Abstract
Small heat shock proteins (sHSPs) are usually upregulated in plants under diverse environmental stresses. These proteins have been suggested to function as molecular chaperones to safeguard other proteins from stress-induced damage. The ripening of pepper (Capsicum annuum L.) fruit involves important phenotypic, physiological, and biochemical changes, which have associated endogenous physiological nitro-oxidative stress, but they can also be significantly affected by environmental conditions, such as temperature. Based on the available pepper genome, a total of 41 sHSP genes were identified in this work, and their distributions in the 12 pepper chromosomes were determined. Among these genes, only 19 sHSP genes were found in the transcriptome (RNA-Seq) of sweet pepper fruits reported previously. This study aims to analyze how these 19 sHSP genes present in the transcriptome of sweet pepper fruits are modulated during ripening and after treatment of fruits with nitric oxide (NO) gas. The time-course expression analysis of these genes during fruit ripening showed that 6 genes were upregulated; another 7 genes were downregulated, whereas 6 genes were not significantly affected. Furthermore, NO treatment triggered the upregulation of 7 sHSP genes and the downregulation of 3 sHSP genes, whereas 9 genes were unchanged. These data indicate the diversification of sHSP genes in pepper plants and, considering that sHSPs are important in stress tolerance, the observed changes in sHSP expression support that pepper fruit ripening has an associated process of physiological nitro-oxidative stress, such as it was previously proposed.
Keywords: fruit ripening, HSP20 family, nitric oxide, pepper, small heat shock proteins
1. Introduction
Environmental heat stress has a negative impact on the growth, development, and productivity of crop plants and is considered a serious threat [1,2], where the metabolism of reactive oxygen and nitrogen species (ROS and RNS, respectively) is usually involved as a response mechanism [3,4,5,6,7,8,9]. Consequently, the plant-adaptive response to high temperatures is very important for plant development and also for food security worldwide. Under heat stress conditions, the heat-responsive genes, mainly those encoding heat shock proteins (HSPs), are switched on [10,11,12,13].
Plant HSPs are grouped into five main families based on the molecular weight and sequence homology, including HSP100s, HSP90s, HSP70s, HSP60s, and HSP20s [14]. Among all the five conserved families, HSP20s, a group of small HSPs (sHSPs) with molecular sizes ranging from 15 to 42 kDa, is the most prevalent and abundant family induced by heat stress in many higher plants [15,16,17,18]. These proteins function as molecular chaperones to safeguard other proteins from stress-induced damage. A common signature of the sHSPs is the alfa-crystallin domain (ACD) with around 90 amino acids that form a seven-stranded β-sandwich. This core is flanked by a variable N-terminal domain (NTD) with fewer than 85 amino acids and by a short C-terminal extension (CTE). The sHSPs are present in the cytosol and different organelles, including chloroplast, mitochondrion, endoplasmic reticulum, and peroxisome. Therefore, they bear the necessary and specific targeting signal either in the N- or C-terminal regions to lead them to diverse organelles. Usually, the majority of the sHSPs form multi-subunit oligomers in their native state [17,19,20,21].
Nitric oxide (NO) is a radical molecule that can exert regulatory functions of protein through posttranslational modifications, mainly S-nitrosation, tyrosine nitration, and metal nitrosylation [22,23,24,25], or through interaction with phytohormones, hydrogen peroxide, hydrogen sulfide, and melatonin, among others [26,27,28,29,30]. Furthermore, NO applied exogenously using either NO donors, gas or nanomaterials, has been shown to trigger beneficial effects on plants under physiological and stress conditions [31,32,33,34,35]. In the case of climacteric and non-climacteric fruits, NO has been shown to modulate the ripening process as well as to extend the postharvest storage [36,37].
Pepper (Capsicum annuum L.) plants have a significant economical relevance since their fruits constitute a group of horticultural products that are among the most consumed worldwide either fresh or processed. This relevance is related to their significant nutritional potentiality since pepper fruits are a source of vitamins (A, C, E, and B6), β-carotene, minerals, folic acid, and fiber [38,39]. Experimental data supporting the ripening of pepper fruits is associated to physiological nitro-oxidative stress [40,41], where the ROS metabolism is significantly regulated [42,43,44,45] and modulated by NO [46,47,48,49,50]. In a previous study, Guo and colleagues [51] analyzed the pepper CaHSP20 genes family, focusing on their heat (40 °C, 2 h)-induced expression in various plant tissues, including root, stem, leaf, and flower. With this previous information, the present study focused on the analysis of the sHSP genes in the sweet pepper fruit using the RNAseq transcriptome obtained previously [46], and also on how the exogenous NO gas treatment could modulate their expression.
2. Results
2.1. sHSP Genes/Proteins from Pepper: Sequence, Structure, and Phylogenetic Analysis
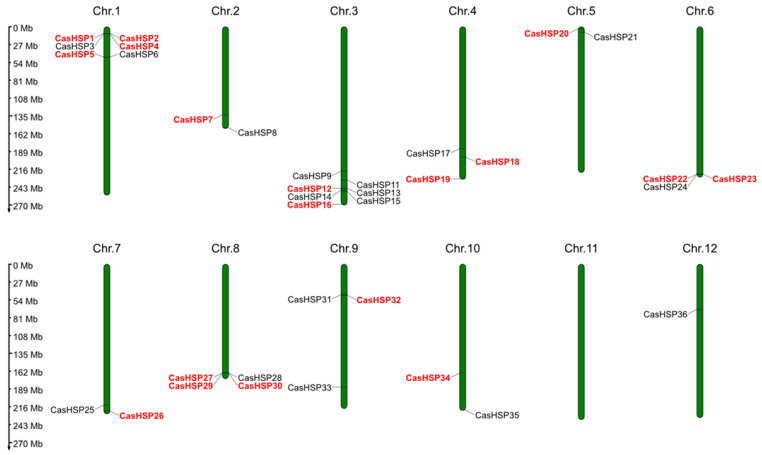
sHSPs are a type of molecular chaperone with high diversity that are widespread in higher plants [17,52]. They are involved in protein homeostasis by binding proteins in non-native conformations, avoiding the irreversible aggregation of unfolded proteins. In this study, a total of 41 sHSP genes were identified and characterized in the pepper genome, which was named CasHSP1 to CasHSP41 based on their chromosomal location (Table 1). These 41 CasHSP genes were distributed across 12 pepper chromosomes (Figure 1). Thus, chromosome (Chr.) 3 contains 8 genes followed by Chr. 1, which has 7 genes, Chr. 8 with 4 genes, Chrs. 4 and 6 with 3 genes, Chrs. 2, 5 and 7 with 2 genes, and Chr. 12 with 1 gene. The only chromosome that does not contain any genes is Chr. 11. However, in the transcriptome obtained in sweet pepper fruits [46], only 19 sHSP genes, indicated in red in Table 1, were identified, suggesting that they are fruit-specific.
Table 1.
Summary of the 41 small heat shock proteins (sHSPs) genes identified in the pepper (Capsicum annuum L.) genome and some of the properties related to the protein encoded for these genes and their subcellular localization. The nineteen CasHSP genes specifically detected in the sweet pepper fruit transcriptome are highlighted in red.
| Gene Name | Gene ID | Chr. | Genomic Location | Protein ID | Length (aa) | kDa | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| CasHSP1 | 107867615 | 1 | 9708054-9710773 | XP_016569420.2 | 215 | 24.5 | Plastid |
| CasHSP2 | 107867608 | 1 | 9725633-9727173 | NP_001311883.1 | 211 | 24.1 | Cytosol |
| CasHSP3 | 107852753 | 1 | 10229995-10231900 | XP_016553288.2 | 195 | 21.5 | Plastid |
| CasHSP4 | 107852746 | 1 | 10242027-10244276 | XP_016553275.2 | 170 | 18.4 | Plastid |
| CasHSP5 | 107845765 | 1 | 46048175-46056638 | XP_047266663.1 | 200 | 17.7 | Cytosol |
| CasHSP6 | 107874604 | 1 | 46076284-46077623 | XP_016576855.1 | 229 | 25.9 | Plastid/Mitochondrion |
| CasHSP7 | 107861257 | 2 | 133802215-133803014 | XP_016562092.1 | 137 | 15.7 | Cytosol |
| CasHSP8 | 107860709 | 2 | 152930539-152932661 | XP_016561651.1 | 141 | 16.4 | Cytosol |
| CasHSP9 | 107865071 | 3 | 219296047-219302039 | XP_047264613.1 | 159 | 18.5 | Cytosol/Nucleus |
| CasHSP10 | 107863044 | 3 | 232468924-232469818 | XP_047263866.1 | 234 | 25.7 | Plastid |
| CasHSP11 | 107863044 | 3 | 232468924-232469818 | NP_001311776.1 | 235 | 25.7 | Plastid |
| CasHSP12 | 107862598 | 3 | 245438867-245439343 | XP_016563712.2 | 158 | 17.7 | Cytosol |
| CasHSP13 | 107862597 | 3 | 245535719-245536195 | XP_016563710.2 | 158 | 17.7 | Cytosol |
| CasHSP14 | 107862438 | 3 | 248116668-248117246 | XP_016563509.1 | 192 | 19.1 | Extracellular |
| CasHSP15 | 107865962 | 3 | 249658853-249659681 | XP_016567632.1 | 211 | 24.0 | Cytosol |
| CasHSP16 | 107861648 | 3 | 270049897-270050678 | XP_016562442.1 | 148 | 16.5 | Cytosol |
| CasHSP17 | 107869601 | 4 | 185294212-185295359 | XP_016571594.2 | 313 | 35.5 | Cytosol |
| CasHSP18 | 107853496 | 4 | 197561845-197562964 | XP_016553971.2 | 171 | 18.4 | Cytosol |
| CasHSP19 | 107866765 | 4 | 231026649-231027604 | XP_016568214.1 | 153 | 16.7 | Plastid |
| CasHSP20 | 107870139 | 5 | 2638802-2640752 | XP_016572055.1 | 147 | 16.3 | Peroxisome |
| CasHSP21 | 107872316 | 5 | 7720799-7721401 | XP_016574557.1 | 200 | 22.6 | Vacuole |
| CasHSP22 | 107875508 | 6 | 223245724-223246143 | XP_016577738.2 | 139 | 17.8 | Cytosol |
| CasHSP23 | 107875506 | 6 | 223249290-223249754 | XP_016577734.2 | 154 | 17.8 | Cytosol |
| CasHSP24 | 107875505 | 6 | 223271811-223272275 | XP_016577732.1 | 154 | 17.6 | Cytosol |
| CasHSP25 | 107877347 | 7 | 213882206-213884807 | XP_016579494.1 | 251 | 27.6 | Plastid |
| CasHSP26 | 107856376 | 7 | 223234365-223235748 | XP_016556884.1 | 190 | 21.7 | Cytosol |
| CasHSP27 | 107840198 | 8 | 164788707-164789186 | XP_016539463.1 | 159 | 18.2 | Cytosol |
| CasHSP28 | 107879949 | 8 | 164801975-164802454 | XP_016582357.1 | 159 | 18.2 | Cytosol |
| CasHSP29 | 107840199 | 8 | 164826536-164827015 | XP_016539464.1 | 159 | 18.1 | Cytosol |
| CasHSP30 | 107840200 | 8 | 164831732-164832211 | XP_016539465.1 | 159 | 18.1 | Cytosol |
| CasHSP31 | 107842139 | 9 | 45981288-45981734 | XP_016541392.1 | 148 | 17.2 | Cytosol |
| CasHSP32 | 107842861 | 9 | 46468003-46469487 | XP_016542384.1 | 203 | 23.8 | Cytosol |
| CasHSP33 | 107841934 | 9 | 186338719-186339366 | XP_016541270.2 | 215 | 24.3 | Plastid/Mitochondrion |
| CasHSP34 | 107844800 | 10 | 164686556-164687242 | XP_016544631.2 | 228 | 26.5 | Cytosol/Mitochondrion |
| CasHSP35 | 107845199 | 10 | 221623125-221623644 | XP_016544930.1 | 144 | 16.6 | Cytosol/Golgi |
| CasHSP36 | 107850800 | 12 | 68905209-68906648 | XP_016551048.1 | 279 | 31.6 | Cytosol/Nucleus |
| CasHSP37 | 107848440 | Unplaced | 532-1187 | XP_047258842.1 | 254 | 28.4 | Cytosol/Golgi |
| CasHSP38 | 107855006 | Unplaced | 357733-359091 | XP_016555471.1 | 246 | 26.9 | Golgi |
| CasHSP39 | 107855027 | Unplaced | 665247-666808 | XP_016555490.1 | 215 | 24.3 | Cytosol/Golgi |
| CasHSP40 | 124890240 | Unplaced | 3954230-3955587 | XP_047258018.1 | 246 | 26.8 | Golgi/Plastid |
| CasHSP41 | 124890244 | Unplaced | 4261704-4263265 | XP_047258024.1 | 215 | 24.3 | Cytosol/Golgi |
Figure 1.
Chromosomal location of the CasHSP genes identified in pepper (Capsicum annuum L.) genome. The approximate locations of the 41 CasHSP genes among the 12 chromosomes are displayed in each chromosome (Chr.1–Chr.12). The distribution of the 19 CasHSP genes specifically detected in the sweet pepper fruit transcriptome is highlighted in red. Chr., chromosome.
The identified CasHSP proteins, obtained from the nucleotide sequences, show a molecular weight ranging from 15.7 to 35.5 kDa and have a wide cellular distribution with 17 in the cytosol, 5 in the plastid, 1 in the peroxisome, 1 in the cytoskeleton, 1 in extracellular space, 1 in the vacuole, and 1 in the Golgi apparatus. Furthermore, there is dual localization of several sHSPs with 4 in plastid/mitochondrion, 3 in cytosol/nucleus, 2 in cytosol/mitochondrion, 4 in cytosol/Golgi, and 1 in Golgi/plastid.
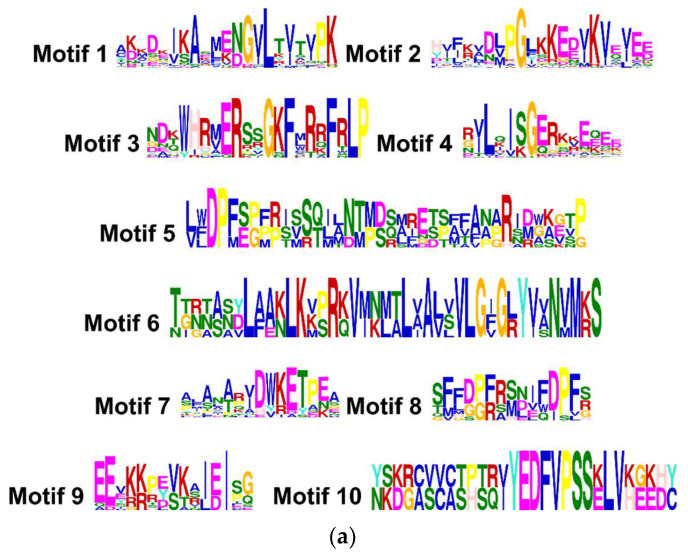
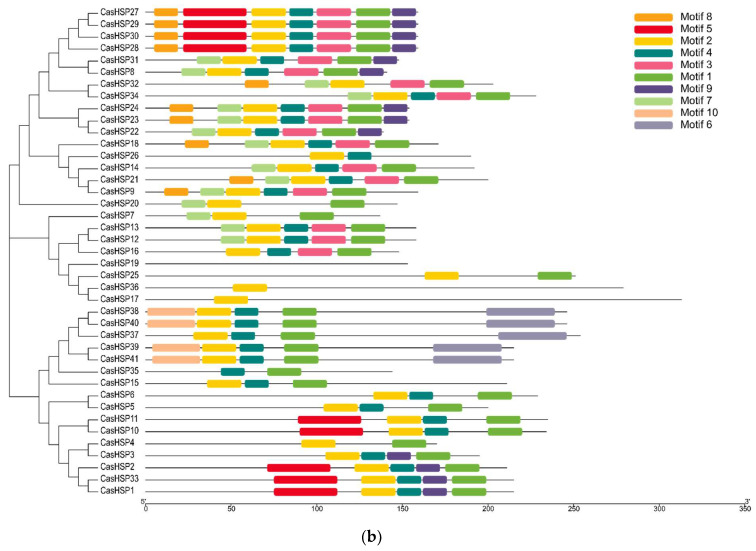
The analysis of the primary structure of the forty-one sHSP proteins revealed a high degree of identity with the families of sHSPs from other plant species, including Arabidopsis, tomato or potato, and allowed discriminating until ten amino acid motifs (Figure 2a). The distribution of these ten conserved motifs in the different CasHSPs from sweet pepper is represented in Figure 2b.
Figure 2.
Identification and position of consensus amino acid motifs for pepper CasHSPs. (a) Amino acids motifs. Ten amino acid motifs with various widths were identified. The height of each amino acid symbol is proportional to the degree of conservation in the consensus sequences depicted in the ten motifs. (b) Distribution of conserved motifs. The distribution of conserved motifs, numbers 1–10, of the forty-one pepper sHSPs, are represented by boxes of different colors. Sequence logos of conserved motifs were created by MEME.
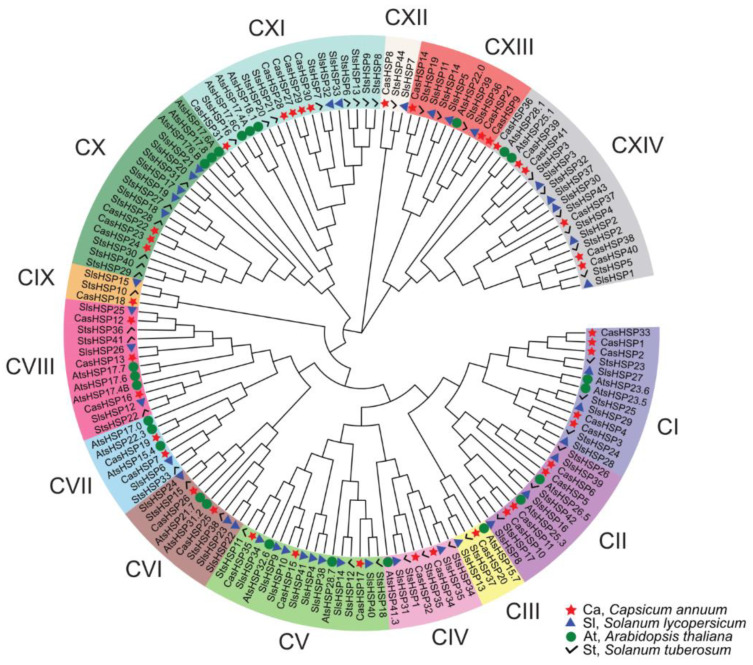
The phylogenetic comparative analysis among the sHSPs from four plant species including sweet pepper (Capsicum annuum), Arabidopsis thaliana, tomato (Solanum lycopersicum), and potato (Solanum tuberosum) allowed the identification of fourteen main sHSP groups, designated as I to XIV and depicted with different colors (Figure 3).
Figure 3.
Phylogenetic analysis of plant small heat shock proteins (sHSPs). Identified CasHSP proteins in the fruit transcriptome are highlighted in red color. The scale bar represents the phylogenetic branch length. Different subgroups of sHSPs are depicted in different colors and designated as I to XIV. Species abbreviations: At (Arabidopsis thaliana), Ca (Capsicum annuum), Sl (Solanum lycopersicum), and St (Solanum tuberosum).
2.2. NO Gas Differentially Modulates sHSP Gene Expression during Pepper Fruit Ripening
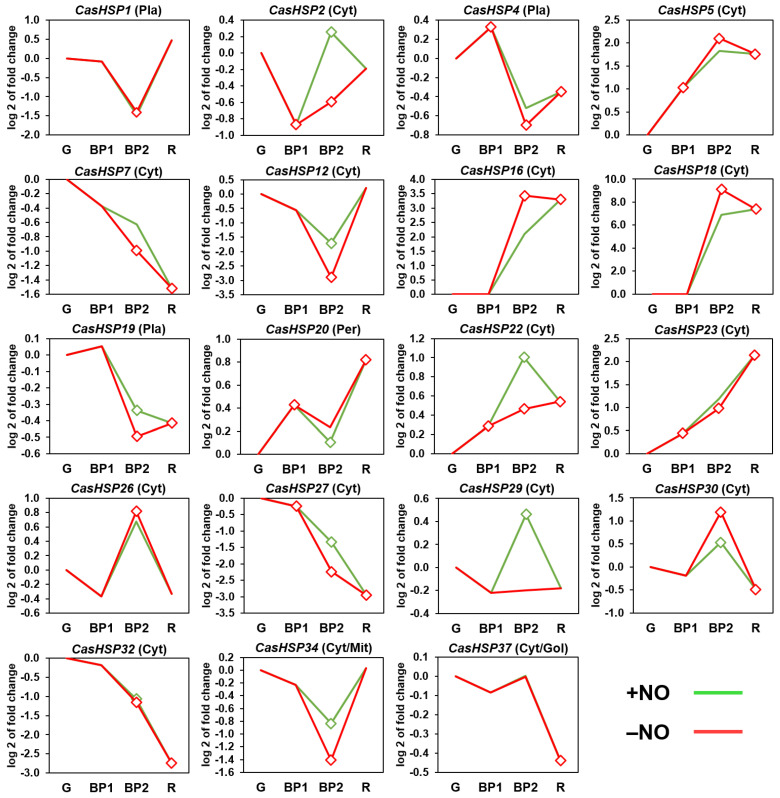
Based on previous analyses of sweet pepper fruits using the experimental design illustrated in Figure S1, three developmental steps were established: green immature (G), breaking point (BP1), and red ripe (R). Likewise, for the exposition to exogenous NO gas, two additional groups were established: fruits treated with 5 ppm NO for 1 h (BP2 + NO) and a parallel group that was not treated with NO (BP2 − NO), which was used as the control. Thus, the expression of the nineteen sHSP genes identified in the sweet pepper fruits was analyzed during ripening as well as the possible effect of NO gas treatment in comparison to untreated fruits. Figure 4 shows the time-course expression analysis of these nineteen sHSP genes by RNA-Seq at different stages of pepper fruit ripening. During ripening from green (G) to red (R) stages, it was possible to distinguish three groups of genes whose expression was increased (CasHSP5, CasHSP16, CasHSP18, CasHSP20, CasHSP22, and CasHSP23), decreased (CasHSP4, CasHSP7, CasHSP19; sHSP27, CasHSP30, CasHSP32, and CasHSP37) or not significantly affected (CasHSP1, CasHSP2, sHSP12, CasHSP26, CasHSP29, and sHSP34).
Figure 4.
Time-course expression analysis of nineteen CasHSP genes (RNA-Seq) under natural ripening conditions and after exogenous NO gas treatment. Samples of sweet pepper fruits at different ripening stages correspond to immature green (G), breaking point 1 (BP1), breaking point 2 with (green line) and without (red line) NO treatment (BP2 + NO and BP2 − NO, respectively) and ripe red (R). Diamonds indicate statistically significant changes in expression levels (p < 0.05) in comparison to immature green (G). Cyt, cytosol. Mit, mitochondrion. Nuc, nucleus. Pla, plastid. Pex, peroxisome.
On the other hand, independently of the modulation during ripening, under NO gas treatment at the BP2 stage, some of these genes were also either positive (CasHSP2, CasHSP7, CasHSP12, CasHSP22, CasHSP27, CasHSP29, and CasHSP34), negatively (CasHSP16, CasHSP18, and CasHSP30) or not significantly affected (sHSP1, sHSP4, sHSP5, CasHSP19, CasHSP20, CasHSP23, CasHSP26, CasHSP32, and CasHSP37) by NO treatment. Overall, a total of 10 CasHSP genes were modulated by NO.
3. Discussion
3.1. Pepper sHSP Genes Are Modulated during Ripening
Fruit ripening is a complex physiological process that involves high coordination among the different metabolic pathways that take place in the different subcellular compartments. Pepper is a non-climacteric fruit, which means that its ripening is independent of the phytohormone ethylene. Previous studies in pepper fruits have shown that the ripening implies significant changes at the transcriptome, proteome, and metabolic levels, and particularly there is a modulation of the metabolism of ROS and RNS [43,47,48,49,50] in a process that can be defined as a “physiological nitro-oxidative stress”, which has been also defined in the animal system as “oxidative eustress [53,54].
In a previous report, a search in the Pepper Genome Database (PGD, http://peppergenome.snu.ac.kr/ (accessed on 28 May 2022), CM334, Zunla-1, a variety which produced hot fruit) allowed to identify 36 genes of the CaHSP20 family. The expression patterns of these genes were evaluated in four different tissues (root, stem, leaf, and flower) from two cultivars called B6 (thermo-sensitive) and R94 (thermo-tolerant) under heat stress. Among these genes, only 7 genes (CaHsp16.4, 18.2a, 18.7, 21.2, 22.0, 25.8, and 25.9) showed higher expression levels in both lines B6 and R94 under heat stress, and the rest of the genes showed what the author called “hysteretic expression” [51]; however, fruits were not analyzed in that work. In the present study, using our transcriptome from California-type sweet pepper fruits, cultivar Melchor, we analyzed the sHSP gene family because, to our knowledge, the information on sHSPs functions during fruit ripening, for either climacteric or non-climacteric fruits, are very limited. In our fruit system, we found 19 sHSP genes, which are 17 fewer than those found in pepper plants of the cultivar Zunla-1 [51]. This difference in the number of genes is notable and could be explained by the fact that our Melchor cultivar corresponds to California-type sweet peppers, while Zunla-1 produces hot peppers. In addition, our transcriptome derives exclusively from the fruits, which suggests that these 19 genes could be more specific to this plant organ.
In tomato (Solanum lycopersicum cv Heinz 1706) plants, 33 sHSP genes have been reported and are mainly related to heat stress response and ripening [55]. In the case of tomato fruit which is a climacteric fruit, it was found that HSP17.7 and sugar can interact to regulate the development of tomato fruit and affect its quality [56]. More recently, it was shown that ethylene and RIPENING INHIBITOR can regulate gene expression of the tomato HSP17.7A and HSP17.7B, which usually are upregulated during the ripening transition of mature green to breaker stage [57]. Another study of the sHSPs during ripening and storage at a low temperature in two tomato cultivars (Micro-Tom and Minitomato) and storage at a low temperature showed a differential response, where sHSPs were induced in Micro-Tom but not in Minitomato fruits. Thus, sHSP17.4-CII and sHSP23.8-M transcripts were strongly accumulated in Micro-Tom fruit, suggesting that these sHSPs could be involved in the mechanism of chilling tolerance in the Micro-Tom cultivar [58]. Moreover, early reports showed that the tomato sHSP21 participates in the protection of photosystem II against oxidative stress as well as promoting the color changes during fruit ripening through the carotene accumulation [59]. Moreover, an accumulation of sHSP during the chromoplast development during tomato ripening was reported [60].
On the other hand, using two pepper cultivars with different sensitivities to temperature, it was observed that the thermo-tolerant cultivar had a higher CaHSP16.4 expression. Likewise, under combined stress of high temperature and drought, this cultivar had higher levels of antioxidant systems. Thus, the authors proposed that the higher expression of CaHSP16.4 favors the scavenger capacity against ROS [61].
Similarly, in pepper plants, it has been shown that the CaHsp25.9 expression increases under heat, salt, and drought stress, which confers tolerance since it was accompanied by a decrease in the accumulation of ROS, an increase of the activity of antioxidant enzymes, and upregulation in the expression of stress-related genes [62]. It should be mentioned that neither CaHsp16.4 nor CaHsp25.9 (named CasHSP8 and CasHSP6, respectively, in Table 1) has been identified in pepper fruits. More recently, the expression analysis of CaHSP18.1a (named CasHPS29 in Table 1 and expressed in fruit) in two pepper lines with different sensitivities to temperature (the thermo-sensitive B6 line and thermo-tolerant R9 line) showed that CaHSP18.1a is induced under heat stress, salt, and drought stress in both lines. Furthermore, the overexpression of CaHSP18.1a in Arabidopsis evidenced that this gene provides an increased resistance under these stresses since this transgenic Arabidopsis plants had also higher activity of some antioxidant enzymes, including catalase, SOD, and ascorbate peroxidase [63].
3.2. NO Modulates the Expression of Nine sHSP Genes during Pepper Ripening
NO is a well-recognized signal molecule that mediated many physiological processes including seed and pollen germination, root development, plant growth, stomatal movement, leaf senescence, flowering, and fruit ripening, and it is also involved in the mechanism of response against multiple adverse abiotic and biotic conditions [64,65,66]. Furthermore, new biotechnological approaches have been started to evaluate its potential application in crops using either NO donors or NO-releasing nanomaterial. In fact, postharvest storage has become one of the most attractive sectors to preserve horticultural products in good conditions and also to avoid losses due to pathogen attacks since NO can activate enzymatic and non-enzymatic antioxidant systems to palliate these negative outcomes [67,68,69].
The effect of exogenous NO gas on the pull of CasHSP genes during sweet pepper fruit ripening is, to our knowledge, unknown, and the provided data indicate that among the 19 identified sHSP genes, a total of 7 genes were upregulated and 3 were downregulated. It implies that, in a certain way, NO seems to restrict the effects triggered by the ripening process since a lower number of genes are influenced due to the treatment with the NO gas. In a previous study using the model plant Arabidopsis thaliana, the exogenous application of nitro-linolenic acid (NO2-Ln) which is a NO donor [6] triggered the induction of different families of HSPs including members of HSP40, HSP60, HSP70, and HSP90 but most of them corresponded to sHSPs such as Hsp17.6II (At5g12020) or Hsp17.6A (At5g12030) [70]. Our data show that in pepper fruit the CasHSP12 was positively regulated by NO. Consequently, our data provide additional evidence of the correlation between sHSPs and the signaling molecule NO which opens new questions to identify the mechanisms involved in this process. However, also, the knowledge of why and how the differential expression of these CasHSPs is orchestrated during ripening and by the NO treatment is a promising avenue that deserves to be investigated in the future.
In this sense, a more specific analysis, for example of the peroxisomal CasHSP20 which encodes for an sHSP of 16.3 kDa provides some additional evidence about its potential function. In Arabidopsis, it was shown that two sHSPs are present in the peroxisomal matrix, specifically, AtsHSP15.7 (At5g37670) and AtsHSP31.2 (At1g06460), that are targeted by a functional peroxisomal targeting signal (PTS1) [SKL>] and a functional PTS2 (RLX5HF), respectively [71]. More recently, in this plant species, it has also been shown the presence of an additional sHSP, exactly the AtsHSP17.6 (At5g12020) which possesses unusual non-canonical PTS1, QKL, 16 amino acids upstream from the C-terminus and functions as a chaperone of the catalase, the main antioxidant enzyme present in peroxisomes which catalyzes the decomposition of H2O2 [72]. Thus, its overexpression confers higher catalase activity and tolerance to abiotic stresses [73]. In this sense, it should be mentioned that catalase of pepper fruit is negatively modulated by two NO-derived posttranslational modifications, S-nitrosation, and tyrosine nitration [50,72]; consequently, it could be suggested that the CaHSP20 (16.3 kDa) may exert this function in pepper peroxisomes whose gene is significantly upregulated during ripening.
4. Materials and Methods
4.1. Identification of the sHSP Family Members
The set of pepper (Capsicum annuum, assembly UCD10Xv1.1) protein sequences was downloaded from the NCBI database (BioProject ID PRJNA376668). To identify the different sHSP candidates, we employed the HMMER3 software [74], which is based on the hidden Markov model (HMM) method. For this purpose, we obtained the alpha-crystallin/HSP20 domain (PF00011) from the Pfam database [75], which was used to search homologous sequences in the pepper proteome. Then, the presence of the conserved domain was confirmed using the SMART tool [76]. Proteins whose molecular weight was outside the range of 15–42 kDa were also rejected. A similar procedure was followed for the identification of sHSP members in Arabidopsis thaliana (assembly TAIR10.1), tomato (Solanum lycopersicum, assembly SL3.1), and potato (Solanum tuberosum, assembly SolTub_3.0).
4.2. Chromosomal Location, Phylogenetic and Conserved Motif Analyses of sHSP Sequences
Information about the chromosomal location of the identified CasHSPs was withdrawn from the NCBI database. The corresponding genetic map was constructed using the MG2C_v2.1 tool [77].
The sHSPs identified in sweet pepper, tomato, potato, and Arabidopsis were used to construct a phylogenetic tree. The alignment of sHSPs was performed using the MUSCLE method [78]. Then, the aligned sequences were subjected to MEGA11 [79] to perform an unrooted maximum likelihood phylogenetic tree with default parameters. Finally, the resulting phylogenetic tree was modified using the online tool Evolview_v3 [80].
Conserved motifs of CasHSPs were discovered using the MEME tool [81] and visualized using TBtools software [82]. The protein localization based on their amino acid sequences was predicted using WoLF PSORT [83].
4.3. Plant Material and Exogenous Nitric Oxide (NO) Gas Treatment
California-type sweet pepper cultivar Melchor fruits were collected from plants grown in plastic-covered greenhouses (Zeraim Iberica/Syngenta Seeds, Ltd., Roquetas de Mar/El Ejido, Almería, Spain). Fruits without any external damage were selected at three developmental stages: green immature (G), breaking point (BP1), and red ripe (R). Once harvested, these selected fruits were placed in black plastic bags and transported to the laboratory at room temperature, washed with distilled water, and kept for 24 h at a low temperature (about 7 °C ± 1 °C). For the analysis of the exogenous NO gas treatment, we set two additional groups: treated fruits with 5 ppm NO for 1 h (BP2 + NO) and another group that was not treated with NO (BP2 − NO) [37,81]. After 3 days, all fruits were chopped into small cubes (5 mm/edge), frozen under liquid nitrogen, and stored at 80 °C until use. Supplementary Figure S1 shows a representative picture of the experimental design followed in this study with the representative phenotypes of sweet pepper fruits at different ripening stages and subjected to NO treatment [46].
4.4. Library Preparation and RNA-Sequencing
All procedures were performed as previously described [46] with minor modifications. Briefly, libraries were prepared using an Illumina protocol and were sequenced on an Illumina NextSeq550 platform using 2 × 75 bp paired-end reads. These reads were pre-processed to remove low-quality sequences. Useful reads were mapped against the set of transcripts available for Capsicum annuum species in the NCBI database (assembly UCD10Xv1.1) using Bowtie2 [84]. Transcript counts were obtained using Samtools [85]. Differential expression analyses were done using DEgenes-Hunter [86]. This R pipeline examined the relative change in expression between the different samples using different algorithms (EdgeR, DESeq2, Limma, and NOISeq) which apply their own normalizations and statistical tests to validate the whole experiment. On the other hand, an analysis of the time course of CasHSP gene expression was performed considering as reference the expression levels found in green fruits (G). Raw data are accessible at the Sequence Read Archive (SRA) repository under the accession PRJNA668052. This reference pepper fruit transcriptome and differentially expressed (DE) genes among the analyzed ripening stages and the NO treatment involved the analysis of 24 biological replicates corresponding to 5 replicates of each stage, except for green fruits that involved 4 replicates.
5. Conclusions and Future Research Prospects
Nineteen sHSP genes were identified in sweet pepper fruits. The proteins encoded by these genes ranged from 16 to 28 kDa in size, and these sHSP proteins shared 10 motifs, but only motifs 6, 8, 9, and 10 were highly conserved. Pepper fruit sHSPs displayed a wide subcellular localization, including cytosol, plastids, mitochondria, nuclei, Golgi, and peroxisomes. Furthermore, 19 sHSP genes were differentially regulated by ripening, but the expression of 10 genes was also modulated by NO. Overall, it was found that sHSPs might be involved in the mechanism of fruit ripening and in the response to NO treatment, thus confirming the situation of stress undergone by fruits once this physiological process is triggered. In fact, the ripening of sweet pepper fruits is characterized to have a very active ROS and RNS metabolism, which is associated with a physiological nitro-oxidative stress. The anti-ripening effect of NO also corroborates that this signal molecule is possibly exerting a relevant role through this family of proteins to modulate a global response against the physiological stress conditions imposed by ripening.
Future research should be aimed at identifying the molecular mechanisms by which NO can modulate specific sHSP gene expression, for example through NO-derived posttranslational modifications of specific transcription factors. However, it will be necessary to explore whether other signal molecules could also achieve these regulatory functions on sHSP. Among these signal molecules, it should be mentioned certain phytohormones (ethylene, abscisic acid, etc.), hydrogen sulfide (H2S), hydrogen peroxide (H2O2), or melatonin [25,27,28,36,87] whose functional network regulates numerous physiological processes, including the ripening of fruits.
Acknowledgments
SG-G acknowledges a contract (BES-2016-078368) from the Ministry of Economy and Competitiveness, Spain. The provision of pepper fruits by Zeraim Iberica/Syngenta Seeds Ltd. (El Ejido, Almería, Spain) is acknowledged, particularly Víctor J. Domínguez, Lidia Martín, and Manuel Solís. The valuable technical assistance of María J. Campos and Carmelo Ruiz-Torres is deeply acknowledged.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12020389/s1, Figure S1: Illustrative picture showing the experimental design used in this study with the representative phenotype of sweet pepper (Capsicum annuum L.) fruits at different stages and treatments.
Author Contributions
S.G.-G. performed bioinformatics analyses. F.J.C. and J.M.P. designed the work, drove and coordinated the tasks. F.J.C. wrote the first draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Sequence Read Archive (SRA) data are available at the following link https://www.ncbi.nlm.nih.gov/sra/PRJNA668052 (accessed on 28 May 2020).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Our research is supported by a European Regional Development Fund co-financed grants from the Ministry of Science and Innovation (PID2019-103924GB-I00) and Junta de Andalucía (P18- 274 FR-1359), Spain.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Kotak S., Larkindale J., Lee U., von Koskull-Döring P., Vierling E., Scharf K.-D. Complexity of the Heat Stress Response in Plants. Curr. Opin. Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Ohama N., Sato H., Shinozaki K., Yamaguchi-Shinozaki K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017;22:53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 3.Corpas F.J., Chaki M., Fernández-Ocaña A., Valderrama R., Palma J.M., Carreras A., Begara-Morales J.C., Airaki M., del Río L.A., Barroso J.B. Metabolism of Reactive Nitrogen Species in Pea Plants under Abiotic Stress Conditions. Plant Cell Physiol. 2008;49:1711–1722. doi: 10.1093/pcp/pcn144. [DOI] [PubMed] [Google Scholar]
- 4.Chaki M., Valderrama R., Fernández-Ocaña A.M., Carreras A., Gómez-Rodríguez M.V., López-Jaramillo J., Begara-Morales J.C., Sánchez-Calvo B., Luque F., Leterrier M., et al. High Temperature Triggers the Metabolism of S-Nitrosothiols in Sunflower Mediating a Process of Nitrosative Stress Which Provokes the Inhibition of Ferredoxin-NADP Reductase by Tyrosine Nitration. Plant Cell Environ. 2011;34:1803–1818. doi: 10.1111/j.1365-3040.2011.02376.x. [DOI] [PubMed] [Google Scholar]
- 5.Ara N., Nakkanong K., Lv W., Yang J., Hu Z., Zhang M. Antioxidant Enzymatic Activities and Gene Expression Associated with Heat Tolerance in the Stems and Roots of Two Cucurbit Species (“Cucurbita maxima” and “Cucurbita moschata”) and Their Interspecific Inbred Line “Maxchata”. Int. J. Mol. Sci. 2013;14:24008–24028. doi: 10.3390/ijms141224008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mata-Pérez C., Sánchez-Calvo B., Begara-Morales J.C., Carreras A., Padilla M.N., Melguizo M., Valderrama R., Corpas F.J., Barroso J.B. Nitro-Linolenic Acid is a Nitric Oxide Donor. Nitric Oxide. 2016;57:57–63. doi: 10.1016/j.niox.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Hu L., Bi A., Hu Z., Amombo E., Li H., Fu J. Antioxidant Metabolism, Photosystem II, and Fatty Acid Composition of Two Tall Fescue Genotypes with Different Heat Tolerance Under High Temperature Stress. Front. Plant Sci. 2018;9:1242. doi: 10.3389/fpls.2018.01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iqbal N., Umar S., Khan N.A., Corpas F.J. Crosstalk between Abscisic Acid and Nitric Oxide under Heat Stress: Exploring New Vantage Points. Plant Cell Rep. 2021;40:1429–1450. doi: 10.1007/s00299-021-02695-4. [DOI] [PubMed] [Google Scholar]
- 9.Rahman M.A., Woo J.H., Song Y., Lee S.-H., Hasan M.M., Azad M.A.K., Lee K.-W. Heat Shock Proteins and Antioxidant Genes Involved in Heat Combined with Drought Stress Responses in Perennial Rye Grass. Life. 2022;12:1426. doi: 10.3390/life12091426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter K., Haslbeck M., Buchner J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Maestri E., Klueva N., Perrotta C., Gulli M., Nguyen H.T., Marmiroli N. Molecular Genetics of Heat Tolerance and Heat Shock Proteins in Cereals. Plant Mol. Biol. 2002;48:667–681. doi: 10.1023/A:1014826730024. [DOI] [PubMed] [Google Scholar]
- 12.Huang B., Xu C. Identification and Characterization of Proteins Associated with Plant Tolerance to Heat Stress. J. Integr. Plant. Biol. 2008;50:1230–1237. doi: 10.1111/j.1744-7909.2008.00735.x. [DOI] [PubMed] [Google Scholar]
- 13.Driedonks N., Xu J., Peters J.L., Park S., Rieu I. Multi-Level Interactions Between Heat Shock Factors, Heat Shock Proteins, and the Redox System Regulate Acclimation to Heat. Front. Plant Sci. 2015;6:999. doi: 10.3389/fpls.2015.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang Y., Lee K., Hoshikawa K., Kang M., Jang S. Molecular Bases of Heat Stress Responses in Vegetable Crops with Focusing on Heat Shock Factors and Heat Shock Proteins. Front. Plant Sci. 2022;13:837152. doi: 10.3389/fpls.2022.837152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddique M., Gernhard S., von Koskull-Döring P., Vierling E., Scharf K.-D. The Plant sHSP Superfamily: Five New Members in Arabidopsis thaliana with Unexpected Properties. Cell Stress Chaperones. 2008;13:183–197. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carra S., Alberti S., Benesch J.L.P., Boelens W., Buchner J., Carver J.A., Cecconi C., Ecroyd H., Gusev N., Hightower L.E., et al. Small Heat Shock Proteins: Multifaceted Proteins with Important Implications for Life. Cell Stress Chaperones. 2019;24:295–308. doi: 10.1007/s12192-019-00979-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters E.R., Vierling E. Plant Small Heat Shock Proteins—Evolutionary and Functional Diversity. New Phytol. 2020;227:24–37. doi: 10.1111/nph.16536. [DOI] [PubMed] [Google Scholar]
- 18.Bourgine B., Guihur A. Heat Shock Signaling in Land Plants: From Plasma Membrane Sensing to the Transcription of Small Heat Shock Proteins. Front. Plant Sci. 2021;12:710801. doi: 10.3389/fpls.2021.710801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters E.R., Aevermann B.D., Sanders-Reed Z. Comparative Analysis of the Small Heat Shock Proteins in Three Angiosperm Genomes Identifies New Subfamilies and Reveals Diverse Evolutionary Patterns. Cell Stress Chaperones. 2008;13:127–142. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waters E.R. The Evolution, Function, Structure, and Expression of the Plant SHSPs. J. Exp. Bot. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- 21.Singh R.K., Muthamilarasan M., Prasad M. SiHSFA2e Regulated Expression of SisHSP21.9 Maintains Chloroplast Proteome Integrity under High Temperature Stress. Cell. Mol. Life. Sci. 2022;79:580. doi: 10.1007/s00018-022-04611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begara-Morales J.C., Chaki M., Sánchez-Calvo B., Mata-Pérez C., Leterrier M., Palma J.M., Barroso J.B., Corpas F.J. Protein Tyrosine Nitration in Pea Roots during Development and Senescence. J. Exp. Bot. 2013;64:1121–1134. doi: 10.1093/jxb/ert006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Begara-Morales J.C., Sánchez-Calvo B., Chaki M., Valderrama R., Mata-Pérez C., López-Jaramillo J., Padilla M.N., Carreras A., Corpas F.J., Barroso J.B. Dual Regulation of Cytosolic Ascorbate Peroxidase (APX) by Tyrosine Nitration and S-Nitrosylation. J. Exp. Bot. 2014;65:527–538. doi: 10.1093/jxb/ert396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corpas F.J., Leterrier M., Begara-Morales J.C., Valderrama R., Chaki M., López-Jaramillo J., Luque F., Palma J.M., Padilla M.N., Sánchez-Calvo B., et al. Inhibition of Peroxisomal Hydroxypyruvate Reductase (HPR1) by Tyrosine Nitration. Biochim. Biophys. Acta. 2013;1830:4981–4989. doi: 10.1016/j.bbagen.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Corpas F.J., González-Gordo S., Palma J.M. Nitric Oxide: A Radical Molecule with Potential Biotechnological Applications in Fruit Ripening. J. Biotechnol. 2020;324:211–219. doi: 10.1016/j.jbiotec.2020.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Asgher M., Per T.S., Masood A., Fatma M., Freschi L., Corpas F.J., Khan N.A. Nitric Oxide Signaling and Its Crosstalk with Other Plant Growth Regulators in Plant Responses to Abiotic Stress. Environ. Sci. Pollut. Res. Int. 2017;24:2273–2285. doi: 10.1007/s11356-016-7947-8. [DOI] [PubMed] [Google Scholar]
- 27.Mishra V., Singh P., Tripathi D.K., Corpas F.J., Singh V.P. Nitric Oxide and Hydrogen Sulfide: An Indispensable Combination for Plant Functioning. Trends Plant Sci. 2021;26:1270–1285. doi: 10.1016/j.tplants.2021.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Corpas F.J., Rodríguez-Ruiz M., Muñoz-Vargas M.A., González-Gordo S., Reiter R.J., Palma J.M. Interactions of Melatonin, Reactive Oxygen Species, and Nitric Oxide during Fruit Ripening: An Update and Prospective View. J. Exp. Bot. 2022;73:5947–5960. doi: 10.1093/jxb/erac128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iqbal N., Umar S., Khan N.A., Corpas F.J. Nitric Oxide and Hydrogen Sulfide Coordinately Reduce Glucose Sensitivity and Decrease Oxidative Stress via Ascorbate-Glutathione Cycle in Heat-Stressed Wheat (Triticum aestivum L.) Plants. Antioxidants. 2021;10:108. doi: 10.3390/antiox10010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Yang J., Zhang H., Cong L., Zhai R., Yang C., Wang Z., Ma F., Xu L. Melatonin Inhibits Ethylene Synthesis via Nitric Oxide Regulation to Delay Postharvest Senescence in Pears. J. Agric. Food Chem. 2019;67:2279–2288. doi: 10.1021/acs.jafc.8b06580. [DOI] [PubMed] [Google Scholar]
- 31.Leshem Y.Y., Wills R.B.H., Ku V.V.-V. Evidence for the Function of the Free Radical Gas—Nitric Oxide (•NO)—As an Endogenous Maturation and Senescence Regulating Factor in Higher Plants. Plant Physiol. Biochem. 1998;36:825–833. doi: 10.1016/S0981-9428(99)80020-5. [DOI] [Google Scholar]
- 32.Correa-Aragunde N., Graziano M., Chevalier C., Lamattina L. Nitric Oxide Modulates the Expression of Cell Cycle Regulatory Genes during Lateral Root Formation in Tomato. J. Exp. Bot. 2006;57:581–588. doi: 10.1093/jxb/erj045. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad P., Ahanger M.A., Alyemeni M.N., Wijaya L., Alam P. Exogenous Application of Nitric Oxide Modulates Osmolyte Metabolism, Antioxidants, Enzymes of Ascorbate-Glutathione Cycle and Promotes Growth under Cadmium Stress in Tomato. Protoplasma. 2018;255:79–93. doi: 10.1007/s00709-017-1132-x. [DOI] [PubMed] [Google Scholar]
- 34.Bhat J.A., Ahmad P., Corpas F.J. Main Nitric Oxide (NO) Hallmarks to Relieve Arsenic Stress in Higher Plants. J. Hazard. Mater. 2021;406:124289. doi: 10.1016/j.jhazmat.2020.124289. [DOI] [PubMed] [Google Scholar]
- 35.Seabra A.B., Silveira N.M., Ribeiro R.V., Pieretti J.C., Barroso J.B., Corpas F.J., Palma J.M., Hancock J.T., Petřivalský M., Gupta K.J., et al. Nitric Oxide-Releasing Nanomaterials: From Basic Research to Potential Biotechnological Applications in Agriculture. New Phytol. 2022;234:1119–1125. doi: 10.1111/nph.18073. [DOI] [PubMed] [Google Scholar]
- 36.Manjunatha G., Gupta K.J., Lokesh V., Mur L.A.J., Neelwarne B. Nitric Oxide Counters Ethylene Effects on Ripening Fruits. Plant Signal. Behav. 2012;7:476–483. doi: 10.4161/psb.19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steelheart C., Alegre M.L., Vera Bahima J., Senn M.E., Simontacchi M., Bartoli C.G., Gergoff Grozeff G.E. Nitric Oxide Improves the Effect of 1-Methylcyclopropene Extending the Tomato (Lycopersicum esculentum L.) Fruit Postharvest Life. Sci. Hortic. 2019;255:193–201. doi: 10.1016/j.scienta.2019.04.035. [DOI] [Google Scholar]
- 38.Kantar M.B., Anderson J.E., Lucht S.A., Mercer K., Bernau V., Case K.A., Le N.C., Frederiksen M.K., DeKeyser H.C., Wong Z.Z., et al. Vitamin Variation in Capsicum Spp. Provides Opportunities to Improve Nutritional Value of Human Diets. PLoS ONE. 2016;11:e0161464. doi: 10.1371/journal.pone.0161464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosa-Martínez E., García-Martínez M.D., Adalid-Martínez A.M., Pereira-Dias L., Casanova C., Soler E., Figàs M.R., Raigón M.D., Plazas M., Soler S., et al. Fruit Composition Profile of Pepper, Tomato and Eggplant Varieties Grown under Uniform Conditions. Food Res. Int. 2021;147:110531. doi: 10.1016/j.foodres.2021.110531. [DOI] [PubMed] [Google Scholar]
- 40.Palma J.M., Sevilla F., Jiménez A., del Río L.A., Corpas F.J., Álvarez de Morales P., Camejo D.M. Physiology of Pepper Fruit and the Metabolism of Antioxidants: Chloroplasts, Mitochondria and Peroxisomes. Ann. Bot. 2015;116:627–636. doi: 10.1093/aob/mcv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corpas F.J., Freschi L., Rodríguez-Ruiz M., Mioto P.T., González-Gordo S., Palma J.M. Nitro-Oxidative Metabolism during Fruit Ripening. J. Exp. Bot. 2018;69:3449–3463. doi: 10.1093/jxb/erx453. [DOI] [PubMed] [Google Scholar]
- 42.Jiménez A., Romojaro F., Gómez J.M., Llanos M.R., Sevilla F. Antioxidant Systems and Their Relationship with the Response of Pepper Fruits to Storage at 20 °C. J. Agric. Food Chem. 2003;51:6293–6299. doi: 10.1021/jf030052i. [DOI] [PubMed] [Google Scholar]
- 43.Chu-Puga Á., González-Gordo S., Rodríguez-Ruiz M., Palma J.M., Corpas F.J. NADPH Oxidase (Rboh) Activity Is Up Regulated during Sweet Pepper (Capsicum annuum L.) Fruit Ripening. Antioxidants. 2019;8:9. doi: 10.3390/antiox8010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corpas F.J., Freschi L., Palma J.M. Advances in Botanical Research. Academic Press; Cambridge, MA, USA: 2022. ROS Metabolism and Ripening of Fleshy Fruits. [DOI] [Google Scholar]
- 45.González-Gordo S., Cañas A., Muñoz-Vargas M.A., Palma J.M., Corpas F.J. Lipoxygenase (LOX) in Sweet and Hot Pepper (Capsicum annuum L.) Fruits during Ripening and under an Enriched Nitric Oxide (NO) Gas Atmosphere. Int. J. Mol. Sci. 2022;23:15211. doi: 10.3390/ijms232315211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.González-Gordo S., Bautista R., Claros M.G., Cañas A., Palma J.M., Corpas F.J. Nitric Oxide-Dependent Regulation of Sweet Pepper Fruit Ripening. J. Exp. Bot. 2019;70:4557–4570. doi: 10.1093/jxb/erz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.González-Gordo S., Rodríguez-Ruiz M., Palma J.M., Corpas F.J. Superoxide Radical Metabolism in Sweet Pepper (Capsicum annuum L.) Fruits Is Regulated by Ripening and by a NO-Enriched Environment. Front. Plant Sci. 2020;11:485. doi: 10.3389/fpls.2020.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.González-Gordo S., Rodríguez-Ruiz M., López-Jaramillo J., Muñoz-Vargas M.A., Palma J.M., Corpas F.J. Nitric Oxide (NO) Differentially Modulates the Ascorbate Peroxidase (APX) Isozymes of Sweet Pepper (Capsicum. annuum L.) Fruits. Antioxidants. 2022;11:765. doi: 10.3390/antiox11040765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodríguez-Ruiz M., Mioto P., Palma J.M., Corpas F.J. S-Nitrosoglutathione Reductase (GSNOR) Activity is Down-Regulated during Pepper (Capsicum annuum L.) Fruit Ripening. Nitric Oxide. 2017;68:51–55. doi: 10.1016/j.niox.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Rodríguez-Ruiz M., González-Gordo S., Cañas A., Campos M.J., Paradela A., Corpas F.J., Palma J.M. Sweet Pepper (Capsicum annuum L.) Fruits Contain an Atypical Peroxisomal Catalase That Is Modulated by Reactive Oxygen and Nitrogen Species. Antioxidants. 2019;8:374. doi: 10.3390/antiox8090374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo M., Liu J.-H., Lu J.-P., Zhai Y.-F., Wang H., Gong Z.-H., Wang S.-B., Lu M.-H. Genome-Wide Analysis of the CaHsp20 Gene Family in Pepper: Comprehensive Sequence and Expression Profile Analysis under Heat Stress. Front. Plant Sci. 2015;6:806. doi: 10.3389/fpls.2015.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J., Gao T., Hu J., Zhao L., Yu C., Ma F. Research Advances in Function and Regulation Mechanisms of Plant Small Heat Shock Proteins (sHSPs) under Environmental Stresses. Sci. Total Environ. 2022;825:154054. doi: 10.1016/j.scitotenv.2022.154054. [DOI] [PubMed] [Google Scholar]
- 53.Sies H. Oxidative Eustress: On Constant Alert for Redox Homeostasis. Redox Biol. 2021;41:101867. doi: 10.1016/j.redox.2021.101867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sies H., Jones D.P. Reactive Oxygen Species (ROS) as Pleiotropic Physiological Signalling Agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 55.Arce D., Spetale F., Krsticevic F., Cacchiarelli P., las Rivas J.D., Ponce S., Pratta G., Tapia E. Regulatory Motifs Found in the Small Heat Shock Protein (sHSP) Gene Family in Tomato. BMC Genom. 2018;19:860. doi: 10.1186/s12864-018-5190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Zhu M., Ren L., Li A., Chen G., Hu Z. The SlFSR Gene Controls Fruit Shelf-Life in Tomato. J. Exp. Bot. 2018;69:2897–2909. doi: 10.1093/jxb/ery116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Upadhyay R.K., Tucker M.L., Mattoo A.K. Ethylene and RIPENING INHIBITOR Modulate Expression of SlHSP17.7A, B Class I Small Heat Shock Protein Genes During Tomato Fruit Ripening. Front. Plant Sci. 2020;11:975. doi: 10.3389/fpls.2020.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ré M.D., Gonzalez C., Escobar M.R., Sossi M.L., Valle E.M., Boggio S.B. Small Heat Shock Proteins and the Postharvest Chilling Tolerance of Tomato Fruit. Physiol. Plant. 2017;159:148–160. doi: 10.1111/ppl.12491. [DOI] [PubMed] [Google Scholar]
- 59.Neta-Sharir I., Isaacson T., Lurie S., Weiss D. Dual Role for Tomato Heat Shock Protein 21: Protecting Photosystem II from Oxidative Stress and Promoting Color Changes during Fruit Maturation. Plant Cell. 2005;17:1829–1838. doi: 10.1105/tpc.105.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence S.D., Cline K., Moore G.A. Chromoplast Development in Ripening Tomato Fruit: Identification of cDNAs for Chromoplast-Targeted Proteins and Characterization of a cDNA Encoding a Plastid-Localized Low-Molecular-Weight Heat Shock Protein. Plant Mol. Biol. 1997;33:483–492. doi: 10.1023/A:1005785321165. [DOI] [PubMed] [Google Scholar]
- 61.Huang L.-J., Cheng G.-X., Khan A., Wei A.-M., Yu Q.-H., Yang S.-B., Luo D.-X., Gong Z.-H. CaHSP16.4, a Small Heat Shock Protein Gene in Pepper, Is Involved in Heat and Drought Tolerance. Protoplasma. 2019;256:39–51. doi: 10.1007/s00709-018-1280-7. [DOI] [PubMed] [Google Scholar]
- 62.Feng X.-H., Zhang H.-X., Ali M., Gai W.-X., Cheng G.-X., Yu Q.-H., Yang S.-B., Li X.-X., Gong Z.-H. A Small Heat Shock Protein CaHsp25.9 Positively Regulates Heat, Salt, and Drought Stress Tolerance in Pepper (Capsicum annuum L.) Plant Physiol. Biochem. 2019;142:151–162. doi: 10.1016/j.plaphy.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y.-L., Liu S., Xiao J.-J., Cheng G.-X., Ul H.S., Gong Z.-H. CaHSP18.1a, a Small Heat Shock Protein from Pepper (Capsicum annuum L.), Positively Responds to Heat, Drought, and Salt Tolerance. Horticulturae. 2021;7:117. doi: 10.3390/horticulturae7050117. [DOI] [Google Scholar]
- 64.Libourel I.G.L., Bethke P.C., de Michele R., Jones R.L. Nitric Oxide Gas Stimulates Germination of Dormant Arabidopsis Seeds: Use of a Flow-through Apparatus for Delivery of Nitric Oxide. Planta. 2006;223:813–820. doi: 10.1007/s00425-005-0117-8. [DOI] [PubMed] [Google Scholar]
- 65.García-Mata C., Lamattina L. Nitric Oxide Induces Stomatal Closure and Enhances the Adaptive Plant Responses against Drought Stress. Plant Physiol. 2001;126:1196–1204. doi: 10.1104/pp.126.3.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corpas F.J., Barroso J.B. Functions of Nitric Oxide (NO) in Roots during Development and under Adverse Stress Conditions. Plants. 2015;4:240–252. doi: 10.3390/plants4020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L., Guo Y., Jia L., Chu H., Zhou S., Chen K., Wu D., Zhao L. Hydrogen Peroxide Acts Upstream of Nitric Oxide in the Heat Shock Pathway in Arabidopsis Seedlings. Plant Physiol. 2014;164:2184–2196. doi: 10.1104/pp.113.229369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh S., Husain T., Kushwaha B.K., Suhel M., Fatima A., Mishra V., Singh S.K., Bhatt J.A., Rai M., Prasad S.M., et al. Regulation of Ascorbate-Glutathione Cycle by Exogenous Nitric Oxide and Hydrogen Peroxide in Soybean Roots under Arsenate Stress. J. Hazard. Mater. 2021;409:123686. doi: 10.1016/j.jhazmat.2020.123686. [DOI] [PubMed] [Google Scholar]
- 69.Prajapati P., Gupta P., Kharwar R.N., Seth C.S. Nitric Oxide Mediated Regulation of Ascorbate-Glutathione Pathway Alleviates Mitotic Aberrations and DNA Damage in Allium cepa L. under Salinity Stress. Int. J. Phytoremediation. 2022:1–12. doi: 10.1080/15226514.2022.2086215. [DOI] [PubMed] [Google Scholar]
- 70.Mata-Pérez C., Sánchez-Calvo B., Padilla M.N., Begara-Morales J.C., Luque F., Melguizo M., Jiménez-Ruiz J., Fierro-Risco J., Peñas-Sanjuán A., Valderrama R., et al. Nitro-Fatty Acids in Plant Signaling: Nitro-Linolenic Acid Induces the Molecular Chaperone Network in Arabidopsis. Plant Physiol. 2016;170:686–701. doi: 10.1104/pp.15.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma Y., Creanga A., Lum L., Beachy P.A. Prevalence of Off-Target Effects in Drosophila RNA Interference Screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- 72.Palma J.M., Mateos R.M., López-Jaramillo J., Rodríguez-Ruiz M., González-Gordo S., Lechuga-Sancho A.M., Corpas F.J. Plant Catalases as NO and H2S Targets. Redox Biol. 2020;34:101525. doi: 10.1016/j.redox.2020.101525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li G., Li J., Hao R., Guo Y. Activation of Catalase Activity by a Peroxisome-Localized Small Heat Shock Protein Hsp17.6CII. J. Genet. Genom. 2017;44:395–404. doi: 10.1016/j.jgg.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 74.Mistry J., Finn R.D., Eddy S.R., Bateman A., Punta M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Letunic I., Khedkar S., Bork P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021;49:D458–D460. doi: 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chao J., Li Z., Sun Y., Aluko O.O., Wu X., Wang Q., Liu G. MG2C: A User-Friendly Online Tool for Drawing Genetic Maps. Mol. Hortic. 2021;1:16. doi: 10.1186/s43897-021-00020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edgar R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Subramanian B., Gao S., Lercher M.J., Hu S., Chen W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019;47:W270–W275. doi: 10.1093/nar/gkz357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bailey T.L., Elkan C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 82.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 83.Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B., Salzberg S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gayte I.G., Moreno R.B., Zonjic P.S., Claros M.G. DEgenes Hunter—A Flexible R Pipeline for Automated RNA-Seq Studies in Organisms without Reference Genome. Genom. Comput. Biol. 2017;3:31. doi: 10.18547/gcb.2017.vol3.iss3.e31. [DOI] [Google Scholar]
- 87.Aghdam M.S., Mukherjee S., Flores F.B., Arnao M.B., Luo Z., Corpas F.J. Functions of Melatonin During Postharvest of Horticultural Crops. Plant Cell Physiol. 2021:pcab175. doi: 10.1093/pcp/pcab175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence Read Archive (SRA) data are available at the following link https://www.ncbi.nlm.nih.gov/sra/PRJNA668052 (accessed on 28 May 2020).