Abstract
Periodontitis is a chronic inflammatory disease affecting oral tissues. Oral epithelial cells represent the primary barrier against bacteria causing the disease. We examined the responses of such cells to an arginine-specific cysteine proteinase (RgpB) produced by a causative agent of periodontal disease, Porphyromonas gingivalis. This protease caused an intracellular calcium transient in an oral epithelial cell line (KB), which was dependent on its enzymatic activity. Since protease-activated receptors (PARs) might mediate such signaling, reverse transcription-PCR was used to characterize the range of these receptors expressed in the KB cells. The cells were found to express PAR-1, PAR-2, and PAR-3, but not PAR-4. In immunohistochemical studies, human gingival epithelial cells were found to express PAR-1, PAR-2, and PAR-3 on their surface, but not PAR-4, indicating that the cell line was an effective model for the in vivo situation. PAR-1 and PAR-2 expression was confirmed in intracellular calcium mobilization assays by treatment of the cells with the relevant receptor agonist peptides. Desensitization experiments strongly indicated that signaling of the effects of RgpB was occurring through PAR-1 and PAR-2. Studies with cells individually transfected with each of these two receptors confirmed that they were both activated by RgpB. Finally, it was shown that, in the oral epithelial cell line, PAR activation by the bacterial protease-stimulated secretion of interleukin-6. This induction of a powerful proinflammatory cytokine suggests a mechanism whereby cysteine proteases from P. gingivalis might mediate inflammatory events associated with periodontal disease on first contact with a primary barrier of cells.
Periodontal diseases include a group of infections affecting the tissues supporting the teeth. Mild clinical conditions involve inflammation of the gingiva, whereas more severe forms, known as periodontitis, are characterized by destruction of periodontal tissues and can lead to tooth loss (47). The major pathogens associated with periodontitis are Porphyromonas gingivalis, Bacteroides forsythus and Actinobacillus actinomycetemcomitans (14). Among these organisms, P. gingivalis, a gram-negative anaerobic bacterium, has been identified as a major etiological agent in the pathogenesis of adult periodontitis in humans (14, 49). It has also been recognized as a virulence agent that initiates progression of periodontitis in primate models of periodontal destruction (16).
In periodontal diseases, bacteria bind to the tooth surface and extend into the gingival sulcus, at the base of which, the gingival epithelium forms a thin junctional epithelium directly attached to the tooth. Plaque bacteria colonize and disrupt the attachment between the junctional epithelium and the tooth during the progression of periodontitis, causing the formation of a periodontal pocket. Thus, the cells of the junctional epithelium form an interface with the subgingival bacteria which is directly exposed to the bacteria and their products. The interaction that occurs between periodontal bacteria and epithelial cells, and the subsequent molecular signals that are generated are of great interest, as they may contribute to the overall effect of the bacteria on the host and the progression of periodontal disease. The periodontal pathogen P. gingivalis has been detected in gingival tissues in vivo (46), indicating that bacteria may pass through the epithelial cell barrier. Certain strains of P. gingivalis have also been found to invade (28) and even replicate (32) within epithelial cells in vitro.
The epithelial cells may contribute to the host's defense as a number of studies have shown that gastrointestinal and uroepithelial cells express proinflammatory cytokines following exposure to invasive and noninvasive bacteria (2, 12, 24, 44). Periodontal bacteria and their products can also elicit signals in epithelial cells and produce a cellular response. Bacteria common in normal oral flora such as Fusobacterium nucleatum, Eikenella corrodens, and A. actinomycetemcomitans were found to induce the production of interleukin-8 (IL-8) in gingival epithelial cells (15, 19, 60). P. gingivalis was found to inhibit IL-8 accumulation (10, 19), and it was determined that IL-8 down-regulation was dependent upon invasion of the epithelial cells by P. gingivalis (5). The invasion efficiency of P. gingivalis is relatively low, and immunohistochemical (IHC) studies have shown that P. gingivalis cells are in close contact with the epithelium in the periodontal pockets (36); therefore, it is of interest to examine whether this bacterium can affect epithelial cells without being inside the cells.
To understand whether a molecular dialogue between epithelial cells and P. gingivalis can occur, the bacterial products affecting extracellular components of the cells and thereby eliciting an intracellular signal leading to a cellular response need to be identified. Proteases produced by P. gingivalis have been shown to act as important pathogenic agents (53). Two types of cysteine proteases, responsible for the so-called trypsin-like activity of the bacterium, have been purified (40, 41): a lysine-specific protease, termed lys-gingipain (Kgp) and an arginine-specific protease referred to as arg-gingipain. The latter is present as three variants: 50-kDa RgpB, 50-kDa RgpAcat, and 95-kDa HRgpA. HRgpA is the high-molecular-mass form of RgpAcat, formed by RgpAcat noncovalently complexed with binding proteins identified as hemagglutinins (43).
RgpB has been found to cleave and activate human protease-activated receptor 2 (PAR-2) on human neutrophils (31). To date, four PARs have been identified: PAR-1, -2, -3, and -4 (reviewed in reference 7). Cleavage within the extracellular domain of a PAR produces a new N terminus that acts as a tethered ligand, binds to the body of the receptor, and leads to its activation. Synthetic peptide agonists corresponding to the tethered ligand of each receptor are able to activate the receptor, with the exception of PAR-3, allowing the cellular responses mediated by the different receptors to be distinguished and studied by treatment of cells with their respective agonist peptides. PAR-1 was the first receptor discovered and is cleaved and activated by thrombin (55), as are PAR-3 and PAR-4, which were discovered more recently (21, 25, 58). Trypsin and mast cell tryptase have been identified as PAR-2 activators (11). Gingival fibroblasts express PAR-1, and activation of this receptor stimulated IL-6 secretion (18). Human keratinocytes were found to express PAR-1 and PAR-2, and their activation induced IL-6, granulocyte-macrophage colony-stimulating factor, and IL-8 secretion (17, 56), indicating that proteases, acting through these receptors, can affect the course of physiological and pathological processes by stimulating the production of such proinflammatory cytokines. KB is an oral epithelial cell line that has been extensively used as a model to study gingival epithelial cells. The first aim of this study was to determine whether the model KB cells express PARs and whether human gingival epithelial cells in situ similarly express the receptors. We then examined if RgpB could cleave and activate these receptors on the KB cells, resulting in the induction of an intracellular signal, and whether the cellular response induced affects the secretion of cytokines by these cells.
MATERIALS AND METHODS
Materials.
KB cells were purchased from the American Type Culture Collection (Manassas, Va.). Chinese hamster ovary (CHO) cells and CHO cells stably expressing human PAR-2 (CHO PAR-2 cells) were a gift from Vanitha Ramakrishnan, COR Pharmaceuticals, San Francisco, Calif.
Fura-2 AM was obtained from Molecular Probes (Eugene, Oreg.). All tissue culture reagents were purchased from Gibco BRL Life Technologies (Melbourne, Australia). PAR-1-activating peptide (TRAP; SFFLRN), PAR-4-activating peptide (TRAP-4′; GYPGQV) and PAR-2-activating peptide (RAP; SLIGKV) were synthesized by Auspep (Parkville, Australia). Ready to Go U Prime Synthesis beads were purchased from Pharmacia (Sydney, Australia). Tissue culture media and medium supplements were obtained from Trace Biosciences and Gibco BRL (Melbourne, Australia). Enzyme-linked immunosorbent assay (ELISA) kits were purchased from Endogen (CSL Biosciences, Melbourne, Australia), and tetramethylbenzidine dihydrochloride was obtained from Sigma (Sydney, Australia). Polyclonal antibodies against PAR-3 and PAR-4 were purchased from Santa Cruz (Santa Cruz, Calif.). The avidin-biotin labeling system used for the IHC procedure was purchased from Vector Laboratories (Burlingame, Calif.). Target buffer for IHC and a monoclonal antibody to vimentin were purchased from Dako (Carpenteria, Calif.). All other materials were purchased from Sigma (Sydney, Australia).
IHC.
Human gingival tissues were obtained from Paul Farber of the Temple University Medical School, Philadelphia, Pa. Tissues were routinely fixed in 10% neutral buffered saline and routinely processed for paraffin embedding. Tissues were then sectioned (5 μm) onto SuperFrost Plus+ slides (Fisher, Pittsburgh, Pa.) and dried overnight. Slides were deparaffinized, hydrated, and processed for routine IHC as previously described (8). Briefly, slides were microwaved in Target buffer, cooled, placed in phosphate-buffered saline (PBS) (pH 7.4), and treated with 3% (vol/vol) H2O2 for 10 min. Slides were processed through an avidin-biotin blocking system according to the manufacturer's instructions (Vector Laboratories) and then placed in PBS. All subsequent reagent incubations and washes were performed at room temperature. Normal blocking serum was placed on all slides for 10 min. After being briefly rinsed in PBS, primary antibodies were placed on slides for 30 min. Polyclonal rabbit anti-human PAR-1 (6, 9) and PAR-2 (8, 9, 50) antibodies have been previously characterized, while polyclonal antibodies to PAR-3 and PAR-4 were purchased from Santa Cruz. The slides were washed, and biotinylated goat anti-rabbit secondary antibodies were placed on the tissue sections for 30 min. After being rinsed in PBS, the avidin-horseradish peroxidase-biotin complex reagent was added for 30 min. Slides were washed and treated twice with chromogen 3,3′-diaminobenzidine for 5 min each, then rinsed in distilled water, and counterstained with hematoxylin. A monoclonal antibody to vimentin (Dako), the widely conserved, ubiquitous, intracellular, filament protein, was utilized as a positive control to demonstrate tissue antigenicity and control reagent quality. The negative controls included replacement of the primary antibody with preimmune serum or with the same species of immunoglobulin G from nonimmune serum.
Purification of the bacterial proteases.
RgpB was purified to homogeneity according to Potempa et al. (41). The amount of active enzyme was determined by active site titration with Phe-Phe-Arg-chloromethylketone (42). RgpB was activated for cellular studies with 10 mM cysteine at 37°C for 10 min in 0.1 M Tris-HCl and 5 mM CaCl2, pH 7.4. Polymyxin B S04 (100 μg/ml) was routinely added to inhibit any cellular stimulation by bacterial lipopolysaccharides. RgpB was inactivated by treatment with 2 μM antipain for 10 min.
Cells and culture conditions.
The adherent cell lines KB, CHO, CHO PAR-2, N1LF, and N1LF PAR-1 were grown in a humidified atmosphere with 5% CO2 at 37°C in the appropriate media. KB cells were grown in Eagle's medium containing Earle's balanced salts and 5% (vol/vol) fetal calf serum (FCS). CHO cells were grown in Dulbecco's modified Eagle medium and Ham's F-12 medium (1:1) supplemented with 10% (vol/vol) FCS. The CHO PAR-2 cells were grown in the same medium as the CHO cells, supplemented with 40 U of G418/ml. N1LF cells are immortalized murine lung myofibroblasts derived from PAR-1-deficient mice that lack functional PAR-1, -2, and -4, and N1LF PAR-1 cells stably express human PAR-1 (3). N1LF cells were grown in Dulbecco's modified Eagle medium containing glucose (1,000 mg/liter), 4 mM l-glutamine, and 10% (vol/vol) heat-inactivated FCS. N1LF PAR-1 cells were grown in the same medium as the N1LF cells, with the addition of 200 μg of hygromycin B/ml. All growth media were supplemented with penicillin (100 U/ml) and streptomycin sulfate (100 μg/ml).
Purification of platelets from human blood.
Platelets were isolated from freshly drawn human blood. Venous blood was anticoagulated by adding 6 volumes of blood to 1 volume of acid-citrate-dextrose (85 mM sodium citrate, 111 mM dextrose, and 71 mM citric acid supplemented with prostacyclin (50 ng/ml) and apyrase (0.67 U/ml). Whole blood was centrifuged at 200 × g for 20 min at room temperature to obtain the supernatant platelet-rich plasma, which was then centrifuged at 730 × g for 10 min at room temperature to sediment the platelets. The platelet pellets were resuspended in CGS (13 mM trisodium citrate, 120 mM sodium chloride, and 30 mM dextrose [pH 7.0], containing 50 ng of prostacyclin/ml) and washed twice. Platelet preparations contained less than 0.1% leukocytes as determined by light microscopy.
Reverse transcription-PCR (RT-PCR) for the determination of expression of PARs.
RNA was isolated from KB cells (grown to 70% confluency) and washed human platelets with TRI reagent, according to the manufacturer's instructions. Total RNA (5 μg) was transcribed to cDNA in the presence of deoxyribosylthymine primer (40 pmol) and diethyl pyrocarbonate-treated Milli-Q H2O to a volume of 33 μl using Ready to Go U Prime Synthesis beads (Pharmacia) according to the manufacturer's instructions at 37°C for 1 h. The reaction mixture was then heated at 65°C for 5 min. The entire RT product was amplified by PCR in a final volume of 100 μl containing the appropriate forward and reverse primers (20 pmol for PAR-2, -3, and -4 primers and 100 pmol for PAR-1 primers; see below) and 2.5 U of Taq polymerase (MBI Fermentas). A control reaction mixture, containing RNA from KB cells or platelets in a reaction mixture that had not undergone RT analysis, was used for the PAR-1 primers as they did not span an intron.
For amplification of PAR-2, PAR-3, and PAR-4 products, PCR was performed for 36 cycles. The first cycle included a denaturation step for 5 min at 95°C. Cycles 2 to 36 had a denaturation step of 1 min at 95°C, 1 min of annealing at 55°C, and 3 min of elongation at 72°C. For PAR-1 amplification, PCR was performed for 34 cycles. The first cycle included a denaturation step for 2 min at 95°C. Cycles 2 to 34 had a denaturation step of 1 min at 95°C, 1 min of annealing at 55°C, and 1 min of elongation at 72°C.
Oligonucleotides for PCR analysis of receptors. (i) PAR-1.
The sense primer was TGTGAACTGATCATGTTTATG. The antisense primer was TTCGTAAGATAAGAGATATGT (55). The resulting PCR product spanned positions 2421 to 3129 (708 bp).
(ii) PAR-2.
The sense primer was GCAGCCTCTCTCTCCTGCAGTGG; the antisense primer was CTTGCATCTGCTTTACAGTGCG (51). The resulting PCR product spanned positions 48 to 1114 (1,066 bp).
(iii) PAR-3.
The sense primer was ATAACGTTTAAGAGACGGGACT; the antisense primer was TAGCAGTAGATGATAAGCACA (48). The resulting PCR product spanned positions 111 to 969 (858 bp).
(iv) PAR-4.
The sense primer was GACGAGAGCGGGAGCACC; the antisense primer was CCCGTAGCACAGCAGCATGG (58). The resulting PCR product spanned positions 195 to 970 (725 bp).
DNA products and molecular weight markers SPP1-EcoRI were separated in a 2% agarose gel, following which the gels were stained with ethidium bromide (5 μg/ml) and visualized under UV light.
Intracellular calcium measurement.
Intracellular calcium levels were measured in KB, CHO, CHO PAR-2, N1LF, and N1LF PAR-1 cells. The cells were grown to 80% confluence and detached from the culture dishes by treatment with nonenzymatic dissociation solution. The cells were prepared for intracellular calcium ion ([Ca2+]i) measurements as described previously (4). Cells were washed and resuspended at 6 × 106 cells/ml in an extracellular medium (EM). The EM consisted of 121 mM NaCl, 5.4 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 6 mM NaHCO3, 5.5 mM glucose, 25 mM HEPES, and 0.1% (wt/vol) bovine serum albumin (pH 7.3). In all subsequent steps, the cells were protected from light.
Cells were loaded with 1 μM Fura-2 AM by occasional shaking for 30 min at room temperature. After centrifugation at 200 × g for 5 min, the cells were resuspended in EM and incubated for 30 min at room temperature with occasional shaking to allow hydrolysis of the intracellular Fura-2 AM and then centrifuged (200 × g for 5 min). The cells were resuspended in EM without bovine serum albumin at 2 × 106 cells/ml for fluorescence measurements. [Ca2+]i was determined using a Perkin-Elmer LS-50B fluorimeter by measuring Fura-2 fluorescence at excitation and emission wavelengths of 340 or 380 and 510 nm, respectively. Loaded cells were maintained at 37°C in stirred plastic cuvettes throughout the experiment. After a stable baseline was established, the agonist was added to cells, and the ratio of fluorescence at the two excitation wavelengths was measured, which is proportional to [Ca2+]i.
ELISA for the expression of IL-6, TNF-α, and IFN-γ by KB cells.
KB cells were grown in six-well plates until 70% confluence and then treated for varying times (as indicated in Results) with RAP, TRAP, thrombin, trypsin, RgpB, or cysteine buffer in medium lacking FCS. Following treatment, the cells were washed with medium and grown in medium containing 5% (vol/vol) FCS for 24 h. The supernatant from the KB cells was then removed, centrifuged, aliquoted, and stored at −80°C. After the medium was harvested, the cells were removed and live cells were counted by the trypan blue exclusion method. At the time of analysis, supernatants were thawed and IL-6, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) concentrations were measured with ELISA kits (Endogen) at room temperature, according to the manufacturer's instructions.
RESULTS
KB cells express PAR-1, -2, and -3.
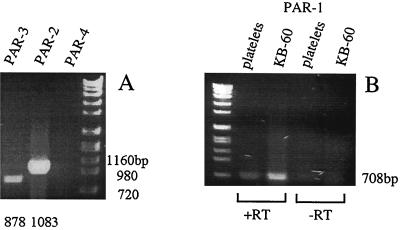
RT-PCR analysis of RNA extracted from KB cells revealed the presence of PAR-1, -2, and -3 mRNA (Fig. 1). Since the PAR-1 primers did not span an intron, additional controls were employed to verify the results for this receptor. RNA isolated from platelets, known to express PAR-1, was used as a positive control to verify the expression of this receptor in KB cells. PAR-1 expression was also confirmed by the fact that only RNA from KB cells that had undergone RT resulted in the appearance of the expected PAR-1 cDNA product (Fig. 1B). Although competitive RT-PCR would be required to determine the quantity of each PAR mRNA in KB cells, based on the intensity of the PAR-2 product, it appears that PAR-2 is expressed at a higher level than PAR-1 and PAR-3. Based on the expression of both PAR-1 and PAR-2 by KB cells, it is apparent that they are able to serve as an effective model for gingival epithelial cells for the studies carried out here.
FIG. 1.
Expression of mRNAs encoding PARs in KB cells. RNA isolated from KB cells or platelets was analyzed for PAR transcripts by RT-PCR as described in Materials and Methods. Since the PAR-1 primers did not span an intron, RNA from platelets was used as a positive control for expression of PAR-1; RNA which had not undergone RT (−RT) was used as a negative control in relation to RNA which had (+RT). (A) KB cells express PAR-2 and PAR-3 (the sizes of the expected products of RT-PCR are shown below the lanes, in comparison to the sizes of selected markers to the right). (B) KB cells and platelets express PAR-1 (the size of the expected product for PAR-1 is shown to the right of panel B).
Human gingival epithelial cells express PAR-1 and PAR-2.
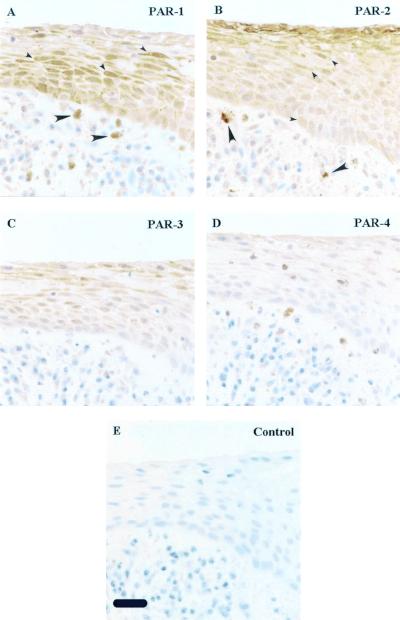
In sections of human gingival tissue, epithelial cells stained strongly for PAR-1 and PAR-2, less strongly for PAR-3, and weakly or not at all for PAR-4 (Fig. 2). Both PAR-1 and PAR-2 are readily apparent in the epithelial layer (Fig. 2). PAR-1 protein expression appears to wane as the cells differentiate to the squamous layer (Fig. 2A), whereas the PAR-2 expression is in contrast to that of PAR-1, the labeling pattern increasing to a maximum in the squamous layer (Fig. 2B). PAR-3 staining is more similar to PAR-1 in distribution, although generally much weaker (Fig. 2C). This indicates that both receptors are strongly expressed in the gingival epithelial cells, although PAR-2 might be expected be the receptor most likely to make first contact with bacteria binding to oral epithelium. Further interesting observations are of PAR-1 and PAR-2 expression in the dermal mast cells and macrophages (Fig. 2) seen in the subendothelial connective tissue. It was previously reported that PAR-1 and PAR-2 are expressed on these cells (1, 9).
FIG. 2.
Expression of PAR-1, -2, -3, and -4 by human gingival epithelium. Serial sections of human gingival tissue were immunolocalized with rabbit polyclonal antibodies against the receptors indicated in the upper right of each picture: PAR-1 (A), PAR-2 (B), PAR-3 (C), PAR-4 (D), and nonimmune rabbit immunoglobulin G (E), using the avidin-horseradish peroxidase-biotin complex system and a hematoxylin counterstain. There was specific staining for both PAR-1 and PAR-2 throughout the gingival epithelium; some examples of positive cells are indicated by small arrowheads (A and B). Macrophages and mast cells scattered in the subepithelial connective tissue were also positive for PAR-1 and PAR-2 (A and B, large arrowheads). Bar (panel E), 25 μm.
RgpB increased [Ca2+]i in KB cells.
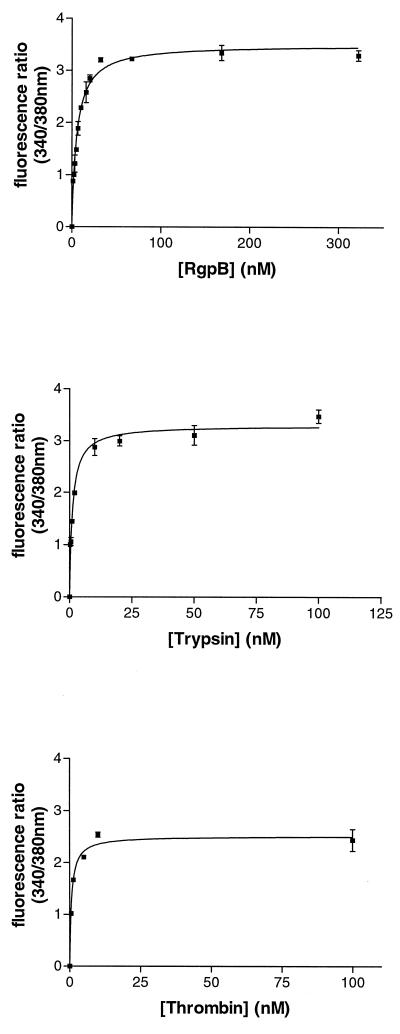
KB cells were tested for a [Ca2+]i response to trypsin, thrombin, and RgpB, and all proteases were found to induce a concentration-dependent increase in [Ca2+]i (Fig. 3). These data yield an enzyme concentration inducing the half-maximal response (50% effective concentration, or EC50) of 0.94, 1.77, and 5.52 nM for thrombin, trypsin, and RgpB, respectively. Trypsin and RgpB appear to elicit a higher maximal mobilization of calcium than thrombin (Fig. 3). Pretreatment of cells with phorbol myristate acetate (an inhibitor of protein kinase C) inhibited the calcium mobilization induced by trypsin, thrombin, or RgpB (data not shown).
FIG. 3.
The [Ca2+]i response of KB cells to different concentrations of RgpB, trypsin, and thrombin. Each data point represents the mean from two traces similar to those shown in the figure.
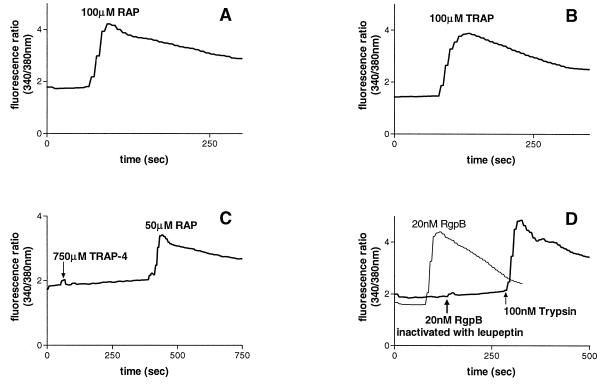
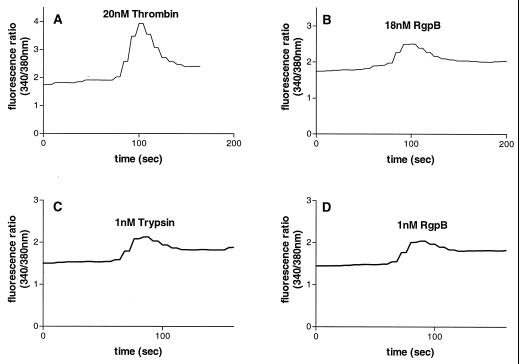
Thrombin is a known activator of PAR-1, -3, and -4 (21, 55, 58) and trypsin can activate PAR-1, -2, and -4 (37, 39, 54, 55, 58). KB cells were found to express PAR-1, -2, and -3; thus, cleavage and activation of these receptors probably mediate the calcium mobilization induced by trypsin and thrombin. In support of this hypothesis is the finding that treatment of KB cells with RAP or TRAP (Fig. 4A and B), but not TRAP-4 (Fig. 4C), caused an increase in [Ca2+]i, verifying the expression of functional PAR-1 and PAR-2 on KB cells and the absence of PAR-4. RgpB, which had been inactivated by antipain, did not induce a calcium response in KB cells (Fig. 4D), indicating that the [Ca2+]i increase induced is due to the proteolytic activity of the enzyme.
FIG. 4.
The [Ca2+]i responses in KB cells to 100 μM RAP (A), 100 μM TRAP (B), 750 μM TRAP-4 (C), and 20 nM RgpB inactivated with antipain, followed by 100 μM trypsin to demonstrate that the cells were capable of mobilizing [Ca2+]i, or 20 nM RgpB alone (D).
The question then addressed was whether the increase in [Ca2+]i induced by RgpB activity is due to cleavage of a PAR on the surface of KB cells. Once a PAR is cleaved by a protease, it cannot be activated a second time by the same or another protease in a short period of time. Desensitization of [Ca2+]i responses to subsequent protease challenges by trypsin, thrombin, and RgpB were therefore examined.
Desensitization profiles of thrombin and trypsin, which are known PAR activators, were first examined. Treatment of KB cells with 100 nM trypsin reduced a subsequent response to 100 nM thrombin by approximately 90% (Fig. 5A), whereas exposure of KB cells to 100 nM thrombin reduced a secondary response to 100 nM trypsin by approximately 20% (Fig. 5B). Pretreatment of KB cells with trypsin can cause activation of PAR-1 and PAR-2, whereas PAR-3 can still be activated by thrombin. It would be expected that exposure of the cells to thrombin causes cleavage of PAR-1 and PAR-3 but not PAR-2, which can then be activated by addition of trypsin. The fact that trypsin desensitizes a subsequent response to thrombin much more efficiently than thrombin desensitizes a secondary response to trypsin suggests that the PAR-2 expression level on KB cells is higher than that of PAR-1 and PAR-3. This suggestion is also supported by the observation that maximal [Ca2+]i responses to trypsin are greater than those to thrombin.
FIG. 5.
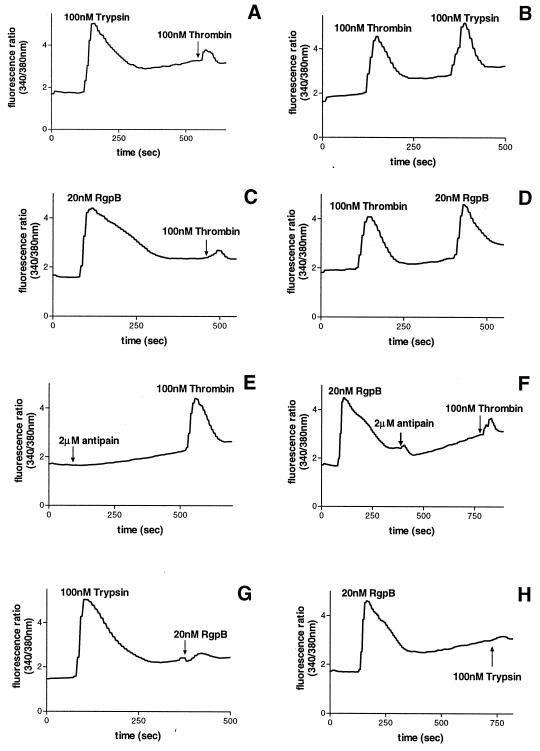
The [Ca2+]i responses in KB cells to 100 nM trypsin followed by 100 nM thrombin (A), 100 nM thrombin followed by 100 nM trypsin (B), 20 nM RgpB followed by 100 nM thrombin (C), 100 nM thrombin followed by 20 nM RgpB (D), 2 μM antipain followed by 100 nM thrombin (E), 20 nM RgpB followed by 2 μM antipain and then 100 nM thrombin (F), 100 nM trypsin followed by 20 nM RgpB (G), and 20 nM RgpB followed by 100 nM trypsin (H).
Similarly, treatment of KB cells with 20 nM RgpB reduced a subsequent response to 100 nM thrombin by approximately 90% (Fig. 5C), whereas exposure of KB cells to 100 nM thrombin reduced a secondary response to 20 nM RgpB only by approximately 10% (Fig. 5D). It was shown that the desensitization of the responses was not due to RgpB inactivating thrombin. This is illustrated by the use of antipain at a concentration known to inhibit RgpB effectively without affecting thrombin (Fig. 5E and F). When added to the cell suspension after the initial activation with RgpB, antipain did not affect the desensitization obtained with thrombin (Fig. 5F). RgpB therefore exhibited a similar ability to trypsin in efficiently desensitizing subsequent thrombin responses.
When KB cells were initially activated with 100 nM trypsin, a subsequent response to 100 nM trypsin was eliminated, and similarly following treatment with 20 nM RgpB, a secondary response to this enzyme was abolished (data not shown). Addition of 100 nM trypsin decreased a subsequent challenge with 20 nM RgpB by 90% (Fig. 5G) and treatment with 20 nM RgpB abolished a secondary response to 100 nM trypsin (Fig. 5H). In each case the cells could still respond to ADP or bradykinin to the same extent as cells that had not been exposed to proteases (data not shown), indicating that the cells were capable of responding to other agonists. Therefore, RgpB effectively desensitizes the calcium mobilization induced by trypsin and vice versa.
These desensitization studies suggest that RgpB and trypsin activate a common receptor(s) on KB cells. Trypsin can activate PAR-1 and PAR-2 on the surface of KB cells by cleaving an arginine residue in their extracellular domains. Since RgpB has a high specificity of cleavage limited to peptide bonds after arginine residues, the calcium response observed is probably due to activation of PAR-1 and PAR-2 by this bacterial enzyme.
RgpB activates cells stably expressing human PAR-1.
To demonstrate that RgpB can cleave and activate human PAR-1, the calcium response elicited by this enzyme was investigated in N1LF PAR-1 cells in comparison to nontransfected N1LF cells (3). Treatment of N1LF PAR-1 cells with 150 μM TRAP (data not shown) or 20 nM thrombin (Fig. 6A) elicited an increase in [Ca2+]i, while nontransfected N1LF cells did not respond to TRAP or to 20 nM thrombin (data not shown). Similarly, trypsin elicited calcium mobilization in N1LF PAR-1 cells but not in control nontransfected N1LF cells (data not shown). RgpB at concentrations as low as 2 nM elicited a Ca2+ increase in N1LF PAR-1 cells, while up to 100 nM RgpB did not induce a calcium response in nontransfected N1LF cells; Fig. 6B illustrates the response of these cells to 18 nM RgpB, followed by bradykinin, to demonstrate that the cells were capable of mobilizing [Ca2+]i.
FIG. 6.
Calcium mobilization in N1LF PAR-1 cells (A and B) or CHO PAR-2 cells (C and D) treated with 20 nM thrombin (A), 18 nM RgpB (B), 1 nM trypsin (C), or 1 nM RgpB (D). The traces are representative of three observed.
RgpB activates cells stably expressing human PAR-2.
We have previously shown that RgpB can activate PAR-2 on human neutrophils (31). The ability of RgpB to activate human PAR-2 was further verified by the fact that the RgpB elicited a calcium response in CHO PAR-2 cells in comparison to nontransfected CHO cells. Treatment of CHO PAR-2 cells with 200 μM RAP (data not shown) or 1 nM trypsin (Fig. 6C) elicited an increase in [Ca2+]i, while nontransfected CHO cells did not respond to RAP or to 1 nM trypsin (data not shown). Concentrations of RgpB as low as 1 nM elicited an increase in [Ca2+]i in CHO PAR-2 cells (Fig. 6D) but did not induce a calcium response in nontransfected CHO cells (data not shown), verifying that RgpB can activate human PAR-2.
Treatment of KB cells with RgpB causes an increase in secretion of IL-6.
Human keratinocytes have been found to express PAR-1 and PAR-2 and their activation with their respective agonist peptides or trypsin, and thrombin was found to induce IL-6 up-regulation (56). We investigated whether treatment with RAP, TRAP, trypsin, thrombin, or RgpB could increase IL-6, TNF-α, and IFN-γ secretion levels in KB cells.
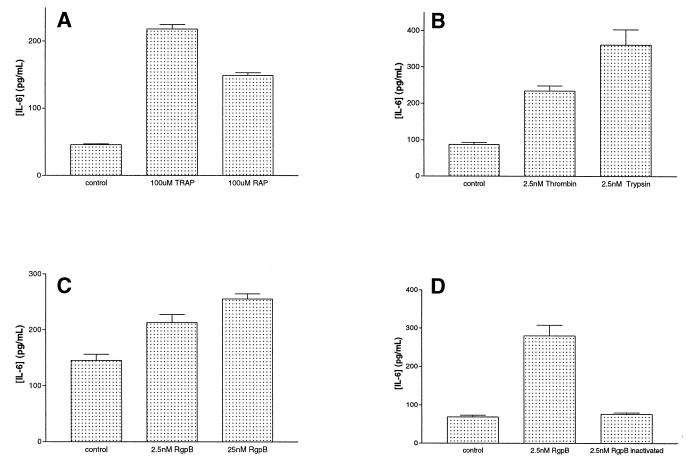
KB cells were stimulated with RAP or TRAP (100 μM) for up to 1 h, and both peptides caused an increase in IL-6 secretion. Preliminary studies showed that maximal concentrations of IL-6 were produced in the supernatant after treatment with RAP or TRAP for 1 h, and this time point was used for further analysis. It was found that TRAP (100 μM) stimulated IL-6 production at levels that were higher than those induced by RAP (100 μM) (Fig. 7A). IL-6 levels in supernatant from cells treated with RAP were found to be 2.5-fold higher than those measured in supernatants from control (nontreated) KB cells, whereas in supernatant from cells treated with TRAP, IL-6 secretion was 4-fold higher than that from untreated cells. RAP activates PAR-2, whereas TRAP activates both PAR-1 and PAR-2, and thus TRAP, by acting through both receptors, may elicit a higher production of IL-6 than RAP.
FIG. 7.
Measurement of IL-6 concentrations by ELISA in culture supernatants from KB cells following treatment for 1 h (A through C) or 15 min (D) with 100 μM TRAP or RAP (A), 2.5 nM thrombin or trypsin (B), 2.5 nM RgpB or 25 nM RgpB (C), or 2.5 nM RgpB or 2.5 nMRgpB inactivated with antipain (D). The results shown in panels A, B, and D represent the mean ± standard error of the mean obtained from three experiments and those shown in panel C are results obtained from five experiments. All results were significantly different (P < 0.001).
KB cells were treated with various concentrations of trypsin or thrombin for up to 1 h, and both proteases induced IL-6 secretion. Treatment of KB cells with inactivated trypsin or thrombin yielded levels of IL-6 secretion that were identical to those of the untreated cells; thus, the cytokine increase observed was dependent on the proteolytic activity of the enzymes.
Treatment of cells for 1 h with trypsin (2.5 nM) was found to cause a higher level of production of IL-6 than with thrombin (2.5 nM) (Fig. 7B). Thrombin and trypsin were found to cause a 2.5- and 3.5-fold increase in IL-6 secretion, respectively, compared to that in untreated cells (Fig. 7B). This is possibly due to the fact that trypsin can activate both PAR-1 and PAR-2 (55, 54, 39, 37), whereas thrombin only activates PAR-1 and the small amount of PAR-3 present.
Treatment of KB cells with RgpB (2.5 nM) for 1 h induced a threefold increase in IL-6 secretion compared to that in untreated cells (Fig. 7C). RgpB that had been inactivated by antipain did not cause an increase in IL-6 production (Fig. 7C), indicating that the IL-6 up-regulation is due to the proteolytic activity of RgpB. Treatment of the cells for a shorter time period (15 min) was required to show the dependence of the increase in IL-6 secretion on the concentration of the protease used (Fig. 7D). Following treatment of KB cells with thrombin, trypsin, or RgpB (2.5 nM) for 1 h, 95% of cells were viable as assessed by trypan blue exclusion. No changes in cell morphology or cell detachment were observed at the concentrations and incubation times used.
TNF-α and IFN-γ levels were measured in supernatants from KB cells following treatment with trypsin, thrombin, or RgpB (2.5 nM) for up to 1 h and were found to be the same as those from untreated control cells. This indicates that the increases in IL-6 levels obtained are not simply due to elevation in cytokine levels generally but are rather a reasonably specific phenomenon.
DISCUSSION
Chronic marginal gingivitis is characterized by gingival inflammation, bleeding, changes in the contour of the gingiva, and increased flow of gingival crevicular fluid (GCF). Hallmarks of periodontitis, which is a progression of gingivitis, are the loss of gingival connective tissue attachment to the teeth, gum regression, and loss of the periodontal ligament, which attaches the tooth to the surrounding alveolar bone. These events lead to alveolar bone resorption and tooth loss. Refractory periodontitis is characterized by its relative resistance to repeated therapeutic procedures aimed at controlling the progression of periodontal tissue destruction.
The gingival epithelium is directly exposed to periodontal bacteria and their products and, by receiving and transmitting signals, plays an important role in the overall dialogue that occurs between pathogens and the host. In this study we showed that PAR-1, -2, and -3, which elicit a number of cellular responses, are expressed in human epidermoid KB cells and that PAR-1, -2, and -3 are also expressed on the surface of human gingival epithelial cells in situ. We then investigated whether an arginine-specific protease from the bacterium P. gingivalis, RgpB, interacted with these receptors. RgpB induced an increase in [Ca2+]i in human KB cells which was dependent upon its proteolytic activity. Several findings strongly support the hypothesis that this increase is mediated by activation of PAR-1 and PAR-2. Treatment of KB cells with RgpB desensitized the [Ca2+]i response to a second challenge with the same enzyme, a phenomenon that is in agreement with the rapid desensitization that PARs undergo after a short period of activation with a protease. Desensitization studies carried out with trypsin showed that when KB cells were activated with this enzyme, a secondary challenge with RgpB was eliminated. Similarly, exposure of KB cells to the bacterial enzyme desensitized the response to a second challenge with trypsin, suggesting that both enzymes activate the same receptors on KB cells.
Pretreatment of KB cells with trypsin drastically reduced a subsequent challenge with thrombin, whereas treatment of the cells with thrombin reduced a subsequent response to trypsin by approximately 10%. Trypsin is known to activate both PAR-1 and PAR-2 (37, 39, 54, 55), while thrombin activates PAR-1 and PAR-3. PAR-1 and PAR-2 are expressed on the surface of KB cells; thus, trypsin would be expected to cleave both PAR-1 and PAR-2 on these cells and thus dramatically reduce a subsequent challenge to thrombin, since only PAR-3 would be available for thrombin cleavage. The finding that the response to thrombin was minimal, subsequent to trypsin treatment, indicates that PAR-1 mediates the majority of the calcium mobilization induced by thrombin and that PAR-3 represents a minimal portion, if any, of the thrombin response. Similarly, O'Brien et al. (38) detected PAR-1, -2, and -3 mRNA, but not PAR-4 mRNA, in human umbilical vein endothelial cells and found that thrombin-induced inositol accumulation in human umbilical vein endothelial cells was reduced by 93% in the presence of PAR-1-blocking antibodies, making a contribution from PAR-3 appear minimal, if not unlikely. O'Brien et al. (38) postulated that PAR-3 might actually be unable to mediate a thrombin response on its own in the absence of PAR-4, a suggestion which has subsequently been supported by work with mouse platelets (35).
PAR-2 expression appears to be higher than PAR-1 expression; therefore, pretreatment with thrombin would result in cleavage of PAR-1 (and PAR-3) alone, leaving PAR-2 intact and available for activation by a secondary treatment with trypsin. The fact that thrombin reduced a secondary response to trypsin to a lesser degree than the desensitization caused by trypsin supports the suggestion that the majority of trypsin-induced calcium mobilization is induced by activation of PAR-2, with only a small percentage of the response due to PAR-1 activation. Like trypsin, RgpB efficiently desensitized subsequent responses to thrombin, but thrombin did not efficiently desensitize responses to RgpB, indicating that RgpB, like trypsin, induces the bulk of its response in KB cells via PAR-2. The suggestion that the majority of RgpB's effects occur through PAR-2 is interesting in the light of the different distribution of PAR-2 and PAR-1 observed in staining of sections of the human gingiva. PAR-2 was predominantly expressed by the superficial cells of the squamous epithelium, while PAR-1 expression appeared to fall off towards this layer. This indicates that not only is RgpB able to activate PAR-2 strongly, but it will also contact PAR-2 more readily in the in vivo situation, making the interaction between these two molecules highly relevant to the process of bacterial pathogenesis and subsequent host response.
Both PAR-1 and PAR-2, expressed on the surface of KB cells, are activated following cleavage after a specific arginine residue in their extracellular domain. RgpB, which is absolutely specific for hydrolysis at Arg-Xaa sites, was found to activate both PAR-1 and PAR-2 in transfected cells stably expressing these receptors. Based on the desensitization studies and the ability of RgpB to activate PAR-1 and PAR-2 on transfected cells, it can be concluded that the bacterial protease activates these receptors on the surface of KB cells, resulting in calcium mobilization. The same observations were made using the high-molecular-weight variant HRgpA (data not shown). It should be noted that, due to the specificity of these proteinases for cleavage after arginine residues, they are unable to activate PAR-3, which has a lysine residue at the cleavage point required to activate the receptor.
Activation of PAR-1 and PAR-2 on KB cells by their respective agonist peptides was found to cause an increase in IL-6 secretion. Similarly, treatment of KB cells with either thrombin or trypsin resulted in IL-6 up-regulation, which was due to the proteolytic activity of the enzymes. Treatment of KB cells with TRAP, which activates both PAR-1 and PAR-2, caused a higher increase in IL-6 levels than with RAP, which activates only PAR-2. Similarly, trypsin, which cleaves both PAR-1 and PAR-2, resulted in a greater stimulation of IL-6 expression than thrombin, which activates PAR-1 alone. Exposure of KB cells to RgpB also resulted in an increase in IL-6 secretion, which was found to be dependent upon the proteolytic activity of the enzyme. RgpB also induced a higher level of stimulation of IL-6 than thrombin, presumably due to the fact that it activates both PAR-1 and PAR-2, like trypsin. The fact that the bacterial enzyme did not induce IL-6 stimulation to the extent seen with trypsin, however, may be due to the fact that the concentration of RgpB used (2.5 nM) was lower than the EC50 of the enzyme for receptor activation in the KB cells (5.5 nM), while trypsin has a lower EC50 (1.8 nM) and thus would be expected to induce a greater response at a concentration of 2.5 nM. During the period of treatment no changes in cell shape or attachment were observed. The up-regulation of IL-6 secretion from KB cells following treatment with RgpB is of particular interest, as IL-6 is a proinflammatory cytokine associated with periodontal disease.
Cytokines are molecules released by cells in their local environment and have numerous properties, including chemoattraction of inflammatory cells. Cytokines that are readily found in periodontium and GCF of patients with periodontal disease are IL-6, IL-8, IL-1, TNF-α, and prostaglandin E2 (47). It has been suggested that these cytokines play significant roles in the pathogenesis of periodontitis (26, 57). In the majority of investigations carried out, IL-6 expression was found to be higher at sites of periodontal inflammation (20). IL-6 levels were found to be increased in diseased gingiva from patients with periodontitis compared to gingiva from periodontally healthy subjects (5, 52). Compared to normal tissue, IL-6 expression appears to be elevated in inflamed tissue from periodontitis sites and is highest in gingivitis sites (33, 59). Studies have indicated that IL-6 levels are higher in GCF from refractory periodontitis patients and may also correlate with progression of disease in these patients (13, 29, 45). IL-6 stimulates plasma cell proliferation and therefore may promote the presence of plasma cells that are readily found in periodontitis lesions (47). IL-6 is secreted by human osteoblasts in response to bone-resorbing agents (30), promotes bone resorption (22), and acts as a potent inducer of osteoclast formation in vitro (27, 34). Thus, IL-6 may contribute to the bone resorption associated with periodontal disease (47).
We found that concentrations of RgpB as low as 1 nM were sufficient to induce an increase in [Ca2+]i in KB cells, indicating that RgpB is very efficient in eliciting an intracellular signal by activating the PARs. Following contact with epithelial cells, invading P. gingivalis bacterial cells cause a transient increase in [Ca2+]i in these cells (23); thus, it may be of interest to determine whether PAR activation by the bacterium's proteases is involved in this process.
The findings presented here provide evidence that PAR-1 and PAR-2 expressed on the surface of epidermoid cells can be activated by the arginine-specific bacterial protease, RgpB. Cleavage of the PARs and the resultant up-regulation of IL-6 secretion by the bacterial protease are likely to contribute to the local inflammatory reaction within the pathological periodontal pocket and the propagation of the chronic inflammatory condition present in periodontal diseases.
ACKNOWLEDGMENTS
We express our gratitude for the excellent histological and immunohistochemical expertise provided by Patti A. Reiser, Brenda M. Hertzog, Norah A. Gumula, and Debbie Polkovich.
We acknowledge the support of the National Health and Medical Research Council (Australia), grant 990199 (to R.N.P.); the Committee of Scientific Research (KBN, Poland), grant 6 PO4A 047 17 (to J.P.); and the National Institutes of Health, grant DE 09761 (to J.T.).
REFERENCES
- 1.Abraham L A, Jenkins A L, Stone S R, Mackie E J. Expression of the thrombin receptor in developing bone and associated tissues. J Bone Miner Res. 1998;13:818–827. doi: 10.1359/jbmr.1998.13.5.818. [DOI] [PubMed] [Google Scholar]
- 2.Agace W, Hedges S, Andersson U, Andersson J, Ceska M, Svanborg C. Selective cytokine production by epithelial cells following exposure to Escherichia coli. Infect Immun. 1993;61:602–609. doi: 10.1128/iai.61.2.602-609.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade-Gordon P, Maryanoff B E, Derian C K, Zhang H C, Addo M F, Darrow A L, Eckardt A J, Hoekstra W J, McComsey D F, Oksenberg D, Reynolds E E, Santulli R J, Scarborough R M, Smith C E, White K B. Design, synthesis, and biological characterization of a peptide-mimetic antagonist for a tethered-ligand receptor. Proc Natl Acad Sci USA. 1999;96:12257–12262. doi: 10.1073/pnas.96.22.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bootman M D, Taylor C W, Berridge M J. The thiol reagent, thimerosal, evokes Ca2+ spikes in HeLa cells by sensitizing the inositol 1,4,5-trisphosphate receptor. J Biol Chem. 1992;267:25113–25119. [PubMed] [Google Scholar]
- 5.Chen C C, Chang K L, Huang J F, Huang J S, Tsai C C. Correlation of interleukin-1 beta, interleukin-6, and periodontitis. Kaohsiung J Med Sci. 1997;13:609–617. [PubMed] [Google Scholar]
- 6.Cheung W-M, D'Andrea M R, Andrade-Gordon P, Damiano B P. Altered vascular injury responses in mice deficient in protease-activated receptor-1. Arterioscler Thromb Vasc Biol. 1999;19:3014–3024. doi: 10.1161/01.atv.19.12.3014. [DOI] [PubMed] [Google Scholar]
- 7.Coughlin S R. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 8.D'Andrea M R, Derian C K, Leturcq D, Baker S M, Brunmark A, Ling P, Darrow A L, Santulli R J, Brass L F, Andrade-Gordon P. Characterization of protease activated receptor (PAR-2) immunoreactivity in normal human tissues. J Histochem Cytochem. 1998;46:1–8. doi: 10.1177/002215549804600204. [DOI] [PubMed] [Google Scholar]
- 9.D'Andrea M R, Rogahn C J, Andrade-Gordon P. Localization of protease-activated receptors -1 and -2 in human mast cells: indications for mast cell amplification cascade. Biotech Histochem. 2000;75:85–90. doi: 10.3109/10520290009064152. [DOI] [PubMed] [Google Scholar]
- 10.Darveau R P, Belton C M, Reife R A, Lamont R J. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dery O, Corvera C U, Steinhoff M, Bunnett N W. Proteinase-activated receptors: novel mechanisms of signaling by serine proteases. Am J Physiol. 1998;274:C1429–C1452. doi: 10.1152/ajpcell.1998.274.6.C1429. [DOI] [PubMed] [Google Scholar]
- 12.Eckmann L, Kagnoff M F, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geivelis M, Turner D W, Pederson E D, Lamberts B L. Measurements of interleukin-6 in gingival crevicular fluid from adults with destructive periodontal disease. J Periodontol. 1993;64:980–983. doi: 10.1902/jop.1993.64.10.980. [DOI] [PubMed] [Google Scholar]
- 14.Haffajee A D, Socransky S S. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 15.Han Y W, Shi W, Huang G T, Kinder Haake S, Park N H, Kuramitsu H, Genco R J. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt S C, Ebersole J, Felton J, Brunsvold M, Kornman K S. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- 17.Hou L, Kapas S, Cruchley A T, Macey M G, Harriott P, Chinni C, Stone S R, Howells G L. Immunolocalization of protease-activated receptor-2 in skin: receptor activation stimulates interleukin-8 secretion by keratinocytes in vitro. Immunology. 1998;94:356–362. doi: 10.1046/j.1365-2567.1998.00528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou L, Ravenall S, Macey M G, Harriott P, Kapas S, Howells G L. Protease-activated receptors and their role in IL-6 and NF-IL-6 expression in human gingival fibroblasts. J Periodontal Res. 1998;33:205–211. doi: 10.1111/j.1600-0765.1998.tb02192.x. [DOI] [PubMed] [Google Scholar]
- 19.Huang G T, Haake S K, Kim J W, Park N H. Differential expression of interleukin-8 and intercellular adhesion molecule-1 by human gingival epithelial cells in response to Actinobacillus actinomycetemcomitans or Porphyromonas gingivalis infection. Oral Microbiol Immunol. 1998;13:301–309. doi: 10.1111/j.1399-302x.1998.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 20.Irwin C R, Myrillas T T. The role of IL-6 in the pathogenesis of periodontal disease. Oral Dis. 1998;4:43–47. doi: 10.1111/j.1601-0825.1998.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara H, Connolly A J, Zeng D, Kahn M L, Zheng Y W, Timmons C, Tram T, Coughlin S R. Protease-activated receptor 3 is a second thrombin receptor in humans. Nature. 1997;386:502–506. doi: 10.1038/386502a0. [DOI] [PubMed] [Google Scholar]
- 22.Ishimi Y, Miyaura C, Jin C H, Akatsu T, Abe E, Nakamura Y, Yamaguchi A, Yoshiki S, Matsuda T, Hirano T. IL-6 is produced by osteoblasts and induces bone resorption. J Immunol. 1990;145:3297–3303. [PubMed] [Google Scholar]
- 23.Izutsu K T, Belton C M, Chan A, Fatherazi S, Kanter J P, Park Y, Lamont R J. Involvement of calcium in interactions between gingival epithelial cells and Porphyromonas gingivalis. FEMS Microbiol Lett. 1996;144:145–150. doi: 10.1111/j.1574-6968.1996.tb08521.x. [DOI] [PubMed] [Google Scholar]
- 24.Jung H C, Eckmann L, Yang S K, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff M F. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Investig. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahn M L, Zheng Y W, Huang W, Bigornia V, Zeng D, Moff S, Farese R V, Jr, Tam C, Coughlin S R. A dual thrombin receptor system for platelet activation. Nature. 1998;394:690–694. doi: 10.1038/29325. [DOI] [PubMed] [Google Scholar]
- 26.Kjeldsen M, Holmstrup P, Bendtzen K. Marginal periodontitis and cytokines: a review of the literature. J Periodontol. 1993;64:1013–1022. doi: 10.1902/jop.1993.64.11.1013. [DOI] [PubMed] [Google Scholar]
- 27.Kurihara N, Bertolini D, Suda T, Akiyama Y, Roodman G D. IL-6 stimulates osteoclast-like multinucleated cell formation in long term human marrow cultures by inducing IL-1 release. J Immunol. 1990;144:4226–4230. [PubMed] [Google Scholar]
- 28.Lamont R J, Chan A, Belton C M, Izutsu K T, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H J, Kang I K, Chung C P, Choi S M. The subgingival microflora and gingival crevicular fluid cytokines in refractory periodontitis. J Clin Periodontol. 1995;22:885–890. doi: 10.1111/j.1600-051x.1995.tb01788.x. [DOI] [PubMed] [Google Scholar]
- 30.Littlewood A J, Russell J, Harvey G R, Hughes D E, Russell R G, Gowen M. The modulation of the expression of IL-6 and its receptor in human osteoblasts in vitro. Endocrinology. 1991;129:1513–1520. doi: 10.1210/endo-129-3-1513. [DOI] [PubMed] [Google Scholar]
- 31.Lourbakos A, Chinni C, Thompson P, Potempa J, Travis J, Mackie E J, Pike R N. Cleavage and activation of proteinase-activated receptor-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis. FEBS Lett. 1998;435:45–48. doi: 10.1016/s0014-5793(98)01036-9. [DOI] [PubMed] [Google Scholar]
- 32.Madianos P N, Papapanou P N, Nannmark U, Dahlen G, Sandros J. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect Immun. 1996;64:660–664. doi: 10.1128/iai.64.2.660-664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuki Y, Yamamoto T, Hara K. Detection of inflammatory cytokine messenger RNA (mRNA)-expressing cells in human inflamed gingiva by combined in situ hybridization and immunohistochemistry. Immunology. 1992;76:42–47. [PMC free article] [PubMed] [Google Scholar]
- 34.Mihara M, Moriya Y, Kishimoto T, Ohsugi Y. Interleukin-6 (IL-6) induces the proliferation of synovial fibroblastic cells in the presence of soluble IL-6 receptor. Br J Rheumatol. 1995;34:321–325. doi: 10.1093/rheumatology/34.4.321. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi-Matsui M, Zheng Y W, Sulciner D J, Weiss E J, Ludeman M J, Coughlin S R. PAR3 is a cofactor for PAR4 activation by thrombin. Nature. 2000;404:609–613. doi: 10.1038/35007085. [DOI] [PubMed] [Google Scholar]
- 36.Noiri Y, Ozaki K, Nakae H, Matsuo T, Ebisu S. An immunohistochemical study on the localization of Porphyromonas gingivalis, Campylobacter rectus and Actinomyces viscosus in human periodontal pockets. J Periodontal Res. 1997;32:598–607. doi: 10.1111/j.1600-0765.1997.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 37.Nystedt S, Emilsson K, Wahlestedt C, Sundelin J. Molecular cloning of a potential proteinase activated receptor. Proc Natl Acad Sci USA. 1994;91:9208–9212. doi: 10.1073/pnas.91.20.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Brien P J, Prevost N, Molino M, Hollinger M K, Woolkalis M J, Woulfe D S, Brass L F. Thrombin responses in human endothelial cells. Contributions from receptors other than PAR1 include the transactivation of PAR2 by thrombin-cleaved PAR1. J Biol Chem. 2000;275:13502–13509. doi: 10.1074/jbc.275.18.13502. [DOI] [PubMed] [Google Scholar]
- 39.Parry M A, Myles T, Tschopp J, Stone S R. Cleavage of the thrombin receptor: identification of potential activators and inactivators. Biochem J. 1996;320:335–341. doi: 10.1042/bj3200335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 41.Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen I B, Enghild J J, Travis J. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273:21648–21657. doi: 10.1074/jbc.273.34.21648. [DOI] [PubMed] [Google Scholar]
- 42.Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 43.Rangarajan M, Aduse-Opoku J, Slaney J M, Young K A, Curtis M A. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol Microbiol. 1997;23:955–965. doi: 10.1046/j.1365-2958.1997.2831647.x. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen S J, Eckmann L, Quayle A J, Shen L, Zhang Y X, Anderson D J, Fierer J, Stephens R S, Kagnoff M F. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Investig. 1997;99:77–87. doi: 10.1172/JCI119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinhardt R A, Masada M P, Kaldahl W B, DuBois L M, Kornman K S, Choi J I, Kalkwarf K L, Allison A C. Gingival fluid IL-1 and IL-6 levels in refractory periodontitis. J Clin Periodontol. 1993;20:225–231. doi: 10.1111/j.1600-051x.1993.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 46.Saglie F R, Marfany A, Camargo P. Intragingival occurrence of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis in active destructive periodontal lesions. J Periodontol. 1988;59:259–265. doi: 10.1902/jop.1988.59.4.259. [DOI] [PubMed] [Google Scholar]
- 47.Schenkein H. The pathogenesis of periodontal diseases. J Periodontol. 1999;70:457–470. doi: 10.1902/jop.1999.70.4.457. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt V A, Nierman W C, Maglott D R, Cupit L D, Moskowitz K A, Wainer J A, Bahou W F. The human proteinase-activated receptor-3 (PAR-3) gene. Identification within a Par gene cluster and characterization in vascular endothelial cells and platelets. J Biol Chem. 1998;273:15061–15068. doi: 10.1074/jbc.273.24.15061. [DOI] [PubMed] [Google Scholar]
- 49.Slots J, Bragd L, Wikstrom M, Dahlen G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol. 1986;13:570–577. doi: 10.1111/j.1600-051x.1986.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 50.Smith-Swintosky V L, Cheo-Isaacs C T, D'Andrea M R, Santulli R J, Darrow A L, Andrade-Gordon P. Protease-activated receptor (PAR-2) is present in the rat hippocampus and is associated with neurodegeneration. J Neurochem. 1997;69:1890–1896. doi: 10.1046/j.1471-4159.1997.69051890.x. [DOI] [PubMed] [Google Scholar]
- 51.Storck J, Kusters B, Vahland M, Morys-Wortmann C, Zimmermann E R. Trypsin induced von Willebrand factor release from human endothelial cells is mediated by PAR-2 activation. Thromb Res. 1996;84:463–473. doi: 10.1016/s0049-3848(96)00214-9. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Takashiba S, Nagai A, Takigawa M, Myoukai F, Kurihara H, Murayama Y. Assessment of interleukin-6 in the pathogenesis of periodontal disease. J Periodontol. 1994;65:147–153. doi: 10.1902/jop.1994.65.2.147. [DOI] [PubMed] [Google Scholar]
- 53.Travis J, Banbula A, Potempa J. The role of bacterial and host proteinases in periodontal disease. Adv Exp Med Biol. 2000;477:455–465. doi: 10.1007/0-306-46826-3_46. [DOI] [PubMed] [Google Scholar]
- 54.Vouret-Craviari V, Grall D, Chambard J C, Rasmussen U B, Pouyssegur J, Van Obberghen-Schilling E. Post-translational and activation-dependent modifications of the G protein-coupled thrombin receptor. J Biol Chem. 1995;270:8367–8372. doi: 10.1074/jbc.270.14.8367. [DOI] [PubMed] [Google Scholar]
- 55.Vu T K, Hung D T, Wheaton V I, Coughlin S R. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 56.Wakita H, Furukawa F, Takigawa M. Thrombin and trypsin induce granulocyte-macrophage colony-stimulating factor and interleukin-6 gene expression in cultured normal human keratinocytes. Proc Assoc Am Physicians. 1997;109:190–207. [PubMed] [Google Scholar]
- 57.Wilson M, Reddi K, Henderson B. Cytokine-inducing components of periodontopathogenic bacteria. J Periodontal Res. 1996;31:393–407. doi: 10.1111/j.1600-0765.1996.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 58.Xu W F, Andersen H, Whitmore T E, Presnell S R, Yee D P, Ching A, Gilbert T, Davie E W, Foster D C. Cloning and characterization of human protease-activated receptor 4. Proc Natl Acad Sci USA. 1998;95:6642–6646. doi: 10.1073/pnas.95.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamazaki K, Nakajima T, Gemmell E, Polak B, Seymour G J, Hara K. IL-4- and IL-6-producing cells in human periodontal disease tissue. J Oral Pathol Med. 1994;23:347–353. doi: 10.1111/j.1600-0714.1994.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 60.Yumoto H, Nakae H, Fujinaka K, Ebisu S, Matsuo T. Interleukin-6 (IL-6) and IL-8 are induced in human oral epithelial cells in response to exposure to periodontopathic Eikenella corrodens. Infect Immun. 1999;67:384–394. doi: 10.1128/iai.67.1.384-394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]