Abstract
Porphyromonas gingivalis, a bacterium associated with active chronic periodontitis lesions, produces several proteolytic enzymes that are thought to be involved in host colonization, perturbation of the immune system, and tissue destruction. The aim of the present study was to investigate the contribution of Arg- and Lys-gingipains produced by P. gingivalis to its growth. Although all of the proteins studied were degraded by P. gingivalis, only human serum albumin and transferrin supported growth during serial transfers in a chemically defined medium (CDM). Growth studies with site-directed gingipain-deficient mutants of P. gingivalis revealed that inactivation of both gingipains prevents growth, whereas inactivation of either Arg- or Lys-gingipain activity extended the doubling times to 33 or 13 h, respectively, compared to 9 h for the parent strain. Growth of the mutants and the parent strain was similar when the CDM was supplemented with a protein hydrolysate instead of human serum albumin. Incubation of resting P. gingivalis ATCC 33277 cells with fluorophore-labeled albumin indicated that the proteolytic fragments generated by the gingipains were internalized by the bacterial cells. Internalization of fluorophore-labeled albumin fragments was reduced or completely inhibited in the proteinase-deficient mutants. Interestingly, gingival crevicular fluid samples from diseased periodontal sites contained low-molecular-mass albumin fragments, whereas samples from healthy sites did not. The critical role of proteinases in the growth of P. gingivalis was further investigated using specific Arg- and Lys-gingipain inhibitors. Adding the inhibitors to CDM containing albumin revealed that leupeptin (Arg-gingipain A and B inhibitor) was more efficient at inhibiting growth than cathepsin B inhibitor II (Lys-gingipain inhibitor). Our study suggests that Arg-gingipains and, to a lesser extent, Lys-gingipain play an important role in the growth of P. gingivalis in a defined medium containing a human protein as the sole carbon and nitrogen source.
Gram-negative anaerobic bacteria play an important role in the initiation and progression of periodontitis. More particularly, Porphyromonas gingivalis has been strongly associated with active chronic periodontitis lesions (24). This bacterial species produces several proteinases that are thought to be involved in host colonization, perturbation of the immune system, and tissue destruction (10, 12, 15). Most of the proteolytic activity exhibited by P. gingivalis is due to Arg- and Lys-gingipain cysteine proteinases (3, 12, 15). Two different genes code for the arginine-X (Arg-gingipain A [rgpA] and B [rgpB])-specific cysteine proteinases, whereas one gene codes for the lysine-X (Lys-gingipain [kgp])-specific cysteine proteinase (3). By cleaving a variety of host proteins, these proteinases may provide small peptides and amino acids that meet the nutritional requirements of P. gingivalis (7, 22) and may thus participate in the pathogenic process of periodontitis. The critical role of P. gingivalis proteinases in pathogenicity is supported by the fact that immunization with purified Arg-gingipain A or B protects against colonization and invasion of a mouse chamber model by P. gingivalis (6, 21). There is also evidence indicating that P. gingivalis proteinases are expressed in periodontal sites, since serum immunoglobulin G responses in patients suffering from periodontitis indicate that P. gingivalis proteinases are important antigens (5, 11).
The gingival crevicular fluid bathing periodontal pockets contains a variety of proteins, including albumin, transferrin, and immunoglobulin G (4, 16). We showed in a previous study (2) that human transferrin may be a source of iron for supporting the growth of P. gingivalis and that cysteine proteinases, and more particularly Lys-gingipain, are critical in the acquisition of iron from this protein. No clear evidence is currently available concerning potential sources of peptides and amino acids for P. gingivalis in subgingival sites or the role played by Arg- and Lys-gingipains in producing these nutrients from human proteins. The aims of the present study were to investigate whether various human proteins could serve as sources of peptides and amino acids for P. gingivalis and to determine the contribution of Arg- and Lys-gingipains to the growth of P. gingivalis in a chemically defined medium (CDM) supplemented with human serum albumin as the sole source of carbon and nitrogen.
Six strains of P. gingivalis were used in the study: ATCC 33277, ATCC 49417, and W50, as well as three proteinase-deficient mutants (KDP129, KDP112, and KDP128) derived from ATCC 33277. The construction of these mutants using suicide plasmids has been previously reported (20, 23). KDP129 is a kgp (Lys-gingipain) mutant, KDP112 is a rgpA rgpB (Arg-gingipains A and B) double mutant, and KDP128 is a rgpA rgpB kgp (Arg- and Lys-gingipains) triple mutant. Bacteria were maintained by weekly transfers on Todd-Hewitt agar plates (BBL Microbiology Systems, Cockeysville, Md.) supplemented with hemin (10 μg/ml), vitamin K (1 μg/ml), and sheep blood (5% [vol/vol]). To prevent the appearance of revertants and to ensure the correct genotype, KDP112 and KDP128 were cultivated in the presence of tetracycline (0.7 μg/ml) and erythromycin (10 μg/ml). All cultures were incubated at 37°C in an anaerobic chamber (N2H2CO2, 75:10:15). Prior to using the mutants in the experiments described below, their phenotypes were confirmed by testing their ability to cleave chromogenic substrates for Arg-gingipain (benzoyl-arginine-p-nitroanilide) and Lys-gingipain (N-p-tosyl-glycine-proline-lysine-p-nitroanilide) as previously described (2).
Growth studies were carried out in the CDM previously described by Milner et al. (19), which contains NaH2PO4 (10 mM), KCl (10 mM), MgCl2 (1.2 mM), ZnCl2 (25 mM), CaCl2 (20 mM), CoCl2 (10 mM), CuCl2 (5 mM), NaMoO4 (0.1 mM), boric acid (5 mM), citric acid (2 mM), α-ketoglutarate (50 mM), hemin (7.5 μM), and vitamin K (3 μM). The medium was adjusted to pH 7 and sterilized by filtration (0.22 μm [pore size]). It was supplemented with 3% of either a protein or a protein hydrolysate. The proteins used were human serum albumin, human transferrin (30% iron saturated), human immunoglobulin G, and calf skin type I soluble collagen. All were obtained from Sigma Chemical Co. (St. Louis, Mo.). The protein hydrolysates used were pancreatic hydrolysate of casein (BBL Microbiology Systems) and proteolytic hydrolysate of chicken egg albumin (Sigma Chemical Co.). The final maximum optical densities at 660 nm (OD660) reached by the cultures were recorded. In terms of dry weight, a value of 1.0 at OD660 in CDM supplemented with either 3% human serum albumin or 3% pancreatic hydrolysate of casein corresponds to 1.1 ± 0.08 or 0.92 ± 0.04 mg/ml, respectively. When growth occurred, the cultures were transferred (1% [vol/vol] inoculum) to the corresponding medium up to four times. Growth of strain ATCC 33277 and the proteinase-deficient mutants (KDP112, KDP129, and KDP128) in CDM containing human serum albumin was followed by recording the OD660 at various times during the incubation. The doubling times were calculated by regression analysis of the OD660 values from the exponential phase of growth.
Mutant KDP129 was used to prepare a purified fraction containing Arg-gingipains (rgpA and rgpB), whereas KDP112 was used to isolate Lys-gingipain (kgp). The use of these mutants facilitated the purification of the gingipains, which are difficult to separate from one another. Bacteria were grown (48 h) in 2 liters of Todd-Hewitt broth containing hemin and vitamin K. The cells were harvested by centrifugation at 8,000 × g for 15 min, washed twice with 50 mM phosphate-buffered saline (PBS; pH 7.2), and resuspended in 50 ml of the same buffer. The suspensions were sonicated (8 min, 60% duty cycle, output 6; Sonifier Cell Disrupter W-350; Branson Sonic Power Co.), and unbroken cells and cellular debris were removed by centrifugation at 8,000 × g for 15 min. Membranes (cytoplasmic and outer) were pelleted by centrifugation of the supernatant at 120,000 × g for 1 h at 4°C and resuspended in 10 ml of 50 mM Tris-HCl (pH 8.2) prior to adding CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} to a final concentration of 0.5%. The membrane suspension was stirred overnight at 4°C and then centrifuged at 120,000 × g for 1 h at 4°C. The supernatant containing the soluble membrane fraction was kept at −20°C until used. Solubilization of the pelleted membranes with 0.5% CHAPS was repeated until no residual Arg- or Lys-gingipain activity could be detected in the soluble fraction. The soluble membrane fractions were pooled and dialyzed (molecular mass cutoff of 6,000 to 8,000 Da) overnight at 4°C against 2 volumes of 50 mM Tris-HCl (pH 7.4) containing 200 mM NaCl. The gingipains were purified by affinity chromatography on arginine-Sepharose 4B (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada) as previously described (8). Arg- and Lys-gingipain activities were quantified using benzoyl-arginine-p-nitroanilide and N-p-tosyl-glycine-proline-lysine-p-nitroanilide, respectively. One unit of enzyme was defined as the amount required to release one nanomole of p-nitroaniline per hour.
Protein degradation by P. gingivalis was tested using bacterial suspensions (OD660 = 2.0 in PBS) prepared from 24-h Todd-Hewitt broth cultures. Mixtures containing bacteria (20 μl), protein (10 μl; 1 mg/ml), PBS (20 μl), and dithiothreitol (10 μl; 10 mM) were incubated at 37°C for 30 min, 1, 2, and 4 h. An equal volume of electrophoresis sample buffer was then added, and the mixtures were boiled prior to analysis by sodium dodecyl sulfate (SDS)–12.5% (11% for collagen analysis) polyacrylamide gel electrophoresis (PAGE) according to the procedure of Laemmli (14). In the case of albumin, transferrin, and immunoglobulin G, undegraded proteins and protein fragments were electrophoretically transferred onto a nitrocellulose membrane and detected using alkaline phosphatase-conjugated goat anti-human albumin antibody (1/8,000 dilution), alkaline phosphatase-conjugated goat anti-human transferrin antibody (1/3,000 dilution), and alkaline phosphatase-conjugated goat anti-human immunoglobulin G antibody (1/3,000 dilution), respectively. The antibodies were obtained from Bethyl Laboratories, Inc. (Montgomery, Tex.). Undegraded proteins and fragments were visualized following development in carbonate buffer containing 0.3 mg of NBT and 0.15 mg of BCIP (5-bromo-4-chloro-3-indolylphosphate) per ml. The degradation of calf skin type I collagen was determined by using 14C-labeled substrate (18). Following incubation with bacterial cells, the collagen was analyzed by SDS-PAGE and autofluorography. Lastly, the degradation of human serum albumin by the Arg- and Lys-gingipain preparations was evaluated as described above except that the bacterial suspensions were replaced by 312 U of enzyme.
The uptake of albumin degradation products by P. gingivalis ATCC 33277 and the three proteinase-deficient mutants was tested using 24-h Todd-Hewitt broth cultures. Self-quenched DQ green bovine serum albumin (Molecular Probes, Eugene, Oreg.) was used. Degradation products emitted maximally at 515 nm following excitation at 505 nm. The assay mixture contained bacteria (100 μl of the culture), PBS (80 μl; O2-free), and DQ albumin (20 μl; 200 μg/ml). Heat-treated P. gingivalis cultures were used as controls. In one experiment, proteinase K (100 μg/ml) was used instead of bacterial cells. Assay mixtures were incubated at 37°C for 3 h under darkness prior to recovering the cells by centrifugation (6,000 × g, 10 min). The fluorescence of the supernatant, which indicated the degree of albumin degradation, was measured using a fluorometer at excitation and emission wavelengths of 490 and 520 nm, respectively. The cell pellet was thoroughly washed twice in PBS and resuspended in 100 μl of 0.05% SDS to lyse the bacteria and allow the internal fluorescence, which corresponded to bovine serum albumin fragments incorporated by P. gingivalis to be measured. The fluorescence results were expressed as relative fluorescence units (RFU).
The effect of cathepsin B inhibitor II (Calbiochem, San Diego, Calif.) and leupeptin (Sigma Chemical Co.), which have been shown to be specific inhibitors of Lys- and Arg-gingipain activities, respectively (2), on the degradation of human serum albumin by P. gingivalis ATCC 33277 was verified by adding them (10 μM) to the assay mixture described above. Their effect on the growth of P. gingivalis ATCC 33277 in CDM containing either human serum albumin or pancreatic hydrolysate of casein was also tested. Twofold serial dilutions of the inhibitors were tested beginning with an initial concentration of 20 μM.
Gingival crevicular fluid samples were obtained from 24 patients attending the dental clinic at Université Laval. The pocket depth of each site was measured using a Michigan 0 periodontal probe. Patients were distributed into three categories: healthy (pocket depth of ≤3 mm), moderate periodontitis (pocket depth of between 4 and 6 mm), and advanced periodontitis (pocket depth of ≥7 mm). Paper strips (3MM; 2 by 8 mm) were inserted into the subgingival sites for 30 s. The strips were then placed in 250 μl of PBS containing a protease inhibitor cocktail with a broad inhibitory spectrum (complete mini #1836153; Roche Diagnostics, Laval, Québec, Canada) and frozen at −20°C. To recover the adsorbed proteins, the samples were thawed rapidly at room temperature and shaken at 4°C for 2 h. The paper strips were then removed, and the samples were kept frozen until used. A 10-μl volume of gingival crevicular fluid sample was mixed with 10 μl of electrophoresis sample buffer and boiled for 10 min. Proteins were separated on 12.5% polyacrylamide gels according to the method of Laemmli (14) and then electrophoretically transferred onto a nitrocellulose membrane. The membranes were incubated with alkaline phosphatase-conjugated goat anti-human albumin antibody (1/8,000 dilution) for 2.5 h. Albumin and albumin fragments were visualized as described above.
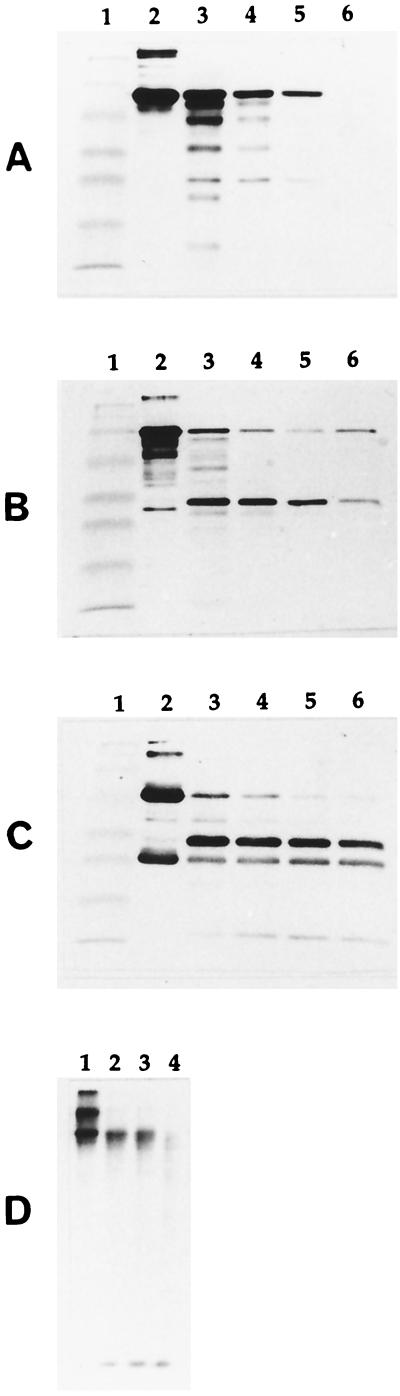
The four proteins selected for the growth studies (albumin, transferrin, immunoglobulin G, and collagen) were first tested for their susceptibility to proteolytic degradation by resting P. gingivalis cells. As shown in Fig. 1, all of the proteins were degraded to various degrees. Albumin and collagen are completely degraded after 4 h. Transferrin and immunoglobulin G appear to be slightly less susceptible since some protein remains undegraded and there is an accumulation of proteolytic fragments. The fact that albumin, transferrin, immunoglobulin G, and collagen are partially or totally degraded by P. gingivalis suggests that they may be potential sources of peptides and amino acids suitable for the growth of this bacterial species. The proteins were individually added to the CDM to determine their ability to support the growth of P. gingivalis ATCC 33277. Human serum albumin and human transferrin supported the growth (final maximal OD660 of >0.8; 48 h of incubation) of P. gingivalis for up to four serial transfers. On the other hand, immunoglobulin G and collagen did not support the growth of P. gingivalis, even after a prolonged incubation period of 72 h (final maximal OD660 of <0.1). These results indicate that the ability of P. gingivalis to hydrolyze proteins does not necessarily imply that these proteins can be used by the bacterium to support its growth.
FIG. 1.
Degradation of human serum albumin (gel A), human transferrin (gel B), human immunoglobulin G (gel C), and calf skin type I collagen (gel D) by P. gingivalis ATCC 33277. Gels A, B, and C: lane 1, molecular mass markers (myosin [206 kDa], β-galactosidase [117 kDa], bovine serum albumin [79 kDa], ovalbumin [48 kDa], carbonic anhydrase [34.7 kDa], soybean trypsin inhibitor [29 kDa], lysozyme [21 kDa]); lane 2, protein without cells; lane 3, protein incubated with cells for 30 min; lane 4, protein incubated with cells for 1 h; lane 5, protein incubated with cells for 2 h; lane 6, protein incubated with cells for 4 h. Gel D: lane 1, protein without cells; lane 2, protein incubated with cells for 30 min; lane 3, protein incubated with cells for 2 h; lane 4, protein incubated with cells for 4 h. Degradation of albumin, transferrin, and immunoglobulin G was determined by SDS-PAGE and Western immunoblotting analysis. Degradation of collagen was determined by SDS-PAGE and autofluorography.
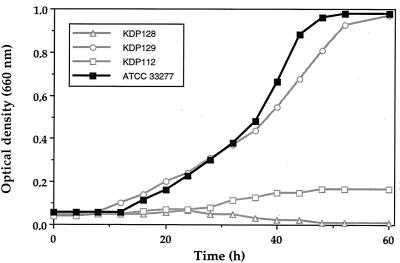
Since albumin is the predominant protein in the gingival crevicular fluid bathing periodontal pockets (4) and also supports the growth of P. gingivalis in a CDM with no other sources of peptides and amino acids, it was selected for further growth studies. The doubling times of three laboratory strains of P. gingivalis (ATCC 33277, ATCC 49417, and W50) in CDM supplemented with serum albumin were 9.2, 9, and 5.4 h, respectively. The fact that strain W50 grew much better may be related to a more efficient degradation of serum albumin, to a lower peptide requirement, or to a more efficient system of peptide transport. Phase-contrast microscopic observations showed that P. gingivalis grew in large cell aggregates in CDM containing serum albumin, whereas isolated or short-chain coccobacilli were observed in Todd-Hewitt broth (data not shown). It is possible that albumin receptors on the cell surface of P. gingivalis could favor the phenomenon of aggregation. Growth curves of P. gingivalis ATCC 33277 and the three proteinase-deficient mutants in CDM containing serum albumin are presented in Fig. 2. The ability to use albumin as a carbon and nitrogen source as well as doubling times varied considerably depending on the missing proteinase activity. The doubling times of mutants KDP129 (kgp) and KDP112 (rgpA rgpB) were 13 and 33 h, respectively, whereas that of the parent strain (ATCC 33277) was 9 h. The final maximum OD660 reached by mutant KDP129 was similar to that of the parent strain. On the other hand, mutant KDP112 grew to a maximum OD660 of 0.16. Mutant KDP128 (kgp rgpA rgpB) did not grow in CDM containing serum albumin, suggesting that proteinase activity is essential for the growth of P. gingivalis. The effect of a deficiency in Arg- and/or Lys-gingipain activity on the growth of P. gingivalis in the presence of a protein hydrolysate rather than serum albumin was investigated. Growth was not affected by the absence of proteinase activities when the CDM was supplemented with either a pancreatic hydrolysate of casein or a proteolytic hydrolysate of egg albumin. These two hydrolysates gave OD660 ranging from 1.3 to 1.6 for all strains (data not shown).
FIG. 2.
Growth curves of P. gingivalis ATCC 33277 and proteinase-deficient mutants KDP112 (rgpA rgpB), KDP129 (kgp), and KDP128 (rgpA rgpB kgp) in a CDM supplemented with human serum albumin.
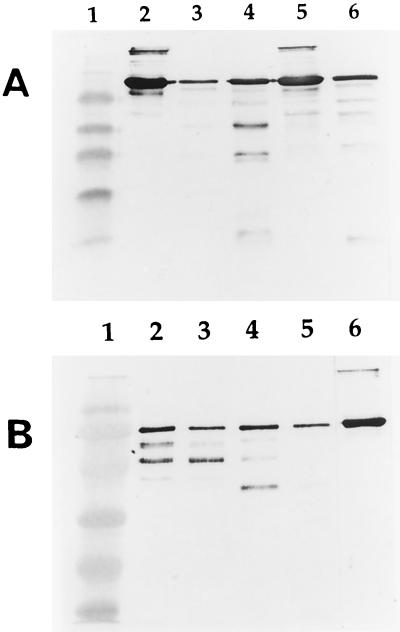
In order to establish a correlation between the growth of P. gingivalis in CDM containing serum albumin and the ability to degrade the protein, strain ATCC 33277 and the proteinase-deficient mutants were incubated with serum albumin for 2 h. Protein degradation was monitored by SDS-PAGE and Western immunoblotting analysis (Fig 3A). Mutant KDP128, which did not grow in CDM supplemented with serum albumin, was unable to degrade the protein. Mutants KDP112 and KDP129 were slightly less effective than the parent strain in hydrolyzing the albumin. Several bands produced by these mutants appeared to have similar molecular masses. The susceptibility of human serum albumin to purified preparations of Arg- and Lys-gingipains is reported in Fig. 3B. After a 2-h incubation, the Lys-gingipain preparation yielded more fragments than the preparation containing Arg-gingipains A and B. Fragments produced by the Arg- and Lys-gingipain preparations differed slightly in their molecular masses. When the incubation was extended to 4 h, a more pronounced degradation of albumin was observed with the Lys-gingipain preparation than the Arg-gingipain A and B preparation.
FIG. 3.
Degradation of human serum albumin by resting P. gingivalis cells (A) and purified preparations of gingipains (B), as determined by SDS-PAGE and Western immunoblotting analysis. Gel A: lane 1, molecular mass markers (myosin [206 kDa], β-galactosidase [117 kDa], bovine serum albumin [79 kDa], ovalbumin [48 kDa], carbonic anhydrase [34.7 kDa], soybean trypsin inhibitor [29 kDa], lysozyme [21 kDa]); lane 2, albumin without cells; lane 3, albumin incubated with ATCC 33277 for 2 h; lane 4, albumin incubated with KDP112 (rgpA rgpB) for 2 h; lane 5, albumin incubated with KDP128 (rgpA rgpB kgp) for 2 h; lane 6, albumin incubated with KDP129 (kgp) for 2 h. Gel B: lane 1, molecular mass markers (myosin [206 kDa], β-galactosidase [117 kDa], bovine serum albumin [79 kDa], ovalbumin [48 kDa], carbonic anhydrase [34.7 kDa], soybean trypsin inhibitor [29 kDa], lysozyme [21 kDa]); lane 2, albumin incubated with the preparation of Arg-gingipains for 2 h; lane 3, albumin incubated with the preparation of Arg-gingipains for 4 h; lane 4, albumin incubated with the preparation of Lys-gingipain for 2 h; lane 5, albumin incubated with the preparation of Lys-gingipain for 4 h; lane 6, albumin alone.
The ability of P. gingivalis to grow in CDM containing serum albumin implies that the bacterium cleaves the protein and assimilates the degradation products, which it then uses as a source of carbon and nitrogen. This was investigated using fluorophore-labeled bovine serum albumin and monitoring the uptake of fluorescence (Table 1). Substantial hydrolysis of the substrate occurred with the parent strain, whereas only partial degradation occurred with mutants KDP112 and KDP129. Mutant KDP128, which is deficient in both Arg- and Lys-gingipain activities, released no fluorescence, indicating that no degradation occurred. When cells incubated with fluorophore-labeled albumin were washed extensively to remove surface-associated fluorescence and lysed with SDS, high levels of intracellular fluorescence were detected with the parent strain. No fluorescence was detected when cells were not lysed, suggesting intracellular uptake rather than cell surface binding of fluorescent peptides. Mutants KDP112, KDP129, and KDP128 internalized approximately 66, 27, and 5%, respectively, of the fluorescence internalized by the parent strain. The fact that mutant KDP112 assimilated more serum bovine albumin-derived peptides than mutant KDP129 but grew poorly compared to this mutant in CDM supplemented with human serum albumin may be related to the different sources of albumin used. It is also possible that the albumin-derived peptides assimilated by mutant KDP112 may not be appropriate to support growth.
TABLE 1.
Uptake of fluorescence from fluorophore-labeled bovine serum albumin by P. gingivalis ATCC 33277, KDP112, KDP129, and KDP128.
| Strain | RFUa
|
|
|---|---|---|
| Supernatantb | Cellsc | |
| ATCC 33277 | 77,240 | 14,370 |
| Control (heat-treated culture) | 1,267 | 0 |
| KDP112 (rgpA rgpB) | 13,213 | 9,474 |
| Control (heat-treated culture) | 1,382 | 0 |
| KDP129 (kgp) | 4,430 | 3,888 |
| Control (heat-treated culture) | 1,286 | 0 |
| KDP128 (rgpA rgpB kgp) | 1,571 | 1,651 |
| Control (heat-treated culture) | 1,326 | 0 |
| Proteinase K | 202,240 | NA |
Mean RFU from two assays.
Following incubation with DQ green bovine serum albumin, the bacteria were removed by centrifugation prior to measuring the fluorescence in the supernatant.
The cells were lysed by adding 0.05% SDS. NA, not applicable.
The importance of proteinases for the growth of P. gingivalis was further investigated using specific Arg- and Lys-gingipain inhibitors. Although the inhibition was not complete, 10 μM leupeptin (Arg-gingipain A and B inhibitor) reduced the degradation of serum albumin by resting P. gingivalis ATCC 33277 cells more effectively than 10 μM cathepsin B inhibitor II (Lys-gingipain inhibitor), as determined by SDS-PAGE and Western immunoblotting analysis (data not shown). The inhibitors were also added to CDM supplemented with serum albumin to investigate their ability to inhibit the growth of P. gingivalis ATCC 33277. Leupeptin was more effective at inhibiting growth than cathepsin B inhibitor II. Inhibition by 5 μM leupeptin exceeded 90%, while inhibition by 20 μM cathepsin B inhibitor II only reached 44%. No inhibition occurred when the CDM was supplemented with pancreatic hydrolysate of casein instead of serum albumin. Inhibition of the growth of P. gingivalis by other proteinase inhibitors (cystatin and bestatin) has been previously reported (1, 13). These inhibitors appear to affect the growth of P. gingivalis by interfering with peptide transport rather than by inactivating gingipain activities.
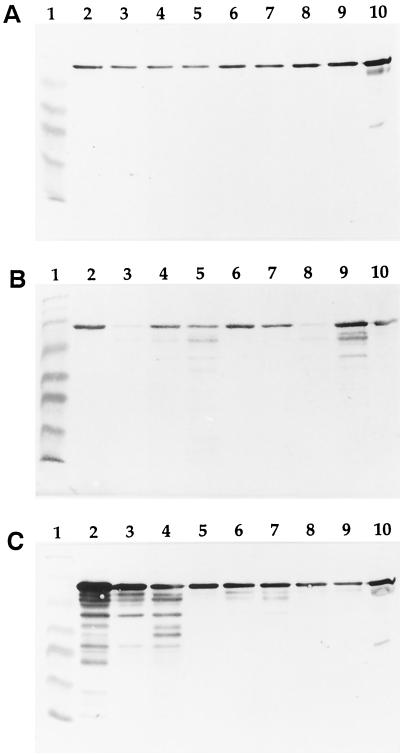
Finally, gingival crevicular fluid samples from patients with various periodontal conditions were analyzed for the presence of serum albumin fragments as an indication of the degradation of this protein in vivo (Fig. 4). Gingival crevicular fluid samples from healthy periodontal sites contained serum albumin but no detectable proteolytic fragments. On the other hand, numerous low-molecular-mass albumin fragments were found in samples from patients with periodontitis, more particularly those from periodontal pockets greater than or equal to 7 mm in depth. The degradation products detected in gingival crevicular fluid may be produced by P. gingivalis, other proteolytic bacteria, or host proteinases. In vivo, the degradation of albumin, in addition to providing peptides, which can serve as carbon and nitrogen sources, may also result in the liberation of heme, which is scavenged and transported by this serum protein.
FIG. 4.
SDS-PAGE Western immunoblotting analysis of gingival crevicular fluid samples for the presence of albumin and albumin fragments. Lanes 1, molecular mass markers (myosin [206 kDa], β-galactosidase [117 kDa], bovine serum albumin [79 kDa], ovalbumin [48 kDa], carbonic anhydrase [34.7 kDa], soybean trypsin inhibitor [29 kDa], lysozyme [21 kDa]); lanes 10, control human serum diluted 1 in 500. Gel A, samples (lanes 2 to 9) from healthy periodontal sites; Gel B, samples (lanes 2 to 9) from periodontal pockets with depths ranging from 4 to 6 mm; Gel C, samples (lanes 2 to 9) from periodontal pockets with depths ranging from 7 to 12 mm.
Previous studies (4, 9) have shown that serum albumin is a key protein in gingival crevicular fluid and may thus be utilized for supporting the growth of asaccharolytic periodontopathogens in subgingival sites. Indeed, increased albumin concentrations in saliva and gingival crevicular fluid during inflammation and periodontal tissue destruction have been reported (4, 9). Evidence suggesting the degradation of albumin in vivo in periodontal pockets was obtained in the present study. The degradation of bovine albumin and hemalbumin by P. gingivalis has been extensively studied by Smalley and Birss (25). They reported the production of protease-stable 55.6-kDa peptides and noted that the total amount of hemalbumin degraded decreased with an increase in the ratio of bound hemin. Heme binding thus appears to protect albumin against proteolysis by P. gingivalis. Heme saturation of albumin in gingival crevicular fluid has not been reported yet.
Our results suggest that Arg-gingipains play a key role in the growth of P. gingivalis in a CDM containing human serum albumin as the sole source of carbon and nitrogen. This is supported by the fact that (i) a mutant deficient in Arg-gingipains did not grow optimally and (ii) a specific Arg-gingipain inhibitor (leupeptin) prevented growth. On the other hand, we found that proteinases are not required for growth in a CDM containing a protein hydrolysate (casein or egg albumin) instead of a whole protein. Shi et al. (23) reported that an Arg-gingipain A and B null mutant could grow in a defined medium containing bovine serum albumin just as well as the wild type. This discrepancy with our results is likely related to the types of albumin (human and bovine) used, which may have different susceptibilities to P. gingivalis gingipains.
Results from the present study have provided a better understanding of the roles of Arg- and Lys-gingipains in the growth of P. gingivalis. Arg-gingipains but not Lys-gingipain appear to be essential when protein sources other than amino acids or low-molecular-weight peptides are provided. We hypothesize that the action of Arg-gingipains generate peptides with C-terminal arginine residues from albumin. Once assimilated by bacteria, an arginine carboxypeptidase previously characterized from P. gingivalis can liberate C-terminal arginine from these peptides (17). The arginine could then be catabolized via an arginine deiminase pathway as in lactic bacteria (26). This pathway involves three enzymes (arginine deiminase, ornithine transcarbamylase, and carbamate kinase) and results in the production of ornithine, ammonia, carbon dioxide, and ATP. Further studies to investigate this pathway in P. gingivalis should be carried out. Since healthy subgingival sites do not appear to contain serum albumin fragments, P. gingivalis proteinases may be even more important in the initiation phase of periodontitis. The local application of gingipain inhibitors might suppress gingipain activities and thus the production of peptides that may be used to support the growth of P. gingivalis. However, it is likely that any periodontal therapy based on the local application of gingipain inhibitors would have only a limited effect on P. gingivalis in the case of advanced periodontitis, a condition where serum albumin fragments are present in large quantity.
ACKNOWLEDGMENTS
This study was supported by grants from the Canadian Institutes of Health Research, FRSQ, Fonds FCAR, and Fonds Emile-Beaulieu.
REFERENCES
- 1.Blankenvoorde M F J, van't Hof W, Walgreen-Weterings E, van Steenbergen T J M, Brand H S, Veerman E C I, Nieuw Amerongen A V. Cystatin and cystatin-derived peptides have antibacterial activity against the pathogen Porphyromonas gingivalis. Biol Chem. 1998;379:1371–1375. [PubMed] [Google Scholar]
- 2.Brochu V, Grenier D, Nakayama K, Mayrand D. Acquisition of iron from human transferrin by Porphyromonas gingivalis: a role for Arg- and Lys-gingipain activities. Oral Microbiol Immunol. 2001;16:79–87. doi: 10.1034/j.1399-302x.2001.016002079.x. [DOI] [PubMed] [Google Scholar]
- 3.Curtis M A, Kuramitsu H K, Lantz M, Macrina F L, Nakayama K, Potempa J, Reynolds E C, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 1999;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 4.Curtis M A, Sterne J A C, Price S J, Griffiths G S, Coulthurst S K, Wilton J M A, Johnson N W. The protein composition of gingival crevicular fluid sampled from male adolescents with no destructive periodontitis: baseline data of a longitudinal study. J Periodontal Res. 1990;25:6–16. doi: 10.1111/j.1600-0765.1990.tb01202.x. [DOI] [PubMed] [Google Scholar]
- 5.Genco C A, Potempa J, Mikolajczyk-Pawlinska J, Travis J. Role of gingipains R in the pathogenesis of Porphyromonas gingivalis—mediated periodontal disease. Clin Infect Dis. 1999;28:456–465. doi: 10.1086/515156. [DOI] [PubMed] [Google Scholar]
- 6.Genco C A, Odusanya B M, Potempa J, Mikolajczyk-Pawlinska J, Travis J. A peptide domain on gingipain R1 which confers immunity against Porphyromonas gingivalis infection in mice. Infect Immun. 1998;66:4108–4114. doi: 10.1128/iai.66.9.4108-4114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gharbia S E, Shah H N. Utilization of aspartate, glutamate and their corresponding peptides by Fusobacterium nucleatum and Porphyromonas gingivalis. Curr Microbiol. 1991;22:159–163. [Google Scholar]
- 8.Grenier, D., P. Gauthier, P. Plamondon, K. Nakayama, and D. Mayrand. Studies on the aminopeptidase activities of Porphyromonas gingivalis. Oral Microbiol. Immunol., in press. [DOI] [PubMed]
- 9.Henskens Y M C, van der Velden U, Veerman E C I, Nieuw Amerongen A V. Protein, albumin and cystatin concentrations in saliva of healthy subjects and of patients with gingivitis or periodontitis. J Periodontal Res. 1993;28:43–48. doi: 10.1111/j.1600-0765.1993.tb01049.x. [DOI] [PubMed] [Google Scholar]
- 10.Holt S, Kesavalu L, Walker S, Genco C A. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 11.Ismaiel M O, Greenman J, Scully J. Serum antibodies against the trypsin-like protease of Bacteroides gingivalis in periodontitis. J Periodontal Res. 1988;23:193–198. doi: 10.1111/j.1600-0765.1988.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 12.Kuramitsu H K. Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol Immunol. 1998;13:263–270. doi: 10.1111/j.1399-302x.1998.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 13.Labbé S, Grenier D, Plamondon P, Uitto V-J, Mayrand D. Effects of dipeptide bestatin on Porphyromonas gingivalis and epithelial cells. J Periodontol. 2001;72:714–721. doi: 10.1902/jop.2001.72.6.714. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makela M, Soderling E, Paunio K, Talonpoika J, Hyyppa T. Protein composition of crevicular fluid before and after treatment. Scand J Dent Res. 1991;99:413–423. doi: 10.1111/j.1600-0722.1991.tb01049.x. [DOI] [PubMed] [Google Scholar]
- 17.Masuda K, Yoshioka M, Hinode D, Nakamura R. Characterization of arginine carboxypeptidase from Porphyromonas gingivalis. J Dent Res. 1999;78:507. doi: 10.1128/IAI.70.4.1807-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayrand D, Grenier D. Detection of collagenase activity in oral bacteria. Can J Microbiol. 1985;31:134–138. doi: 10.1139/m85-026. [DOI] [PubMed] [Google Scholar]
- 19.Milner P, Batten J, Curtis M A. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for α-ketoglutarate. FEMS Microbiol Lett. 1996;140:125–130. doi: 10.1016/0378-1097(96)00159-0. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 21.Page R C, Lantz M, Persson G R, Houston S, Lukehart S, Jeffcoat M, Mancl L, Weinberg A, Braham P, Darveau R. Immunization with Porphyromonas gingivalis cysteine protease attenuates alveolar bone loss. J Dent Res. 1999;78:294. [Google Scholar]
- 22.Shah H N, Gharbia S E. Batch culture and physiological properties. In: Shah H N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 85–103. [Google Scholar]
- 23.Shi Y, Ratnayake D B, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis: construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 24.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol 2000. 1999;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 25.Smalley J W, Birss A J. Albumin and hemalbumin degradation by Porphyromonas gingivalis. Oral Microbiol Immunol. 1997;12:254–258. doi: 10.1111/j.1399-302x.1997.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 26.Vaughan L C, Thomas T D. Arginine metabolism in lactic streptococci. J Bacteriol. 1982;150:1024–1032. doi: 10.1128/jb.150.3.1024-1032.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]