Abstract
Progressive dysfunction and failure of insulin-releasing β-cells are a hallmark of type 2 diabetes (T2D). To study mechanisms of β-cell loss in T2D, we performed islet single-cell RNA sequencing of two obese mouse strains differing in their diabetes susceptibility. With mice on a control diet, we identified six β-cell clusters with similar abundance in both strains. However, after feeding of a diabetogenic diet for 2 days, β-cell cluster composition markedly differed between strains. Islets of diabetes-resistant mice developed into a protective β-cell cluster (Beta4), whereas those of diabetes-prone mice progressed toward stress-related clusters with a strikingly different expression pattern. Interestingly, the protective cluster showed indications of reduced β-cell identity, such as downregulation of GLUT2, GLP1R, and MafA, and in vitro knockdown of GLUT2 in β-cells—mimicking its phenotype—decreased stress response and apoptosis. This might explain enhanced β-cell survival of diabetes-resistant islets. In contrast, β-cells of diabetes-prone mice responded with expression changes indicating metabolic pressure and endoplasmic reticulum stress, presumably leading to later β-cell loss. In conclusion, failure of diabetes-prone mice to adapt gene expression toward a more dedifferentiated state in response to rising blood glucose levels leads to β-cell failure and diabetes development.
Introduction
The development of type 2 diabetes (T2D) is characterized by a gradual decrease of pancreatic islet function accompanied by the disability to secrete sufficient amounts of insulin (1). Currently, no medical treatment can stop or reverse the loss of β-cell mass, resulting in hyperglycemia, severe secondary complications, and premature death. Hence, there is a need to improve our knowledge about the mechanisms driving islet cell failure during the progression from normoglycemia to T2D.
In humans and mice, genetics strongly contribute to islet function and individual T2D risk (2,3). New Zealand Obese (NZO) mice are used as a model for polygenic diabetes with β-cell failure, closely resembling human T2D pathophysiology (4,5). Comparing gene expression signatures in pancreatic islets of diabetes-resistant B6.V-Lepob/ob (OB) and diabetes-prone NZO mice represents an appropriate and valuable strategy for the identification of genetic variants (6,7). Whereas leptin-deficient OB mice are highly insulin resistant but protected from T2D due to adaptive β-cell proliferation, NZO mice exhibit islet cell failure and β-cell apoptosis (8,9). Several diabetes genes have been identified through combining quantitative trait loci (QTL) and islet transcriptome analysis, and their role in β-cell proliferation (Lefty1, Apoa2, Pcp4l1) (7) and cilia function (Kif3a) (10), as well as insulin secretion and β-cell survival (Gjb4) (6), has been demonstrated.
Glucose is a major modulator of β-cell function and fate, and persistent hyperglycemia exerts deleterious effects on insulin secretion and β-cell survival (11,12). The concept of glucotoxicity includes molecular mechanisms such as overstimulation, oxidative stress, and endoplasmic reticulum (ER) stress, which can induce changes in β-cell identity (de- and transdifferentiation) as well as β-cell destruction (13).
Here, we use a synchronized, 2-day glucotoxic stimulus, initiated by dietary carbohydrates after 13 weeks of ketogenic diet, to study early gene expression changes in pancreatic islets of obese mice known to undergo β-cell adaptation (OB mice) or β-cell decompensation (NZO mice) during an extended feeding regimen. Feeding the mice with a fat-enriched carbohydrate-free diet induces severe insulin resistance without affecting islet function, whereas the switch to a carbohydrate-containing diet for 2 days initiates early steps of β-cell dysfunction and failure in NZO islets in a synchronized manner (7,9,10). These early events include decreased expression of Foxo1 and cilia genes and, if extended beyond 2 days, are followed by reduced expression of Pdx1, Nkx6-1, and Mafa; reduced proliferation; and increased apoptosis (10,14). The difference in diabetes susceptibility between NZO and OB mice phenocopies the situation in humans, as there is an overlap of human diabetes susceptibility genes and differentially expressed genes (DEGs) of OB and NZO islets (9).
Recent transcriptomic and functional analyses of single pancreatic endocrine cells have been instrumental in enhancing our understanding of islet cell fate and dysfunction during diabetes (15–18). By using single-cell RNA sequencing (scRNA-seq) (17) combined with diverse bioinformatics analyses, we aim to illuminate the heterogeneity and plasticity of pancreatic islet cells and to decipher the underlying adaptive mechanisms that enable OB islets to escape β-cell loss under metabolic stress conditions.
Research Design and Methods
Animal Studies
Five-week-old male NZO (NZO/HIBomDife) and B6.V-Lepob/ob (OB) mice from our own breeding (German Institute of Human Nutrition Potsdam-Rehbrücke [DIfE]; ethics approval no. 2347-33-2019) were fed a carbohydrate-free high-fat diet (−CH) (C 1057/89; Altromin) for 13 weeks (until 18 weeks of age), followed by random allocation into two diet intervention groups: 2 days of either −CH or a carbohydrate-rich high-fat diet (+CH) (self-made with 40% carbohydrates) (7) (Supplementary Fig. 2A).
Immunofluorescence Staining
Pancreata of OB and NZO mice (−CH and +CH groups, n = 3) were prepared for immunohistochemistry as previously described (7). Pancreatic sections were stained for somatostatin (1:1,000, MA5-16987; Invitrogen), insulin (1:50,000, I2018; Sigma-Aldrich), and glucagon (1:2,000, A0565; Dako). Primary antibodies were detected with fluorophore-labeled secondary antibodies at a dilution of 1:200 (anti-rat Alexa Fluor 546, anti-mouse Alexa Fluor 488, anti-rabbit Alexa Fluor 647; Invitrogen) for 1 h at room temperature. Images of 10–15 randomly selected islets per section were documented with a confocal microscope (TCS SP8 X; Leica Microsystems) and analyzed with ImageJ (19). In total, >6,000 islet cells were evaluated per group.
Isolation of Pancreatic Islets
Islet were isolated through standard collagenase digestion (7).
In Vitro Detection of Cellular Stress, Apoptosis, and Proliferation
One day after seeding, INS-1 823/13 cells were transduced with adenovirus expressing scrambled control shRNA (sh-scrmb) or sh-Slc2a2 (targeting both rat and mouse Slc2a2 isoforms; multiplicity of infection [MOI] 5–20) overnight. Afterward, they were incubated with regular INS-1 medium or INS-1 medium containing 30 mmol/L glucose and 0.2 mmol/L palmitate/BSA for 2–3 days. Finally, cells were lysed in radioimmunoprecipitation assay buffer and ER stress and apoptosis markers were detected via immunoblotting (6,10) (Supplementary Table 1).
Islets from C57BL/6J mice were isolated, dispersed via accutase digestion, seeded onto coverslips coated with fibronectin (F1141; Sigma-Aldrich) and extracellular matrix (E1270; Sigma-Aldrich), and cultured in RPMI medium (P04-16500; PAN Biotech UK) for 24 h. Dispersed islets were transduced with sh-scrmb or sh-Slc2a2 (MOI 20; 24 h) before incubation with regular RPMI medium or glucolipotoxic medium (RPMI medium containing 30 mmol/L glucose and 0.2 mmol/L palmitate/BSA) for 2 days. For prevention of overgrowth with fibroblasts, FibrOut (7-15174; CHI Scientific) was added to media. Finally, cells were lysed in QIAzol and RNA isolated and reverse transcribed and ER stress markers detected via quantitative RT-PCR.
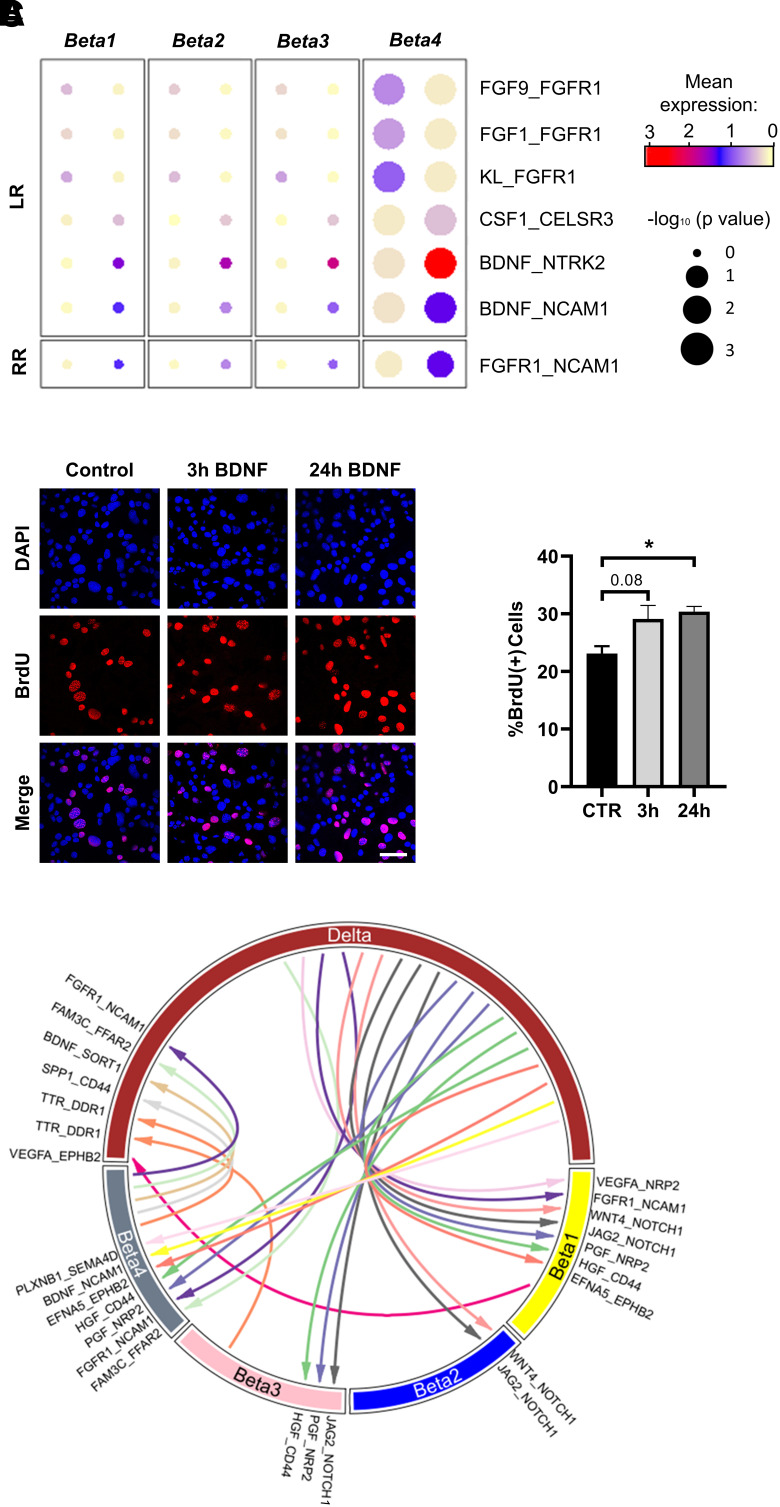
Proliferation assays were performed with MIN6 cells cultured in serum-free MIN6 medium with FGF-1 (4034-50; BioVision), FGF-9 (273-F9-025; R&D Systems), or BDNF (B-250; Alomone Labs) for 3 and 24 h. BrdU labeling (100 μmol/L, 2 h) was detected via immunofluorescence staining and analyzed with ImageJ.
Single-Cell Suspension and Single-Cell Libraries
For scRNA-seq, islets from three mice per condition were pooled (34 islets per animal) and dissociated into single-cell suspension. The cell suspension was immediately used for scRNA-seq library preparation with use of the Chromium Single Cell 3′ Reagent Kit, version 3 (PN-1000075; 10× Genomics) according to the manufacturer’s instructions. Libraries were pooled and sequenced on a NovaSeq 6000 (Illumina) with an average read depth of 60,000–90,000 reads/cell.
Preprocessing of scRNA-seq Data and Identification of Doublet-Like Cell Clusters
Preprocessing was performed with the Cell Ranger pipeline (version 3.0.2; 10× Genomics), as well as python3 and Scanpy (version 1.4.4) (20). mm10 (Ensemble release 97) was used as reference genome. Cells with a mitochondrial content <20% and read counts between 1,000 and 150,000 were selected for further analysis. Lastly, we set a threshold of 500 expressed genes for live cells. An overview of cell numbers at each respective filtering step is shown in Table 1.
Table 1.
Number of cells remaining after several steps of quality control
| Start | Mt in % <20 | Min count >1,000 | Max count <150,000 | Genes/cell >500 | Doublets | |
|---|---|---|---|---|---|---|
| All samples | 34,695 | 21,402 | 20,757 | 20,491 | 19,952 | 19,440 |
| OB −CH | 10,829 | 5,409 | 5,259 | 5,198 | 5,014 | 4,885 |
| OB +CH | 7,022 | 4,189 | 4,189 | 4,146 | 3,872 | 3,787 |
| NZO −CH | 6,634 | 4,393 | 4,224 | 4,083 | 4,061 | 3,964 |
| NZO +CH | 10,210 | 7,411 | 7,085 | 7,064 | 7,005 | 6,804 |
Cells failing quality control were sequentially subtracted from the total number of cells before quality control (Start). Shown are the number of cells remaining after each step. Doublets, exclusion of cell doublets; Genes/cell, number of genes detected per cell; Mt in %, mitochondrial content in percent; Max count, maximum number of reads per cell; Min count, minimum number of reads per cell.
For detection and removal of data reflecting expression of doublet cells, scrublet (21) (version 0.2.1) was applied independently for each study group (Supplementary Fig. 1).
Normalization of scRNA-seq Data
Normalization of scRNA-seq data was performed in independent steps: 1) normalizing the number of reads to the number of expressed genes with the R package scran (22) (version 1.16), 2) log-scaling of data, and 3) via batch correction with scanorama (version 1.4) (23). scanorama used 3,000 highly variable genes, detected per batch and merged afterward (scanpy.pp.highly_variable_genes). If not stated otherwise, subsequent analysis was based on those highly variable genes.
Clustering, Embedding, and Cell Type Annotation
Preliminary clustering of cell types was based on the batch-corrected whole expression profile of single cells as suggested by Büttner et al. (24). For embedding, a neighborhood graph for single cells was calculated, as k-nearest neighbor (KNN) graph, based on the first 50 principal components and considering the nearest 15 neighbors (17). Next, Louvain-based clustering was applied as implemented by Blondel et al. (25) (version 0.6.1). Variant resolutions were used for several different clusters to appropriately detect strong and weak changes in single cell gene expression profiles. Clusters were annotated based on gene expression levels of typical marker genes and hormones (Supplementary Table 2), from Baron et al. (26) with adaptations from Martens et al. (27). Plotting of cell clusters was done via UMAP (28).
Detection of DEGs
DEGs were detected with the MAST R package (29) (version 1.12) and P values corrected for multiple testing through calculation of the false discovery rate (FDR). Genes with an FDR <0.01 and a coefficient >0.2 were defined as differentially expressed. Since input data were already log-transformed, the output was not comparable with classic log (fold change) values. Thresholds similar to those described by others (17) were used for these log-transformed input data.
Gene set enrichment for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms was analyzed with Database for Annotation, Visualization and Integrated Discovery (DAVID) (30).
Network analysis of DEGs between Beta1 cells of OB and NZO mice was performed via Ingenuity Pathway Analysis (QIAGEN) (31). Differentially expressed cilia-associated genes were identified by overlapping with 5,266 orthologs of cilia-annotated genes in the Cildb database (version 3.0) (32,33).
Calculation of Cell Cycle State
For estimation of the cell cycle state of single cells, an S and G2/M phase score with a threshold of 0.25 was used, based on the data of Macosko et al. (34). The scores were calculated as previously described (35), using the scanpy function tl.score_genes_cell_cycle.
β-Cell Embryonic Scores
β-Cell embryonic scores were calculated (17) based on the GSE87375 data set (36). Expression data from cells between E17.5 and P0 were used to define embryonic β-cells, and P60 was used for mature β-cells. DEGs between those two groups were calculated with log (fold change) > 0.25 and FDR < 0.01. In taking into account only the top 500 genes, scores were assigned with the tl.score_genes scanpy function.
Estimation of β-Cell Trajectories
β-Cell trajectories were calculated with slingshot (version 1.4) (37) using only β-cell–related clusters as input (Beta1, Beta2, Beta3, Beta4, Beta5, BetaP, BG, BDG). To calculate strain-specific effects related to the +CH diet, the data set was split into OB (−CH and +CH) and NZO (−CH and +CH) subsets. The Beta1 cluster was used as root node for both sets to arrange cells in pseudotime and obtain diet effects. For confirmation of the observed pseudotime, randomly selected cells with all DEGs were used to generate heat maps based on the trajectories Beta1-Beta2-Beta3-Beta4 (H1) or Beta1-Beta4-Beta2-Beta3 (H2). Finally, 1,000 permutations were performed, and which trajectory the genes correlated with significantly was determined.
Weighted Gene Correlation Network Analysis
A gene coexpression network was created from normalized and batch-corrected transcriptome data of all Beta4 cells through following the best practice workflow for weighted gene correlation network analysis (WGCNA) (38) (R package, version 1.69) (power = 3, minClusterSize = 30, and deepSplit = 2). Hub genes were defined by geneModuleMembership >0.70 among the top 10 connected genes within a module. Graphical representation was created with the R package igraph (version 1.2.6).
Detection of Differentially Coexpressed Genes Between β-Cell Clusters
The code provided by Tesson et al. (39) was used to detect differential coexpression between the Beta4 and Beta1–3 clusters. For detection of a variety of differences, a power value of 3 and a minclustersize of 30 were used.
Estimation of Cell-Cell Communication Using CellPhoneDB
Cell-cell communication was predicted with use of CellPhoneDB (version 2.0) (40). The normalized and batch-corrected gene expression matrix, as well as the cell type clustering depicted in Fig. 2B, were required to identify the most probable interactions. Interactions were visualized using the R packages ComplexHeatmap (version 2.4.3) and circlize (version 0.4.12).
Figure 2.
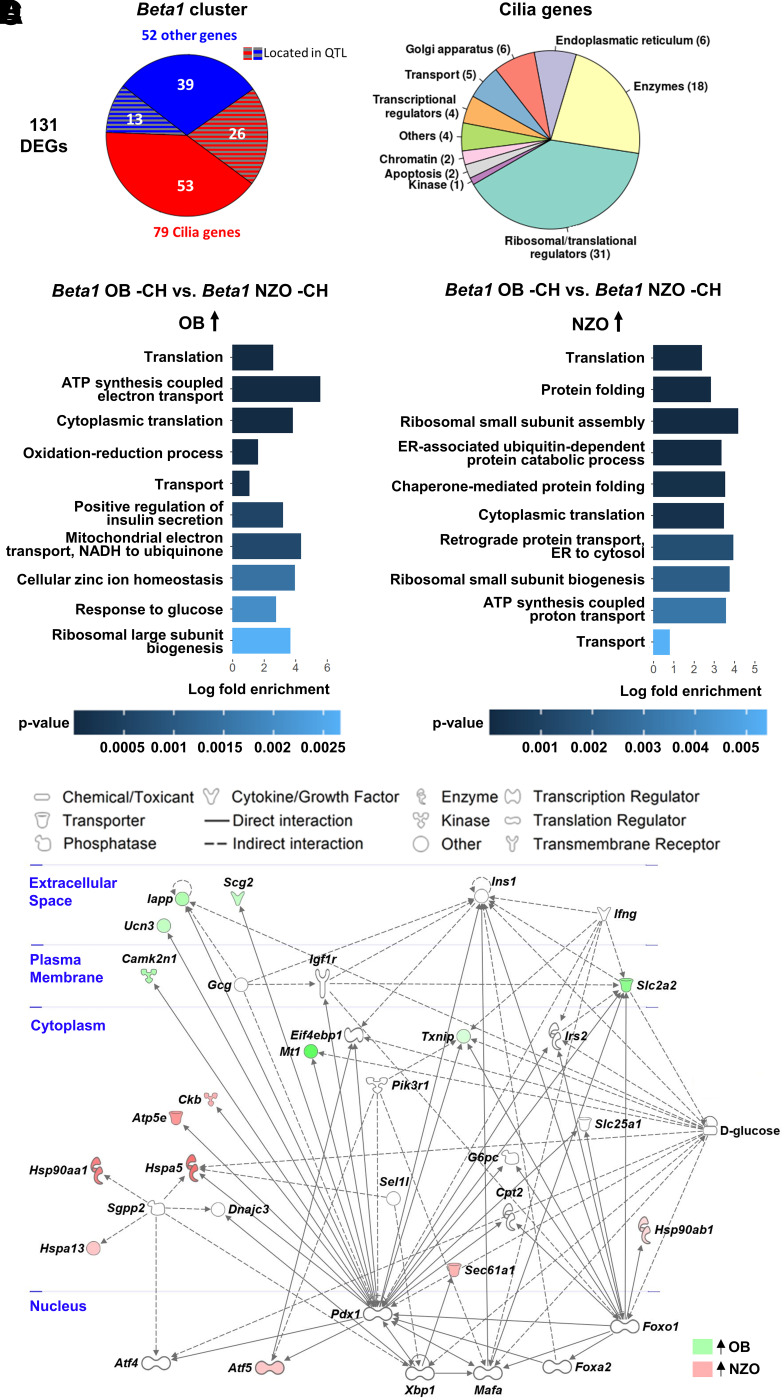
Differential expression of cilia-annotated and PDX1-regulated genes between OB- and NZO mice in the carbohydrate-free β-cell cluster Beta1. A: Annotation of the 131 genes differentially expressed between Beta1 cells of OB and NZO mice by cellular location and process. The DEGs were separated into cilia-annotated genes and remaining genes based on the Cildb database. B and C: Top 10 regulated GO terms (biological process) of DEGs from the comparison of OB −CH Beta1 vs. NZO −CH Beta1 cells. Shown are biological processes upregulated in OB (B) or NZO (C) mice. Absolute log (fold change) > 0.2; FDR < 0.01. D: Network analysis of 131 genes differentially expressed between Beta1 cells of OB and NZO −CH mice. Data were analyzed via Ingenuity Pathway Analysis. Cellular localization of gene products is shown in blue.
Sources of Human Genome-Wide Association Study Genes and Mouse Linkage Studies
The NHGRI-EBI GWAS Catalog (41) was screened for variants associated with T2D, with addition of signals detected in a large metastudy (42). QTLs of C57BL/6J and NZO mice were extracted from https://146.107.176.32/QTL-DZD-Cross/ (43).
Detection and Classification of Mouse Single Nucleotide Polymorphisms
Sequence information and mutations in NZO mice were downloaded from the Wellcome Sanger Institute (44). We calculated effects of genes harboring a missense variant using Protein Variation Effect Analyzer (PROVEAN) (45) and SIFT (46).
Statistics and Plotting
Statistical significance was analyzed with Student t test, one-way ANOVA, or two-way ANOVA through comparison of the test groups with the appropriate control group. Data are presented as mean ± SEM, with significance indicated by asterisks in figures (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). Where applicable, the number of replicates (n) is stated in figure legends. If not stated otherwise, enrichment analysis was performed with Fisher exact test. We created plots with R, version 3.6, using the RCircos (47,48), DiagrammeR, and gplots packages.
Data and Resources
All relevant data generated or analyzed during this study are included in this manuscript. Raw scRNA-seq data were deposited under gene accession no. GSE159211 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159211). Further data sets used or analyzed during the current study are available from the corresponding author on reasonable request. Resources are available on request.
Results
Different Islet Cell Composition in Diabetes-Resistant OB and Diabetes-Susceptible NZO Mice
In contrast to previous bulk RNA-sequencing studies (7,10), we investigated the contribution of different islet cell subtypes from OB and NZO mice fed a −CH or +CH diet for 2 days (10) (Supplementary Fig. 2A). The animals reacted to the diet regimen as expected, with higher blood glucose levels and plasma insulin levels on +CH diet. In line with previous reports (7,9), NZO islets were smaller than their diet-matched OB counterparts and contained significantly fewer β- and more α-cells (Supplementary Results and Supplementary Figs. 2B–F and 3A–D). Subsequently, islets from all four groups (OB −CH, OB +CH, NZO −CH, NZO +CH) were isolated, dispersed, and subjected to scRNA-seq analysis.
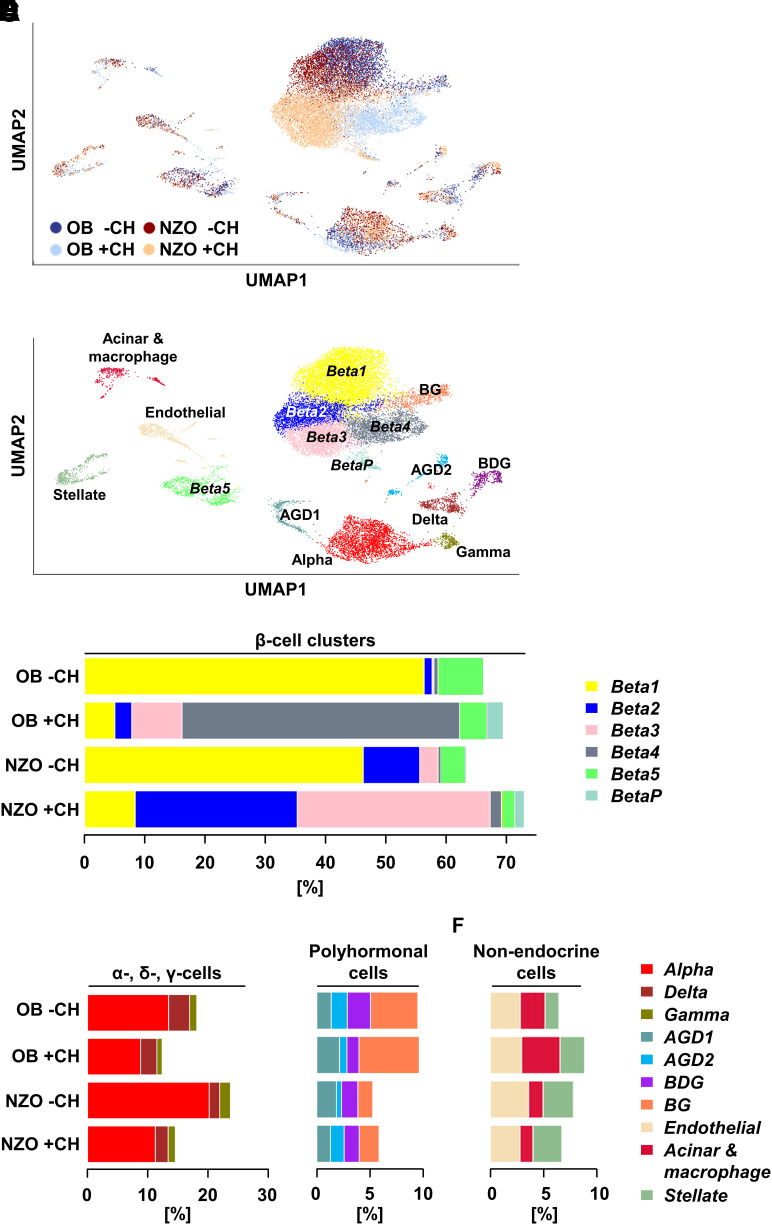
In total, 19,440 cells remained following quality control (Table 1). While the mean number of expressed genes increased in OB +CH compared with −CH islets, NZO +CH islets expressed the lowest average number of genes (Supplementary Fig. 4A). Next, cells from all four groups were subjected to unbiased clustering analysis and resulting clusters assigned to cell types based on known marker genes (Fig. 1A and B and Supplementary Table 2). Among the identified populations were six β-cell clusters and one α-, γ-, and δ-cell cluster, plus several polyhormonal (AGD1, AGD2, BDG, BG; abbreviations based on marker gene expression for α- [A], β- [B], γ- [G], and δ- [D] cells) and nonendocrine (acinar and macrophage, endothelial, and stellate cell) clusters (Fig. 1B). As expected, the majority of cells (63–73%) were β-cells (Fig. 1C and Supplementary Table 3). Notably, both OB and NZO mice on −CH diet showed a comparable β-cell cluster composition, with most β-cells belonging to the Beta1 cluster (56.3 and 46.2%, respectively). In addition, NZO −CH islets exhibited a considerable percentage of Beta2 and Beta3 cells and OB −CH islets a higher number of Beta5 β-cells (Fig. 1C and Supplementary Table 3). Beta5 cells expressed a very low number of genes (Supplementary Fig. 4B) and could represent low-complexity libraries.
Figure 1.
Changes in β-cell subpopulations in response to a 2-day diabetogenic stimulus markedly differ in diabetes-resistant OB and diabetes-prone NZO mice. A: UMAP plot of all 19,440 islet cells from OB −CH (blue dots), OB +CH (lilac dots), NZO −CH (orange dots), and NZO +CH (red dots) mice. Islet cells from n = 3 mice per group were pooled and used for scRNA-Seq. B: UMAP plot of all cells from the four groups with indicated cell clusters. Multihormonal cell clusters were designated by the presence of markers for α-cells (A), β-cells (B), γ-cells (G), and/or δ-cells (D). C: Bar plots showing the percentage of cells belonging to a specific β-cell cluster in OB and NZO islets under −CH or +CH feeding. Percentages of α-, δ-, and γ-cells (D) as well as polyhormonal (E) and nonendocrine (F) cells in pancreatic islets of the indicated groups.
Drastic changes in cluster distribution were observed in +CH-fed mice, in which β-cell composition changed at the expense of a greatly diminished Beta1 cluster (Fig. 1C). The β-cell cluster signature between OB +CH and NZO +CH diverged; most OB +CH β-cells ended up in Beta4, with a lower percentage in Beta3. In contrast, NZO +CH β-cells were allocated into Beta2 and Beta3 clusters at roughly equal proportions (Fig. 1C). These results suggested that β-cell adaptation versus failure was mainly presented as gene expression changes between Beta1 and Beta2–4 clusters. Because NZO mice proceed to develop stark β-cell defects after 16–32 days on a +CH diet (9), the shift from Beta1 to Beta2/3 likely represents failure to adapt to the diabetogenic stimulus. Alternatively, OB β-cells handle elevated blood glucose levels by switching to a Beta4 cell subtype. Furthermore, a cluster of proliferating β-cells, designated BetaP (where P is proliferation), was identified based on proliferation marker genes (Mki67, Pcna) and G2/M cell cycle phase genes (Supplementary Fig. 4C–E). BetaP was strongly induced by 2-day +CH feeding, particularly in OB +CH (2.6%) compared with NZO +CH (1.6%) islets (Fig. 1C and Supplementary Table 3). Whereas NZO islets contained more α- and slightly more γ-cells than OB islets, the percentage of the δ-cell cluster was highest in OB islets (Fig. 1D). Additionally, OB islets exhibited more polyhormonal cells than NZO islets (Fig. 1E), which could indicate that some OB endocrine cells can proceed toward a developmentally more immature, polyhormonal state. This could protect them from the glucotoxic effects of the +CH diet, as opposed to NZO islets, which appear less capable of doing so. Concerning nonendocrine cells, OB islets contained more acinar cells and macrophages than NZO islets (Fig. 1F).
Predisposition of NZO Beta1 Cells to Metabolic Stress
In line with previous studies, we hypothesized that the changes in NZO islets are maladaptive, leading to functional decline and cell death, while changes in OB islets generally promote cell survival and proliferation. Therefore, we addressed the question of why NZO β-cells on a +CH diet progress toward Beta2/Beta3 rather than the more protective Beta4 cluster. If genetic predisposition of NZO mice played a role, transcriptomic differences between OB and NZO β-cells might be manifest in the Beta1 cluster that is common to both strains on −CH diet. While OB and NZO Beta1 gene expression showed 98.48% (8,490 genes) overlap, 131 genes (1.52%) were differentially expressed (Supplementary Table 4). We recently showed enrichment of cilia-annotated genes in DEGs of OB and NZO islets (10), and 79 of the 131 Beta1 DEGs were annotated to cilia (Fisher exact test P < 0.00001; odds ratio 2.5) (Supplementary Table 4), 33 of which were more abundant in OB and 46 in NZO β-cells. Using linkage analysis from B6-NZO crosses (43), we located 13 of 52 noncilia genes and 26 of the 79 cilia-annotated genes to a metabolically related QTL (Supplementary Table 4), suggesting that these genes are relevant for the divergent development of β-cell clusters during the progression from −CH to +CH (Fig. 2A). The cilia-related genes are involved in different cellular processes (Fig. 2A and Supplementary Table 4); several encode ribosomal proteins (18 Rpl and 13 Rps genes), pointing at altered ribosomal biogenesis and protein synthesis, which, among other processes, is needed during cellular proliferation (49,50). Other genes implicated in β-cell function and survival, such as Isl1 (51–53) and Rgs2 (54), were also more highly expressed in OB Beta1 cells (Supplementary Table 4).
Interestingly, OB Beta1 β-cells expressed more genes linked to positive regulation of insulin secretion (Glp1r, Slc30a8, Isl1, Ucn3), translation (Rpl26, Rps5), oxidation-reduction process (Ndufa7, Ero1lb), and transport (Naca, Slc2a2) (Fig. 2B and Supplementary Table 5), implying a more metabolically active state. At the same time, genes specific for NZO Beta1 β-cells were enriched in pathways linked to protein folding (Canx, Hsp90aa1, Calr) and ER-associated ubiquitin-dependent protein catabolic process (Vcp, Ubxn4, Sec61b) (Fig. 2C and Supplementary Table 5), indicating that these β-cells were already undergoing protein misfolding or oxidative stress. Finally, network analysis by Ingenuity Pathway Analysis demonstrated that targets of the key β-cell transcription factor Pdx1 were more abundant in OB cells (Supplementary Results and Fig. 2D). Altogether, the OB-specific gene expression pattern of Beta1 cells pointed toward active, functional β-cells, whereas that of NZO Beta1 cells suggested an ongoing response to metabolic and ER stress.
β-Cell Clusters of Diabetes-Prone Mice Are Less Capable of Mitigating Effects of Diabetogenic Diet
The data above indicated that β-cells of OB and NZO mice have different capacities to react to a diabetogenic diet. To test the assumption that Beta1 mainly changes into the Beta4 cluster in OB +CH, but into Beta2 and Beta3 in NZO +CH islets (Fig. 1C), we estimated cell-to-cell distances based on similarity of gene expression using slingshot (37). Since these cell-to-cell distances were comparable with pseudotime calculations, we refer to them as such below.
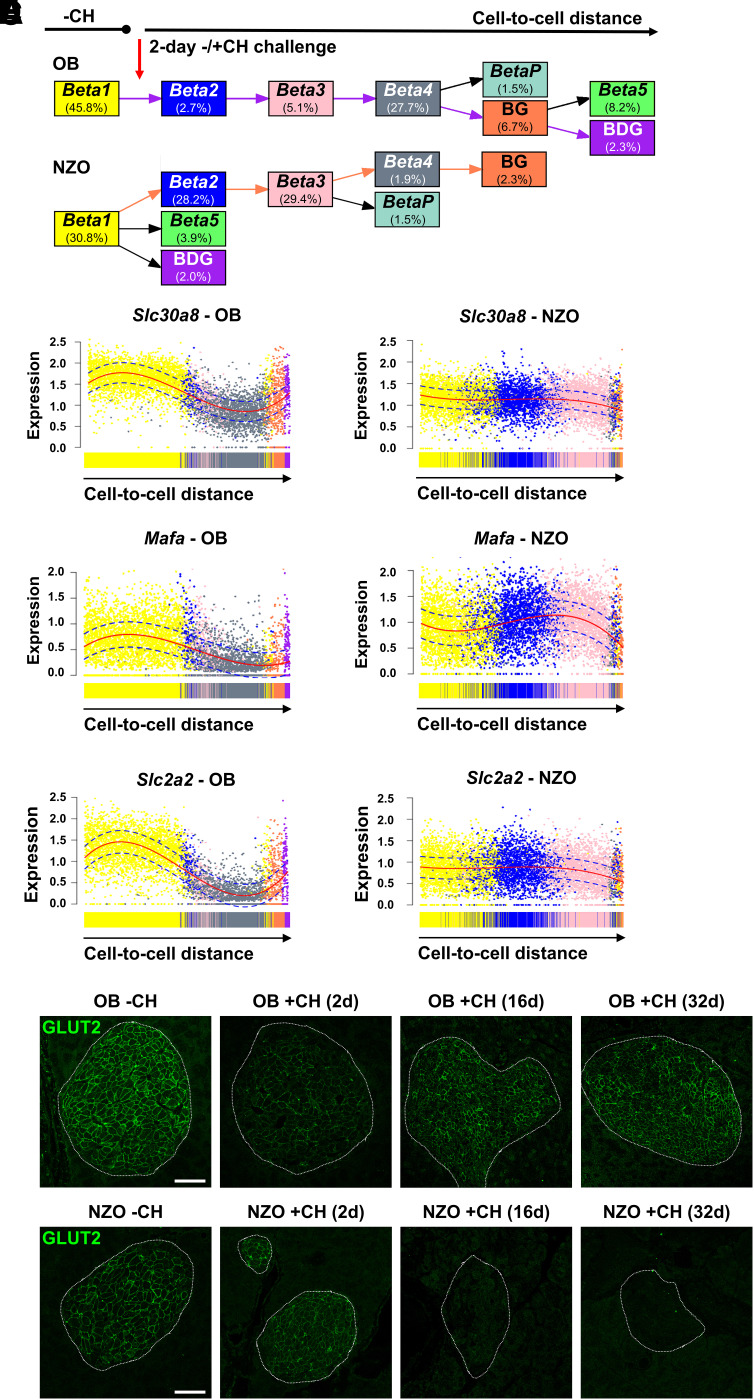
To assess cell fate changes triggered by +CH diet, we defined the predominant cluster in −CH-fed mice (Beta1) as the starting point for the algorithm. Principal pseudotime ordering of the major β-cell clusters was confirmed based on DEGs (Supplementary Fig. 5A), with the most likely lineage progressing from Beta1, via Beta2 and Beta3, to the Beta4 cluster (Supplementary Fig. 5B and C). This lineage was found in both OB and NZO cells. However, while a large proportion of OB β-cells develops into Beta4 (27.7%) with few cells remaining in Beta2/3 (2.7 and 5.1%, respectively), the majority of NZO β-cells fall into clusters Beta2/3 (28.2 and 29.4%) with only few reaching Beta4 (1.9%) (Fig. 3A). Our analysis further predicted that OB Beta4 cells could develop into the proliferation cluster BetaP as well as multihormonal (BG and BDG) or Beta5 cells (Fig. 3A). For NZO islets, percentages of these populations were generally lower, and the algorithm predicted alternative branching points coming from Beta1 and Beta3 instead of Beta4 (Fig. 3A). Together, these factors indicated that similar lineage transitions occurred in β-cells of both strains, but adaptation of NZO +CH β-cell gene expression beyond Beta2/3 was stalled or impeded compared with OB +CH β-cells, a potential underlying cause for the maladaptive response of NZO islets to carbohydrate feeding.
Figure 3.
Trajectories of β-cell clusters in OB and NZO in response to 2-day carbohydrate feeding. A: Pseudotime analysis using slingshot shows predicted trajectories of OB and NZO β-cells based on cell-to-cell distance. The percentage of cells in the different clusters is depicted in the respective boxes. The trajectory from Beta1 to BDG in OB is highlighted with purple arrows and the Beta1 to BG trajectory in NZO with orange arrows. B–D: Expression levels of Slc30a8 (B), Mafa (C), and Slc2a2 (D) along those trajectories highlighted in A. Dots represent scaled expression levels of individual β-cells color coded according to their cluster allocation. Solid red lines depict fitted curves, and dotted blue lines depict CIs. E and F: Representative images of GLUT2 (green) staining in pancreatic sections of OB (E) and NZO (F) mice fed a −CH or +CH diet for 2, 16, or 32 days. After 16 and 32 days, GLUT2-positive signals were nearly undetectable in NZO islets, whereas a transient reduction in OB islets at 2 days with +CH was reversed at the later time points. Scale bar, 50 µm. d, days.
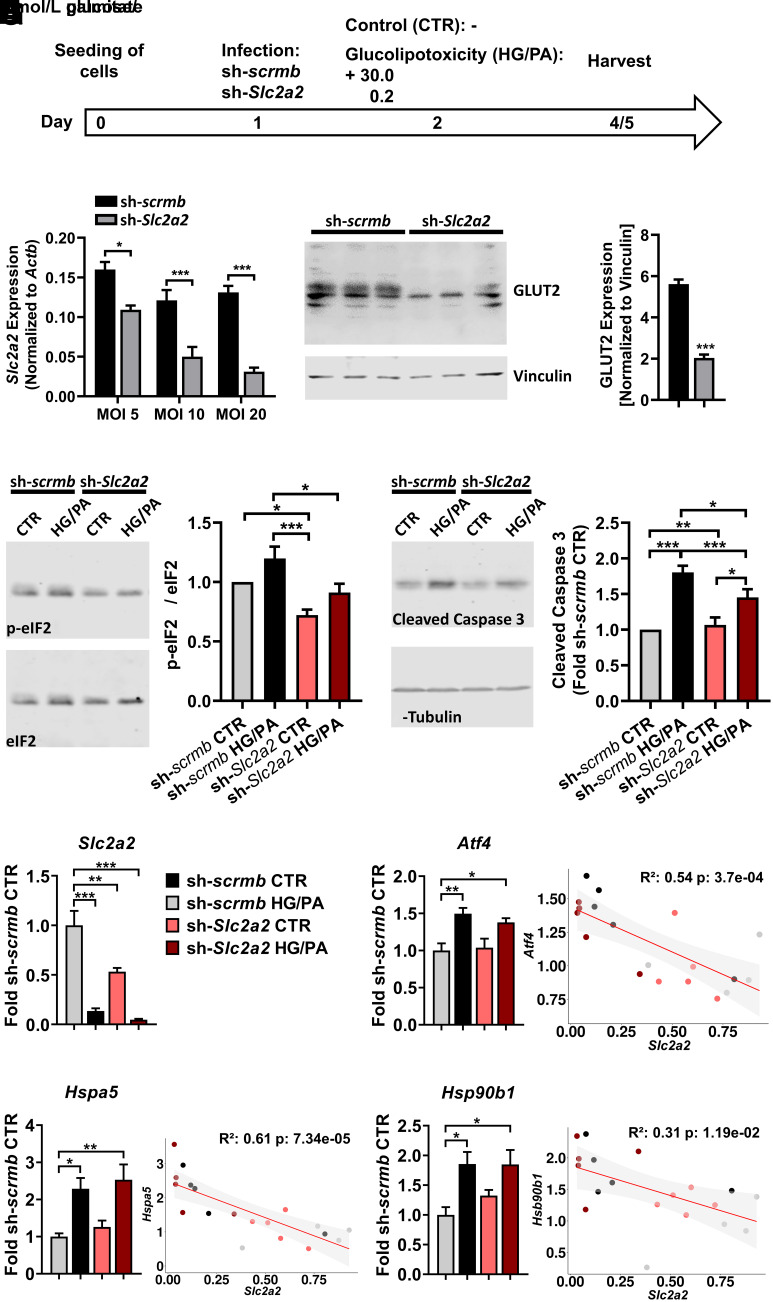
Indeed, dynamic expression changes of several important β-cell and ER stress response genes along the pseudotime lineage indicated that NZO β-cells (Beta2/3) were less adept than OB β-cells (Beta4) at compensating for the increased carbohydrate intake on +CH diet (Supplementary Results, Fig. 3B and C, and Supplementary Fig. 6A–E). For example, Slc2a2/GLUT2 fell drastically during the OB Beta1 to Beta4 cluster transition (Fig. 3D and Supplementary Fig. 6E). Immunohistochemical staining confirmed a substantial, transient reduction of GLUT2 signal in OB +CH versus −CH islets (Fig. 3E), which partially recovered at later time points (16 and 32 days, +CH) (7,9), whereas GLUT2 decreased successively in NZO islets (Fig. 3F). For testing of whether short-term downregulation of GLUT2 provides temporary protection from the toxic effects of glucose overload and thereby protects β-cells from ER stress and apoptosis, its expression was suppressed in INS-1 cells (Fig. 4A–C). While treatment with palmitate and glucose increased the ER stress marker phosphorylated (phospho)-eIF2α in sh-scrmb–transduced cells, this effect was abolished in sh-Slc2a2–infected cells (Fig. 4D), indicating that short-term loss of GLUT2 expression reduces ER stress. In addition, control cells exhibited elevated activation of caspase 3 when treated with glucolipotoxic medium, an effect that was also lower in sh-Slc2a2–transduced cells (Fig. 4E). Because insulinoma cells are partially dedifferentiated and may already express lower levels of Slc2a2 as primary β-cells, we sought to validate our hypothesis of a protective effect due to temporary GLUT2 reduction in dispersed islets using the above conditions. Both treatment with glucolipotoxic medium for 2 days and adenoviral Slc2a2 downregulation resulted in upregulation of Atf4, Hspa5, and Hsp90b1. Thus, lower levels of Slc2a2 correlate with higher levels of these stress response genes (Fig. 4F–I), suggesting that following Slc2a2 downregulation, metabolic stress is resolved successfully, presumably due to lower glucose uptake.
Figure 4.
Downregulation of GLUT2 in β-cells ameliorates glucolipotoxic effects. A: INS-1 823/13 (INS-1) cells or dispersed C57BL/6J islets were infected with adenovirus containing scrambled (sh-scrmb) or GLUT2-specific (sh-Slc2a2) shRNA and incubated without (CTR) or with 30 mmol/L glucose and 0.2 mmol/L palmitate (HG/PA) for 2–3 days. B: Successful downregulation of GLUT2 mRNA at different viral loads (n = 4) (one-way ANOVA). C: Representative Western blot of GLUT2 in INS-1 cells infected with sh-scrmb or sh-Slc2a2 (MOI 20) (left panel) and quantification of GLUT2 levels (right panel) (n = 3, Welch t test). D: Representative immunoblots of phospho-eIF2α and total eIF2α in sh-scrmb or sh-Slc2a2–infected INS-1 cells (MOI 20) (left panel) and quantification of phospho-eIF2α levels relative to total eIF2α (right panel) (n = 8, two-way ANOVA). E: Representative immunoblot of cleaved caspase 3 in sh-scrmb or sh-Slc2a2–infected INS-1 cells (MOI 20) (left panel) and quantification of cleaved caspase 3 levels relative to α-tubulin (right panel) (n = 12, two-way ANOVA). F–I: Quantification of gene expression via quantitative PCR in dispersed C57BL/6J islets infected with sh-scrmb or sh-Slc2a2 (MOI 20). Slc2a2 expression (F) was correlated with levels of Atf4 (G), Hspa5 (H), and Hsp90b1 (I). Data are presented as mean ± SEM (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). p-, phosphorylated.
Increased Cell Surface Marker Gene Expression in β-Cells of Diabetes-Resistant Mice on a Diabetogenic Diet
Dysregulated expression of receptors, transporters, channels, and cell adhesion molecules cause β-cell defects (55). We asked whether the maladaptive responses to diabetogenic diet in NZO islets were accompanied by genes coding for cell surface proteins. In the α-, β-, δ-, and γ-cell clusters, 874 of 3,702 predicted islet cell surface genes (55) were detected (mean normalized expression ±0.1). Average expression levels of cell surface protein–encoding transcripts were comparable between β-cell clusters, except for the OB +CH-enriched Beta4, which exhibited much higher expression (Supplementary Fig. 7B). For α- and δ-cells, the highest content of surface marker genes was found in the OB +CH cluster, while there was no difference between groups in γ-cells (Supplementary Fig. 7C).
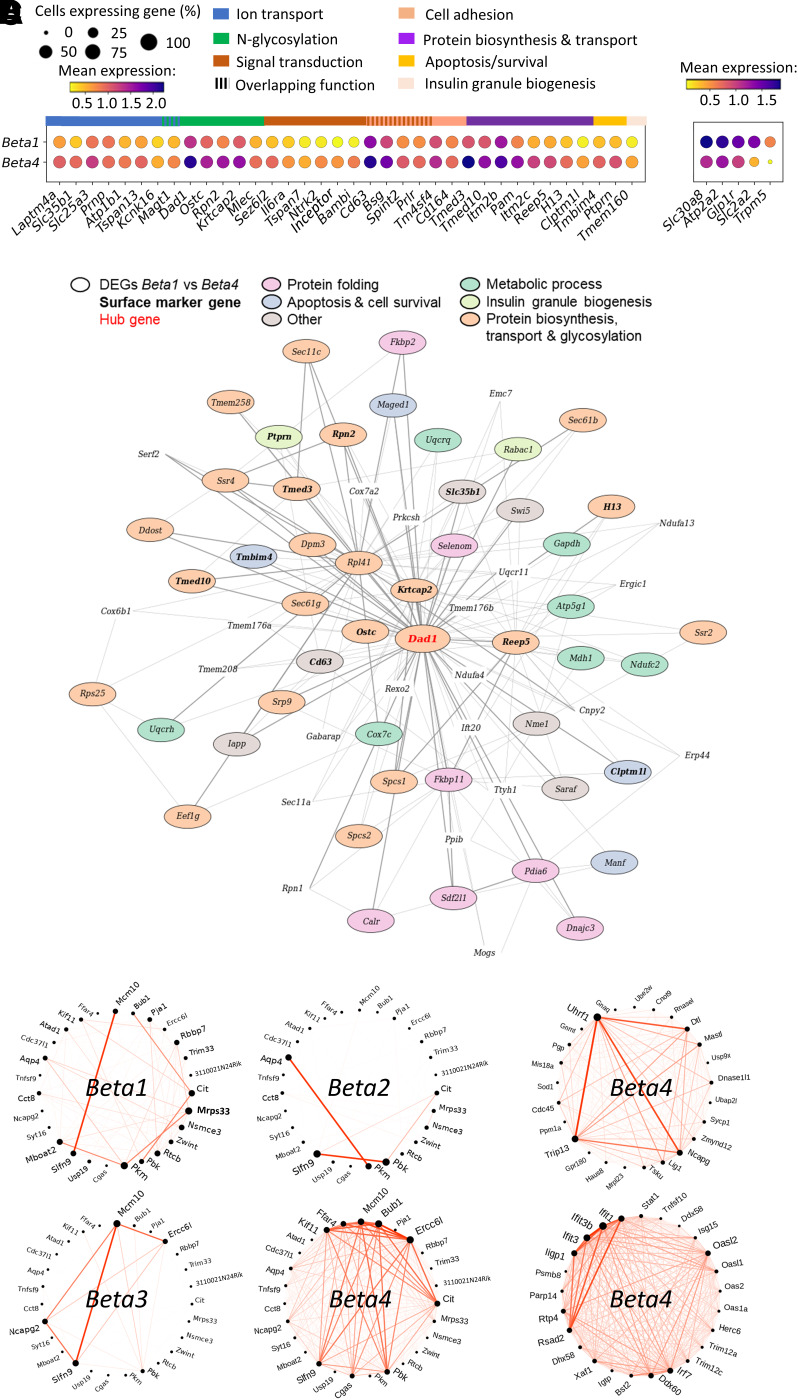
In our further analysis, we focused on the Beta4 cluster because we hypothesized that its expression pattern participates in the protective adaptations of diabetes-resistant mice. Of 41 cell surface marker genes differentially expressed between Beta1 and Beta4, 36 were more highly and 5 less highly expressed in Beta4 cells (Fig. 5A). The upregulated genes were associated with ion transport (e.g., Atp1b1), N-glycosylation (e.g., Dad1), signal transduction (e.g., Il6ra), cell adhesion (e.g., Bsg), and protein biosynthesis, processing, and transport (e.g., Tmed10). The downregulated genes (Slc2a2, Scl30a8, Glp1r, Trpm5, Atp2a2) play essential roles for β-cell function, confirming a more dedifferentiated phenotype of Beta4 (Supplementary Fig. 7A). A temporary reduction of these genes in the Beta4 cluster might participate in protecting β-cells from glucotoxicity, as shown for Slc2a2/GLUT2 (Fig. 4).
Figure 5.
Diet- and strain-dependent differences in cell surface marker gene expression and coexpression analysis. A: Dot plot representing the number of cells (dot size) and mean gene expression levels in the Beta1 and Beta4 clusters. Shown are genes differentially expressed between the two clusters and either up- or downregulated in Beta4. Genes were sorted with respect to their main function, as indicated with horizontal bars. B: Coexpression relationships of Beta4 cells according to WGCNA. Shown are the top 70 genes of module M2, including the M2 hub gene Dad1 (highlighted in red). The main function of genes differentially expressed between Beta1 and Beta4 are indicated with colored circles; those DEGs that are surface markers are highlighted in bold. C and D: Circos plots showing DiffCoEx of the Beta4-specific modules M3 (C), M6 (D, upper panel), and M10 (D, lower panel) as detected with DiffCoEx. Coexpressed genes are connected via red lines, with the thickness reflecting the degree of coexpression. The Beta4 cluster (C and D) exhibits the highest number of connections in comparison with the other (Beta1–3) clusters (C).
To determine coexpression relationships in Beta4 cells and to identify important gene modules and hub genes, we conducted WGCNA. Overall, 18 modules were identified by the clustering algorithm (Supplementary Table 7). Module M2 contained many genes related to protein processing in ER, protein export, N-glycan biosynthesis, lysosome, and proteasome, with the antiapoptotic oligosaccharyl transferase subunit Dad1 as a hub gene (Fig. 5B and Supplementary Table 7). Among the 70 most connected genes in module M2, 69% were upregulated in Beta4 compared with Beta1 cells (Fig. 5B [circled]) and 13 genes encoded cell-surface proteins (Fig. 5B [highlighted in bold]). The majority of these DEGs were involved in metabolic processes (e.g., Ndufc2), protein biosynthesis (Eef1g), transport (Sec61b) and folding (Dnajc3), and glycosylation (Ddost). Thus, promoting protein biosynthesis and stability through co- or posttranslational N-glycosylation seems to be a key characteristic of the Beta4 cluster. Next, we applied DiffCoEx in combination with WGCNA to detect differentially coexpressed genes (DiffCoEx) unique for one of the 18 Beta4-specific modules. Three modules (M3, M6, M10) exhibited coexpressed genes (Supplementary Fig. 8A), which were specifically and highly connected in Beta4, with limited connections in Beta1–3 (Fig. 5C and Supplementary Fig. 8B and C). In M3, the strongest DiffCoEx were linked to cell proliferation (Mcm10, Bub1, Ercc6l, Cit, Kif11, and Pbk) (Fig. 5C). Candidate genes in M6 were related to DNA repair (Uhrf1, Trip13, Lig1, Dtl, and Mastl) and in M10 to interferon signaling (Ifit1, Ifit3b, Ifit3, and Irf7) (Fig. 5D).
Taken together, these data indicate that OB β-cells—by developing a Beta4 cluster expression pattern—adapt to increased insulin demand and promote survival on a carbohydrate-rich diet via an upregulation of ER chaperones, N-glycosylation, and cell adhesion genes, including many cell surface proteins.
Enhanced Cell-Cell Communication in β-Cells From Diabetes-Resistant Mice
To investigate whether cell-cell communication plays a role in the protective phenotype of OB islets, we analyzed potential ligand-receptor interactions by comparing the distribution of receptor and ligand gene expression within and across cell types using CellPhoneDB (40). A set of ligands (FGF1, FGF9, KL) identified as partners of FGFR1 was more likely to interact specifically in the Beta4 cluster (Fig. 6A), indicating that Beta4 cells could potentially promote survival and proliferation in an autocrine and/or paracrine manner. Furthermore, Beta4 gene expression for receptors activated by CSF1 (Celsr3) and BDNF (Ntrk2, Ncam1) was higher, including a putative receptor-receptor interaction between FGFR1 and NCAM1, implicating processes like cell survival, adhesion, and cell-cell communication (Fig. 6A). We next tested whether FGF1, FGF9, and BDNF have an effect on β-cell proliferation by treating MIN6 cells with these ligands. BDNF (Fig. 6B) increased β-cell proliferation, shown via higher BrdU incorporation into ligand-treated cells after 24 h; for FGF9, this effect did not reach statistical significance (Supplementary Fig. 9A and C). In contrast, FGF1 did not affect cell division (Supplementary Fig. 9B and D). These data suggest that elevated expression of BDNF receptors contributes to prevention of β-cell failure by increasing proliferation in OB +CH-enriched Beta4 cells.
Figure 6.
Analysis of islet cell-cell communication based on ligand and receptor gene expression. A: Dot plots representing potential ligand-receptor (LR) and receptor-receptor (RR) interactions in Beta1–4 cells and upregulated in Beta4. B: Induction of proliferation in MIN6 β-cells via exposure to BDNF. Serum-starved MIN6 cells were treated with BDNF for 3 or 24 h. Subsequently, BrdU was added for 2 h, followed by fixation and immunostaining with a BrdU antibody (left panel). Quantification of BrdU-positive MIN6 cells shown as percentage of DAPI-positive nuclei (right panel) (n = 3). C: Circle plot depicting potential intercellular communication (based on ligand-receptor gene expression) in δ-cells and β-cell clusters Beta1–4. Arrow directions are from ligand to receptor. Nonunique interactions more likely to occur between δ-cells and all four β-cell clusters are not shown. CTR, control.
Lastly, genes important for cell-cell communication exhibited different expression patterns between β-cell clusters and other endocrine cell types. Putting a focus on unique ligand-receptor interactions shared only between specific β-cell clusters and δ-cells (Supplementary Table 8), more significant interactions were predicted between δ-cell ligands and their respective receptors in Beta1 and Beta4 cells (Fig. 6C). Of note, SST signaling was likely comparable between β-cell clusters because the most abundant receptor Sstr3 was expressed at similar levels in Beta1–4 (Supplementary Fig. 9E). Finally, only few interactions were seen between δ-cells and the diabetes-prone Beta2 and Beta3 clusters (Fig. 6C), suggesting that impaired cell-cell communication is part of the diabetes susceptibility of NZO islets following a diabetogenic diet. Similarly, more ligand-receptor interactions were observed between Beta4 and the dual hormonal BG cells, whereas fewer were seen between BG and the other β-cell clusters (Supplementary Fig. 9F).
Expression Changes in Mouse Islets Are Associated With Human Diabetes Risk Genes
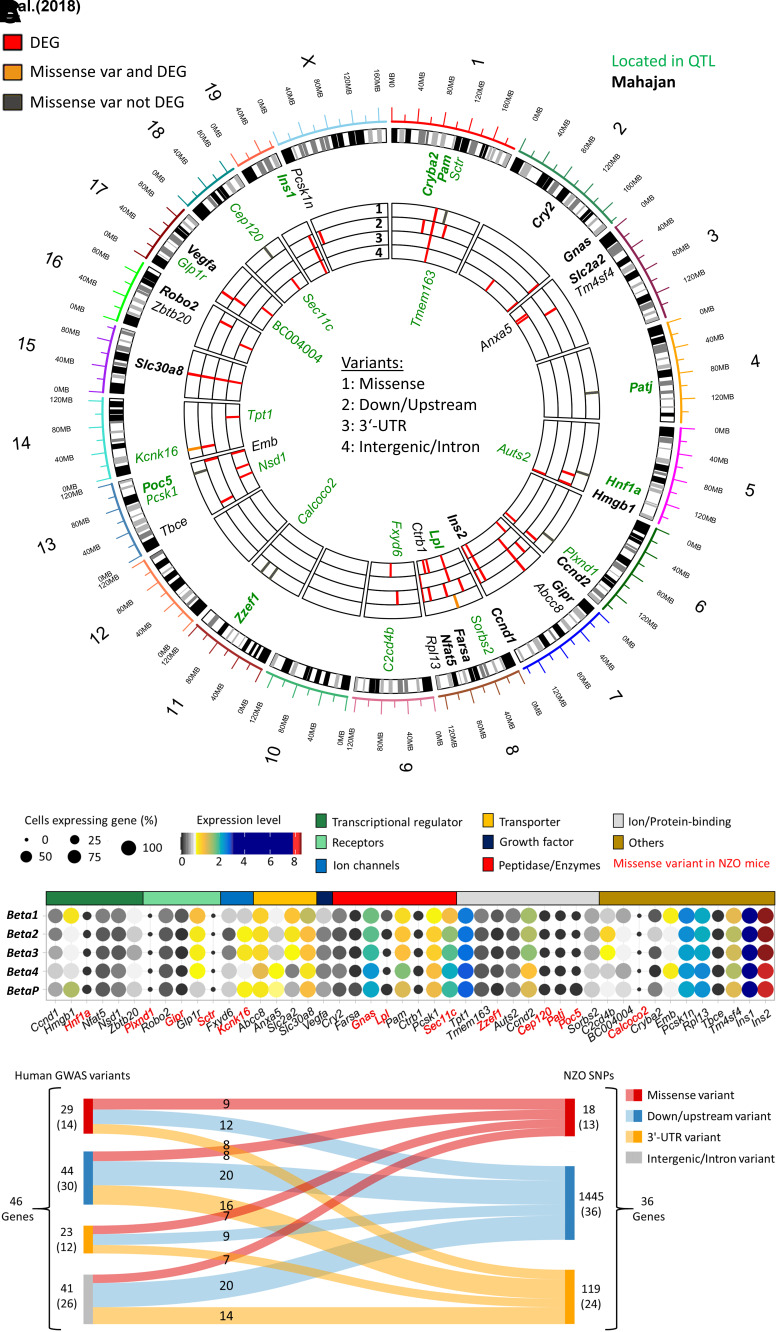
Finally, we assessed the relevance of our observations from diabetes-susceptible NZO mice for human diabetes (T2D). DEGs from our β-cell clusters were compared with a data set of human genes and loci associated with diabetes susceptibility. Overall, 38 of 665 genome-wide association study (GWAS) genes from the NHGRI-EBI GWAS Catalog (41) (odds ratio 2.1, P = 8.5e−5) corresponded to DEGs detected in our scRNA-seq data of mouse β-cells, which included 21 of 255 genes (odds ratio 3.1, P = 2.13e−5) from the meta-analysis by Mahajan et al. (42) (Fig. 7A). This does not only represent a significant enrichment; the majority of genes (23 of 38) were also located in B6/NZO diabetes-related QTLs (43), further illustrating their relevance for metabolic disease and validity of our analysis. Additionally, eight human diabetes genes (Sctr, Patj, Hnf1a, Plxnd1, Zzef1, Calcoco2, Poc5, Cept120 [identified with GWAS]) feature a missense variant in NZO mice but are not differentially expressed according to the scRNA-seq data set, indicating that NZO β-cells might express proteins with an altered structure or function (Fig. 7A).
Figure 7.
Comparison of human T2D GWAS genes with NZO SNPs and mouse transcriptome. A: Circos plot of mouse scRNA-seq β-cell cluster DEGs compared with human genes and loci associated with diabetes susceptibility. B: Expression and functional annotation of T2D GWAS genes in the different β-cell clusters. Dot color and size represent expression level and cells expressing the gene. Genes marked in red harbor a missense variant in NZO mice. C: Mapping of human GWAS variants into NZO variants by variant consequence, focusing only on regulatory (down/upstream, 3′-UTR) and missense variants. var, variant.
Next, we were interested in the classification and distribution of GWAS genes within the β-cell clusters. Among them were six transcriptional regulators, which we hypothesized were promising candidates explaining differences between β-cell clusters and trajectory development. Three were expressed at similarly high levels in Beta1 and Beta4 (Nfat5, Nsd1, Zbtb20), one at higher levels in Beta4 (Ccnd1) and one in BetaP (Hmgb1). Hnf1a was not differentially expressed, but has a missense variant in NZO mice (Fig. 7B), while Zbtb20 was located in a QTL for pancreatic insulin. Together, these transcriptional regulators could in part mediate the differences in OB and NZO β-cell responses to a diabetogenic stimulus.
To assess whether the human diabetes gene variants were conserved in NZO mice, we mapped the 46 diabetes-related GWAS genes to NZO single nucleotide polymorphisms (SNPs). Focusing only on regulatory (down/upstream, 3′-untranslated region [UTR]) and missense variants, we found 13 genes with missense mutations in NZO, of which 9 also have a similar alteration in humans (Fig. 7C). In addition, we found a large number of NZO variants (119 SNPs in 24 genes) in the 3′-UTR, which might affect RNA stability. Altogether, our analysis points to multiple genetic factors, partially shared between mice and humans, as a possible underlying cause for differences in β-cell fates in diabetes-susceptible and -resistant mice, mimicking early T2D in humans.
Discussion
In the current study, we used diabetes-resistant and -susceptible mouse models and evaluated their islet single-cell transcriptome to illuminate the transition from health to diabetes. We observed islet cell–specific alterations and various trajectories of β-cells in diabetes-prone or -resistant mice in response to a short diabetogenic challenge. This study supported conceptual findings from earlier transcriptome analyses, such as the discovery of diabetes suppressor genes in OB (Lefty, Apoa2, Pcp4l1) and diabetes genes in NZO (Ifi202b, Kif2a) (7,9,10), while enabling us to find new gene expression signatures at much increased resolution.
In our analysis, we mainly focused on β-cells, hypothesizing that OB mice are protected from diabetes due to an altered expression of cilia and antiapoptotic genes and a higher degree of maturation in the absence of carbohydrates. In addition, OB mice adapt to carbohydrate feeding via activation of genes promoting protein biosynthesis and folding, encoding for cell surface proteins and for cell-cell communication. At the same time, genes essential for β-cell function, like Slc2a2, Scl30a8, and Glp1r, were downregulated after feeding of a carbohydrate-containing diet. We believe that—at least via suppression of Slc2a2/GLUT2—β-cells protect themselves from glucotoxic conditions because INS-1 cells showed a lower degree of ER stress (phospho-eIF2α) and a reduced induction of apoptosis (cleaved caspase 3) after Slc2a2 knockdown. In contrast, NZO β-cells showed indications of metabolic pressure, which might participate in the later loss of β-cell function.
Interestingly, the cell cluster pattern of OB and NZO islets on the −CH diet was very similar. Despite higher blood glucose in NZO mice, both strains mainly exhibited β-cells of the Beta1 cluster. However, these cells responded differently to a rise in blood glucose on the +CH diet. While OB Beta1 cells transition to OB +CH-enriched Beta4 cells, NZO Beta1 cells progress into Beta2/3. We propose that the 131 DEGs of OB versus NZO Beta1 cells participate in their divergent development. NZO Beta1 cells showed indications of elevated protein misfolding and oxidative stress, which could expose them to reduced plasticity and prevent an appropriate adaptation to rising blood glucose levels. Conversely, a higher degree of maturity of OB Beta1 cells, reflected by increased expression of Glp1r, Ucn3, Slc2a2, and other PDX1 target genes (shown via IPA), appears to be a key determinant for their diabetes resistance. We propose that if the most differentiated β-cells are able to mount a higher initial response to metabolic changes (e.g., high blood glucose), OB Beta1 cells are better adapted to rising blood glucose levels.
According to pseudotime analysis, Beta1 cells attempt to take a path via Beta2 and Beta3 to become Beta4 cells in both mouse strains. However, the Beta4 cluster is more prominent in OB than in NZO islets, indicating that the transition to Beta4 occurs faster or to a higher degree in OB mice, reducing their diabetes susceptibility. For example, a notable upregulation of N-glycosylation–related genes in the OB-enriched Beta4 cluster might participate in their diabetes resistance, as N-glycosylation defects are linked to T2D pathogenesis (56,57). At the same time, important β-cell genes were downregulated, such as Mafa, Slc30a8, and Slc2a2. These alterations might protect OB β-cells from glucotoxicity, as shown in GLUT2 knockdown experiments where palmitate/glucose treatment showed attenuated effects on ER stress and the induction of apoptosis. This response is expected, since GLUT2 is the main glucose transporter in mouse β-cells and its reduction decreases glucose uptake and metabolism. Importantly, downregulation of GLUT2 in OB +CH mice is transient, since permanent loss contributes to diabetes (58). The downregulation of several typical β-cell genes in OB islets could also indicate that their β-cells transition to a state of reduced β-cell identity, thereby preventing apoptosis (59) and/or allowing proliferation. Recently, Whitticar and Nunemaker proposed an intervention that reduces overactive glucokinase, which restored pulsatility and functionality to β-cells in prediabetic mouse islets (60). Their observations indicate that such strategies mimic a physiologic response occurring in individuals without diabetes, which is or progressively becomes dysfunctional in patients with diabetes.
OB islets carry more cilia and express higher amounts of cilia genes than NZO, and cilia disassemble after 2 days of +CH feeding. This effect is accompanied by an induction of islet cell proliferation (10), a process coinciding with functional immaturity (61). Interestingly, 79 cilia-annotated genes were among the 131 DEGs of the Beta1 cluster in OB. This could indicate that functional cilia dynamics are essential for the capacity of Beta1 cells to develop into the OB-enriched cluster Beta4, of which a small fraction develops into proliferation cluster BetaP. In support of the ability of Beta4 cells to transition to a proliferative state, many MYC target genes related to ribosome function were upregulated. Together, this helps clarify why the proliferative capacity of NZO β-cells was markedly impaired following +CH in comparisons with OB β-cells (7), i.e., due to a lack of or delayed dedifferentiation toward Beta4 and BetaP. The reduction in mature β-cell expression signature in our study is transient and likely allows cells to reenter the cell cycle (transition to BetaP) while maintaining β-cell properties. It differs from pathologic β-cell dedifferentiation, including β- to α-cell conversion, that is observed during T2D progression (62–64).
Finally, the evaluation of cell-to-cell cross talk indicated that Beta1 and Beta4 cells in particular communicate with δ-cells, for instance, via the ephrin ligand EFNA5 (Beta4) and its receptor EPHB2 (δ-cells). Targeting of ephrin receptors by small molecules increased glucose-stimulated insulin secretion (65). Another noteworthy cell-cell signaling event could occur via binding of the ligand semaphorin 4D (SEMA4D) to the plexin receptor PLXNB1, which is important for tissue homeostasis and morphogenesis (66). Furthermore, BDNF receptors are highly expressed in OB-enriched Beta4 cells and treatment of MIN6 β-cells with BDNF increased proliferation, implicating it in the adaptation of OB mice to increased β-cell demand. Altogether, our analyses showed that elevated islet cell-cell communication contributes to diabetes resistance of OB mice.
The NZO mouse is a well-established model of polygenic diabetes (4), sharing many similarities with human T2D. OB mice (on a C57BL/6J background) with their monogenic defect in leptin are comparable with respect to hyperphagia, reduced thermogenesis, and physical inactivity (all slightly more pronounced in OB) (67) but maintain functional islet mass despite insulin resistance and do not develop T2D. Like OB mice, NZO show an element of dysregulated leptin signaling, as they develop peripheral leptin resistance and defective transport of leptin across the blood-brain barrier, suggesting that with progressive age and duration of HFD feeding, defects in leptin signaling in NZO become more comparable with OB mice (68). Altogether, that makes these two strains a good combination for identifying diabetes risk genes. A limitation of the inbred NZO strain is genetic homogeneity, which means that it only reflects a subpopulation of all possible human diabetes susceptibility genes. However, several diabetes-related genes discovered in NZO mice overlapped with human diabetes GWAS genes (9). Furthermore, we detected an enrichment of human T2D genes among our DEGs (Fig. 7), demonstrating the relevance of our mouse model for human T2D.
In conclusion, our intensive scRNA-seq–based analysis of islets from diabetes-resistant and -susceptible mice has helped to clarify the picture of the heterogeneous responses of islet cells exposed to diabetogenic conditions. In diabetes-prone mice, the failure to adapt gene expression leads to higher metabolic stress and β-cell failure. In diabetes-resistant mice, exposure to a diabetogenic diet induces transcriptional processes that might participate in β-cell protection. This includes activation of genes involved in N-glycosylation, cell adhesion, cell surface markers, and indications for elevated cell-cell communication.
Article Information
Acknowledgments. The skillful technical assistance of Anett Helms (A.S. laboratory) is gratefully acknowledged.
Funding. The study was supported by the German Ministry of Education and Research (BMBF: DZD grant 82DZD00302) and the Brandenburg State.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. P.G. performed data analysis. T.S. performed experiments and data analysis and wrote the manuscript. M.Sta. conceived and designed the experiments and participated in the drafting of the manuscript. M.Ste. performed experiments. M.B. contributed to the acquisition and analysis of data. P.M.S. performed experiments. M.J. performed data analysis. E.Z. performed data analysis. H.A. performed experiments and data analysis. S.R.B. critically edited and revised the manuscript. F.J.T. performed data analysis. H.L. performed study conception and critically edited and revised the manuscript. A.S. performed study conception and design and wrote the manuscript. All authors subsequently edited the manuscript and approved its final version for publication. A.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented orally at the 2022 Diabetes Congress of the German Diabetes Association (DDG), Berlin, Germany, 25–28 May 2022.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.20080700.
P.G. and T.S. contributed equally.
References
- 1. DeFronzo RA, Ferrannini E, Groop L, et al. Type 2 diabetes mellitus. Nat Rev Dis Primers 2015;1:15019. [DOI] [PubMed] [Google Scholar]
- 2. Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature 2016;536:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joost HG, Schürmann A. The genetic basis of obesity-associated type 2 diabetes (diabesity) in polygenic mouse models. Mamm Genome 2014;25:401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kleinert M, Clemmensen C, Hofmann SM, et al. Animal models of obesity and diabetes mellitus. Nat Rev Endocrinol 2018;14:140–162 [DOI] [PubMed] [Google Scholar]
- 5. Jürgens HS, Neschen S, Ortmann S, et al. Development of diabetes in obese, insulin-resistant mice: essential role of dietary carbohydrate in beta cell destruction. Diabetologia 2007;50:1481–1489 [DOI] [PubMed] [Google Scholar]
- 6. Gässler A, Quiclet C, Kluth O, et al. Overexpression of Gjb4 impairs cell proliferation and insulin secretion in primary islet cells. Mol Metab 2020;41:101042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kluth O, Matzke D, Kamitz A, et al. Identification of four mouse diabetes candidate genes altering β-cell proliferation. PLoS Genet 2015;11:e1005506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kluge R, Scherneck S, Schürmann A, Joost HG. Pathophysiology and genetics of obesity and diabetes in the New Zealand obese mouse: a model of the human metabolic syndrome. Methods Mol Biol 2012;933:59–73 [DOI] [PubMed] [Google Scholar]
- 9. Kluth O, Matzke D, Schulze G, Schwenk RW, Joost HG, Schürmann A. Differential transcriptome analysis of diabetes-resistant and -sensitive mouse islets reveals significant overlap with human diabetes susceptibility genes. Diabetes 2014;63:4230–4238 [DOI] [PubMed] [Google Scholar]
- 10. Kluth O, Stadion M, Gottmann P, et al. Decreased expression of cilia genes in pancreatic islets as a risk factor for type 2 diabetes in mice and humans. Cell Rep 2019;26:3027–3036.e3 [DOI] [PubMed] [Google Scholar]
- 11. Bensellam M, Laybutt DR, Jonas JC. The molecular mechanisms of pancreatic β-cell glucotoxicity: recent findings and future research directions. Mol Cell Endocrinol 2012;364:1–27 [DOI] [PubMed] [Google Scholar]
- 12. Rossetti L, Giaccari A, DeFronzo RA. Glucose toxicity. Diabetes Care 1990;13:610–630 [DOI] [PubMed] [Google Scholar]
- 13. Bensellam M, Jonas JC, Laybutt DR. Mechanisms of β-cell dedifferentiation in diabetes: recent findings and future research directions. J Endocrinol 2018;236:R109–R143 [DOI] [PubMed] [Google Scholar]
- 14. Kluth O, Mirhashemi F, Scherneck S, et al. Dissociation of lipotoxicity and glucotoxicity in a mouse model of obesity associated diabetes: role of forkhead box O1 (FOXO1) in glucose-induced beta cell failure. Diabetologia 2011;54:605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Camunas-Soler J, Dai XQ, Hang Y, et al. patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metab 2020;31:1017–1031.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krentz NAJ, Lee MYY, Xu EE, et al. Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Reports 2018;11:1551–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sachs S, Bastidas-Ponce A, Tritschler S, et al. Targeted pharmacological therapy restores β-cell function for diabetes remission. Nat Metab 2020;2:192–209 [DOI] [PubMed] [Google Scholar]
- 18. Wigger L, Barovic M, Brunner AD, et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat Metab 2021;3:1017–1031 [DOI] [PubMed] [Google Scholar]
- 19. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 2019;8:281–291.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000 Res 2016;5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hie B, Bryson B, Berger B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat Biotechnol 2019;37:685–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Büttner M, Miao Z, Wolf FA, Teichmann SA, Theis FJ. A test metric for assessing single-cell RNA-seq batch correction. Nat Methods 2019;16:43–49 [DOI] [PubMed] [Google Scholar]
- 25. Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech 2008;2008:P10008 [Google Scholar]
- 26. Baron M, Veres A, Wolock SL, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst 2016;3:346–360.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martens GA, Jiang L, Hellemans KH, et al. Clusters of conserved beta cell marker genes for assessment of beta cell phenotype. PLoS One 2011;6:e24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018;37:38–44 [DOI] [PubMed] [Google Scholar]
- 29. Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnaiz O, Cohen J, Tassin AM, Koll F. Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia 2014;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arnaiz O, Malinowska A, Klotz C, et al. Cildb: a knowledgebase for centrosomes and cilia. Database (Oxford) 2009;2009:bap022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015;33:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu WL, Zhang YW, Feng Y, Li LC, Yang L, Xu CR. Deciphering pancreatic islet β cell and α cell maturation pathways and characteristic features at the single-cell level. Cell Metab 2017;25:1194–1205.e4 [DOI] [PubMed] [Google Scholar]
- 37. Street K, Risso D, Fletcher RB, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 2018;19:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tesson BM, Breitling R, Jansen RC. DiffCoEx: a simple and sensitive method to find differentially coexpressed gene modules. BMC Bioinformatics 2010;11:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–1506 [DOI] [PubMed] [Google Scholar]
- 41. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47(D1):D1005–D1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vogel H, Kamitz A, Hallahan N, et al. A collective diabetes cross in combination with a computational framework to dissect the genetics of human obesity and Type 2 diabetes. Hum Mol Genet 2018;27:3099–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keane TM, Goodstadt L, Danecek P, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011;477:289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015;31:2745–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40:W452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2019 [Google Scholar]
- 48. Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics 2013;14:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thomson E, Ferreira-Cerca S, Hurt E. Eukaryotic ribosome biogenesis at a glance. J Cell Sci 2013;126:4815–4821 [DOI] [PubMed] [Google Scholar]
- 50. Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology 2004;39:129–138 [DOI] [PubMed] [Google Scholar]
- 51. Ediger BN, Du A, Liu J, et al. Islet-1 Is essential for pancreatic β-cell function. Diabetes 2014;63:4206–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wade AK, Liu Y, Bethea MM, Toren E, Tse HM, Hunter CS. LIM-domain transcription complexes interact with ring-finger ubiquitin ligases and thereby impact islet β-cell function. J Biol Chem 2019;294:11728–11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang H, Wang WP, Guo T, et al. The LIM-homeodomain protein ISL1 activates insulin gene promoter directly through synergy with BETA2. J Mol Biol 2009;392:566–577 [DOI] [PubMed] [Google Scholar]
- 54. Dong H, Zhang Y, Wang J, et al. Regulator of G protein signaling 2 is a key regulator of pancreatic β-cell mass and function. Cell Death Dis 2017;8:e2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stützer I, Esterházy D, Stoffel M. The pancreatic beta cell surface proteome. Diabetologia 2012;55:1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohtsubo K, Chen MZ, Olefsky JM, Marth JD. Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat Med 2011;17:1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ohtsubo K, Takamatsu S, Minowa MT, Yoshida A, Takeuchi M, Marth JD. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 2005;123:1307–1321 [DOI] [PubMed] [Google Scholar]
- 58. Guillam MT, Hümmler E, Schaerer E, et al. Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2. Nat Genet 1997;17:327–330 [DOI] [PubMed] [Google Scholar]
- 59. Moin ASM, Butler AE. Alterations in beta cell identity in type 1 and type 2 diabetes. Curr Diab Rep 2019;19:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Whitticar NB, Nunemaker CS. Reducing glucokinase activity to enhance insulin secretion: a counterintuitive theory to preserve cellular function and glucose homeostasis. Front Endocrinol (Lausanne) 2020;11:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Puri S, Roy N, Russ HA, et al. Replication confers β cell immaturity. Nat Commun 2018;9:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 2012;150:1223–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spijker HS, Song H, Ellenbroek JH, et al. Loss of β-cell identity occurs in type 2 diabetes and is associated with islet amyloid deposits. Diabetes 2015;64:2928–2938 [DOI] [PubMed] [Google Scholar]
- 64. Lee K, Chan JY, Liang C, et al. XBP1 maintains beta cell identity, represses beta-to-alpha cell transdifferentiation and protects against diabetic beta cell failure during metabolic stress in mice. Diabetologia 2022;65:984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jain R, Jain D, Liu Q, et al. Pharmacological inhibition of Eph receptors enhances glucose-stimulated insulin secretion from mouse and human pancreatic islets. Diabetologia 2013;56:1350–1355 [DOI] [PubMed] [Google Scholar]
- 66. Janssen BJ, Robinson RA, Pérez-Brangulí F, et al. Structural basis of semaphorin-plexin signalling. Nature 2010;467:1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jürgens HS, Schürmann A, Kluge R, et al. Hyperphagia, lower body temperature, and reduced running wheel activity precede development of morbid obesity in New Zealand obese mice. Physiol Genomics 2006;25:234–241 [DOI] [PubMed] [Google Scholar]
- 68. Hileman SM, Pierroz DD, Masuzaki H, et al. Characterizaton of short isoforms of the leptin receptor in rat cerebral microvessels and of brain uptake of leptin in mouse models of obesity. Endocrinology 2002;143:775–783 [DOI] [PubMed] [Google Scholar]