine and cytokine antibody array was used to detect proteins in CM from ADR-treated HS-5 cells. CM from NS-treated HS-5 cells was a control. B, quantitative results of significantly increased proteins are represented in the bar chart. C, qRT–PCR analysis of activin A, BTC, FGF10, EG-VEGF, 6Ckine, and GH mRNA levels in HS-5 cells after 200 μg/l ADR, 20 μg/l IDA, and 400 μg/l Ara-C exposures relative to pretreatment transcript amounts. D, analysis of signaling pathway. Top 20 most significantly enriched KEGG pathways from data of chemokine and cytokine antibody array. E, GO functional enrichment of the differential proteins. F, CCL27, GH, and TNFS8 of the differentially secreted proteins are presented in their cytokine–cytokine receptor interaction pathway (GPR2, GH, and SF8) according to GO database. Activin is shown in activin R-Smad signaling pathway and GF in RTK and downstream effectors including Ras, Raf, and PI3K–Akt signaling pathways. ADR, adriamycin; Ara-C, cytarabine; BTC, betacellulin; CM, conditioned medium; EG-VEGF, endocrine gland–derived vascular endothelial growth factor; FGF10, fibroblast growth factor-10; GH, growth hormone; GO, Gene Ontology; IDA, idarubicin; KEGG, Kyoto Encyclopedia of Genes and Genomes; NS, normal saline; qRT–PCR, quantitative RT–PCR; RTK, receptor tyrosine kinase.
Table 1.
The fold change of differential proteins from damaged HS-5 cells
| Protein symbol | Fold change | p |
|---|---|---|
| 6Ckine | 1.566 | 0.032 |
| Activin A | 3.631 | 0.002 |
| Activin C | 1.567 | 0.008 |
| Activin RII A/B | 1.404 | 0.009 |
| Activin RIIA | 1.590 | 0.029 |
| AgRP | 1.512 | 0.025 |
| ALCAM | 1.780 | 0.025 |
| Angiopoietin-like 1 | 1.576 | 0.017 |
| Angiostatin | 1.284 | 0.032 |
| AR (Amphiregulin) | 1.434 | 0.046 |
| BMP-3 | 1.314 | 0.042 |
| BMP-15 | 1.670 | 0.027 |
| BTC | 1.527 | 0.028 |
| Cardiotrophin-1/CT-1 | 1.444 | 0.002 |
| CCR9 | 1.558 | 0.016 |
| CD30 ligand/TNFSF8 | 2.011 | 0.021 |
| CD40/TNFRSF5 | 1.466 | 0.006 |
| Chordin-like 1 | 1.393 | 0.025 |
| CNTF R alpha | 1.728 | 0.030 |
| Coagulation factor III/tissue factor | 1.779 | 0.005 |
| CRIM 1 | 1.434 | 0.021 |
| Cripto-1 | 1.641 | 0.006 |
| CTACK/CCL27 | 1.485 | 0.014 |
| CTLA-4/CD152 | 1.427 | 0.014 |
| CXCR2/IL-8 RB | 1.489 | 0.017 |
| DR6/TNFRSF21 | 1.995 | 0.001 |
| EG-VEGF/PK1 | 1.556 | 0.028 |
| ENA-78 | 1.394 | 0.040 |
| Endoglin/CD105 | 1.537 | 0.019 |
| Endothelin | 1.489 | 0.045 |
| Erythropoietin | 1.640 | 0.020 |
| FGF-BP | 1.473 | 0.009 |
| FGF R5 | 1.467 | 0.035 |
| FGF-10/KGF-2 | 1.698 | 0.041 |
| FGF-11 | 1.657 | 0.001 |
| FGF-13 1B | 1.339 | 0.036 |
| FGF-16 | 1.476 | 0.032 |
| FGF-20 | 1.579 | 0.003 |
| Follistatin | 1.238 | 0.046 |
| Frizzled-1 | 1.666 | 0.019 |
| GH | 1.775 | 0.031 |
| HVEM/TNFRSF14 | 1.435 | 0.036 |
| IL-12 p70 | 1.231 | 0.046 |
| NrCAM | 0.875 | 0.038 |
We then performed signaling pathway and GO Term analyses of the chemokine and cytokine antibody array data through http://amigo.geneontology.org/ and http//www.genome.jp/networks (Datas S3 and S4). This pathway analysis showed that proteins upregulated in response to DNA damage operated through cytokine–cytokine receptor interactions and the ALK1, FGFR2, TGF-β, and PI3K–AKT pathways (Fig. 3D). Enrichment analysis revealed that these differential proteins were significantly enriched in top 20 GO terms, mainly associated with GF activity, receptor binding, and regulation of cell proliferation (Fig. 3E). The cytokine–cytokine receptor interaction pathway was the dominant pathway influenced by CCL27, GH, and TNFS8. Activin involved in cell functions was presented in activin R-Smad signaling pathway and GF in the receptor tyrosine kinase and downstream effectors, including Ras, Raf, and PI3K–AKT signaling pathways (Fig. 3F). These results suggest that proteins increased by DNA damage activate the downstream signaling pathways including MAPK, PI3K–AKT, and FGFR2.
DNA damage induces inflammatory microenvironment in BM through NF-κB signaling, and suppressing NF-κB attenuate the proliferation effects of BMSCs
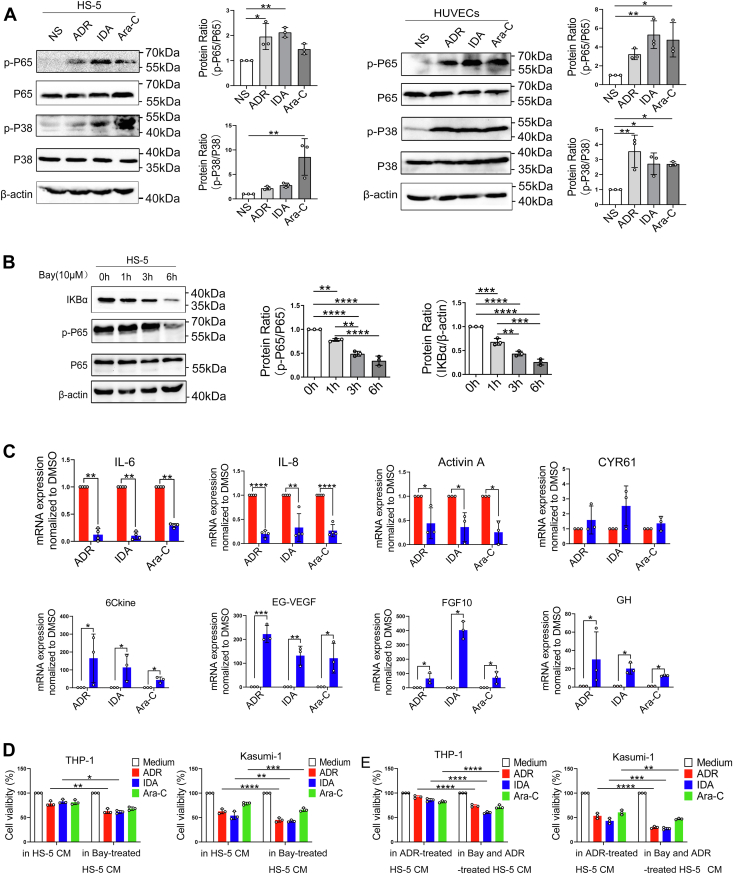
Previous report suggests the key role of NF-κB in DNA damage with apoptosis and senescence mechanisms (20, 21, 22). NF-κB and P38 MAPK signaling pathways are also involved in stress-associated induction of inflammatory networks including upregulation of IL-6 and IL-8 (23, 24, 25). We therefore analyzed changes of the NF-κB and P38 MAPK signaling pathways in HS-5 and HUVEC cells treated with ADR, IDA, or Ara-C. We found that the phosphorylation levels of NF-κB P65 and P38 MAPK were significantly increased compared with controls (Fig. 4A). To determine whether DNA damage secretory proteins are associated with activated NF-κB, we further inhibited NF-κB with Bay11-7082 (Fig. 4B) and measured mRNA levels of inflammatory factors encoding the relevant proteins in HS-5 cells. Interestingly, suppressing NF-κB in HS-5 cells with Bay11-7082 significantly decreased IL-6, IL-8, and activin A mRNA levels but no significant change in CYR61 mRNA levels (Fig. 4C, up). However, the mRNA levels of 6Ckine, EG-VEGF, FGF10, and GH were markedly increased (Fig. 4C, down) suggesting these factors might not be directly regulated by NF-κB response to DNA damage. These data present a probable role of NF-κB signaling in IL-6 and IL-8 inflammatory networks in damaged BMSCs.
Figure 4.
Drug resistance associated with damaged bone marrow stromal cells is mediated by NF-κB signaling.A, HS-5 and HUVEC cells were exposed to 200 μg/l ADR, 20 μg/l IDA, and 400 μg/l Ara-C for 24 h, and cell lysates were prepared and analyzed for phosphorylation of NF-κB P65 and p-P38 through Western blotting. Quantification of three replicate experiments is shown. B, HS-5 cells were treated with NF-κB inhibitor Bay11-7082 or dimethyl sufoxide (DMSO) as a vehicle control for the indicated intervals, and cell lysates were prepared to determine protein levels of p-P65 and IKBα. Representative images are shown, and blots were quantified using β-actin as a loading control. C, HS-5 cells were treated with 10 μmol/l Bay11-7082 combined with 200 μg/l ADR, 20 μg/l IDA, and 400 μg/l Ara-C for 24 h. mRNA levels of IL-6, IL-8, activin A, CYR61, 6Ckine, EG-VEGF, FGF10, and GH were assessed by qRT–PCR. D, THP-1 and Kasumi-1 cells were cultured with CM from HS-5 cells treated with Bay11-7082 and treated with 100 μg/l ADR, 5 μg/l IDA, and 200 μg/l Ara-C for 72 h. CM from NS-treated HS-5 was a control. E, THP-1 and Kasumi-1 cells were cultured with CM from HS-5 cells treated with Bay11-7082 combined with ADR and treated with 100 μg/l ADR, 5 μg/l IDA, and 200 μg/l Ara-C for 72 h. CM from 200 μg/l ADR-treated HS-5 was a control. CCK-8 analysis was performed to assess percent viable cells. Values are the mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p< 0.001, and ∗∗∗∗p< 0.0001. ADR, adriamycin; Ara-C, cytarabine; CCK-8, Cell Counting Kit-8; CM, conditioned medium; EG-VEGF, endocrine gland–derived vascular endothelial growth factor; FGF10, fibroblast growth factor-10; GH, growth hormone; HUVEC, human umbilical vein endothelial cell; IDA, idarubicin; IL, interleukin; qRT–PCR, quantitative RT–PCR.
To precisely determine the effect of NF-κB on the survival of AML cells, HS-5 cells were treated with Bay11-7082 or Bay11-7082 combined with ADR prior to collection of CM. Adding these CM to THP-1 and Kasumi-1 cells significantly reduced the proportion of surviving cells compared with CM from an equal number of untreated HS-5 cells (Fig. 4, D and E). These data indicate that NF-κB inhibition in BMSCs reduces the prosurvival effect of AML cells.
FGF10–FGFR2 signaling promotes survival of AML cell lines by activating P38 MAPK, AKT, ERK1/2, and STAT3 signaling pathways
To validate factors contributing to AML cell line survival after DNA damage of HS-5 cells, we used human recombinant proteins and/or neutralizing antibodies in vitro to test their effect on apoptosis, proliferation, or drug sensitivity of AML cell lines. Interestingly, among several factors studied, adding FGF10 resulted in a significant increase of surviving THP-1 cells following IDA treatment for 72 h (Fig. 5A). Besides, proliferation of THP-1 cells was slightly increased by increased concentrations of BTC, but apoptosis was not significantly decreased (Fig. S4, A and B). The proportion of surviving THP-1 cells was slightly increased at increased concentrations of BTC after treatment with IDA compared with controls (Fig. S4C). In addition, activin A stimulation did not significantly increase the sensitivity of Kasumi-1 cells to ADR and IDA (Fig. S5, B and C). And, activin A had no effect on proliferation of Kasumi-1 cells (Fig. S5A), and neutralization of activin A with antibody did not inhibit the protective role of damaged HS-5 cells (Fig. S5, D–F). The similar results were found in Kasumi-1 cells after incubation with EG-VEGF (Fig. S6, C and D). EG-VEGF also had no effect on the apoptosis and proliferation of THP-1 cells (Fig. S6, A and B). These data suggest a primary role of FGF10 in resistance of AML cells to antileukemia drugs induced by damaged BMSCs.
Figure 5.
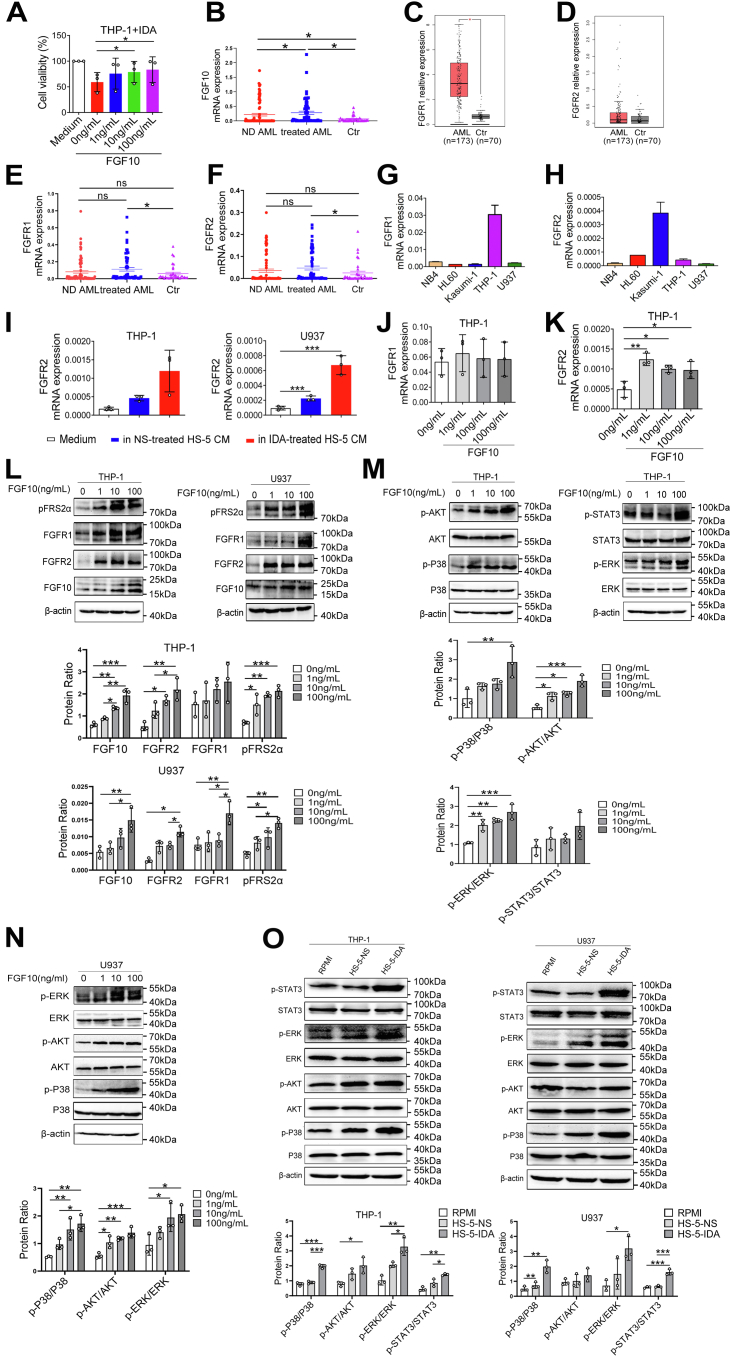
FGF10–FGFR2 signaling promotes AML cell lines survival by activating P38 MAPK, AKT, ERK1/2, and STAT3 signaling pathways.A, THP-1 cells were treated with different concentrations of FGF10 followed by 5 μg/l IDA for 72 h. CCK-8 was used to detect percent viable cells. B, quantitative mRNA level of FGF10 in newly diagnosed (ND) AML patients, AML patients after chemotherapy, and controls (Ctr). C and D, the expression levels of FGFR1 ad FGFR2 in AML patients (n = 173) and Ctr (n = 70) from TCGA database. Quantitative mRNA levels of FGFR1 (E) and FGFR2 (F) in ND AML, AML patients after chemotherapy, and Ctr. Quantitative mRNA levels of FGFR1 (G) and FGFR2 (H) in AML cell lines. I, THP-1 and U937 cells were cultured with CM from 20 μg/l IDA-treated or NS-treated HS-5 cells for 48 h, and FGFR2 mRNA levels were determined by qRT–PCR. J and K, qRT–PCR analyses of FGFR1 and FGFR2 mRNA expressions in THP-1 cells after stimulated with different concentrations of FGF10 for 24 h. L, THP-1 and U937 cells were starved in serum-free media overnight and treated with 1, 10, or 100 ng/ml FGF10 for 1 h. Cell lysates were prepared to detect levels of FGF10, FGFR1, FGFR2, and pFRS2α proteins by Western blotting. β-actin is a loading control. M, Western blotting was used to detect p-P38, p-AKT (S473), p-ERK1/2 (T202/Y204), and p-STAT3 (Tyr705) in THP-1 cells after stimulation with FGF10 for 1 h. N, Western blotting was used to detect p-P38, p-AKT (S473), and p-ERK1/2 (T202/Y204) in U937 cells after stimulation with FGF10 for 1 h. O, THP-1 and U937 cells were cultured with CM from 20 μg/l IDA-treated and NS-treated HS-5 cells for 6 h followed by Western blotting analyses of phosphorylation levels of P38 MAPK, AKT, ERK1/2, and STAT3 in cell lysates. Blots from (L–O) were quantified by measuring the density of all bands. And anti–total-antibody and anti-β-actin were loading controls. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p< 0.001. AML, acute myeloid leukemia; CCK-8, Cell Counting Kit-8; CM, conditioned media; ERK1/2, extracellular signal–regulated kinase 1/2; FGF10, fibroblast growth factor-10; FGFR2, fibroblast growth factor receptor 2; IDA, idarubicin; MAPK, mitogen-activated protein kinase; NS, normal saline; qRT–PCR, quantitative RT–PCR; STAT3, signal transducer and activator of transcription 3; TCGA, The Cancer Genome Atlas.
Recent data suggest FGF10 promotes neoplastic cell proliferation, migration, and invasion by activating FGFRs and triggering the STAT1/P21, MAPK, PLC-γ, and PI3K pathways (26, 27). In the present study, we found that the mRNA levels of FGF10 were increased in newly diagnosed (ND) AML and chemotreated AML patients, more obviously in the latter (Fig. 5B). Using Gene Expression Profiling Interactive Analysis (GEPIA), we compared the mRNA expressions of FGFR1 and FGFR2 between AML patients and normal controls. As shown in Figure 5, C and D, the mRNA levels of FGFR1 and FGFR2 were higher in AML patients than in controls. Besides, the mRNA levels of FGFR1 and FGFR2 were significantly increased in AML patients after chemotherapy (Fig. 5, E and F). We then detected FGFR mRNA levels in five AML cell lines. Highest levels of FGFR1 transcript were found in THP-1 cells (Fig. 5G). FGFR2 transcript levels were high in Kasumi-1, HL60, and THP-1 cells (Fig. 5H). To determine whether FGF10 increased survival of AML cell lines through cognate receptors, we assessed effects of exogenous FGF10 stimulation on FGFR1 and FGFR2 expressions in THP-1 and U937 cells by qRT–PCR and Western blotting. We found a small increase in mRNA levels of FGFR2 (Fig. 5K). No significant increase of FGFR1 mRNA was found (Fig. 5J). The protein levels of FGFR2 and FGFR1 were significantly increased in THP-1 and U937 cells, especially after exposure to 100 ng/ml FGF10 (Fig. 5L). Phosphorylation of FRS2α, a downstream substrate of the FGFR2, was also increased by adding FGF10 indicating that FGF10 secreted by damaged HS-5 cells activates FGFR2 signaling. In FGF10-stimulated conditions, the levels of FGFR2 downstream molecules, including P38 MAPK, AKT, ERK1/2, and STAT3 phosphorylation, were significantly increased in THP-1 and U937 cells (Fig. 5, M and N). Moreover, CM from HS-5 cells representing all the secretory proteins also significantly upregulated FGFR2 mRNA levels in THP-1 and U937 cells, more significantly after culture with CM from IDA-treated HS-5 cells (Fig. 5I). Meanwhile, CM from IDA-treated HS-5 cells significantly increased the P38, ERK1/2, AKT, and STAT3 phosphorylation levels in THP-1 and U937 cells (Fig. 5O). These data suggest that FGF10 exposure increases FGFR2 and activates downstream P38 MAPK, AKT, ERK1/2, and STAT3 signaling pathways, which increase survival of AML cell lines.
FGFR inhibition or genetic knockdown of FGFR2 attenuates FGF10-induced promotion of survival of AML cells
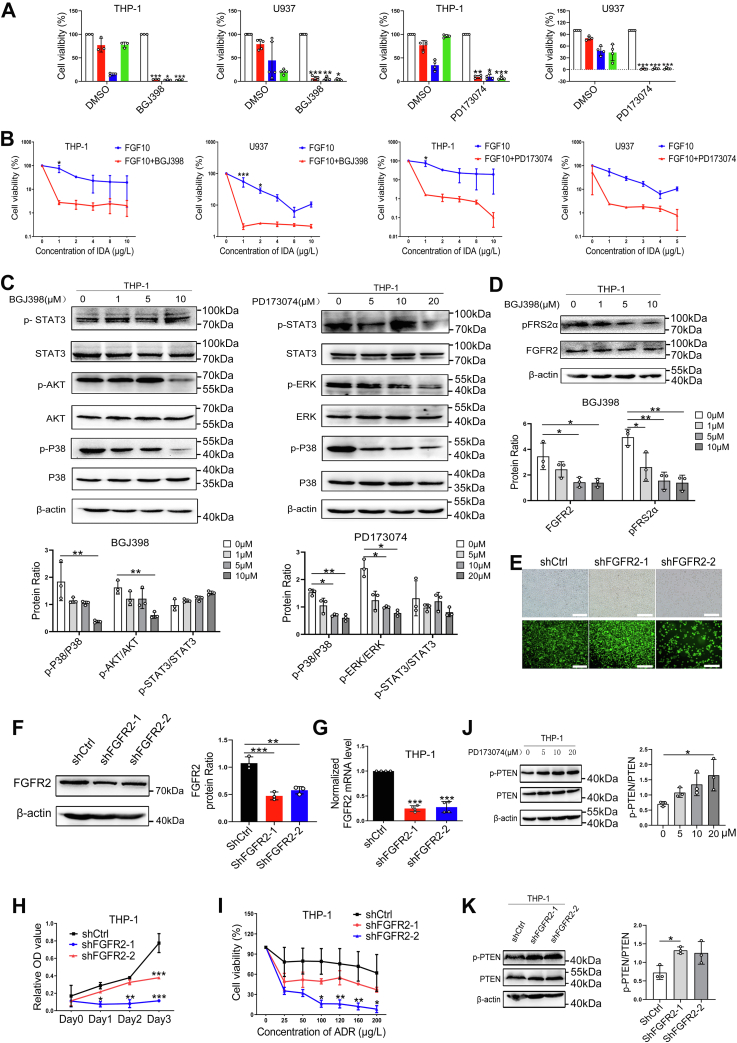
FGF10 is not only an autocrine signaling protein for stroma but also promotes tumor growth through paracrine signaling (28, 29, 30). We hypothesized that FGFR inhibition blocks FGF10-mediated paracrine protection of leukemia cells in BMM. To evaluate the relative effect of FGFR inhibition on the growth of AML cells, THP-1 and U937 cells were pretreated with selective FGFR inhibitors BGJ398 and PD173074 prior to exposure to ADR, IDA, and Ara-C for 72 h. Inhibition of FGFR significantly increased killing of THP-1 and U937 cells by ADR, IDA, and Ara-C (Fig. 6A). Adding FGF10 to the medium containing BGJ398 or PD173074 did not reverse IDA resistance of these AML cell lines (Fig. 6B). Treatment of THP-1 cells with BGJ398 or PD173074 decreased phosphorylation levels of FRS2α, P38 MAPK, ERK1/2, and AKT (Fig. 6, C and D).
Figure 6.
Inhibition of paracrine FGF10–FGFR2 signaling blocks FGF10-promoted survival of AML cell lines.A, THP-1 and U937 cells were treated with FGFR inhibitors BGJ398 and PD173074 at the indicated concentration for 1 h followed by treatment with 100 μg/l ADR, 5 μg/l IDA, and 200 μg/l Ara-C for 72 h, and percent viable cells were analyzed by CCK-8 assay. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p< 0.001 versus DMSO. B, THP-1 and U937 cells were incubated with 10 ng/ml FGF10 for 1 h before treated with 10 μmol/l BGJ398 or 20 μmol/l PD173074 combined with indicated concentrations of IDA. CCK-8 analysis was used to detect percent surviving cells. C, Western blotting was used to quantify levels of P38 MAPK, AKT, ERK1/2, and STAT3 in cell lysates of THP-1 cells after treatment with BGJ398 and PD173074 at the indicated concentration for 1 h. D, representative blots of FGFR2 protein and phosphorylation of FRS2α were shown. Quantitative analysis was performed and relativized to β-actin. E, THP-1 cells were transfected with FGFR2 shRNA and shCtrl lentivirus for 72 h. Efficiency of infection was determined by fluorescence microscope. The scale bar represents 150 μm. Cells were then collected for qRT–PCR and Western blotting analyses to detect FGFR2 protein (F) and mRNA levels (G). CCK-8 analysis was performed to detect the proliferation (H) and percent viable THP-1-shRNAFGFR2 after treatment with ADR at the indicated concentrations (I). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p< 0.001 versus THP-1-shCtrl. Western blotting analysis of PTEN protein and phosphorylation levels in cell lysates of THP-1 cells treated with 20 μmol/l PD173074 (J) or transfected with shFGFR2 (K). Anti–total-PTEN and anti-β-actin were loading controls. ADR, adriamycin; AML, acute myeloid leukemia; Ara-C, cytarabine; CCK-8, Cell Counting Kit-8; DMSO, dimethyl sulfoxide; ERK1/2, extracellular signal–regulated kinase 1/2; FGF10, fibroblast growth factor-10; FGFR2, fibroblast growth factor receptor 2; IDA, idarubicin; MAPK, mitogen-activated protein kinase; PTEN, phosphatase and tensin homolog; qRT–PCR, quantitative RT–PCR; STAT3, signal transducer and activator of transcription 3.
To confirm the decreased protection of AML cells is specific for FGFR2 inhibition, we generated a THP-1 clone expressing shRNA specific to FGFR2 (THP-1-shRNAFGFR2; Fig. 6E). Data from qRT–PCR and Western blotting showed that mRNA and protein levels of FGFR2 were significantly decreased in THP-1-shRNAFGFR2 compared with THP-1-shRNACtrl (Fig. 6, F and G). FGFR2 silencing significantly inhibited THP-1 cell growth (Fig. 6H) and increased sensitivity of THP-1 cells to ADR (Fig. 6I).
Phosphatase and tensin homolog (PTEN) is an important negative feedback regulator of PI3K–AKT signaling pathway blocking activation of the MAPK and PI3K–AKT signaling pathway (31, 32). When we studied effects of FGFR inhibition on protein levels of PTEN in THP-1 cells, we found the phosphorylation level of PTEN was slightly increased in PD173074-treated group and THP-1-shRNAFGFR2 compared with controls (Fig. 6, J and K).
Cytotoxic stress induces FGF10 secretion through β-catenin in damaged BMSCs
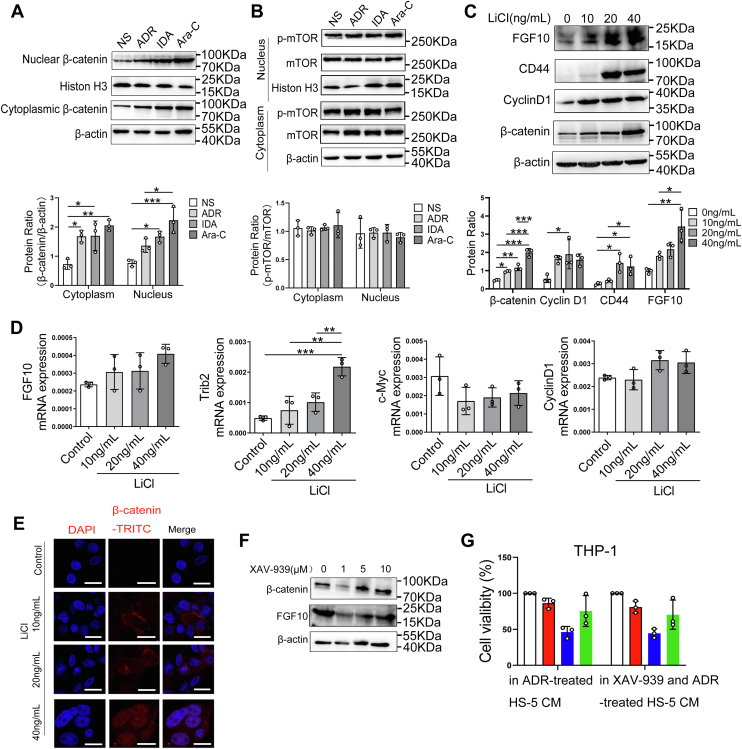
To determine the mechanistic link between DNA damage and FGF10 in BMSCs, we then measured the protein level of FGF10 in damaged HS-5 cells. The result indicated that FGF10 transcription levels were significantly elevated in HUVECs and HS-5 cells after drug exposure (Fig. S7, A and B). In addition, the immunofluorescence images confirmed that higher FGF10 levels were associated with higher γ-H2AX in HS-5 cells after drug treatment (Fig. S7C). These results suggest that cytotoxic damage induces the expression of FGF10 protein in BMSCs. Recent data suggest that β-catenin or mammalian target of rapamycin (mTOR) signaling pathways regulate FGF10 expression (33). We further tested whether DNA damage–induced FGF10 in HS-5 cells is mediated by β-catenin or mTOR signaling pathways. Levels of β-catenin were significantly increased in cytoplasm and nuclei of HS-5 cells treated with ADR, IDA, or Ara-C compared with controls (Fig. 7A). However, there was no significant change of total and phosphorylated mTOR protein (Fig. 7B). We next investigated whether activated β-catenin is the pivotal regulator of FGF10 in damaged HS-5 cells. Interestingly, levels of FGF10, β-catenin, and downstream target CyclinD1 and CD44 proteins were significantly increased in lithium chloride (LiCl)-treated HS-5 cells compared with controls by Western blotting (Fig. 7C). Furthermore, levels of β-catenin protein were also significantly increased in LiCl-treated HS-5 cells detected by immune fluorescence (Fig. 7E). The mRNA levels of Trib2 and FGF10 were slightly increased in HS-5 cells after treated with LiCl by qRT–PCR. However, no significant change of c-Myc and CyclinD1 mRNA expressions was found (Fig. 7D). Next, we examined the effect of β-catenin inhibition on FGF10 expression in HS-5 cells using Wnt signaling inhibitor XAV-939. Treatment of HS-5 cells with 1 μM XAV-939 resulted in significant reduction of FGF10 protein levels (Fig. 7F). However, adding these medium to THP-1 cells did not significantly reduce the proportion of surviving cells compared with controls (Fig. 7G).
Figure 7.
Cytotoxic stress induces FGF10 secretion by damaged bone marrow stromal cells through β-catenin.A and B, HS-5 cells were respectively treated with 200 μg/l ADR, 10 μg/l IDA, and 400 μg/l for 24 h, and protein levels of β-catenin and p-mTOR in cytoplasm and nuclei lysates were assessed by Western blotting. Western blots from (A and B) were quantified, and data are shown normalized to cells treated with NS. C, HS-5 cells were exposed to β-catenin activator (LiCl) at the indicated concentrations for 24 h. Western blotting was used to detect protein levels of FGF10, β-catenin, and downstream target CyclinD1, CD44. Quantitative analysis of three replicate experiments was performed and shown relativized to β-actin. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p< 0.001. D, the mRNA levels of FGF10 and β-catenin downstream target c-Myc, CyclinD1, and Trib2 in LiCl-treated HS-5 cells were assessed by qRT–PCR. ∗∗p < 0.01 and ∗∗∗p< 0.001 versus control. E, LiCl-treated HS-5 cells were probed with antibodies recognizing β-catenin (red and pink signals) by immunofluorescence, and nuclei were counterstained with DAPI (blue). The scale bar represents 20 μm. F, HS-5 cells were treated with Wnt signaling inhibitor XAV-939 at the indicated concentration for 1 h, and levels of β-catenin and FGF10 protein were quantified by Western blotting. G, CM was collected from HS-5 cells after treatment with 200 μg/l ADR or 1 μM XAV-939 combined with 200 μg/l ADR, and percent viable THP-1 cells in the CM after treated with 100 μg/l ADR, 5 μg/l IDA, and 200 μg/l Ara-C for 72 h were determined by CCK-8 analysis. ADR, adriamycin; Ara-C, cytarabine; CCK-8, Cell Counting Kit-8; CM, conditioned media; DAPI, 4′, 6-diamidino-2-phenylindole; FGF10, fibroblast growth factor-10; IDA, idarubicin; LiCl, lithium chloride; mTOR, mammalian target of rapamycin; NS, normal saline; qRT–PCR, quantitative RT–PCR.
Discussion
The BMM has been reported to protect leukemia cells from the effects of both antileukemia drugs and targeted kinase inhibitors (34, 35), but their mechanisms are poorly understood. Until recently, stromal protection of leukemia cells was thought to be largely mediated by secreted cytokines or through direct contact. In solid tumor, nonspecific treatments involving ionizing radiation and genotoxic drugs are not entirely restricted to neoplastic cells and can also induce genotoxic stress in benign cells (23). In the present study, we demonstrate that antileukemia drugs result in an obvious highly expression of γ-H2AX and PARP1 in HS-5 cells, and HS-5 cells undergo substantial transcriptional rewiring of DNA damage after drug exposure, revealing a damaged microenvironment induced by chemotherapy in AML. In an HS-5 CM culture and direct coculture model, we show an increase in AML cell survival and a reduction in cell apoptosis. These data highlight a prosurvival effect of damaged BMSCs on leukemia cells related to involvement of secreted factors and cell–cell interactions.
As reported, genotoxic-induced damage to the tumor microenvironment elicits a secretory response and subsequent senescence-related secretory phenotypes (18, 19). The composition of damage response program is complex and mainly includes a diverse spectrum of proinflammatory cytokines such as IL-6 and IL-8, extracellular matrix–altering protease, proneurogenic factors, angiogenic GFs, and epithelial mitogens, including agonists for the epidermal growth factor receptor (36, 37, 38). In a mouse model of lymphoma, Luke et al. (14) demonstrated that the thymus released IL-6 and TIMP-1, creating a “chemo-resistant niche” that promotes the survival of residual tumor. Yu Sun et al. (13) identified a spectrum of secreted proteins derived from fibroblasts including WNT16B. Here, using an ex vivo functional screen, we show that ADR-damaged HS-5 cells produce several of these proteins, including activin A, BTC, FGF10, EG-VEGF, 6Ckine, and GH rather than IL-6, IL-8, and CYR61. Exactly how these released proteins protect AML cell lines is uncertain.
Recent studies have identified FGFs as an important mediator in organogenesis, development, and homeostasis (39). FGF10, a member of FGF7/10/22 subfamily, which is specially expressed in the mesenchymal cells, mediates mesenchymal to epithelial signaling and induces migration and invasion of cancer cells by binding and activating to their specific FGFRs (40, 41). FGFRs are tyrosine kinase receptors including FGFR1, FGFR2, FGFR3, and FGFR4. FGFR2 has the highest affinity for FGF10 (42, 43). FGF10–FGFR2 signaling promotes proliferation and inhibits apoptosis of tumor cells through activation of STAT1/P21, MAPK, PLC-γ, and PI3K pathways (26, 27). Recently, overexpression of FGF10 has been found to be associated with poor prognosis and related to FLT3 and NPM1 mutations in AML (44). Consistent with this report, we found significant increased FGF10 expression in primary AML marrow samples and identified a central role of FGF10 in AML chemoresistance among detected secreted proteins in damaged BMSCs. By analyzing the data of ND AML patients from The Cancer Genome Atlas (TCGA) database, we found FGFR1 and FGFR2 are significantly elevated in AML patients. Similar to these observations, FGFR1 and FGFR2 were elevated in primary AML marrow samples after chemotherapy and differentially expressed in AML cell lines. We also showed that FGFR2 was activated in AML cells in the CM from damaged HS-5 cells. Moreover, exposure of AML cells to recombinant FGF10 also induced FGFR2 expression. FGFR inhibition by small-molecule inhibitors or knockdown of FGFR2 in AML cells significantly increase the sensitivity to antileukemia drugs, which was even not regained after exposure to recombinant FGF10. Mechanistically, FGF10–FGFR2 signaling exerts its effect through activation of several downstream pathways, including P38 MAPK, AKT, and ERK1/2 pathways. Thus, our work reveals that FGF10 secreted by damaged BMSC-mediated AML chemoresistance in a paracrine manner through activation of FGFR2, P38 MAPK, AKT, and ERK1/2 signaling and blockage of FGF10–FGFR2 signaling in AML cells can reverse chemoresistance.
The FGF10–FGFR2 is a receptor tyrosine kinase signaling pathway that has been shown to drive tumor growth and mediate resistance to anticancer therapies. Little is known about function and prognostic value of FGF10–FGFR2 signaling in AML. Currently, numerous FGFR inhibitors, including PD173074, BGJ398, dovitinib, erdafitinib, and ponatinib blocking the tyrosine kinase domain of FGFRs are undergoing clinical trials for cancer treatment (45, 46, 47, 48, 49). A recent phase I clinical study by Meric-Bernstam et al. (50) reported that futibatinib, a highly selective FGFR inhibitor, showed clinical activity and a tolerable safety profile in patients with advanced solid tumors. Another phase II pivotal study revealed that FGFR inhibitor erdafitinib significantly increased the objective response rate of patients with FGFR2- and FGFR3-altered urothelial cancer (51). The oncogenic role of aberrant FGFR signaling and its sensitivity to FGFR inhibition in preclinical trials have provided a strong confidence to discover and develop FGFR-targeted therapies in cancer. The FGF10–FGFR2 signaling transduction relies on MAPK and PI3K–AKT–mTOR, and blockade of FGFR2 signaling results in inhibition of P38 MAPK, AKT, and ERK1/2 in this study. Recently, a high degree of synergism between FGFR inhibitors and PI3K has been reported in preclinical models (52). So far, several phase I or II trials of MAPK–ERK inhibition and AKT inhibitor have also been reported in AML (53, 54). Based on the recent reports, we believe that inhibition of FGF10–FGFR2 signaling pathway could be a promising therapeutic avenue to delay or stop leukemia progression.
Canonical WNT signaling activation in the BMM is implicated in the pathogenesis of AML (55). Here, we identify an increased level of β-catenin protein in damaged HS-5 cells, and high levels of FGF10 produced by damaged BMSCs might be regulated by β-catenin consistent with its role as an upstream mediator of FGF10 (33, 56).
There are several important limitations to our study. For example, the BMM is complex, and we studied only two components represented by cell lines. Moreover, we studied these cells in liquid culture rather than the complex 3-dimensional structural context of the BMM. Another limitation is that we studied AML cell lines rather than AML cells from patients. Also, we did not validate our observations in patients with AML resistant to the antileukemia drugs. As in all laboratory experiments, we report associations and correlations, which suggest which should not be assumed to be cause and effect.
Conclusions
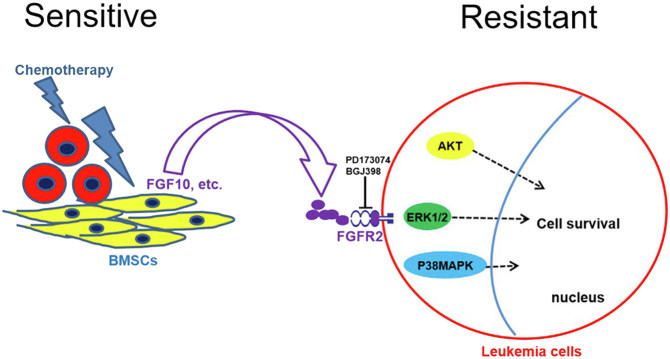
In summary, our studies explore mechanisms by which DNA damage to BMSCs promotes resistance of AML cell lines to killing by antileukemia drugs. This is a novel way to view how resistance to antileukemia drugs develops. Targeting pathways we identified could be a strategy to overcome drug resistance in patients with AML (Fig. 8).
Figure 8.
Model of drug-resistance effects of bone marrow stromal cells (BMSCs) in response to antileukemia drugs. Antileukemia drugs activate a DNA damage response (DDR) in BMSCs and induce diverse proteins that promote survival of AML cell lines via progrowth signaling pathways. Drug-induced β-catenin activation in damaged BMSCs regulates FGF10 secretion–promoting survival of AML cell lines by activating FGFR2, P38 MAPK, AKT, and ERK1/2 signaling pathways. Blocking FGF10–FGFR2 signaling with PD173074 and BGJ398 reverses the prosurvival effects of BMSCs on AML cell lines by inhibiting P38 MAPK, AKT, and ERK1/2 signaling pathways. AML, acute myeloid leukemia; ERK1/2, extracellular signal–regulated kinase 1/2; FGF10, fibroblast growth factor-10; FGFR2, fibroblast growth factor receptor 2; MAPK, mitogen-activated protein kinase.
Experimental procedures
Drugs, antibodies, and reagents
ADR, IDA, and Ara-C were dissolved in NS and diluted in RPMI1640 medium immediately before use. Primary antibodies to β-actin, phospho-Histone H2AX (Ser139) (clone JBW301), NF-κB P65, p-P65, IKBα, STAT3, p-STAT3 (Tyr705), PTEN, p-PTEN, ERK1/2, p-ERK1/2 (T202/Y204), P38, p-P38, AKT, p-AKT (S473), mTOR, p-mTOR, FGFR1, FGFR2, pFRS2α, CyclinD1, and CD44 were purchased from Cell Signaling Technology. PARP1 and FGF10 were from Santa Cruz Biotechnology. β-catenin were from Sigma–Aldrich, β-tubulin was from Abways, and histone H3 and c-MYC were from Immunoway. All secondary antibodies were obtained from ZSGB-BIO. The NF-κB inhibitor Bay11-7082 was purchased from Selleckchem and solubilized at 100 mM stock solution in dimethyl sulfoxide (Sigma) and stored at −20 °C. The selective FGFR inhibitor (BGJ398 and PD173074) and Wnt signaling inhibitor (XAV-939) were also purchased from Selleckchem and dissolved in dimethyl sulfoxide to a concentration of 10 and 50 mM, respectively, and diluted in RPMI1640 medium to a working concentration immediately before use. The β-catenin activator LiCl was from Sigma–Aldrich. Recombinant human BTC, activin A, and activin A βA subunit antibody were purchased from R&D Systems. Recombinant human EG-VEGF and FGF10 were purchased from Peprotech.
Cell cultures
Human AML cell lines THP-1, NB4, Kasumi-1, HL-60 and U937, and HUVECs were obtained from Shanghai Institutes for Biological Sciences of China. The human BMSC line HS-5 was kindly provided by Dr Feiyang Liu (High Magnetic Field Laboratory, Chinese Academy of Sciences) and Dr Ellen Weisberg (Dana-Farber Cancer Institute) (57). HL-60 cells were cultured under Iscove's modified Dulbecco's medium containing 20% FBS (Gibco) and 1% penicillin–streptomycin. Other cells were all maintained in RPMI1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin in an incubator at 37 °C and 5% CO2.
Patient samples
BM samples were collected from 81 ND AML and 59 AML patients after chemotherapy at Qilu Hospital of Shandong University. Normal BM samples were obtained from 35 healthy honors at Qilu Hospital of Shandong University. Mononuclear cells were obtained from BM by density-gradient centrifugation with Ficoll–Hypaque (Sigma–Aldrich). The studies in this work are abide by the Declaration of Helsinki principles. All patients and subjects gave their written informed consent for use of BM, and study protocols were approved by the Medical Ethics Committee of the Affiliated Qilu Hospital of Shandong University (Jinan, China).
Treatment with DNA-damaging agents
For DNA damage, HS-5 cells or HUVECs were grown until 80% confluence and treated with NS, 200 μg/l ADR, 20 μg/l IDA, or 400 μg/l Ara-C for 24 h. After treatment, cells were rinsed thrice with PBS and incubated for 3 days in RPMI1640 with 0.5% FBS medium. The supernatants from cells were harvested, concentrated, and either stored frozen at −80 °C or applied immediately.
Coculture HS-5 cells with AML cell lines
HS-5 cells were mixed with Kasumi-1 cells at ratios of 0:1, 0.2:1, 0.5:1, 1:1, 2:1, and 4:1 preseeded 1 day before and treated with 100 μg/l ADR. The cultures were incubated for 48 h after which cocultured Kasumi-1 cells were harvested for further apoptosis analysis. Kasumi-1 cells were cultured alone and treated with ADR in parallel as a control.
Human chemokine and cytokine antibody arrays
Human antibody array L-507 (AAH-BLG-1; RayBiotech) was used according to the manufacturer’s instructions. In brief, supernatants from NS-treated and ADR-treated HS-5 cells were collected and incubated on antibody-coated membranes for 24 h at 4 °C with gentle shaking. Detection of cytokines was performed and visualized with a charge-coupled device camera system (FUSION SL; Peqlab). Mean chemoluminescence of all positive-control spots per membrane was calculated, and the intensity of the chemoluminescent signal was normalized to the internal positive control.
Treatment of recombinant cytokines or neutralizing antibodies
AML cell lines were starved in serum-free medium overnight to synchronize them, and culture medium containing different concentrations of candidate cytokines or neutralizing antibodies was then added. Cultures were continued for 6 days, and cell proliferation was analyzed. After overnight starvation, AML cell lines were cultured in medium with different concentrations of candidate cytokines or neutralizing antibodies combined with antileukemia drugs for 48 or 72 h. Cell apoptosis and sensitivity to drugs were assayed.
Treatment of small-molecule inhibitor
HS-5 cells were treated with 10 μmol/l Bay11-7082 for 1, 3, and 6 h, and Western blotting was used to quantify protein levels of NF-κB P65, p-P65, and IKBa. THP-1 and U937 cells were treated with BGJ398 and PD173074 for 1 h, cell lysates were prepared, and changes in FGFR1, FGFR2, downstream molecules pFRS2α and p-P38 MAPK, p-AKT, p-ERK1/2, and p-STAT3 were quantified. XAV-939 was used to inhibit β-catenin. HS-5 cells were treated for 1 h, and Western blotting was used to quantify protein levels of β-catenin and downstream targets CyclinD1, CD44, and FGF10.
Lentivirus infection
The FGFR2-shRNA (shFGFR2) and control-shRNA (shCtrl) lentivirus were purchased from GeneChem Co, Ltd. The following are the target sequences of FGFR2-RNAi: shFGF2-1: GAA TGA AGA ACA CGA CCA A; shFGFR2-2: CCC TGT TTG ATA GAG TAT A. THP-1 cells were transfected with shFGFR2 and shCtrl lentivirus, and infection efficiency was quantified using a fluorescence microscope after 72 h. Puromycin was used to stabilize infected cells. Western blotting and qRT–PCR were performed to quantify protein and mRNA levels of FGFR2 to verify efficiency of infection.
Cell proliferation and viability assays
For cell proliferation assays, THP-1 and NB4 cells were cultured with CM from ADR-treated and NS-treated HS-5 cells for 6 days, and RPMI1640 medium containing 0.5% FBS was used as a control. THP-1 cells transfected with shFGFR2 and shCtrl lentivirus were cultured with complete medium for 3 days. For cell viability assays, THP-1, NB4, Kasumi-1 and HL-60 cells were seeded in CM from ADR-treated and NS-treated HS-5 cells or RPMI1640 medium with 0.5% FBS followed by treatment with ADR or IDA and cultured for 72 h. THP-1 and Kasumi-1 cells were cultured in CM from HS-5 cells treated with Bay11-7082 or Bay11-7082 combined with ADR and treated with a certain concentration of ADR, IDA, or Ara-C for 72 h. THP-1 and U937 cells were treated with BGJ398 or PD173074 for 1 h followed by treatment with ADR, IDA, and Ara-C for 72 h. THP-1 cells transfected with shFGFR2 and shCtrl lentivirus were cultured in complete medium and treated with indicated concentrations of ADR for 72 h. THP-1 cells were cultured in CM from HS-5 cells treated with XAV-939 or XAV-939 combined with ADR and treated with ADR, IDA, or Ara-C for 72 h. Cell lines were incubated with 10 μl Cell Counting Kit-8 (Dojindo) at 37 °C for 4 h. Absorbance measured at 450 and 650 nm was used as a reference wavelength. Cell viability was expressed by the relative absorbance value in treated samples compared with controls after correcting for background absorbance. Samples were performed in triplicates.
Apoptosis assay
The apoptosis assay was performed using annexin V–APC/propidium iodide apoptosis detection kit (BestBio) according to the manufacturer’s instructions. Fluorescence of at least 10,000 cells was determined on a flow cytometer (Beckman Coulter) to determine percent apoptotic cells.
TCGA RNA-Seq
The publicly available RNA-Seq data of 173 AML patients were downloaded from TCGA database. This database contains patients with previously untreated AML, and all patients had been diagnosed according to the National Comprehensive Cancer Network guidelines. The RNA-Seq was analyzed using GEPIA.
qRT–PCR
Total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription was performed using M-MLV reverse transcriptase complementary DNA Synthesis Kit (Takara). qPCR was done in triplicate using SYBR Green Real-time PCR Master Mix kit (Toyobo) on LightCycler 480 II Real-time PCR system (Hoffmann-La Roche) using standard settings: 95 °C (10 min) and 40 cycles of 95 °C (20 s) and 60 °C (1 min). The primers for real-time RT–PCR are displayed in Table 2. Melting curve analyses was applied to guarantee amplification specificity. mRNA levels were expressed relative to GAPDH levels. Relative expression ratio was calculated as the fold change relative to control (2−ΔΔCT).
Table 2.
The primers for qRT–PCR
| Name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| IL-6 | ACTCACCTCTTCAGAACGAATTG | CCATCTTTGGAAGGTTCAGGTTG |
| IL-8 | ACTGAGAGTGATTGAGAGTGGAC | AACCCTCTGCACCCAGTTTC |
| CYR61 | GGTCAAAGTTACCGGGCAGT | GGAGGCATCGAATCCCAGC |
| Activin A | AGCCATATAGCAGGCACGTC | GAGGTTGGCAAAGGGGCTAT |
| 6Ckine | CCTTGCCACACTCTTTCTCCC | CAAGGAAGAGGTGGGGTGTA |
| EG-VEGF | AGGTCCCCTTCTTCAGGAAACG | TCCAGGCTGTGCTCAGGAAAAG |
| FGF10 | CAGTAGAAATCGGAGTTGTTGCC | TGAGCCATAGAGTTTCCCCTTC |
| GH | AGCAACGTCTATGACCTCCTAA | CAGGAATGTCTCGACCTTGT |
| BTC | GCCCCAAGCAGTACAAGCAT | GCCCCAGCATAGCCTTCATC |
| FGFR1 | CCCGTAGCTCCATATTGGACA | TTTGCCATTTTTCAACCAGCG |
| FGFR2 | CGCTGGTGAGGATAACAACACG | TGGAAGTTCATACTCGGAGACCC |
RNA-Seq and bioinformatic analysis
The dataset included six HS-5 cell samples, treated with NS, and 20 μg/l IDA for 24 h in three biological replicates. Total RNA was extracted from cells using TRIzol reagent (Invitrogen), followed by sample integrity, quality, and purity examination. Library construction and sequencing were performed by LC-BIO Technology company (Hangzhou). The sequencing was detected by Illumina NovaSeq 6000. Differentially expressed mRNAs were selected according to fold change >2 or fold change <0.5 and p value <0.05 by edge R or DESeq2. The differentially expressed genes were subjected to GO enrichment and Kyoto Encyclopedia of Genes and Genomes enrichment analysis.
Western blotting
Cells were harvested by centrifugation, washed two times with PBS, and solubilized in radioimmunoprecipitation assay lysis buffer (Beyotime) containing protease inhibitor compound (Beyotime). Cell lysates (30 μg) were fractionated by SDS-PAGE and transferred onto nitrocellulose membranes (Millipore). Membranes were blocked in 5% nonfat milk protein for 1 h and incubated in appropriate primary antibodies overnight at 4 °C followed by horseradish peroxidase–conjugated antimouse, anti-rabbit, or antigoat immunoglobulin G at 24 °C for 1 h. After washing, signals were detected and analyzed by FluorChem E Chemiluminescent imaging system (ProteinSimple).
Immunofluorescence analysis
For detection of DNA damage, HS-5 cells grown on coverslips (Marienfeld-Superior) were treated with NS, 200 μg/l ADR, 20 μg/l IDA, and 400 μg/l Ara-C for 24 h. HS-5 cells were treated with different concentrations of LiCl for 24 h to investigate effects of β-catenin pathway on FGF10 level. Treated cells were rinsed and fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 before immune staining and then incubated in normal goat serum. Primary rabbit monoclonal anti–phospho-Histone H2AX (Ser139), anti-FGF10, anti–β-catenin, and secondary antibody Alexa Fluor 647 goat anti-rabbit immunoglobulin G (H + L) were sequentially applied. Nuclei were counterstained with 2 μg/ml of 4′,6-diamidino-2-phenylindole, and coverslips were mounted onto glass slides. Image was acquired by laser scanning confocal microscope (UltraVIEW Vox; PerkinElmer).
Statistical analysis
The SAS (China Burning Union EcHO19), version 8 and GraphPad Prism 8.0 software (Graphpad) were used to analyze data, which are reported as mean ± SD. Statistical analyses were performed on raw data for each group by one-way ANOVA, two-tailed Student’s t or Kruskal–Wallis tests. p Values <0.05 was considered statistically significant.
Data availability
Public datasets can be found here: The TCGA database/GEPIA. All other data are available from the corresponding author upon reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by grants from the Distinguished Taishan National Scholars in Climbing Plan (grant no.: tspd20210321), the National Nature Science Foundation of China (grant nos.: 81370662, 81770159, and 82070160), the Major Research plan of the National Natural Science Foundation of China (grant no.: 91942306), the key program of Natural Science Foundation of Shandong Province (grant no.: ZR2020KH016), the Fundamental Research Funds for the Central Universities (grant no.: 2022JC012), and the Clinical Practical new Technology and Development Fund of Qilu Hospital, Shandong University (grant no.: 2019-5), and the Independently Cultivate Innovative Teams of Jinan, Shandong Province (grant no.: 2021GXRC050).
Author contributions
J. Y. conceptualization; N. L. formal analysis; S. Y. investigation; Y. L. resources; Y. W. and T. L. data curation; S. Y. writing–original draft; D. M. and R. P. G. writing–review & editing; J. Z., F. L., and C. J. supervision; C. J. project administration.
Funding and additional information
R. P. G. acknowledges support from the National Institute of Health Research Biomedical Research Centre funding scheme.
Edited by Donita Brady
Supporting information
References
- 1.Dohner H., Weisdorf D.J., Bloomfield C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y., Hu F., Li J.Y., Nie R.C., Chen S.L., Cai Y.Y., et al. Systematic construction and validation of a metabolic risk model for prognostic prediction in acute myelogenous leukemia. Front. Oncol. 2020;10:540. doi: 10.3389/fonc.2020.00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisfeld A.K., Kohlschmidt J., Mims A., Nicolet D., Walker C.J., Blachly J.S., et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia. 2020;34:3215–3227. doi: 10.1038/s41375-020-0872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar B., Garcia M., Weng L., Jung X., Murakami J.L., Hu X., et al. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia. 2018;32:575–587. doi: 10.1038/leu.2017.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Usmani S., Sivagnanalingam U., Tkachenko O., Nunez L., Shand J.C., Mullen C.A. Support of acute lymphoblastic leukemia cells by nonmalignant bone marrow stromal cells. Oncol. Lett. 2019;17:5039–5049. doi: 10.3892/ol.2019.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciciarello M., Corradi G., Forte D., Cavo M., Curti A. Emerging bone marrow microenvironment-driven mechanisms of drug resistance in acute myeloid leukemia: tangle or chance? Cancers (Basel) 2021;13:5319. doi: 10.3390/cancers13215319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacamo R., Chen Y., Wang Z., Ma W., Zhang M., Spaeth E.L., et al. Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-kappaB mediates chemoresistance. Blood. 2014;123:2691–2702. doi: 10.1182/blood-2013-06-511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L., Zhao L., Man J., Liu B. CXCL2 benefits acute myeloid leukemia cells in hypoxia. Int. J. Lab. Hematol. 2021;43:1085–1092. doi: 10.1111/ijlh.13512. [DOI] [PubMed] [Google Scholar]
- 9.Koschmieder S., Chatain N. Role of inflammation in the biology of myeloproliferative neoplasms. Blood Rev. 2020;42 doi: 10.1016/j.blre.2020.100711. [DOI] [PubMed] [Google Scholar]
- 10.Long X., Yu Y., Perlaky L., Man T.K., Redell M.S. Stromal CYR61 confers resistance to mitoxantrone via spleen tyrosine kinase activation in human acute myeloid leukaemia. Br. J. Haematol. 2015;170:704–718. doi: 10.1111/bjh.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J., Li Y., Liu S., Jiang Y., Ma J., Wan L., et al. CXCL8 derived from mesenchymal stromal cells supports survival and proliferation of acute myeloid leukemia cells through the PI3K/AKT pathway. FASEB J. 2019;33:4755–4764. doi: 10.1096/fj.201801931R. [DOI] [PubMed] [Google Scholar]
- 12.Lama-Sherpa T.D., Shevde L.A. An emerging regulatory role for the tumor microenvironment in the DNA damage response to double-strand breaks. Mol. Cancer Res. 2020;18:185–193. doi: 10.1158/1541-7786.MCR-19-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y., Campisi J., Higano C., Beer T.M., Porter P., Coleman I., et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat. Med. 2012;18:1359–1368. doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert L.A., Hemann M.T. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–366. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acharyya S., Oskarsson T., Vanharanta S., Malladi S., Kim J., Morris P.G., et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruet-Hennequart S., Prendergast A.M., Shaw G., Barry F.P., Carty M.P. Doxorubicin induces the DNA damage response in cultured human mesenchymal stem cells. Int. J. Hematol. 2012;96:649–656. doi: 10.1007/s12185-012-1196-5. [DOI] [PubMed] [Google Scholar]
- 17.Gynn L.E., Anderson E., Robinson G., Wexler S.A., Upstill-Goddard G., Cox C., et al. Primary mesenchymal stromal cells in co-culture with leukaemic HL-60 cells are sensitised to cytarabine-induced genotoxicity, while leukaemic cells are protected. Mutagenesis. 2021;36:419–428. doi: 10.1093/mutage/geab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuilman T., Peeper D.S. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 19.Fumagalli M., d'Adda di Fagagna F. SASPense and DDRama in cancer and ageing. Nat. Cell Biol. 2009;11:921–923. doi: 10.1038/ncb0809-921. [DOI] [PubMed] [Google Scholar]
- 20.He Y., Pasupala N., Zhi H., Dorjbal B., Hussain I., Shih H.M., et al. NF-kappaB-induced R-loop accumulation and DNA damage select for nucleotide excision repair deficiencies in adult T cell leukemia. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2005568118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smale S.T. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol. Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 22.Chien Y., Scuoppo C., Wang X., Fang X., Balgley B., Bolden J.E., et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McConkey D.J., Choi W., Marquis L., Martin F., Williams M.B., Shah J., et al. Role of epithelial-to-mesenchymal transition (EMT) in drug sensitivity and metastasis in bladder cancer. Cancer Metastasis Rev. 2009;28:335–344. doi: 10.1007/s10555-009-9194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuilman T., Michaloglou C., Vredeveld L.C., Douma S., van Doorn R., Desmet C.J., et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 25.Acosta J.C., O'Loghlen A., Banito A., Guijarro M.V., Augert A., Raguz S., et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 26.Koyama N., Hayashi T., Ohno K., Siu L., Gresik E.W., Kashimata M. Signaling pathways activated by epidermal growth factor receptor or fibroblast growth factor receptor differentially regulate branching morphogenesis in fetal mouse submandibular glands. Dev. Growth Differ. 2008;50:565–576. doi: 10.1111/j.1440-169x.2008.01053.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakao Y., Mitsuyasu T., Kawano S., Nakamura N., Kanda S., Nakamura S. Fibroblast growth factors 7 and 10 are involved in ameloblastoma proliferation via the mitogen-activated protein kinase pathway. Int. J. Oncol. 2013;43:1377–1384. doi: 10.3892/ijo.2013.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehira M., Kikuchi T., Santoso A., Tode N., Hirano T., Ohkouchi S., et al. Human marrow stromal cells downsize the stem cell fraction of lung cancers by fibroblast growth factor 10. Mol. Cell Biol. 2014;34:2848–2856. doi: 10.1128/MCB.00871-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner N., Grose R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 30.Ndlovu R., Deng L.C., Wu J., Li X.K., Zhang J.S. Fibroblast growth factor 10 in pancreas development and pancreatic cancer. Front. Genet. 2018;9:482. doi: 10.3389/fgene.2018.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campion T.J., 3rd, Sheikh I.S., Smit R.D., Iffland P.H., 2nd, Chen J., Junker I.P., et al. Viral expression of constitutively active AKT3 induces CST axonal sprouting and regeneration, but also promotes seizures. Exp. Neurol. 2021;349 doi: 10.1016/j.expneurol.2021.113961. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y., Fang B., Feng X., Jiang Y., Zeng Y., Jiang J. Mechanism of IDH1-R132H mutation in T cell acute lymphoblastic leukemia mouse model via the Notch1 pathway. Tissue Cell. 2022;74 doi: 10.1016/j.tice.2021.101674. [DOI] [PubMed] [Google Scholar]
- 33.Hertzler-Schaefer K., Mathew G., Somani A.K., Tholpady S., Kadakia M.P., Chen Y., et al. Pten loss induces autocrine FGF signaling to promote skin tumorigenesis. Cell Rep. 2014;6:818–826. doi: 10.1016/j.celrep.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang Y.T., Hernandez D., Alonso S., Gao M., Su M., Ghiaur G., et al. Role of CYP3A4 in bone marrow microenvironment-mediated protection of FLT3/ITD AML from tyrosine kinase inhibitors. Blood Adv. 2019;3:908–916. doi: 10.1182/bloodadvances.2018022921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Battula V.L., Le P.M., Sun J.C., Nguyen K., Yuan B., Zhou X., et al. AML-induced osteogenic differentiation in mesenchymal stromal cells supports leukemia growth. JCI Insight. 2017;2 doi: 10.1172/jci.insight.90036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bavik C., Coleman I., Dean J.P., Knudsen B., Plymate S., Nelson P.S. The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer Res. 2006;66:794–802. doi: 10.1158/0008-5472.CAN-05-1716. [DOI] [PubMed] [Google Scholar]
- 37.Coppe J.P., Patil C.K., Rodier F., Sun Y., Munoz D.P., Goldstein J., et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coppe J.P., Kauser K., Campisi J., Beausejour C.M. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J. Biol. Chem. 2006;281:29568–29574. doi: 10.1074/jbc.M603307200. [DOI] [PubMed] [Google Scholar]
- 39.Agrawal S., Maity S., AlRaawi Z., Al-Ameer M., Kumar T.K.S. Targeting drugs against fibroblast growth factor(s)-induced cell signaling. Curr. Drug Targets. 2021;22:214–240. doi: 10.2174/1389450121999201012201926. [DOI] [PubMed] [Google Scholar]
- 40.Watson J., Francavilla C. Regulation of FGF10 signaling in development and disease. Front. Genet. 2018;9:500. doi: 10.3389/fgene.2018.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh N., Ornitz D.M. Functional evolutionary history of the mouse Fgf gene family. Dev. Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- 42.Clayton N.S., Grose R.P. Emerging roles of fibroblast growth factor 10 in cancer. Front. Genet. 2018;9:499. doi: 10.3389/fgene.2018.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Jiang X., Shi M., Yang Z., Chen Z., Hua X., et al. Vascular adventitial fibroblasts-derived FGF10 promotes vascular smooth muscle cells proliferation and migration in vitro and the neointima formation in vivo. J. Inflamm. Res. 2021;14:2207–2223. doi: 10.2147/JIR.S305204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling Y., Du Q. FGF10/FGF17 as prognostic and drug response markers in acute myeloid leukemia. Curr. Res. Transl. Med. 2021;70 doi: 10.1016/j.retram.2021.103316. [DOI] [PubMed] [Google Scholar]
- 45.Touat M., Ileana E., Postel-Vinay S., Andre F., Soria J.C. Targeting FGFR signaling in cancer. Clin. Cancer Res. 2015;21:2684–2694. doi: 10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 46.Sobhani N., Fassl A., Mondani G., Generali D., Otto T. Targeting aberrant FGFR signaling to overcome CDK4/6 inhibitor resistance in breast cancer. Cells. 2021;10:293. doi: 10.3390/cells10020293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renhowe P.A., Pecchi S., Shafer C.M., Machajewski T.D., Jazan E.M., Taylor C., et al. Design, structure-activity relationships and in vivo characterization of 4-amino-3-benzimidazol-2-ylhydroquinolin-2-ones: a novel class of receptor tyrosine kinase inhibitors. J. Med. Chem. 2009;52:278–292. doi: 10.1021/jm800790t. [DOI] [PubMed] [Google Scholar]
- 48.Ahn D.H., Uson Junior P.L.S., Masci P., Kosiorek H., Halfdanarson T.R., Mody K., et al. A pilot study of Pan-FGFR inhibitor ponatinib in patients with FGFR-altered advanced cholangiocarcinoma. Invest. New Drugs. 2022;40:134–141. doi: 10.1007/s10637-021-01170-x. [DOI] [PubMed] [Google Scholar]
- 49.Javle M., Roychowdhury S., Kelley R.K., Sadeghi S., Macarulla T., Weiss K.H., et al. Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: Mature results from a multicentre, open-label, single-arm, phase 2 study. Lancet Gastroenterol. Hepatol. 2021;6:803–815. doi: 10.1016/S2468-1253(21)00196-5. [DOI] [PubMed] [Google Scholar]
- 50.Meric-Bernstam F., Bahleda R., Hierro C., Sanson M., Bridgewater J., Arkenau H.T., et al. Futibatinib, an irreversible FGFR1-4 inhibitor, in patients with advanced solid tumors harboring FGF/FGFR aberrations: a phase I dose-expansion study. Cancer Discov. 2022;12:402–415. doi: 10.1158/2159-8290.CD-21-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loriot Y., Necchi A., Park S.H., Garcia-Donas J., Huddart R., Burgess E., et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 2019;381:338–348. doi: 10.1056/NEJMoa1817323. [DOI] [PubMed] [Google Scholar]
- 52.Packer L.M., Geng X., Bonazzi V.F., Ju R.J., Mahon C.E., Cummings M.C., et al. PI3K inhibitors synergize with FGFR inhibitors to enhance antitumor responses in FGFR2(mutant) endometrial cancers. Mol. Cancer Ther. 2017;16:637–648. doi: 10.1158/1535-7163.MCT-16-0415. [DOI] [PubMed] [Google Scholar]
- 53.Maiti A., Naqvi K., Kadia T.M., Borthakur G., Takahashi K., Bose P., et al. Phase II trial of MEK inhibitor binimetinib (MEK162) in RAS-mutant acute myeloid leukemia. Clin. Lymphoma Myeloma Leuk. 2019;19:142–148.e141. doi: 10.1016/j.clml.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Tong F.K., Chow S., Hedley D. Pharmacodynamic monitoring of BAY 43-9006 (Sorafenib) in phase I clinical trials involving solid tumor and AML/MDS patients, using flow cytometry to monitor activation of the ERK pathway in peripheral blood cells. Cytometry B Clin. Cytom. 2006;70:107–114. doi: 10.1002/cyto.b.20092. [DOI] [PubMed] [Google Scholar]
- 55.Stoddart A., Wang J., Hu C., Fernald A.A., Davis E.M., Cheng J.X., et al. Inhibition of WNT signaling in the bone marrow niche prevents the development of MDS in the Apc(del/+) MDS mouse model. Blood. 2017;129:2959–2970. doi: 10.1182/blood-2016-08-736454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen E.D., Wang Z., Lepore J.J., Lu M.M., Taketo M.M., Epstein D.J., et al. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J. Clin. Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weisberg E., Wright R.D., McMillin D.W., Mitsiades C., Ray A., Barrett R., et al. Stromal-mediated protection of tyrosine kinase inhibitor-treated BCR-ABL-expressing leukemia cells. Mol. Cancer Ther. 2008;7:1121–1129. doi: 10.1158/1535-7163.MCT-07-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Public datasets can be found here: The TCGA database/GEPIA. All other data are available from the corresponding author upon reasonable request.