Abstract
A model system mimicking Staphylococcus aureus bacteremia was developed by growth in serum under microaerobic conditions. Eight genes induced by growth in serum were identified, including an antimicrobial peptide biosynthesis locus, amino acid biosynthetic loci, and genes encoding putative surface proteins. Nine independent insertions were found in the major lysine biosynthesis operon, which encodes eight genes, is repressed by lysine in vitro, and is expressed in vivo.
Staphylococcus aureus is a highly adaptable human pathogen in which differential gene expression is known to occur in response to environmental conditions, both in vitro (1, 8, 17) and in vivo (9). Previous reports have demonstrated that in vitro conditions can be used to mimic those in vivo, for example, the use of cell culture extracts and mammalian cell cultures (13, 17). In this study, a model system of growth in serum has been established and characterized.
Growth of S. aureus in serum.
S. aureus 8325-4 (12) was grown in both brain heart infusion (BHI) broth and pig serum (Sigma) under aerobic and microaerobic conditions (8% O2–5% CO2–87% N2). Microaerobic growth in serum resulted in a higher growth rate and yield than did aerobic growth (optical densities at 600 nm [OD600], 6.1 and 4.2, respectively; 7 h) (results not shown). Growth in human serum produced trends identical to those seen in pig serum (results not shown). BHI gave a higher growth yield than did serum and, in contrast to the results for serum, in BHI aerobic growth was found to be optimal (OD600, 7.9 [microaerobic] and 10.2 [aerobic]; 7 h) (results not shown).
Identification of serum-expressed genes (seg).
Genes specifically induced in serum versus BHI were identified by replica plating Tn917 insertion libraries (19) on serum agar and BHI agar, both containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (80 μg/ml). Following incubation at 37°C (microaerobic for serum and aerobic for BHI), colonies that were blue on serum and white on BHI were selected and rescreened. Twenty-three clones with increased LacZ activity on serum were selected and further characterized. No growth defects on serum or BHI were observed for any of the clones.
Identification of seg loci.
Following marker rescue cloning and DNA sequencing (19), transposon insertion sites were identified. In total, nine different seg genes were identified (Table 1). The genes insertionally inactivated in mutants seg5, seg7, and seg37 are all likely to encode surface proteins. Thus, differentially expressed surface proteins that are produced by S. aureus in serum may have a role in virulence.
TABLE 1.
Characterization of serum-expressed genes (seg)
| Classification | Mutant(s) sibling[s] | Similar gene (EMBL accession no.) | Identitya | Ratio of β-galactosidase activityb | Putative function |
|---|---|---|---|---|---|
| Biosynthesis | seg1 (seg21), seg13, seg14, seg25 (seg50), seg45 (seg48) | lysC (AAD36585.1)c | 39 (157/395) | 75 | Lysine biosynthesis, peptidoglycan biosynthesis |
| seg22 (seg30, seg33) | Putative lysC control region (AAD36585.1)c | 39 (157/395) | 77 | Lysine biosynthesis , peptidoglycan biosynthesis | |
| seg24, seg26 | asd (AAC07674.1)d | 49 (164/333) | 70 | Lysine biosynthesis, peptidoglycan biosynthesis | |
| seg10 | dapA (AAB98232.1)e | 41 (119/285) | 80 | Lysine biosynthesis, peptidoglycan biosynthesis | |
| seg8 | yjcI (CAB13044.1)f | 47 (174/365) | 45 | Methionine biosynthesis | |
| seg29, seg43 | gltA (CAB13728.1)f | 52 (792/1513) | 105 | Glutamate synthesis | |
| Surface protein | seg7 | sai-1 (BAA97049.1)g | 91 (268/293) | 50 | 29-kDa cell surface protein |
| seg37 (seg39, seg40) | sitC (CAA67571.1)h | 76 (235/309) | 68 | Lipoprotein, putative adhesin | |
| Antimicrobial peptide | seg35 | gdhM (AAB61135.1)i | 51 (23/45) | 7 | Lantibiotic gallidermin precursor |
| No database match | seg5 | ? | No significant homologyj | 58 | Unknown |
Percentage (number of identical amino acids).
Expressed as the ratio of β-galactosidase activity of stationary-phase bacteria cultured in serum versus BHI.
Thermotoga maritima.
Aquifex aeolicus.
Methanococcus jannaschii.
B. subtilis.
S. aureus.
Staphylococcus epidermidis.
Staphylococcus gallinarum.
Identity score with no significant database match.
In seg35 the transposon has been inserted within a putative lantibiotic precursor-encoding gene (Table 1). Lantibiotics are a group of antibiotic peptides which are produced by and primarily act on gram-positive bacteria (15). A signature-tagged mutagenesis study of S. aureus also identified a lantibiotic precursor-encoding gene in which a mutation led to significant attenuation in a mouse abscess model and in a 50% lethal dose assay (3).
A gene showing homology to the glutamate synthase large-subunit gene (gltA) was identified in two independent transposon insertion mutants, seg29 and seg43 (Table 1). Glutamate synthase is involved in the incorporation of ammonium ions into organic compounds, an important step in the production of amino acids (16).
The four remaining seg genes are all putatively involved in the biosynthesis of the aspartate family amino acids, lysine (Lys), methionine (Met), threonine (Thr), and isoleucine (Ile) (Table 1). lysC and asd encode the “common-pathway” enzymes aspartokinase II and aspartate semialdehyde dehydrogenase, respectively, which are involved in the synthesis of all four aspartate family amino acids. The other two enzymes have a role in only lysine (dihydrodipicolinate synthase; dapA) or methionine (cystathionine γ-synthase; yjcI) biosynthesis (Table 1). Lysine is a particularly important amino acid in S. aureus, being required not only as a building block for proteins but also as a component of the cell wall peptidoglycan. Interestingly, genes encoding lysine biosynthetic enzymes (lysC, asd, dapA, dapB, and lysA) have been identified not only in other in vitro screens (8, 17) but also as mutations (asd, ykuQ, and lysA) resulting in attenuation in vivo (3, 10).
Analysis of lysC, asd, and dapA genes in S. aureus.
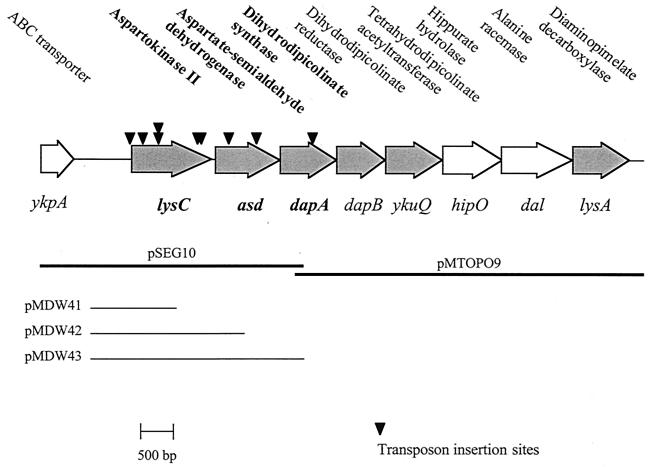
Nine independent transposon insertions were identified in genes involved in the biosynthesis of lysine and the other aspartate family amino acids. The entire dap locus was sequenced from S. aureus strain 8325-4, leading to the identification of a putative eight-gene transcription unit termed the dap operon (Fig. 1). Protein homology suggests that the dap operon contains six genes involved in the biosynthesis of lysine. The dapB, ykuQ, and lysA genes encode dihydrodipicolinate reductase, tetrahydrodipicolinate succinylase, and diaminopimelate decarboxylase, respectively. Notably, the dap genes are in the same order as they appear in the biosynthetic pathway (Fig. 1) (17).
FIG. 1.
Diagrammatic representation of the dap operon of S. aureus, showing the sequenced regions cloned in pSEG10 and pMTOPO9 and those cloned to generate the lacZ transcriptional fusion strains. Shaded genes are putatively involved in lysine biosynthesis. Tn917 insertions from left to right: aspartokinase II (lysC) control-leader region (putative)—seg22 (siblings seg30 and seg33); aspartokinase II (lysC)—seg14, seg25 (bottom, sibling seg50), seg45 (top, sibling seg48), seg13, and seg1 (sibling seg21); aspartate semialdehyde dehydrogenase (asd)—seg24 and seg26; and dihydrodipicolinate synthase (dapA)—seg10. ABC, ATP-binding cassette.
The first of the two remaining genes of the dap operon, hipO, encodes hippurate hydrolase. This enzyme acts to cleave benzoylglycine (hippuric acid) into the constituent products, benzoic acid and glycine (4). The second gene, dal, encodes alanine racemase, which interconverts l-alanine and d-alanine, providing d-alanine for bacterial cell wall synthesis (18). The identification of alanine racemase for the dap operon confirmed the prediction that S. aureus, like Escherichia coli and Bacillus subtilis, possesses two alanine racemase isozymes (7).
Amino acid requirements of selected seg mutants.
A chemically defined medium (19) was adapted to allow the analysis of potential aspartate family amino acid auxotrophy (results not shown). These studies revealed that a mutation in lysC, asd, or dapA leads to lysine auxotrophy. Interestingly, while a mutation within the common pathway enzyme gene asd leads to an additional requirement for methionine and threonine, a lysC mutation does not. The phenotype of the lysC mutant can be explained only by the presence of multiple aspartokinase isozymes (as in B. subtilis) able to rescue the methionine and threonine biosynthesis functions of the lysC-encoded isozyme, but not lysine biosynthesis. The presence in S. aureus of a likely second aspartokinase isozyme homologous to aspartokinase III (yclM) of B. subtilis (44% over 171 amino acids) was confirmed by BLAST analysis of S. aureus databases (results not shown). Additionally, due to the genetic organization of the dap operon, where a lysC mutation is polar on asd, it is likely that a further promoter upstream of asd drives transcription independently of lysC.
Analysis of the expression and regulation of lysC, asd, and dapA.
Reporter gene fusions MDW41 (lysC::lacZ), MDW42 (asd::lacZ), and MDW43 (dapA::lacZ), containing the fragments shown in Fig. 1, were cloned as BamHI-EcoRI PCR fragments into similarly digested pAZ106 (6). Recombinant plasmids were introduced into S. aureus RN4220 by electroporation (14), and the resulting chromosomal fusions were then transduced into S. aureus 8325-4 by phage transduction (12) and verified by Southern blot analysis (results not shown). The fusion strains all contained an intact copy of the dap operon. LacZ activity was measured as previously described (5).
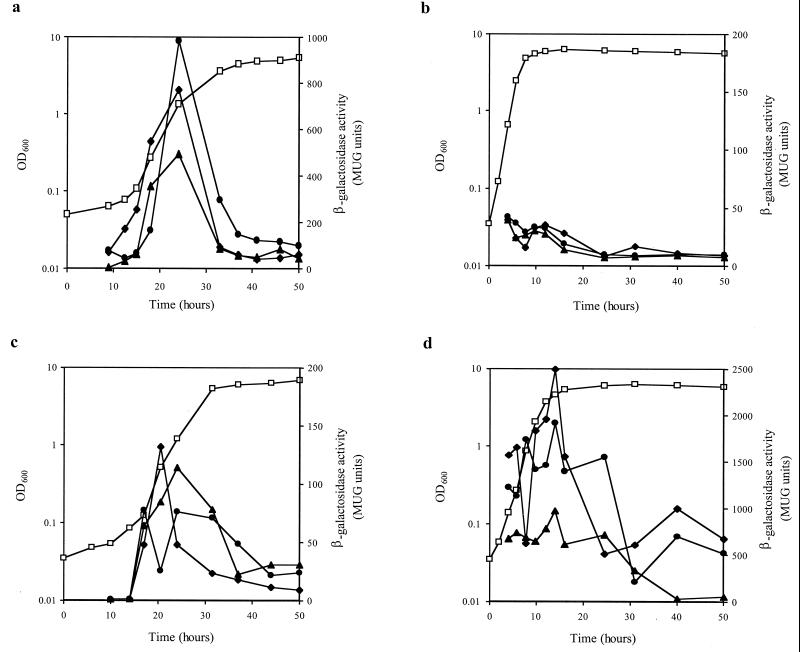
Using the defined medium (19), the effects of Lys, Met, Thr, and Ile on the expression of the lacZ fusions were tested. Without any aspartate family amino acids, all fusion strains show a decreased growth rate but similar final yields compared to the results obtained with medium containing all four amino acids (Fig. 2a and b). All three fusions showed similar expression kinetics, being maximally expressed during the exponential phase and repressed >5-fold by Lys (Fig. 2). The addition of Met, Thr, and Ile had no effect in the absence of Lys (Fig. 2a and d), although the presence of all four aspartate family amino acids led to an almost complete lack of expression of any of the three fusions (Fig. 2b). These results suggest that serum lacks sufficient available Lys, leading to induction of the synthesis of the dap operon.
FIG. 2.
Expression of lysC, asd, and dapA in different chemically defined media (CDM). Shown is analysis of transcription (closed symbols) of lysC::lacZ (MDW41) (diamonds), asd::lacZ (MDW42) (circles), and dapA::lacZ (MDW43) (triangles). Basal CDM was prepared without the aspartate family amino acids (lysine [Lys], methionine [Met], threonine [Thr], and isoleucine [Ile]). Supplements to 100 mg of Lys and Met per liter and to 150 mg of Thr and Ile per liter were added as follows: a, none; b, Lys, Met, Thr, and Ile; c, Lys; and d, Met, Thr, and Ile. A representative growth curve (MDW41) is shown on each graph (OD600 [open squares]). MUG, methylumbelliferyl-β-d-galactoside.
In vivo analysis of lysC, asd, and dapA.
Using reverse transcription PCR and a murine pyelonephritis model (11), lysC, asd, and dapA were all shown to be expressed in vivo (results not shown). Three murine infection models (mouse abscess [2], pyelonephritis, and wound infection [11]) were used to investigate the role of lysC, asd, and dapA in vivo. In all three models, there was no significant difference between the number of cells recovered from the host following a 7-day infection for any of the mutant strains and the number recovered for the wild-type strain (results not shown). However, Lys biosynthetic components, including asd, have been shown to have roles in pathogenesis, as they have been identified during signature-tagged mutagenesis screening of S. aureus using bacteremia models of infection (3, 10).
The serum model is useful for the identification of genes which may contribute to the establishment (surface protein genes) and the persistence (biosynthetic genes) of S. aureus in the bloodstream. The serum screen is a simple and complementary approach to both signature-tagged mutagenesis and in vivo expression technology (3, 9, 10) and may allow environmental parameters important in the host to be elucidated. Further study of the role and regulation of the genes identified by these techniques will shed light on the complex processes involved in the ability of S. aureus to cause disease.
Nucleotide sequence accession number.
The sequence determined for the entire dap locus from S. aureus strain 8325-4 has been deposited in GenBank under accession number AF306669.
Acknowledgments
We thank Martin Burnham (Glaxo SmithKline) and E. Ingham (University of Leeds, Leeds, United Kingdom) for help with in vivo models and useful discussions.
We are grateful to the Staphylococcus aureus Genome Sequencing project and to B. A. Roe, Y. Qian, A. Dorman, F. Z. Najar, S. Clifton, and J. Iandolo, who received funding from NIH and the Merck Genome Research Institute for preliminary sequence data. Sequence data were also obtained from The Institute for Genomic Research website (http://www.tigr.org) with support from NIH and the Merck Genome Research Institute. This research program was supported by the BBSRC (to M.D.W), the Royal Society (to S.J.F.), and Glaxo SmithKline.
REFERENCES
- 1.Chan P F, Foster S J. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus8325-4. Microbiology. 1998;144:2469–2479. doi: 10.1099/00221287-144-9-2469. [DOI] [PubMed] [Google Scholar]
- 2.Chan P F, Foster S J, Ingham E, Clements M O. The Staphylococcus aureus alternative sigma factor ςBcontrols the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coulter S N, Schwan W R, Ng E Y W, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Staphylococcus aureusgenetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 4.Hani E K, Chan V L. Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase (hippuricase) gene in Escherichia coli. J Bacteriol. 1995;177:2396–2402. doi: 10.1128/jb.177.9.2396-2402.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horsburgh M J, Ingham E, Foster S J. In Staphylococcus aureus, Fur is an interactive regulator with PerR and is necessary for oxidative stress resistance through positive regulation of catalase, iron homeostasis, and virulence. J Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kemp E H, Sammons R L, Moir A, Sun D, Setlow P. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis168. J Bacteriol. 1991;173:4646–4652. doi: 10.1128/jb.173.15.4646-4652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullik I, Jenni R, Berger-Bachi B. Sequence of the putative alanine racemase operon in Staphylococcus aureus: insertional interruption of this operon reduces d-alanine substitution of lipoteichoic acids and autolysis. Gene. 1998;219:9–17. doi: 10.1016/s0378-1119(98)00404-1. [DOI] [PubMed] [Google Scholar]
- 8.Lammers A, Kruijt E, van de Kuijt C, Nuijten P J M, Smith H E. Identification of Staphylococcus aureus genes expressed during growth in milk: a useful model for selection of genes important in bovine mastitis? Microbiology. 2000;146:981–987. doi: 10.1099/00221287-146-4-981. [DOI] [PubMed] [Google Scholar]
- 9.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivoexpression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 10.Mei J, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureusvirulence genes in a murine model of bacteremia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 11.Nicholas R O, Li T, McDevitt D, Marra A, Sucoloski S, Demarsh P L, Gentry D R. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect Immun. 1999;67:3667–3669. doi: 10.1128/iai.67.7.3667-3669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 13.Renzoni A, Cossart P, Dramsi S. PrfA, the transcriptional activator of virulence genes, is up regulated during interaction of Listeria monocytogeneswith mammalian cells and in eukaryotic cell extracts. Mol Microbiol. 1999;34:552–561. doi: 10.1046/j.1365-2958.1999.01621.x. [DOI] [PubMed] [Google Scholar]
- 14.Schenk S, Laddaga R A. Improved methods for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 15.Schnell N, Entian K-D, Schneider U, Gotz F, Zahner H, Kellner R, Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with 4 sulfide-rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 16.Schreier H J. Biosynthesis of glutamine and glutamate and the assimilation of ammonia. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 281–298. [Google Scholar]
- 17.Vriesema A J M, Beekhuizen H, Hamdi M, Soufan A, Lammers A, Willekens B, Bakker O, Welten A G A, Veltrop M H A M, van de Gevel J S, Dankert J, Zaat S A J. Altered gene expression in Staphylococcus aureusupon interaction with human endothelial cells. Infect Immun. 2000;68:1765–1772. doi: 10.1128/iai.68.4.1765-1772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserman S A, Daub E, Grisafi P, Botstein D, Walsh C T. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence, enzyme purification, and characterization. Biochemistry. 1984;23:5182–5187. doi: 10.1021/bi00317a015. [DOI] [PubMed] [Google Scholar]
- 19.Watson S P, Antonio M, Foster S J. Isolation and characterisation of Staphylococcus aureusstarvation-induced, stationary-phase mutants defective in survival or recovery. Microbiology. 1998;144:3159–3169. doi: 10.1099/00221287-144-11-3159. [DOI] [PubMed] [Google Scholar]