Abstract
Opioid drugs have analgesic properties used to treat chronic and post-surgical pain due to descending pain modulation. The use of opioids is often associated with adverse effects or clinical issues. This study aimed to evaluate the toxicity of opioids by exposing the neuroblastoma cell line (SH-SY5Y) to 0, 1, 10, and 100 µM oxycodone and naloxone for 24 h. Analyses were carried out to evaluate cell cytotoxicity, identification of cell death, DNA damage, superoxide dismutase (SOD), glutathione S-transferase (GST), and acetylcholinesterase (AChE) activities, in addition to molecular docking. Oxycodone and naloxone exposure did not alter the SH-SY5Y cell viability. The exposure to 100 µM oxycodone and naloxone significantly increased the cells’ DNA damage score compared to the control group. Naloxone exposure significantly inhibited AChE, GST, and SOD activities, while oxycodone did not alter these enzymes’ activities. Molecular docking showed that naloxone and oxycodone interact with different amino acids in the studied enzymes, which may explain the differences in enzymatic inhibition. Naloxone altered the antioxidant defenses of SH-SY5Y cells, which may have caused DNA damage 24 h after the exposure. On the other hand, more studies are necessary to explain how oxycodone causes DNA damage.
Keywords: opioids, molecular docking, antioxidant enzymes
1. Introduction
Opioids have been used for thousands of years to treat pain. Opium extracted from poppy seeds is the oldest opioid used in medicine, described in the third century BC [1]. Opium was responsible, in the second half of the 19th century, for the Opium War, an international conflict between China and England, because of the ban on opium consumption by China [2].
Opioids are first-pass metabolism drugs. These drugs can be administered orally, intrathecally, intravenously, subcutaneously, or epidurally [3]. Generally, opioids are metabolized in the liver and eliminated by the kidneys [4]. For instance, naloxone is deactivated in the liver 30–60 min after intravenous administration, and the inactive metabolite (6-beta-naloxol) is excreted by the urine [5]. Likewise, oxycodone is biotransformed into noxicodone (inactive metabolite) and oxymorphine (active metabolite) in the liver [6].
Among the recommended opioids sold worldwide, fentanyl, oxycodone (OxyContin®), hydrocodone (Vicodin®), codeine, and morphine stand out [7]. Although the Opioid Prescription Guideline recommends the use of opioids for limited periods, chronic pain patients make use of these drugs for long periods [8,9]. Adverse reactions after opioid treatment are usually mood alterations, psychological and psychosocial changes, and physical dependence [10,11]. Neurotoxic effects can be observed with opioid chronic use, such as cognitive dysfunction, neurodegeneration, neuroinflammation, brain abscesses, ischemic strokes, embolism, aneurysms, and leukoencephalopathy [12,13]. In 2020, 91,799 drug overdose deaths occurred in the United States, and opioids were involved in 68,630 of these deaths (74.8% of all drugs) [14].
The in vitro acute toxic effect of oxycodone and naloxone is underexplored. Due to the increase in opioid prescriptions, it is pivotal to evaluate the cellular toxicity of these drugs. Therefore, this study aimed to evaluate the toxic effects of oxycodone and naloxone in a neuroblastoma cell line (SH-SY5Y); thus, analyses were carried out to evaluate cell viability, genotoxicity, and the activity of antioxidant enzymes, in addition to molecular docking.
2. Results
2.1. Cytotoxicity
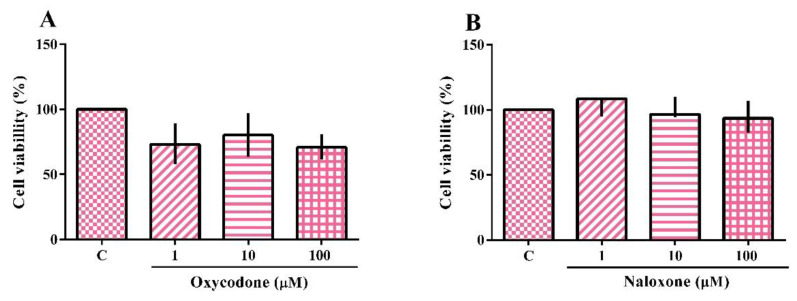
The cytotoxicity of the neuroblastoma cell line (SH-SY5Y) exposed to oxycodone and naloxone is shown in Figure 1A,B, respectively. The Kruskal–Wallis test showed no treatment effect. In fact, neither opioid drug induced a statistically significant change in cell viability. This test was also performed with the oxycodone vehicle (methanol 3.3%), which did not alter the cell viability.
Figure 1.
Cytotoxicity analysis of a neuroblastoma cell line (SH-SY5Y) exposed for 24 h to oxycodone (A) and naloxone (B). The results were analyzed by the Kruskal–Wallis test followed by Dunn’s post-test and presented as the median ± interquartile interval (n = 3–4). C: control, nonexposed cells.
2.2. Identification of Apoptotic and Necrotic Cells
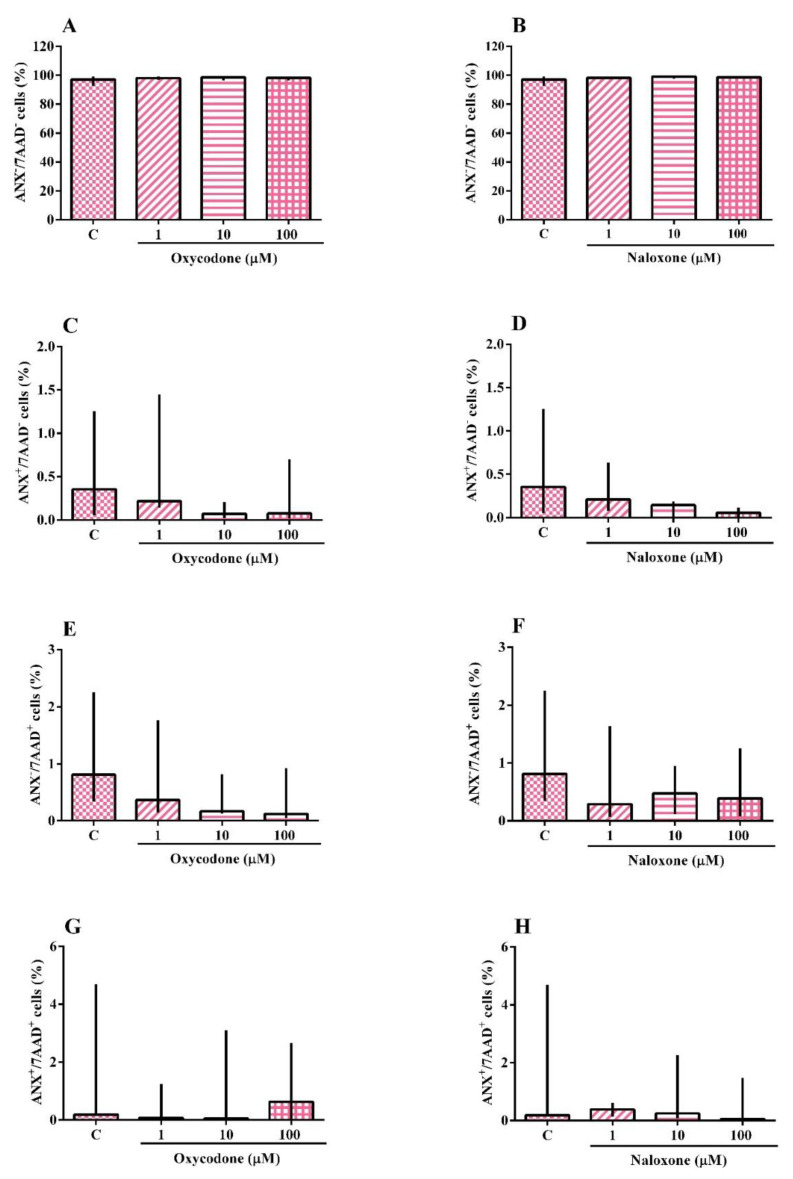
The analysis of death by apoptosis and/or necrosis of the SH-SY5Y cell line exposed to oxycodone or naloxone is shown in Figure 2A–H. The Kruskal–Wallis test showed no effect of the treatment. SH-SY5Y cells exposed to oxycodone or naloxone did not have apoptotic or necrotic processes. The test was also performed with the oxycodone vehicle (methanol 3.3%), which did not induce an apoptotic or necrotic process.
Figure 2.
Identification of cell death by necrosis and apoptosis of a neuroblastoma cell line (SH-SY5Y) exposed for 24 h to oxycodone and naloxone. Viable cells (ANX−/7AAD−) (A,B), apoptotic cells (ANX+/7AAD−) (C,D), necrotic cells (ANX−/7AAD+) (E,F), and late apoptotic and/or necrotic cells (ANX+/7AAD+) (G,H). The results were analyzed by the Kruskal–Wallis test followed by Dunn’s post-test and presented as median ± interquartile interval (n = 3–5). C: control, nonexposed cells.
2.3. DNA Damage
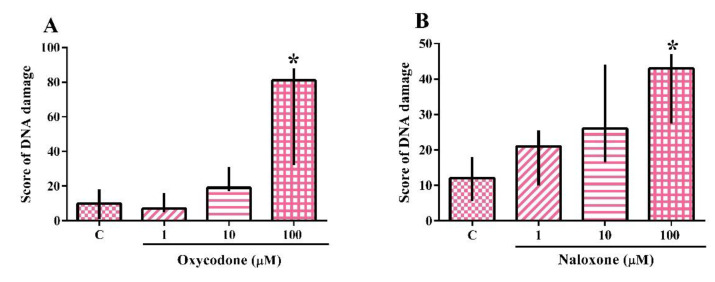
The analysis of the genotoxicity assay performed on the SH-SY5Y cell line exposed to oxycodone or naloxone is shown in Figure 3A,B. The Kruskal–Wallis test demonstrated the effect of oxycodone (H(5) = 8.744; p = 0.0064) and naloxone (H(4) = 8.016; p = 0.0457) exposure. The exposure to 100 µM oxycodone and naloxone caused a statistically significant increase in the score of DNA damage when compared to the nonexposed cells. The test was also performed with the oxycodone vehicle (methanol 3.3%), which did not cause DNA damage. The positive control, methyl methane sulfonate, caused a DNA damage score of 341.3, demonstrating the reliability of the test.
Figure 3.
Genotoxicity analysis of a neuroblastoma cell line (SH-SY5Y) exposed for 24 h to oxycodone (A) and naloxone (B). The results were analyzed by the Kruskal–Wallis test followed by Dunn’s post-test and presented as median ± interquartile interval (n = 3–5). C: control, nonexposed cells. * Statistically different from C.
2.4. Biochemical Analysis
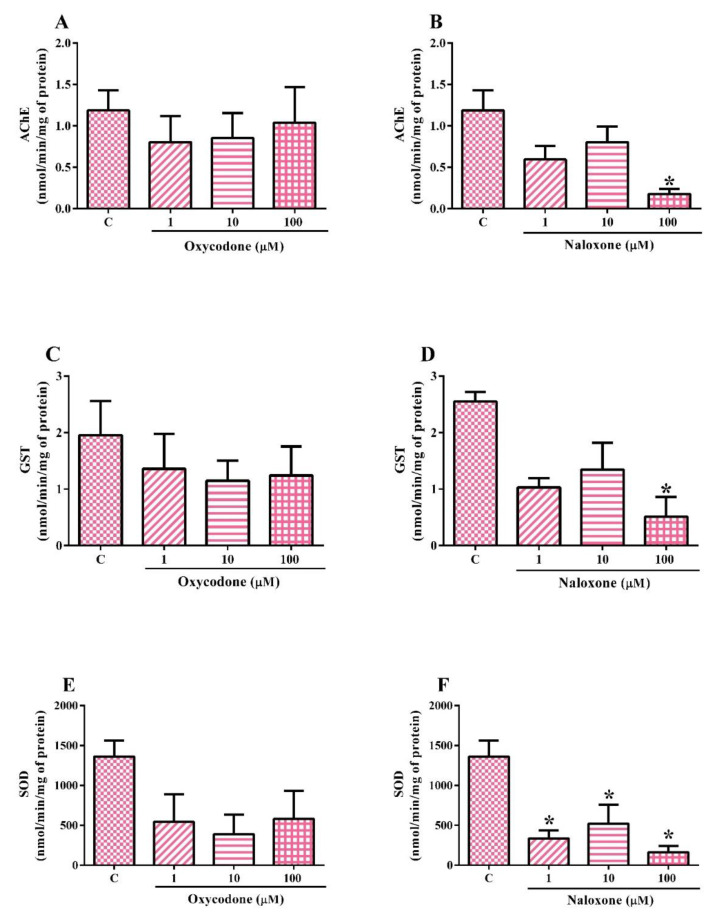
The biochemical analyses performed on the SH-SY5Y cell line exposed to oxycodone or naloxone are shown in Figure 4A–F. The tests were also performed with the oxycodone vehicle control (methanol 3.3%), which did not alter enzyme activities. One-way ANOVA revealed no effect of oxycodone exposure on acetylcholinesterase (AChE) activity (Figure 4A). On the other hand, one-way ANOVA revealed the effect of naloxone exposure (Figure 4B) on AChE activity (F(3,8) = 5.641; p = 0.0225). SH-SY5Y cells exposed to 100 µM naloxone had a statistically significant AChE activity inhibition (85%) when compared to nonexposed cells. Although 1 and 10 µM naloxone inhibited the AChE activity by ~40%, there was no statistically significant difference compared to control cells.
Figure 4.
Biochemical analysis of a neuroblastoma cell line (SH-SY5Y) exposed for 24 h to oxycodone (A,C,E) and naloxone (B,D,F). The results were analyzed by one-way ANOVA followed by Dunnett’s post-test and presented as the mean ± SEM (n = 2–3). C: control, nonexposed cells. * Statistically different from C.
One-way ANOVA showed no effect of oxycodone treatment on glutathione S-transferase (GST) activity (Figure 4C). Albeit not statistically significant, the exposure to oxycodone (1, 10, and 100 µM) inhibited the GST activity by ~40% compared to control cells. On the other hand, the one-way ANOVA showed the effect of naloxone exposure (Figure 4D) on GST activity (F(3,4) = 7.364; p = 0.0417). The exposure to 100 µM naloxone induced a statistically significant inhibition of GST activity (~80%) compared to control cells. Although 1 and 10 µM naloxone inhibited the GST activity by 55%, there was no statistically significant difference compared to control cells.
One-way ANOVA showed no effect of oxycodone treatment on superoxide dismutase (SOD) activity (Figure 4E). Although not statistically significant, the exposure to oxycodone (1, 10, and 100 µM) caused approximately 29% SOD activity inhibition compared to control cells. The one-way ANOVA showed the effect of naloxone exposure (Figure 4D) on SOD activity (F(3,7) = 8.528; p = 0.0098). The exposure to naloxone (1, 10, and 100 µM) induced a statistically significant inhibition of SOD activity compared to control cells.
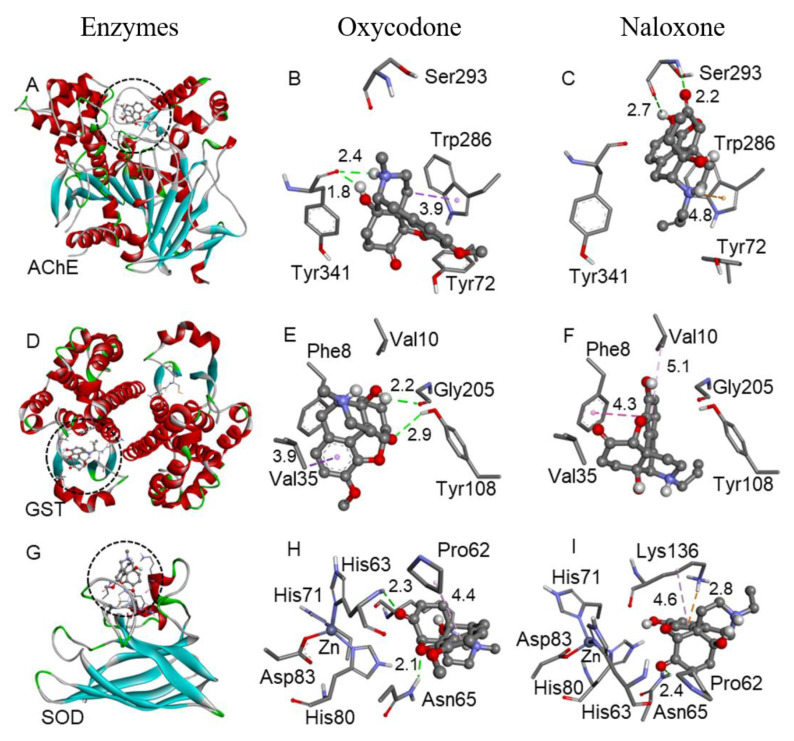
2.5. Molecular Docking
To verify the binding pose and interactions among oxycodone and naloxone with AChE, GST, and SOD enzymes, molecular docking simulations were performed (Figure 5). The predicted binding free energy (ΔG) suggested that the interactions were thermodynamically favorable, i.e., the compounds can interact in the enzyme active sites. The ΔG of oxycodone with the AChE, GST, and SOD was −7.1, −6.7, and −6.3 kcal/mol, respectively. Similarly, the ΔG of naloxone was −7.1 (AChE), −6.5 (GST), and −6.1 (SOD) kcal/mol. Despite the structural similarity between oxycodone and naloxone, the molecules showed different binding poses with the studied enzymes, suggesting that the phenol and allylamine groups from naloxone and the ether and methylamine moieties from oxycodone could interfere in the molecules’ conformation and interactions.
Figure 5.
Binding poses of naloxone and oxycodone with the AChE (A–C), GST (D–F), and SOD (G–I) enzymes. Oxycodone (B,E,H) and naloxone (C,F,I) are represented by the ball-and-stick model. Only the main residues involved in the interactions are shown in the stick model. The interaction distances are in Å, with the H-bonds (green), electrostatic (orange), and hydrophobic interactions (purple) represented by dotted lines.
3. Discussion
The analgesic ladder of the World Health Organization determines the most appropriate form of opioid use, staggering the type of pain and opioid of choice [15]. However, opioids are frequently used incorrectly either by self-medication or by inappropriate drug prescription. Moon and Chun [16] reported three clinical cases of patients who used fentanyl without a correct prescription. The inadequate use of opioids leads to side-effects, such as alterations in pyramidal physiology [17], apnea, hypoxia, respiratory depression [11,18], constipation, nausea, vomiting, and reflux [19,20]. In this study, we observed that oxycodone and naloxone did not alter cell viability. However, exposure of cells to oxycodone and naloxone caused enzymatic inhibition and DNA damage.
The cell viability assay was carried out using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). In this assay, the metabolically active cells transform the water-soluble dye MTT into an insoluble formazan [21]. In our study, there was no change in cell viability of cells exposed to 1, 10, and 100 µM oxycodone or naloxone for 24 h. In addition, neither necrosis nor apoptosis was observed after oxycodone and naloxone exposure. In contrast, Kokki et al. [22] showed that at higher concentrations (500–2000 µM), morphine and oxycodone decreased the SH-SY5Y cell viability after 48 h of exposure. Another study using drugs from the same pharmacological class showed that neuroblastoma cells exposed to tramadol (~600 µM) and tapentadol (~243 µM) for 48 h had a reduction in cell viability [23]. Moreover, Lin et al. [24] demonstrated the induction of apoptosis in 60% of cells exposed to 4000 µM of morphine for 48 h.
In this study, the exposure to naloxone inhibited the AChE activity. AChE hydrolyzes acetylcholine neurotransmitters in the cholinergic synapses; the inhibition of this enzyme leads to the interruption of acetylcholine degradation, which in turn hyperstimulates neuronal activity [25]. The inhibition of rodent AChE by opioid drugs, such as naloxone, has already been reported in the literature [25]. Motel et al. [26] suggested the use of naloxone as an adjuvant in the treatment of Alzheimer’s disease. However, inhibiting other enzymes must be considered; for instance, in this study, naloxone exposure also inhibited SOD and GST activity.
The enzyme GST is part of the antioxidant system, performing electrophilic detoxification [27]. Animal model studies demonstrated the inhibition of hepatic GST by morphine [28,29]. SOD enzyme catalyzes the degradation of superoxide anion [30]. Sadat-Shirazi et al. [31] demonstrated the inhibition of SOD activity in the prefrontal cortex of opioid abusers. SOD and GST inhibition increases reactive oxygen species and consequent oxidative stress [32,33]. As demonstrated by Ma et al. [32] and Salarian et al. [33], oxidative stress is related to addiction, dependence, and tolerance. The increase in reactive species is one of the factors linked to hyperalgesia. This factor requires ingesting a larger dose or a more potent drug to reach the desired analgesia threshold [34,35]. Therefore, the development of physiological and chemical dependence related to opioids can occur [17,36].
Even though naloxone and oxycodone have similar steric conformations, they exhibit distinct interactions with the enzymes (AChE, GST, and SOD), as demonstrated by molecular docking. Oxycodone and naloxone bind in the same region of the enzymes but interact with different amino acids. According to molecular docking, naloxone has more stable interactions. This may contribute to the different results of AChE, SOD, and GST inhibition observed in this study. Oxycodone and naloxone bind in the AChE peripheral anionic site (PAS). While oxycodone interacts with Trp286 (hydrophobic interaction) and Tyr341 (H-bonds) residues, naloxone interacts with Ser293 (H-bond) and Trp286 (electrostatic interaction). These types of interactions were observed with other AChE inhibitors [37]. Regarding GST, oxycodone and naloxone interact in the enzyme hydrophobic substrate-binding site (H-site). While oxycodone interacts with Val35 (hydrophobic interaction), Tyr108 (H-bond), and Gly205 (H-bond) residues, naloxone interacts with Val10 and Phe8 (hydrophobic interaction). In the SOD, the compounds bind near the zinc site. Oxycodone interacts with the Pro62 (hydrophobic interaction), Asn65 (H-bond), and His63 (H-bond) residues, and naloxone interacts with Lys136 (hydrophobic and electrostatic interactions).
The significant inhibition of the antioxidant system by naloxone may explain the DNA damage observed in this study; however, more studies are necessary to explain the oxycodone causing DNA damage. Sandoval-Sierra et al. [38] observed an increase in DNA methylation after 24 h opioid treatment. Another study by Tsujikawa et al. [39] demonstrated that DNA damage is a likely mechanism for morphine-induced P53 upregulation, probably through activation of the kappa-opioid receptor, which might lead to immune suppression.
In addition to the differences pointed out above, oxycodone and naloxone have different physiological action mechanisms. Oxycodone has an agonist action; in this way, the drug inhibits the pain, via the descending pathway, blocking the nociception [22]. On the other hand, naloxone has an antagonist action, inhibiting the agonists’ action, binding in the receptor, and displacing the drug that performs agonist action [40]. The different actions of these two drugs can activate distinct pathways in the cells, which may explain the results found in this work; however, more studies are needed to better explain these alterations.
4. Materials and Methods
4.1. Neuroblastoma Cell Line
The cell line SH-SY5Y (neuroblastoma) was acquired at the cell bank of Rio de Janeiro, Brazil. SH-SY5Ycells are neuroblastic-type cells, i.e., are characterized as immature nerve cells [41].
4.2. Cell Cultivation
Cells were cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 (DMEM-F12) (Sigma-Aldrich®—USA) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich®—USA) and 1% penicillin/streptomycin (Sigma-Aldrich®—USA). SH-SY5Y cells were kept in a 75 cm2 culture flask at 37 °C in a humidified atmosphere with 5% CO2. The medium was replaced every 2–3 days.
4.3. Oxycodone and Naloxone Exposure
SH-SY5Y cells were plated into six- and 96-well plates at a density of 1 × 105, 2.5 × 105, or 1 × 106 cells/well depending on the analysis performed. After 24 h, the complete DMEM-F12 medium was replaced by 0, 1, 10, and 100 µM oxycodone (Sigma-Aldrich®—St. Louis, MO, USA) or naloxone (Cayman Chemical Company®—Ann Arbor, MI, USA) for 24 h. Cells were also exposed to 3.3% methanol the oxycodone vehicle. Naloxone was prepared in DMEM-F12 medium.
4.4. Cytotoxicity Assay
After oxycodone and naloxone exposure, the cell medium was replaced by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, 1 mg/mL of FBS-free DMEM-F12) (Invitrogen®—Waltham, MA, USA) solution and the plates were incubated at 37 °C. After 3 h of incubation, the MTT solution was replaced by 100 µL of dimethyl sulfoxide (DMSO; Êxodo Cientifica®—Sumaré, SP, Brazil) to dissolve the formazan crystals. Absorbance was measured spectrophotometrically at a wavelength of 595 nm [42]. Results were expressed as percentages of the control.
4.5. Identification of Apoptotic and Necrotic Cells
After oxycodone and naloxone exposure, the cell medium was collected, and the cells were washed twice with phosphate-buffered saline (PBS, Sigma-Aldrich®—St. Louis, MO, USA) and harvested using trypsin (Sigma-Aldrich®—St. Louis, MO, USA). The cell suspensions were centrifuged at 446× g to remove the culture medium. After centrifugation, the cell pellets were resuspended with 250 µL of binding buffer (BD Biosciences®—New Jersey, NJ, USA), to which 3 µL of FITC-conjugated Annexin V (Invitrogen®—USA) and 5 µL of 7 aminoactinomycin D (Invitrogen®— Waltham, Massachusetts, USA) were added, and the cell suspensions were incubated in the dark for 15 min. A FACS Canto II flow cytometer (Becton Dickinson—Franklin Lakes, NJ, USA) with FITC and PERCP channels was used to evaluate the cells. Analyses were performed using Infinicyt software version 6.0. The results were expressed as percentages of the control [43].
4.6. DNA Damage
The DNA damage was analyzed using the alkaline comet assay according to Singh et al. [44] with modifications by Ferraro et al. [45]. After oxycodone and naloxone, the cell medium was collected, and the wells were washed two times with PBS and harvested by trypsin. All samples were submitted to viability analysis by the Trypan Blue (Sigma-Aldrich®—St. Louis, MO, USA) dye exclusion technique, and only samples with more than 80% viability were used in the comet assay. The cells were resuspended in low-melting-point agarose (Kasvi®—São José dos Pinhais, PR, Brazil) (0.5% w/v in PBS) and then deposited on microscope slides precoated with 1.5% agarose. After solidification under refrigeration at 4 °C for 10 min, the slides were immersed in freshly prepared lysis solution (2.5 M NaCl (Dinâmica®— Indaiatuba, SP, Brazil), 100 mM ethane-1,2-diyldinitrilo tetra acetic acid (EDTA; Neon®—Suzano, SP, Brazil), 10 mM Tris-HCl (Synth®—São Paulo, SP, Brazil), 1% N-lauryl sarcosinate (Synth®—São Paulo, SP, Brazil), 1% Triton X-100 (Synth®—São Paulo, SP, Brazil), and 10% DMSO) overnight. The slides were then transferred to a horizontal electrophoresis vat, filled with freshly prepared buffer solution (200 mM EDTA and 10 M NaOH (Dinâmica®—Indaiatuba, SP, Brazil), pH > 13) at 4 °C for 25 min for nucleoid unfolding, and then electrophoresed at 300 mA and 1 V/cm for another 25 min. After the end of this step, the slides were neutralized with Tris-HCl buffer solution (4.85%, pH 7.5) in three baths of 5 min each, and finally fixed in 100% ethanol (Alphatec®—Carlsbad, CA, USA). The DNA damage was analyzed using an epifluorescence microscope at 400× magnification after staining the slides with 40 µL of ethidium bromide (Sigma-Aldrich®—St. Louis, MO, USA) solution (0.02 g/mL). A total of 100 nucleoids per slide were visually evaluated using the “tail” size as a parameter, assuming values from 0 to 4 (0: no apparent damage and 4: maximum apparent damage). A total score was calculated from the 100 nucleoids, which assumes a value spectrum ranging from 0 to 400 [46]. To demonstrate the reliability of the test, positive control was performed with 0.5 mM methyl methane sulfonate (Sigma-Aldrich®—St. Louis, MO, USA).
4.7. Biochemical Analyses
4.7.1. Sample Preparation
After oxycodone and naloxone exposure, the cell medium was collected, and the cells were washed two times with PBS and harvested by trypsin. Samples were centrifuged at 1200 rpm, the supernatant was discarded, and the pellet was washed two times with PBS. The samples were resuspended in PBS, and the aliquots were stored at −80 °C until the biochemical analyses.
4.7.2. Acetylcholinesterase
AChE activity was measured as described by Ellman et al. [47]. Briefly, in a 96-well plate, 25 µL of the sample (prepared as described in Section 4.7.1), 0.75 mM 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB; Sigma-Aldrich®—St. Louis, MO, USA), and 10 mM acetylthiocholine (Sigma-Aldrich®—St. Louis, MO, USA) were added. The plate was incubated for 5 min at 37 °C, and the absorbance was measured at 405 nm once a minute for five minutes. The activity was expressed as nmol/min/mg of protein.
4.7.3. Glutathione S-Transferase
The GST activity was evaluated using the method described by Keen et al. [48]. Briefly, in a 96-well plate, 20 µL of the sample (prepared as described in Section 4.7.1), 3.0 mM 1-chloro-2,4-dinitrobenzene (CDNB; Sigma-Aldrich®—St. Louis, MO, USA), 3.0 mM reduced glutathione (GSH; Sigma-Aldrich®—St. Louis, MO, USA), and 0.1 M phosphate buffer (pH 6.5) were added. Absorbance was measured at 340 nm once a minute for 4 min. The activity was expressed as nmol/min/mg of protein.
4.7.4. Superoxide Dismutase
The SOD activity was performed according to the method proposed by Gao et al. [49]. In a 96-well plate, 40 µL of the sample (prepared as described in Section 4.7.1), 1 M Tris (Sigma-Aldrich®—USA), 5 mM EDTA (VWR Life Science®—Denver, CO, USA) buffer, pH 8.0, and 15 mM pyrogallic acid (Labsynth®—Br, Diadema, SP, Brazil) were added. After 30 min of incubation at room temperature, 1 N hydrochloric acid (Sigma-Aldrich®—St. Louis, MO, USA) was added to stop the reaction. The absorbance was measured spectrophotometrically at 440 nm. The results were expressed as U of SOD/mg of protein.
4.7.5. Protein Quantification
Total protein was quantified as described by Bradford [50]. Briefly, 10 µL of the sample (prepared as described in Section 4.7.1) and 250 µL of Bradford reagent (Thermo Scientific®—Waltham, MA, USA) were pipetted into the 96-well plate. A standard curve of bovine serum albumin (BSA) (Sigma-Aldrich®—USA) was prepared (0–500 µg BSA/mL). The absorbance was measured spectrophotometrically at 595 nm.
4.8. Molecular Docking
Computational docking studies were performed using AutoDock Vina 1.1.1 [51] based on previous studies [52,53,54], with an exhaustiveness of 20. The AChE, GST, and SOD enzymes were obtained from the Protein Data Bank (PDB ID: 4EY6, 6GSS, and 2C9V, respectively), and the grid box was centered on the active site (AChE: −13.25, −46.76, 33.52 with size 30 × 30 × 30 Å; GST: 11.44, 4.81, 26.31 with size 20 × 20 × 20 Å; SOD: 22.35, −15.74, 15.49 with size 20 × 20 × 20 Å). The ligands (oxycodone and naloxone) were created using Avogadro 1.1.1 software [55] followed by semi-empirical PM6 geometry optimization using the MOPAC2016 program [56]. The amine group of the ligands was considered protonated at physiological pH, as previously reported for morphine [57]. The files were prepared for docking using AutoDock Tools 4.2 [58], in which the compounds were considered flexible and the enzymes rigid. Accelrys discovery studio visualizer [59] was used in the analysis of the results.
4.9. Statistical Analyses
For all tests, at least, three independent experiments were carried out. The data were statistically analyzed using the Prisma Graphpad software, version 6.0, using the Kruskal–Wallis test followed by the Dunn’s test and presented as the median ± interquartile interval (cell viability, identification of apoptotic and necrotic cells, and DNA damage) or one-way ANOVA followed by the Dunnett test and presented as the mean ± SEM (biochemical analyses). Results were considered statistically different when p < 0.05.
5. Conclusions
In conclusion, we can observe that, in addition to the chronic toxicity caused by using opioids, 24 h of exposure to naloxone or oxycodone was also harmful, causing toxic effects to the cells, such as inhibition of enzymes of the antioxidant system and DNA damage without altering cell viability. Interestingly, the naloxone exposure caused more pronounced toxic effects on the SH-SY5Y cells than oxycodone.
Author Contributions
Conceptualization, L.S.L., A.C.I. and C.S.O.; investigation, L.S.L., N.d.S.d.C., M.E.P., M.E.A.G., W.A., P.A.N. and C.S.O. writing—review and editing, L.S.L., N.d.S.d.C., W.A., P.A.N., M.M.C., A.C.I. and C.S.O.; visualization, C.S.O.; supervision, A.C.I. and C.S.O. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Trescot A.M., Datta S., Lee M., Hansen H. Opioid pharmacology. Pain Physician. 2008;11:133–153. doi: 10.36076/ppj.2008/11/S133. [DOI] [PubMed] [Google Scholar]
- 2.Chan P.C.W. China’s Approaches to International Law since the Opium War. Leiden J. Int. Law. 2014;27:859–892. doi: 10.1017/S0922156514000399. [DOI] [Google Scholar]
- 3.Pathan H., Williams J. Basic opioid pharmacology: An update. Br. J. Pain. 2012;6:11–16. doi: 10.1177/2049463712438493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huddart R., Clarke M., Altman R.B., Klein T.E. PharmGKB summary: Oxycodone pathway, pharmacokinetics. Pharmacogenet. Genom. 2018;28:230. doi: 10.1097/FPC.0000000000000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith H.S. Opioid Metabolism. Mayo Clin Proc. 2009;84:613–624. doi: 10.1016/S0025-6196(11)60750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira G.H., Camargo M.M.A. Opiáceos e Opioides. In: OGA S., Camargo M.M.A., Batistuzzo J.A.O., editors. Fundamentos da Toxicologia. 4th ed. Atheneu; São Paulo, Brazil: 2014. [Google Scholar]
- 7.National Institute on Drug Abuse—NIDA Naloxone Drug Facts. [(accessed on 13 June 2022)];2022 January; Available online: https://nida.nih.gov/publications/drugfacts/naloxone.
- 8.Dowell D., Haegerich T.M., Chou R. CDC Guideline for Prescribing Opioids for Chronic Pain—United States, 2016. Recomm. Rep. 2016;65:1–49. doi: 10.15585/mmwr.rr6501e1. [DOI] [PubMed] [Google Scholar]
- 9.Tyan P., Carey E.T. Physiological Response to Opioids. Clin. Obstet. Gynecol. 2019;62:11–21. doi: 10.1097/GRF.0000000000000421. [DOI] [PubMed] [Google Scholar]
- 10.Chahl L.A. Opioids—Mechanisms of action. Exp. Clin. Pharmacol. 1996;19:63–65. [Google Scholar]
- 11.Parkin S., Neale J., Brown C., Campbell A.N.C., Castillo F., Jones J.D., Strang J., Comer S.D. Opioid overdose reversals using naloxone in New York City by people who use opioids: Implications for public health and overdose harm reduction approaches from a qualitative study. Int. J. Drug Policy. 2020;79:102751. doi: 10.1016/j.drugpo.2020.102751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motaghinejad M.S.M., Motaghinejad O., Shabab B., Asadighaleni M., Fatima S. The effect of various morphine weaning regimens on the sequelae of opioid tolerance involving physical dependency, anxiety and hippocampus cell neurodegeneration in rats. Fundam. Clin. Pharmacol. 2015;29:299–309. doi: 10.1111/fcp.12121. [DOI] [PubMed] [Google Scholar]
- 13.Bates D.D., Gallagher K., Yu H., Uyeda J., Murakami A.M., Setty B.N., Anderson S.W., Clement M.O. Acute radiologic manifestations of America’s opioid epidemic. RadioGraphics. 2018;3:109–123. doi: 10.1148/rg.2018170114. [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention (CDC) Death Rate Maps & Graphs, Drug Overdose Deaths Remain High. [(accessed on 25 June 2022)];2022 June 2; Available online: https://www.cdc.gov/drugoverdose/deaths/index.html.
- 15.Anekar A.A., Cascella M. StatPearls [Internet] StatPearls Publishing; Treasure Island, FL, USA: 2022. WHO Analgesic Ladder. [PubMed] [Google Scholar]
- 16.Moon J.M., Chun B.J. Fentanyl Intoxication Caused by Abuse of Transdermal Fentanyl. J. Emer. Med. 2011;40:37–40. doi: 10.1016/j.jemermed.2007.10.075. [DOI] [PubMed] [Google Scholar]
- 17.Pujol C.N., Paasche C., Laprevote V., Trojak B., Vidailhet P., Bacon E., Lalanne L. Cognitive effects of labeled addictolytic medications. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;81:306–332. doi: 10.1016/j.pnpbp.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Marshansky S., Mayer P., Rizzo D., Baltzan M., Denis R., Lavigne G.J. Sleep, chronic pain, and opioid risk for apnea. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;87:234–244. doi: 10.1016/j.pnpbp.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Ortiz V., García-Campos M., Sáez-González E., Delpozo P., Garrigues V. A concise review of opioid-induced esophageal dysfunction: Is this a new clinical entity? Dis. Esophagus. 2018;31:1–6. doi: 10.1093/dote/doy003. [DOI] [PubMed] [Google Scholar]
- 20.Whitman Z., O’neil D.H.R. Gastric disorders: Modifications of gastric content, antacids and drugs influencing gastric secretions and motility. Anaesth. Intensive Care Med. 2018;19:25–29. doi: 10.1016/j.mpaic.2017.10.011. [DOI] [Google Scholar]
- 21.Rai Y., Pathak R., Kumari N., Sah D.K., Pandey S., Kalra N., Soni R., Dwarakanath B.S., Bhatt A.N. Mitochondrial biogenesis and metabolic hyperactivation limits the application of MTT assay in the estimation of radiation induced growth inhibition. Sci. Rep. 2018;8:1531. doi: 10.1038/s41598-018-19930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokki M., Pesonen M., Vehviläinen P., Litmala O., Pasanen M., Kokki H. Cytotoxicity of oxycodone and morphine in human neuroblastoma and mouse motoneuronal cells: A comparative approach. Drugs R&D. 2016;16:155–163. doi: 10.1007/s40268-016-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faria J., Barbosa J., Queirós O., Moreira R., Carvalho F., Dinis-Oliveira R.J. Comparative study of the neurotoxicological effects of tramadol and tapentadol in SH-SY5Y cells. Toxicology. 2016;359:1–10. doi: 10.1016/j.tox.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Lin X., Wang Y.J., Li Q., Hou Y.Y., Hong M.H., Cao Y.L., Chi Z.Q., Liu J.G. Chronic high-dose morphine treatment promotes SH-SY5Y cell apoptosis via c-Jun N-terminal kinase-mediated activation of mitochondria-dependent pathway. FEBS J. 2009;276:2022–2036. doi: 10.1111/j.1742-4658.2009.06938.x. [DOI] [PubMed] [Google Scholar]
- 25.Aramjoo H., Riahi-Zanjani B., Farkhondeh T., Forouzanfar F., Sadeghi M. Modulatory effect of opioid administration on the activity of cholinesterase enzyme: A systematic review of mice/rat models. Environ. Sci. Pollut. Res. 2021;28:52675–52688. doi: 10.1007/s11356-021-16044-1. [DOI] [PubMed] [Google Scholar]
- 26.Motel W.C., Coop A., Cunningham C.W. Cholinergic modulation by opioid receptor ligands: Potential application to Alzheimer’s disease. Mini-Rev. Med. Chem. 2013;13:456–466. doi: 10.2174/138955713804999784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lannutti F., Marrone A., Re N. Binding of GSH conjugates to π-GST: A cross-docking approach. J. Mol. Graph. Model. 2012;32:9–18. doi: 10.1016/j.jmgm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Payabvash S., Beheshtian A., Salmasi A.H., Kiumehr S., Ghahremani M.H., Tavangar S.M., Sabzevari O., Dehpour A.R. Chronic morphine treatment induces oxidant and apoptotic damage in the mice liver. Life Sci. 2006;79:972–980. doi: 10.1016/j.lfs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Samarghandian S., Afshari R., Farkhondeh T. Effect of long-term treatment of morphine on enzymes, oxidative stress indices and antioxidant status in male rat liver. Int. J. Clin. Exp. Med. 2014;7:1449. [PMC free article] [PubMed] [Google Scholar]
- 30.Hashemi S.A., Karami M., Bathaie S.Z. Saffron carotenoids change the superoxide dismutase activity in breast cancer: In vitro, in vivo and in silico studies. Int. J. Biol. Macromol. 2020;158:845–853. doi: 10.1016/j.ijbiomac.2020.04.063. [DOI] [PubMed] [Google Scholar]
- 31.Sadat-Shirazi M.S., Zarrindast M.R., Ashabi G. Oxidative stress enzymes are changed in opioid abusers and multidrug abusers. J. Clin. Neurosci. 2020;72:365–369. doi: 10.1016/j.jocn.2019.12.064. [DOI] [PubMed] [Google Scholar]
- 32.Ma J., Yuan X., Qu H., Zhang J., Wang D., Sun X., Zheng Q. The role of reactive oxygen species in morphine addiction of SH-SY5Y cells. Life Sci. 2015;124:128–135. doi: 10.1016/j.lfs.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Salarian A., Kadkhodaee M., Zahmatkesh M., Seifi B., Bakhshi E., Akhondzadeh S., Adeli S., Askari H., Arbabi M. Opioid use disorder induces oxidative stress and inflammation: The attenuating effect of methadone maintenance treatment. Iran. J. Psychiatry. 2018;13:46. [PMC free article] [PubMed] [Google Scholar]
- 34.Khomula E.V., Araldi D., Bonet I.J., Levine J.D. Opioid-induced hyperalgesic priming in single nociceptors. J. Neurosci. 2021;41:31–46. doi: 10.1523/JNEUROSCI.2160-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobore T.O. Towards a Comprehensive Theory of Non-Cancer Acute and Chronic Pain Management: The Critical Role of Reactive Oxygen and Nitrogen Species in Pain, and Opioid Dependence, Addiction, Hyperalgesia, and Tolerance. Adv. Redox Res. 2021;2:100003. doi: 10.1016/j.arres.2021.100003. [DOI] [Google Scholar]
- 36.Hearing M. Prefrontal-accumbens opioid plasticity: Implications for relapse and dependence. Pharmacol. Res. 2019;139:158–165. doi: 10.1016/j.phrs.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckroat T.J., Manross D.L., Cowan S.C. Merged tacrine-based, multitarget-directed acetylcholinesterase inhibitors 2015–present: Synthesis and biological activity. Int. J. Mol. Sci. 2020;21:5965. doi: 10.3390/ijms21175965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandoval-Sierra J.V., García F.I.S., Brooks J.H., Derefinko K.J., Mozhui K. Effect of short-term prescription opioids on DNA methylation of the OPRM1 promoter. Clin. Epigenetics. 2020;12:1–14. doi: 10.1186/s13148-020-00868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsujikawa H., Shoda T., Mizota T., Fukuda K. Morphine induces DNA damage and P53 activation in CD3+ T cells. Biochim. Biophys. Acta—Gen. Subj. 2009;1790:793–799. doi: 10.1016/j.bbagen.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Tylleskar I., Skarra S., Skulberg A.K., Dale O. The pharmacokinetic interaction between nasally administered naloxone and the opioid remifentanil in human volunteers. Eur. J. Clin. Pharmacol. 2021;77:1901–1908. doi: 10.1007/s00228-021-03190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campos C.S., Gradowski F.C.N.T., Almeida B.P.F., França J.N., Santos R.B., Cavalli L.R., Elifio-Esposito S. An overview of neuroblastoma cell lineage phenotypes and in vitro models. Exp. Biol. Med. 2020;245:1637–1647. doi: 10.1177/1535370220949237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sumantran V.N. Cancer Cell Culture. Springer; Berlin/Heidelberg, Germany: 2011. Cellular chemosensitivity assays: An overview; pp. 219–236. [DOI] [PubMed] [Google Scholar]
- 43.Vermes I., Haanen C., Reutelingsperger C. Flow cytometry of apoptotic cell death. J. Immunol. Methods. 2000;243:167–190. doi: 10.1016/S0022-1759(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 44.Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 45.Ferraro M.V.M., Fenocchio A.S., Mantovani M.S., Ribeiro C.O., Cestari M.M. Mutagenic effects of lead (Pb II) on the fish H. malabaricus as evaluated using the comet assay, piscine micronucleus and chromosome aberrations tests. Genet. Mol. Biol. 2004;27:103–107. doi: 10.1590/S1415-47572004000100017. [DOI] [Google Scholar]
- 46.Collins A.R., Ai-Guo M., Duthie S.J. The kinetics of repair of oxidative DNA damage (strand breaks and oxidised pyrimidines) in human cells. Mutat. Res. /DNA Repair. 1995;336:69–77. doi: 10.1016/0921-8777(94)00043-6. [DOI] [PubMed] [Google Scholar]
- 47.Ellman G.L., Courtney K.D., Andres V., Jr., Featherstone R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 48.Keen J.H., Habig W.H., Jakoby W.B. Mechanism for the several activities of the glutathione S-transferases. J. Biol. Chem. 1976;251:6183–6188. doi: 10.1016/S0021-9258(20)81842-0. [DOI] [PubMed] [Google Scholar]
- 49.Gao R., Yuan Z., Zhao Z., Gao X. Mechanism of pyrogallol autoxidation and determination of superoxide dismutase enzyme activity. Bioelect. Bioenerg. 1998;45:41–45. doi: 10.1016/S0302-4598(98)00072-5. [DOI] [Google Scholar]
- 50.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 51.Trott O., Olson A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arya A., Azarmehr N., Mansourian M., Doustimotlagh A.H. Inactivation of the superoxide dismutase by malondialdehyde in the nonalcoholic fatty liver disease: A combined molecular docking approach to clinical studies. Arch. Physiol. Biochem. 2021;127:557–564. doi: 10.1080/13813455.2019.1659827. [DOI] [PubMed] [Google Scholar]
- 53.Kobzar O.L., Shulha Y.V., Buldenko V.M., Mrug G.P., Kolotylo M.V., Stanko O.V., Onysko P.P., Vovk I. Alkyl and aryl α-ketophosphonate derivatives as photoactive compounds targeting glutathione S -transferases. Phosphorus Sulfur Silicon Relat. Ele. 2021;96:672–678. doi: 10.1080/10426507.2021.1901703. [DOI] [Google Scholar]
- 54.Silva F.D., Nogara P.A., Ochoa-Rodríguez E., Nunez-Figueredo Y., Wong-Guerra M., Rosemberg D.B., Rocha J.B.T. Molecular docking and in vitro evaluation of a new hybrid molecule (JM-20) on cholinesterase activity from different sources. Biochimie. 2020;168:297–306. doi: 10.1016/j.biochi.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 55.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart J.J.P. Stewart Computational Chemistry—MOPAC Home Page. 2016. [(accessed on 13 June 2022)]. Available online: http://openmopac.net/home.html.
- 57.Møllendal H., Balcells D., Eisenstein O., Syversen L., Suissa M.R. Conformational complexity of morphine and morphinum in the gas phase and in water. A DFT and MP2 study. RSC Adv. 2014;4:24729–24735. doi: 10.1039/C4RA02992E. [DOI] [Google Scholar]
- 58.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 59.BIOVIA Discovery Studio Accelrys Discovery Studio [WWW Document] 2017. [(accessed on 13 June 2022)]. Available online: https://discover.3ds.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.