Abstract
Dysregulation of hepatic VLDL secretion contributes to the pathogenesis of metabolic diseases, such as nonalcoholic fatty liver disease (NAFLD) and hyperlipidemia. Accumulating evidence has suggested that long noncoding RNAs (lncRNAs) had malfunctioning roles in the pathogenesis of NAFLD. However, the function of lncRNAs in controlling hepatic VLDL secretion remains largely unillustrated. Here, we identified a novel lncRNA, lncRNA regulator of hyperlipidemia (lncRHL), which was liver-enriched, downregulated on high-fat diet feeding, and inhibited by oleic acid treatment in primary hepatocytes. With genetic manipulation in mice and primary hepatocytes, depletion of lncRHL induces hepatic VLDL secretion accompanied by decreased hepatic lipid contents. Conversely, lncRHL restoration reduces VLDL secretion with increased lipid deposition in hepatocytes. Mechanistic analyses indicate that lncRHL binds directly to heterogeneous nuclear ribonuclear protein U (hnRNPU), and thereby enhances its stability, and that hnRNPU can transcriptional activate Bmal1, leading to inhibition of VLDL secretion in hepatocytes. lncRHL deficiency accelerates the protein degradation of hnRNPU and suppresses the transcription of Bmal1, which in turn activates VLDL secretion in hepatocytes. With results taken together, we conclude that lncRHL is a novel suppressor of hepatic VLDL secretion. Activating the lncRHL/hnRNPU/BMAL1/MTTP axis represents a potential strategy for the maintenance of intrahepatic and plasma lipid homeostasis.
Introduction
As a central metabolic organ, the liver plays an important role in maintaining lipid metabolism. Under normal physiological conditions, hepatic lipid homeostasis is regulated by the interplay between the delivery of lipids to liver and hepatic uptake, synthesis, oxidation, and secretion of lipids (1). Dysregulated hepatic lipid homeostasis causes excessive accumulation of lipids in liver, which results in hepatic steatosis and contributes to the pathogenesis of nonalcoholic fatty liver disease (NAFLD). NAFLD has been associated with hyperlipidemia, obesity, insulin resistance, and hyperglycemia (2). In NAFLD, hyperlipidemia is manifested as increased serum triglyceride and LDL levels and decreased HDL levels, all of which are key risk factors for cardiovascular disease (CVD) (1). Previous studies have demonstrated that CVD is the leading causes of mortality in patient with NAFLD (3,4). The characteristic mixed hyperlipidemia phenotype observed in NAFLD patients is a consequence of increased secretion of triacylglycerol (TAG)-rich VLDL particles (5). VLDL particles are formed in the endoplasmic reticulum, where apolipoprotein (apo)B100 is lipidated in a process catalyzed by the enzyme microsomal triglyceride transfer protein (MTTP) (6–8). Consequently, APOB100 and MTTP are key components in hepatic VLDL secretion and in maintaining hepatic lipid homeostasis. As such, hepatic steatosis, secondary to compromised triglyceride export, is common in patients with genetic defects in the ApoB or Mttp gene (9,10).
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides, which do not have protein-coding potential. lncRNAs are emerging regulators that are involved in gene expression and diverse physiological and pathological processes. Accumulating studies have revealed that lncRNAs participate in lipid metabolism by influencing the expression of key genes, networks, and pathways that are involved in cholesterol and triglyceride biosynthesis, cholesterol transport, lipid uptake, and efflux (11–16). For example, lncLSTR, a liver-specific triglyceride regulator, maintains systemic lipid homeostasis via a TDP-43/FXR/apoC2-dependent pathway in liver (13). Another lncRNA, lncRNA suppressor of hepatic gluconeogenesis and lipogenesis (lncSHGL), which is reduced in human liver with steatosis, has been shown to suppress hepatic gluconeogenesis and lipogenesis by recruiting heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) to enhance calmodulin mRNAs translation in an insulin- and calcium-independent manner (17). However, the roles of lncRNAs involved in hepatic VLDL secretion during NAFLD progression remain elusive. Therefore, to further characterize new lncRNAs that regulate lipid metabolism is of great importance. Particularly, identifying new lncRNAs that regulate hepatic VLDL secretion will shed light on the pathogenesis of NAFLD.
In this study, we identified a novel lncRNA, lncRNA regulator of hyperlipidemia (lncRHL), as an important regulator participating in hepatic VLDL secretion both in vivo and in vitro, and revealed that lncRHL interacts with hnRNPU to form a ribonucleoprotein complex. Deficiency of lncRHL leads to an accelerate protein degradation of hnRNPU without affecting its mRNA level, which results in a suppressed expression of Bmal1 and its downstream genes associated with hepatic VLDL secretion. Our findings illustrate a novel protein-lncRNA regulatory network in hepatic lipid metabolism, revealing a new molecular mechanism underlying the intrahepatic and plasma lipid homeostasis, thereby providing an opportunity for further understanding of the pathogenesis of NAFLD and dyslipidemia.
Research Design and Methods
Experimental Animals
C57BL/6J mice used in this study were purchased from Beijing Vital River Laboratory Animal Technology (Beijing, China). The mice were housed in a pathogen-free barrier facility of the Nanjing Medical University in 12-h light/dark cycles at 25°C with free access to food and water. All animal protocols were approved by the institutional animal care and use committee of Nanjing Medical University.
High-Throughput RNA Sequencing
Eight-week-old wild-type male C57BL/6J mice (n = 6) were maintained on a normal diet (ND) or high-fat diet (HFD) (60% kcal from fat) (D12492; Research Diets, New Brunswick, NJ). After 23 weeks of HFD administration, mice were sacrificed under 4-h fasting condition and livers were collected for high-throughput RNA sequencing. We constructed sequencing libraries from the fragmented cDNA using NEBNext Ultra DNA Library Prep Kit according to the manufacturer’s instructions (E7370; New England Biolabs, Ipswich, MA). Paired-end 150-bp sequencing was performed on HiSeq X Ten by Annoroad Gene Technology Company (China). We used a cutoff of normalized array values (fold change >2 or <0.5) to enrich for transcripts present in ND and HFD. RNA-sequencing (RNA-seq) data of this study have been deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (GSE157482).
5′ and 3′ Rapid Amplification of cDNA Ends
For identification of the full-length sequence of lncRHL, 5′ and 3′ rapid amplification of cDNA ends (RACE) experiments were performed with the GeneRacer Kit (Invitrogen) according to the manufacturer’s protocol. The gene-specific primers used for RACE analysis are listed in Supplementary Table 4.
Cell Culture and Transfection
Hepa1-6 cells were purchased from ATCC and maintained in high-glucose DMEM (Life Technologies) containing 10% FBS (Gibco) and 1% penicillin-streptomycin. Mouse primary hepatocytes were isolated from 6- to 8-week-old C57BL/6J mice as previously described (18). Primary hepatocytes (2 × 105 cells) were cultured in low-glucose DMEM supplemented with 10% FBS (Gibco) at 37°C and 5% CO2. For knockdown experiments, primary hepatocytes at ∼70% confluence were transfected with negative control Smart Silencer (ssNC) or lncRHL Smart Silencer (sslncRHL) (100 nmol/L) (Ri-boBio, Guangzhou, China) by Lipofectamine 2000 (Smart Silencer: Lipofectamine 2000 = 1: 0.5) after 4 h isolation. sslncRHL was a mixture of three siRNAs and three antisense oligo nucleotides. The target sequences of siRNAs are listed in Supplementary Table 4. Cells and supernatant were collected at 48 h posttransfection.
Adenovirus Infection
Adenoviruses, of either scrambled (Ad-scramle) or shRNA for mouse lncRHL (Ad-shlncRHL), were constructed with use of the Ad-Track Adenoviral Vector System (StrataGene) and purified as previously described (19). Adenoviruses of shRNA sequence specifically targeted for lncRHL and MTTP (Ad-shMTTP) are listed in Supplementary Table 4. C57BL/6J mice 6–8 weeks old were injected with 2.5 × 1011 recombinant adenoviral particles of Ad-shlncRHL, Ad-shMTTP, or Ad-Scramble through tail veins. After injection, mice were fed an ND or HFD for 7 days and the mice were sacrificed after a 16-h fast. Primary mouse hepatocytes were isolated from C57BL/6J mice and treated with adenovirus (6 × 109 particles/well) for 6 h. Cells and supernatant were then collected at 48 h postinfection.
Quantitative RT-PCR and Western Blot
RNA isolation, cDNA synthesis, and quantitative PCR were performed as previously described (19). The sequences of the specific primers used in this study are listed in Supplementary Table 4. Western blot analysis was performed as previously described (19). The antibodies used in this study are listed in Supplementary Table 5. Protein signals were detected by incubation with horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch), followed by enhanced chemiluminescent detection reagent (Pierce).
RNA Pull-down and Mass Spectrometry Assays
RNA pull-down and mass spectrometry assays were performed via a method previously described (20). Briefly, we prepared lncRHL sense or antisense RNA using in vitro transcription of appropriately linearized plasmid templates (cat. no. AM1344; Invitrogen). Then biotin-labeled RNA were synthesized and used for RNA pull-down assay according to the manufacturer’s instructions (cat. no. 20164, Magnetic RNA-Protein Pull-Down Kit; Thermo Fisher Scientific). RNA-associated proteins were eluted, followed by SDS-PAGE and silver staining, and then subjected to liquid chromatography–mass spectrometry analysis.
RNA Immunoprecipitation Assay
For RNA immunoprecipitation (RIP) experiments, nuclear lysates were isolated from primary hepatocytes and treated with 300 units/mL RNase inhibitor, followed by incubation with mouse anti-hnRNPU antibody or mouse IgG for 3 h at 4°C with continuous rotation. Antibody/protein/RNA complexes were immunoprecipitated with protein A/G beads (B23202; Bimake.com). RNA associated with hnRNPU were extracted with Trizol and analyzed with quantitative RT-PCR (RT-qPCR) as previously described (21).
Nuclear and Cytosolic RNA Extraction
Fresh liver tissue (∼100 mg) was homogenized with use of a glass Dounce homogenizer on ice with 1 mL extraction buffer containing 20 mmol/L HEPES, 1 mmol/L EDTA, 250 mmol/L sucrose, 10 mg/mL leupeptin, 10 mg/mL aprotinin, 2 mmol/L dithiothreitol, 100 μmol/L NaVO4, 100 μmol/L phenylmethylsulfonyl fluoride (PMSF), and complete protease inhibitor cocktail (11697498001; Roche). Separation of nuclear and cytosolic RNA fractions was performed as published previously (22).
Lipid Extraction and Analysis
Liver lipids were extracted from 100–200 mg frozen liver samples with the method of Folch et al. (23). TAG and total cholesterol (TC) levels were measured with a TAG assay kit and TC assay kit (Wako, Richmond, VA) and normalized to sample weight as previously described (24). Plasma TAG, free fatty acid, TC, LDL, and HDL levels were measured with an enzymatic kit (Wako). Intracellular TAG content in cultured primary hepatocytes was measured with a TAG enzymatic assay kit (Applygen Technologies, Beijing, China) and normalized to total protein amount.
VLDL Secretion in Mice and Primary Hepatocytes
For in vivo VLDL secretion in mice, 6- to 8-week-old C57BL/6J male mice were infected with 2.5 × 1011 recombinant adenovirus particles of Ad-Scramble or Ad-shlncRHL through tail vein injection for 7 days. After 8 h fasting, mouse were injected with the lipase inhibitor tyloxapol (Triton WR-1339, 500 mg/kg i.v.; Sigma-Aldrich) and 20 mL blood was collected from the retro-orbital plexus in heparinized tubes at the indicate time point. The collected blood samples were used for TAG measurements, and the VLDL-TAG production rate was calculated from the slope of the plasma TAG versus time curve as previously described (24). For in vitro VLDL secretion experiments, after 4-h attachment, primary mouse hepatocytes isolated from 6- to 8-week-old C57BL/6J mice were transfected with ssNC or sslncRHL for 6 h. After that, those primary hepatocytes were incubated for 6 h with DMEM containing 0.4 mmol/L oleic acid/0.5% fatty acid–free BSA. Cells were then washed with PBS and incubated for a further 16 h with serum-free and phenol red–free DMEM; then the cells and medium were collected and assayed for TAG with an enzymatic TAG assay kit (Wako) and mouse VLDL ELISA kit (Jiancheng Bioengineering Research Institute, Jiangsu, China), respectively, as previously described (19).
Protein Stability and In Vivo Ubiquitination Assay
Protein stability of hnRNPU in primary hepatocytes or Hepa1-6 cells was performed with a cycloheximide (CHX)-based protein chase experiment as previously described (25). For in vivo ubiquitination assay of hnRNPU, primary hepatocytes were transfected with sslncRHL or infected with recombinant adenovirus particles expressing full-length lncRHL (Ad-lncRHL) for 12 h and then transfected with HA-ubiquitin (2 mg). After 18 h, cells were treated with 10 μmol/L MG132 for 4 h, harvested, and lysed in immunoprecipitation (IP) buffer. Total cell lysate (1 mg) was used for IP with 50 µL protein A/G beads and anti-hnRNPU antibody. The immunoprecipitates or total cell lysates were analyzed by Western blot with indicated antibodies.
RNA-Fluorescence In Situ Hybridization
RNA-fluorescence in situ hybridization (RNA-FISH) for lncRHL was performed on primary mouse hepatocytes. Cell slides fixed with 4% paraformaldehyde were treated with several buffers in RNA-FISH Kit (Ri-boBio, Guangzhou, China). After denaturing at 37°C for 5 min, probe mixture was hybridized in a cell slide overnight in darkness at 37 °C. CY3-labeled nucleic acid probes were targeting lncRHL. After washing, the signal was observed under confocal microscope. Detailed procedures were followed according to the manufacturer’s protocol.
Cell Apoptosis Assay
After transfection with sslncRHL or ssNC for 48 h, primary hepatocytes were collected for the detection of cell apoptosis. The cell apoptosis of primary hepatocytes was measured with use of Annexin V-FITC/PI Apoptosis Detection Kit (MedChemExpress, Shanghai, China), and the apoptotic rate was evaluated with FACS. TUNEL analysis was performed with TUNEL Apoptosis Detection Kit (MedChemExpress) according to the manufacturer protocol (26).
Statistical Analysis
Statistical analysis was determined with two-tailed unpaired Student t test for single comparisons and ANOVA followed by Bonferroni-Dunn for multiple comparisons. All statistical analyses were performed with GraphPad Prism 7.0 software (GraphPad Software, San Diego, CA). Data are expressed as mean ± SEM. Differences were considered significant when P was <0.05 (*P < 0.05; **P < 0.01; ***P < 0.001).
Data and Resource Availability
RNA-seq data were deposited in NCBI’s GEO with accession no. GSE157482. All other data are available from the corresponding author on reasonable request.
Results
Identification of a Liver-Enriched lncRNA Implicated in Lipid Metabolism
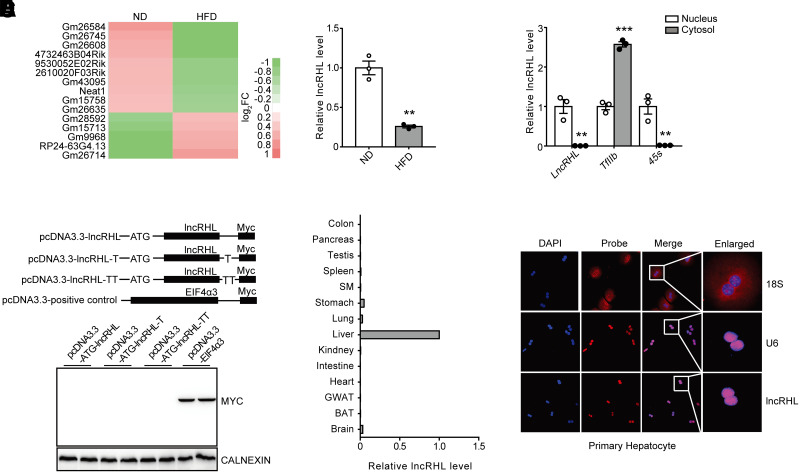
For comprehensive investigation of the lncRNAs expression profile in the liver of HFD and ND mice, specifically those involved in hepatic lipid metabolism, the Illumina HiSeq X Ten platform was used to obtain a comprehensive view of lncRNAs in liver. HFD for 23 weeks resulted in significantly higher liver fat accumulation compared with that of mice fed normal chow. HFD-fed mice showed a significant increase of hepatic TAG and TC levels both in serum (Supplementary Fig. 1A) and liver (Supplementary Fig. 1B) compared with ND. An average of 84.1 million high-quality paired-end sequencing reads was obtained for each sample (range 74.5–93.7 million). Thus, an average of 93.6% of the total reads (range 91.29–95.91%) could be mapped to the mouse genome. As a result, a total of 886 genes (614 annotated lncRNAs and 272 unknown lncRNAs) with sufficiently high expression levels were identified (summarized reads information shown in Supplementary Table 1). Specifically, we found that there were 77 lncRNAs with induction folds of >2 or <0.5 in response to ND (Supplementary Table 2). We next performed a cluster analysis by setting another three criteria to further screen these lncRNAs: annotation as authentic lncRNAs in Ensembl, relatively high abundance in the mouse liver, and relatively large fold change. Fifteen uncharacterized lncRNAs were then identified as dysregulated candidates in the livers from HFD mice (Fig. 1A). For confirmation of these results, an independent RT-qPCR assay was performed. Consistent with our RNA-seq results, the hepatic expression of Gm26584 was significantly reduced (Fig. 1B) while Gm26714 and Gm9968 were induced by HFD (Supplementary Fig. 1C).
Figure 1.
Identification and characterization of lncRHL in the liver of mice. A: The cluster heat map showing differentially expressed hepatic lncRNAs from RNA-seq data in mice liver after ND or HFD for 23 weeks. B: RT-qPCR analysis of relative lncRHL expression in liver of C57BL/6J mice fed with ND or HFD for 23 weeks (n = 3/group). C: Characterization of noncoding property of lncRHL. The full-length lncRHL was cloned into the eukaryotic expression vector pcDNA3.3-Myc with an N-terminal codon ATG in the three expression patterns. These plasmids were then transfected into human embryonic kidney (HEK)293A cells separately. After 48 h posttransfection, Western blot analysis was performed to detect the expression of Myc-tagged protein, EIF4α3, with Myc tag severed as a positive control (n = 2/group). D: Relative expression of lncRHL in mice tissues (three samples pooled together per tissue). Total RNA was subjected to RT-qPCR quantification and level of lncRHL in tissues was expressed relative to liver. E: Relative expression of lncRHL, TFIIb, and 45S in the isolated cytosol and nucleus fraction of primary hepatocytes. Total RNA was subjected to RT-qPCR quantification, and levels of lncRHL, TFIIb, and 45S in cytosol were expressed relative to nucleus. TFIIb and 45S were used as markers of the cytosol and nucleus, respectively (n = 3/group). F: RNA-FISH analysis in mouse primary hepatocytes. lncRHL subcellular localization in mouse primary hepatocytes was assessed with laser confocal microscopy. Red, lncRHL or small nuclear (sn)RNA U6 or 18S (snRNA U6 and 18S were selected as a positive control for nucleus and cytosol marker, respectively); blue, DAPI staining. Magnification: ×400. All values are presented as mean ± SEM. BAT, brown adipose tissue; FC, fold change; GWAT, gonadal white adipose tissue. SM, skeletal muscle. *P < 0.05; **P < 0.01; ***P < 0.001.
We named Gm26584 as lncRHL (lncRNA regulator of hyperlipidemia) based on its function characterized in this report. RACE assay revealed a full length of 3,923 nt for lncRHL (Supplementary Fig. 2A), slightly longer than that annotated in Ensembl (Supplementary Fig. 2B). Coding Potential Calculator (CPC) (http://cpc.gao-lab.org/) (27) analysis indicated a lack of coding potential for this transcript (Supplementary Fig. 2C). Furthermore, the lncRHL expression vector failed to produce any protein when transfected into 293T cells (Fig. 1C). RT-qPCR analysis indicated that lncRHL was specifically enriched in the mouse liver (Fig. 1D) and mainly distributed in the nucleus fraction of liver (Fig. 1E). The result of RNA-FISH further confirmed that lncRHL was localized in the nucleus of primary hepatocytes and Hepa1-6 cells (Fig. 1F and Supplementary Fig. 2D). We further validated that lncRHL was downregulated in primary hepatocytes with oleic acid treatment (Supplementary Fig. 2E).
These data indicate that lncRHL is a liver-enriched and metabolic stimuli–regulated lncRNA, suggesting that it may serve as a potential regulator of hepatic lipid metabolism.
Liver-Specific Knockdown of lncRHL Induces Hyperlipidemia in Mice
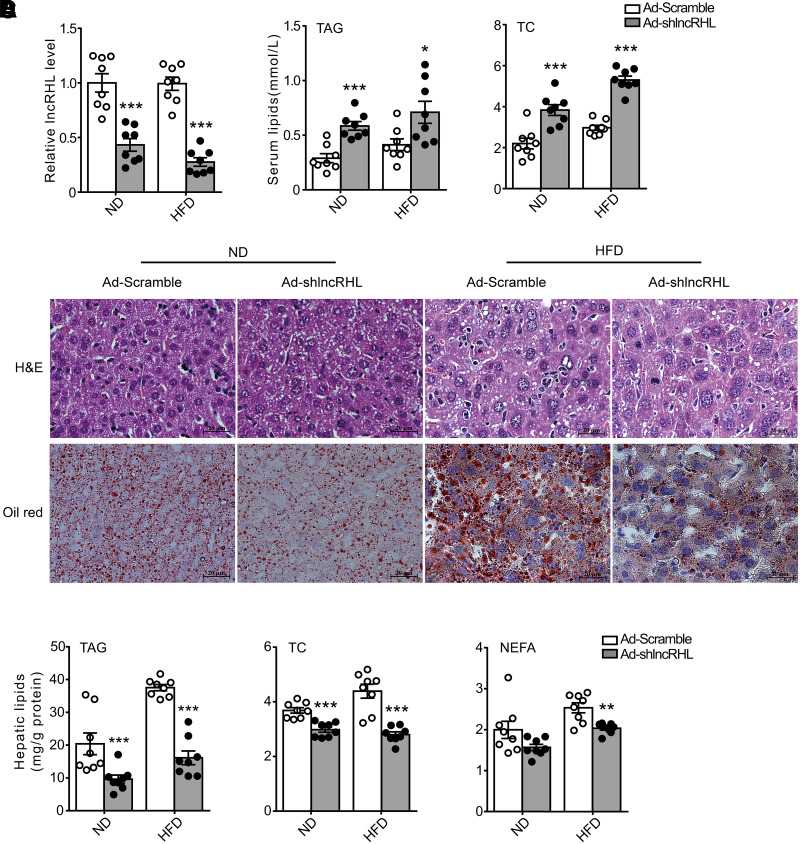
To directly address the physiological role of lncRHL in hepatic lipid metabolism, we specifically knocked down lncRHL in C57BL/6J mice liver using adenoviral shRNA. The RT-qPCR results indicated that tail vein injection of Ad-lncRHL shRNA effectively silenced the expression level of lncRHL by >50% in mouse liver compared with Ad-Scramble in both ND and HFD conditions (Fig. 2A). lncRHL knockdown in vivo did not markedly affect body weight but increased liver weight correlated with increased gonadal fat (Table 1). Interestingly, the knockdown of lncRHL significantly increased serum level of TAG, TC (Fig. 2B), nonesterified fatty acid (NEFA), and LDL cholesterol but decreases HDL cholesterol as compared with that of control mice under both ND and HFD conditions (Supplementary Fig. 3). In contrast, Oil Red O and hematoxylin-eosin staining revealed a dramatic reduction in lipid droplets accumulation in the liver of lncRHL knockdown mice (Fig. 2C). These results were further confirmed by reduced hepatic TAG, TC, and NEFA levels in lncRHL knockdown mice (Fig. 2D). These data suggest that liver-specific knockdown of lncRHL induces hyperlipidemia accompanied by lower hepatic lipid contents in vivo.
Figure 2.
Hepatic knockdown of lncRHL induces hyperlipidemia in mice. C57BL/6 J male mice 6–8 weeks old were infected with 2.5 × 1011 recombinant adenovirus particles of Ad-Scramble or Ad-shlncRHL via tail vein injection and fed with ND or HFD for 7 days. Mice were sacrificed and the tissues and plasma were collected for analysis after 16 h fasting (n = 8/group). A: Knockdown efficiency of lncRHL in the liver of C57BL/6J mice. The relative expression levels of lncRHL in the liver of mice infected with Ad-shlncRHL were verified with RT-qPCR analysis and are expressed relative to those of control mice infected with Ad-Scramble. B: Knockdown of lncRHL increases serum TAG (left) and TC (right) levels in mice. Total TAG and cholesterol levels in serum of control (Ad-Scramble) or lncRHL knockdown (Ad-shlncRHL) mice were measured with enzymatic kits (n = 8/group). C: Representative images of hematoxylin-eosin (H&E) and Oil Red O staining of liver sections from control (Ad-Scramble) or lncRHL knockdown (Ad-shlncRHL) mice (n = 3/group), scale bar = 20 μm. D: lncRHL knockdown decreases hepatic TAG, TC, and NEFA levels in mice. Total hepatic TAG, cholesterol, and free fatty acids were extracted from the livers of control (Ad-Scramble) and lncRHL knockdown (Ad-shlncRHL) mice and measured using enzymatic kits (n = 8/group). All values are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Table 1.
Metabolic parameters of mice with or without knockdown of lncRHL fed an ND or HFD for 7 days
| ND | HFD | |||||
|---|---|---|---|---|---|---|
| Ad-Scramble (n = 8) | Ad-shlncRHL (n = 8) | P * | Ad-Scramble (n = 8) | Ad-shlncRHL (n = 8) | P * | |
| Body weight (g) | 18.5 ± 0.71 | 20.24 ± 0.69 | 0.1011 | 21.17 ± 0.94 | 21.47 ± 1.11 | 0.84 |
| Glucose (mmol/L) | 4.44 ± 0.74 | 4.844 ± 0.52 | 0.6571 | 4.14 ± 0.65 | 3.4 ± 0.26 | 0.31 |
| Liver (g) | 1.03 ± 0.08 | 1.417 ± 0.09 | 0.0052** | 1.116 ± 0.035 | 1.49 ± 0.07 | 0.0004*** |
| Liver/body weight (%) | 5.58 ± 0.36 | 7.04 ± 0.44 | 0.0218* | 5.30 ± 0.13 | 7.05 ± 0.47 | 0.0029** |
| gWAT (g) | 0.097 ± 0.024 | 0.182 ± 0.026 | 0.0324* | 0.23 ± 0.02 | 0.27 ± 0.03 | 0.31 |
| gWAT/body weight (%) | 0.502 ± 0.12 | 0.873 ± 0.11 | 0.0356* | 1.05 ± 0.08 | 1.24 ± 0.12 | 0.21 |
Data are means ± SEM. C57BL/6J mice 6-8 weeks old were infected with 2.5 × 1011 recombinant adenoviral particles of Ad-shlncRHL or Ad-Scramble through tail vein injection. After that, mice were fed an ND or HFD for 7 days and the mice were sacrificed after a 16-h fast. gWAT, gonadal white adipose tissue. P values are the results of t test.
P < 0.05;
P < 0.01;
P < 0.001.
Knockdown of lncRHL Enhances Hepatic VLDL Secretion
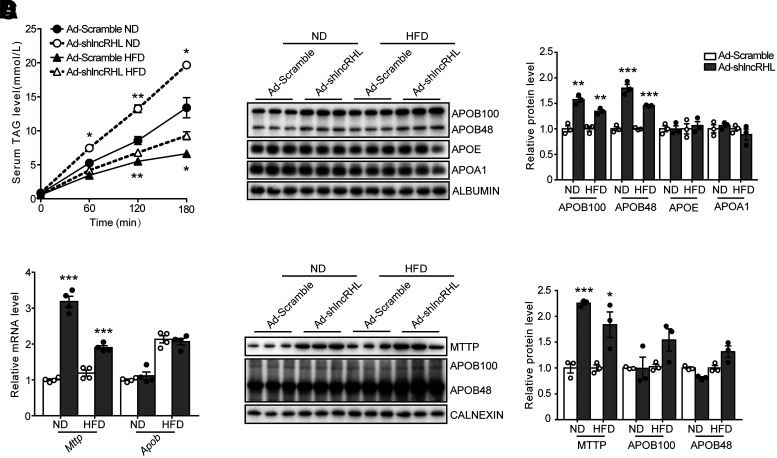
It is therefore interesting to investigate the cause of hyperlipidemia in lncRHL knockdown mice. The RT-qPCR results indicated that liver-specific knockdown of lncRHL did not affect expression level of genes involved in fatty acid synthesis, β-oxidation, or TAG synthesis or hydrolysis in liver of these mice (Supplementary Fig. 4A). The protein level of proprotein convertase subtilisin/kexin type 9 (PCSK9) and lipoprotein lipase (LPL), key regulators of plasma lipid metabolism, remained unchanged in plasma, suggesting that lncRHL knockdown did not influence lipid clearance (Supplementary Fig. 4B). Of note, in lncRHL knockdown mice, the tyloxapol-induced TAG increment was ∼30% and 25% higher than those of control mice under ND and HFD condition, respectively (Fig. 3A). Consistently, plasma APOB100 and APOB48 levels were significantly increased in lncRHL knockdown mice, but APOE, APOA-I, and ALBUMIN were not influenced (Fig. 3B). Unlike plasma APOB level, hepatic expression of APOB did not show any change in the liver of lncRHL knockdown mice, whereas the mRNA and protein level of Mttp were induced approximately two- to threefold in the liver of lncRHL knockdown mice, respectively (Fig. 3C and D). These results revealed that the manifestation of hyperlipidemia observed in mice with liver-specific knockdown of lncRHL was due to enhanced hepatic TAG-VLDL secretion.
Figure 3.
Knockdown of lncRHL promotes hepatic VLDL secretion in vivo. A: VLDL-TAG secretion increases in lncRHL knockdown C57BL/6J mice. After infected with Ad-Scramble or Ad-shlncRHL for 7 days, mice were fasted for 8 h and injected with 500 mg/kg body wt tyloxapol (Triton WR-1339) via tail veins. Blood samples were collected from the retro-orbital plexus at the indicated time point posttreatment. Serum TAG levels was measured with enzymatic kit (n = 3/group). B: lncRHL knockdown results in the induction of serum apoB proteins. Representative Western blot (left) and quantitative analysis (right) of apoB-100/48, apoE, apoA1, and albumin protein levels in the serum of control (Ad-Scramble) and lncRHL knockdown (Ad-shlncRHL) mice (n = 3/group). C: Knockdown of lncRHL increases mRNA level of Mttp in mice liver. RT-qPCR analysis of relative mRNA levels of Mttp and ApoB in the livers of control (Ad-Scramble) and lncRHL knockdown (Ad-shlncRHL) mice (n = 4/group). D: Knockdown of lncRHL increases hepatic protein level of MTTP in mice. Representative Western blot (left) and quantitative analysis (right) of MTTP and apoB100/48 protein levels in the livers of control (Ad-Scramble) and lncRHL knockdown (Ad-shlncRHL) mice (n = 3/group). All values are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
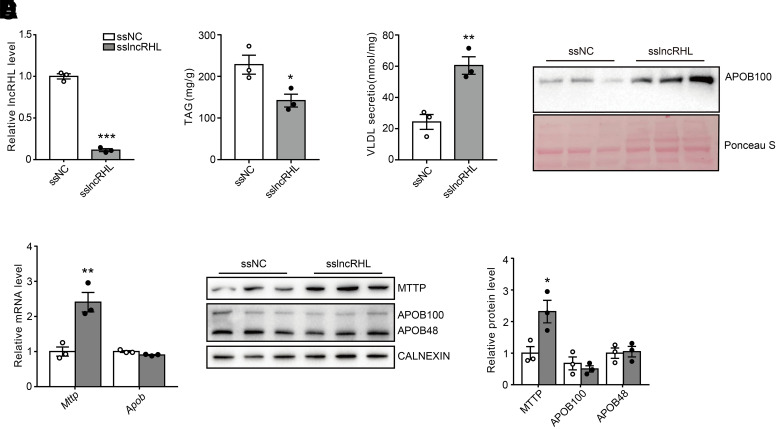
For confirmation that the increased VLDL-TAG in the lncRHL knockdown mice was cell autonomous and not dependent on factors extrinsic to the liver such as free fatty acids, we knocked down lncRHL with sslncRHL in primary hepatocytes to measure intracellular and secreted TAG (Fig. 4A). As shown in Fig. 4B, the intracellular levels of TAG were decreased by ∼37% in hepatocytes following lncRHL knockdown, whereas the secreted VLDL-TAG in medium was ∼1.48-fold higher than that of control cells with treatment of 0.4 mmol/L oleic acid (60.42 ± 9.73 vs. 24.29 ± 8.28 nmol/mg protein, respectively) (Fig. 4C). Accordingly, knockdown lncRHL increases the content of APOB100 in the culture medium of primary hepatocytes (Fig. 4D). In addition, the knockdown of lncRHL increased mRNA and protein levels of MTTP in primary hepatocytes, although apoB expression was unchanged (Fig. 4E and F). Consistent with the observations in mice liver, the mRNA levels of genes involved in lipogenesis, lipolysis, and fatty acid β-oxidation such as Srebp-1c, Acc1, Fasn, and Atgl remained unchanged in lncRHL-knockdown hepatocytes. These data suggest that the inhibition of lncRHL resulted directly in increased VLDL-TAG secretion from hepatocytes rather than TAG synthesis or hydrolysis (Supplementary Fig. 5A). Furthermore, the results of Annexin V-FITC/PI Apoptosis Detection Kit analysis and TUNEL staining showed that the percentage of apoptotic cells in primary hepatocytes was ∼11% after 48-h culture in vitro. There was no difference in cell mortality between lncRHL knockdown primary hepatocytes and control cells (Supplementary Fig. 5B and C).
Figure 4.
Knockdown of lncRHL increases VLDL secretion in vitro. A: Knockdown efficiency of lncRHL in primary hepatocytes (n = 3/group). The relative expression of lncRHL in primary hepatocytes transfected with sslncRHL was verified with RT-qPCR analysis and expressed relative to that of control hepatocytes transfected with ssNC. B: Decreased intracellular TAG levels in primary hepatocytes with knockdown of lncRHL (n = 3/group). The intracellular TAG levels were measured with TAG assay kit and normalized to total protein. Values represent mean ± SEM of triplicates. C: Increased VLDL secretion in primary hepatocytes with knockdown of lncRHL (n = 3/group). The secreted VLDL in the medium was measured with ELISA kit and normalized to total protein. Values represent mean ± SEM of triplicates. D: Increased protein levels of apoB in culture medium of primary hepatocytes with knockdown of lncRHL (n = 3/group). Representative Western blot of apoB protein levels in culture medium of primary hepatocytes transfected with ssNC or sslncRHL. Lower panel show Ponceau S staining as loading control. E: Knockdown of lncRHL increases mRNA level of Mttp in primary hepatocytes. RT-qPCR analysis of relative mRNA levels of Mttp and ApoB in the primary hepatocytes transfected with ssNC or sslncRHL (n = 3/group). F: Knockdown of lncRHL increases protein level of MTTP in primary hepatocytes. Representative Western blot (left) and quantitative analysis (right) of MTTP and apoB100/48 protein levels in the primary hepatocytes transfected with ssNC or sslncRHL (n = 3/group). Error bars are ±SEM. These experiments were repeated three times with similar results. *P < 0.05; **P < 0.01; ***P < 0.001.
To determine the role of MTTP in lncRHL-regulated VLDL secretion, we further knockdown lncRHL in primary hepatocytes isolated from mice injected with Ad-shMTTP. We found that knockdown lncRHL–induced increases in the mRNA level of Mttp were significantly abolished by depletion of MTTP in primary hepatocytes, and no changes in the mRNA level of Apob (Supplementary Fig. 6A). Importantly, the decreased intracellular TAG levels (Supplementary Fig. 6B) and increased VLDL secretion (Supplementary Fig. 6C) as well as apoB content in culture medium (Supplementary Fig. 6D) and protein level of MTTP (Supplementary Fig. 6E) were also restored by depletion of MTTP in lncRHL knockdown primary hepatocytes. These results indicate that the depletion of Mttp significantly attenuated the effects of lncRHL knockdown on VLDL secretion and hepatic lipid accumulation in primary hepatocytes.
lncRHL Interacts With hnRNPU and Modulates Its Stability
lncRNAs generally coordinate with protein partners to exert their specific function. To uncover the molecular mechanism of lncRHL function, we sought to identify protein partners that directly bind to lncRHL using an RNA pull-down assay by incubating full-length lncRHL or its antisense (negative control) RNA in lysates of primary hepatocytes. The proteins retrieved by the RNA pull-down in lncRHL sense line were subjected to SDS-PAGE and visualized with sliver staining followed by mass spectrometry analysis (Supplementary Fig. 7A). A total of 151 candidates were identified, including 16 nucleus proteins and 47 cytosolic proteins (Supplementary Table 3). The results of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that the interacting proteins of lncRHL were mainly involved in metabolic pathways (Supplementary Fig. 7B).
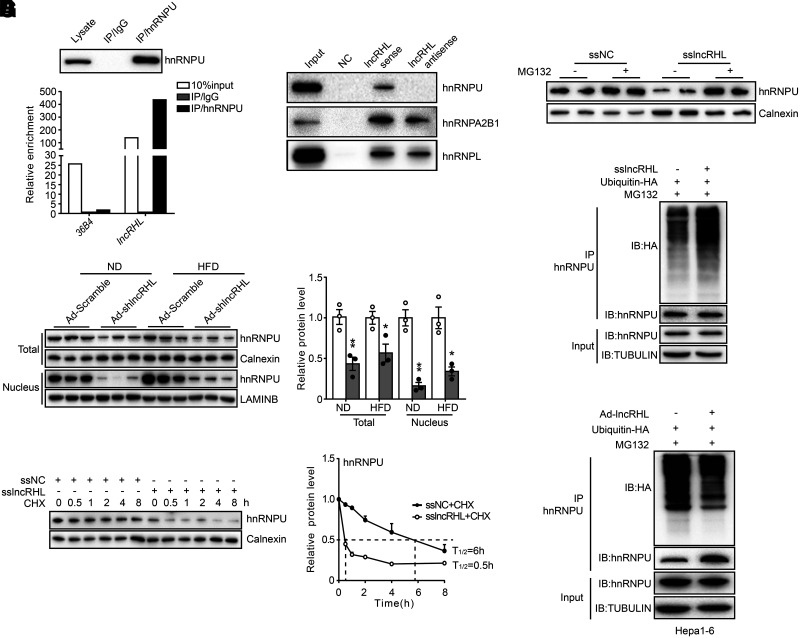
Among them, we noticed that the heterogenous nuclear ribonucleoprotein (hnRNP) family members were enriched (Supplementary Fig. 7C). Interestingly, we found multiple putative hnRNPU binding motifs within lncRHL with RBPmap (28) using default parameters with high stringency levels (Supplementary Fig. 7D), suggesting a possible interaction between hnRNPU and lncRHL. RIP assay was subsequently performed with nuclear extracts from primary hepatocytes using hnRNPU antibodies, and the results demonstrated that lncRHL was strongly associated with the hnRNPU (Fig. 5A). In parallel with RIP experiments, RNA pull-down assay further confirmed lncRHL specifically interacted with hnRNPU but not hnRNPA2B1 or hnRNPL (Fig. 5B). These findings suggested that lncRHL and hnRNPU form a ribonucleoprotein complex.
Figure 5.
lncRHL interacts with hnRNPU and modulates its stability. A: Association between endogenous lncRHL and hnRNPU in primary hepatocytes. RIP enrichment was assessed as RNA associated with hnRNPU relative to IgG control by Western blot (upper) or RT-qPCR (lower). 36B4 served as negative control of RIP. B: lncRHL and hnRNPU specifically interact in vitro. Western blot for niotin-lncRHL RNA pull-down shows specific interaction between lncRHL and hnRNPU but not for hnRNPA2B1 or hnRNPL in primary hepatocytes. Biotin-lncRHL RNA and biotin-lncRHL antisense RNA (negative control) were synthesized by in vitro transcription. Lysate used as a positive loading control. C: Knockdown of lncRHL decreases hnRNPU protein level in mice liver. Representative Western blot (left) and quantitative (right) analysis of hnRNPU protein levels in total lysate and nucleus fraction of the livers from control (Ad-Scramble) and lncRHL knockdown (Ad-shlncRHL) mice (n = 3/group). D: Knockdown of lncRHL decreases hnRNPU protein stability in primary hepatocytes. Western blot (left) analysis of hnRNPU protein levels in primary hepatocytes transfected with ssNC or sslncRHL and treated with CHX (100 ng/mL) for the indicated time, respectively. hnRNPU protein half-life (T1/2) (lower) was analyzed with ImageJ software. Values represent mean ± SEM of triplicate samples. E: MG132 treatment reverses reduction of hnRNPU caused by knockdown of lncRHL in vitro. Western blot analysis of endogenous hnRNPU protein level in the presence or absence of MG132 after transfection with ssNC or sslncRHL in primary hepatocytes. F: Knockdown of lncRHL induces hnRNPU ubiquitination in primary hepatocytes. Endogenous hnRNPU was immunoprecipitated with anti-hnRNPU antibody in primary hepatocytes cotransfected with HA-Ub and ssNC or sslncRHL, respectively. The immunoprecipitates were subjected to Western blot, and hnRNPU ubiquitination was detected with anti-HA antibody. G: Overexpression of lncRHL reduces hnRNPU ubiquitination in Hepa1-6 cells. Endogenous hnRNPU was immunoprecipitated with anti-hnRNPU antibody in Hepa1-6 cells transfected with HA-Ub after overexpression of lncRHL. The immunoprecipitates were subjected to Western blot, and hnRNPU ubiquitination was detected with anti-HA antibody. All values are presented as mean ± SEM. These experiments were repeated three times with similar results. IB, immunoblot. *P < 0.05; **P < 0.01; ***P < 0.001.
Interestingly, liver-specific knockdown of lncRHL reduced hnRNPU protein levels, especially in nucleus fractions (Fig. 5C), without affecting its mRNA level (Supplementary Fig. 8A). More importantly, the half-life of endogenous hnRNPU protein was markedly shortened in lncRHL knockdown hepatocytes treated with CHX (Fig. 5D). Concomitantly, the reduced protein level of hnRNPU was restored by MG132, a proteasome inhibitor (Fig. 5E). These results indicated that lncRHL may regulate hnRNPU protein level at the posttranslational level.
Furthermore, the ubiquitination and co-IP assays revealed that MG132 treatment and lncRHL knockdown led to a robust increase in endogenous hnRNPU ubiquitination in primary hepatocytes (Fig. 5F). Conversely, hnRNPU ubiquitination level was markedly reduced in lncRHL-overexpressed hepatocytes (Fig. 5G).
Taken together, these results support that lncRHL physically interacts with hnRNPU and regulates its protein stability by influencing the proteasome-mediated ubiquitination of hnRNPU.
lncRHL Regulates the Transcriptional Activity of hnRNPU/BMAL1 Axis
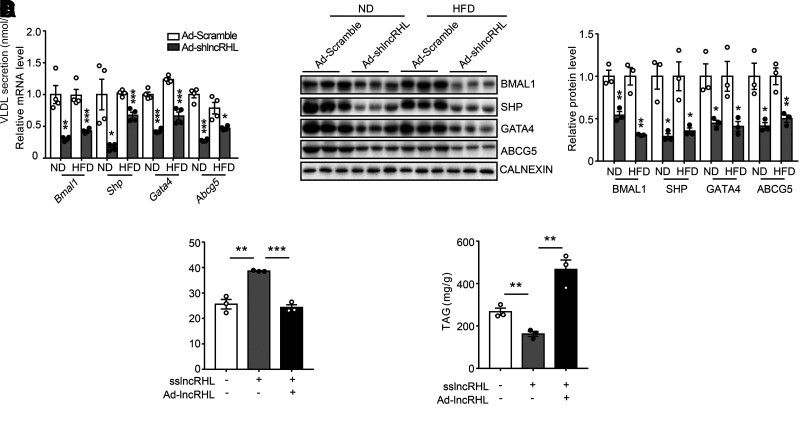
We next sought to explore the downstream targets of lncRHL/hnRNPU ribonucleoprotein complex in lipid metabolism. Recent hnRNPU chromatin IP with massively parallel DNA sequencing (ChIP-seq) identified 5,285 peaks in AML12 hepatocytes. Of these peaks, at least four were located within Bmal1 gene body (29). In addition, using the electrophoretic mobility shift assay also confirmed the binding of 3′-flanking region of Bmal1 gene promoter by SAF-A/hnRNPU protein complex (30). Together, the results of these experiments supported the transcriptional regulation of hnRNPU on Bmal1. Global and hepatocytes-specific ablation of BMAL1 induces hyperlipidemia and enhances atherosclerosis through upregulation of MTTP and downregulation of SHP (31). It is therefore interesting to confirm the role of hnRNPU in the regulation of Bmal1 in hepatocytes. As expected, knockdown of hnRNPU significantly decreased mRNA and protein levels of BMAL1 in primary hepatocytes (Supplementary Fig. 8B and 8C). In agreement, we observed significantly reduced BMAL1 expression accompanied by decreased protein levels of hnRNPU in the liver of lncRHL knockdown mice (Fig. 6A and B). Furthermore, lncRHL deficiency reduced liver mRNA and protein levels of ABCG5, SHP, and GATA4, correlating with the changes in BMAL1 levels (Fig. 6A and B). These results indicated that lncRHL knockdown in liver exerts its functional influence at least partially by repressing the transcriptional activity of hnRNPU/BMAL1 axis.
Figure 6.
Knockdown of lncRHL interrupts transcriptional activity of hnRNPU/BAML1 axis. A: Knockdown of lncRHL reduces mRNA levels of downstream targets of hnRNPU in mice liver. RT-qPCR analysis of mRNA levels of Bmal1, Shp, Gata4, and Abcg5 in the liver of mice injected with Ad-Scramble or Ad-shlncRHL (n = 4/group). The same liver samples as used in Figure 2. B: Knockdown of lncRHL reduces protein levels of downstream targets of hnRNPU in mice liver. Representative Western blot (left) and quantitative (right) analysis of BMAL1, SHP, GATA4, and ABCG5 protein level in the livers from control (Ad-Scramble) and lncRHL knockdown (Ad-shlncRHL) mice (n = 3/group). C: Overexpression of lncRHL inhibits VLDL secretion in primary hepatocytes with knockdown of lncRHL. The secreted VLDL in the medium was measured with ELISA kit and normalized to total protein (n = 3/group). D: Overexpression of lncRHL increases intracellular TAG level in primary hepatocytes with knockdown of lncRHL. The intracellular TAG level was measured with TAG assay kit and normalized to total protein (n = 3/group). All values are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. These experiments were repeated three times with similar results.
In vitro studies further confirmed that manipulation of lncRHL did not affect hnRNPU mRNA levels (Supplementary Fig. 9A), whereas the protein levels of hnRNPU were significantly decreased on lncRHL knockdown and were restored in response to lncRHL overexpression in primary hepatocytes (Supplementary Fig. 9B). Accordingly, the mRNA levels of Bmal1, Shp, and Gata4 were significantly reduced, and Mttp was increased in lncRHL knockdown hepatocytes. Moreover, lncRHL overexpression reversed their levels (Supplementary Fig. 9A and 9B). In agreement with this, silencing of lncRHL in primary hepatocytes resulted in a significant induction of VLDL secretion (Fig. 6C), associated with reduced intracellular TAG content (Fig. 6D). Importantly, the secreted VLDL and intracellular TAG levels in these lncRHL knockdown primary hepatocytes were recovered with exogenous expression of lncRHL following Ad-lncRHL infection (Fig. 6C and D).
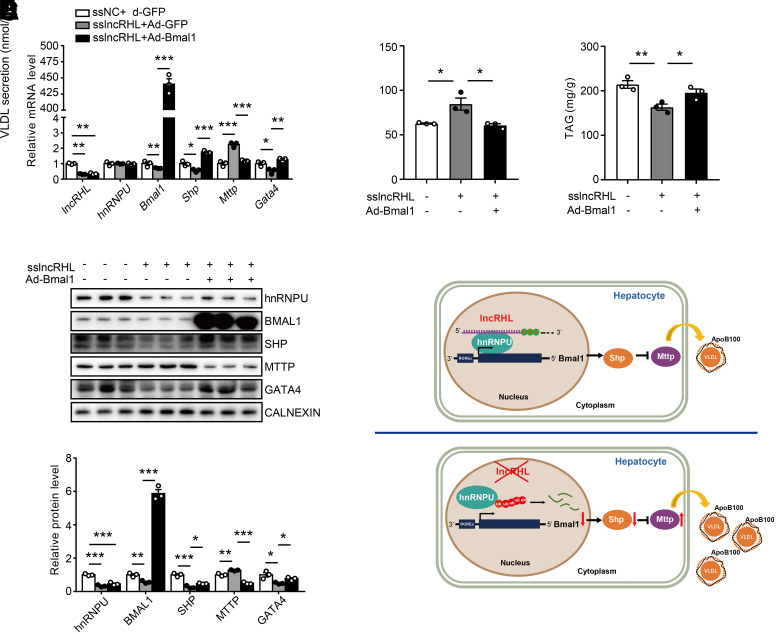
We overexpressed Bma1 to further determine the effect of Bmal1 on the enhanced VLDL secretion in lncRHL knockdown primary hepatocytes. Again, mRNA and protein levels of Bmal1, Shp, and Gata4 were decreased and Mttp were increased in lncRHL knockdown hepatocytes, whereas restoring Bmal1 reverted Shp, Gata4, and Mttp mRNA and protein expression (Fig. 7A and B). Importantly, the increased VLDL secretion and decreased intracellular TAG levels were also restored by overexpression of BMAL1 in lncRHL knockdown primary hepatocytes (Fig. 7C and D). Collectively, these results demonstrate that the deficiency of lncRHL promotes VLDL secretion by attenuating the transcriptional activity of hnRNPU/BMAL1/MTTP axis.
Figure 7.
Overexpression of BMAL1 rescues VLDL secretion in primary hepatocytes with knockdown of lncRHL. A: Overexpression of Bmal1 restores mRNA levels of Shp, Mttp, and Gata4 in primary hepatocytes with knockdown of lncRHL. After transfected with ssNC or sslncRHL for 12 h, the primary hepatocytes were infected with adenoviruses expressing Bmal1 (Ad-Bmal1) or GFP (Ad-GFP). The mRNA levels of lncRHL, hnRNPU, Bmal1, Shp, Mttp, and Gata4 were assessed with RT-qPCR (n = 3/group). B: Overexpression of BMAL1 restores protein levels of SHP, MTTP, and GATA4 in primary hepatocytes with knockdown of lncRHL. Western blot (upper) and quantitative analysis (lower) of hnRNPU, BMAL1, SHP, MTTP, and GATA4 protein levels. C: Overexpression of BMAL1 inhibits VLDL secretion in primary hepatocytes with knockdown of lncRHL. The secreted VLDL in the medium was measured with ELISA kit and normalized to total protein (n = 3/group). D: Overexpression of BMAL1 increases intracellular TAG level in primary hepatocytes with knockdown of lncRHL. The intracellular TAG level was measured with TAG assay kit and normalized to total protein (n = 3/group). E: A working model of lncRHL regulates hepatic VLDL secretion via hnRNPU/BMAL1/MTTP axis in mouse liver. All values are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. These experiments were repeated three times with similar results.
Discussion
The liver is an essential metabolic organ, and metabolic function of liver is controlled by nutritional and hormonal signaling (32). NAFLD is the most common liver disease due to overnutrition and obesity. The presence of hyperlipidemia was reported in 20%–80% of cases of NAFLD (33). The increased serum TAG, cholesterol, and LDL are known risk factors for CVD. Therefore, identifying new regulatory factors participating in hepatic VLDL secretion is crucial for understanding the pathogenesis of NAFLD and dyslipidemia.
Recent studies show an emerging role of lncRNAs in hepatic lipid metabolism (34). lncRNAs are expressed in a more tissue-specific manner than protein-coding genes (35). In this study, lncRHL was identified as a novel lncRNA specifically enriched in liver and downregulated in HFD-fed mice as well as in oleic acid–treated primary hepatocytes. Gain- and loss-of-function studies revealed that lncRHL suppressed hepatic VLDL secretion without affecting lipogenesis and fatty acid oxidation. These findings strongly suggest that expression of lncRHL is regulated by nutritional signaling to meet the requirements for metabolic homeostasis and participates in the pathogenesis of NAFLD and dyslipidemia.
Compared with mRNAs, a great proportion of lncRNAs are localized in the nucleus. Depending on nucleus localization, lncRNAs often function by partnering with RNA binding proteins (RBPs) to form ribonucleoprotein complexes, which is critical for lncRNAs to exert their function as gene regulators (36,37). In this study we found that lncRHL was mainly distributed in nucleus and directly interacted with hnRNPU, an RNA binding protein participating in mRNA stabilization and transcriptional or translational regulation of downstream targets (38) to form ribonucleoprotein complex. Interestingly, depletion of lncRHL decreased the hnRNPU protein levels without affecting its transcription. Through in vivo and in vitro studies, lncRHL-regulated hnRNPU protein levels most likely resulted from posttranslational regulation because knockdown of lncRHL promoted proteasome-mediated ubiquitination and degradation of hnRNPU, whereas overexpression of lncRHL stabilized the protein levels of hnRNPU. Therefore, the interaction between lncRHL and hnRNPU is essential and responsible for protein stability and nuclear retention of hnRNPU, just as lncRNA DINO regulates p53 stability (39).
Recently, increasing evidence has indicated a tight connection between circadian rhythms and energy metabolism. Synchronizing of a physiological circadian rhythm is crucial for metabolic health and an important strategy for treatment of obesity and NAFLD (40–42). BMAL1 was initially identified as an essential component of the molecular circadian clock in mammals (43). Beside the role in circadian regulation, an influence of BAML1 on lipid metabolism has been reported (31,44,45). Depletion of Bmal1 induces hyperlipidemia by enhancing VLDL assembly and secretion with upregulation of Mttp and downregulation of Shp. Meanwhile, ablation of Bmal1 inhibits cholesterol secretion to bile acid with downregulation of Abcg5/8 and Gata4 (31). In agreement, we found that knockdown of lncRHL significantly decreased mRNA and protein levels of BMAL1, accompanied by induced VLDL secretion in vivo and in vitro. Consistent with previous observation, downregulation of Baml1 resulted in reduced Shp expression and increased Mttp expression. The expression patterns of Gata4, Abcg5, and Abcg8, downstream targets of BMAL1, were also regulated accordingly. Importantly, restoring Bmal1 reversed the induced VLDL secretion and expression of downstream targets in lncRHL knockdown primary hepatocytes. Thus, our findings suggest that BMAL1 is the downstream target of lncRHL in the regulation of hepatic VLDL secretion.
As a member of the hnRNPU subfamily, hnRNPU mainly localized in the nucleus, where it is able to bind pre-mRNA through RGG box in the glycine-rich COOH terminus (46). Previous studies showed transcriptional regulation of hnRNPU on Bmal1 (29,30). Indeed, knockdown of hnRNPU significantly impaired Bmal1 expression in primary hepatocytes. The critical role of lncRHL in mediating the transcriptional function of hnRNPU/BMAL1/MTTP axis was illustrated by shRNA knockdown studies. We found that shRNA-mediated depletion of lncRHL markedly reduced hnRNPU protein level and BMAL1 expression, which in turn impaired the transcriptional regulation effects of BMAL1 on gene expression of Mttp, Shp, Gata4, Abcg5, and Abcg8 in hepatocytes. In contrast, restoring lncRHL recovers the protein level of hnRNPU and reverted the expression level of BMAL1 as well as downstream targets of BMAL1. These studies raise the possibility that lncRHL may engage the BMAL1 expression through its association with hnRNPU. Recently, hnRNPU has been shown to facilitate the assembly of brown fat lncRNA 1 (Blnc1) with transcription factors EBF2 and ZBTB7B to form ribonucleoprotein transcriptional complexes to stimulate the thermogenic gene expression in adipose tissue (47–49). This indicates that lncRHL recruiting hnRNPU to regulate the transcription efficiency of other mRNAs besides Bmal1 is also possible. Further identification of the targets of the lncRHL/hnRNPU axis is of great interest. Further work will be needed to clarify whether lncRHL may also regulate the hepatic lipid metabolism via other mechanisms beyond hnRNPU/BMAL1/MTTP pathway.
Taken together, the results of our study provide the first evidence that lncRHL is involved in the regulation of hepatic VLDL-TAG secretion by modulating hnRNPU/BMAL1/MTTP axis. Specifically, the expression levels of lncRHL were strongly downregulated on HFD treatment, indicating that lncRHL plays a major role in the development of HFD-induced hyperlipidemia. The reduction of lncRHL leads to enhanced protein degradation of hnRNPU, which further reduces BMAL1 expression and affects the transcription of its downstream target genes, thus increasing the VLDL secretion and development of hyperlipidemia (Fig. 7E). VLDL secretion is a hepatic-specific defense against the excessive liver TAG accumulation that occurs in nutritional overload or metabolism. Decreased levels of lncRHL in liver may therefore represent a potential risk factor for CVD. These results highlight a novel molecular mechanism of hepatic lipid secretion regulated by lncRNA, thereby providing an opportunity for further understanding of the pathogenesis of the development of NAFLD and dyslipidemia.
Article Information
Acknowledgments. The authors are grateful to Dr. Chang Liu (China Pharmaceutical University) for kindly providing adenoviruses Ad-Bmal1and Ad-GFP, Dr. Zheng Chen (Harbin Institute of Technology) for kindly providing adenoviruses Ad-shlncRHL and Ad-Scramble, Dr. Tong-Jin Zhao (Fudan University) for critical reading for the manuscript, and members of the J.Z.L. laboratory for technical assistance and helpful discussion.
Funding. This work was supported by grants from the National Key R&D Program of China (grant 2018YFA0506904 to J.Z.L.), the National Natural Science Foundation of China (grants 32130050, 31771307, 91957115, and 81471079 to J.Z.L. and 81770859 to Q.W.), and the Jiangsu Provincial Innovation Team Program Foundation.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. X.S. and Y.Z. designed the experiments. X.S., Y.Z., X.J., B.L., Y.W., Y.H., X.Z., J.Y., R.Z., and D.Q. performed the experiments. X.S., Y.Z., Q.W., and J.Z.L. analyzed data. H.Z., Q.W., and J.Z.L. contributed to discussion and wrote the manuscript. All authors approved the final manuscript. Q.W. and J.Z.L. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.20126396.
X.S. and Y.Z. contributed equally.
References
- 1. Meex RCR, Watt MJ. Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance. Nat Rev Endocrinol 2017;13:509–520 [DOI] [PubMed] [Google Scholar]
- 2. Katsiki N, Mikhailidis DP, Mantzoros CS. Non-alcoholic fatty liver disease and dyslipidemia: an update. Metabolism 2016;65:1109–1123 [DOI] [PubMed] [Google Scholar]
- 3. Ong JP, Pitts A, Younossi ZM. Increased overall mortality and liver-related mortality in non-alcoholic fatty liver disease. J Hepatol 2008;49:608–612 [DOI] [PubMed] [Google Scholar]
- 4. Söderberg C, Stål P, Askling J, et al. Decreased survival of subjects with elevated liver function tests during a 28-year follow-up. Hepatology 2010;51:595–602 [DOI] [PubMed] [Google Scholar]
- 5. Stols-Gonçalves D, Hovingh GK, Nieuwdorp M, Holleboom AG. NAFLD and atherosclerosis: two sides of the same dysmetabolic coin? Trends Endocrinol Metab 2019;30:891–902 [DOI] [PubMed] [Google Scholar]
- 6. Olofsson SO, Stillemark-Billton P, Asp L. Intracellular assembly of VLDL: two major steps in separate cell compartments. Trends Cardiovasc Med 2000;10:338–345 [DOI] [PubMed] [Google Scholar]
- 7. Gusarova V, Brodsky JL, Fisher EA. Apolipoprotein B100 exit from the endoplasmic reticulum (ER) is COPII-dependent, and its lipidation to very low density lipoprotein occurs post-ER. J Biol Chem 2003;278:48051–48058 [DOI] [PubMed] [Google Scholar]
- 8. Olofsson SOJB, Borèn J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med 2005;258:395–410 [DOI] [PubMed] [Google Scholar]
- 9. Segrest JP, Jones MK, Dashti N. N-terminal domain of apolipoprotein B has structural homology to lipovitellin and microsomal triglyceride transfer protein: a “lipid pocket” model for self-assembly of apob-containing lipoprotein particles. J Lipid Res 1999;40:1401–1416 [PubMed] [Google Scholar]
- 10. Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res 2003;44:22–32 [DOI] [PubMed] [Google Scholar]
- 11. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Halley P, Kadakkuzha BM, Faghihi MA, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep 2014;6:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li P, Ruan X, Yang L, et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 2015;21:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qin W, Li X, Xie L, et al. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res 2016;44:6423–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47–62 [DOI] [PubMed] [Google Scholar]
- 16. Muret K, Désert C, Lagoutte L, et al. Long noncoding RNAs in lipid metabolism: literature review and conservation analysis across species. BMC Genomics 2019;20:882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J, Yang W, Chen Z, et al. Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes 2018;67:581–593 [DOI] [PubMed] [Google Scholar]
- 18. Klaunig JE, Goldblatt PJ, Hinton DE, Lipsky MM, Chacko J, Trump BF. Mouse liver cell culture. I. Hepatocyte isolation. In Vitro 1981;17:913–925 [DOI] [PubMed] [Google Scholar]
- 19. Wang X, Guo M, Wang Q, et al. The patatin-like phospholipase domain containing protein 7 facilitates VLDL secretion by modulating ApoE stability. Hepatology 2020;72:1569–1585 [DOI] [PubMed] [Google Scholar]
- 20. Zhang E, He X, Zhang C, et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol 2018;19:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alvarez-Dominguez JR, Bai Z, Xu D, et al. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab 2015;21:764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Firmin FF, Oger F, Gheeraert C, et al. The RBM14/CoAA-interacting, long intergenic non-coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARγ. Sci Rep 2017;7:14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497–509 [PubMed] [Google Scholar]
- 24. Li JZ, Huang Y, Karaman R, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest 2012;122:4130–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chan SC, Lin SC, Li P. Regulation of Cidea protein stability by the ubiquitin-mediated proteasomal degradation pathway. Biochem J 2007;408:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu YL, He Y, Shi JJ, Zheng TX, Lin XJ, Lin X. Microcystin-LR promotes necroptosis in primary mouse hepatocytes by overproducing reactive oxygen species. Toxicol Appl Pharmacol 2019;377:114626. [DOI] [PubMed] [Google Scholar]
- 27. Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 2007;35:W345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paz I, Kosti I, Ares M Jr, Cline M, Mandel-Gutfreund Y. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res 2014;42:W361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan H, Lv P, Huo X, et al. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res 2018;28:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Onishi Y, Hanai S, Ohno T, Hara Y, Ishida N. Rhythmic SAF-A binding underlies circadian transcription of the Bmal1 gene. Mol Cell Biol 2008;28:3477–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan X, Bradfield CA, Hussain MM. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat Commun 2016;7:13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rui L. Energy metabolism in the liver. Compr Physiol 2014;4:177–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Souza MR, Diniz MdeF, Medeiros-Filho JE, Araújo MS. Metabolic syndrome and risk factors for non-alcoholic fatty liver disease. Arq Gastroenterol 2012;49:89–96 [DOI] [PubMed] [Google Scholar]
- 34. van Solingen C. Scacalossi KR, Moore KJ. Long noncoding RNAs in lipid metabolism. Curr Opin Lipidol 2018;29:224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7–21 [DOI] [PubMed] [Google Scholar]
- 36. He RZ, Luo DX, Mo YY. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis 2019;6:6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet 2016;135:851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmitt AM, Garcia JT, Hung T, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet 2016;48:1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weger BD, Gobet C, David FPA, et al. Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc Natl Acad Sci U S A 2021;118:e2015803118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shi D, Chen J, Wang J, et al. Circadian clock genes in the metabolism of non-alcoholic fatty liver disease. Front Physiol 2019;10:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ray S, Valekunja UK, Stangherlin A, et al. Circadian rhythms in the absence of the clock gene Bmal1. Science 2020;367:800–806 [DOI] [PubMed] [Google Scholar]
- 43. Bunger MK, Wilsbacher LD, Moran SM, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 2000;103:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anea CB, Zhang M, Stepp DW, et al. Vascular disease in mice with a dysfunctional circadian clock. Circulation 2009;119:1510–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paschos GK, Ibrahim S, Song WL, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med 2012;18:1768–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 2010;430:379–392 [DOI] [PubMed] [Google Scholar]
- 47. Zhao XY, Li S, DelProposto JL, et al. The long noncoding RNA Blnc1 orchestrates homeostatic adipose tissue remodeling to preserve metabolic health. Mol Metab 2018;14:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li S, Mi L, Yu L, et al. Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc Natl Acad Sci U S A 2017;114:E7111–E7120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mi L, Zhao XY, Li S, Yang G, Lin JD. Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol Metab 2016;6:101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]