Abstract
Protein kinases and phosphatases catalyze the phosphorylation and dephosphorylation of their protein substrates, respectively, and these are important mechanisms in cellular signal transduction. The rice blast fungus Magnaporthe oryzae possesses 6 protein phosphatases of type 2C class, including MoPtc1, 2, 5, 6, 7 and 8. However, only very little is known about the roles of these phosphatases in filamentous fungi. Here in, we deployed genetics and molecular biology techniques to identify, characterize and establish the roles of MoPtc5 and MoPtc7 in M. oryzae development and pathogenicity. We found that during pathogen-host interaction, MoPTC7 is differentially expressed. Double deletion of MoPTC7 and MoPTC5 suppressed the fungal vegetative growth, altered its cell wall integrity and reduced its virulence. The two genes were found indispensable for stress tolerance in the phytopathogen. We also demonstrated that disruption of any of the two genes highly affected appressorium turgor generation and Mps1 and Osm1 phosphorylation levels. Lastly, we demonstrated that both MoPtc5 and MoPtc7 are localized to mitochondria of different cellular compartments in the blast fungus. Taken together, our study revealed synergistic coordination of M. oryzae development and pathogenesis by the type 2C protein phosphatases.
Keywords: blast fungus, protein phosphatases, phosphorylation, pathogenicity
1. Introduction
Magnaporthe oryzae is a filamentous fungus that causes the blast disease of rice which results in severe losses of global rice production. Due to the economic significance and experimental tractability of M. oryzae, it is considered a perfect pathosystem for studying host-pathogen interactions [1]. Infection wise, M. oryzae is similar to other important pathogens of cereals [2,3]. Hence there is possibility of establishing at least a broad-spectrum drug that targets common disease determinants in the fungi for effective disease management. The rice blast fungus causes up to 15% of the potential worldwide rice harvest, and this is adequate food for more than sixty million people [4]. M. oryzae is highly destructive to rice partly because of its ability to adopt different cycles (polycyclic pathogen) within a growing season [5]. M. oryzae infects rice plants at all stages of development and attacks the leaves, stems, nodes and panicles [2]. For the infection cycle, conidia are dispersed by wind onto the host leaf surfaces where each conidium develops a germ tube, the tip of which swells into an infection structure called appressorium [6]. Mature appressorium then accumulates turgor pressure (up to about 8 MPa) that mechanically ruptures the leaf cuticle to enable the pathogen enter the plant cells [7]. Under favorable environmental conditions, lesions occur within 3–4 days of infection, after which new conidia are produced to start a new disease cycle [3]. Previously, some studies revealed that a group of signaling pathways regulate the pathogenicity of M. oryzae, including MAPK and cAMP pathways which affect conidiation as well as appressorium formation [8,9].
Cellular processes such as cell growth and differentiation, pH regulation, cell cycle, morphogenesis and apoptosis are largely regulated via phosphorylation and dephosphorylation of proteins by kinase and phosphatase groups of enzymes [10]. Protein phosphorylation constitutes a very essential mechanism by which higher plants and animals regulate cellular processes [11]. In particular, signal transductions are transmitted through phosphorylation and dephosphorylation of some enzymes and regulatory proteins. Protein phosphatases of serine/threonine family are classified into PPM and PPP superfamilies. Members of the latter superfamily have both regulatory and catalytic subunits [12], while members of the former are monomeric enzymes [13].Generally, both groups are share common inhibitors; and PP2C phosphatase requires Mg2+ or Mn2+ as coenzymes [14].
Within the PP2C family, Ptc1 is identified in Saccharomyces cerevisiae to be involved in the regulation of MAPK pathways [15]. In S. cerevisiae, Ptc1 regulates stress response pathways [15,16]. Furthermore, Ptc1 was shown to mediate tRNA splicing, organelles distribution, inheritance, cation homeostasis and sporulation in budding yeast [17]. In S. cerevisiae, Ptc2 proteins share 60% identity, and both proteins are structurally different from Ptc1 as each of them possesses a 170 residues extension at its carboxyl terminus, which allows them achieve maximum activity [16]. Protein phosphatases play vital roles in preserving cell viability when the cells are exposed to agents that compromise DNA integrity, thereby maintaining genome stability [18]. in S. cerevisiae, cell cycle is partly regulated by synergistic effects of Ptc2 and Ptc3 on dephosphorylation of Thr-169 in Cdc28 kinase [19]. Ptc4 is localized in the cytoplasm in S. cerevisae and its overexpression significantly reduced the phosphorylation activity of Hog1 [20]. Ptc2 and Ptc4 participate in cell wall integrity, and possibly limit the hyperphosphorylation of CWI pathway [21].
There is limited information on the functions of Ptc5 and Ptc7 in filamentous fungi like M. oryzae, but in budding yeast, these proteins have been shown to regulate pyruvate dehydrogenase activity by dephosphorylating Pda1, an E1 α-subunit of the PDH complex in mitochondria [22]. Ptc7 regulates the PDH activity by dephosphorylating the canonical citrate synthase of the tricarboxylic acid cycle CIT1, thereby maintaining its proper dimerization during its activity. Deletion of AP2C1 and PP2C5 generated plant mutants with abnormal physiologic and phenotypic characteristics [23]. More so, both Ptc5 and Ptc7 were previously demonstrated to be involved in rapamycin-induced stress resistance [24]. Therefore, understanding the relationship between these proteins (Ptc5 and Ptc7) and the pathogenesis and development of M. oryzae would be critical in handling the menace of the rice blast infection, thereby strengthening the global food security. As such, we employed gene deletion approach to investigate the functions of MoPtc5 and MoPtc7 in relation to the pathogenesis and development of M. oryzae. Our findings indicated that deletion of MoPTC5 and MoPTC7 perturbs the fungal vegetative growth, conidiation, conidiophores development, pathogenicity and Mps1 and Osm1 phosphorylation levels. Our results also reveal that the type 2C protein phosphatases are required for multiple stress tolerance in the rice blast pathogen.
2. Materials and Methods
2.1. Strains and Culture Conditions
Guy11 strain of M. oryzae was used in this study as the WT (wild type), and the various mutant strains were generated from the Guy11 background. Complete media was used to culture the fungal strains [25]. Liquid complete media (CM) were used for the growth of mycelia for nucleic acids extractions and protoplast preparation. TB3 medium (200 g sucrose, 3 g casamino acids, 3 g yeast extract and 7 g agar in 1 L of distilled water) was used for transformation, growth and screening of transformants.
Rice bran media (RBM: 40 g of rice bran in 1 L of ddH2O) was used for induction of conidiation and the fungal cultures were incubated in the dark at 28 °C for 10 days, and then exposed to light condition for another 3 days. Conidia were harvested, washed with sterile double distilled water (ddH2O), filtered and counted under a light microscope with the help of a hemocytometer. For growth assays, we cultured the strains on complete media (CM: 6 g yeast extract, 6 g casamino acid, 10 g sucrose in 1 L of water), Starch yeast media (SYM: 10 g starch extract, 2 g yeast extract, 3 g sucrose) and RBM.
2.2. Targeted Gene Deletion and Complementation
For MoPP2C gene deletion, homologous recombination approach was adopted. The flanking regions of the phosphatase genes were amplified using the appropriate primers (Table S1). The PCR products were then ligated with hygromycin phosphotransferase (hph) gene by overlap PCR. M. oryzae protoplasts were prepared, into which the above constructs were transformed following the protocols previously reported [26]. A TB3 medium was used for the screening of transformants in the presence of hygromycin B (Roche Applied Science, Penzberg, Germany). Appropriate mutants were subjected to Southern blot (elaborated in Section 2.5 below) for confirmation.
For complementation assay, complementation vectors were designed by amplifying full length ORFs (and their respective promoters) of the deleted genes and ligating them with GFP. These products were then cloned into PKNTG plasmid, respectively, after digestion with a mixture of KpnI and HindIII restriction enzymes. The recombinant DNA construct was sequenced to ensure successful cloning. The MoPTC5 and MoPTC7 constructs were respectively transformed into the protoplasts of ΔMoptc5 and ΔMoptc7 mutants and the transformants screened by PCR using the appropriate primers. For confirmation, GFP signals were analyzed by confocal microscopy (Nikon, Tokyo, Japan).
2.3. Infection Assays, Cuticle Penetration and Incipient Cytorrhysis Assay
To perform rice infection assay, we prepared conidia suspensions (5 × 104 spores/mL) from Guy11, ΔMoptc5, ΔMoptc7, ΔMoptc7ΔMoptc5, ΔMoptc5_C, and ΔMoptc7_C strains and sprayed them on 3-week-old blast susceptible rice seedling (Oryza sativa, CO39). The seedlings were then incubated in the dark at 28 °C for 24 h in a humid growth chamber, and later subjected to 12/12 h of light/dark photoperiod. Leaves were then collected from the seedlings for observation of disease lesions at 7 days post infection (dpi). For barley infection assay, 10 days old barley leaves were subjected to same treatments and incubated under same conditions. Leaves from the infected plants were collected for observation at 7 days.
To study the cuticle penetration potential, we dropped 10 μL of conidia suspensions (5 × 104 spores/mL) from the various strains onto 10 days old barley leaves, and incubated the leaves at 28 °C for 30 h, 48 h and 60 h under humid condition. A confocal microscope (Nikon, Japan) was then used to check for cuticle penetration and growths of invasive hyphae.
The turgor pressures in the appressoria of the strains were analyzed by incipient cytorrhysis assay on hydrophobic coverslips after 24 h of treatments with glycerol solutions (1, 2, and 3 M). The number of collapsed appressoria was counted under a light microscope.
2.4. Microscopy
While a confocal microscope was used for investigation of subcellular localizations, light microscopy using Olympus DP80 (Tokyo, Japan) was utilized for conidiophores formation assay, cytorryhsis incipient assay, invasive hyphae development and appressorium penetration and glycogen degradation.
2.5. Southern Blot Analysis, Extraction of Total RNA, qRT-PCR and Gene Expressions
For southern blot assay, we extracted DNA from the fungal hyphae following cetyl trimethyl ammonium bromide (CTAB) method [25]. Restriction digestion, gel electrophoresis, blotting, probe labeling, ligation and hybridization were conducted using a commercial kit (Roche), following the manufacturer protocol. Briefly, the gels were immersed in alkaline buffer for 15 min with mild shaking to depurinate the fractioned DNA and denature them by soaking in the gel blotting solution (0.4 M NaOH, 0.6 M NaCl) for 30 min. The gels were then moved to neutralization buffer (1.5 M NaCl, 0.5 M Tris -HCl pH 7.5) for 30 min before capillary blotting on hybond-N (GE-healthcare). The gel blots were achieved by putting the inverted gels on the sheet of filter papers. The hybond–N membrane was then placed on the gels and covered with filter papers. The filter papers were being regularly changed after getting wet for 2 days. The membrane was UV cross-linked immediately probed or wrapped for storage in saran wrap.
To examine the gene transcription profiles in planta at different growth stages, we sprayed Guy11 conidia on 3 weeks old CO-39 rice, the leaf samples were then collected at different time points (4 h, 8 h, 12 h, 24 h, 48 h and 72 h). Next, we extracted total RNA using an RNA extraction kit (Eastep super RNA) and cDNA was prepared following the protocols described in the manual (accurate biology, Hunan, China). qPCR was performed where actin gene was used as an internal control. Mean Ct values were normalized as previously described [27].
For analysis of the expressions of conidiation responsive and chitinase encoding genes in the various strains, we inoculated the strains in liquid CM placed in an incubator shaker at 28 °C, 110 rpm for 3 days. After this period, samples were collected and total RNA extracted using a kit (Eastep super RNA). cDNA were prepared following the aforementioned protocols. qPCR was conducted and tubulin gene was used as an internal control. Mean Ct values were normalized as previously described [27]. The primers used are listed in Table S1.
2.6. Bioinformatics Analysis
Protein sequence alignment was performed using DNAMAN software. The protein sequences were obtained from NCBI (National Center for Biotechnology Information) database by BLAST analysis of the protein sequences of PP2C homologs in S. cerevisiae. The identified homologs in M. oryzae were confirmed by BLASTp search at the fungi and oomycetes genomics resources database (http://fungidb.org/fungidb/, accessed on 10 January 2022). For domain prediction and phylogenetic analysis, amino acid sequences of MoPP2c and their respective orthologs were fed into Pfam based domain prediction analysis software and the various domains identified, while phylogenetic analyses were performed using MEGA x software, adopting Neighbor-Joining method and 1000 bootstrap.
2.7. Cell Wall Integrity, Osmotic and Oxidative Stresses and Cell Wall Thickness Assays
Sodium dodecyl sulphate (SDS, 0.01%), Calcofluor White (CFW, 200 µg/mL) and Congo Red (CR, 200 µg/mL) were supplemented into CM agar, respectively, on which the fungal strains were grown for analysis of cell wall stress sensitivity. For osmotic stress assay, CM media was supplemented with sodium chloride (NaCl, 1 M), potassium chloride (KCl, 1 M) and sorbitol (1 M). For oxidative stress assay, the CM was supplemented with hydrogen peroxides (5 mM and 10 mM H2O2). The strains were cultured and their colony diameters measured at 10 dpi. Inhibition rates were calculated as previously described [28]. For cell wall thickness assay, we inoculated the strains in liquid CM and incubated under constant agitation for three days. Next, mycelia were harvested and dried. Transmission electron micrographs of the transverse section of the cell walls were taken by scanning electron microscopy (SEM).
2.8. Western Blot Assay
The fungal mycelia were used for extraction of total proteins as previously described [29]. The extracted proteins were subjected to SDS-PAGE and fixed on PDVF (polyvinylidene fluoride) membranes, and the proteins of interest were spotted by antiphospho p44/42 and P42/44 primary antibodies. HRP-Rabbit were used as conjugated secondary antibodies. A western blot detection Kit (Advansta, San Jose, CA, USA) was used for signals visualization using an imaging system (Tanon 5200). Pictures were processed using imaging software.
2.9. Subcellular Localization Assays
One Step Cloning Kit (Vazyme, Nanjing, China) was used for the construction of MoPtc5 and MoPtc7-GFP and RFP vectors. The vectors were transformed into the WT background, respectively. The transformants were screened by GFP and RFP fluorescence signal analyses at different developmental stages in planta, by confocal microscopy. Images were processed using NIS-viewer software.
3. Results
3.1. Phylogenetic Analysis of MoPtc5 and MoPtc7
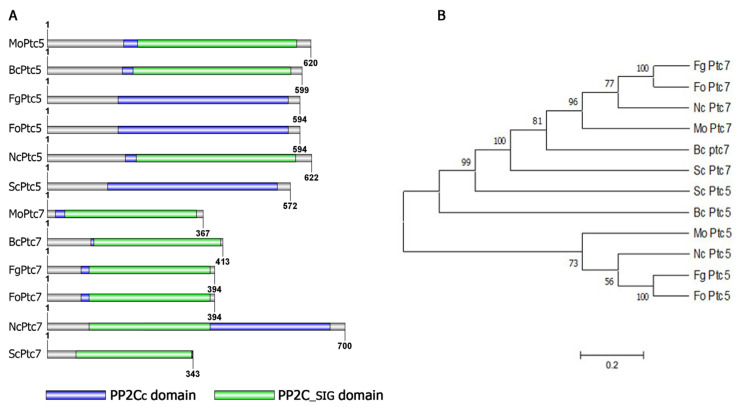
The putative homologs of Ptc5 and Ptc7 were identified in M. oryzae by BLASTp search at the fungi and oomycetes database using S. cerevisiae Ptc5 and Ptc7 sequences as queries. The BLASTp hits identified two copies of PP2C genes MGG_03154 and MGG_00166 in the rice blast genome, which were subsequently named MoPTC5 and MoPTC7, respectively. Protein structure and domain prediction analysis showed that MoPtc5 protein contains a single PP2C domain while MoPtc7 has a PP2C and PP2C-SIG domains (Figure 1A). Phylogenetic analysis revealed that MoPtc5 shared a close homology with its orthologs in Neurospora crassa and Fusarium oxyporum while MoPtc7 has close homology with its orthologs in Botrytis cinera and N. crassa (Figure 1B).
Figure 1.
Domain architecture and phylogenetic analysis of Ptc5 and Ptc7 in different fungi. (A) Domain architecture of Ptc5 and Ptc7 orthologs in in different organisms. (B) A phylogenetic tree generated using MEGA X software and a UPGMA test with 1000 Boot straps replications was conducted. Fg: Fusarium gamineum; Fo: Fusarium oxyporum; Nc: Neurospora crassa; Mo: Magnaporthe oryzae; Bc: Botytis cinera; and Sc: Saccharomyces cerevisae.
3.2. MoPtc5 and MoPtc7 Are Differentially Expressed during Pathogen-Host Interaction
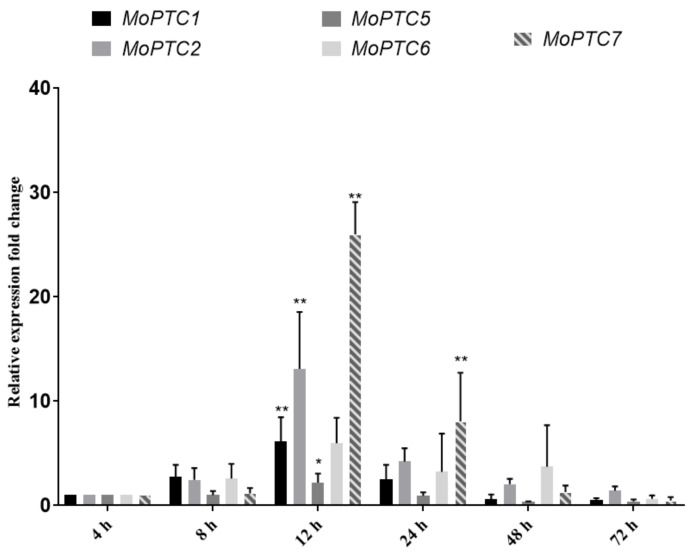
To gain a clue on the importance of MoPtc5 and MoPtc7 in M. oryzae virulence, we first decided to check the expression patterns of all the type 2C genes (MoPTC1, MoPTC2, MoPTC5, MoPTC6 and MoPTC7) in Guy11 during host infection, at different time points. To achieve this, we harvested conidia from the WT (Guy11) strain and sprayed them on 21-day-old rice seedlings. We then quantified the type 2C genes expression levels at 8, 12, 24, 48 and 72 h. We found that, with the exception of MoPTC6, all the type 2C genes were significantly expressed at 12 hpi (hours post inoculation), while only MoPTC7 was significantly expressed at 24 hpi (Figure 2). These results show that MoPtc5 and MoPtc7 could be involved in the development and virulence of M. oryzae.
Figure 2.
Transcriptional expression patterns of M. oryzae type 2C genes during interaction with host. Quantitative realtime (q-PCR) was used to check the genes expressions at the indicated time points. The 4 h time point was taken as background control with tubulin gene serving as internal control. The experiments were repeated three times with three technical replicates each. Error bars represent standard deviations while asterisks show significant differences at different p-values (* p < 0.1; ** p < 0.01). The data were analyzed by one-way ANOVA using Tukey’s multiple-comparison test.
3.3. MoPtc5 and MoPtc7 Redundantly Contribute to Vegetative Growth of M. oryzae
In order to examine the roles played by MoPtc5 and MoPtc7 in the vegetative growth of the fungal pathogen, we generated ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants by homologous recombination approach (Figure S1). The mutants (plus the controls) were then cultured on CM, SYM and RBM and incubated for 10 days at 28 °C. Compared to the controls, the colony diameters of ΔMoptc5 and ΔMoptc7 on CM and RBM were not significantly different, while ΔMoptc5 revealed significant difference on SYM (Figure 3A,B). In contrast, a significant reduction was observed in the growth of the double deletion mutant ΔMoptc7ΔMoptc5 on both SYM and RBM media. These suggest the redundant roles of MoPtc5 and MoPtc7 in the vegetative growth of the blast fungus.
Figure 3.
Synergistic roles of MoPtc5 and MoPtc7 in M. oryzae vegetative growth of. (A) The growths of the various strains on different media plates. CM, SYM and RBM were used for the growth of the fungal mycelia and incubated in the dark for 10 days at 28 °C. (B) Bar graphs showing the colony diameters of the various strains on the indicated media. Asterisks show significant differences at different p-values (* p < 0.1; ** p < 0.01) while error bars represent standard deviations from three independent replicates. The data were analyzed by one-way ANOVA using Tukey’s multiple-comparison test.
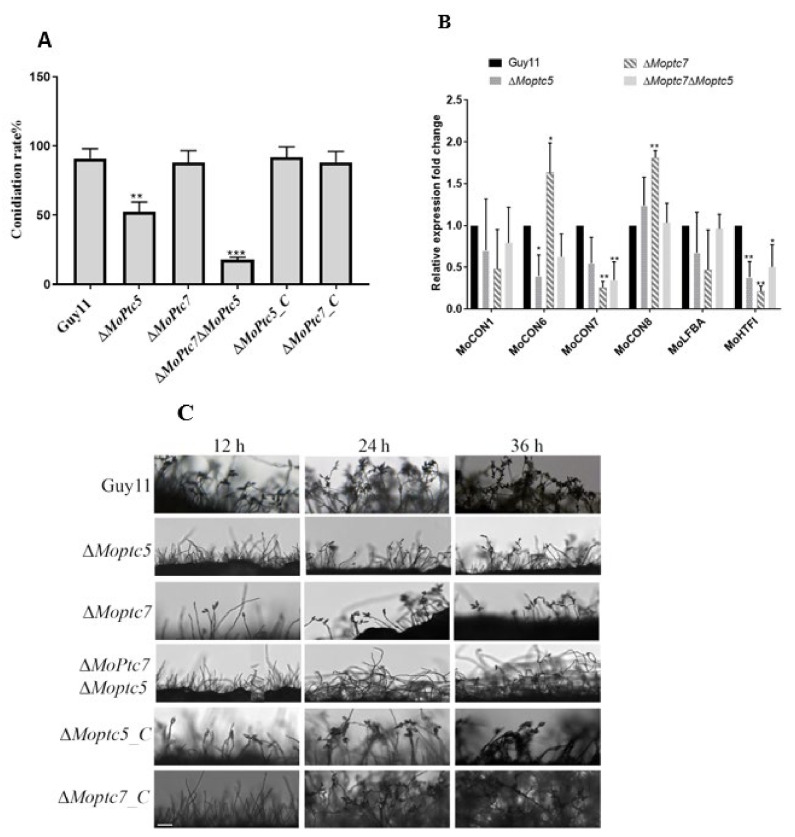
3.4. MoPtc5 Is Involved in Asexual Reproduction of the Rice Blast Fungus
To understand the roles of MoPtc5 and MoPtc7 in M. oryzae conidiation, we cultured the controls and mutant strains on RBM for 10 days period, and monitored conidia development. From our results, we found that ΔMoptc7 produced almost similar quantity of conidia as the WT. However, conidia production in the ΔMoptc5 and ΔMoptc7ΔMoptc5 mutants was significantly reduced compared to Guy11 (Figure 4A). Since conidia are always borne on conidiophores, we decided to check for conidiophore formation of the ΔMoptc5 and ΔMoptc7ΔMoptc5 mutants at 12 h, 24 h and 36 h time points. The results indicated that the number of conidiophores in both ΔMoptc5 and ΔMoptc7ΔMoptc5 mutants were highly reduced especially at 12 h and 24 h compared to the controls (Figure 4B). We further conducted qPCR to check the expressions of some conidiation-responsive genes, including MoCON1, MoCON6, MoCON7, MoCON8, MoFLBA and MoHTFI. We found that the expressions of MoCON1, MoCON6, MoCON7 and MoHTFI were downregulated in ΔMoptc5 and ΔMoptc7ΔMoptc5 mutants (Figure 4C). Put together, we conclude here that MoPtc5 is important for M. oryzae asexual reproduction.
Figure 4.
MoPTC5 deletion causes reduction in conidiation and conidiophores formation of M. oryzae. (A) Conidia production of the strains grown on RBM for 10 days period. (B) Conidiophores formation from Guy11, ΔMoptc5, and ΔMoptc7ΔMoptc5 strains on RBM. ΔMoptc7ΔMoptc5 double mutant shows highly reduced conidiophores formation. (C) Relative expressions of genes involved in M. oryzae conidiation. The various strains were inoculated in liquid CM and incubated at 28 °C under constant shaking at 110 rpm for 3 days, after which total RNAs were extracted for qPCR analysis. Tubulin gene was used as a control. Results are means obtained from three independent replicates. Error bars represent standard deviations while asterisks show significant differences at different p-values (*, p < 0.1; **, p < 0.01; ***, p < 0.001)). The data were analyzed by one-way ANOVA using Tukey’s multiple-comparison test.
3.5. MoPtc5 and MoPtc7 Are Involved in Cell Wall Stress Resistance and Dephosphorylation of MoMps1
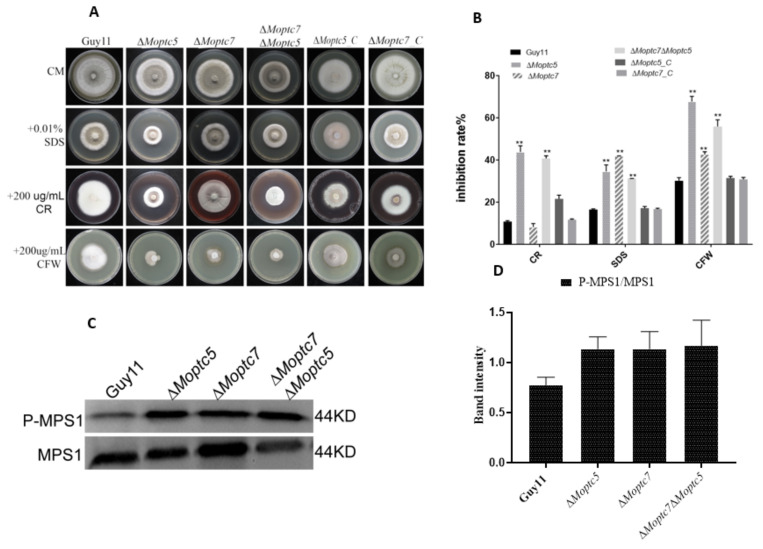
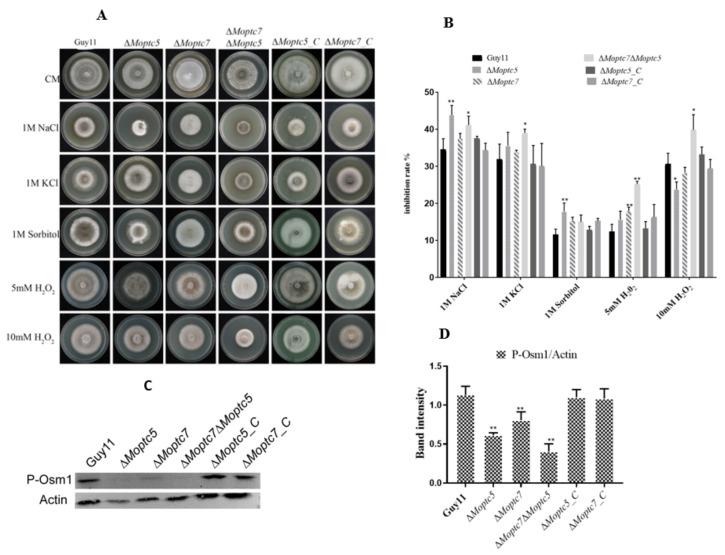
To analyse the effects of cell wall stress on ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants, we supplemented sodium dodecyl sulphate (SDS, 0.01%), Calcofluor White (CFW, 200 µg/mL) and Congo red (CR, 200 µg/mL) to separate CM growth media. We then cultured the fungal strains on these media and incubated for 10 days in the dark at 28 °C, after which the colony diameters of the cultures were measured. Growth inhibition rates were calculated as earlier stated (Materials and Methods). We found that the growths of ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 were affected towards cell wall stress agents (Figure 5A), however ΔMoptc5 and ΔMoptc7ΔMoptc5 were highly inhibited on media containing SDS, CR and CFW, while ΔMoptc7 significantly inhibited only by SDS and CFW (Figure 5B), these results suggesting the importance of MoPtc5 and MoPtc7 in stress resistance. Since MoMps1 phosphorylation level is required for M. oryzae cell wall integrity and the two type 2C proteins are protein phosphatases [9], we quantified MoMps1 phosphorylation level in the fungal samples by western blot. We recorded an increase in MoMps1 phosphorylation in ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants, although the differences were not significant when compared to Guy11 (Figure 5C,D). We therefore conclude that MoPtc5 and MoPtc7 are involved in stress resistance and MoMps1 dephosphorylation in the rice blast fungus.
Figure 5.
MoPtc5 and MoPtc7 are involved in stress resistance in the blast fungus. (A) Colony diameters of the strains were measured 10 days post inoculation on CM media containing cell wall-perturbing agents. (B) Growth inhibition rates of the mutants due to the effects of the cell wall stressing substances (Inhibition rate = (the diameter of untreated strain − the diameter of treated strain)/(the diameter of untreated strain) × 100%. Three independent replicates were involved. Error bars represent standard deviations while asterisks show significant differences at different p-values (** p < 0.01). The data were analyzed by one-way ANOVA using Tukey’s multiple-comparison test. (C) MoMps1 phosphorylation level in ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants. (D) Comparison of band intensities for MoMps1 phosphorylation levels in the various strains as calculated using imaging software. Despite the obvious increase in the phosphorylation levels, the differences were not significant compared to the wild type.
3.6. MoPtc5 and MoPtc7 May Mediate Chitin Synthesis to Confer Resistance to Cell Wall Stress in the Blast Fungus
Considering our earlier finding that showed the involvement of MoPtc5 and MoPtc7 cell wall stress resistance, we decided to investigate whether the two proteins contribute to the fungal cell wall thickness. The results from transmission electron microscopy indicated no significant difference in the cell wall thickness of the WT, ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 strains (Figure 6A,B). Since the cell wall integrity is intact, we reasoned that the reduction of mycelial growth on media containing cell wall perturbing agents could have been due to altered chitin synthesis. As such, qPCR assay was performed to check the expression levels of the chitin synthase encoding genes MoCHS1, MoCHS2, MoCHS3, MoCHS4, MoCHS5, MoCHS6 and MoCHS7 in the various strains. We found that the expression of MoCHS2 was upregulated in ΔMoptc7 and ΔMoptc7ΔMoptc5, while MoCHS7 was highly expressed in ΔMoptc7. However, all of these genes were downregulated in ΔMoptc5 mutant, though with no significant difference (Figure 6C). These results suggest an alteration in chitin content due to MoPTC5 and MoPTC7 deletions.
Figure 6.
Deletion of MoPTC5 and MoPTC7 does not affect cell wall thickness. (A) Transverse sections of hyphae from Guy11, ΔMoptc5, ΔMoptc7 and ΔMoptc5ΔMoptc7 strains following transmission electron microscopy. (B) Graphs indicating differences in hyphal cell wall thickness between the mutants and Guy11. (C) The transcript levels of chitin encoding genes using actin gene as internal control. Three replicates were involved. Error bars represent standard deviations while asterisks show significant differences at different p-values (* p < 0.1; ** p < 0.01). The data were analyzed by one-way ANOVA using Tukey’s multiple-comparison test.
3.7. MoPtc5 and MoPtc7 Are Required for Osmotic and Oxidative Stress Tolerance
To check the possible involvement of MoPTC5 and MoPTC7 in M. oryzae tolerance to osmotic and oxidative stresses, we inoculated mycelia plugs from cultures of Guy11, ΔMoptc5, ΔMoptc7, ΔMoptc5ΔMoptc7 and complementation strains were on media supplemented with 1 M NaCl, 1 M KCl, 1 M Sorbitol and 5 and 10 mM H2O2 and their colony growths measured. Our results revealed that the growths of ΔMoptc5 and ΔMoptc5ΔMoptc7 mutants were significantly inhibited by 1 M NaCl while the double mutant ΔMoptc5ΔMoptc7 was highly inhibited by both 5 mM H2O2 and 10 mM H2O2 (Figure 7A,B). MoOsm1 is a crucial member of the osmoregulation pathway. A study previously demonstrated that deletion of MoOSM1 caused hypersensitivity to oxidative stress-inducing agents [30]. To establish its relationship with MoPtc5 and ΔMoptc7ΔMoptc5, we performed western blot assay of MoOsm1 in ΔMoptc5, ΔMoptc7, ΔMoptc7ΔMoptc5 and wild type strains. Our results support that the expression of MoOsm1 protein is strongly attenuated in the absence of MoPtc7 and MoPtc5 (Figure 7C,D).
Figure 7.
MoPtc5 and ΔMoptc7ΔMoptc5 are indispensable for tolerance to osmotic and oxidative stresses in the blast fungus. (A) Colony diameters of the various strains were measured 10 days after inoculation on media containing 1 M NaCl, 1 M KCl, 1 M Sorbitol, 5 mM, 10 mM H2O2. (B) Inhibition rate of the oxidative and osmotic stressors. Inhibition rate = (the diameter of untreated strain − the diameter of treated strain)/(the diameter of untreated strain × 100%). There were three replicates. Error bars represent standard deviations while asterisks show significant differences at different p-values (* p < 0.1; ** p < 0.01). The data were analyzed by one-way ANOVA using Tukey’s multiple-comparison test. (C) Osm1 protein abundance in ΔMoptc5, ΔMoptc7, ΔMoptc7ΔMoptc5 mutants. (D) Comparison of band intensities for Osm1/Actin phosphorylation levels in the various strains as calculated using imaging software.
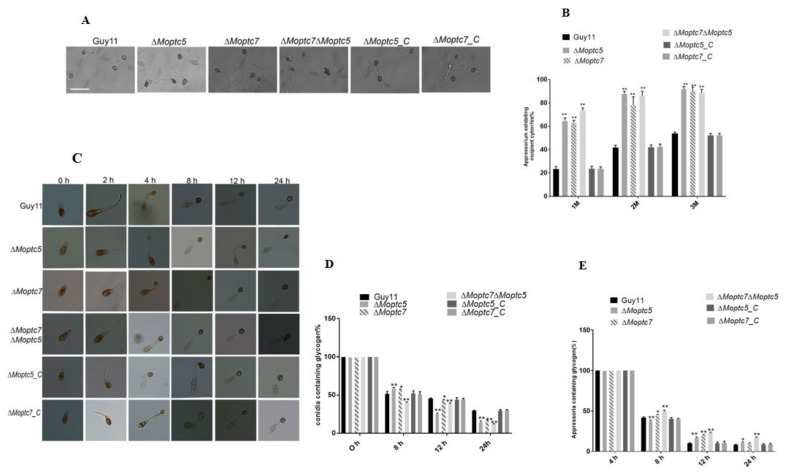
3.8. MoPtc5 and MoPtc7 Are Responsible for Appressorium Turgor Generation and Degradation of Glycogen in M. oryzae
The involvement of MoPtc5 and MoPtc7 in appressorium-mediated host penetration was also tested by measuring appressorium turgor in ΔMoptc5, ΔMoptc7, ΔMoptc5ΔMoptc7, ΔMoptc5_C, ΔMoptc7_C and Guy11 after induction of appressorium formation on artificial hydrophobic surfaces. They were then incubated for 24 h, after which they were treated with 1 M, 2 M and 3 M glycerol. The number of collapsed appressoria were counted under a light microscope. The results showed that most of the appressoria produced by ΔMoptc5, ΔMoptc7 and ΔMoptc5ΔMoptc7 were collapsed (Figure 8A,B). To examine glycogen degradation, the conidia and appressoria of the strains were stained with KI/I2 and glycogen degradation was checked at 2 h, 4 h, 8 h, 12 h and 24 h time points. We found delayed glycogen degradation from conidia to appressoria formation in the mutants (Figure 8C–E).
Figure 8.
Deletion of MoPTC5 and MoPTC7 delayed glycogen mobilization from conidia to appressoria and reduced appressorium turgor pressure. (A,B) represent the percentages of appressorium exhibiting incipient cytorryhsis after being treated with glycerol. (C) Mobilization of glycogen in the mutants and Guy11 at different time points. (D,E) Micrographs showing percentage of conidia and appressoria containing glycogen.
3.9. MoPtc5 and MoPtc7 Are Involved in M. oryzae Pathogenicity
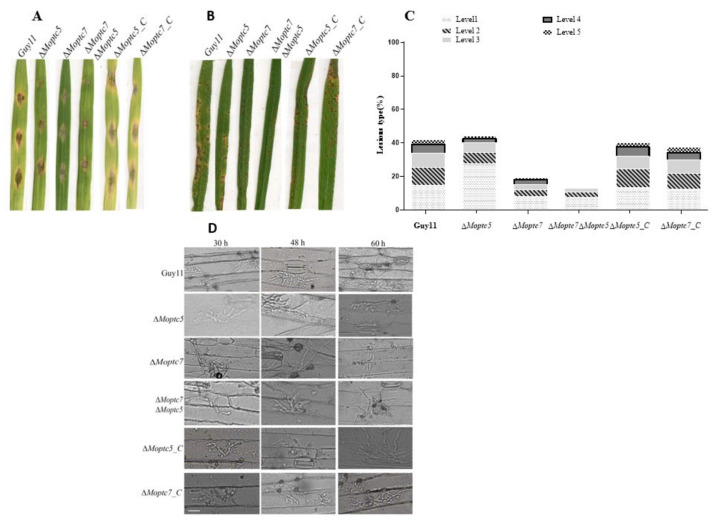
We analyzed the virulence of ΔMoptc5, ΔMoptc7, ΔMoptc7ΔMoptc5, and Guy11 strains by placing drops of their conidia suspensions on 10 days old barley leaves. At the 7th day of the inoculation, the virulence of the mutants were obviously reduced compared to Guy11 and the complemented strains (Figure 9A). We further harvested spores from these strains to infect 3 weeks old CO39 rice seedlings. The results showed slightly reduced pathogenicity for the mutant strains (Figure 9B,C). To further confirm these results, we investigated the cuticle penetration potential of the mutants on barley leaves at different time points. We found that the ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants had weak cuticle penetrating capacity at 30 h and 48 h time points relative to Guy11 (Figure 9D), which is consistent with their defects in appressoria formation earlier observed. These results reveal the importance of MoPtc5 and MoPtc7 in the pathogenicity of M. oryzae.
Figure 9.
MoPtc5 and MoPtc7 play important roles in M. oryzae virulence. (A) Pathogenicity assay for Guy11, ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 on barley leaves using drops of conidia suspension. (B) The pathogenicity of ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants in comparison to Guy11 using conidia sprays on 3 weeks old barley leaves. (C) Extents of lesions development as blast symptoms caused by the indicated strains, Level 1 (uniform dark brown pinpoint lesions without visible centers), Level 2 (small lesions with distinct centers surrounded by a dark brown margin, 1 mm in diameter), Level 3 (small eyespot lesions approximately 2 mm in length with tan centers surrounded by dark brown margins), Level 4 (intermediate size eyespot lesions, approximately 3–4 mm in length), Level 5 (large eyespot lesions approximately 5 mm in length). (D) Cuticle penetration defects of the mutants.
3.10. Subcellular Localization of MoPtc5 and MoPtc7 Genes
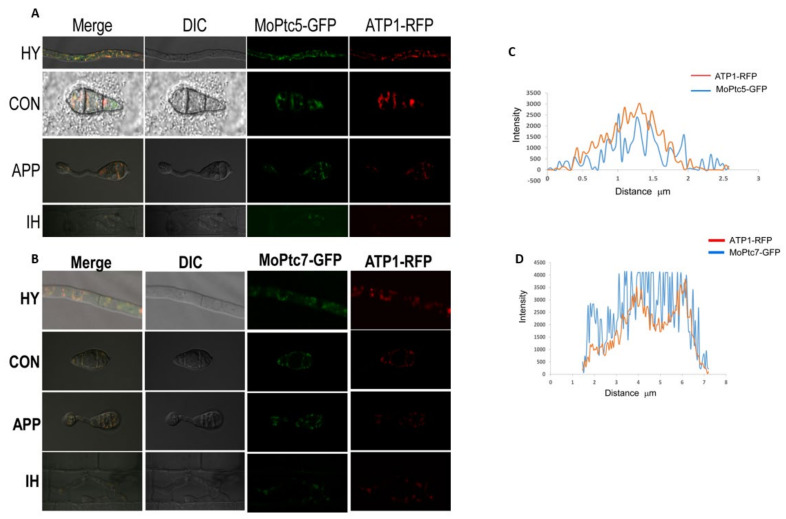
To evaluate the location of MoPtc5 and MoPtc7 in this fungus, we amplified MoPtc5 and MoPtc7 fragments from Guy11 genomic DNA and cloned the two separate fragments in the KpnI/HindIII site of pKNTG plasmid which is fused with GFP. We transformed these constructs separately into the protoplasts of their respective mutants. It was observed that both MoPtc5 and MoPtc7 are localized in mitochondria at all developmental stages. To confirm the mitochondria localization, we cotransformed ATP1 vectors fused with Red fluorescent protein (RFP) and MoPtc5-GFP, MoPtc7-GFP vectors in Guy11 protoplast. The transformants were selected using hygromycin and geneticin antibiotics. We screened the transformants by PCR method and fluorescence microscopy and observed that a colocalization of MoPtc5-GFPand MoPtc7-GFP with the mitochondria marker in hyphae, conidia and in appressorium (Figure 10).
Figure 10.
Localization of MoPtc5 and MoPtc7 in rice blast fungus. (A,B) displays co-localization of MoPtc5, MoPtc7 in hyphae, conidia, appressorium and in plantae as it co-localizes with ATP1_RFP (Red Fluorescence Protein) mitochondria marker. (C,D) graphs representing intensity fluorescence and distance for MoPtc5 and MoPtc7 co-localization. The pictures were taken by a NikonA1 confocal microscope (Scale bar = 20 µm).
4. Discussion
Type 2C protein phosphatases perform various functions in cell biology and signal pathways in eukaryotes. However, their roles in filamentous fungi are poorly known. In this study, we analyzed the functions of MoPtc5 and MoPtc7 to unveil their individual and combined roles in the development and pathogenesis of Magnaporthe oryzae. We showed through phylogenetic analysis that MoPtc5 shares high percentage similarity to S. cerevisiae, F. graminearum and F. oxysporum while MoPtc7 is closest to F. graminearum and B. cinerea (Figure 1B). We also showed the presence of PP2C and PP2C_SIG domains in MoPtc5 and MoPtc7 proteins (Figure 1A). The PP2C and PP2C_SIG domains are conserved in all type 2C protein phosphatases. In this study, we demonstrated that both MoPtc5 and MoPtc7 proteins are localized to mitochondria in M. oryzae at different development stages, including hyphae, conidia, appressoria and in infected plants.
Transcriptomic analyses demonstrated that MoPtc5 and MoPtc7 are differentially expressed at 12 hpi, while at 24 hpi MoPtc7 is up-regulated. These results suggest that MoPtc7 is more active in planta than MoPtc5. Furthermore, we also established that MoPTC5 and MoPTC7 genes do not influence M. oryzae vegetative growth. However, double deletion of both genes in a single mutant resulted in significantly reduced M. oryzae mycelial growth on SYM and RBM. These findings are consistent with that reported by [28]. Which revealed that double deletion of MoPtc5 and MoPtc7 disrupted the growth of S. cerevisiae. This simply shows the redundant functions of the two proteins in the vegetative growth of the fungus.
Conidia formation is very crucial during the lifetime and epidemics of M. oryzae, since conidia serve as primary inoculums during host infection [7]. The M. oryzae mutant lacking MoPTC5 gene showed slightly reduced conidiation rate while deletion of MoPTC7 had no adverse defect on conidiation. However, double knockout of the two genes resulted in a mutant that displayed significant impairment in conidiation rate. These results are consistent with the observed down-regulation of conidiation responsive genes and poor conidiophore developments in the double mutant strain. Therefore, the protein phosphatases might play overlapping roles in asexual reproduction in M. oryzae.
Fungal cell wall is a very powerful structure that acts as a barrier that maintains cellular integrity; it is involved in signal transduction, and plays a key function in selective permeability of the cells [31]. In addition, cell wall is important for response to external stresses [32]. In this study, we examined the influence of some cell wall-damaging agents including Calcofluor white (CFW), SDS and Congo red (CR) on ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants. Our results indicate that deletion of MoPTC5 as well as the double deletion of MoPTC7 and MoPTC5 resulted in increased CFW and CR. However, these observations and previous findings on the type 2C protein phosphatases in S. cerevisiae [16] revealed the essential function of MoPtc5 and MoPtc7 in cell wall integrity. In addition, double deletion of MoPTC5 and MoPTC7 has no effect on the fungal cell wall thickness, but altered the expressions of the chitinase-encoding genes MoCHS2 and MoCHS7 which became up-regulated in the double deletion mutant ΔMoptc7ΔMoptc5. Taken together, these results suggest that MoPtc5 and MoPtc7 synergistically modulate chitin biosynthesis in M. oryzae.
Reactive oxygen species (ROS) production and homeostasis are important for general fungal development [33]. In this research work, the double mutant ΔMoptc7ΔMoptc5 was significantly more sensitive to different concentrations of hydrogen peroxide (5 mM H2O2 and 10 mM H2O2) than ΔMoptc7 and ΔMoptc5 mutants. Based on these results we speculated that MoPtc5 and MoPtc7 might play important roles in the fungal tolerance to oxidative stress. A previous study demonstrated that these type 2C phosphatases are required for osmotic stress tolerance in yeast and in higher eukaryotes [16]. In M. oryzae, lacking both proteins showed higher NaCl and KCl sensitivities. Therefore, we reasoned that the type 2C phosphatase gene might redundantly confer tolerance to osmotic stress in M. oryzae.
More so, we observed reduction in the fungal pathogenicity due to single as well as double deletions of the type 2C genes in M. oryzae. ΔMoptc7 and ΔMoptc5 mutants’ virulence in rice and barley was slightly reduced. However, double deletion of MoPTC7 and MoPTC5 resulted in highest reduction in M. oryzae pathogenicity. We reasoned that MoPtc5 and MoPtc7 might be playing a synergetic role in rice blast disease morphogenesis. Hence, we concluded that MoPtc5 and MoPtc7 may play redundant roles in M. oryzae pathogenicity.
Glycerol accumulation in an appressorium generates turgor pressure that is used to physically break the host cuticle for penetration [34]. A previous study indicated that maintenance of turgor pressure is necessary for M. oryzae penetration into the host [31]. We therefore tried to investigate whether the observed reduction in the pathogenicity of the mutants was due to reduced turgor generation in their appressoria. Our results showed that ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants had attenuated appressorium turgor pressure. Furthermore, glycogen degradation and transportation from conidia to mature appressoria are key biochemical processes that promote appressorial efficiency and cuticle penetration [35]. We found here that double deletion of MoPTC7 and MoPTC5 delayed the degradation and transport of glycogen from conidia to mature appressoria. As such, we speculated that the type 2C phosphatases MoPtc5 and MoPtc7 are positively involved in pathogenesis in M. oryzae by regulating glycogen trafficking pathways.
In summary, the type 2C protein phosphatases MoPtc5 and MoPtc7 in M. oryzae play overlapping roles in vegetative growth, conidiation, stress tolerance, pathogenicity, and MoMps1 phosphorylation. The findings unveil additional targets for the control and management of the globally devastating blast disease of rice.
Acknowledgments
I’m grateful for the Fujian government scholarship. I would like also to acknowledge Stefan Olsson for the discussion to improve the project.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9010001/s1, Figure S1, Southern blot for confirmation for ΔMoptc5, ΔMoptc7 and ΔMoptc7ΔMoptc5 mutants; Table S1, List of primers used in this project.
Author Contributions
Z.W., W.T. and J.B. designed this project, all authors contributed to conducting the experiments, data analysis and writing the draft and confirmation of the final manuscript for publication. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors confirmed no conflict of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. U1805232, No. 32272513), Fujian Provincial Science and Technology Key Project (2022NZ030014), the Natural Science Foundation of Fujian Province (No. 2022J01129) and the Program of Fujian Key Laboratory for Monitoring and Integrated Management of Crop Pests (MIMCP-202101).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Talbot N. Exploring the Biology of Magnaporthe grisea, on the Trail of a Cereal Killer. Annu. Rev. Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- 2.Dean R.A., Talbot N.J., Ebbole D.J., Farman M.L., Mitchell T.K., Orbach M.J., Thon M., Kulkarni R., Xu J.-R., Pan H. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 3.Ebbole D.J. Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 2007;45:437–456. doi: 10.1146/annurev.phyto.45.062806.094346. [DOI] [PubMed] [Google Scholar]
- 4.Cooper A.J., Jeitner T.M. Central role of glutamate metabolism in the maintenance of nitrogen homeostasis in normal and hyperammonemic brain. Biomolecules. 2016;6:16. doi: 10.3390/biom6020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim K.S., Lee Y.-H. Gene expression profiling during conidiation in the rice blast pathogen Magnaporthe oryzae. PLoS ONE. 2012;7:e43202. doi: 10.1371/journal.pone.0043202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker S.L., Talbot N.J. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 2001;39:385–417. doi: 10.1146/annurev.phyto.39.1.385. [DOI] [PubMed] [Google Scholar]
- 7.Howard R.J., Ferrari M.A., Roach D.H., Money N.P. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA. 1991;88:11281–11284. doi: 10.1073/pnas.88.24.11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1-and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/S1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 9.Xu J.-R., Hamer J.E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 10.Seshacharyulu P., Pandey P., Datta K., Batra S.K. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 2013;335:9–18. doi: 10.1016/j.canlet.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez P.L. Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol. Biol. 1998;38:919–927. doi: 10.1023/A:1006054607850. [DOI] [PubMed] [Google Scholar]
- 12.Ariño J., Velázquez D., Casamayor A. Ser/Thr protein phosphatases in fungi: Structure, regulation and function. Microb. Cell. 2019;6:217. doi: 10.15698/mic2019.05.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X. Defining the Molecular Function of Mitochondrial Phosphatases. The University of Wisconsin-Madison; Madison, WI, USA: 2017. [Google Scholar]
- 14.Tasdelen I., van Beekum O., Gorbenko O., Fleskens V., van den Broek N.J., Koppen A., Hamers N., Berger R., Coffer P.J., Brenkman A.B. The serine/threonine phosphatase PPM1B (PP2Cβ) selectively modulates PPARγ activity. Biochem. J. 2013;451:45–53. doi: 10.1042/BJ20121113. [DOI] [PubMed] [Google Scholar]
- 15.Warmka J., Hanneman J., Lee J., Amin D., Ota I. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 2001;21:51–60. doi: 10.1128/MCB.21.1.51-60.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ariño J., Casamayor A., González A. Type 2C protein phosphatases in fungi. Eukaryot. Cell. 2011;10:21–33. doi: 10.1128/EC.00249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Y., Walker L., Novick P., Ferro-Novick S. Ptc1p regulates cortical ER inheritance via Slt2p. EMBO J. 2006;25:4413–4422. doi: 10.1038/sj.emboj.7601319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leroy C., Lee S.E., Vaze M.B., Ochsenbien F., Guerois R., Haber J.E., Marsolier-Kergoat M.-C. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell. 2003;11:827–835. doi: 10.1016/S1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 19.Cheng M., Olivier P., Diehl J.A., Fero M., Roussel M.F., Roberts J.M., Sherr C.J. The p21Cip1 and p27Kip1 CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shitamukai A., Hirata D., Sonobe S., Miyakawa T. Evidence for antagonistic regulation of cell growth by the calcineurin and high osmolarity glycerol pathways in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:3651–3661. doi: 10.1074/jbc.M306098200. [DOI] [PubMed] [Google Scholar]
- 21.Markovich S., Yekutiel A., Shalit I., Shadkchan Y., Osherov N. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2004;48:3871–3876. doi: 10.1128/AAC.48.10.3871-3876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause-Buchholz U., Gey U., Wünschmann J., Becker S., Rödel G. YIL042c and YOR090c encode the kinase and phosphatase of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. FEBS Lett. 2006;580:2553–2560. doi: 10.1016/j.febslet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Brock A.K., Willmann R., Kolb D., Grefen L., Lajunen H.M., Bethke G., Lee J., Nürnberger T., Gust A.A. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 2010;153:1098–1111. doi: 10.1104/pp.110.156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González A., Ruiz A., Casamayor A., Arino J. Normal function of the yeast TOR pathway requires the type 2C protein phosphatase Ptc1. Mol. Cell. Biol. 2009;29:2876–2888. doi: 10.1128/MCB.01740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talbot N.J., Ebbole D.J., Hamer J.E. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Z., Xue C., Peng Y., Katan T., Kistler H.C., Xu J.-R. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 2002;15:1119–1127. doi: 10.1094/MPMI.2002.15.11.1119. [DOI] [PubMed] [Google Scholar]
- 27.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Aron O., Wang M., Lin L., Batool W., Lin B., Shabbir A., Wang Z., Tang W. MoGLN2 is important for vegetative growth, conidiogenesis, maintenance of cell wall integrity and pathogenesis of Magnaporthe oryzae. J. Fungi. 2021;7:463. doi: 10.3390/jof7060463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q., Liu H., Du D., Yu Y., Ma C., Jiao F., Yao H., Lu C., Zhang W. Identification of novel laminin-and fibronectin-binding proteins by far-western blot: Capturing the adhesins of Streptococcus suis type 2. Front. Cell. Infect. Microbiol. 2015;5:82. doi: 10.3389/fcimb.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Qian B., Gao C., Huang S., Cai Y., Zhang H., Zheng X., Wang P., Zhang Z. The putative protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate the development and pathogenicity in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2016;29:496–507. doi: 10.1094/MPMI-11-15-0259-R. [DOI] [PubMed] [Google Scholar]
- 31.Lin L., Cao J., Du A., An Q., Chen X., Yuan S., Batool W., Shabbir A., Zhang D., Wang Z. eIF3k Domain-Containing Protein Regulates Conidiogenesis, Appressorium Turgor, Virulence, Stress Tolerance, and Physiological and Pathogenic Development of Magnaporthe oryzae Oryzae. Front. Plant Sci. 2021;12:748120. doi: 10.3389/fpls.2021.748120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novaković L., Guo T., Bacic A., Sampathkumar A., Johnson K.L. Hitting the wall—Sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants. 2018;7:89. doi: 10.3390/plants7040089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Chen Y., Li B., Chen T., Tian S. Reactive oxygen species: A generalist in regulating development and pathogenicity of phytopathogenic fungi. Comput. Struct. Biotechnol. J. 2020;18:3344–3349. doi: 10.1016/j.csbj.2020.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang H.-X., Miller L.A., Hartman G.L. Melanin-independent accumulation of turgor pressure in appressoria of Phakopsora pachyrhizi. Phytopathology. 2014;104:977–984. doi: 10.1094/PHYTO-12-13-0335-R. [DOI] [PubMed] [Google Scholar]
- 35.Zhang S.-R., Hao Z.-M., Wang L.-H., Shen S., Cao Z.-Y., Xin Y.-Y., Hou M.-L., Gu S.-Q., Han J.-M., Dong J.-G. StRas2 regulates morphogenesis, conidiation and appressorium development in Setosphaeria turcica. Microbiol. Res. 2012;167:478–486. doi: 10.1016/j.micres.2012.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.