Abstract
Superoxide dismutases convert superoxide anions to molecular oxygen and hydrogen peroxide, which, in turn, is metabolized by catalases and/or peroxidases. These enzymes constitute one of the major defense mechanisms of cells against oxidative stress and hence play a role in the pathogenesis of certain bacteria. We previously demonstrated that group B streptococci (GBS) possess a single Mn-cofactored superoxide dismutase (SodA). To analyze the role of this enzyme in the pathogenicity of GBS, we constructed a sodA-disrupted mutant of Streptococcus agalactiae NEM316 by allelic exchange. This mutant was subsequently cis complemented by integration into the chromosome of pAT113/Sp harboring the wild-type sodA gene. The SOD specific activity detected by gel analysis in cell extracts confirmed that active SODs were present in the parental and complemented strains but absent in the sodA mutant. The growth rates of these strains in standing cultures were comparable, but the sodA mutant was extremely susceptible to the oxidative stress generated by addition of paraquat or hydrogen peroxide to the culture medium and exhibited a higher mutation frequency in the presence of rifampin. In mouse bone marrow-derived macrophages, the sodA mutant showed an increased susceptibility to bacterial killing by macrophages. In a mouse infection model, after intravenous injection the survival of the sodA mutant in the blood and the brain was markedly reduced in comparison to that of the parental and complemented strains whereas only minor effects on survival in the liver and the spleen were observed. These results suggest that SodA plays a role in GBS pathogenesis.
Streptococcus agalactiae is a leading cause of invasive infections (septicemia, meningitis, and pneumonia) in neonates and a serious cause of mortality or morbidity in immunocompromised adults (38). In addition, this bacterium is considered one of the major causes of bovine intramammary infections, in particular in North America (24), and could be responsible for meningitis in fish (47). Newborns are usually colonized during delivery by the strain present in the vaginal flora of the mother (17). The main route of infection is assumed to be aspiration of the vaginal contents or the amniotic fluid containing group B streptococci (GBS) by the neonate during parturition, resulting in subsequent colonization of the respiratory epithelium (2). Pneumonia results from local infections, whereas sepsis and meningitis may be due to the spread of bacteria followed by systemic infection. The humoral and cellular inflammatory responses that contribute to the clearance of S. agalactiae in the host are the opsonization of the bacteria with specific antibodies or with complement, followed by phagocytosis by macrophages or neutrophils (6, 17, 30). Opsonin-independent phagocytosis mediated by CR3 receptor has also been reported in GBS infection (1). However, the functionality of the phagocytic cells also seems to be important in the pathogenesis of GBS infection in neonates (6). An important killing mechanism of professional phagocytes involves the production of highly microbicidal reactive oxygen metabolites during the so-called oxidative burst, which is generally induced by the engulfment of the bacteria (28). Reactive oxygen intermediates, including superoxide anions (O2.−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH.), have many deleterious effects on living organisms. They are known to cause severe damage to DNA, RNA, proteins, and lipids (28). Oxidative bacterial killing by phagocytic cells (polymorphonuclear neutrophils [PMN] and macrophages) involves an NADPH oxidase, which assembles in the phagosomal membrane and converts oxygen to superoxide when the bacteria are ingested (39). Superoxide by itself exhibits little toxicity toward bacteria, presumably because, as a negatively charged ion, it requires a transport function to permeate the phospholipid bilayer of biological membranes (41). However, superoxide produced by phagocytic cells acts as a precursor of hydrogen peroxide, which, as an uncharged molecule, is freely permeable through biological membranes (22). Once in the bacterial cytoplasm, H2O2 can react (Fenton reaction) with reduced iron or copper ions to generate hydroxyl radicals (OH.) that cause cellular damage such as lipid peroxidation, protein oxidation, and DNA strand breaks (22). Bacteria can make use of five enzymatic mechanisms to detoxify oxygen radicals; these mechanisms involve superoxide dismutase (SOD), catalase, NADH oxidase, alkyl hydroperoxide reductase, and glutathione reductase.
SODs convert the superoxide anions (O2.−) to molecular oxygen (O2) and hydrogen peroxide (H2O2), which, in turn, is metabolized by catalases and/or peroxidases (3). SODs are metalloenzymes that are classified into three types depending on the metal cofactor utilized: Cu/Zn-SOD (SodC), Mn-SOD (SodA), and Fe-SOD (SodB). Cu/Zn-SODs are essentially found in eukaryotes; however, several gram-negative bacteria containing Cu/Zn-SOD have recently been reported (7, 26, 40, 48). In contrast, Mn-SODs are present in prokaryotes and in the mitochondria of eukaryotic cells whereas Fe-SODs are present in prokaryotes and in the chloroplasts of eukaryotic cells (23). Prokaryotes might possess several types of SODs (10, 23), but all streptococci tested thus far appear to synthesize only as Mn-SOD (29, 32, 37). In addition to the Mn-SOD, the presence of a Fe-SOD in Streptococcus pneumoniae has been recently reported (49), but examination of the S. pneumoniae genome did not reveal the occurrence of a sodB-like gene (our data not shown). SODs constitute one of the major defense mechanisms of cells against oxidative stress and hence play a role in the pathogenesis of numerous bacteria (e.g., Campylobacter jejuni, Shigella flexneri, Salmonella enterica serovar typhimurium, Yersinia enterocolitica, Neisseria meningitidis, Haemophilus influenzae, Nocardia asteroides, and Streptococcus pneumoniae) by impairing the oxygen-dependent microbicidal mechanisms of the phagocytes (4, 20, 31, 34, 45, 48, 49).
S. agalactiae is a facultative anaerobe, which, like all streptococci, lacks catalase. The absence of this enzyme in this bacterial genus suggests that SOD could play an important role against oxidative stress, affecting both the survival and, consequently, the virulence of the bacteria. We previously cloned and analyzed the expression of the sodA gene of S. agalactiae (21). We report here that SodA plays a role in GBS pathogenesis.
MATERIALS AND METHODS
Bacterial strains, growth, and media.
The bacterial strains used in this study are listed in Table 1. S. agalactiae NEM316, responsible for a fatal septicemia, belongs to capsular serotype III. Escherichia coli DH5α was used for cloning experiments. S. agalactiae was cultured in brain heart infusion (BHI) broth or agar, and E. coli was cultured on tryptic soy medium (Difco Laboratories, Detroit, Mich.) at 37°C. Unless otherwise specified, antibiotics were used at the following concentrations: for E. coli, ampicillin, 100 μg/ml; erythromycin, 150 μg/ml; kanamycin, 50 μg/ml; and spectinomycin, 60 μg/ml; for S. agalactiae, erythromycin, 10 μg/ml; kanamycin, 1,000 μg/ml; rifampin, 40 μg/ml; and spectinomycin, 250 μg/ml. S. agalactiae liquid cultures were grown in standing filled flasks. Growth rates of strains were determined by measuring the optical density at 600 nm (OD600) in BHI broth.
TABLE 1.
Bacterial strains and plasmids used in this study
| Straina or plasmid | Relevant propertiesb | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | recA1 gyrA (Nal) Δ(lacIZYA-argF) (Φ80dlacΔlacZM15) | Gibco-BRL |
| HB101 | F−, hsd-20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 (Str) xyl-5 mtl-I supE44 | 9 |
| S. agalactiae | ||
| NEM316 | Serotype III isolated from neonatal blood culture (early onset disease) | 21 |
| NEM1640 | NEM316 sodAΩaphA-3, Km | This work |
| NEM1641 | NEM1640::pAT113/SpΩsodA, Km, Sp | This work |
| NEM1871 | NEM316 cpsDΩaphA-3, Km | This work |
| Plasmids | ||
| pRK24 | Ap, Tc, Tra+, Mob+ (IncP) | 42 |
| pG+host5 | Em; ColE1 replicon, thermosensitive derivative of pGK12, MCS pBluescript | 8 |
| pAT113/Sp | Sp, Mob+ (IncP); oriR pACYC184, attTn1545, lacZα+, MCS pUC19 | 11 |
| pTCV-int | Em, Km, Mob+ (IncP); a pTCV-erm derivative with a 1.8-kb EcoRI-PstI fragment carrying PdltRΩintTn1545 | Submittedc |
Bacteria were grown in BHI broth or agar.
Ap, Cm, Em, Km, Nal, Sp, Str, and Tc indicate resistance to ampicillin, chloramphenicol, erythromycin, kanamycin, nalidixic acid, spectinomycin, streptomycin, and tetracycline, respectively. attTn and MCS indicate transposon attachment site and multiple-cloning site, respectively.
C. Poyart, M.-C. Lamy, C. Boumaila, F. Fiedler, and P. Trieu-Cuot, submitted for publication.
General DNA techniques.
Genomic streptococcal DNA was isolated as previously described (33). Standard recombinant DNA techniques were used for nucleic acid preparation and analysis (36). Plasmid DNA preparation were isolated with Nucleospin Plasmid (Macherey Nagel, Düren, Germany). The oligonucleotides used in this study are listed in Table 2, and PCRs were carried out with Pfu polymerase as described by the manufacturer (Stratagene, La Jolla, Calif.). Amplification products were purified on Sephadex S-400 columns (Pharmacia, Uppsala, Sweden) and sequenced with an ABI 310 automated DNA sequencer, using the ABI PRISM dye terminator cycle sequencing kit (Perkin-Elmer, Applied Biosystems, Roissy, France). Electrocompetent cells of S. agalactiae were prepared as described previously (15).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a | Target | Match positionb |
|---|---|---|---|
| SOD1 | CGGAATTCGGATTATCTCTTTGATTTAGCC | sodA locus | 1729–1750 |
| SOD2 | AAAGGTACCTGTTGCTGCTGCTGTG | sodA locus | 2210–2227 |
| SOD3 | AGAGGATCCAATTATGGAAGGTAAG | sodA locus | 2309–2327 |
| SOD4 | AACCTGCAGTCGTCGTTGAATAGG | sodA locus | 2731–2747 |
| SOD5 | CGGAATTCAAGATGTCTTACTTGGCAGG | sodA locus | 1406–1426 |
| CAPE | AAGAATTCTTACGCTAAGTTTTCACG | cpsD | |
| CAPK | GGGGTACCTATAATAGCACCCGTGAT | cpsD | |
| CAPB | GGGGATCCAGAAAAGATGGTGGACCGGC | cpsD | |
| CAPP | GGCTGCAGATGACCACCACTTGAACC | cpsD | |
| KanK | GGGGTACCTTTAAATACTGTAG | aphA-3 | |
| KanB | TCTGGATCCTAAAACAATTCATCC | aphA-3 | |
| attRin | GGGATATATCAACGGTGG | pAT113/Sp | |
| attRout | GATAAGTCCAGTTTTTATGCGG | pAT113/Sp | |
| attLin | CCTTCTCGTTCGGAGGAAATCC | pAT113/Sp | |
| attLout | TTCTGACAGCTAAGACATGAGG | pAT113/Sp |
The restriction sites included in the oligonucleotides for subsequent cloning of the amplified fragments are indicated in bold type.
Numbering is according to the nucleotide sequence of the sodA locus (21).
Construction of bacterial strains.
We previously showed that the sodA gene of S. agalactiae was transcribed monocistronically as an 800-base mRNA (21). Although in this peculiar case insertional inactivation of sodA by a single crossover would not have generated transcriptional polar effects, we decided to inactivate this gene by inserting a resistance cassette devoid of promoter and terminator following a double-crossover event to construct a genetically stable mutant. A similar strategy was used to inactivate the cpsD gene, which is essential for type III capsule expression in GBS (35). To construct S. agalactiae strains NEM1640 and NEM1871, we inserted, in the same direction of transcription, the promoterless and terminatorless kanamycin resistance cassette aphA-3 (44) within DNA segments internal to sodA and cpsD, respectively. This was done by ligating, after digestion with the appropriate enzymes, the amplicons SOD1-SOD2, KanK-KanB, and SOD3-SOD4 (NEM1640 construction) or CAPE-CAPK, KanK-KanB, and CAPB-CAPP (NEM1871 construction). The corresponding EcoRI-PstI fragments were cloned into pG+host5, and the resulting recombinant vectors were introduced into NEM316 by electroporation. The double-crossover events leading to the expected gene replacements were screened and obtained as described previously (8). In NEM1640 (NEM316 ΔsodA), the aphA-3 cassette is transcribed from the promoter PsodA previously characterized (21). Southern analysis of restriction enzyme-digested DNA revealed that in both strains, insertion of the kanamycin resistance cassette occurred at the expected location and that the host chromosome was devoid of sequences related to pG+host5 (data not shown).
The pair of oligonucleotides SOD5-SOD4 was used to amplify the sodA gene associated with its promoter. The resulting fragment was digested with BamHI and PstI and inserted into the integrative vector pAT113/Sp (11) to give pAT113/SpΩsodA. This vector was conjugatively transferred from the mobilizing strain HB101/pRK24 to S. agalactiae NEM1640/pTCV-int to restore the SodA activity in this mutant strain. The plasmid insertion site was characterized by inverted PCR in three integrants harboring a single copy of pAT113/SpΩsodA inserted at different loci. This was done by using ligated Sau3A-digested chromosomal DNA as template in PCRs carried out with the primer pairs attRin plus attRout and attLin plus attLout to characterize the right and left chromosome-plasmid junction fragments, respectively (attL and attR were previously arbitrarily defined [43]). Sequence analysis of the three insertion sites revealed that in neither case was the integrative vector inserted within a putative coding sequence (data not shown). The complemented strain NEM1641 was chosen for further studies because no transcript running through the corresponding vector integration site in NEM316 was detected by Northern blot analysis and reverse transcriptase PCR (data not shown).
Insertional inactivation of sodA with the kanamycin resistance cassette by a double crossover constitutes a genetically irreversible event, like the insertion of the functional sodA gene in the chromosome of the sodA mutant. These strategies were chosen to construct genetically stable strains in order to avoid the use of antibiotics during the long-term animal experiments.
Protein extraction and SOD activity assay.
Crude cell lysates of GBS strains were prepared as described previously (21). Then 50 μg of total proteins was loaded onto a 10% polyacrylamide gel run under nondenaturing conditions, and the gel was stained for SOD activity by the method of Beauchamp and Fridovich (5).
Oxygen free-radical resistance assay.
Sensitivities to paraquat (methyl viologen) and hydrogen peroxide (H2O2) were determined as follows. To assess the sensitivity of strains to oxidative-stress-generating agents, aliquots of overnight cultures of S. agalactiae strains were inoculated (1:100) into prewarmed BHI broth containing 10 mM paraquat (Sigma-Aldrich Chemical Co.), and the cells were incubated aerobically without agitation (standing culture) at 37°C. The growth was monitored by assessing the OD600 hourly for 11 h. Sensitivity to H2O2 was determined by measuring cell survival after exposure to 20 mM H2O2. Overnight cultures were diluted (1:100) into prewarmed fresh BHI broth and grown at 37°C to an OD600 of 0.6 (mid-exponential phase). At this point, the culture were sampled and exposed to 20 mM H2O2. Cells were left in contact with H2O2 at 37°C for 30, 60, or 120 min. Samples of the cultures were drawn and diluted in BHI, and appropriate dilutions were plated on BHI agar. Colonies were counted after 24 h, and the cell survival was expressed as the percentage of the original CFU. All experiments were performed at least in triplicate.
Spontaneous mutation rates.
Mutagenesis, as measured by the emergence of rifampin resistance, was determined for the wild-type strain, NEM316; the sodA mutant, NEM1640; and the complemented strain, NEM1641. These strains, cultivated aerobically without agitation in BHI broth at 37°C, were collected in the mid-log phase of growth, and appropriate dilutions of the respective cultures were plated on BHI agar devoid of antibiotic, to enumerate total CFU, or containing rifampin, to enumerate rifampin-resistant CFU. The mutation rate was calculated by dividing total rifampin-resistant CFU by total CFU per milliliter of culture. For each strain, the mean mutation frequency was calculated from three independent experiments.
Cell culture techniques, macrophage survival assay, and determination of the oxidative burst.
Bone marrow-derived macrophages (BMMs) from C57 BL/6 mice were precultured and infected essentially as described previously (16). Briefly, cells were obtained by seeding 1 × 105 to 2 × 105 BMMs from 6- to 10-week-old C57BL/6 female mice per 35-mm tissue culture dish. Cells were grown in RPMI 1640 medium containing NaHCO3 (2 g/liter) and supplemented with 10% heat-inactivated fetal calf serum, 10% L-cell-conditioned supernatant (a source of colony-stimulating factor-1), and 2 mM l-glutamine. On day 4 after seeding, the adherent cells were rinsed twice with Hank's balanced salt solution containing 10 mM HEPES and refed with fresh medium. The medium was then changed once on day 5, and the infection experiment was performed on day 6 or 7.
Cell monolayers were infected with streptococci (bacterium-to-cell ratio, 10:1) in RPMI 1640 medium supplemented as described above with NaHCO3, fetal calf serum, L-cell-conditioned supernatant, and l-glutamine. For bacterial infection, the mixture was incubated for 15 min at 4°C and then for 30 min at 37°C. After this incubation, the cells were washed three times with phosphate-buffered saline (PBS)-Ca2+, and fresh medium containing gentamicin (100 μg/ml) was added to kill extracellular bacteria (time zero of the assay). To quantify the intracellular streptococci at different times of postinfection, the supernatants were removed and the cells were washed three times with PBS-Ca2+ buffer and then lysed with Triton X-100 to a final concentration of 0.1% (vol/vol). Serial dilutions of lysate from each well were plated onto BHI agar. The number of CFU was determined after 24 h incubation at 37°C. Three independent assays in triplicate were carried out for each bacterial strain. Double fluorescence labeling of F-actin and bacteria was performed as described previously (25) using β-phalloidin coupled to Oregon Green 488 (Molecular Probes, Eugene, Oreg.) and a rabbit polyclonal anti-GBS antibody revealed with an anti-immunoglobulin G coupled to Alexa 546 (Molecular Probes), respectively. Images were scanned on a Zeiss LSM 510 confocal microscope.
The oxidative burst of BMMs was measured as described previously (12) by using dihydrorhodamine 123 (DHR 123; Sigma-Aldrich Chemical Co.), a compound which becomes fluorescent on oxidation to rhodamine by reactive oxygen species produced during the respiratory burst of macrophages. After 90 min of infection, DHR 123 was added to infected BMMs at a final concentration of 10 μg/ml and the mixture was incubated for an additional 30 min in the dark at 37°C. Macrophages were washed three times in PBS-Ca2+ buffer, scraped, and resuspended in 0.5 ml of the same buffer, and the fluorescence intensities of 10,000 cells were recorded by flow cytometry. Uninfected macrophages and BMMs treated with opsonized zymosan were incubated with DHR 123 as previously described and used as negative and positive controls, respectively.
Electron microscopy.
For transmission electron microscopy (TEM), infected macrophages (100 bacteria per macrophage) were fixed for 1 h at room temperature in cacodylate buffer containing 2.5% glutaraldehyde and 0.1 M sucrose and washed three times with this buffer. The cells were then fixed for 1 h at room temperature with 1% (wt/vol) osmium tetraoxide in cacodylate buffer, washed three times with cacodylate buffer, dehydrated through a graded series of acetone, embedded in an Epon resin, thinly sectioned, and finally stained with uranyl acetate and lead citrate. Sections were examined at calibrated magnifications with a JEOL transmission electron microscope.
Mouse virulence assays.
Pathogen-free ICR female Swiss mice (Janvier, Le Geneset St-Isle, France) (6 to 8 weeks old) were used in this study. Groups of 10 mice were inoculated intravenously (i.v.) in the tail vein with 6 × 106 CFU of S. agalactiae NEM316, NEM1640, or NEM1641. The mortality and clinical symptoms (progressively starry coat, hunched posture, lethargy, circle syndrome, and then moribund) were observed over a 14-day period. For estimation of bacterial numbers in organ homogenates, groups of four mice were inoculated i.v. with 106 bacteria diluted in 0.9% NaCl. Bacterial numbers in homogenates of spleen, liver, brain, and blood were determined at various intervals by plating on BHI agar plates supplemented, when possible, with the appropriate antibiotic(s). To ensure the genetic stability of the mutant and complemented strain, 10 selected colonies of each strain were characterized by PCR in every experiment for the presence of the kanamycin cassette within sodA in NEM1640 and NEM1641 (primers SOD1-KanB) and for the presence of the functional sodA gene in the complemented strain NEM1641 (universal −20 forward and −40 reverse pUC primers).
Mice were deeply anesthetized with ketamine (10 μg/g) and xylazine (13 μg/g) administered by intramuscular injection or killed by cervical dislocation in accordance with the policies of the Animal Welfare Committee of the Faculté Necker (Paris). Each experiment was performed in triplicate.
RESULTS AND DISCUSSION
Construction of a sodA mutant of S. agalactiae and complementation.
The NEM1640 mutant (NEM316 ΔsodA) was constructed by inserting a promoterless and terminatorless aphA-3 gene conferring resistance to kanamycin into the sodA gene through a double-recombination event. The chromosomal sodA gene was replaced by the disrupted sodA gene through homologous recombination using the thermosensitive shuttle vector pG+host5. The appropriate gene disruption was confirmed by Southern blotting and PCR analysis (data not shown). The absence of SOD activity in the mutant NEM1640 was confirmed by analyzing whole-cell crude protein extracts by nondenaturing polyacrylamide gel electrophoresis with specific staining for SOD activity (Fig. 1, lane 2). These results confirmed the inactivation of sodA by allelic exchange in NEM1640.
FIG. 1.

SOD activity gel. Crude cell extracts (50 μg) of S. agalactiae were loaded onto a nondenaturing 10% polyacrylamide gel stained for SOD activity. Lanes 1, S. agalactiae NEM316; 2, sodA mutant NEM1640; 3, sodA-complemented mutant NEM1641.
For complementation of the SodA mutation in NEM1640, the integrative vector pAT113/SpΩsodA, containing the entire sodA gene and its promoter region, was introduced into S. agalactiae NEM1640 as described in Materials and Methods. One complemented strain, designated NEM1641 (NEM1640::pAT113/SpΩsodA) was selected for further studies, and complementation of the gene defect was confirmed by SOD activity assay (Fig. 1, lane 3).
Role of S. agalactiae SodA in oxidative stress resistance.
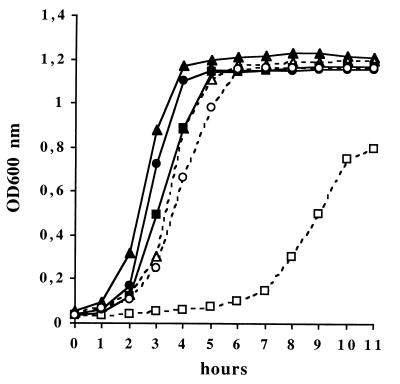
The growth of the wild-type strain, NEM316, the sodA mutant, NEM1640, and the complemented strain, NEM1641, was compared under various oxidative stress conditions. When the parental and complemented strains were cultivated aerobically without agitation in BHI broth at 37°C, their growth was similar but that of the sodA mutant was slightly delayed, reaching the stationary phase with a 1-h lag (Fig. 2). No difference was observed when these strains were grown under 5% CO2 atmosphere (data not shown). By contrast, when 10 mM paraquat was added to the culture medium, the growth of the mutant was strongly delayed, such that it reached the stationary phase with a 4-h lag and a maximum OD600 value inferior to that of the parental strain (Fig. 2). Bacterial growth was fully restored in the complemented strain NEM1641.
FIG. 2.
Growth curves of S. agalactiae NEM316, the sodA mutant, NEM1640, and the complemented strain, NEM1641, in BHI broth at 37°C under aerobic conditions with and without paraquat (10 mM). Strains and growth conditions are represented as follows: wild-type strain without (▴) or with 10 mM paraquat (▵); sodA mutant without (■) or with 10 mM paraquat (□); sodA-complemented mutant without (●) or with 10 mM paraquat (○). The results shown are representative of at least three independent experiments showing less than 10% variation.
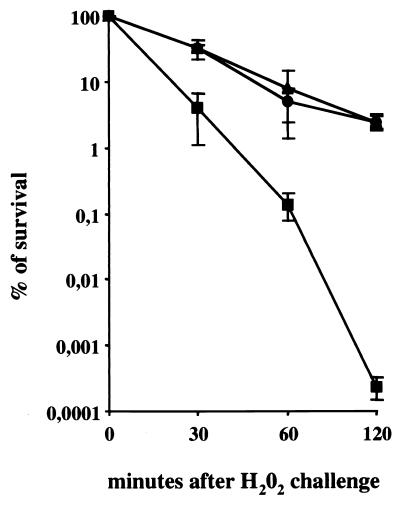
An E. coli sodA sodB double mutant exhibits an increased sensitivity to H2O2 which is thought to be due to Fenton-mediated killing (10). We therefore assessed the viability of NEM316, NEM1640, and NEM1641 in BHI broth containing 20 mM H2O2. The survival of the wild-type strain, NEM316, and the complemented strain, NEM1641, was greater by 4 orders of magnitude than that of the sodA mutant after 2 h of exposure to H2O2 (Fig. 3). The cellular toxicity of H2O2 is partly due to its ability to cause DNA damage mediated by the Fenton reaction, and SOD is protective against oxidative DNA damage that may result in gene mutations (19). Accordingly, comparison of the mutation frequencies to rifampin revealed that the sodA mutant, NEM1640, exhibited a significantly higher rate of spontaneous mutation (4.15 × 10−7 ± 1.2 × 10−7) than did the wild-type strain, NEM316 (2.58 × 10−8 ± 0.96 × 10−8), and the complemented strain, NEM1641 (3.45 × 10−8 ± 1.2 × 10−8).
FIG. 3.
Sensitivity of S. agalactiae to H2O2. The wild-type strain, NEM316 (▴), the sodA mutant, NEM1640 (■), and the complemented strain, NEM1641 (●) were grown and treated with H2O2 as described in Materials and Methods. Exponential-phase cells were exposed to 20 mM H2O2 for 30, 60, or 120 min at 37°C. Viability was determined by plating on BHI agar. Error bars represent the standard deviations of three independent experiments.
Taken together, these results indicate that SodA plays an essential role in conferring protection against oxidative stress in S. agalactiae. Similar effects have been observed in sodA mutants of other facultative anaerobic gram-positive cocci such as Streptococcus mutans, S. pneumoniae, and Lactococcus lactis (29, 37, 49). The ability to grow under aerated conditions indicates that an alternate protection mechanism operates against oxidative damage in these bacteria and, because streptococci and lactococci lack catalase, NADH oxidase, alkyl hydroperoxide reductase, and glutathione reductase could play this role.
SodA is required for intracellular survival of S. agalactiae in macrophages.
To analyze the role of Mn-SOD in the defense of S. agalactiae against the microbicidal mechanisms of macrophages, bacterial survival of the parental strain, the sodA mutant, and the complemented strain was studied in BMMs. Cell monolayers were infected with the wild-type strain, NEM316, the sodA mutant, NEM1640, and the sodA-complemented mutant, NEM1641, and the number of intracellular bacteria was estimated on cell lysates at selected intervals by quantitative plating on BHI agar (Fig. 4). The uptake was similar for the three strains since 2 × 104 CFU of GBS was recovered from monolayers of 105 macrophages in all cases at time zero of the assay. The NEM316 parental strain was able to survive inside the macrophages and was maintained at a level of 103 CFU during the course of the assay (8 h), whereas the number of recoverable sodA mutant organisms declined steadily to zero after 7 h of infection (Fig. 4). Survival of the complemented mutant was restored and was similar to that of the parental strain (Fig. 4). Moreover, survival of the unencapsulated strain, NEM1871, in the BMMs was similar to that of the wild-type strain (Fig. 4), which demonstrates that the capsule is not necessary for intramacrophagic survival, as already shown with peritoneal macrophages (14).
FIG. 4.
Growth of S. agalactiae in macrophages. BMMs were cultured in vitro and exposed to S. agalactiae NEM316 (▴), the sodA mutant, NEM1640 (■), and the complemented strain, NEM1641 (●), or the cpsD mutant, NEM1871 (⧫). Error bars represent the standard deviation of three independent experiments done in triplicate for each strain studied.
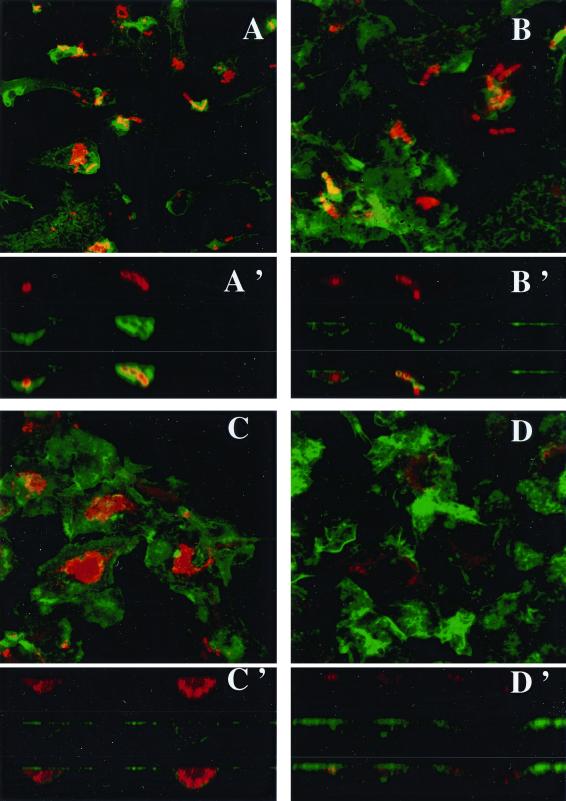
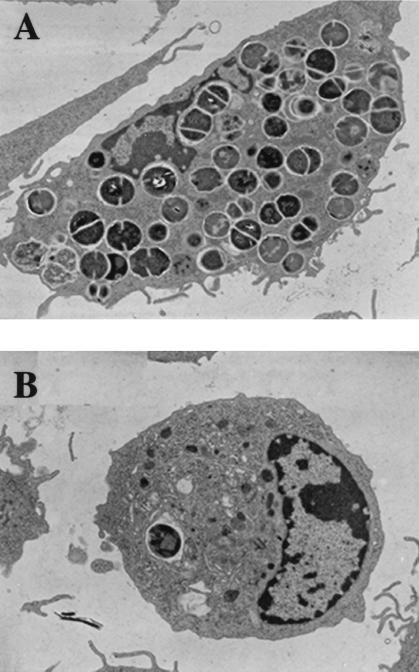
The time course of invasion was also studied qualitatively by confocal microscopy and TEM. The pictures obtained by confocal microscopy after double staining with an anti-GBS antibody, to visualize the bacteria, and with β-phalloidin, to visualize the F-actin, showed the rapid uptake process regardless of the strain (Fig. 5A and B). This uptake was associated with polymerization of F-actin, since the F-actin sheets associated with bacteria and are indicated by the overlapping of green and red light (orange-yellow) on xz sections (Fig. 5A′ and B′). After 3 h of infection, many intracellular bacteria could be detected with the wild-type strain (Fig. 5C and C′) whereas only degraded bacteria were visualized with the sodA mutant (Fig. 5D and D′). These results were confirmed by TEM. After 3 h of infection, many intracellular NEM316 streptococci were present inside the macrophages (Fig. 6A) whereas very few of the sodA mutant bacteria were seen (Fig. 6B). Taken together, these results suggest that the sodA mutant is highly susceptible to the bactericidal activity of macrophages.
FIG. 5.
Fluorescent confocal microscopy of BMMs infected (100 bacteria per cell) with S. agalactiae NEM316 (A and C) or the sodA mutant, NEM1640 (B and D). F-actin was stained with β-phalloidin (green). Bacteria were labeled with anti-S. agalactiae antibodies (red). F-actin sheets associated with bacteria are indicated by the overlapping of green and red light (orange-yellow). After 30 min of infection, characteristic chains are observed and the bacterial uptake is similar for both strains (A and B). Images reconstructed from confocal xz sections show that bacterial phagocytosis for both strains is associated with actin polymerization (A' and B'). After 3 h of infection, bacterial clusters are observed with the wild-type strain (C) whereas only bacterial degradation products are labeled in the mutant (D). Images reconstructed from confocal xz sections demonstrate the intracellular localization of bacteria (C'). Magnification, ×130.
FIG. 6.
Transmission electron micrographs of BMMs infected (100 bacteria per cell) with S. agalactiae NEM316 (A) or the sodA mutant, NEM1640 (B). Samples were taken 3 h postinfection. Magnification, ×4,558.
Previous studies have shown that GBS are able to enter and persist efficiently in macrophages (14, 46). Intracellular localization of GBS in macrophages could protect these bacteria from the microbicidal activity of neutrophils, the main effector cells against GBS infection, and from the action of antibiotics. Entry of GBS into macrophages probably involves phagocytosis and receptor-mediated endocytosis as the two principal mechanisms (46). Our confocal micrographs, which visualize rearrangements of the cellular actin microfilament system, confirmed that phagocytosis is one of the mechanisms of entry of GBS into the macrophages (Fig. 5A, A′, C, and C′). Long-term survival of GBS in macrophages has been reported; however, the mechanisms of survival have not been well identified. Several mechanisms, such as resistance to the oxidative burst, inhibition of phagolysosomal fusion, resistance to lysosomal enzymes, and attenuation of phagolysosomal acidification, have been reported to be used by pathogenic bacteria to evade intraphagosomal killing (18). Inhibition of phagolysosomal fusion and impairment of the protein kinase C-dependent signal transduction pathway may contribute to the intracellular survival of GBS (14, 46). However, a recent report revealed that following ingestion by BMMs, nonopsonized GBS fail to trigger or elicit a weak oxidative burst, a feature which may account for their intramacrophage survival (12). Our data are consistent with this report since our measurement of the oxidative burst by flow cytometry revealed that 5% of the uninfected BMMs emitted a fluorescent signal whereas infection with the wild-type strain, NEM316, and the sodA mutant, NEM1640, resulted in 13 and 15% of cells emitting fluorescence, respectively. In the positive control, 68% of the cells treated with opsonized zymosan were fluorescent, which indicates that the BMMs are able to generate an oxidative burst. Since our results demonstrate that sodA is essential for the survival of nonopsonized GBS in murine BMMs, we cannot exclude the possibility that the sodA mutant is more susceptible to an oxygen-independent killing mechanism. However, we do not favor this hypothesis because the only function thus far known for the corresponding encoded SodA protein is the detoxification of oxygen radicals. Accordingly, we demonstrated that the sodA mutant is extremely susceptible to oxidative stress but remains as resistant as the wild-type strain to lyzozyme, a cell wall-degradative enzyme, or to the cationic peptide colistin, which mimics the activity of defensins (data not shown). We therefore hypothesize that the sodA gene enables S. agalactiae to survive in the phagosome against the basal production of reactive oxygen species by the murine macrophages or to a weak and short-lived oxidative burst triggered by phagocytosis.
SodA contributes to the virulence of S. agalactiae in the mouse.
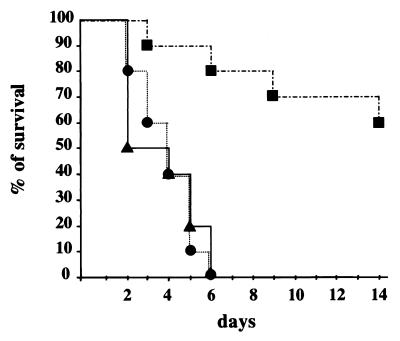
We studied the role of SodA in the virulence of S. agalactiae by infecting Swiss mice i.v. with the wild-type strain, the sodA mutant, and the sodA-complemented mutant. Over a period of 15 days, we monitored the clinical symptoms and mortality of mice infected i.v. with 6 × 106 bacteria (Fig. 7). At 1 day postinfection, all mice infected with the wild-type strain, NEM316, were symptomatic, and 50% were dead within 2 days. In contrast, no animal infected with the sodA mutant, NEM1640, showed symptoms until 2 days postinfection and the first death was recorded by day 3. Within 6 days, all mice infected with NEM316 died, while only two deaths were recorded with NEM1640 and two moribund mice subsequently died by days 9 and 14. When mice were challenged with the sodA-complemented mutant, NEM1641, data similar to those obtained with the wild-type strain, NEM316, were observed (Fig. 7).
FIG. 7.
Mortality curves in mice infected with S. agalactiae NEM316 (▴), the sodA mutant, NEM1640 (■); or the complemented strain, NEM1641 (●). Mice (10 per group) were inoculated i.v. with 6 × 106 bacteria.
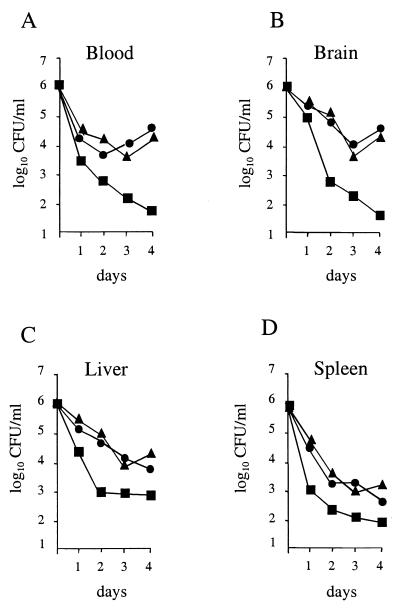
To further investigate the nature of impaired virulence, the bacterial survival of these strains in the blood and in organs (spleen, liver, and brain) of mice infected i.v. with a sublethal dose of 106 bacteria was monitored over a 4-day period. The results are illustrated in Fig. 8. We found that the numbers of the wild-type strain, NEM316, slowly declined in the spleen and liver during the early phase of infection, reaching 103 to 104 after 4 days of infection, while the sodA mutant NEM1640 was more rapidly eliminated in both organs. Survival of the sodA-complemented mutant, NEM1641, was very similar to that of the parental strain in the liver and spleen (Fig. 8C and D, respectively). Interestingly, the survival of the sodA mutant in the blood and the brain was also markedly reduced, with a 2-log-unit difference by days 2 to 4, in comparison to the parental strain (Fig. 8A and B). The growth of the complemented mutant was restored and was comparable to that of the wild-type strain. These results indicate that SodA might play a role at the early phase of infection by enabling a high level of bacteremia, which probably favors the brain invasion and the persistence of bacteria in the spleen and liver. The spleen and liver contain mainly resident macrophages that are associated with a poor oxidative activity compared to that of the PMNs, which constitute the main host cellular defense against microbes in the bloodstream and cerebrospinal fluid. Thus, the fact that the sodA mutant is more rapidly eliminated from the blood and brain than from the liver and spleen is consistent with its impaired ability to resist oxidative killing.
FIG. 8.
Mouse virulence assays. Growth of S. agalactiae NEM316 (▴), the sodA mutant, NEM1640 (■), or the complemented strain, NEM1641 (●) was monitored in the blood (A), brain (B), liver (C), and spleen (D) of mice inoculated i.v. with 106 bacteria. Means of bacterial counts in four organs per time point are shown (standard deviation, ≤0.25).
SodA contributes differently to the virulence of numerous bacteria. For gram-positive bacteria, a sodA mutant of S. pneumoniae was shown to be less virulent in a mouse intranasal infection model (49) whereas in Staphylococcus aureus, no difference was observed between a wild-type strain and a mutant strain in a mouse abscess model (13). For gram-negative bacteria, inactivation of sodA in Y. enterocolitica resulted in a marked reduction in virulence in an i.v. mouse infection model (34); in contrast, the virulence of a sodA mutant of S. enterica serovar typhimurium was only weakly attenuated in the same mouse infection model (45).
In conclusion, we have shown here that SodA plays a role in the pathogenicity of GBS. We have demonstrated that this enzyme is essential for the protection of S. agalactiae against oxidative stress and for survival in murine macrophages and is necessary to maintain a high level of bacteremia in a mouse infection model. Extension of these results to human pathogenesis will necessitate the use of an animal model that more closely simulates the neonatal disease (neonatal rat or mice model) and/or the use of human phagocytic cells such as alveolar macrophages or blood PMNs. In addition, it would be of obvious interest to study the contribution of SodA to the survival of GBS in human PMNs isolated from patients with chronic granulomatous disease, which are unable to mediate oxidative killing because of their defect in NADPH oxidase activity (27).
ACKNOWLEDGMENTS
We thank S. Nair, G. Milon, B. Deschamp, and F. Heffron for critical reading of the manuscript and for helpful discussions; P. Berche for his interest in this work and constant support; E. Eugène for technical help with confocal microscopy; and E. Schneider for fluorescence-activated cell sorter experiments.
M. Baptista was supported by Programa Praxis XXI of the fundaçao para a Ciênca e Tecnologia, Portugal. This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Pasteur Institute, and the University of Paris V.
REFERENCES
- 1.Antal J M, Cunningham J V, Goodrum K J. Opsonin-independent phagocytosis of group B streptococcus: role of complement receptor type three. Infect Immun. 1992;60:1114–1121. doi: 10.1128/iai.60.3.1114-1121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker C J, Edwards M S. Group B streptococcal infections. In: Remington J S, Klein J O, editors. Infectious diseases of the fetus and newborn infant. Philadelphia, Pa: The W. B. Saunders Co; 1990. pp. 743–811. [Google Scholar]
- 3.Bannister J V, Bannister W H, Rotilio G. Aspects of the structure, function and applications of superoxide dismutase. Crit Rev Biochem. 1987;22:111–180. doi: 10.3109/10409238709083738. [DOI] [PubMed] [Google Scholar]
- 4.Beaman L, Beaman B L. Monoclonal antibodies demonstrate that superoxide dismutase contributes to protection of Nocardia asteroides within the intact host. Infect Immun. 1992;58:3122–3128. doi: 10.1128/iai.58.9.3122-3128.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauchamp C, Fridovitch I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 6.Becker I D, Robinson O M, Bazan T S, Lopez-Osuna M, Kretschmer R R. Bactericidal capacity of newborn phagocytes against group B beta-hemolytic streptococci. Infect Immun. 1981;34:535–539. doi: 10.1128/iai.34.2.535-539.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benov L T, Fridovich I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J Biol Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 8.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 10.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celli J, Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol. 1998;28:103–117. doi: 10.1046/j.1365-2958.1998.00778.x. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Q, Carlson B, Pillai S, Eby R, Edwards L, Olmsted S B, Cleary P. Antibody against surface-bound C5a peptidase is opsonic and initiates macrophage killing of group B streptococci. Infect Immun. 2001;69:2302–2308. doi: 10.1128/IAI.69.4.2302-2308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements M O, Watson S P, Foster S J. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181:3898–3903. doi: 10.1128/jb.181.13.3898-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornacchione P, Scaringi L, Fettucciari K, Rosati E, Sabatini R, Orefici G, von Hunolstein C, Modesti A, Modica A, Minelli F, Marconi P. Group B streptococci persist inside macrophages. Immunology. 1998;93:86–95. doi: 10.1046/j.1365-2567.1998.00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz-Rodz A, Gilmore M S. High efficiency introduction of plasmid DNA into glycin treated Enterococcus faecalis. Mol Gen Genet. 1990;224:152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- 16.de Chastellier C, Berche P. Fate of Listeria monocytogenes in murine macrophages: evidence for stimulation killing and survival of intracellular bacteria. Infect Immun. 1994;62:543–553. doi: 10.1128/iai.62.2.543-553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards M S, Baker C J. Streptococcus agalactiae (group B Streptococcus) In: Mandell G L, Bennett J E, Dolin R, editors. Mandell, Douglas and Bennett's principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone, Inc.; 1995. pp. 1835–1845. [Google Scholar]
- 18.Falkow S, Isberg R R, Portnoy D A. The interaction of bacteria with mammalian cells. Annu Rev Cell Biol. 1992;8:333–363. doi: 10.1146/annurev.cb.08.110192.002001. [DOI] [PubMed] [Google Scholar]
- 19.Farr S B, D'Ari R, Touati D. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc Natl Acad Sci USA. 1986;83:8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzon V L, Arondel J, Sansonetti P J. Contribution of superoxide dismutase and catalase activities to Shigella flexneri pathogenesis. Infect Immun. 1990;58:529–535. doi: 10.1128/iai.58.2.529-535.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaillot O, Poyart C, Berche P, Trieu-Cuot P. Molecular characterization and expression analysis of the superoxide dismutase gene from Streptococcus agalactiae. Gene. 1997;204:213–218. doi: 10.1016/s0378-1119(97)00548-9. [DOI] [PubMed] [Google Scholar]
- 22.Halliwell B, Gutteridge J M. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 23.Hassan H M. Microbial superoxide dismutases. Adv Genet. 1989;26:65–97. doi: 10.1016/s0065-2660(08)60223-0. [DOI] [PubMed] [Google Scholar]
- 24.Keefe G P. Streptococcus agalactiae mastitis: a review. Can Vet J. 1997;38:429–437. [PMC free article] [PubMed] [Google Scholar]
- 25.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 26.Langford P R, Loynds B M, Kroll J S. Copper-zinc superoxide dismutase in Haemophilus species. J Gen Microbiol. 1992;138:517–522. doi: 10.1099/00221287-138-3-517. [DOI] [PubMed] [Google Scholar]
- 27.Meischl C, Roos D. The molecular basis of chronic granulomatous disease. Springer Semin Immunopathol. 1998;19:417–434. doi: 10.1007/BF00792600. [DOI] [PubMed] [Google Scholar]
- 28.Miller R A, Britigan B E. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama K. Nucleotide sequence of Streptococcus mutans superoxide dismutase gene and isolation of insertion mutants. J Bacteriol. 1992;174:4928–4934. doi: 10.1128/jb.174.15.4928-4934.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noel G J, Katz S L, Edelson P J. The role of C3 in mediating binding and ingestion of group B Streptococcus serotype III by murine macrophages. Pediatr Res. 1991;30:118–123. doi: 10.1203/00006450-199107000-00023. [DOI] [PubMed] [Google Scholar]
- 31.Pesci E C, Cottle D L, Pickett C L. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect Immun. 1994;62:2687–2694. doi: 10.1128/iai.62.7.2687-2694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poyart C, Berche P, Trieu-Cuot P. Characterization of superoxide dismutase genes from Gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett. 1995;131:41–45. doi: 10.1016/0378-1097(95)00232-t. [DOI] [PubMed] [Google Scholar]
- 33.Poyart-Salmeron C, Carlier C, Trieu-Cuot P, Courtieu A, Courvalin P. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet. 1990;335:1422–1426. doi: 10.1016/0140-6736(90)91447-i. [DOI] [PubMed] [Google Scholar]
- 34.Roggenkamp A, Bittner T, Leitritz L, Sing A, Heesemann J. Contribution of the Mn-cofactored superoxide dismutase (SodA) to the virulence of Yersinia enterocolitica serotype O8. Infect Immun. 1997;65:4705–4710. doi: 10.1128/iai.65.11.4705-4710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubens C E, Heggen L M, Haft R F, Wessels M R. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol Microbiol. 1993;8:843–855. doi: 10.1111/j.1365-2958.1993.tb01631.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanders J W, Leenhouts K J, Haandrikman A J, Venema G, Kok J. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol. 1995;177:5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuchat A. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin Microbiol Rev. 1998;11:497–513. doi: 10.1128/cmr.11.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal A W. The electron transport chain of the microbicidal oxidase of phagocytic cells and its involvement in the molecular pathology of chronic granulomatous disease. J Clin Investig. 1989;83:1785–1793. doi: 10.1172/JCI114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St. John G, Steinman H M. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi M A, Asada K. Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch Biochem Biophys. 1983;226:558–566. doi: 10.1016/0003-9861(83)90325-9. [DOI] [PubMed] [Google Scholar]
- 42.Thomas C, Smith C. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 43.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from Gram-positive bacteria. Gene. 1991;106:21–27. doi: 10.1016/0378-1119(91)90561-o. [DOI] [PubMed] [Google Scholar]
- 44.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 45.Tsolis R M, Baumler A J, Heffron F. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect Immun. 1995;63:1739–1744. doi: 10.1128/iai.63.5.1739-1744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valentin-Weigand P, Benkel P, Rohde M, Chhatwal G S. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect Immun. 1996;64:2467–2473. doi: 10.1128/iai.64.7.2467-2473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandamme P, Devriese L A, Pot B, Kersters K, Melin P. Streptococcus difficile is a nonhemolytic group B, type Ib Streptococcus. Int J Syst Bacteriol. 1997;47:81–85. doi: 10.1099/00207713-47-1-81. [DOI] [PubMed] [Google Scholar]
- 48.Wilks K E, Dunn K L, Farrant J L, Reddin K M, Gorringe A R, Langford P R, Kroll J S. Periplasmic superoxide dismutase in meningococcal pathogenicity. Infect Immun. 1998;66:213–217. doi: 10.1128/iai.66.1.213-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yesilkaya H, Kadioglu A, Gingles N, Alexander J E, Mitchell T J, Andrew P W. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect Immun. 2000;68:2819–2826. doi: 10.1128/iai.68.5.2819-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]