Abstract
The potential utility of microRNAs (miRNAs) as diagnostic or prognostic biomarkers, as well as therapeutic targets, for chronic kidney disease (CKD) has been advocated. However, studies evaluating the expression profile of the same miRNA signatures in CKD report contradictory findings. This review aimed to characterize miRNAs associated with CKD and/or measures of kidney function and kidney damage in the general population, and also in high-risk subgroups, including people with hypertension (HTN), diabetes mellitus (DM) and human immunodeficiency virus (HIV) infection. Medline via PubMed, Scopus, Web of Science, and EBSCOhost databases were searched to identify relevant studies published in English or French languages on or before 30 September 2022. A total of 75 studies fulfilled the eligibility criteria: CKD (n = 18), diabetic kidney disease (DKD) (n = 51) and HTN-associated CKD (n = 6), with no study reporting on miRNA profiles in people with HIV-associated nephropathy. In individuals with CKD, miR-126 and miR-223 were consistently downregulated, whilst in DKD, miR-21 and miR-29b were consistently upregulated and miR-30e and let-7a were consistently downregulated in at least three studies. These findings suggest that these miRNAs may be involved in the pathogenesis of CKD and therefore invites further research to explore their clinical utility for CKD prevention and control.
Keywords: chronic kidney disease, diabetic kidney disease, hypertension-associated CKD, micro-RNAs
1. Introduction
The incidence of chronic kidney disease (CKD) is on the rise globally, and it is expected that CKD will be the fifth leading cause of death by 2045 [1]. This is partly attributable to the high burden of diabetes mellitus (DM) and hypertension (HTN), which are the leading causes of CKD, as well as other causes, including human immunodeficiency virus (HIV) infection and advanced age [2]. Chronic kidney disease is described as a silent condition due to a lack of obvious clinical symptoms, particularly in its early stages. As a result, most affected individuals are unaware of their disease status, and are often only detected at an advanced stage of the disease [3]. Furthermore, CKD is an independent risk factor for cardiovascular disease (CVD), and individuals with CKD are more likely to die of CVDs than progress to end-stage kidney disease (ESKD) [4]. Early diagnosis of CKD and effective screening of high-risk individuals is critical to mitigate disease progression and substantially reduce related poor health outcomes [5].
The indirect measurement of glomerular filtration rate (GFR) by clearance of exogenous filtration markers remains the reference standard method for determining kidney function. However, this method is complex, lengthy, expensive, invasive, and as such, not ideal for routine practice or research purposes [6]. As a result, endogenous filtration markers such as serum creatinine and cystatin C are used to estimate GFR (eGFR), and kidney function in clinical practice. However, serum creatinine can be affected by factors independent of glomerular filtration such as muscle mass, age and gender, whereas the measurement of cystatin C is complex, expensive and has not been standardized [7]. Furthermore, the predictive equations for eGFR are biased and imprecise, translating into overestimation of GFR and underdiagnosis of CKD, particularly in black Africans [8,9]. Although, albuminuria is a well-established marker of kidney damage used to define stages 1 and 2 of CKD where the level of GFR is above 60 mL/min/1.73 m2, it has limited predictive ability and specificity for early detection of CKD [10]. Kidney biopsies can be used to confirm a diagnosis, but this option comes with significant risk and possibility for complications, and therefore is not ideal for routine practice or research purposes [11]. Put together, these diagnostic challenges highlight the need for more accurate, minimally invasive, highly sensitive and specific, and readily available biomarkers that will improve the diagnosis/prognosis of CKD.
Research into microRNAs (miRNAs) as potential biomarkers of disease diagnosis and prognosis, as well as therapeutic targets, has gained traction over the last 10 years [12]. MiRNAs are a class of small non-coding RNAs, whose main function is regulating gene expression by degrading messenger RNA (mRNA) or inhibiting mRNA translation into functional proteins [13]. They play an important role in various cellular regulatory processes, such as differentiation, proliferation, development and apoptosis [13], and are also involved in the development and normal functioning of the kidneys [14]. Although they were initially considered to be intracellular gene regulators, emerging evidence suggests that a number of miRNAs are also detectable in biological fluids, such as urine, plasma, serum, and saliva in highly stable forms [15]. Previous studies found that these extracellular miRNAs presented unique patterns in pathological conditions and suggested that they may be utilized as potential diagnostic and prognostic biomarkers [16,17,18,19]. There has been a growing interest in exploring the role of extracellular miRNAs in the development and progression of CKD [19,20,21,22]. However, most findings describing miRNA expression in various biological fluids from CKD patients are inconsistent.
As such, the main purposes of this review were: (1) to identify all reported miRNAs associated with CKD and/or measures of kidney function and kidney damage in the general population, as well as in high-risk subgroups (HTN, DM and HIV-infected), and (2) to explore the specific expression patterns of the identified miRNAs in prevalent CKD. We also aimed to explore (3) whether the expression patterns of the identified miRNAs differed depending on the human sample type used and/or 4) whether the expression profile of the identified miRNAs differed depending on the stage of CKD.
2. Materials and Methods
2.1. Protocol and Registration
This review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The protocol for this systematic review was registered on the PROSPERO database (Registration No. CRD42021270028), and detailed methods outlining the steps followed in conducting the systematic review have been previously published [23].
2.2. Search Strategy
A comprehensive and systematic search of Medline via PubMed, Scopus, Web of Science, and EBSCOhost databases was conducted to identify eligible studies, published in English or French languages on or before 30 September 2022, without a starting date. The search strategy made use of keywords and phrases such as “microRNAs, miRNA, miRNAs, chronic kidney disease, CKD, chronic kidney injury, chronic renal disease, chronic renal injury, renal failure, end-stage renal disease, diabetic kidney disease, diabetic nephropathy, hypertensive nephrosclerosis, chronic kidney failure, chronic renal failure, end-stage renal failure, HIV-associated nephropathy, HIVAN, HIV-associated renal disease, HIV-associated kidney disease, serum creatinine, serum cystatin C, estimated glomerular filtration rate, urinary albumin excretion rate (UAER), urinary albumin-to-creatinine ratio (UACR), urinary albumin” in combination with Boolean operators (AND/OR/NOT) (refer to Additional files: Tables S1–S4). Furthermore, we manually scanned reference lists of the included studies for additional studies.
2.3. Data Collection
Two authors (DDM and DMM) independently conducted the database searches and screened studies by title, abstract and full text to identify those meeting the inclusion criteria, as shown in Figure 1. Disagreements encountered were resolved through discussions or consultation with a third author (CG). Studies were included if they: (i) were original articles reporting on miRNAs associated with prevalent CKD and/or measures of kidney function (serum creatinine, serum cystatin C, eGFR) or kidney damage (urinary albumin excretion rate, albumin-to-creatinine ratio, urinary protein) in the general adult population and/or high-risk subgroups (HTN, DM, HIV-infection), ii) written in English and French languages, (iii) clearly described the type of sample in which miRNA analysis was done, methods used for miRNA detection and quantification, as well as the normalization control used, (iv) with clearly defined cases and controls. Studies were excluded if they were: (i) conducted in animal or cell models, (ii) qualitative in nature (reviews, case reports, newspaper articles, editorials, commentaries, book chapters), or (iii) pre-prints or unpublished research.
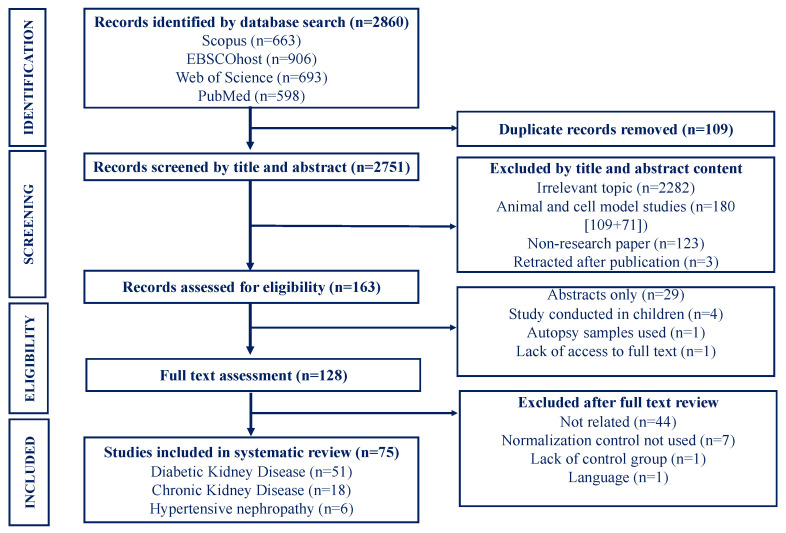
Figure 1.
Selection process for studies included.
2.4. Data Extraction, Assessment, and Synthesis
The following data were independently extracted by two reviewers (DDM and DMM) from the eligible studies: publication details (first author, year of publication, country); study details [design, sample size, demographics (age, sex)]; disease outcome [CKD (of unspecified cause), diabetic kidney disease (DKD)/HTN-associated CKD/HIV-associated nephropathy (HIVAN) or ESKD]; population [general or high-risk subgroups (HTN, DM and HIV)]; participant clinical characteristics [body mass index (BMI), C-reactive protein (CRP), smoking status, alcohol consumption, lipid profile (low-density lipoprotein (LDL), high-density lipoprotein (HDL), triglycerides and total cholesterol)]; clinical outcomes (diagnostic criteria, classification/staging, medication status); CKD diagnostic criteria (eGFR or proteinuria/albuminuria and eGFR equation used); miRNA analysis [sample type, molecular techniques, inclusion of screening and validation cohorts, expression pattern (upregulated or downregulated) and normalization control used]. Any inconsistencies or disagreements were resolved by discussions or consultation with a third author (CG). Furthermore, we assessed the quality of studies using the Newcastle-Ottawa Quality Assessment Scale for observational studies (NOS) tool [24]. The assessment was done based on a critical appraisal of three domains, namely: (i) participant group selection, (ii) how comparable the groups are, and (iii) determination of the exposure of interest. The quality of evidence was then assessed using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) framework [25]. Given that very few studies investigated the association of the same miRNA with CKD risk or markers of kidney function or damage in the same sample type, making use of different normalization controls and miRNA quantification techniques as well as variabilities in disease outcome measures, and attempting to pool studies was meaningless. We, therefore, instead opted for a narrative synthesis of evidence.
3. Results
3.1. Search Results
We obtained a total of 2860 related citations (663 from Scopus, 906 from EBSCOhost, 693 from Web of Science and 598 from PubMed) from database searches. Of these, 2732 citations were excluded for various reasons (Figure 1). The remaining 128 articles were further assessed for eligibility and 53 studies were subsequently excluded from the review because they were irrelevant to our review (n = 44); they did not report on the quantitative reverse transcription polymerase chain reaction (RT-qPCR) normalization controls used (n = 7), did not include a control group (n = 1) and did not meet our reporting language restrictions (n = 1). Ultimately, 75 studies fulfilled the eligibility criteria and were retained for the systematic review (Figure 1). The eligible studies were classified according to disease outcome, as follows: CKD (n = 18), DKD (n = 51) and HTN-associated CKD (6). Database searches did not return studies reporting on miRNA profiles in humans with HIVAN.
3.2. Characteristics of Included Studies
Table 1, Table 2 and Table 3 detail the main characteristics of studies that were included in the systematic review according to disease outcome. Table 1 summarizes the 18 studies that quantified miRNA expression patterns in CKD compared to controls, whilst Table 2 is a summary of the 51 studies that quantified miRNA expression patterns in DKD relative to controls. A summary of the six studies that quantified miRNA expression patterns in individuals with HTN-associated CKD compared to controls is shown in Table 3. All 75 studies were published between 2013 and 2022, and from diverse geographical locations, including China (n = 26), the United States of America (n = 6), Spain (n = 4), Egypt (n = 6), South Africa (n = 1), Germany (n = 3), Italy (n = 2), Austria (n = 2), Japan (n = 3), Iran (n = 3), Belgium (n = 1), Sweden (n = 1), Turkey (n = 1), Poland (n = 1), Bahrain (n = 2), Brazil (n = 2), United Kingdom (n = 1), India (n = 2), Romania (n = 2), France (n = 1), Canada (n = 1), Ireland (n = 1), Republic of Korea (n = 1), Netherlands (n = 1) and Malaysia (n = 1). The design of most studies was either case-control or cross-sectional, with study participant numbers ranging between 28 to 1385 in CKD, 11 to 1018 in DKD and 30 to 150 in HTN-associated CKD. The included studies used varying diagnostic methods to classify kidney disease, with 20% of the studies using only eGFR to classify CKD, 42% defined CKD by the level of albuminuria alone, whilst 34% of the studies used both albuminuria and eGFR to classify CKD. The remaining 4% of studies were not clear on the methods used for CKD classification. For estimation of GFR, most studies used the Modification of Diet in Renal Disease Study (MDRD) equation [26] (44% of the studies) and Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation [27] (38% of the studies). Of those included, 26% of the studies further validated their findings in a separate cohort [CKD (n = 3), DKD (n = 14) and HTN with CKD (n = 1)].
Table 1.
Characteristics of studies evaluating microRNA expression patterns in chronic kidney disease.
| Study | Country | Study Population [Cases] | Study Population [Control] | Quantification Method | Sample Type | microRNAs | Upregulated | Downregulated |
|---|---|---|---|---|---|---|---|---|
| Carmona, 2020 [28] | Spain | 45 | 10 | RT-qPCR | Serum | miR-126-3p, miR-191-5p, miR-223-3p, miR-363-3p, miR-495-3p | - | miR-126-3p, miR-191-5p, miR-223-3p |
| Chen, 2013 [29] | United States of America | 110 | 8 | RT-qPCR | Serum | miR-125b, miR-145, miR-155 | - | miR-125b, miR-145, miR-155 |
| Donderski, 2021 [30] | Poland | 45 | 17 | RT-qPCR | urine, serum | miR-155-5p, miR-214-3p, miR-200a-5p, miR-29a-5p, miR-21-5p, miR-93-5p, miR-196a-5p | Urine—miR-29-5p, miR-21-5p, miR-196a-5p. Serum—miR-155-5p, miR-214-3p and miR-200a-5p |
Urine—miR-155-5p, miR-214-5p, miR-200a-5p, miR-93-5p |
| Eckersten, 2017 [31] | Sweden | 30 | 18 | RT-qPCR | Serum | miR-155 | miR-155 | - |
| Fourdinier, 2019 [21] | Belgium | 601 | 31 | RT-qPCR | Serum | miR-223, miR-126 | - | miR-223, miR-126 |
| Fujii, 2019 [22] | Japan | 395 | 118 | RT-qPCR | Serum | miR-17, miR-21, miR-150 | - | - |
| Fujii, 2019 [32] | Japan | 229 | 1156 | RT-qPCR | Serum | miR-126, miR-197, miR-223 | - | miR-126, miR-197, miR-223 |
| Fujii, 2021 [33] | Japan | 29 | 140 | RT-qPCR | Serum | miR-126, miR-197, miR-21, miR-150, miR-17 |
- | - |
| Lange, 2019 [34] | Germany | 41 | 5 | RT-qPCR | urine exosomes | miR-21-5p, miR-30a-5p, miR-92a-3p | miR-21 | - |
| Li, 2020 [35] | China | 116 | 127 | RT-qPCR | Serum | miR-155 | miR-155 | - |
| Liu, 2020 [36] | China | 110 | 35 | NGS, RT-qPCR | serum | miR-483-5p, miR-363-3p | miR-483-5p | miR-363-3p |
| Motshwari, 2021 [37] | South Africa | 171 | 740 | NGS, RT-qPCR | whole blood | miR-novel- chr1_36178, miR-novel-chr2_55842, miR-novel-chr7_76196, miR-novel-chr5_67265, miR-novel-chr13_13519, and miR-novel-chr15_18383 | All novel miRNAs | - |
| Muralidharan, 2017 [38] | United States of America | 19 | 9 | Microarray, RT-qPCR | plasma and urine exosomes | Urine—miR-1281, miR-1825, miR-130a-3p, let-7a-5p Plasma—miR-1825p miR-1281, miR-423 |
Urine—miR-1825, miR-1281. Plasma—miR-1825, miR-1281, miR-144-5p, miR-548ap-5p | Urine—miR-4525. Plasma—miR-423-5p, miR-3648 |
| Rudnicki, 2016 [39] | Austria | 20 | 52 | RT-qPCR | Kidney biopsy | miR-30d, miR-140-3p, miR-532-3p, miR-194, miR-190, miR-204, miR-206 | miR-206, miR-532-3 | - |
| Sayilar, 2016 [40] | Turkey | 30 | 15 | RT-qPCR | plasma, urine | miR-21, miR-124, miR-192, miR-195, miR-451 | Urine—miR-124 Plasma—miR-195, miR-451 | Urine—miR-195, miR-451 |
| Shang, 2017 [41] | China | 208 | 37 | RT-qPCR | serum | miR-92a, miR-126, miR-155, miR-483 | miR-92a | - |
| Trojanowicz, 2019 [42] | Germany | 48 | 23 | RT-qPCR | serum | miR-421 | miR-421 | - |
| Ulbing, 2017 [43] | Austria | 137 | 36 | RT-qPCR | serum | miR-223-3p, miR-93-5p, miR-142-3p, miR-146a-5p | - | miR-223-3p, miR-93-5p, miR-142-3p |
Table 2.
Characteristics of studies evaluating microRNA expression patterns in diabetic kidney disease.
| Study | Country | Study Population (n) | Quantification Method | microRNAs | Sample Type | Upregulated | Downregulated | ||
|---|---|---|---|---|---|---|---|---|---|
| Healthy | Normoalbuminuria | Diabetic Kidney Disease | |||||||
| Abdelsalam, 2020 [44] | Egypt | 30 | 30 | 60 | RT-qPCR | miR-451 | plasma | miR-451 | - |
| urine | - | miR-451 | |||||||
| Abdou, 2022 [45] | Egypt | 20 | 20 | 40 | RT-qPCR | miR-152-3p | serum | miR-152-3p | - |
| Akhbari, 2018 [46] | Iran | 22 | 21 | 40 | RT-qPCR | miR-93 | serum | - | miR-93 |
| Akhbari, 2019 [47] | Iran | 22 | - | 61 | RT-qPCR | miR-155 | cell-free serum | - | miR-155 |
| Al-kafaji, 2016 [48] | Bahrain | 50 | 52 | 50 | RT-qPCR | miR-126 | peripheral whole blood | - | miR-126 |
| Al-kafaji, 2018 [49] | Bahrain | 30 | 30 | 25 | RT-qPCR | miR-377, miR-192 | whole blood | miR-377 | miR-192 |
| Argyropoulos, 2013 [50] | United States of America | - | 10 | 30 | RT-qPCR | 27 microRNAs | urine | miR-214-3p, miR-92b-5p, miR-765, miR-429, miR-373-5p, miR-1913, miR-638 | miR-323b-5p, miR-221-3p, miR-524-5p, miR-188-3p |
| Assmann, 2019 [51] | Brazil | 20 | 33 | 54 | RT-qPCR | miR-16-5p, miR-21-3p, miR-29a-3p, miR-378a-5p, miR-503-5p | plasma | miR-21-3p, miR-378a-5p | miR-16-5p, miR-29a-3p |
| Barutta, 2013 [52] | Italy | 10 | 12 | 12 | RT-qPCR | miR-130a, miR-424, miR-155, miR-145 | urine exosomes | miR-145, miR-130a | miR-424, miR-155 |
| Beltrami, 2018 [53] | United Kingdom | 61 | 62 | 109 | MicroRNA array, RT-qPCR | miR-126-3p, miR-155-5p, miR-29b-3p | urine | miR-126-3p, miR-155-5p, miR-29b-3p | - |
| Cardenas-Gonzalez, 2017 [54] | United States of America | 93 | 71 | 132 | RT-qPCR, miRNA in situ hybridization | miR-1915-3p, miR-2861, miR-4532, miR-4536-3p, miR-6747-3p | urine | miR-4536-3p, miR-6747-3p | miR-1915-3p, miR-2861, miR-4532 |
| Conserva, 2019 [55] | Italy | 20 | - | 37 | Microarray, RT-qPCR | miR-27b-3p, miR-1228-3p | kidney biopsy, cell-free urine | - | miR-27b-3p, miR-1228-3p |
| Delić, 2016 [56] | Germany | 14 | 14 | 13 | Microarray, RT-qPCR | miR-320c, miR-6068 | urine exosomes | miR-320c, miR-6068 | - |
| Dieter, 2019 [57] | Brazil | - | 17 | 23 | RT-qPCR | miR-15a-5p, miR-30e-5p | plasma | - | miR-30e-5p |
| urine | - | miR-30e-5p | |||||||
| Eissa, 2016 [58] | Egypt | 56 | 60 | 116 | MicroRNA array, RT-qPCR | miR-15b, miR-34a, miR-636 | urine pellets, exosomes | miR-15b, miR-34a, miR-636 | - |
| Eissa, 2016b [59] | Egypt | 54 | 56 | 110 | RT-qPCR | miR-133b, miR-342, miR-30a | urine exosomes | miR-133b, miR-342, miR-30a | - |
| Florijn, 2019 [60] | Netherlands | 12 | - | 33 | RT-qPCR | miR-1, miR-21, miR-29a, miR-126, miR-132, miR-145, miR-152, miR-212, miR-223, miR-574, miR-660 | plasma endothelial vesicles | miR-21, miR-126 | - |
| Plasma | miR-126 | ||||||||
| high density lipoprotein fraction | - | miR-132 | |||||||
| Apo-2 | miR-126, miR-145, miR-660 | - | |||||||
| Fouad, 2020 [61] | Egypt | 100 | 120 | 120 | RT-qPCR | miR-21 | plasma | miR-21 | - |
| Guo, 2017 [62] | China | 45 | 33 | 42 | Microarray, RT-qPCR | miR-29c | plasma | miR-29c | - |
| urine | - | miR-29c | |||||||
| kidney tissue | - | miR-29c | |||||||
| Han, 2021 [63] | China | - | 5 | 6 | Microarray, RT-qPCR | miR-95-3p, miR-185-5p, miR-1246, miR-631 | urine sediment | miR-95-3p, miR-185-5p, miR-1246, miR-631 | - |
| He, 2014 [64] | China | 6 | - | 6 | Microarray hybridisation, RT-qPCR | miR-15a, miR-17, miR-21, miR-30b, miR-126, miR-135a, miR-192, miR-377, miR-34a, miR-194-1, miR-205, miR-215 | serum | miR-15a, miR-17, miR-21, miR-30b, miR-126, miR-135a, miR-192, miR-377 | miR-34a, miR-194-1, miR-205, miR-215 |
| kidney tissue | miR-135a | - | |||||||
| Hong, 2021 [65] | China | 36 | 36 | 51 | Microarray, RT-qPCR | miR-193a-3p, miR-320c, miR-27a-3p | plasma | miR-193a-3p, miR-320c | - |
| Jia, 2016 [66] | China | 10 | 30 | 50 | RT-qPCR | miR-192, miR-194, miR-215 | urine extracellular vesicles | miR-192, miR-194, miR-215 | - |
| Khokhar, 2021 [67] | India | 36 | 38 | 35 | RT-qPCR | miR-21-5p | whole blood | miR-21-5p | - |
| Lin, 2021 [68] | China | 30 | 36 | 32 | RT-qPCR | miR-638 | serum | miR-638 | |
| Liu, 2021 [69] | China | 180 | 64 | 116 | RT-qPCR | miR-29a | serum | miR-29a | |
| Ma, 2016 [70] | China | 127 | 157 | 307 | RT-qPCR | miR-192 | serum | - | miR-192 |
| Milas, 2018 [71] | Romania | 11 | 26 | 42 | RT-qPCR | miR-21, miR-124, miR-192 | urine | miR-21, miR-124 | miR-192 |
| Monjezi, 2022 [72] | Iran | 30 | 31 | RT-qPCR | miR-124-3p | peripheral blood mononuclear cells | miR-124-3p | ||
| Motawi, 2018 [73] | Egypt | 25 | 25 | 25 | RT-qPCR | miR-130b | serum | - | miR-130b |
| Park, 2022 [74] | Republic of Korea | 7 | - | 12 | NGS | miR-320b, miR-30d-5p, miR-30e-3p, miR-30c-5p, miR-190a-5p, miR-29c-5p, miR-98-3p, miR-331-3p, let-7a-3p, miR-106b-3p, miR-30b-5p, miR-99b-5p, let-7f-1-3p | plasma and urine extracellular vesicles | miR-320b | miR-30d-5p, miR-30e-3p, miR-30c-5p, miR-190a-5p, miR-29c-5p, miR-98-3p, miR-331-3p, let-7a-3p, miR-106b-3p, miR-30b-5p, miR-99b-5p, let-7f-1-3p |
| Peng, 2013 [75] | China | - | 41 | 42 | RT-qPCR | miR-29a, miR-29b, miR-29c | urine supernatant | miR-29a | - |
| Petrica, 2019 [76] | Romania | 11 | 36 | 81 | RT-qPCR | miR-125a, miR-126, miR-146a, miR-21p, miR-124, miR-192 | serum | miR-192, miR-21p | miR-124, miR-125a, miR-126, miR-146 |
| urine | miR-21p, miR-124, miR-125a, miR-126 | miR-192, miR-146a | |||||||
| Pezzolesi, 2015 [77] | United States of America | - | 40 | 76 | RT-qPCR | let-7b-5p, let-7c-5p, miR-21-5p, miR-29a-3p, miR-29c-3p | plasma | let-7b-5p, miR-21-5p | let-7c-5p, miR-29a-3p |
| Prabu, 2019 [78] | India | 40 | 40 | 80 | RT-qPCR | let-7i-5p, miR-135b-5p, miR-15b-3p, miR-197-3p, miR-24-3p, miR-27b-3p | urine exosomes | let-7i-5p, miR-24-3p, miR-27b-3p, miR-30a-5p | miR-15b-3p |
| Regmi, 2019 [79] | China | 25 | 50 | 42 | RT-qPCR | miR-20a, miR-99b, miR-122-5p, miR-486-5p | serum | miR-99b, miR-122 | miR-20a, miR-486 |
| Ren, 2019 [80] | China | 280 | 273 | 465 | RT-qPCR | miR-154-5p | serum | miR-154-5p | - |
| Ren, 2020 [81] | China | - | 136 | 254 | RT-qPCR | miR-154-5p | serum | miR-154-5p | - |
| Roux, 2018 [82] | France | - | 73 | 73 | NGS, RT-qPCR | miR-362-5p, miR-152-3p, miR-196b-5p, miR-140-3p | plasma | miR-152-3p | - |
| Rovira-Llopis, 2018 [83] | Spain | 24 | 13 | 13 | RT-qPCR | miR-31 | serum | - | miR-31 |
| Shao, 2017 [84] | China | 195 | 186 | 309 | RT-qPCR | miR-217 | serum | miR-217 | - |
| Sham, 2022 [85] | Malaysia | - | 15 | 26 | miS-cript miRNA qPCR array, RT-qPCR | miR-874-3p, miR-101-3p, miR-145-5p | serum | miR-874-3p, miR-101-3p | |
| Su, 2020 [86] | China | 20 | - | 20 | MicroRNA array, RT-qPCR | miR-140-5p | peripheral blood, kidney tissue | - | miR-140-5p |
| Wang, 2019 [19] | China | 40 | 40 | 66 | MicroRNA array, qPCR | miR-27a-3p, miR-30e, miR-33b, miR-50, miR-125b-5p, miR-150-5p, miR-155-5p, miR-296, miR-320e, miR-328, miR-484, miR-487, miR-550a-5p, miR-590-5p, miR-744, miR-885-5p, miR-933. miR-3196, let-7a-5p, let-7c-5p | plasma | miR-125b-5p, miR- 484, miR-550 | miR-30e, miR-155-5p, miR-320, let-7a-5p, miR-150-5p, miR-3196 |
| Xiao, 2017 [87] | China | 35 | - | 140 | Real time PCR | miR-9 | serum | miR-9 | - |
| Xie, 2017 [88] | China | - | 35 | 5 | MicroRNA array, qPCR | miR-362-3p, miR-877-3p, miR-15a-5p, miR-150-5p |
urine exosomes | miR-362-3p, miR-877-3p, miR-150-5p | miR-15a-5p |
| Zang, 2019 [89] | Ireland | 18 | 30 | 36 | MicroRNA arrays, RT-PCR | miR-21-5p, let-7e-5p, miR-23b-3p, miR-30b-5p, miR-125b-5p | urine sediment exosome | miR-21-5p | miR-30b-5p |
| Zhang, 2017 [90] | China | 28 | 30 | 27 | Microarray, qPCR | miR-223-3p, miR-106b-5p, miR-103a-3p, miR-126-3p, miR-27a-3p, miR-29a-3p, miR-29c-3p, miR-425-5p, miR-93-5p, miR-1249-5p, miR-2276-3p, miR-1225-5p, miR-345-3p, miR-3679-5p, miR-4281, miR-4442 | plasma | - | miR-223-3p |
| Zhang, 2020 [91] | China | - | 30 | 30 | RT-qPCR | miR-135a-5p | serum | miR-135a-5p | - |
| Zhao, 2020 [92] | China | - | 17 | 17 | MicroRNA arrays, qRT-PCR | miR-4491, miR-2117, miR-4507, miR-5088-5p, miR-1587, miR-219a-3p, miR-5091, miR-498, miR-4687-3p, miR-516b-5p, mir-4534, miR-1275, miR-5007-3p, miR-4516 | urine exosomes | miR-4687-3p, miR-4534, miR-5007-3p | - |
| Zhou, 2013 [93] | China | 62 | 104 | 108 | MicroRNA microarrays, real time RT-PCR | let-7a, let-7d, let-7f, miR-4429, miR-363 | whole blood | - | let-7a |
Table 3.
Characteristics of studies evaluating microRNA expression patterns in hypertension-associated chronic kidney disease.
| Study | Country | Study Population | Quantification Method | Sample Type | microRNAs | Upregulated | Downregulated | ||
|---|---|---|---|---|---|---|---|---|---|
| Healthy | Hypertensive | Hypertensive CKD | |||||||
| Berillo, 2020 [94] | Canada | 15 | 31 | 16 | Hi-seq, RT-qPCR | platelet-poor plasma | let-7g-5p, miR-191-5p | - | let-7g-5p, miR-191-5p |
| Huang, 2018 [95] | China | 0 | 50 | 100 | RT-qPCR | plasma | miR-29a | miR-29a | - |
| Huang, 2020 [96] | China | 0 | 50 | 100 | RT-qPCR | plasma | miR-29b | miR-29b | - |
| Nandakumar, 2017 [97] | United States of America | - | 15 | 15 | NGS | whole blood | miR-17-5p, miR-130a-3p, miR-15b-5p, miR-106b-3p, miR-106a-5p, miR-16-5p, miR-181a-5p, miR-1285-3p, miR-15a-5p, miR-29c-5p, miR-345-5p, miR-142-3p, miR-339-3p, miR-210-3p | - | miR-17-5p, miR-15a-5p, miR-15b-5p, miR-16-5 |
| Perez-Hernandez, 2018 [98] | Spain | 20 | 28 | 24 | NGS, RT-qPCR | Urinary exosomes | miR-146a and miR-335 | - | miR-146a |
| Perez-Hernandez, 2021 [99] | Spain | 15 | 56 | 61 | NGS, RT-qPCR | plasma and urine exosomes | miR-143-3p, miR-126-3p, miR-26a-5p, miR-144-3p, miR-191-5p, miR-220a-3p, miR-222-3p, miR-423-5p |
Plasma exosome—miR-191-5p | Plasma exosome—miR-222-3p, miR-26a-5p, miR-126-3p |
Of the included studies, 29% performed initial miRNA expression discovery using next-generation sequencing (NGS) techniques [CKD (n = 2); DKD (n = 2), HTN with CKD (n = 4)] and microarrays [CKD (n = 1); DKD (n = 15)], followed by a validation step using a PCR-based technique. PCR-based techniques were used for the quantification of miRNAs in instances where the miRNA was already identified by NGS and microarrays in previous studies. A wide range of normalization techniques were employed by the studies included in this review, with 31% using an exogenous spike-in control containing non-mammalian synthetic miRNAs such as Caenorhabditis elegans-miR-39 (cel-miR-39) (n = 22) and 52% of the studies used endogenous controls such as small non-human ubiquitous miRNA (U6) (n = 28) or miR-16 (n = 8). Of the included studies, 15% of the studies used more than one normalization control [CKD (n = 4), DKD (n = 5) and in HTN-associated CKD (n = 2)]. The sample types in which miRNA expression levels were determined varied widely across studies, with 36% conducted in serum [CKD (n = 13) and DKD (n = 17)], 21% in plasma [CKD (n = 2), DKD (n = 10) and HTN-associated CKD (n = 4)], 1% in plasma endothelial vesicle [DKD (n = 1)], 8% in whole blood [CKD (n = 1), DKD (n = 4), and HTN-associated CKD (n = 1)], 19% in urine [CKD (n = 2), and DKD (n = 12)], 19% in urinary exosomes [CKD (n = 2), DKD (n = 8) and HTN-associated CKD (n = 2)], 7% in kidney tissue biopsy [CKD (n = 1) and DKD (n = 4)] and 1% peripheral blood mononuclear cells [DKD (n = 1)]. Only 17% of the studies quantified miRNA expression in two or more sample types, and this included two studies in CKD, 10 in DKD and one in HTN-associated CKD.
The expression patterns of 288 miRNAs were investigated across the 75 studies included in this review. Of the 288 miRNAs, 67 miRNAs were evaluated in populations with prevalent CKD, with 53 miRNAs found to be dysregulated (25 downregulated and 28 upregulated). Of the 193 miRNAs evaluated in populations with DKD, 155 miRNAs were found to be dysregulated (67 downregulated and 88 upregulated), whilst 13 (10 downregulated and 3 upregulated) of the 28 miRNAs evaluated in populations with HTN-associated CKD were dysregulated. The dysregulation discussed below refers only to miRNAs evaluated in three or more studies, of which HTN-associated CKD had none.
3.3. Dysregulated miRNAs in CKD
Of the 53 differentially expressed miRNAs in individuals with CKD, four miRNAs (miR-126, miR-223, miR-155 and miR-21) were reported in at least three studies (Table 4). Of these, serum miR-126 [21,28,32] and serum miR-223 [21,28,32,43] were consistently downregulated in individuals with CKD across studies. Inconsistent expression patterns were observed for miR-155, two studies found the miRNA to be downregulated in serum and urine, [29,30] and two studies found it upregulated in serum [30,31] in CKD, with one study reporting no difference [41] in the expression of miR-155 in both urine and serum, in individuals with and without CKD. Of the studies reporting on the expression pattern of miR-21 in CKD, one study showed a downregulation in this miRNA in serum sample [30], whilst two studies evaluated miR-21 in urine and urine exosomes and found it upregulated [30,34], and one study found no difference in urine and plasma samples between the case and control groups [40].
Table 4.
MicroRNAs reported in at least three studies in chronic kidney disease subtypes.
| MicroRNA | Study | Sample Type | Expression Pattern |
|---|---|---|---|
| CHRONIC KIDNEY DISEASE | |||
| miR-126 | Carmona, 2020 [28] | Serum | Downregulated |
| Fourdinier, 2019 [21] | Serum | Downregulated | |
| Fujii, 2019b [32] | Serum | Downregulated | |
| Shang, 2017 [41] | Serum, urine | No difference | |
| miR-223 | Carmona, 2020 [28] | Serum | Downregulated |
| Fourdinier, 2019 [21] | Serum | Downregulated | |
| Fujii, 2019b [32] | Serum | Downregulated | |
| Ulbing, 2017 [43] | Serum | Downregulated | |
| miR-155 | Chen, 2013 [29] | Serum | Downregulated |
| Donderski, 2021 [30] | Urine | Downregulated | |
| Serum | Upregulated | ||
| Eckersten, 2017 [31] | Serum | Upregulated | |
| Shang, 2017 [41] | Serum, urine | No difference | |
| miR-21 | Donderski, 2021 [30] | Urine | Upregulated |
| Serum | Downregulated | ||
| Lange, 2019 [34] | Urine exosomes | Upregulated | |
| Sayilar, 2016 [40] | Urine, plasma | No difference | |
| DIABETIC KIDNEY DISEASE | |||
| miR-155 | Akhbari, 2019 [47] | Cell-free serum | Downregulated |
| Barutta, 2013 [49] | Urinary exosomes | Downregulated in microalbuminuria | |
| Beltrami, 2018 [53] | Urine | Upregulated | |
| Wang, 2019 [19] | Plasma | Downregulated | |
| miR-126 | Al-kafaji, 2016 [48] | Whole blood | Downregulated |
| Beltrami, 2018 [53] | Urine | Upregulated | |
| Florijn, 2019 [60] | Plasma exosomal vesicles | Upregulated | |
| Plasma | Upregulated | ||
| Plasma Ago | Upregulated | ||
| Petrica, 2019 [76] | Urine | Upregulated | |
| Serum | Downregulated | ||
| He, 2014 [64] | Serum | Upregulated | |
| miR-192 | Al-kafaji, 2018 [49] | Whole blood | Downregulated |
| Jia, 2016 [66] | Urine extracellular vesicles | Upregulated in microalbuminuria and downregulated in macro albuminuria | |
| Ma, 2016 [70] | Serum | Downregulated | |
| Milas, 2018 [71] | Urine | Downregulated | |
| Petrica, 2019 [76] | Urine | Upregulated | |
| Serum | Upregulated | ||
| He, 2014 [64] | Serum | Upregulated | |
| miR-21 | Assmann, 2019 [51] | Plasma | Upregulated in macroalbuminuria |
| Florijn, 2019 [60] | Plasma exosomal vesicles | Upregulated | |
| Plasma | No difference | ||
| Fouad, 2020 [61] | Plasma | Upregulated | |
| Khokhar, 2021 [67] | Whole blood | Upregulated | |
| Milas, 2018 [71] | Urine | Upregulated | |
| Petrica, 2019 [76] | Serum | Upregulated | |
| Urine | Upregulated | ||
| Pezzolesi, 2015 [77] | Plasma | Upregulated in rapid progressors to ESKD | |
| Zang, 2019 [89] | Urinary exosomes | Upregulated | |
| He, 2014 [64] | Serum | Upregulated | |
| miR-29b | Beltrami, 2018 [53] | Urine | Upregulated |
| Peng, 2013 [75] | Urine supernatant | No difference | |
| Argyropoulos, 2013 [50] | Urine | Upregulated | |
| miR-15a-5p | He, 2014 [64] | Serum | Upregulated |
| Xie, 2017 [88] | Urinary exosomes | No difference | |
| Dieter, 2019 [57] | Urine and plasma | No difference | |
| miR-29a | Assmann, 2019 [51] | Plasma | Downregulated in macroalbuminuria |
| Peng, 2013 [75] | Urine supernatant | Upregulated | |
| Pezzolesi, 2015 [77] | Plasma | Downregulated in fast progressors to ESKD | |
| Liu, 2021 [69] | Serum | Upregulated | |
| miR-29c | Guo, 2017 [62] | Plasma | Upregulated |
| Urine sediments | Downregulated | ||
| Kidney tissue | Downregulated | ||
| Pezzolesi, 2015 [77] | Plasma | No difference | |
| Peng, 2013 [75] | Urine supernatant | No difference | |
| miR-124 | Milas, 2018 [71] | Urine | Upregulated |
| Monjezi, 2022 [72] | Peripheral blood mononuclear cells | downregulated | |
| Petrica, 2019 [76] | Serum | Downregulated | |
| Urine | Upregulated | ||
| Let-7a | Park, 2022 [74] | Plasma | Downregulated |
| Urinary extracellular vesicles | Downregulated | ||
| Wang, 2019 [19] | Plasma | Downregulated | |
| Zhou, 2013 [93] | Whole blood | Downregulated | |
| miR-30e | Dieter, 2019 [57] | Plasma | Downregulated |
| Urine | Downregulated | ||
| Park, 2022 [74] | Plasma | Downregulated | |
| Urinary extracellular vesicles | Downregulated | ||
| Wang, 2019 [19] | Plasma | Downregulated | |
| miR-30b | He, 2014 [64] | Serum | Upregulated |
| Park, 2022 [74] | Plasma | Downregulated | |
| Urinary extracellular vesicles | Downregulated | ||
| Zang, 2019 [89] | Urine sediment exosome | Downregulated | |
3.4. Dysregulated miRNAs in DKD
One hundred and ninety-three miRNAs were differentially expressed in DKD, and of these, 12 miRNAs (miR-155, miR-126, miR-192, miR-21, miR-29b, miR-15a-5p, miR-29a, miR-29c, miR-124, let-7a, miR-30e and miR-30b) were reported in at least three studies. miR-21 [51,60,61,64,67,71,76,77,89] and miR-29b [50,53], were consistently upregulated whereas let-7a [19,74,93] and miR-30e [19,57,74] were consistently downregulated in individuals with DKD across studies (Table 5). Although discordant results were observed for miR-155, miR-126 and miR-192, they were commonly studied in at least four different studies. miR-155 was downregulated in DKD in three different studies [19,47,52], with one study reporting upregulation [53]. In the five studies where miR-126 expression was investigated in serum, three different studies showed consistent upregulation of the miRNA [53,60,64], whilst two studies reported downregulation [48,76], in individuals with DKD and controls. Three studies reported downregulation of miR-192 [49,70,71], whilst three other studies [64,66,76] observed upregulation of this miRNA in individuals with DKD compared to controls.
Table 5.
Association of miRNAs with kidney disease outcome.
| Study | microRNA | Adjustment | Effect Estimate [OR (95%CI)] | Outcome |
|---|---|---|---|---|
| Fujii, 2019 [22] | miR-17 | sex, age, proteinuria, body mass index, systolic blood pressure, triglyceride, blood glucose, smoking status, alcohol consumption, exercise habit, and medication for non-communicable diseases |
0.42 (0.24 to 0.75); p = 0.004 | CKD |
| miR-21 | 0.47 (0.26 to 0.85); p = 0.01 | |||
| miR-150 | 0.49 (0.27 to 0.88); p = 0.02 | |||
| Fujii, 2019b [32] | miR-126 | age, sex, blood glucose, body mass index, systolic blood pressure, smoking status, alcohol consumption, relocation frequency, degree of housing damage, current housing environment, and psychological condition | 0.67 (0.45 to 0.98); p = 0.04 | CKD |
| miR-197 | 0.67 (0.46 to 0.99); p = 0.05 | |||
| miR-223 | 0.53 (0.35 to 0.79); p = 0.002 | |||
| Fujii, 2021 [33] | miR-126 | Sex, age, body mass index, blood glucose levels, systolic blood pressure, smoking status, alcohol intake, habitual exercise, proteinuria and baseline eGFR or blood urea nitrogen | 3.85 (1.01 to 16.8); p = 0.05 | CKD |
| Huang, 2018 [95] | miR-29a | age, sex, SBP, fasting blood-glucose, body mass index, glomerular filtration rate, triglyceride, C-reactive protein, and TGF-β1 | 1.11 (1.08 to 1.37); p = 0.002 | Proteinuria |
| Huang, 2020 [96] | miR-29b | age, gender, SBP, fasting blood-glucose, body mass index, glomerular filtration rate, low density lipoprotein cholesterol, C-reactive protein and TGF-β1 | 0.55 (0.35 to 0.79); p < 0.001 | Albuminuria |
| Motshwari, 2021 [37] | miR-novel-chr2_55842 | age, gender, smoking status, drinking status, HTN, and DM status | 1.65 (1.33 to 2.05); p < 0.0001 | CKD |
| miR-novel-chr7_76196 | 4.89 (2.48 to 9.64); p < 0.0001 | |||
| miR-novel-chr5_67265 | 1.37 (1.17 to 1.60); p < 0.0001 | |||
| miR-novel-chr13_13519 | 1.79 (1.40 to 2.28); p < 0.0001 | |||
| miR-novel-chr1_36178 | 1.22 (1.10 to 1.37); p < 0.0001 | |||
| miR-novel-chr15_18383 | 1.44 (1.09 to 1.89); p = 0.009 | |||
| Al-kafaji, 2016 [48] | miR-126 | age, gender, BMI and blood pressure, fasting glucose, HbA1c, triglyceride, and LDL | 0.51 (0.37 to 0.71); p = 0.002 | DKD |
| 0.78 (0.70 to 0.95); p = 0.04 | Microalbuminuria | |||
| 0.43 (0.30 to 0.70); p = 0.03 | Macroalbuminuria | |||
| Al-kafaji, 2018 [49] | miR-377 | age, sex, BMI, HbA1c, mean blood pressure, LDL, triglyceride and total cholesterol | 1.12 (0.98 to 1.22); p = 0.018 | DKD |
| Cardenas-Gonzalez, 2017 [54] | miR-4536-3p | Not reported | 3.03 (1.95 to 4.72) | DKD |
| Pezzolesi, 2015 [77] | let-7b-5p | Sex, age, HbA1c, duration of type 1 diabetes | 2.51 (1.42 to 4.43); p = 0.002 | ESKD |
| miR-21-5p | 6.33 (1.75 to 22.92); p = 0.005 | |||
| let-7c-5p | 0.23 (0.10 to 0.52); p = 0.0004 | |||
| miR-29a-3p | 0.38 (0.20–0.74); p = 0.004 |
3.5. MicroRNAs Associated with Kidney Disease Subgroups
Studies reporting on the independent associations of various miRNAs with kidney disease outcomes and/or markers of kidney function and kidney damage are summarized in Table 5 and Table 6, as well as in Figure 2. In the general population, miR-17 [22], miR-21 [22], miR-150 [22], miR-197 [32] and miR-223 [32] were inversely associated with prevalent CKD, whilst miR-novel-chr2_55842, miR-novel-chr7_76196, miR-novel-chr5_67265, miR-novel-chr13_13519, miR-novel-chr1_36178 and miR-novel-chr15_18383 [37] were positively associated with prevalent CKD. In individuals with DM, miR-377 [49] was inversely associated with DKD, whereas miR-4536-3p [54] was positively associated with DKD. MicroRNAs, let-7b-5p, miR-21-5p were positively associated with progression to ESKD, whereas let-7c-5p, and miR-29a-3p were inversely associated with progression to ESKD in those with DM [77]. In the general population, miR-126 was inversely associated with the risk of CKD [32,33], whereas in individuals with DM it was inversely associated with DKD, microalbuminuria and macroalbuminuria [48]. In individuals with HTN-associated CKD, miR-29a [95] and miR-29b [96] were positively associated with albuminuria.
Table 6.
Association of miRNAs with kidney function and damage.
| Study | microRNA | Adjustment | Unstandardized/Standardized β-Coefficient (95%CI) | Outcome |
|---|---|---|---|---|
| Chen, 2013 [29] | miR-125b | Not reported | Not reported | eGFR |
| miR-145 | ||||
| miR-155 | ||||
| Donderski, 2021 [30] | miR-155-5p | Not reported | 0.32; p = 0.042 | eGFR |
| Fourdinier, 2019 [21] | miR-223 | age, body mass index, diabetes, urea, calcium, phosphate, parathyroid hormone, platelet count, cholesterol, and low-density lipoprotein | 0.02 (0.01 to 0.03); p < 0.0001 | eGFR |
| miR-126 | hypertension, body mass index, diabetes, urea, phosphate, parathyroid hormone, proteinuria, cholesterol, and low-density lipoprotein | 0.00 (0.000 to 0.001); p = 0.002 | eGFR | |
| Fujii, 2019 [22] | miR-17 | sex, age, proteinuria, body mass index, systolic blood pressure, triglyceride, blood glucose, smoking status, alcohol consumption, exercise habit, and medication for non-communicable diseases | 0.121; p = 0.004 | eGFR |
| miR-21 | 0.134; p = 0.002 | |||
| miR-150 | 0.123; p = 0.004 | |||
| Fujii, 2021 [33] | miR-126 | age, sex, smoking habits, alcohol intake, habitual exercise, BMI, SBP, glucose levels, proteinuria, and baseline eGFR | −3.18; p = 0.04 | eGFR |
| Motshwari, 2021 [37] |
miR-novel-chr2_55842 | age, gender, smoking status, drinking status, hypertension, and diabetes mellitus status | −2.70 (−4.82 to −0.57); p = 0.013 | eGFR |
| miR-novel-chr7_76196 | −7.39 (−14.05 to −0.72); p = 0.030 | |||
| Shang, 2017 [41] | miR-92a | age, sex, smoking, diabetes mellitus, coronary artery disease, and hyperlipidaemia | −0.684; p < 0.001 −0.548; p < 0.001 |
eGFR |
| Berillo, 2020 [94] | let-7g-5p | age, urinary albumin creatinine ratio, carotid distensibility, neutrophil and lymphocyte fractions, neutrophil number and neutrophil-to-lymphocyte ratio | 0.41; p < 0.001 | eGFR |
| miR-191-5p | 0.30; p < 0.014 | |||
| Eissa, 2016 [58] | miR-15b | Not reported | 0.452 (0.000 to 0.000); p < 0.001 | UACR |
| miR-34a | −0.914 (0.000 to 0.000); p < 0.03 | |||
| miR-636 | 0.889 (0.000 to 0.000); p < 0.02 | |||
| Eissa, 2016b [59] | miR-133b | Not reported | 0.4 (0.395 to 1.855); p < 0.01 | eGFR |
| Ma, 2016 [70] | miR-192 | Age, duration, body mass index, systolic and diastolic blood pressure, fasting blood glucose, postprandial blood glucose, HbA1C, fasting insulin, postprandial insulin, fasting C peptides, prandial C peptides, blood urea nitrogen, creatinine, low- and high-density lipoprotein cholesterol, triglycerides, cholesterol, TGF-β1, and fibronectin |
Not reported | UACR |
| Milas, 2018 [71] | miR-21 | lipid profile, HbA1c, and high-sensitive C-reactive protein | −0.007 (−0.011 to −0.003); p = 0.0001 | eGFR |
| miR-124 | −0.007 (−0.011 to −0.003); p = 0.0001 | |||
| miR-192 | 0.005 (0.002 to 0.008); p = 0.0001 | |||
| miR-21 | −0.0005 (−0.0007 to −0.0002); p = 0.0001 | UACR | ||
| miR-124 | −0.0005 (−0.0007 to −0.0002); p = 0.0001 | |||
| Xiao, 2017 [87] | miR-9 | pigment epithelium-derived factor, vascular endothelial growth factor, low-density lipoprotein cholesterol, total cholesterol, fibrinogen, HbA1c, insulin resistance | 0.431; p = 0.023 | UAER |
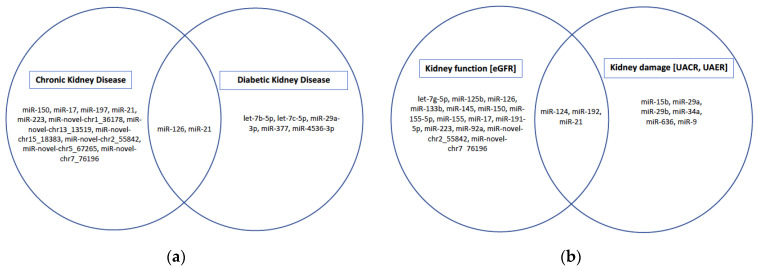
Figure 2.
Associations between miRNAs and CKD subgroups and markers of kidney function and damage. (a) Shows miRNAs independently associated with CKD in general population and in high-risk individuals with HTN and DM. (b) miRNAs associated with markers of kidney function and or kidney damage in general population and in high-risk individuals with HTN and DM.
4. Discussion
Recent studies have demonstrated that miRNAs are key mediators in the pathophysiology of CKD, suggesting that circulating miRNAs have potential utility as alternative markers for early detection and progression of CKD, as well as monitoring treatment responses. Circulating and urinary miRNAs are ideal minimally- or non-invasive biomarkers because they are stable in body fluids and exosomes and can be detected using validated techniques for quantification. However, miRNA profile studies in humans have shown contradictory results, with few miRNAs being consistently dysregulated across studies. We performed a systematic review of studies that evaluated miRNA expressions in CKD in the general population, and high-risk subgroups (individuals with HTN and DM). We found that two miRNAs (miR-126 and miR-223) were consistently downregulated in the general population with CKD, whilst miR-21 and miR-29b were consistently upregulated and let-7a-3p and miR-30e were consistently downregulated in individuals with DKD, in whole blood, plasma, serum, urine, or urinary exosomes. Although showing inconsistent data, miR-155, miR-192, miR-15a-5p, miR-29a, miR-29c were also commonly quantified in the studies included in this review. Of note, only a few studies quantified miRNA expression in individuals with HTN-associated CKD, and reported inconsistent findings and none in HIVAN.
MiR-126 is endothelial cell-specific and promotes vascular integrity and angiogenesis via regulation of vascular endothelial growth factor (VEGF) signalling and, as a result, inhibits vascular inflammation [100]. MiR-126, which was downregulated in CKD in the general population [21,28,32] and individuals with DM [48,76] and HTN [99] when quantified in serum and whole blood samples, was inversely associated with prevalent CKD [32,33] and positively associated with eGFR [21,33]. A prospective study showed that lower levels of miR-126 were associated with an increased risk of developing CKD and rapid decline in kidney function over a period of five years [33]. Zhou et al. also demonstrated that miR-126 has an atheroprotective role, as it increases vascular smooth muscle cells (VSMCs) turnover, thereby regulating the contractile phenotype of VSMC [101]. Taken together, the downregulation of miR-126 may result in vascular dysfunction, which is very common in early-stage CKD. MiR-126 may therefore be a potential biomarker for the early identification of CKD and a potential target for the prevention or treatment of CKD-related vascular complications. Contrary to these findings, a few studies reported upregulated expression of miR-126 individuals with DKD [53,60,64,76] when quantified in urine, plasma as well as serum. It is plausible that this may be a compensatory mechanism resulting in increased release of miR-126 when microvascular endothelial cells are exposed to stressful conditions [102]. Indeed, Beltrami and colleagues used in vitro analyses and showed that miR-126 is released from glomerular endothelial cells in response to DKD-related cytokines [53].
Similarly to miR-126, miR-223 also plays a role in the regulation of VSMC proliferation [103]. This miRNA was consistently downregulated in the general population with CKD in serum samples [21,28,32,43]. Moreover, lower levels of this miRNA were associated with lower levels of eGFR [32], and prevalent CKD [21]. Although commonly considered to be involved in inflammatory pathways, evidence also suggests a protective role of miR-223 in VSMCs, through the inhibition of calcification by the regulation of interleukin-6 (IL-6)/signal transducer and activator of transcription 3 (STAT3) [63]. These findings imply that increased levels of miR-223 may improve kidney function, and thus may serve as a therapeutic strategy to improve CKD outcomes in the general population.
The involvement of miR-21 in kidney fibrosis is well established, although the mechanisms involved have not been completely clarified. miR-21 acts as a pro-fibrotic factor, and its upregulation induces kidney fibrosis through TGF-β signalling pathway regulation [104]. Consistent upregulation of miR-21 in individuals with DKD [51,61,64,67,71,76,77,89] as well as its inverse association with eGFR [71] and positive association with albuminuria [71] have been reported. Moreover, increased levels of miR-21 were associated with rapid progression to ESKD over a 10-year follow-up period [77]. Correspondingly, in vitro and in vivo knockdown of miR-21 ameliorated DKD by reducing albuminuria, kidney inflammation, podocyte loss and interstitial fibrosis, suggesting its value as a potential therapeutic target against DKD progression [105]. Although the observed findings imply that increased expression of miR-21 may be associated with the development and progression of DKD, the data were inconclusive in the case of CKD. In the general population with CKD, contrasting results were reported, with miR-21 upregulated in urine [30] and urine exosomes [34] but downregulated in serum samples [30]. Moreover, Fujii and colleagues found that increased levels of miR-21 were positively associated with eGFR and inversely associated with the risk of CKD in the general population [22]. Donderski et al. (2021) explained that the lower levels of miR-21 detected in serum samples of individuals with CKD may be as a result of suppression caused by increased fibrosis and TGF–β activity [30]. This is in line with the findings by Sun et al. (2018), using a murine kidney fibrosis model, they observed that miR-21 is the main regulator of fibroblasts activation through an auto-regulatory loop between miR-21 and programmed cell death protein 4 and activated protein 1, therefore suggesting that miR-21 may act as pro- or anti-fibrotic depending on the cell type [106]. It has been suggested that identification of the cellular source of miRNAs would be helpful instead of the biofluid sample to link the miRNAs to the specific disease process [107].
The miR-29 family has been well studied with regard to the TGF-β signalling pathway [104]. Two studies included in our review reported that miR-29b was upregulated in the urine samples of individuals with DKD [50,53], although one study reported no difference in the expression level of miR-29b when quantified in urine supernatant [75]. The lack of difference could be explained by the relatively lower abundance of miR-29b in urine supernatant sample reported in this study [75]. In individuals with HTN, increased expression of miR-29b was positively associated with albuminuria and inversely associated with kidney function [96]. However, these findings are contrary to previous studies that have reported on the protective role of this miRNA in DKD. Chen et al. (2014) showed that knockdown of miR-29b in diabetic mice was associated with increased albuminuria and TGFβ mediated fibrosis whereas overexpression of miR-29b attenuated kidney fibrosis in DKD through the regulation of TGFβ1/Smad3 pathway [108]. The upregulated expression of miR-29b in DKD observed in the included studies in our review may be due to the compensatory release of miR-29b. Beltrami and colleagues used in vitro analyses and observed increased release of miR-29b from glomerular endothelial cells in response to DKD-related cytokines [53]. Regarding the expression profile of miR-29a and miR-29c, contradictory results were observed when quantified in various samples of individuals with DKD. miR-29a was downregulated in plasma samples of individuals with severe DKD [51] and inversely associated with rapid progression to ESKD over a 10-year follow-up period [77], suggesting that this miRNA may have a protective effect against the progression of DKD. However, when quantified in urine supernatant, upregulated expression of miR-29a was observed in individuals with DKD [75]. Studies have previously highlighted that urine supernatant miRNAs inversely reflect intracellular miRNAs [109], which could be the possible reason for the discrepancy, however, tissue expression of miR-29a was not analyzed in this study [75]. On the other hand, Guo et al. (2017) [62] analyzed the expression of miR-29c in three different samples of individuals with DKD relative to those without, and found downregulated expression in urinary sediments and kidney tissues but upregulated expression in plasma. Cui and Cui (2020) found that relative to blood, urinary miRNAs highly reflected kidney tissue miRNAs and suggested that urine should be a better sample for kidney miRNAs analysis [110].
MicroRNAs miR-30e and let-7a were consistently downregulated in individuals with DKD relative to those without DKD, suggesting that increased expression of these miRNAs may have protective effects in the kidney and therefore may serve as possible diagnostic and prognostic markers of DKD. Accordingly, previous evidence suggests that the let-7 family of miRNAs may be a negative regulator of kidney fibrosis in DKD [111]. Muralidharan and colleagues [38] validated the expression of let-7a in an Alb/TGFβ mouse model of CKD and found that the miRNA was significantly downregulated further suggesting its possible role in the development or progression of CKD. MicroRNAs in the miR-30 family are highly enriched in kidney podocytes cells where they are involved in regulatory roles and are essential for structural and functional homeostasis [112]. In vitro and in vivo experimental studies showed that the expression of miR-30e was significantly decreased in those with DKD whereas overexpression of miR-30e was protective against the development of kidney fibrosis in DKD suggestive the potential role of this miRNA as a therapeutic target [113].
The miR-155 was commonly analyzed in the studies included in this review. Experimental studies have shown that suppressing miR-155 expression in DKD mice protects against kidney damage, attenuates hyperglycaemia-induced kidney damage and downregulates IL-17 expression by enhancing the suppression of cytokine signalling 1 (SOCS1) [114]. In line with these, upregulated expression of miR-155 was observed in the general population with CKD [30,31] and individuals with DKD 53. However, downregulated expression of miR-155 was commonly reported in individuals with DKD [19,47,52], as well as in the general population with CKD [29,30]. Furthermore, Donderski and colleagues reported on the positive association of miR-155 and eGFR [30]. These findings suggest that miR-155 may also play a role in the development of DKD. Wang et al. (2018) demonstrated that miR-155 is involved in the regulation of the autophagic process in DKD through the regulation of a signalling loop p53/miR-155-5p/Sirt1 and may therefore serve as a therapeutic strategy for DKD [115].
miR-192 has been shown to have a protective effect against kidney fibrosis. Downregulated expression of miR-192 was observed in individuals with DKD [49,70,71], and reduced levels of miR-192 were positively associated with kidney function [71] and inversely associated with kidney damage [70,71], suggesting that miR-192 levels may decrease with the increasing level of albuminuria and a decline in kidney function. Consistently, in vivo studies have shown that loss of miR-192 was associated with the development and progression of DKD through exacerbation of kidney fibrosis by enhancing TGF-β1 signalling pathway [116]. These findings suggest that reduced expression of miR-192 in the early stage may be associated with the development of DKD and therefore may serve as an early indicator of DKD. However, contrary to these findings, increased expression of miR-192 in individuals with DKD relative to controls has been observed in a few studies [64,76]. Jia and colleagues reported that the expression of miR-192 was increased during the early stages of DKD and decreased in the advanced stages of DKD [66]. They further showed that miR-192 was positively correlated with albuminuria and TGF-β1 levels [66], suggesting a profibrotic role of this miRNA. Jenkins et al. (2012) highlighted that miR-192 is pleiotropic, involved in multiple important roles in the kidney, and its role as an antifibrotic or profibrotic factor may be cell dependent [117].
This review provides an overview of miRNA dysregulation in CKD, including diabetic and hypertensive-related CKD in humans. It also highlights miRNAs that are associated with CKD and its clinical indicators. A few miRNAs showed consistent expression patterns in CKD relative to controls, whilst most of the frequently studied miRNAs showed contradictory findings. These discrepancies may be partly explained by the technical and methodological inter-study variabilities, such as the use of different biological samples, sample handling and processing procedures, miRNA extraction protocols, detection and quantification techniques, and normalizing controls [118]. Although the majority of included studies quantified miRNA expression in blood, recent evidence points to the superiority of urine miRNAs to serum/plasma miRNAs for CKD diagnosis with the non-invasive nature in which urine samples are collected, adding to its preference [119]. Another challenge is the lack of a standardized normalization control for miRNA expression studies. Although a wide range of endogenous and exogenous miRNAs are employed as normalizers, emerging evidence suggests that the use of synthetic spike in controls such as cel-miR-39 is preferable [120]. Therefore, to be able to identify reliable miRNA biomarkers, research findings need to be reproducible and comparable between studies. This can be achieved when normalization controls have been validated, and there is a standardization of robust protocols for sample processing and extraction [121].
Strength and Limitations
The main strength of this review is its comprehensive report of miRNAs dysregulated in CKD, their association with CKD as well as clinical markers of CKD in the general population as well as in high-risk individuals with HTN and/or DM for the very first time. The review also provides a list of miRNAs that have been frequently studied in diverse geographical areas and showed consistent expression patterns across studies in CKD and DKD and therefore are worthy of future research.
The studies included in the review had their own shortcomings and, as such, impacted the review’s overall quality of evidence. The most important limitation of the review was the inability to perform a pooled meta-analysis of our studies due to various factors, including insufficient raw data on fold changes or relative expression of miRNAs, technical and methodological variabilities between studies, such as the use of different biological samples, normalization control used, and different miRNA quantification techniques. Moreover, due to insufficient data, we could not report on the expression patterns of miRNAs across different stages of CKD. The language restriction on the inclusion criteria may have excluded other relevant studies, thus biasing our findings. Additionally, there were differences in the classification of CKD, wide sample size ranges, variability in participant demographic factors such as age, sex proportion, and race, as well as environmental and regional factors between studies.
5. Conclusions
MiRNAs detected in biofluids are promising as potential biomarkers of disease diagnosis and therapeutic targets for future clinical applications. However, understanding their role in CKD pathophysiology and how their expression pattern is regulated is still in its infancy. As such, further research is required to fully elucidate their roles before any extrapolation for clinical use. Prevention and early detection of CKD has been a topic of interest for many researchers and clinicians in the field. This review highlights several dysregulated miRNAs that were frequently studied and showed consistent findings across studies in CKD (miR-126 and miR-223) or DKD (miR-21, miR-29b, let-7a and miR-30e) with a potential for clinical application in CKD diagnosis/prognosis in the future. This consistent alteration of miRNAs with CKD/DKD and their stability and detectability in bodily fluids suggests that these miRNAs are promising potential non-invasive or minimally invasive diagnostic markers for early detection and therapeutic targets of CKD/DKD and warrant further scrutiny in future investigations. Besides these specific miRNAs, miR-155, miR-192, miR-15a-5p, miR-29a and miR-29c, despite their inconsistent expression patterns reported in different studies, were commonly dysregulated in CKD and/or DKD, and therefore may also play an important role in CKD. As such, their further exploration is warranted. Furthermore, it may be worthwhile for future studies to focus on identifying target genes and pathways of these frequently studied miRNAs, to get a complete understanding of their role in the development and progression of CKD, as well as to assess their potential value as diagnostic markers or therapeutic targets.
Acknowledgments
The authors would like to thank Michelle Snyders, the research librarian at the Cape Peninsula University of Technology, for her assistance with accessing databases and advice on the search strategy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24021792/s1.
Author Contributions
Study conceptualization, D.D.M., D.M.M. and C.G.; research protocol development D.D.M., D.M.M. and C.G.; online database search and selection of studies, D.D.M. and D.M.M.; data extraction and quality assessment, D.D.M. and D.M.M. and C.G. as the third author in cases of disagreement; writing – original draft preparation, D.D.M.; writing- reviewing and editing, D.D.M., D.M.M. and C.G.; critical reviewing of the manuscript, C.G., A.P.K., T.E.M. and R.T.E.; supervision, C.G. and T.E.M.; funding acquisition, T.E.M., A.P.K. and R.T.E. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated during this study is available upon reasonable request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Foreman K.J., Marquez N., Dolgert A., Fukutaki K., Fullman N., McGaughey M., Pletcher M.A., Smith A.E., Tang K., Yuan C.W., et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: Reference and alternative scenarios for 2016–2140 for 195 countries and territories. Lancet. 2018;392:2052–2090. doi: 10.1016/S0140-6736(18)31694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD Chronic Kidney Disease Collaboration Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395:709–733. doi: 10.1016/S0140-6736(20)30045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Essue B.M., Jha V., John O., Knight J., Jan S. Universal health coverage and chronic kidney disease in India. Bull. World Health Organ. 2018;96:442. doi: 10.2471/BLT.18.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oguntola S.O., Hassan M.O., Duarte R., Vachiat A., Manga P., Naicker S. Atherosclerotic vascular disease is more prevalent among black ESKD patients on long-term CAPD in South Africa. BMC Nephrol. 2019;20:1–10. doi: 10.1186/s12882-019-1583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li P., Garcia-Garcia G., Lui S.-F., Andreoli S., Fung W., Hradsky A., Kumaraswami L., Liakopoulos V., Rakhimova Z., Saadi G., et al. Kidney health for everyone everywhere–from prevention to detection and equitable access to care. Br. J. Med. Biol. Res. 2020;53:1–10. doi: 10.1590/1414-431X20209614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Giacoman S., Madero M. Biomarkers in chronic kidney disease, from kidney function to kidney damage. World J. Nephrol. 2015;4:57–73. doi: 10.5527/wjn.v4.i1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inker L.A., Eneanya N.D., Coresh J., Tighiouart H., Wang D., Sang Y., Crews D.C., Doria A., Estrella M.M., Froissart M., et al. New creatinine-and cystatin C–based equations to estimate GFR without race. N. Engl. J. Med. 2021;385:1737–1749. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukabau J.B., Yayo E., Gnionsahé A., Monnet D., Pottel H., Cavalier E., Nkodila A., Makulo J.R.R., Mokoli V.M., Lepira F.B., et al. Performance of creatinine-or cystatin C–based equations to estimate glomerular filtration rate in sub-Saharan African populations. Kidney Int. 2019;95:1181–1189. doi: 10.1016/j.kint.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 9.Fabian J., Kalyesubula R., Mkandawire J., Hansen C.H., Nitsch D., Musenge E., Nakanga W.P., Prynn J.E., Dreyer G., Snyman T., et al. Measurement of kidney function in Malawi, South Africa, and Uganda: A multicentre cohort study. Lancet Glob. Health. 2022;10:e1159–e1169. doi: 10.1016/S2214-109X(22)00239-X. [DOI] [PubMed] [Google Scholar]
- 10.Al-Rubeaan K., Siddiqui K., Al-Ghonaim M.A., Youssef A.M., Al-Sharqawi A.H., AlNaqeb D. Assessment of the diagnostic value of different biomarkers in relation to various stages of diabetic nephropathy in type 2 diabetic patients. Sci. Rep. 2017;7:2684. doi: 10.1038/s41598-017-02421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin A., Tonelli M., Bonventre J., Coresh J., Donner J.A., Fogo A.B., Fox C.S., Gansevoort R.T., Heerspink H.J.L., Jardine M., et al. Global kidney health 2017 and beyond: A roadmap for closing gaps in care, research, and policy. Lancet. 2017;390:1888–1917. doi: 10.1016/S0140-6736(17)30788-2. [DOI] [PubMed] [Google Scholar]
- 12.Bonneau E., Neveu B., Kostantin E., Tsongalis G.J., De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2019;30:114–127. [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatt K., Mi Q.S., Dong Z. MicroRNAs in kidneys: Biogenesis, regulation, and pathophysiological roles. Am. J. Physiol.-Ren. Physiol. 2011;300:602–610. doi: 10.1152/ajprenal.00727.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X., Xie D., Zhao Q., You Z.H. MicroRNAs and complex diseases: From experimental results to computational models. Brief Bioinform. 2019;20:515–539. doi: 10.1093/bib/bbx130. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H., Wang G., Zhou X., Song X., Gao H., Ma C., Chang H., Li H., Liu F.F., Lu J., et al. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed. Pharm. 2016;83:792–797. doi: 10.1016/j.biopha.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 18.Amiri A., Tehran M.M., Asemi Z., Shafiee A., Hajighadimi S., Moradizarmehri S., Mirzaei H.R., Mirzaei H. Role of resveratrol in modulating microRNAs in human diseases: From cancer to inflammatory disorder. Curr Med. Chem. 2019;26:360–376. doi: 10.2174/0929867326666191212102407. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Wang G., Liang Y., Zhou X. Expression profiling and clinical significance of plasma microRNAs in diabetic nephropathy. J. Diabetes Res. 2019;2019:5204394. doi: 10.1155/2019/5204394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Usa K., Wang F., Liu P., Geurts A.M., Li J., Williams A.M., Regner K.R., Kong Y., Liu H., et al. MicroRNA-214-3p in the kidney contributes to the development of hypertension. J. Am. Soc. Nephrol. 2018;29:2518–2528. doi: 10.1681/ASN.2018020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fourdinier O., Schepers E., Metzinger-Le Meuth V., Glorieux G., Liabeuf S., Verbeke F., Vanholder R., Brigant B., Pletinck A., Diouf M., et al. Serum levels of miR-126 and miR-223 and outcomes in chronic kidney disease patients. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-41101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii R., Yamada H., Munetsuna E., Yamazaki M., Ohashi K., Ishikawa H., Maeda K., Hagiwara C., Ando Y., Hashimoto S., et al. Associations of Circulating MicroRNAs (miR-17, miR-21, and miR-150) and Chronic Kidney Disease in a Japanese Population. J. Epidemiol. 2019;30:177–182. doi: 10.2188/jea.JE20180233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motshwari D.D., Matshazi D.M., Erasmus R., Kengne A.P., Matsha T.E., George C. MicroRNAs associated with chronic kidney disease in the general population and high-risk subgroups: Protocol for a systematic review and meta-analysis. BMJ Open. 2022;12:e057500. doi: 10.1136/bmjopen-2021-057500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 25.Guyatt G., Oxman A.D., Akl E.A., Kunz R., Vist G., Brozek J., Norris S., Falck-Ytter Y., Glasziou P., DeBeer H., et al. GRADE guidelines: 1. Introduction—GRADE evidence profiles and summary of findings tables. J. Clin. Epidemiol. 2011;64:383–394. doi: 10.1016/j.jclinepi.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Levey A.S., Coresh J., Greene T., Marsh J., Stevens L.A., Kusek J.W., Van Lente F., Chronic Kidney Disease Epidemiology Collaboration Expressing the Modification of Diet in Renal Disease Study equation for estimating glomerular filtration rate with standardized serum creatinine values. Clin. Chem. 2007;53:766–772. doi: 10.1373/clinchem.2006.077180. [DOI] [PubMed] [Google Scholar]
- 27.Levey A.S., Stevens L.A., Schmid C.H., Zhang Y., Castro I.I.I.A.F., Feldman H.I., Kusek J.W., Eggers P., Van Lente F., Greene T., et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carmona A., Guerrero F., Jimenez M.J., Ariza F., Agüera M.L., Obrero T., Noci V., Muñoz-Castañeda J.R., Rodríguez M., Soriano S., et al. Inflammation, Senescence and MicroRNAs in Chronic Kidney Disease. Front. Cell Dev. Biol. 2020;8:739. doi: 10.3389/fcell.2020.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen N.X., Kiattisunthorn K., O’Neill K.D., Chen X., Moorthi R.N., Gattone V.H., Allen M.R., Moe S.M. Decreased microRNA is involved in the vascular remodeling abnormalities in chronic kidney disease (CKD) PLoS ONE. 2013;8:e64558. doi: 10.1371/journal.pone.0064558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donderski R., Szczepanek J., Naruszewicz N., Naruszewicz R., Tretyn A., Skoczylas-Makowska N., Tyloch J., Odrowąż-Sypniewska G., Manitius J. Analysis of profibrogenic microRNAs (miRNAs) expression in urine and serum of chronic kidney disease (CKD) stage 1-4 patients and their relationship with proteinuria and kidney function. Int. Urol. Nephrol. 2021;54:937–947. doi: 10.1007/s11255-021-02928-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckersten D., Tsatsanis C., Giwercman A., Bruun L., Pihlsgård M., Christensson A. MicroRNA-155 and Anti-Müllerian Hormone: New Potential Markers of Subfertility in Men with Chronic Kidney Disease. Nephron. Extra. 2017;7:33–41. doi: 10.1159/000458711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujii R., Yamada H., Yamazaki M., Munetsuna E., Ando Y., Ohashi K., Ishikawa H., Shimoda H., Sakata K., Ogawa A., et al. Circulating microRNAs (miR-126, miR-197, and miR-223) are associated with chronic kidney disease among elderly survivors of the Great East Japan Earthquake. BMC Nephrol. 2019;20:474. doi: 10.1186/s12882-019-1651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii R., Yamada H., Tsuboi Y., Ando Y., Munetsuna E., Yamazaki M., Ohashi K., Ishikawa H., Ishihara Y., Hashimoto S., et al. Association between circulating microRNAs and changes in kidney function: A five-year prospective study among Japanese adults without CKD. Clin. Chim. Acta. 2021;521:97–103. doi: 10.1016/j.cca.2021.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Lange T., Artelt N., Kindt F., Stracke S., Rettig R., Lendeckel U., Chadjichristos C.E., Kavvadas P., Chatziantoniou C., Endlich K., et al. MiR-21 is up-regulated in urinary exosomes of chronic kidney disease patients and after glomerular injury. J. Cell Mol. Med. 2019;23:4839–4843. doi: 10.1111/jcmm.14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Qiu F.X., Tian F., Shi X.Z., Gao A.Q., Song L., Liu J. Changes of miR-155 expression in serum of uremic patients before and after treatment and risk factors analysis. Exp. Ther. Med. 2020;20:3352–3360. doi: 10.3892/etm.2020.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Wang W., Bai Y., Zhang H., Zhang S., He L., Zhou W., Zhang D., Xu J. Identification of a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for chronic kidney disease using next-generation sequencing. J. Int. Med. Res. 2020;48:300060520969481. doi: 10.1177/0300060520969481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motshwari D.D., George C., Matshazi D.M., Weale C.J., Davids S.F.G., Erasmus R.T., Kengne A.P., Matsha T.E. Novel Whole Blood MicroRNAs Predicting Chronic Kidney Disease in South Africans with Hypertension and Diabetes Mellitus. Appl. Sci. 2021;11:7674. doi: 10.3390/app11167674. [DOI] [Google Scholar]
- 38.Muralidharan J., Ramezani A., Hubal M., Knoblach S., Shrivastav S., Karandish S., Scott R., Maxwell N., Ozturk S., Beddhu S., et al. Extracellular microRNA signature in chronic kidney disease. Am. J. Physiol.-Ren. Physiol. 2017;312:F982–F991. doi: 10.1152/ajprenal.00569.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudnicki M., Perco P., DHaene B., Leierer J., Heinzel A., Mühlberger I., Schweibert N., Sunzenauer J., Regele H., Kronbichler A., et al. Renal microRNA- and RNA-profiles in progressive chronic kidney disease. Eur. J. Clin. Investig. 2016;46:213–226. doi: 10.1111/eci.12585. [DOI] [PubMed] [Google Scholar]
- 40.Sayilar E.I., Gullulu M., Tuncel E., Peynirci H., Alemdar A., Tunca B., Egeli U., Cecener G., Bayindir M., Cosgun G. Biomarker Potential of Urine miR-451 at Different Stages of Diabetic Nephropathy. J. Diabetes Metab. 2016;7:650. doi: 10.4172/2155-6156.1000650. [DOI] [Google Scholar]
- 41.Shang F., Wang S.-C., Hsu C.-Y., Miao Y., Martin M., Yin Y., Wu C.C., Wang Y.T., Wu G., Chien S., et al. MicroRNA-92a Mediates Endothelial Dysfunction in CKD. J. Am. Soc. Nephrol. 2017;28:3251–3261. doi: 10.1681/ASN.2016111215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trojanowicz B., Imdahl T., Ulrich C., Fiedler R., Girndt M. Circulating miR-421 Targeting Leucocytic Angiotensin Converting Enzyme 2 Is Elevated in Patients with Chronic Kidney Disease. Nephron. 2019;141:61–74. doi: 10.1159/000493805. [DOI] [PubMed] [Google Scholar]
- 43.Ulbing M., Kirsch A.H., Leber B., Lemesch S., Münzker J., Schweighofer N., Hofer D., Trummer O., Rosenkranz A.R., Müller H., et al. MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone. 2017;95:115–123. doi: 10.1016/j.bone.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdelsalam M., Wahab A.M., El Sayed Zaki M., Motawea M. MicroRNA-451 as an Early Predictor of Chronic Kidney Disease in Diabetic Nephropathy. Int. J. Nephrol. 2020;2020:1–7. doi: 10.1155/2020/8075376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdou A.E., Anani H.A.A., Ibrahim H.F., Ebrahem E.E., Seliem N., Youssef E.M.I., Ghoraba N.M., Hassan A.S., Ramadan M.A.A., Mahmoud E., et al. Urinary IgG, serum CX3CL1 and miRNA-152-3p: As predictors of nephropathy in Egyptian type 2 diabetic patients. Tissue Barriers. 2022;10:1994823. doi: 10.1080/21688370.2021.1994823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akhbari M., Biglari A., Shahrabi-Farahani M., Khalili M., Bandarian F. Expression Level of Circulating miR-93 in Serum of Patients with Diabetic Nephropathy. TURKISH J. Endocrinol. Metab. 2018;22:78–84. doi: 10.25179/tjem.2018-59661. [DOI] [PubMed] [Google Scholar]
- 47.Akhbari M., Khalili M., Shahrabi-Farahani M., Biglari A., Bandarian F. Expression Level of Circulating Cell Free miR-155 Gene in Serum of Patients with Diabetic Nephropathy. Clin. Lab. 2019;65:190209. doi: 10.7754/Clin.Lab.2019.190209. [DOI] [PubMed] [Google Scholar]
- 48.Al-Kafaji G., Al-Mahroos G., Al-Muhtaresh H.A., Skrypnyk C., Sabry M.A., Ramadan A.R. Decreased expression of circulating microRNA-126 in patients with type 2 diabetic nephropathy: A potential blood-based biomarker. Exp. Ther. Med. 2016;12:815–822. doi: 10.3892/etm.2016.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Kafaji G., Al-Muhtaresh H.A. Expression of microRNA-377 and microRNA-192 and their potential as blood-based biomarkers for early detection of type 2 diabetic nephropathy. Mol. Med. Rep. 2018;18:1171–1180. doi: 10.3892/mmr.2018.9040. [DOI] [PubMed] [Google Scholar]
- 50.Argyropoulos C., Wang K., McClarty S., Huang D., Bernardo J., Ellis D., Orchard T., Galas D., Johnson J. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS ONE. 2013;8:e54662. doi: 10.1371/annotation/37e647d5-1781-4edf-86a8-e3b533c32ad9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Assmann T.S., Recamonde-Mendoza M., Costa A.R., Puñales M., Tschiedel B., Canani L.H., Bauer A.C., Crispim D. Circulating miRNAs in diabetic kidney disease: Case–control study and in silico analyses. Acta Diabetol. 2019;56:55–65. doi: 10.1007/s00592-018-1216-x. [DOI] [PubMed] [Google Scholar]
- 52.Barutta F., Tricarico M., Corbelli A., Annaratone L., Pinach S., Grimaldi S., Bruno G., Cimino D., Taverna D., Deregibus M.C., et al. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS ONE. 2013;8:e73798. doi: 10.1371/journal.pone.0073798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beltrami C., Simpson K., Jesky M., Wonnacott A., Carrington C., Holmans P., Newbury L., Jenkins R., Ashdown T., Dayan C., et al. Association of Elevated Urinary miR-126, miR-155, and miR-29b with Diabetic Kidney Disease. Am. J. Pathol. 2018;188:1982–1992. doi: 10.1016/j.ajpath.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Cardenas-Gonzalez M., Srivastava A., Pavkovic M., Bijol V., Rennke H.G., Stillman I.E., Zhang X., Parikh S., Rovin B.H., Afkarian M., et al. Identification, confirmation, and replication of novel urinary microrna biomarkers in lupus nephritis and diabetic nephropathy. Clin. Chem. 2017;63:1515–1526. doi: 10.1373/clinchem.2017.274175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conserva F., Barozzino M., Pesce F., Divella C., Oranger A., Papale M., Sallustio F., Simone S., Laviola L., Giorgino F., et al. Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of Kidney Fibrosis in Diabetic Nephropathy. Sci. Rep. 2019;9:11357. doi: 10.1038/s41598-019-47778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delić D., Eisele C., Schmid R., Baum P., Wiech F., Gerl M., Zimdahl H., Pullen S.S., Urquhart R. Urinary Exosomal miRNA Signature in Type II Diabetic Nephropathy Patients. PLoS ONE. 2016;11:e0150154. doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dieter C., Assmann T.S., Costa A.R., Canani L.H., de Souza B.M., Bauer A.C., Crispim D. MiR-30e-5p and MiR-15a-5p Expressions in Plasma and Urine of Type 1 Diabetic Patients With Diabetic Kidney Disease. Front. Genet. 2019;10:563. doi: 10.3389/fgene.2019.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eissa S., Matboli M., Aboushahba R., Bekhet M.M., Soliman Y. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J. Diabetes Complicat. 2016;30:1585–1592. doi: 10.1016/j.jdiacomp.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 59.Eissa S., Matboli M., Bekhet M.M. Clinical verification of a novel urinary microRNA panal: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed. Pharmacother. 2016;83:92–99. doi: 10.1016/j.biopha.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 60.Florijn B.W., Duijs J.M.G.J., Levels J.H., Dallinga-Thie G.M., Wang Y., Boing A.N., Yuana Y., Stam W., Limpens R.W.A.L., Au Y.W., et al. Diabetic Nephropathy Alters the Distribution of Circulating Angiogenic MicroRNAs Among Extracellular Vesicles, HDL, and Ago-2. Diabetes. 2019;68:2287–2300. doi: 10.2337/db18-1360. [DOI] [PubMed] [Google Scholar]
- 61.Fouad M., Salem I., Elhefnawy K., Raafat N., Faisal A. MicroRNA-21 as an Early Marker of Nephropathy in Patients with Type 1 Diabetes. Indian J. Nephrol. 2020;30:21–25. doi: 10.4103/ijn.IJN_80_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo J., Li J., Zhao J., Yang S., Wang L., Cheng G., Liu D., Xiao J., Liu Z., Zhao Z. MiRNA-29c regulates the expression of inflammatory cytokines in diabetic nephropathy by targeting tristetraprolin. Sci. Rep. 2017;7:2314. doi: 10.1038/s41598-017-01027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han Q., Zhang Y., Jiao T., Li Q., Ding X., Zhang D., Cai G., Zhu H. Urinary sediment microRNAs can be used as potential noninvasive biomarkers for diagnosis, reflecting the severity and prognosis of diabetic nephropathy. Nutr. Diabetes. 2021;11:24. doi: 10.1038/s41387-021-00166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He F., Peng F., Xia X., Zhao C., Luo Q., Guan W., Li Z., Yu X., Huang F. MiR-135a promotes renal fibrosis in diabetic nephropathy by regulating TRPC1. Diabetologia. 2014;57:1726–1736. doi: 10.1007/s00125-014-3282-0. [DOI] [PubMed] [Google Scholar]
- 65.Hong Y., Wang J., Zhang L., Sun W., Xu X., Zhang K. Plasma miR-193a-3p can be a potential biomarker for the diagnosis of diabetic nephropathy. Ann. Clin. Biochem. 2021;58:141–148. doi: 10.1177/0004563220983851. [DOI] [PubMed] [Google Scholar]
- 66.Jia Y., Guan M., Zheng Z., Zhang Q., Tang C., Xu W., Xiao Z., Wang L., Xue Y. miRNAs in Urine Extracellular Vesicles as Predictors of Early-Stage Diabetic Nephropathy. J. Diabetes Res. 2016;2016:7932765. doi: 10.1155/2016/7932765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khokhar M., Roy D., Bajpai N.K., Bohra G.K., Yadav D., Sharma P., Purohit P. Metformin mediates MicroRNA-21 regulated circulating matrix metalloproteinase-9 in diabetic nephropathy: An in-silico and clinical study. Arch. Physiol. Biochem. 2021:1–11. doi: 10.1080/13813455.2021.1922457. [DOI] [PubMed] [Google Scholar]
- 68.Lin M., Song D., Zhang S., Li P. Dysregulation of miR-638 in diabetic nephropathy and its role in inflammatory response. Diabetol. Metab. Syndr. 2021;13:122. doi: 10.1186/s13098-021-00744-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X., Liu S., Luo D., Huang S., Wang F., Zhang B., Chen Y., Zheng L., Lu J., Li S. Involvement of Circulating Exosomal MicroRNAs in Jian-Pi-Yi-Shen Formula Protection Against Adenine-Induced Chronic Kidney Disease. Front. Pharmacol. 2021;11:622658. doi: 10.3389/fphar.2020.622658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma X., Lu C., Lv C., Wu C., Wang Q. The Expression of miR-192 and Its Significance in Diabetic Nephropathy Patients with Different Urine Albumin Creatinine Ratio. J. Diabetes Res. 2016;2016:6789402. doi: 10.1155/2016/6789402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milas O., Gadalean F., Vlad A., Dumitrascu V., Gluhovschi C., Gluhovschi G., Velciov S., Popescu R., Bob F., Matusz P. Deregulated profiles of urinary microRNAs may explain podocyte injury and proximal tubule dysfunction in normoalbuminuric patients with type 2 diabetes mellitus. J. Investig. Med. Off. Publ. Am. Fed. Clin. Res. 2018;66:747–754. doi: 10.1136/jim-2017-000556. [DOI] [PubMed] [Google Scholar]
- 72.Monjezi A., Khedri A., Zakerkish M., Mohammadzadeh G. Resistin, TNF-α, and microRNA 124-3p expressions in peripheral blood mononuclear cells are associated with diabetic nephropathy. Int. J. Diabetes Dev. Ctries. 2022;42:62–69. doi: 10.1007/s13410-021-00966-0. [DOI] [Google Scholar]
- 73.Motawi T.K., Shehata N.I., ElNokeety M.M., El-Emady Y.F. Potential serum biomarkers for early detection of diabetic nephropathy. Diabetes Res. Clin. Pract. 2018;136:150–158. doi: 10.1016/j.diabres.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 74.Park S., Kim O.-H., Lee K., Park I.B., Kim N.H., Moon S., Im J., Sharma S.P., Oh B.C., Nam S., et al. Plasma and urinary extracellular vesicle microRNAs and their related pathways in diabetic kidney disease. Genomics. 2022;114:110407. doi: 10.1016/j.ygeno.2022.110407. [DOI] [PubMed] [Google Scholar]
- 75.Peng H., Zhong M., Zhao W., Wang C., Zhang J., Liu X., Li Y., Paudel S.D., Wang Q., Lou T. Urinary miR-29 correlates with albuminuria and carotid intima-media thickness in type 2 diabetes patients. PLoS ONE. 2013;8:e82607. doi: 10.1371/journal.pone.0082607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petrica L., Milas O., Vlad M., Vlad A., Gadalean F., Dumitrascu V., Velciov S., Gluhovschi C., Bob F., Ursoniu S., et al. Interleukins and miRNAs intervene in the early stages of diabetic kidney disease in Type 2 diabetes mellitus patients. Biomark Med. 2019;13:1577–1588. doi: 10.2217/bmm-2019-0124. [DOI] [PubMed] [Google Scholar]
- 77.Pezzolesi M.G., Satake E., McDonnell K.P., Major M., Smiles A.M., Krolewski A.S. Circulating TGF-β1-Regulated miRNAs and the Risk of Rapid Progression to ESRD in Type 1 Diabetes. Diabetes. 2015;64:3285–3293. doi: 10.2337/db15-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prabu P., Rome S., Sathishkumar C., Gastebois C., Meugnier E., Mohan V., Balasubramanyam M. MicroRNAs from urinary extracellular vesicles are non-invasive early biomarkers of diabetic nephropathy in type 2 diabetes patients with the “Asian Indian phenotype”. Diabetes Metab. 2019;45:276–285. doi: 10.1016/j.diabet.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 79.Regmi A., Liu G., Zhong X., Hu S., Ma R., Gou L., Zafar M.I., Chen L. Evaluation of Serum microRNAs in Patients with Diabetic Kidney Disease: A Nested Case-Controlled Study and Bioinformatics Analysis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019;25:1699–1708. doi: 10.12659/MSM.913265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren H.W., Ma X.Y., Shao Y., Han J.Y., Yang M., Wang Q.Y. Correlation Between Serum miR-154-5p and Osteocalcin in Males and Postmenopausal Females of Type 2 Diabetes With Different Urinary Albumin Creatinine Ratios. Front. Endocrinol. 2019;10:542. doi: 10.3389/fendo.2019.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren H., Wu C., Shao Y., Liu S., Zhou Y., Wang Q. Correlation between serum miR-154-5p and urinary albumin excretion rates in patients with type 2 diabetes mellitus: A cross-sectional cohort study. Front. Med. 2020;14:642–650. doi: 10.1007/s11684-019-0719-3. [DOI] [PubMed] [Google Scholar]
- 82.Roux M., Perret C., Feigerlova E., Mohand Oumoussa B., Saulnier P.-J., Proust C., Trégouët D.A., Hadjadj S. Plasma levels of hsa-miR-152-3p are associated with diabetic nephropathy in patients with type 2 diabetes. Nephrol. Dial. Transpl. Off. Publ. Eur. Dial. Transpl. Assoc.-Eur. Ren. Assoc. 2018;33:2201–2207. doi: 10.1093/ndt/gfx367. [DOI] [PubMed] [Google Scholar]
- 83.Rovira-Llopis S., Escribano-Lopez I., Diaz-Morales N., Iannantuoni F., Lopez-Domenech S., Andújar I., Jover A., Pantoja J., Pallardo L.M., Bañuls C., et al. Downregulation of miR-31 in Diabetic Nephropathy and its Relationship with Inflammation. Cell Physiol. Biochem. Int. J. Exp. Cell Physiol. Biochem. Pharmacol. 2018;50:1005–1014. doi: 10.1159/000494485. [DOI] [PubMed] [Google Scholar]
- 84.Shao Y., Ren H., Lv C., Ma X., Wu C., Wang Q. Changes of serum Mir-217 and the correlation with the severity in type 2 diabetes patients with different stages of diabetic kidney disease. Endocrine. 2017;55:130–138. doi: 10.1007/s12020-016-1069-4. [DOI] [PubMed] [Google Scholar]
- 85.Sham S.Y.Z., Ng C.T., Azwar S., Yip W.K., Abdullah M., Thevandran K., Osman M., Seow H.F. Circulating miRNAs in Type 2 Diabetic Patients with and without Albuminuria in Malaysia. Kidney Blood Press Res. 2022;47:81–93. doi: 10.1159/000518866. [DOI] [PubMed] [Google Scholar]
- 86.Su J., Ren J., Chen H., Liu B. MicroRNA-140-5p ameliorates the high glucose-induced apoptosis and inflammation through suppressing TLR4/NF-κB signaling pathway in human renal tubular epithelial cells. BioSci. Rep. 2020;40:BSR20192384. doi: 10.1042/BSR20192384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiao Y., Guo S., Zhang Y., Bian Z., Jia L., Hu Y., Chen J., Yin C., Li N., Zhang D., et al. Diabetic nephropathy: Serum miR-9 confers a poor prognosis in and is associated with level changes of vascular endothelial growth factor and pigment epithelium-derived factor. Biotechnol. Lett. 2017;39:1583–1590. doi: 10.1007/s10529-017-2390-6. [DOI] [PubMed] [Google Scholar]
- 88.Xie J.X., Fan X., Drummond C.A., Majumder R., Xie Y., Chen T., Liu L., Haller S.T., Brewster P.S., Dworkin L.D., et al. MicroRNA profiling in kidney disease: Plasma versus plasma-derived exosomes. Gene. 2017;627:1–8. doi: 10.1016/j.gene.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zang J., Maxwell A.P., Simpson D.A., McKay G.J. Differential Expression of Urinary Exosomal MicroRNAs miR-21-5p and miR-30b-5p in Individuals with Diabetic Kidney Disease. Sci. Rep. 2019;9:10900. doi: 10.1038/s41598-019-47504-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang L., Li R., He J., Yang Q., Wu Y., Huang J., Wu B. Co-expression analysis among microRNAs, long non-coding RNAs, and messenger RNAs to understand the pathogenesis and progression of diabetic kidney disease at the genetic level. Methods. 2017;124:46–56. doi: 10.1016/j.ymeth.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang J., Zhang L., Zha D.Q., Wu X.Y. Inhibition of miRNA-135a-5p ameliorates TGF-beta 1-induced human renal fibrosis by targeting SIRT1 in diabetic nephropathy. Int. J. Mol. Med. 2020;46:1063–1073. doi: 10.3892/ijmm.2020.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao Y., Shen A., Guo F., Song Y., Jing N., Ding X., Pan M., Zhang H., Wang J., Wu L. Urinary Exosomal MiRNA-4534 as a Novel Diagnostic Biomarker for Diabetic Kidney Disease. Front. Endocrinol. 2020;11:590. doi: 10.3389/fendo.2020.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou J., Peng R., Li T., Luo X., Peng H., Zha H., Yin P., Wen L., Zhang Z. A potentially functional polymorphism in the regulatory region of let-7a-2 is associated with an increased risk for diabetic nephropathy. Gene. 2013;527:456–461. doi: 10.1016/j.gene.2013.06.088. [DOI] [PubMed] [Google Scholar]
- 94.Berillo O., Huo K.-G., Fraulob-Aquino J.C., Richer C., Briet M., Boutouyrie P., Lipman M.L., Sinnett D., Paradis P., Schiffrin E.L. Circulating let-7g-5p and miR-191-5p are independent predictors of chronic kidney disease in hypertensive patients. Am. J. Hypertens. 2020;33:505–513. doi: 10.1093/ajh/hpaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang Y.-Q., Huang C., Li J., Zhang B., Feng Y.-Q. The association of miR-29a with proteinuria in essential hypertension. J. Hum. Hypertens. 2018;32:775–780. doi: 10.1038/s41371-018-0097-3. [DOI] [PubMed] [Google Scholar]
- 96.Huang C., Huang Y.-Q. The correlation of circulating miR-29b and inflammatory markers with albuminuria in hypertensive patients. Clin. Exp. Hypertens. 2020;42:743–747. doi: 10.1080/10641963.2020.1790585. [DOI] [PubMed] [Google Scholar]
- 97.Nandakumar P., Tin A., Grove M.L., Ma J., Boerwinkle E., Coresh J., Chakravarti A. MicroRNAs in the miR-17 and miR-15 families are downregulated in chronic kidney disease with hypertension. PLoS ONE. 2017;12:e0176734. doi: 10.1371/journal.pone.0176734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perez-Hernandez J., Olivares D., Forner M.J., Ortega A., Solaz E., Martinez F., Chaves F.J., Redon J., Cortes R. Urinary exosome miR-146a is a potential marker of albuminuria in essential hypertension. J. Transl. Med. 2018;16:228. doi: 10.1186/s12967-018-1604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Perez-Hernandez J., Riffo-Campos A.L., Ortega A., Martinez-Arroyo O., Perez-Gil D., Olivares D., Solaz E., Martinez F., Martínez-Hervás S., Chaves F.J., et al. Urinary- and Plasma-Derived Exosomes Reveal a Distinct MicroRNA Signature Associated With Albuminuria in Hypertension. Hypertension. 2021;77:960–971. doi: 10.1161/HYPERTENSIONAHA.120.16598. [DOI] [PubMed] [Google Scholar]
- 100.Fish J.E., Santoro M.M., Morton S.U., Yu S., Yeh R.-F., Wythe J.D., Ivey K.N., Bruneau B.G., Stainier D.Y.R., Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou J., Li Y.-S., Nguyen P., Wang K.-C., Weiss A., Kuo Y.-C., Chiu J.-J., Shyy J.Y., Chien S. Regulation of vascular smooth muscle cell turnover by endothelial cell–secreted microRNA-126: Role of shear stress. Circ. Res. 2013;113:40–51. doi: 10.1161/CIRCRESAHA.113.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Prattichizzo F., Giuliani A., Ceka A., Rippo M.R., Bonfigli A.R., Testa R., Procopio A.D., Olivieri F. Epigenetic mechanisms of endothelial dysfunction in type 2 diabetes. Clin. Epigenetics. 2015;7:56. doi: 10.1186/s13148-015-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Metzinger-Le Meuth V., Burtey S., Maitrias P., Massy Z.A., Metzinger L. microRNAs in the pathophysiology of CKD-MBD: Biomarkers and innovative drugs. BBA-Mol. Basis Dis. 2017;1863:337–345. doi: 10.1016/j.bbadis.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 104.Panizo S., Martínez-Arias L., Alonso-Montes C., Cannata P., Martín-Carro B., Fernández-Martín J.L., Naves-Díaz M., Carrillo-López N., Cannata-Andía J.B. Fibrosis in Chronic Kidney Disease: Pathogenesis and Consequences. Int. J. Mol. Sci. 2021;22:408. doi: 10.3390/ijms22010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kölling M., Kaucsar T., Schauerte C., Hübner A., Dettling A., Park J.-K., Busch M., Wulff X., Meier M., Scherf K., et al. Therapeutic miR-21 Silencing Ameliorates Diabetic Kidney Disease in Mice. Mol. Ther. 2017;25:165–180. doi: 10.1016/j.ymthe.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun Q., Miao J., Luo J., Yuan Q., Cao H., Su W., Zhou Y., Jiang L., Fang L., Dai C. The feedback loop between miR-21, PDCD4 and AP-1 functions as a driving force for renal fibrogenesis. J. Cell Sci. 2018;131:jcs202317. doi: 10.1242/jcs.202317. [DOI] [PubMed] [Google Scholar]
- 107.Sun I.O., Lerman L.O. Urinary microRNA in kidney disease: Utility and roles. Am. J. Physiol.-Ren. Physiol. 2019;316:F785–F793. doi: 10.1152/ajprenal.00368.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen H.-Y., Zhong X., Huang X.R., Meng X.-M., You Y., Chung A.C.K., Lan H.Y. MicroRNA-29b Inhibits Diabetic Nephropathy in db/db Mice. Mol. Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zen K., Zhang C. Circulating microRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 110.Cui C., Cui Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020;10:5644. doi: 10.1038/s41598-020-62534-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sakuma H., Hagiwara S., Kantharidis P., Gohda T., Suzuki Y. Potential Targeting of Renal Fibrosis in Diabetic Kidney Disease Using MicroRNAs. Front. Pharmacol. 2020;11:587689. doi: 10.3389/fphar.2020.587689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo L., Tan K., Luo Q., Bai X. Dihydromyricetin promotes autophagy and attenuates renal interstitial fibrosis by regulating miR-155-5p/PTEN signaling in diabetic nephropathy. Bosn. J. Basic Med. Sci. 2020;20:372–380. doi: 10.17305/bjbms.2019.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao D., Jia J., Shao H. miR-30e targets GLIPR-2 to modulate diabetic nephropathy: In vitro and in vivo experiments. J. Mol. Endocrinol. 2017;59:181–190. doi: 10.1530/JME-17-0083. [DOI] [PubMed] [Google Scholar]
- 114.Lin X., You Y., Wang J., Qin Y., Huang P., Yang F. MicroRNA-155 deficiency promotes nephrin acetylation and attenuates renal damage in hyperglycemia-induced nephropathy. Inflammation. 2015;38:546–554. doi: 10.1007/s10753-014-9961-7. [DOI] [PubMed] [Google Scholar]
- 115.Wang Y., Zheng Z.J., Jia Y.J., Yang Y.L., Xue Y.M. Role of p53/miR-155-5p/sirt1 loop in renal tubular injury of diabetic kidney disease. J. Transl. Med. 2018;16:146. doi: 10.1186/s12967-018-1486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Krupa A., Jenkins R., Luo D.D., Lewis A., Phillips A., Fraser D. Loss of MicroRNA-192 Promotes Fibrogenesis in Diabetic Nephropathy. J. Am. Soc. Nephrol. 2010;21:438–447. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jenkins R.H., Martin J., Phillips A.O., Bowen T., Fraser D.J. Pleiotropy of microRNA-192 in the kidney. Biochem. Soc. Trans. 2012;40:762–767. doi: 10.1042/BST20120085. [DOI] [PubMed] [Google Scholar]
- 118.Witwer K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015;61:56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 119.Li J., Ma L., Yu H., Yao Y., Xu Z., Lin W., Wang L., Wang X., Yang H. MicroRNAs as Potential Biomarkers for the Diagnosis of Chronic Kidney Disease: A Systematic Review and Meta-Analysis. Front. Med. 2021;8:782561. doi: 10.3389/fmed.2021.782561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roberts T.C., Coenen-Stass A.M.L., Wood M.J.A. Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS ONE. 2014;9:e89237. doi: 10.1371/journal.pone.0089237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zampetaki A., Mayr M. Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb. Haemost. 2012;108:592–598. doi: 10.1160/TH12-02-0097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study is available upon reasonable request from the corresponding author.