Abstract
Background
Recent reports suggested that circulating exosomal microRNAs (exomiRs) may serve as non-invasive prediction biomarkers in gastrointestinal (GI) cancers, yet their clinicopathological and prognostic values need to be more clarified. Hence, the present meta-analysis was aimed to quantitatively assess the evidence regarding the association between circulating exomiRs and prognosis in GI cancer patients.
Methods
A comprehensive search was carried out in prominent literature databases, including PubMed, ISI Web of Science, Scopus, and Embase. Odds ratios (ORs) or hazard ratios (HRs) with 95% confidence intervals (CIs) were gathered to evaluate the strength of the association. The quality assessment was investigated through the Newcastle-Ottawa Scale (NOS) and publication bias via Eggers’ test and funnel plots.
Results
A total of 47 studies, comprising of 4881 patients, were considered eligible for this meta-analysis. Both up-regulated and down-regulated circulating exomiRs are significantly associated with differentiation (HR = 1.353, P = 0.015; HR = 1.504, P = 0.016), TNM stage (HR = 2.058, P < 0.001; HR = 2.745, P < 0.001), lymph node metastasis (HR = 1.527, P = 0.004; HR = 2.009, P = 0.002), distant metastasis (HR = 2.006, P < 0.001; HR = 2.799, P = 0.002), worse overall survival (OS) (HR = 2.053, P < 0.001; HR = 1.789, P = 0.001) and poorer disease/relapse/progression-free survival (DFS/RFS/PFS) (HR = 2.086, P < 0.001; HR = 1.607, P = 0.001) in GI cancer patients, respectively. In addition, subgroup analyses based on seven subcategories indicated the robustness of the association. The majority of findings were lack of publication bias except for the association between up-regulated exomiRs and OS or DFS/RFS/PFS and for the down-regulated exomiRs and TNM stage.
Conclusion
This study supports that up- and down-regulated circulating exomiRs are associated with poorer survival outcomes and could be served as potential prognostic biomarkers in GI cancers. Given the limitations of the current findings, such as significant heterogeneity, more investigations are needed to fully clarify the exomiRs prognostic role.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-023-02851-8.
Keywords: Circulating exosomal microRNAs (exomiRs), Gastrointestinal cancers, Prognostic value, Clinicopathological characteristics, Meta-analysis
Introduction
Gastrointestinal (GI) cancers, which include esophageal, gastric, pancreas, colorectal, rectal, and hepatocellular carcinomas, are the leading cause of cancer-related deaths, with incidence rates varying among industrialized and developing countries [1–4]. As reported in 2022, the number of new GI cancer cases and deaths were estimated at 343,040 and 171,920 in the USA, respectively [3]. Although the mortality rate of GI cancers has decreased over the last decade owing to multidisciplinary treatment approaches, the global burden of the disease remains considerable, with a notable unfavorable prognosis. The primary factors causing inferior survival outcomes for these individuals are late-stage diagnosis, inadequate prognostic biomarkers, metastasis and recurrence progression, and therapeutic resistance [5, 6].
The extent burden of GI cancers resulted in the planning of innovative molecular–omics landscapes [5–7]. Historically, tissue biopsy, as the standard method, still provides insight into cancer diagnosis and prognosis. However, this approach is invasive, costly, and associated with some complications, including a partial snapshot of the whole tumor, inaccessibility of tumor tissue in terms of anatomic location, biopsy sampling errors, and inter-observer variability in some GI tissues [8, 9]. Therefore, an accurate non-invasive detection technique is urgently warranted to completely elucidate the characteristics of the tumor, allow for early detection of cancer, and precisely evaluate the efficacy of treatment approaches. Compared to traditional tissue biopsy, blood-based or liquid biopsy as the minimally invasive tools provide close monitoring to identify cancer-associated changes and predict prognosis and acquired resistance or disease recurrence before the appearance of clinical symptoms [10–15].
Exosomes have received much attention in recent years as circulating biomarkers for cancer. Exosomes are cell-secreted nano-sized membrane (30 to 100 nm) vesicles involved in intercellular communication and various pathological features of cancers, including invasion, angiogenesis, immune response modulation, and inflammation [16, 17]. Extracellular vesicles (EVs), which may be recovered from a variety of physiological fluids, often reflect the genetic makeup of the cancer cells that originally made them up [19]. Importantly, microRNAs (miRs) in EVs or exomiRs obtained from blood samples demonstrate the specificity of tumors, indicating that exomiRs may be potential indicators for the diagnosis and prognosis of many malignancies as well as in the age of tailored anticancer therapy [17, 18, 20]. MiRs are endogenous non-coding RNAs subtype with 18–22 nucleotides which mainly modulate cellular gene expression, mostly at the post-transcriptional level. They are involved in many physiological cellular processes, including differentiation, proliferation, and apoptosis [18–21]. Recently, many investigations have reported that aberrant expression of miRs is closely associated with the progression of cancers [22, 23], suggesting miRs as putative biomarkers for diagnosis and prognosis in various tumors, such as colorectal cancer, non-small cell lung carcinoma (NSCLC), and glioma [24–26]. Non-exosomal and exosomal miRNAs (exomiRs) in body fluids are considered as stable "tumor-specific" circulating biomarkers in early diagnosis, prognosis, and screening of various cancer types, including colorectal, esophagus, and hepatocellular cancers [10, 11, 27, 28].
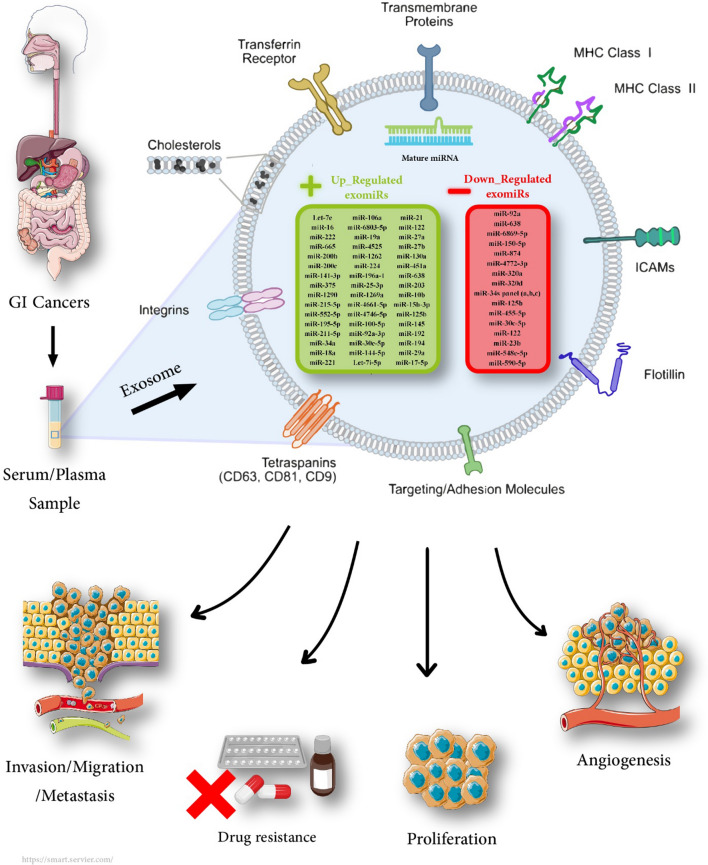
This comprehensive systematic review and meta-analysis was designed to verify the prognostic significance of circulating exomiRs in patients with GI cancers and propose their potential as a non-invasive prognostic tool for monitoring mortality, focusing on clinicopathological outcomes. Deregulated circulating exosomal microRNA(s) in gastrointestinal cancers are shown in Fig. 1.
Fig. 1.
Deregulated circulating exosomal microRNA(s) in gastrointestinal cancers
Material and method
Protocol and registration
This systematic review and meta-analysis has been registered in the PROSPERO International prospective register of systematic reviews (http://www.crd.york.ac.uk/PROSPERO), with the registration number: PROSPERO CRD42017057129; available at https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=57129. In addition, the protocol of the current review has been published in the Systematic Reviews Journal [29].
Eligibility criteria
The research question has been developed using PFO; “P” as Population, “F” as Prognostic Factors (or models of interest), and “O” as Outcome [30]. The following criteria were incorporated into this systematic review based on the PFO components:
i) Observational studies (case-control, cross-sectional, and cohort studies) assessed the association between circulating exomiRs and GI cancers.
ii) Studies published in English with available full texts.
iii) Studies with human GI cancers, including upper and lower GI and hepatopancreatic biliary.
iv) Studies assessing the association between the circulating exomiRs expression and the prognostic values consisting of overall survival (OS), disease/relapse/progression-free survival (DFS/RFS/PFS), and/or clinicopathological characteristics of GI cancers.
v) Hazard ratios (HRs) and 95% confidence intervals (CIs) provided in the article, or availability of data to calculate HRs with 95% CIs.
The investigations meeting the following criteria were excluded:
i) Reviews, meta-analysis, commentaries, case reports, case series studies.
ii) In vitro and in vivo studies.
iii) Studies not related to the topic of the interest (e.g., when the studies evaluated the other solid cancers).
iv) Studies with insufficient and useless data or with unavailable full text.
v) Studies in which cases received anti-cancer treatment (i.e., chemotherapy and/or radiation therapy) before the biopsy.
Information sources
The literature was searched in electronic databases, comprising of Web of Science, PubMed/MEDLINE, Scopus, and Embase until 31st July 2017 and updated on 7th September 2022. Besides, the references of included papers were assessed. Hand searching was performed in key journals that rely on search in Scopus.
Search strategy
This study was designed based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (PRISMA 2020) [31]. The following main keywords were used to carry out the search strategy in the mentioned databases: (Neoplasms OR Cancer OR Carcinoma OR Tumor) AND (“Gastrointestinal Tracts” OR “GI Tract” OR “Digestive Tract”) AND (“extracellular vesicles” OR microvesicle OR “Shedding Microvesicles” OR exosomes). The search syntax was adopted in other databases. The search strategy has been fully presented in Additional file 3: Table S1.
Selection process
Studies were selected in three phases. Phase 1: duplicated studies were deleted using both EndNote software (version X9.3.3, Thomson Reuters, Philadelphia, USA) and hand searching. Phase 2: two authors (E.Gh. and F.T.) independently screened all records by title and abstract. Phase 3: the same authors independently assessed the full text of each potentially eligible study. Any disagreement was resolved through consensus and then checked by a third author (M.R.).
Data collection process
The data of each eligible study’ was extracted by two authors (E.Gh. and F.T.) independently. The obtained data were entered into a “Data Extraction Form” created by Microsoft Excel for quality assessment and data synthesis. A consensus method was applied between the two reviewers to finalize the validity of all collected data and was then checked by a third author (M.R.).
Data items
Data extracted from all eligible papers have consisted of the following items: author’s name, publication year, country, type of exomiRs, type of cancer, expression of exomiRs, detection method, exosome extraction method, sample size, sample type, age, gender, number of patients with up- and down-regulation of circulating exomiRs, clinicopathological parameters (gender, TNM stage of disease, tumor differentiation, lymph node metastasis, distant metastasis), survival data (HR with corresponding 95% CI for OS and DFS/RFS/PFS), cut-off value, and median or mean follow-up times.
Study quality assessment
All studies reporting the prognostic values of exomiRs were included in the meta-analysis and assessed according to Newcastle–Ottawa Scale (NOS) tool by two independent investigators (E.Gh. and F.T.). NOS comprises three sections: selection, comparability, and exposure or outcome, with a score ranging from 0 to 9 [32]. This scoring includes four stars for the selection section, two stars for comparability, and three stars for exposure or outcomes. The result of the quality assessment is divided into three categories good, fair, and poor. Moreover, discrepancies between the two authors were resolved by consensus and were then checked by a third author (M.R.).
Effect measures and synthesis methods
Comprehensive Meta-Analysis (CMA) software version 2.2.064 was applied to perform all statistical analyses. The HRs and corresponding 95% CIs were recorded for all survival data, including OS and DFS/RFS/PFS. HRs were extracted from both multivariate and univariate statistical tests by preferring information from multivariate statistics if available. For studies without providing HR, we calculated HRs by the Kaplan–Meier curves using the method presented by Parmar et al. [33]. In this regard, survival data were extracted from Kaplan-Meyer curves by the software GetData Graph Digitizer (http://getdata-graph-digitizer.com/). Additionally, the combined odds ratios (ORs) and 95% CIs were applied to evaluate the associations between exomiRs expression and clinicopathological features, including gender (male vs. female), TNM stage (III/IV vs. I/II), tumor differentiation (poor vs. moderate/well), lymph node metastasis (positive vs. negative) and distant metastasis (positive vs. negative). A pooled HR/OR larger than one reflected a worse clinical prognostic outcome in GI cancer patients. Heterogeneity among studies was assessed through Cochran’s Q statistic and the I2 index. While an I2 of over 50% and/or P < 0.05 indicated a large degree of heterogeneity and a random effect model was used, the fixed effect model was utilized in the absence of heterogeneity (I2 ≤ 50% or P > 0.05). Afterward, subgroup analyses were employed for prognostic outcomes to recognize possible causes of heterogeneity and evaluate the prognostic importance of various subgroups. The level of significance was set at P < 0.05.
Publication bias assessment
Funnel plots were applied to graphically investigate the potential publication bias. In addition, Egger’s test was conducted to statistically assess the publication bias [34].
Results
Study selection
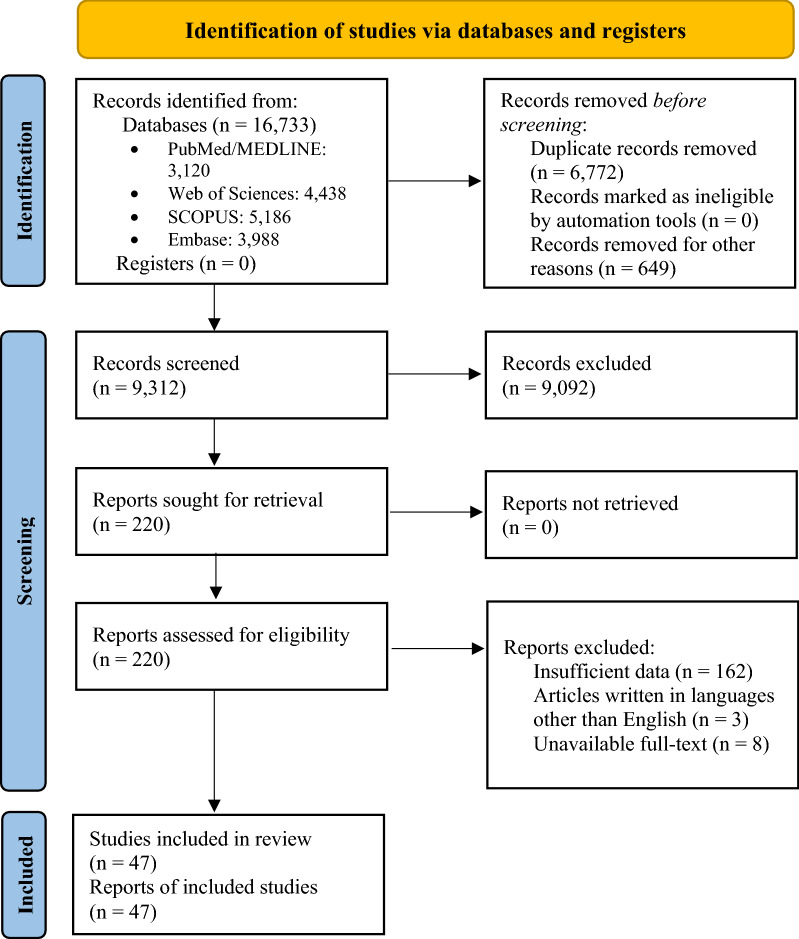
The preliminary search contained 16,733 references, of which 3120, 4438, 5186, and 3988 papers were retrieved from PubMed/MEDLINE, Web of Science, Scopus, and Embase databases, respectively, published from inception to 7th September 2022. Subsequently, the resulting references were imported to the EndNote reference manager to remove duplicate articles (n = 6772). The review articles were excluded (n = 649) before screening studies. Of the remained articles (n = 9312), 9092 papers were excluded following the subsequent screening of titles and abstracts according to eligibility criteria. As a result, 220 eligible studies remained and entered the next phase. The full text of the remaining studies was evaluated, and of these, 173 were excluded according to the exclusion criteria and unavailable full text. Finally, 47 studies were included in the qualitative synthesis. A flowchart of the search process for eligible studies is shown in Fig. 2.
Fig. 2.
Flow-chart for the search strategy according to the Preferred Reporting Items for Systematics Reviews and Meta-Analyses (PRISMA) guideline
Study characteristics
All the enrolled studies were written in English and published between 2015 and 2022, with sample sizes ranging from 4 to 326 patients (Table 1). Geographically, the majority of the papers (n = 31) were conducted in China, whereas the remaining papers (n = 16) were carried out in other countries (Japan, Egypt, Korea, Spain, Germany, Norway, and Taiwan). In addition, a large number of cases were male in most of the studies. Of 47 studies, 32 reported circulating exomiRs with high aberrant expression, and 16 reported low expression (Additional file 4: Table S2). There have been various circulating exomiRs, leading to high heterogeneity in our study. Concerning the types of cancer, the two most commonly evaluated GI tract carcinomas were colorectal cancer (CRC) (n = 16) and hepatocellular carcinoma (HCC) (n = 16), followed by gastric cancer (GC) (n = 8), pancreatic ductal adenocarcinoma (PDAC) (n = 3), hepatoblastoma (HB) (n = 2), pancreatic cancer (PC) (n = 1), and locally-advanced rectal cancer (LARC) (n = 1). Moreover, the exomiRs were derived from either serum (n = 32) or plasma (n = 15). Most of the articles (n = 38) used quantitative real-time PCR (qRT-PCR) for the detection of exomiR expression; 7 studies employed reverse transcription quantitative PCR (RT-qPCR), and 3 applied RNA sequencing. There have been multiple exosome isolation methods; 28 studies employed extraction kit, and 19 papers applied ultracentrifuge (UC). Besides, 33 studies evaluated the relationship between OS and the expression of circulating exomiRs, while 25 studies investigated the prognostic value of the circulating exomiRs on DFS/RFS.
Table 1.
Main characteristics of the studies included in the meta-analysis
| Authors, publication year | Country, Duration (year) | Exosomal miR(s) | Cancer type | Detection method | Exosome extraction method | sample | Sample size (cancer) | mean/ median age (year) | Gender cancer (M/F) | TNM Stage | Outcome | Hazard ratio (HR) | Follow up (month) | Spec % | Sen % | AUC (95% CI) | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-regulationa | |||||||||||||||||
| Soeda N 2019 [35] | Japan 2006–2015 | miR-21 | GC | qRT-PCR | UC | plasma | 129 | 68 | 90/39 | II—III | OS, RFS |
OS: R-Multi/ R-Uni RFS: R-Multi/ R-Uni |
40.8 | 76.8 | 61.6 | NA | good |
| Sun L 2020 [36] | China NA | miR-122 | CRC | qRT-PCR | UC | serum | 85 | 65 | 46/39 | I—IV | OS | R-Multi/ R-Uni | 60 | NA | NA | NA | poor |
| Liu X 2018 [37] | China NA | miR-27a, miR-130a | CRC | qRT-PCR | Exosome isolation kit | plasma | 130 | 64 | 80/50 | I—IV | OS | R-Uni | 60 |
90.9 100 |
81.8 69.3 |
0.866 (0.774–0.957) 0.816 (0.730–0.901) |
poor |
| Wu L. 2020 [38] | China 2018–2019 |
miR-21 miR-210 |
PC | RT-qPCR | Exosome isolation kit | serum | 30 | 60 |
20/10 23/7 |
I—IV | NA | NA | NA |
90.0 90.0 |
80.0 83.0 |
0.869 0.823 |
good |
| Takahasi K 2018 [39] | Japan 2013- 2017 | miR-451a | PDAC | qRT-PCR | UC | plasma | 50 |
High expression group: 66 Low expression group: 69 |
28/22 | I—II | OS, DFS |
OS: R-Multi/ R-Uni DFS: R-Multi/ R-Uni |
30 (range: 5.5–54) |
70.8 | 69.2 | NA | good |
| Tsukamoto M 2017 [40] | Japan 2002- 2012 | miR-21 | CRC | RT-qPCR | UC | plasma | 326 | NA | NA | I—IV | OS, DFS |
OS: R-Multi DFS: R-Multi |
55 | NA | NA | NA | poor |
| Yokota Y 2021 [41] | Japan 2012–2015 | miR-638 | HCC | RT-qPCR | Exosome isolation kit | serum | 54 |
High expression group: 73 Low expression group: 70 |
35/19 | NA | DFS | R-Multi/ R-Uni | 24 | NA | NA | NA | good |
| Takano Y 2017 [42] | Japan 2005- 2012 | miR-203 | CRC | RT-qPCR | UC | serum | 240 | NA | 147/93 | I—IV | OS, DFS |
OS: R-Multi/ R-Uni DFS: R-Multi/ R-Uni |
54 | NA | NA | NA | poor |
| Tian X 2019 [43] | China 2009–2012 |
miR-21 miR-10b |
HCC | qRT-PCR | ExoQuick kit | serum | 124 | 48 | 115/9 | NA | DFS | R-Multi/ R-Uni | NA | NA | NA | NA | poor |
| Wei S.C. 2020 [44] | China NA | miR-15b-3p | GC | qRT-PCR | UC | serum | 108 | 62 | 80/28 | I—IV | OS | E | NA | 80.6 | 74.1 | 0.820 (0.763–0.876) | good |
| Xue X. F. 2019 [45] | China 2015–2017 |
miR-122 miR‐125b miR‐145 miR‐192 miR‐194 miR‐29a miR‐17‐5p miR‐106a |
HCC | qRT-PCR | Exosome isolation kit | serum | 80 | 59 | 61/19 | I—IV | OS | E | 24 | NA | NA |
0.746 (0.650–0.842) 0.650 (0.526–0.774) 0.535 (0.422–0.649) 0.752 (0.658–0.846) 0.738 (0.638–0.838) 0.703 (0.597–0.809) 0.850 (0.764–0.936) 0.704 (0.534–0.873) |
good |
| Yagi T 2019 [46] | Japan NA | miR-125b | CRC | qRT-PCR | UC | plasma | 6 | 65 | 4,2 | III—IV | PFS | R-Multi/ R-Uni | 12 | NA | NA | NA | good |
| Yan S. S. 2018 [47] | China NA | miR-6803-5p | CRC | qRT-PCR | Exosome isolation kit | serum | 168 | 55 | 100/68 | I—IV | OS, DFS |
OS: R-Multi DFS: R-Multi |
60 | NA | NA | 0.739 | poor |
| Matsumura T. 2015 [48] | Japan 1992–2007 | miR-19a | CRC | qRT-PCR | UC | serum | 209 | 65 | 80/129 | I—IV | OS, DFS |
OS: R-Multi/ R-Uni DFS: R-Multi/ R-Uni |
55.68 | NA | NA | NA | poor |
| Kawamura S. 2019 [49] | Japan 2013- 2018 |
miR-4525 miR-451a miR-21 |
PDAC | qRT-PCR | UC | plasma | 55 |
miR-4525: 66 miR-451a: 69 miR-21: 70 |
33/22 | I—II | OS, DFS |
OS: R-Multi/ R-Uni DFS: R-Multi/ R-Uni |
60 |
86.4 77.3 72.7 |
81.8 72.7 72.7 |
NA | poor |
| Abd El Gwad A. 2018 [50] | Egypt 2015–2016 | miR-1262 | HCC | qRT-PCR | UC | serum | 60 | 58 | 43/17 | N/A | RFS | E | 44 | 80.0 | 95.0 | 0.847 | good |
| Cui Y. 2019 [51] | China 2014–2016 | miR-224 | HCC | qRT-PCR | Exosome isolation kit | serum | 89 | 59 | 43/46 | N/A | OS | E | NA | NA | NA | 0.910 (0.840–0.980) | fair |
| Feng C. 2019 [52] | China NA | miR-196a-1 | GC | qRT-PCR | Exosome isolation kit | plasma | 86 | NA | NA | I—IV | OS | E | NA | NA | NA | NA | poor |
| Liu W. B. 2016 [53] | China 2008–2013 | miR-21 | HB | qRT-PCR | ExoQuick kit | serum | 32 | NA | 17/15 | I—IV | DFS | R-Multi/ R-Uni | 42 | NA | NA | 0.861 (0.752–0.935) | poor |
| Cho H. J. 2020 [54] | Korea 2014–2018 |
miR-25-3p miR-1269a miR-4661-5p miR-4746-5p |
HCC |
Cohoort1: NGS-RNA-Seq Cohort 2&3: qRT-PCR |
ExoQuick kit | serum | 72 | 55 | 58/14 | I—IV | NA | NA | NA | 71.4 | 38.0 | 0.704 (0.629–0.772) | good |
| Han J. Y. 2020 [55] | China 2015–2017 |
hsa-miR-100-5p hsa-miR-92a-3p hsa-miR-30e-5p hsa-miR-144-5p hsa-let-7i-5p has-miR-16 |
CRC | qRT-PCR | Exosome isolation kit | plasma | 139 | 60 | 77/62 | III—IV | OS | R-Uni | NA | NA | NA |
0.637 (0.545 -0.729) 0.659 (0.568–0.751) 0.694 (0.606–0.782) 0.791 (0.718 -0.864) 0.650 (0.559–0.742) 0.721 (0.638–0.804) 0.746 (0.664–0.827) 0.778 (0.702–0.854) |
good |
| de Miguel-Pérez D 2020 [56] | Spain NA |
miR-92a miR-222 |
CRC | qRT-PCR | UC | serum | 44 | 55 | 30/14 | NA | OS, DFS/PFS |
miR-92a OS: R-Uni DFS/PFS: R-Multi/ R-Uni miR-222 OS: R-Multi/ R-Uni |
27 | NA | NA |
0.951 (0.900–1.000) 0.896 (0.810–0.980) |
good |
| Qu Z. 2017 [57] | China NA | miR-665 | HCC | qRT-PCR | ExoQuick kit | serum | 30 | 60 | 12/18 | I—IV | OS | E | NA | NA | NA | NA | poor |
| Reese M. 2020 [58] | Germany 2015–2018 |
miR-200b miR-200c |
PDAC | qRT-PCR | UC | serum | 56 | 60 | 36/20 | I—IV | OS |
miR-200b OS: R-Multi/ R-Uni miR-200c OS: R-Multi/ R-Uni |
13 | NA | NA |
0.790 (0.680–0.890) 0.670 (0.550–0.790) |
good |
| Meltzer S. 2019 [59] | Norway NA |
miR-141-3p miR-375 |
LARC | qRT-PCR | Exosome isolation kit | plasma | 64 | 60 | 36/28 | NA | OS, PFS |
OS: R-Uni PFS: R-Uni |
65 (range 4–66) | NA | NA | NA | good |
| Wang Q. 2021 [60] | China NA | miR-1290 | HCC | qRT-PCR | Exosome isolation kit/UC | serum | 49 | 55 | 37/12 | I—IV | NA | NA | NA | NA | NA | NA | poor |
| Zhang Y. 2021 [60] | China NA | miR-215-5p | GC | qRT-PCR | ExoQuick kit | serum | 118 | 60 | 70/48 | I—IV | OS, DFS |
OS: R-Multi/ R-Uni DFS: E |
NA | 97.1 | 68.6 | 0.866 | good |
| Zhu L. 2022 [61] | China 2020–2021 | miR-552-5p | GC | qRT-PCR | UC | plasma | 30 | 56 | 16/14 | I—IV | NA | NA | NA | NA | NA | NA | good |
| Yang J. 2022 [62] | China NA |
miR-195-5p miR-211-5p |
GC | RT-qPCR | UC | plasma | 108 |
62.4 ± 8.9 62.2 ± 9.4 |
80/28 79/27 |
I—IV | NA | NA | NA | NA | NA |
0.745 (0.584–0.906) 0.798 (0.656–0.940) |
good |
| Chen S. 2022 [63] |
China NA |
miR-34a | HCC | qRT-PCR | UC | serum | 60 | 55 | 60/60 | I—IV | OS | R-Multi/ R-Uni | 6–60 | 51.7 | 78.3 | 0.664 (0.572–0.747) | good |
| Huang C. Y. 2021[64] |
China 2017–2018 |
let-7e miR-18a miR-27b miR-221 miR-20b miR-652 |
HCC | RT-qPCR | UC | plasma | 40 | 68 | 13/7 | NA | OS, DFS |
OS: R-Uni DFS: R-Uni |
80 | NA | NA | NA | poor |
| Hao Y. J. 2022 [65] | Taiwan 2019–2020 | miR-21 | CRC | qRT-PCR | Exosome isolation kit | plasma | 113 | 65 | 77/36 | I—IV | DFS, PFS | E | NA | NA | NA | NA | poor |
| Down-regulationa | |||||||||||||||||
| Soeda N 2019 [35] | Japan 2006–2015 | miR-92a | GC | qRT-PCR | UC | plasma | 129 | 68 | 90/39 | II—III | OS, RFS |
OS: R-Multi/ R-Uni RFS: E |
40.8 | 60.7 | 63.0 | NA | good |
| Yan S. 2017 [66] | China 2008–2014 | miR-638 | CRC | qRT-PCR | Exosome isolation kit | serum | 192 | 58 | 108/84 | I—IV | OS, DFS |
OS: R-Multi/ R-Uni DFS: E |
47 | NA | NA | NA | poor |
| Yan S. S. 2018 [67] | China 2012–2014 | miR-6869-5p | CRC | qRT-PCR | Exosome Isolation Kit | serum | 142 | 59 | 85/57 | I—IV | OS | R-Multi/ R-Uni | 36 | NA | NA | NA | good |
| Zou S. L. 2019 [68] | China NA | miR-150-5p | CRC | qRT-PCR | ExoQuick kit | serum | 133 | 60 | 49/84 | I—IV | OS, DFS |
OS: R-Multi/ R-Uni DFS: E |
60 | 76.1 | 81.0 | 0.870 | poor |
| Zhang N. 2020 [69] | China NA | miR-874 | CRC | RT-qPCR | Exosome Isolation kit | serum | 125 | 60 | 76/49 | I—IV | OS | R-Multi/ R-Uni | NA | 78.6 | 80.8 | 0.818 | poor |
| Liu C. 2016 [70] | China 2006–2011 | miR-4772-3p | CRC |
qRT-PCR RNA-seq |
ExoQuick kit | serum | 84 | 57 | 53/31 | II—III | OS | R-Multi | 51 (range: 45–64) | 77.1 | 78.6 | 0.720 (0.590–0.850) | poor |
| Hao X. 2020 [71] | China 2012–2013 | miR-320a | HCC | qRT-PCR | ExoQuick kit | serum | 104 | 60 | 77/27 | I—IV | OS, DFS |
OS: R-Multi/ R-Uni DFS: E |
NA | 80.0 | 77.9 | 0.860 | good |
| Jiao C. W. 2017 [72] | China 2007–2015 |
miR-34 s panel miR-34a miR-34b miR-34c |
HB | qRT-PCR | ExoQuick kit | serum | 89 | NA | 52/37 | I—IV | DFS | R-Multi/ R-Uni | 54 | NA | NA |
0.831(0.071–0.371) 0.813(0.023–0.296) 0.837(0.004–0.342) |
good |
| Li W. 2020 [73] | China NA | miR-320d | HCC | qRT-PCR | Exosome isolation kit | serum | 110 | 60 | 98/12 | I—IV | OS, DFS |
OS: R-Multi DFS: E |
NA | NA | NA | 0.869 | good |
| Liu W. F. 2017 [74] | China 2012 | miR-125b | HCC | qRT-PCR | ExoQuick kit | serum | 128 | 50 | 110/18 | I—III | OS | R-Multi/ R-Uni | 2.9–52.4 | 53.4 | 82.5 | 0.702 (0.602–0.802) | good |
| Sheng L. Q. 2020 [75] | China 2017–2018 |
miR-455-5p miR-30c-5p |
HCC | exomiRs sequencing | Exosome isolation kit | plasma |
cohort1:24 cohort2:27 |
NA |
cohort1:36/8 cohort2:8/4 |
N/A | OS | E | NA | NA | NA | NA | poor |
| Shi M. 2018 [76] | China 2008–2011 | miR-638 | HCC | qRT-PCR | Exosome isolation kit | serum | 126 | 65 | 70/56 | I—IV | OS | R-Multi/ R-Uni | 81.5 | NA | NA | NA | good |
| Suehiro T. 2018 [76] | Japan 2006–2013 | miR-122 | HCC | qRT-PCR | ExoQuick kit | serum | 75 | 73 | 49/26 | NA | DFS | R-Multi | 47 | NA | NA | NA | poor |
| Kumata Y. 2018 [77] | Japan 2006–2013 | miR-23b | GC | qRT-PCR | UC | plasma | 232 | NA | 165/67 | I—IV | OS, DFS |
OS: R-Multi/ R-Uni DFS: R-Multi/ R-Uni |
45.6 (range: 4.8–127.2) | NA | NA | NA | poor |
| Peng Z. Y. 2018 [78] | China 2008–2014 | miR-548c-5p | CRC | qRT-PCR | Exosome isolation kit | serum | 108 | 58 | 61/47 | I—IV | OS | R-Multi/ R-Uni | 44 | NA | NA | NA | poor |
| Zheng G-D. 2021 [79] | China 2008–2011 | miR-590-5p | GC | qRT-PCR | UC | serum | 168 | 60 | 117/51 | I—IV | OS | R-Multi/ R-Uni | 64.2 (range: 43.3–92) | 86.0 | 63.7 | 0.810 (0.751–0.860) | good |
GC Gastric Cancer, CRC Colorectal Cancer, LARC Locally Advanced Rectal Cancer, HCC Hepatocellular Carcinoma, HB Hepatoblastoma, PC Pancreatic Cancer, PDAC Pancreatic Ductal Adenocarcinoma, UC Ultracentrifuge, microRNA miR, NA Not Applicable, OS Overall Survival, DFS Disease Free Survival, PFS Progression Free Survival, RFS Relapse Free Survival, E Estimated, R-Multi Reported Multivariate, R-Uni Reported Univariate, qRT-PCR quantitative real-time PCR, RT-qPCR Reverse transcription quantitative PCR, Sen Sensitivity, Spec Specificity, AUC Area Under Curve.
aThe up- and down-regulation of exomiR(s) has been reported in comparison with the control group
Quality assessment in studies
According to the NOS quality assessment tool, the majority of the included studies (n = 28) had good quality. The remaining studies (n = 18) had poor quality, and only one study had fair quality. A summary of quality assessment for all eligible studies has been shown in Fig. 3.
Fig. 3.
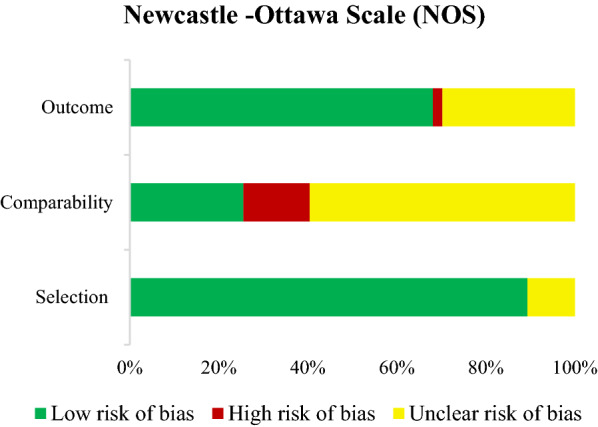
Summary of quality assessment
Meta-analysis
Prognostic accuracy and subgroup analyses
A total of 34 studies included in the present meta-analysis assessed the association between deregulated exomiRs (n = 52) and OS in individuals suffering from GI cancers. Twenty-four studies provided data regarding exomiRs deregulation (n = 29) and DFS/RFS/PFS.
Deregulated exomiRs and the overall survival (OS)
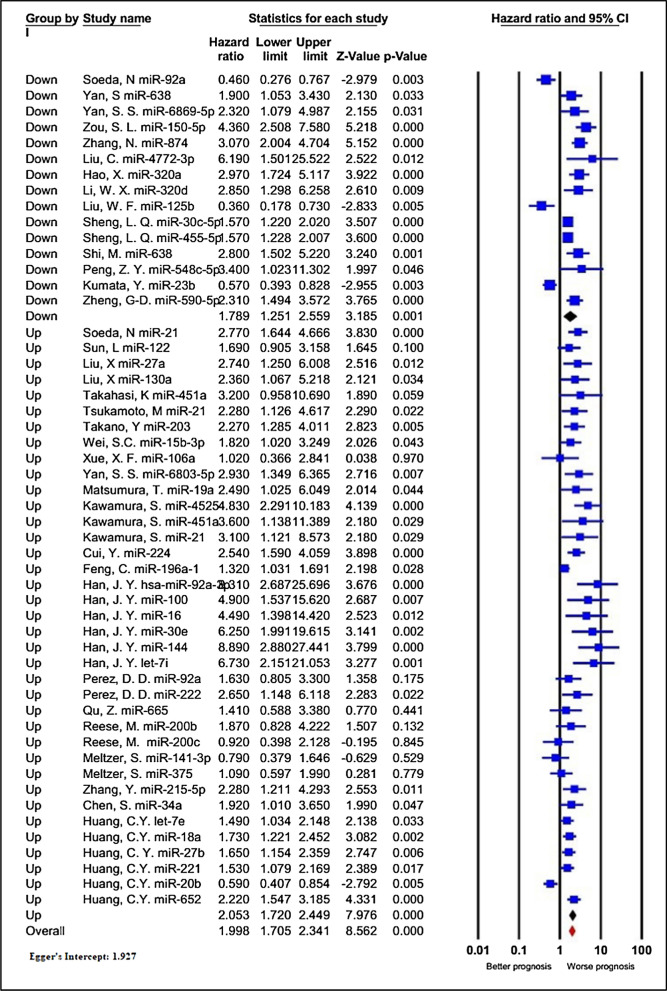
Thirty-four studies containing 3833 patients investigated the impact of exomiRs deregulation on OS in patients with GI cancers. As shown in Table 2, since statistical heterogeneity was identified among the investigations (I2 = 76.564%, P < 0.001), a random-effect model was applied to estimate the combined HR. Patients with deregulated exomiRs displayed a statistically significant decrease in OS (pooled HR = 1.998, 95% CI 1.705–2.341, P < 0.001, Fig. 4).
Table 2.
The results of meta-analyses for the association between deregulated exomiRs and overall survival (OS), and disease/relapse/progression-free survival (DFS/RFS/PFS) in patients with GI cancers
| Study groups | Included exomiRs | Test of association | Test of heterogeneity | ||
|---|---|---|---|---|---|
| HR (95% CI) | P-value | I2% | P-value | ||
| OS | |||||
| All studies | 52 | 1.998 (1.705–2.341) | < 0.001 | 76.564 | < 0.001 |
| The type of exomiRs deregulation | |||||
| Up-regulation | 37 | 2.053 (1.720–2.449) | < 0.001 | 66.043 | < 0.001 |
| Down-regulation | 15 | 1.789 (1.251–2.559) | 0.001 | 87.290 | < 0.001 |
| Type of cancer | |||||
| Colorectal | 21 | 2.928 (2.417–3.547) | < 0.001 | 19.747 | 0.204 |
| Gastric | 7 | 1.353 (0.827–2.214) | 0.229 | 88.803 | < 0.001 |
| Hepatocellular | 16 | 1.582 (1.264–1.979) | < 0.001 | 76.367 | < 0.001 |
| Pancreatic | 6 | 2.514 (1.478–4.279) | 0.001 | 48.243 | 0.085 |
| locally advanced rectal | 2 | 0.958 (0.601–1.525) | 0.856 | 0.000 | 0.506 |
| Sample size | |||||
| ≥ 100 | 29 | 2.306 (1.789–2.973) | < 0.001 | 81.157 | < 0.001 |
| < 100 | 23 | 1.659 (1.359–2.025) | < 0.001 | 65.182 | < 0.001 |
| Sample type | |||||
| Plasma | 27 | 1.890 (1.497–2.386) | < 0.001 | 82.109 | < 0.001 |
| Serum | 25 | 2.125 (1.752–2.578) | < 0.001 | 51.047 | 0.002 |
| Survival analysis | |||||
| Direct | 44 | 2.114 (1.719–2.601) | < 0.001 | 79.360 | < 0.001 |
| Indirect | 8 | 1.559 (1.368–1.776) | < 0.001 | 1.170 | 0.420 |
| NOS score | |||||
| ≥ 7 | 25 | 2.165 (1.577–2.972) | < 0.001 | 78.093 | < 0.001 |
| < 7 | 27 | 1.850 (1.534–2.231) | < 0.001 | 74.863 | < 0.001 |
| Ethnicity | |||||
| Asian | 46 | 2.085 (1.746–2.489) | < 0.001 | 78.377 | < 0.001 |
| Caucasian | 6 | 1.327 (0.934–1.886) | 0.115 | 25.255 | 0.245 |
| DFS/RFS/PFS | |||||
| All studies | 29 | 1.920 (1.641–2.245) | < 0.001 | 55.871 | < 0.001 |
| Deregulation | |||||
| Up-regulation | 21 | 2.086 (1.725–2.522) | < 0.001 | 41.988 | 0.023 |
| Down-regulation | 8 | 1.607 (1.218–2.122) | 0.001 | 72.320 | 0.001 |
| Type of cancer | |||||
| Colorectal | 9 | 2.105 (1.736–2.554) | < 0.001 | 0.000 | 0.734 |
| Gastric | 4 | 1.408 (0.781–2.539) | 0.256 | 86.773 | < 0.001 |
| Hepatocellular | 10 | 1.923 (1.651–2.239) | < 0.001 | 0.000 | 0.740 |
| Pancreatic | 4 | 3.195 (2.019–5.056) | 0.000 | 0.000 | 0.839 |
| Locally advanced rectal | 2 | 1.034 (0.758–1.410) | 0.834 | 0.000 | 0.567 |
| Sample size | |||||
| ≥ 100 | 15 | 1.911 (1.520–2.402) | < 0.001 | 64.800 | < 0.001 |
| < 100 | 14 | 1.890 (1.506–2.373) | < 0.001 | 45.085 | 0.034 |
| Sample type | |||||
| Plasma | 13 | 1.819 (1.317–2.510) | < 0.001 | 74.592 | < 0.001 |
| Serum | 16 | 1.906 (1.680–2.163) | < 0.001 | 0.000 | 0.633 |
| Survival analysis | |||||
| Direct | 20 | 2.019 (1.594–2.558) | < 0.001 | 67.219 | < 0.001 |
| Indirect | 9 | 1.809 (1.552–2.109) | < 0.001 | 0.000 | 0.725 |
| NOS score | |||||
| ≥ 7 | 14 | 1.803 (1.498–2.170) | < 0.001 | 41.843 | 0.050 |
| < 7 | 15 | 2.062 (1.571–2.706) | < 0.001 | 65.905 | < 0.001 |
| Ethnicity | |||||
| Asian | 26 | 1.998 (1.705–2.341) | < 0.001 | 50.425 | 0.002 |
| Caucasian | 3 | 1.109 (0.800–1.538) | 0.535 | 10.141 | 0.329 |
Fig. 4.
Forest plot of the association between exomiRs deregulation and overall survival in patients with GI cancers, stratified by the type of exomiRs deregulation (down-regulated exomiRs and up-regulated exomiRs)
Additionally, to figure out the overall findings’ robustness, subgroup analysis for OS data was conducted according to seven subcategories, including the type of exomiRs deregulation (up or down), type of cancer, sample size, different data extraction methods (direct or indirect), NOS score, and ethnicity (Table 2). Stratified analysis by the different types of exomiRs deregulation indicated a poorer OS for 37 up-regulated exomiRs (HR = 2.053, 95% CI 1.720–2.449, P < 0.001; I2 = 66.043%, P < 0.001), compared to 15 down-regulated exomiRs (HR = 1.789, 95% CI 1.251–2.559, P = 0.001; I2 = 87.290%, P < 0.001). Regarding the cancer types, deregulation of exomiRs was closely associated with inferior OS in cases with CRC (HR = 2.928, 95% CI 2.417–3.547, P < 0.001, I2 = 19.747%, P = 0.204), HCC (HR = 1.582, 95% CI 1.264–1.979, P < 0.001, I2 = 76.367%, P < 0.001), and PDAC (HR = 2.514, 95% CI 1.478–4.279, P = 0.001, I2 = 48.243%, P = 0.085). However, GC (HR = 1.353, 95% CI 0.827–2.214, P = 0.229, I2 = 88.803%, P < 0.001) and LARC (HR = 0.958, 95% CI 0.601–1.525, P = 0.856, I2 = 0.000%, P = 0.506) did not show such association. When the investigations were stratified based on the sample size, a more considerable relationship was identified between worse OS and large sample sizes (≥ 100, HR = 2.306, 95% CI 1.789–2.973, P < 0.001; I2 = 81.157%, P < 0.001) compared to small sample sizes (< 100, HR = 1.659, 95% CI 1.359–2.025, P < 0.001; I2 = 65.182%, P < 0.001). Subgroup analysis of OS for exomiRs de-regulation in plasma demonstrated considerable association (HR = 1.890, 95% CI 1.497–2.386, P < 0.001; I2 = 82.109%, P < 0.001), like serum (HR = 2.125, 95% CI 1.752–2.578, P < 0.001; I2 = 51.047%, P = 0.002). In subgroup analysis stratified by different data extraction methods, exomiRs deregulation revealed a more significant connection with the worse OS in the HR presented in the articles (HR = 2.114, 95% CI 1.719–2.601, P < 0.001; I2 = 79.360%, P < 0.001) than that calculated from the survival curves (HR = 1.559, 95% CI 1.368–1.776, P < 0.001; I2 = 1.170%, P = 0.420). When categorized by quality assessment, deregulated exomiRs was related to poorer OS in both high- (HR = 2.165, 95% CI 1.577–2.972, P < 0.001; I2 = 78.093%, P < 0.001) and low-quality publications (HR = 1.850, 95% CI 1.534–2.231, P < 0.001; I2 = 74.863%, P < 0.001) (Additional file 1: Figure S1). Finally, in subgroup analysis stratified by ethnicity, deregulation of exomiRs revealed a significant connection with the worse OS in the articles published in Asian countries (HR = 2.085, 95% CI 1.746–2.489, P < 0.001; I2 = 78.377%, P < 0.001) unlike articles from Caucasian countries with an HR of 1.327 (95% CI 0.934–1.886, P = 0.115; I2 = 25.255%, P = 0.245).
Deregulated exomiRs and the disease/relapse/progression-free survival (DFS/RFS/PFS)
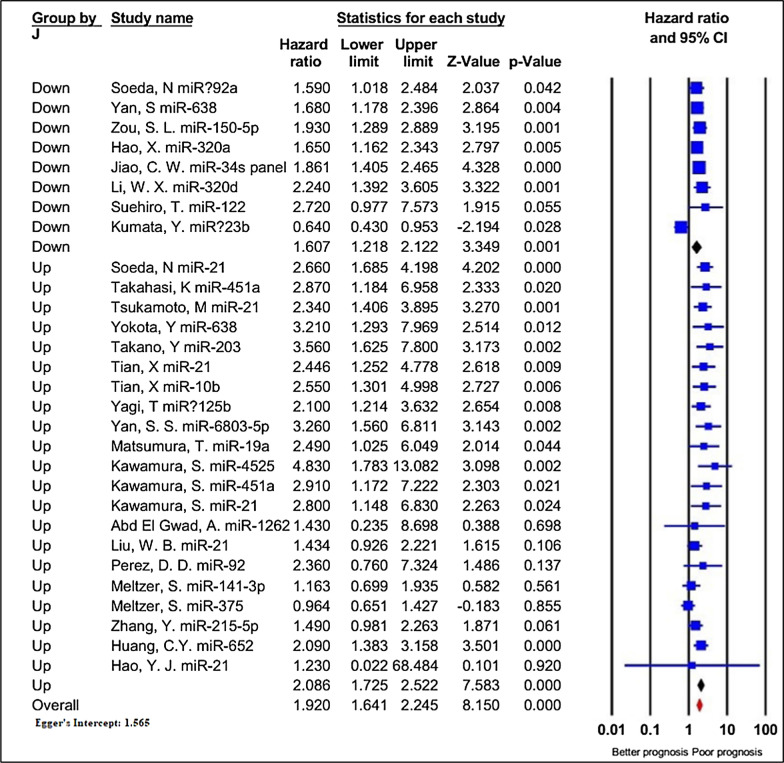
As indicated in Table 2, twenty-nine studies containing 2767 patients reported the data regarding DFS/RFS/PFS, and relatively significant heterogeneity was identified among these investigations (I2 = 55.871%, P < 0.001). The pooled results through a random-effects model revealed a significant link between deregulated exomiRs and poorer DFS/RFS/PFS in patients (HR = 1.920, 95% CI 1.641–2.245, P < 0.001, Fig. 5).
Fig. 5.
Forest plot of the association between exomiRs deregulation and disease/relapse/progression-free survival (DFS/RFS/PFS) in patients with GI cancers, stratified by the type of exomiRs deregulation (down-regulated exomiRs and up-regulated exomiRs)
Following that, subgroup analyses were performed to further investigate the potential predictive significance of the SMYD family members according to the same seven categories utilized for OS (Table 2). The results demonstrated a remarkable connection between deregulated exomiRs and worse DFS/RFS/PFS in all the stratified analyses performed, except patients with GC (HR = 1.408, 95% CI 0.781–2.539, P = 0.256; I2 = 86.773%, P < 0.001), LARC (HR = 1.034, 95% CI 0.758–1.410, P = 0.834; I2 = 0.000%, P = 0.567), and articles published in Caucasian (HR = 1.109, 95% CI 0.800–1.538, P = 0.535; I2 = 10.141%, P = 0.329) (Table 2).
Association between deregulated exomiRs and clinicopathological characteristics
Gender
The relationship between up-regulated exomiRs and gender was evaluated in 27 studies with 2479 patients, while down-regulated exomiRs were reported in 13 studies with 1736 patients. As shown in Additional file 5: Table S3, pooled results from fixed-effects framework (I2 = 19.601%, P = 0.182) indicated that up-regulated exomiRs did not associate with gender (OR = 0.992, 95% CI 0.826–1.190, P = 0.927), as in down-regulated exomiRs (OR = 0.986, 95% CI 0.796–1.222, P = 0.900; I2 = 0.000%, P = 0.826).
TNM stage
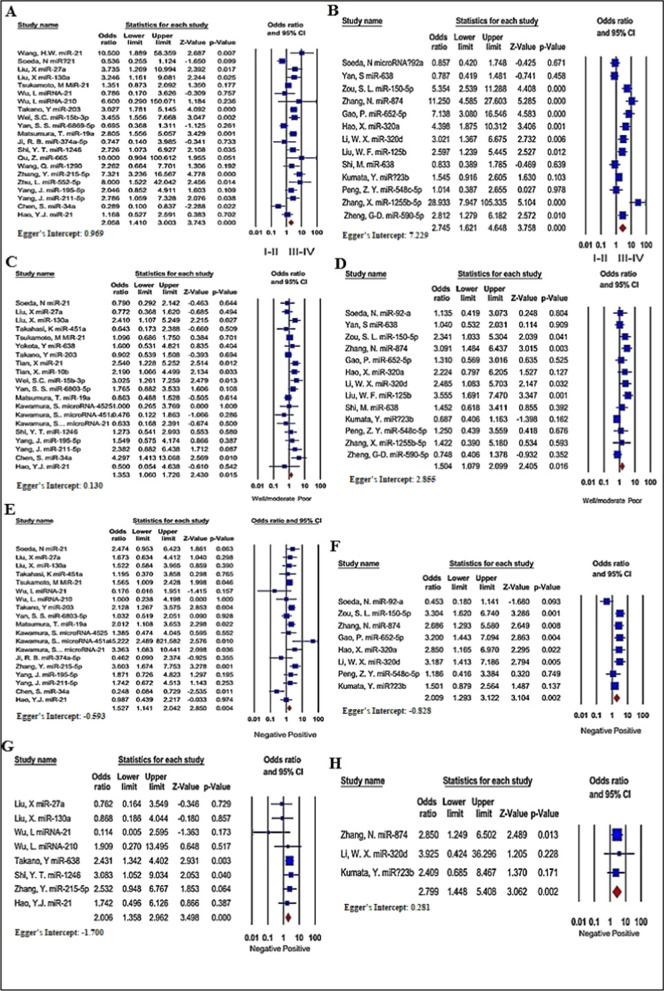
Data from 21 and 13 investigations comprising of 2288 and 1735 patients were collected and pooled to reveal a connection between up- and down-regulated exomiRs and TNM stage, respectively. Based on the random-effect framework (I2 = 71.244%, P < 0.001), it was found that the GI cancer patients with up-regulated exomiRs tended towards the advanced TNM stage (OR = 2.058, 95% CI 1.410–3.003, P < 0.001, Fig. 6A, Additional file 5: Table S3). In addition, the pooled OR indicated that down-regulated exomiRs were directly correlated with a higher TNM stage (HR = 2.745, 95% CI 1.621–4.648, P < 0.001, Fig. 6B, Additional file 5: Table S3).
Fig. 6.
Forest plot of the association between up-regulated A, C, E, and G or down-regulated B, D, F, and H exomiRs and clinicopathological characteristics in patients with GI cancers. TNM stage A, B, Differentiation C, D, Lymph node metastasis E, F, Distant metastasis G, H
Differentiation
A total of 20 studies 2,425 patients, evaluated a possible connection between up-regulated exomiRs and differentiation (Fig. 6C and Additional file 5: Table S3). The pooled OR through a random effects model (I2 = 37.862%, P = 0.045) found statistically significant results (OR = 1.353, 95% CI: 1.060–1.726, P = 0.015). Analysis based on 13 studies through a random-effects model (I2 = 55.128%, P = 0.008) showed that the down-regulated exomiRs correspondent with poorly-differentiated cancer cells (OR = 1.504, 95% CI: 1.079–2.099, P = 0.016; Fig. 6D and Additional file 5: Table S3).
Lymph node metastasis
A total of 19 studies consisting of 2121 cases focused on the dependability between the up-regulated exomiRs and LNM. The overall pooled HR, under a random-effect model (I2 = 47.603%, P = 0.011), indicated that up-regulated exomiRs had a statistically significant relationship with LNM (OR = 1.527, 95% CI 1.141–2.042, P = 0.004; Fig. 6E, Additional file 5: Table S3). Afterward, a clear association was identified between down-regulated exomiRs and LNM (OR = 2.009, 95% CI 1.293–3.122, P = 0.002; Fig. 5F, Additional file 5: Table S3) with significant heterogeneity (I2 = 60.459%, P = 0.013).
Distant metastasis
The relationship between up-regulated exomiRs and distant metastasis was demonstrated in eight studies with 876 cases, while down-regulated exomiRs were reported in three studies with 467 patients. Statistically, non-significant heterogeneity was observed for high expression exomiRs (I2 = 2.450%, P = 0.411) and low expression exomiRs (I2 = 0.000%, P = 0.930); consequently, a fixed-effect model was employed to combine the results. The findings showed that cases with down-regulated exomiRs were more likely to develop distant metastasis (OR = 2.799, 95% CI 1.448–5.408, P = 0.002; Fig. 6H, Additional file 5: Table S3), as in up-regulated exomiRs (OR = 2.006, 95% CI 1.358–2.962, P < 0.001; Fig. 6G, Additional file 5: Table S3).
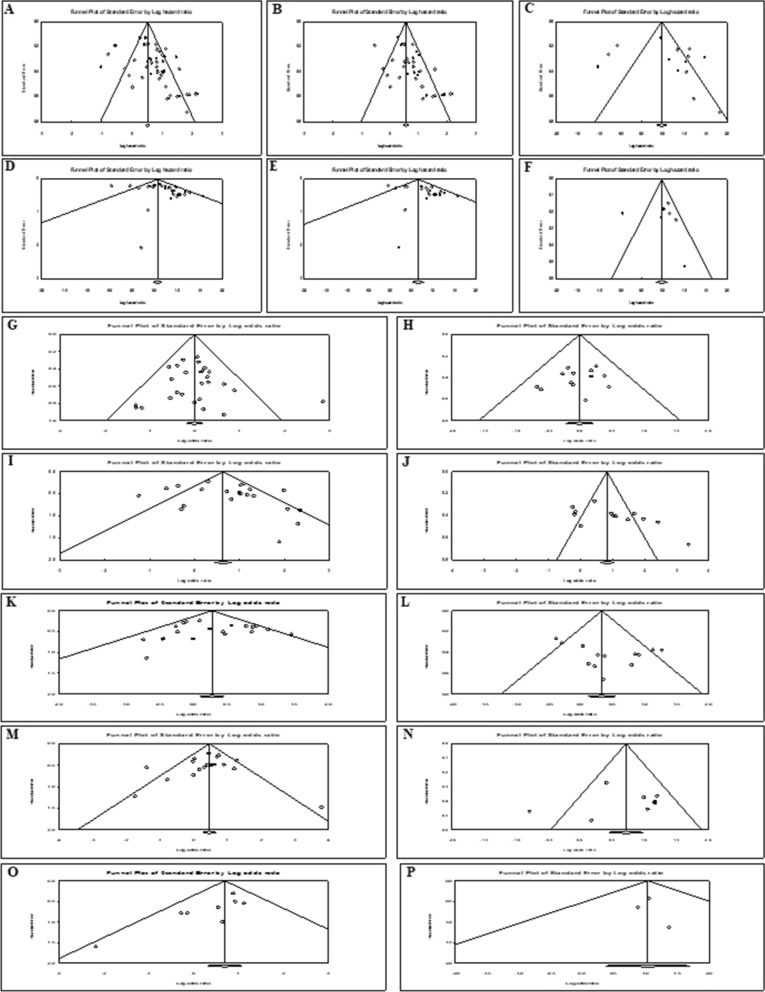
Publication Bias
Funnel plot (Fig. 7) and Egger’s test (Additional file 5: Table S3) were also conducted to identify potential publication bias of all analyses. The P-values for Egger’s test, measuring the asymmetry of performed analyses, indicated statistically non-significant publication bias for all analyses except for OS (overall deregulated exomiRs (P = 0.002) and up-regulated exomiRs (P < 0.001), DFS/RFS/PFS (overall deregulated exomiRs (P = 0.021) and up-regulated exomiRs (P = 0.023), TNM stage (down-regulated exomiRs (P = 0.027) and distant metastasis (up-regulated exomiRs (P = 0.026). The funnel plots, displaying potential publication bias, are shown in Fig. 7.
Fig. 7.
Funnel plot of the publication bias. A the association between overall exomiRs deregulation and OS. B, C the association between up-regulated B and down-regulated C exomiRs and OS. D the association between overall exomiRs deregulation and disease/relapse/progression-free survival (DFS/RFS/PFS). E, F the association between up-regulated E and down-regulated F exomiRs and DFS/RFS/PFS. G–P the association between up-regulated and down-regulated exomiRs and clinicopathological characteristics, including gender G, H, TNM stage I, J, Differentiation K, L, Lymph node metastasis M, N, Distant metastasis O, P, respectively
Association between circulating exomiR-21 and prognostic/clinicopathological characteristics
To analyze the same types of circulating exomiRs in GI cancers, we considered the exomiR-21 as the most overexpressed miRNA in GI tumors. The results of our analysis indicated a statistically significant association between the exomiR-21 and overall survival (HR = 2.655, 95% CI 1.802–3.912, P < 0.001), disease/relapse/progression-free survival (HR = 2.127, 95% CI 1.674–2.703, P < 0.001), and Lymph node metastasis (HR = 1.568, 95% CI 1.118–2.198, P = 0.009) ( Additional file 2: Figure S2). Furthermore, no association was found between circulating exomiR-21 and other clinicopathological characteristics.
Discussion
Exploring novel prognostic biomarkers is crucial for improving management, therapy, and prognosis in patients with GI cancers. Compelling evidence has cleared that deregulated exomiRs, as the hallmarks of most human cancers, regulate a plethora of biological pathways, including maintaining proliferative potential, escaping suppressors, repelling cell death, and triggering invasion metastasis, drug resistance, and angiogenesis [80].
Recently, serum exomiRs were applied as feasible and non-invasive serological biomarkers for several cancers, including GI cancers [41, 48, 81–88]. Therefore, in this systematic review and meta-analysis, we aimed to evaluate the prognostic capacity of circulating exomiRs GI cancers. The four principal databases (Web of Science, PubMed/MEDLINE, Scopus, and Embase) were searched to ensure the identification of all relevant studies. To the best of our knowledge, this is the first comprehensive meta-analysis comprising a wide range of circulating exomiRs with 4881 participants to provide strong evidence regarding the potential clinical applicability of circulating exomiRs in GI cancers. Notably, we attempted to clarify study heterogeneity and publication bias.
In general, we combined the data regarding the deregulation of all exomiRs in our meta-analysis to perceive their potential roles to predict the prognosis. The results emerging from this meta-analysis advocated that deregulated exomiRs were significantly associated with reduced OS (HR = 1.998) and dismal DFS/RFS/PFS (HR = 1.920), supporting the statement that the aberrant expression of exomiRs as minimally invasive biomarkers can predict prognostic outcomes in GI cancers patients. The majority of subcategories, including the type of exomiRs deregulation (up or down), the majority of cancer types (colorectal, hepatocellular, and pancreatic cancers), sample size (≥ 100 or < 100), various data extraction methods (direct or indirect), sample type (plasma or serum), and NOS score (high or low-quality publications), were also strongly associated with poor survival outcomes (OS or DFS/RFS/PFS, strengthening the prognosis. Only some types of cancers (gastric cancer and locally advanced rectal cancer) and Caucasian ethnicity did not show a link with poor survival outcomes. Based on obtained findings from OS, CRC (HR = 2.928), HCC (HR = 1.582), and PDAC (HR = 2.514) are likely to have a significant association with poor prognosis while other cancers of the GI tract, including GC (HR = 1.353, P = 0.229) and LARC (HR = 0.958, P = 0.857) did not show such association. Even the association between DFS/RFS/PFS and the same types of cancers including CRC (HR = 2.105), HCC (HR = 1.923), and PDAC (HR = 3.195) remained significant. This finding highlights the similarity of results related to different types of prognostic reports (OS vs. DFS/RFS/PFS) and GI cancers. Moreover, we did not find any significant association between survival findings and LARC or GC. It is assumed that the assorted outcomes and the insufficient number of included studies might have affected these findings [89, 90]. It was reported that variability in outcomes prediction and categorization of LARC is due to the diversity of tumors and the complexity of diagnostic and prognostic tools in clinical practice [91–93].
We observed partially high heterogeneity in most of our prognostic findings, which is mainly due to the considerable variability within circulating exomiRs, type of cancer, and follow-up duration of the included studies. Moreover, publication bias was only detected between OS and overall deregulated exomiRs (P = 0.002) or up-regulated exomiRs (P < 0.001), as well as DFS and overall deregulated exomiRs (P = 0.021) or up-regulated exomiRs (P = 0.023).
The clinical significance of circulating exomiRs with abnormally elevated or reduced expression in various malignancies has been highlighted by an increasing body of research [79, 98, 99]. We examined 47 studies on 62 distinct exomiRs in GI malignancies in the current research, including 50 exomiRs with high expression, 16 with low expression, and 4 with both low and high expression (miR-122, 638, 125b, 92a). All circulating exomiRs were categorized into two main subgroups as either up- or down-regulated exomiRs. According to our findings, aberrant expression of exomiRs, in terms of both up and down-regulation, are strongly associated with inferior prognostic outcomes, including OS and DFS/RFS/PFS, as well as clinicopathological characteristics, including differentiation, TNM stage, lymph node metastasis, and distant metastasis. However, we did not identify any association between up or down-regulated circulating exomiRs and gender. Similar to our findings, a prior meta-analysis has demonstrated that aberrant exomiRs expression, in terms of both up and down-regulation, is correlated with a worse prognosis in colorectal cancer [23]. The above findings propose that developing the panel of exomiRs could be utilized as valuable biomarkers in GI cancer prognosis.
Among the various up-regulated miRs, miR-21 has been identified to be the most overexpressed miRNA in tumors, which may affect the development of cancer via different signaling cascades [94]. Overexpression of miR-21 was recognized as a prognostic and diagnostic biomarker in various cancers [95–99]. Furthermore, it was studied that high expression of miR-21 was correlated with low OS in glioma patients [100]. Our findings, which indicated a statistically significant correlation between the exomiR-21 and OS (HR = 2.655, P < 0.001), DFS/RFS/PFS (HR = 2.127, P < 0.001), and Lymph node metastasis (HR = 1.568, P = 0.009), are consistent with these findings. Similarly, overexpression of miR-222 can enhance tumorigenesis, migration, and invasion properties in breast cancer (BC) and thyroid cancer [101, 102], along with its association with worse OS and DFS/RFS/PFS in glioma and NSCLC [103, 104]. Besides, high expression of the miR-200 family plays a significant role in tumorigenesis and metastasis in ovarian cancer and endometrial adenocarcinoma [105, 106] and is associated with shorter OS in breast cancer [107]. Regarding the results of our meta-analysis and other investigations, overexpression of some specific exomiRs has the chance to become predictors of long-term survival and metastasis in the patients with GI cancers.
Regarding down-regulated miRs, a low level of miR-320 is correlated with advanced stage, LNM, and poor OS in BC patients [108]. It was reported that miR-34 is expressed at low levels in BC, NSCLC, and bladder cancer and its down-regulation is correlated with recurrence, metastasis, and poor survival outcomes [109–111]. Furthermore, the down-regulation of miR-30c was correlated with poor prognostic outcomes in patients with BC [112, 113]. Thus, our findings revealed that up- or down-regulated exomiRs can be considered new biomarkers in predicting clinical outcomes in GI cancers.
Moreover, the expression of some circulating exomiRs could be either increased or decreased based on the cancer type. Among them, miR-122, a tumor suppressor miR, could regulate metastasis of HCC [114], and circulating miR-122 was up-regulated in BC, NSCLC [115, 116], which is associated with distant metastasis and lowered OS and PFS, considered as a prognostic factor [115]. Other groups of researchers, however, have shown that miR-122 expression is down-regulated in various cancer cells, including bladder and colon tumors [105]. These investigations support our findings, which show that several circulating exomiRs (miR-122, 638, 125b, and 92a) are members of both expression-high and expression-low subgroups in GI cancers. Although our large meta-analysis sheds light on the pathological characteristics and prognostic potentials of GI circulating exomiRs in clinical practice, several limitations should be considered when interpreting the results of the current study. Firstly, while all relevant studies were included in this study, relatively high heterogeneity was observed when analyzing the overall findings for OS or DFS/RFS/PFS. Therefore, subgroup analysis was performed by considering some subcategories to find the factors that caused the heterogeneity. Regarding this consideration, we did not underestimate the variability in patients’ clinicopathological characteristics. Therefore, variations in population and research methodology might affect the heterogeneity. Secondly, the publication bias was identified for the relationship between OS or DFS/RFS/PFS and circulating exomiRs with high expression, affecting the validity of prognostic findings. Third, additional bias may possibly have arisen from (i) the number of investigations for some GI cancers was insufficient; (ii) all articles published in English; and (iii) most of the included publications were performed in Asian ethnicity, possibly leading to selection bias. Finally, some statistical errors may affect the credibility of findings in our meta-analysis, including (i) the indirect estimation of HR and 95% CI via the Kaplan–Meier curves in some studies; and (ii) using univariate analysis information instead of multivariate analysis for some studies without providing the statistical methodology. Despite these limitations, this meta-analysis suggests that circulating exomiRs would be useful as new prognostic biomarkers in GI cancers. However, multi-parameter and large-scale clinical studies with a strong methodology are required to implement exomiRs as biomarkers in the prognosis of GI cancers robustly.
Conclusions
In conclusion, our study demonstrated the widest meta-analysis conducted on the prognostic importance of circulating exomiRs in GI cancers. Our results indicated that up- and down-regulated circulating exomiRs might serve as effective indicators of inferior survival outcomes in patients with gastrointestinal malignancies. In addition, exomiR dysregulation is related to advanced clinical stage, poor differentiation, and tumor spread in GI carcinomas. The current review advocates using a combined panel of circulating exomiRs for better risk stratification and clinical outcomes prediction in GI cancer patients, which could compensate for the unreliability of individual exomiRs in estimating prognosis. We envision our results would bring the attention of researchers and clinicians to the significance of deregulated circulating exomiRs as prognostic biomarkers that may aid better prediction.
Supplementary Information
Additional file 1: Figure S1. Forest plot of the association between NOS and overall survival (A), disease/relapse/progression-free survival (B).
Additional file 2: Figure S2. Forest plot of the association between exomiR-21 and overall survival (A), disease/relapse/progression-free survival (B), and clinicopathological characteristics in patients with GI cancers. Lymph node metastasis (C), Differentiation (D), Distant metastasis (E), TNM stage (F), Gender (G).
Additional file 3: Table S1. Search strategy of electronical databases.
Additional file 4: Table S2. Significantly dysregulated circulating exomiR(s) in GI cancer patients.
Additional file 5: Table S3. Summary of meta-analyses for the association between exomiRs deregulation and clinicopathologic features in patients with GI cancers.
Acknowledgements
Not applicable.
Author contributions
ZM, RGh, SB, MR, EGh, and FT contributed to the concept and study design. The search syntax was provided by KT. The search strategy was developed by EGh, and FT. Data screening and selecting phases were performed by EGh, FT, and MS. Quality assessment was performed by EGh, FT, and MR. Data extraction and preparing the draft of the manuscript were performed by EGh, FT, and MR. All extracted data were analyzed by MR. Moreover, WCSC critically revised and edited the manuscript. Besides, ZM, RG, and MR were responsible for reviewing the manuscript and editing the final manuscript. All authors read and approved the final manuscript.
Funding
The funding source is a college institute Iran University of Medical Sciences (IUMS) and has sponsored the project financially and approved (Grant Number #30805).
Availability of data and materials
All recorded data from the data extraction process were available on request to the extent that they were not included in the systematic review article.
Declarations
Ethics approval and consent to participate
As this systematic review was only based on published data already in the public domain, ethical approval is not required. The findings of this systematic review have been disseminated through an international peer-reviewed journal publication and several scientific conference presentations. To the best of our knowledge, there are no systematic reviews that have specifically looked at the frequency of exosome encapsulated miRNA(s) in the circulating blood of patients with GI cancers.
Consent for publication
Not applicable.
Competing interests
There are no competing interests that the authors declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Elmira Gheytanchi and Fatemeh Tajik contributed equally to this manuscript as co-first authors
Professor Zahra Madjd as the first corresponding author, Dr Roya Ghods as a second and Dr Mahdieh Razmi as a third corresponding authors have contributed to this work
Contributor Information
Mahdieh Razmi, Email: Razmi.mahdieh@ut.ac.ir, Email: Razmi_mahdyeh@yahoo.com.
Roya Ghods, Email: ghods.ro@iums.ac.ir, Email: rghods77@yahoo.com.
Zahra Madjd, Email: Zahra.madjd@yahoo.com, Email: majdjabari.z@iums.ac.ir.
References
- 1.Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159(1):335–49.e15. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76(2):182–188. doi: 10.1111/his.13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics 2022. CA A Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 4.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 5.Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159(1):335–49. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bijlsma MF, Sadanandam A, Tan P, Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2017;14(6):333–342. doi: 10.1038/nrgastro.2017.33. [DOI] [PubMed] [Google Scholar]
- 7.Abyadeh M, Meyfour A, Gupta V, Zabet Moghaddam M, Fitzhenry MJ, Shahbazian S, et al. Recent advances of functional proteomics in gastrointestinal cancers- a path towards the identification of candidate diagnostic, prognostic, and therapeutic molecular biomarkers. Int J Mol Sci. 2020;21:22. doi: 10.3390/ijms21228532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melton SD, Genta RM, Souza RF. Biomarkers and molecular diagnosis of gastrointestinal and pancreatic neoplasms. Nat Rev Gastroenterol Hepatol. 2010;7(11):620–628. doi: 10.1038/nrgastro.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto Y, Kikuchi D, Nagami Y, Nonaka K, Tsuji Y, Fujimoto A, et al. Management of adverse events related to endoscopic resection of upper gastrointestinal neoplasms: review of the literature and recommendations from experts. Dig Endosc. 2019;31:4–20. doi: 10.1111/den.13388. [DOI] [PubMed] [Google Scholar]
- 10.Alves dos Santos K, Clemente dos Santos IC, Santos Silva C, Gomes Ribeiro H, de Farias Domingos I, Nogueira Silbiger V. Circulating exosomal miRNAs as biomarkers for the diagnosis and prognosis of colorectal cancer. Int J Mol Sci. 2021;22(1):346. doi: 10.3390/ijms22010346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danac JMC, Uy AGG, Garcia RL. Exosomal microRNAs in colorectal cancer: overcoming barriers of the metastatic cascade. Int J Mol Med. 2021;47(6):1–16. doi: 10.3892/ijmm.2021.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francavilla A, Turoczi S, Tarallo S, Vodicka P, Pardini B, Naccarati A. Exosomal microRNAs and other non-coding RNAs as colorectal cancer biomarkers: a review. Mutagenesis. 2020;35(3):243–260. doi: 10.1093/mutage/gez038. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Jiang W, Gan Y, Zhou W. The application of exosomal microRNAs in the treatment of pancreatic cancer and its research progress. Pancreas. 2021;50(1):12–16. doi: 10.1097/MPA.0000000000001713. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, et al. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7(1):1–9. doi: 10.1038/s41598-017-04386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi T, Lombaert IM, Hauser BR, Patel VN, Hoffman MP. Exosomal MicroRNA transport from salivary mesenchyme regulates epithelial progenitor expansion during organogenesis. Dev Cell. 2017;40(1):95–103. doi: 10.1016/j.devcel.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer. 2020;147(10):2934–2947. doi: 10.1002/ijc.33111. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Zhou WB, Zhou J, Wei Y, Wang HM, Liu XD, et al. Circulating exosomal microRNAs as novel potential detection biomarkers in pancreatic cancer. Oncol Lett. 2020;20(2):1432–1440. doi: 10.3892/ol.2020.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Ling M, Xue J, Dai X, Sun Q, Chen C, et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics. 2018;8(19):5419–5433. doi: 10.7150/thno.27876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inamura K, Ishikawa Y. MicroRNA in lung cancer: novel biomarkers and potential tools for treatment. J Clin Med. 2016;5:3. doi: 10.3390/jcm5030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Z, Ni L, Yu W, Yu Z, Chen D, Zhang E, et al. MicroRNA-451a is associated with cell proliferation, migration and apoptosis in renal cell carcinoma. Mol Med Rep. 2015;11(3):2248–2254. doi: 10.3892/mmr.2014.2957. [DOI] [PubMed] [Google Scholar]
- 21.Mahabady MK, Mirzaei S, Saebfar H, Gholami MH, Zabolian A, Hushmandi K, et al. Noncoding RNAs and their therapeutics in paclitaxel chemotherapy: mechanisms of initiation, progression, and drug sensitivity. J Cell Physiol. 2022;237(5):2309–2344. doi: 10.1002/jcp.30751. [DOI] [PubMed] [Google Scholar]
- 22.Lamichhane SR, Thachil T, Gee H, Milic N. Circulating microRNAs as prognostic molecular biomarkers in human head and neck cancer: a systematic review and meta-analysis. Dis Markers. 2019 doi: 10.1155/2019/8632018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Guo H, Yang Y, Zhang Y, Liu H. A meta-analysis on the prognosis of exosomal miRNAs in all solid tumor patients. Medicine. 2019;98:16. doi: 10.1097/MD.0000000000015335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017;8(8):13048. doi: 10.18632/oncotarget.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi R, Li Y, Wang FL, Miao G, Qi RM, Zhao YY. MicroRNAs as diagnostic and prognostic biomarkers in colorectal cancer. World J Gastrointest Oncol. 2016;8(4):330–340. doi: 10.4251/wjgo.v8.i4.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan F, Qing Q, Pan Q, Hu M, Yu H, Yue X. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol. 2018;41(1):25–33. doi: 10.1007/s13402-017-0355-3. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119(6):1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 28.Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–538. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gheytanchi E, Madjd Z, Janani L, Rasti A, Ghods R, Atyabi F, et al. Exosomal microRNAs as potential circulating biomarkers in gastrointestinal tract cancers: a systematic review protocol. Syst Rev. 2017;6(1):1–6. doi: 10.1186/s13643-017-0624-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munn Z, Stern C, Aromataris E, Lockwood C, Jordan Z. What kind of systematic review should I conduct? a proposed typology and guidance for systematic reviewers in the medical and health sciences. BMC Med Res Methodol. 2018;18(1):5. doi: 10.1186/s12874-017-0468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ. 2021;372:n160. doi: 10.1136/bmj.n160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stang A. Critical evaluation of the newcastle-ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 33.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soeda N, Iinuma H, Suzuki Y, Tsukahara D, Midorikawa H, Igarashi Y, et al. Plasma exosome-encapsulated microRNA-21 and microRNA-92a are promising biomarkers for the prediction of peritoneal recurrence in patients with gastric cancer. Oncol Lett. 2019;18(5):4467–4480. doi: 10.3892/ol.2019.10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Liu X, Pan B, Hu X, Zhu Y, Su Y, et al. Serum exosomal miR-122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer. 2020;11(3):630–637. doi: 10.7150/jca.33022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, et al. Circulating exosomal miR-27a and miR-130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2018;27(7):746–754. doi: 10.1158/1055-9965.EPI-18-0067. [DOI] [PubMed] [Google Scholar]
- 38.Wu L, Zhou WB, Zhou J, Wei Y, Wang HM, Liu XD, et al. Circulating exosomal microRNAs as novel potential detection biomarkers in pancreatic cancer. Oncol Lett. 2020;20(2):1432–1440. doi: 10.3892/ol.2020.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, et al. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25(2):155–161. doi: 10.1002/jhbp.524. [DOI] [PubMed] [Google Scholar]
- 40.Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y. Circulating exosomal MicroRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology. 2017;92(6):360–370. doi: 10.1159/000463387. [DOI] [PubMed] [Google Scholar]
- 41.Yokota Y, Noda T, Okumura Y, Kobayashi S, Iwagami Y, Yamada D, et al. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci. 2021;112(3):1275–1288. doi: 10.1111/cas.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8(45):78598–78613. doi: 10.18632/oncotarget.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ, et al. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics. 2019;9(7):1965–1979. doi: 10.7150/thno.30958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei S, Peng L, Yang J, Sang H, Jin D, Li X, et al. Exosomal transfer of miR-15b-3p enhances tumorigenesis and malignant transformation through the DYNLT1/caspase-3/caspase-9 signaling pathway in gastric cancer. J Exp Clin Cancer Res. 2020;39(1):32. doi: 10.1186/s13046-019-1511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue XF, Zhao YB, Wang XN, Qin L, Hu RK. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120(1):135–142. doi: 10.1002/jcb.27165. [DOI] [PubMed] [Google Scholar]
- 46.Yagi T, Iinuma H, Hayama T, Matsuda K, Nozawa K, Tsukamoto M, et al. Plasma exosomal microRNA-125b as a monitoring biomarker of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients. Mol Clin Oncol. 2019;11(4):416–424. doi: 10.3892/mco.2019.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan SS, Jiang Y, Liang CH, Cheng M, Jin CW, Duan QH, et al. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J Cell Biochem. 2018;119(5):4113–4119. doi: 10.1002/jcb.26609. [DOI] [PubMed] [Google Scholar]
- 48.Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawamura S, Iinuma H, Wada K, Takahashi K, Minezaki S, Kainuma M, et al. Exosome-encapsulated microRNA-4525, microRNA-451a and microRNA-21 in portal vein blood is a high-sensitive liquid biomarker for the selection of high-risk pancreatic ductal adenocarcinoma patients. J Hepatobiliary Pancreat Sci. 2019;26(2):63–72. doi: 10.1002/jhbp.601. [DOI] [PubMed] [Google Scholar]
- 50.Abd El Gwad A, Matboli M, El-Tawdi A, Habib EK, Shehata H, Ibrahim D, et al. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J Cell Biochem. 2018;119(10):8600–10. doi: 10.1002/jcb.27109. [DOI] [PubMed] [Google Scholar]
- 51.Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, et al. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol. 2019;25(15):1890–1898. doi: 10.3748/wjg.v25.i15.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng C, She JJ, Chen XB, Zhang QC, Zhang X, Wang YS, et al. Exosomal miR-196a-1 promotes gastric cancer cell invasion and metastasis by targeting SFRP1. Nanomedicine. 2019;14(19):2579–2593. doi: 10.2217/nnm-2019-0053. [DOI] [PubMed] [Google Scholar]
- 53.Liu WB, Chen S, Liu B. Diagnostic and prognostic values of serum exosomal microRNA-21 in children with hepatoblastoma: a Chinese population-based study. Pediatr Surg Int. 2016;32(11):1059–1065. doi: 10.1007/s00383-016-3960-8. [DOI] [PubMed] [Google Scholar]
- 54.Cho HJ, Baek GO, Seo CW, Ahn HR, Sung S, Son JA, et al. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020;9(15):5459–5472. doi: 10.1002/cam4.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han JY, Sun W, Liu R, Zhou Z, Zhang HY, Chen X, et al. Plasma exosomal miRNA expression profile as oxaliplatin-based chemoresistant biomarkers in colorectal adenocarcinoma. Front Oncol. 2020 doi: 10.3389/fonc.2020.01495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Miguel PD, Rodriguez Martínez A, Ortigosa Palomo A, Delgado Ureña M, Garcia Puche JL, Robles Remacho A, et al. Extracellular vesicle-miRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci Rep. 2020;10(1):3974. doi: 10.1038/s41598-020-60212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu Z, Wu J, Wu J, Ji A, Qiang G, Jiang Y, et al. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(46):80666–80678. doi: 10.18632/oncotarget.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reese M, Flammang I, Yang Z, Dhayat SA. Potential of exosomal microRNA-200b as liquid biopsy marker in pancreatic ductal adenocarcinoma. Cancers. 2020;12:1. doi: 10.3390/cancers12010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meltzer S, Bjørnetrø T, Lyckander LG, Flatmark K, Dueland S, Samiappan R, et al. Circulating exosomal miR-141-3p and miR-375 in metastatic progression of rectal cancer. Transl Oncol. 2019;12(8):1038–1044. doi: 10.1016/j.tranon.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang YF, Huang FC, Xu N, Wang J, Li D, Yin L. Overexpression of serum extracellular vesicle micro-RNA-215–5p is associated with early tumor recurrence and poor prognosis of gastric cancer. Clinics. 2021 doi: 10.6061/clinics/2021/e2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu LY, Zhang SS, Chen SD, Wu HJ, Jiang MJ, Liu AQ. Exosomal miR-552-5p promotes tumorigenesis and disease progression via the PTEN/TOB1 axis in gastric cancer. J Cancer. 2022;13(3):890–905. doi: 10.7150/jca.66903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang JJ, Li X, Wei SC, Peng L, Sang HM, Jin DC, et al. Evaluation of the diagnostic potential of a plasma exosomal miRNAs panel for gastric cancer. Front Oncol. 2021 doi: 10.3389/fonc.2021.683465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen S, Mao Y, Chen W, Liu C, Wu H, Zhang J, et al. Serum exosomal miR-34a as a potential biomarker for the diagnosis and prognostic of hepatocellular carcinoma. J Cancer. 2022;13(5):1410–1417. doi: 10.7150/jca.57205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang CY, Tang SY, Shen D, Li XZ, Liang L, Ding YQ, et al. Circulating plasma exosomal miRNA profiles serve as potential metastasis-related biomarkers for hepatocellular carcinoma. Oncol Lett. 2021;21:2. doi: 10.3892/ol.2021.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hao YJ, Yang CY, Chen MH, Chang LW, Lin CP, Lo LC, et al. Potential values of circulating microRNA-21 to predict early recurrence in patients with colorectal cancer after treatments. J Clin Med. 2022;11:9. doi: 10.3390/jcm11092400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan S, Dang G, Zhang X, Jin C, Qin L, Wang Y, et al. Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget. 2017;8(42):72220–72226. doi: 10.18632/oncotarget.19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan SS, Liu GY, Jin CW, Wang ZF, Duan QH, Xu J, et al. MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-B signaling pathway in colorectal cancer. J Cell Physiol. 2018;233(9):6660–6668. doi: 10.1002/jcp.26316. [DOI] [PubMed] [Google Scholar]
- 68.Zou SL, Chen YL, Ge ZZ, Qu YY, Cao Y, Kang ZX. Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomarker. 2019;26(1):69–77. doi: 10.3233/CBM-190156. [DOI] [PubMed] [Google Scholar]
- 69.Zhang N, Zhang PP, Huang JJ, Wang ZY, Zhang ZH, Yuan JZ, et al. Reduced serum exosomal miR-874 expression predicts poor prognosis in colorectal cancer. Eur Rev Med Pharmacol Sci. 2020;24(2):664–672. doi: 10.26355/eurrev_202001_20043. [DOI] [PubMed] [Google Scholar]
- 70.Liu C, Eng C, Shen J, Lu Y, Takata Y, Mehdizadeh A, et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget. 2016;7(46):76250–76260. doi: 10.18632/oncotarget.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao X, Xin R, Dong W. Decreased serum exosomal miR-320a expression is an unfavorable prognostic factor in patients with hepatocellular carcinoma. J Int Med Res. 2020;48(4):300060519896144. doi: 10.1177/0300060519896144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiao CW, Jiao XH, Zhu AZ, Ge JT, Xu XQ. Exosomal miR-34s panel as potential novel diagnostic and prognostic biomarker in patients with hepatoblastoma. J Pediatr Surg. 2017;52(4):618–624. doi: 10.1016/j.jpedsurg.2016.09.070. [DOI] [PubMed] [Google Scholar]
- 73.Li W, Ding X, Wang S, Xu L, Yin T, Han S, et al. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J Clin Lab Anal. 2020;34(6):e23239. doi: 10.1002/jcla.23239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu WF, Hu J, Zhou KQ, Chen FY, Wang Z, Liao BY, et al. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843–3851. doi: 10.2147/OTT.S140062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheng LQ, Li JR, Qin H, Liu L, Zhang D, Zhang Q, et al. Blood exosomal micro ribonucleic acid profiling reveals the complexity of hepatocellular carcinoma and identifies potential biomarkers for differential diagnosis. World J Gastrointest Oncol. 2020;12(10):1195–1208. doi: 10.4251/wjgo.v12.i10.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suehiro T, Miyaaki H, Kanda Y, Shibata H, Honda T, Ozawa E, et al. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett. 2018;16(3):3267–3273. doi: 10.3892/ol.2018.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kumata Y, Iinuma H, Suzuki Y, Tsukahara D, Midorikawa H, Igarashi Y, et al. Exosome-encapsulated microRNA-23b as a minimally invasive liquid biomarker for the prediction of recurrence and prognosis of gastric cancer patients in each tumor stage. Oncol Rep. 2018;40(1):319–330. doi: 10.3892/or.2018.6418. [DOI] [PubMed] [Google Scholar]
- 78.Peng ZY, Gu RH, Yan B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J Cell Biochem. 2018 doi: 10.1002/jcb.27291. [DOI] [PubMed] [Google Scholar]
- 79.Zheng GD, Xu ZY, Hu C, Lv H, Xie HX, Huang T, et al. Exosomal miR-590-5p in serum as a biomarker for the diagnosis and prognosis of gastric cancer. Front Mol Biosci. 2021;8:636566. doi: 10.3389/fmolb.2021.636566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nik Mohamed Kamal N, Shahidan WNS. Non-exosomal and exosomal circulatory micrornas: which are more valid as biomarkers? Front Pharmacol. 2019;10:1500. doi: 10.3389/fphar.2019.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal. 2015;2015:657086. doi: 10.1155/2015/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zöller M. Pancreatic cancer diagnosis by free and exosomal miRNA. World J Gastrointest Pathophysiol. 2013;4(4):74–90. doi: 10.4291/wjgp.v4.i4.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219. doi: 10.1186/1477-7819-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119(6):1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 87.Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47(9):e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE. 2012;7(10):e46874. doi: 10.1371/journal.pone.0046874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tokunaga M, Kurokawa Y, Machida R, Sato Y, Takiguchi S, Doki Y, et al. Impact of postoperative complications on survival outcomes in patients with gastric cancer: exploratory analysis of a randomized controlled JCOG1001 trial. Gastric Cancer. 2021;24(1):214–223. doi: 10.1007/s10120-020-01102-3. [DOI] [PubMed] [Google Scholar]
- 90.Osseis M, Esposito F, Lim C, Doussot A, Lahat E, Fuentes L, et al. Impact of postoperative complications on long-term survival following surgery for T4 colorectal cancer. BMC Surg. 2018;18(1):87. doi: 10.1186/s12893-018-0419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kokelaar RF, Evans MD, Davies M, Harris DA, Beynon J. Locally advanced rectal cancer: management challenges. Onco Targets Ther. 2016;9:6265–6272. doi: 10.2147/OTT.S100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Apffelstaedt JP, Driscoll DL, Spellman JE, Velez AF, Gibbs JF, Karakousis CP. Complications and outcome of external hemipelvectomy in the management of pelvic tumors. Ann Surg Oncol. 1996;3(3):304–309. doi: 10.1007/BF02306287. [DOI] [PubMed] [Google Scholar]
- 93.Nacion AJD, Park YY, Kim NK. Contemporary management of locally advanced rectal cancer: resolving issues, controversies and shifting paradigms. Chin J Cancer Res. 2018;30(1):131–46. doi: 10.21147/j.issn.1000-9604.2018.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 95.Sun M, Song J, Zhou Z, Zhu R, Jin H, Ji Y, et al. Comparison of serum microRNA21 and Tumor markers in diagnosis of early non-small cell lung cancer. Dis Markers. 2016;2016:3823121. doi: 10.1155/2016/3823121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5(4):395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen J, Wang X. MicroRNA-21 in breast cancer: diagnostic and prognostic potential. Clin Transl Oncol. 2014;16(3):225–233. doi: 10.1007/s12094-013-1132-z. [DOI] [PubMed] [Google Scholar]
- 98.Qu J, Yang J, Chen M, Cui L, Wang T, Gao W, et al. MicroRNA-21 as a diagnostic marker for hepatocellular carcinoma: A systematic review and meta-analysis. Pak J Med Sci. 2019;35(5):1466–1471. doi: 10.12669/pjms.35.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Y, Xing QF, Liu XQ, Guo ZJ, Li CY, Sun G. MiR-122 targets VEGFC in bladder cancer to inhibit tumor growth and angiogenesis. Am J Transl Res. 2016;8(7):3056–3066. [PMC free article] [PubMed] [Google Scholar]
- 100.Li C, Sun J, Xiang Q, Liang Y, Zhao N, Zhang Z, et al. Prognostic role of microRNA-21 expression in gliomas: a meta-analysis. J Neurooncol. 2016;130(1):11–17. doi: 10.1007/s11060-016-2233-7. [DOI] [PubMed] [Google Scholar]
- 101.Chatterjee A, Jana S, Chatterjee S, Wastall LM, Mandal G, Nargis N, et al. MicroRNA-222 reprogrammed cancer-associated fibroblasts enhance growth and metastasis of breast cancer. Br J Cancer. 2019;121(8):679–689. doi: 10.1038/s41416-019-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang Y, Yu S, Cao S, Yin Y, Hong S, Guan H, et al. MicroRNA-222 promotes invasion and metastasis of papillary thyroid cancer through targeting protein phosphatase 2 regulatory subunit B alpha expression. Thyroid. 2018;28(9):1162–1173. doi: 10.1089/thy.2017.0665. [DOI] [PubMed] [Google Scholar]
- 103.Zhang R, Pang B, Xin T, Guo H, Xing Y, Xu S, et al. Plasma miR-221/222 family as novel descriptive and prognostic biomarkers for glioma. Mol Neurobiol. 2016;53(3):1452–1460. doi: 10.1007/s12035-014-9079-9. [DOI] [PubMed] [Google Scholar]
- 104.Ulivi P, Petracci E, Marisi G, Baglivo S, Chiari R, Billi M, et al. Prognostic role of circulating miRNAs in early-stage non-small cell lung cancer. J Clin Med. 2019;8:2. doi: 10.3390/jcm8020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi PW, Ng SW. The functions of microRNA-200 family in ovarian cancer: beyond epithelial-mesenchymal transition. Int J Mol Sci. 2017;18:6. doi: 10.3390/ijms18061207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Snowdon J, Zhang X, Childs T, Tron VA, Feilotter H. The microRNA-200 family is up-regulated in endometrial carcinoma. PLoS ONE. 2011;6(8):e22828. doi: 10.1371/journal.pone.0022828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thi Chung Duong T, Nguyen THN, Thi Ngoc Nguyen T, Huynh LH, Ngo HP, Thi Nguyen H. Diagnostic and prognostic value of miR-200 family in breast cancer: a meta-analysis and systematic review. Cancer Epidemiol. 2022;77:102097. doi: 10.1016/j.canep.2022.102097. [DOI] [PubMed] [Google Scholar]
- 108.Luo L, Yang R, Zhao S, Chen Y, Hong S, Wang K, et al. Decreased miR-320 expression is associated with breast cancer progression, cell migration, and invasiveness via targeting aquaporin 1. Acta Biochim Biophys Sin. 2018;50(5):473–480. doi: 10.1093/abbs/gmy023. [DOI] [PubMed] [Google Scholar]
- 109.Zhang L, Wang L, Dong D, Wang Z, Ji W, Yu M, et al. MiR-34b/c-5p and the neurokinin-1 receptor regulate breast cancer cell proliferation and apoptosis. Cell Prolif. 2019;52(1):e12527. doi: 10.1111/cpr.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tafsiri E, Darbouy M, Shadmehr MB, Zagryazhskaya A, Alizadeh J, Karimipoor M. Expression of miRNAs in non-small-cell lung carcinomas and their association with clinicopathological features. Tumour Biol. 2015;36(3):1603–1612. doi: 10.1007/s13277-014-2755-6. [DOI] [PubMed] [Google Scholar]
- 111.Juracek J, Stanik M, Vesela P, Radova L, Dolezel J, Svoboda M, et al. Tumor expression of miR-34a-3p is an independent predictor of recurrence in non-muscle-invasive bladder cancer and promising additional factor to improve predictive value of EORTC nomogram. Urol Oncol. 2019;37(3):184.e1–7. doi: 10.1016/j.urolonc.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 112.Rodríguez-González FG, Sieuwerts AM, Smid M, Look MP, Meijer-van Gelder ME, de Weerd V, et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2011;127(1):43–51. doi: 10.1007/s10549-010-0940-x. [DOI] [PubMed] [Google Scholar]
- 113.Han W, Cui H, Liang J, Su X. Role of microRNA-30c in cancer progression. J Cancer. 2020;11(9):2593–2601. doi: 10.7150/jca.38449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49(5):1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 115.Saleh AA, Soliman SE, Habib MSE, Gohar SF, Abo-Zeid GS. Potential value of circulatory microRNA122 gene expression as a prognostic and metastatic prediction marker for breast cancer. Mol Biol Rep. 2019;46(3):2809–2818. doi: 10.1007/s11033-019-04727-5. [DOI] [PubMed] [Google Scholar]
- 116.Fan X, Wu X. MicroRNA-122-5p promotes the development of non-small cell lung cancer via downregulating p53 and activating PI3K-AKT pathway. J buon. 2019;24(1):273–279. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Forest plot of the association between NOS and overall survival (A), disease/relapse/progression-free survival (B).
Additional file 2: Figure S2. Forest plot of the association between exomiR-21 and overall survival (A), disease/relapse/progression-free survival (B), and clinicopathological characteristics in patients with GI cancers. Lymph node metastasis (C), Differentiation (D), Distant metastasis (E), TNM stage (F), Gender (G).
Additional file 3: Table S1. Search strategy of electronical databases.
Additional file 4: Table S2. Significantly dysregulated circulating exomiR(s) in GI cancer patients.
Additional file 5: Table S3. Summary of meta-analyses for the association between exomiRs deregulation and clinicopathologic features in patients with GI cancers.
Data Availability Statement
All recorded data from the data extraction process were available on request to the extent that they were not included in the systematic review article.