Abstract
This nationwide study aimed to investigate the molecular characteristics of metallo-β-lactamase (MBL)-producing Pseudomonas aeruginosa in Serbia, underlying resistance mechanisms, the genetic context of detected MBL genes, and the clonal relationship between isolates harboring genes-encoding MBL. Overall, 320/5334 isolates collected from 2018 to 2021 were identified as P. aeruginosa. Carbapenem-resistant P. aeruginosa (CRPA) were screened for the presence of blaVIM, blaIMP, and blaNDM, genes whereas MBL-positive isolates were tested for the presence of the blaCTX-M-2, blaPER, blaTEM, blaSHV, blaVEB, and blaGES. Multilocus sequence typing and phylogenomic analysis were performed for P. aeruginosa-producing MBL. The majority of the P. aeruginosa isolates were recovered from the lower respiratory tract (n = 120; 37.5%) and wound specimens (n = 108; 33.75%). CRPA isolates accounted for 43.1% (n = 138) of the tested isolates, 31 out of them being blaNDM-1-positive (22.5%). The colistin resistance rate was 0.3%. MLST analysis revealed the occurrence of ST235 (n = 25) and ST654 (n = 6), mostly confined to Serbia. The distribution of beta-lactamase-encoding genes in these isolates suggested clonal dissemination and possible recombination: ST235/blaNDM-1, ST235/blaNDM-1/blaPER-1, ST654/blaNDM-1, ST654/blaNDM-1/blaPER-1, and ST654/blaNDM-1/blaGES-5. High-risk clones ST235 and ST654 identified for the first time in Serbia, are important vectors of acquired MBL and ESBL and their associated multidrug resistance phenotypes represent a cause for considerable concern.
Keywords: high-risk clones, carbapenem-resistant Pseudomonas aeruginosa, blaNDM, blaPER, blaGES, MLST, WGS
1. Introduction
Pseudomonas aeruginosa is an important opportunistic human pathogen involved in a variety of community and hospital-acquired infections, ranging from respiratory tract infections, endocarditis, and urinary tract infections to septicemia [1]. P. aeruginosa has been recognized as a common cause of ventilator-associated pneumonia, particularly in intensive care units (ICU) [2]. High adaptability, genetic plasticity, and versatile and ubiquitous features make P. aeruginosa a very concerning and difficult-to-treat pathogen. In addition, the treatment of P. aeruginosa infections has become a great challenge due to its intrinsic resistance to many antimicrobial agents and the ability to easily acquire antibiotic resistance through mutational changes or the acquisition of resistance genes via horizontal gene transfer [3]. P. aeruginosa is one of the six leading mortality-causing pathogens, which together were responsible for 929,000 deaths attributable to antimicrobial resistance (AMR) and 3.57 million deaths associated with AMR in 2019 globally [4]. Thus, multidrug-resistant P. aeruginosa (MDR-PA) and extensively drug-resistant P. aeruginosa (XDR-PA) strains are becoming major clinical threats worldwide due to their association with high mortality, long hospital stays, and increased costs [5]. The World Health Organization (WHO) has listed carbapenem-resistant P. aeruginosa (CRPA) as one of the three bacterial species in which there is a critical need for the development of new antibiotics to treat infections [6]. Likewise, P. aeruginosa is included in the group ESKAPE, an acronym introduced to designate a group of bacteria that escape the action of antibiotics: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species [7]. Data from the annual report of Central Asian and European Surveillance of Antimicrobial Resistance (CAESAR) showed that CRPA isolates accounted for 55% of invasive P. aeruginosa in Serbia in 2020 [8]. Carbapenem resistance in P. aeruginosa has shown to be multifactorial, including (i) acquisition of carbapenemase-encoding genes through horizontal gene transfer [9], (ii) deficiency or repression of the porin (OprD) for carbapenems [10], (iii) overexpression of mexAB-oprM efflux pump [11], and (iv) overexpression of the chromosomal gene (ampC) encoding the P. aeruginosa intrinsic cephalosporinase [12]. However, the acquired carbapenemase-encoding genes need special attention because horizontal gene transfer through mobile genetic elements, such as transposons, plasmids, and integrative and conjugative elements, could accelerate the dissemination of the CRPA. Indeed, acquired extended-spectrum β-lactamases (ESBLs) located on mobile genetic elements, such as serine-β-lactamases of molecular classes A and D and metallo-β-lactamases (MBLs) of class B, are important resistance mechanisms in P. aeruginosa [13]. Considering the role of P. aeruginosa in the development of nosocomial infections, the analysis of the genetic context of the MBL genes is essential in identifying associations with other genes conferring antimicrobial resistance and revealing potential routes of dissemination. The first report on the P. aeruginosa producing MBL in Serbia and the Balkan region was the detection of isolates carrying blaNDM-1 gene in 2011 [14].
Epidemiological studies based on multilocus sequence typing (MLST) may help to understand and identify circulating MDR P. aeruginosa strains worldwide. Previously published reports showed that P. aeruginosa sequence type (ST) 235 is the predominant global clinical lineage [15]. Besides ST235, most of the carbapenemase-producing P. aeruginosa reported worldwide belong to ubiquitous high-risk clones, such as ST111, ST175, and ST654 [16,17]. However, in Serbia, there is a lack of data regarding circulating STs and the genomic epidemiology of CRPA isolates.
To address the prevalence and underlying mechanisms of antibiotic resistance and the epidemiology of P. aeruginosa, the objectives of the current study were (i) to determine the antimicrobial resistance profile of P. aeruginosa isolated from hospitalized patients across Serbia, (ii) to investigate the prevalence of P. aeruginosa producing MBLs, (iii) to analyze the genetic context of detected MBL genes and (iv) to investigate the clonal relationship between isolates harboring genes encoding MBL.
2. Results
Out of 5334 collected bacterial isolates, 320 (6%) corresponded to P. aeruginosa. The isolates were recovered from hospitalized patients with an average age of 59.72 years within the age range of 2 months and 88 years old. The clinical samples included lower respiratory tract samples (n = 120; 37.5%), wound specimens (n = 108; 33.75%), urine (n = 71; 22.2%), and blood (n = 21; 6.6%). Most of the P. aeruginosa isolates were obtained from the ICU (n = 178; 55.6%). Detailed information concerning specimen types, patient characteristics, and hospital units is listed in Table 1.
Table 1.
Demographic and clinical characteristics of Pseudomonas aeruginosa-infected study patients.
| Characteristic | No. (%) |
|---|---|
| Gender | |
| Male gender | 172 (53.8) |
| Female gender | 148 (46.2) |
| Admission ward | |
| Intensive care unit | 126 (39.4) |
| Thoracic surgery | 3 (0.9) |
| Orthopedic surgery | 3 (0.9) |
| Plastic surgery | 6 (1.9) |
| Cardiovascular surgery | 39 (12.2) |
| Neurosurgery | 10 (3.1) |
| General surgery | 29 (9.1) |
| Urology surgery | 3 (0.9) |
| Internal wards | 52 (16.3) |
| Department for COVID-19-positive patients | 23 (7.2) |
| Geriatric department | 19 (5.9) |
| Department of Oncology | 7 (2.2) |
| Type of specimen | |
| Tracheal aspirate | 74 (23.1) |
| Bronchoalveolar lavage | 26 (8.1) |
| Sputum | 20 (6.3) |
| Wound specimen | 108 (33.8) |
| Blood | 21 (6.6) |
| Urine | 71 (22.2) |
| Comorbidity | |
| Malignancy | 36 (11.2) |
| Chronic venous insufficiency | 18 (5.6) |
| Heart insufficiency | 18 (5.6) |
| Diabetes | 23 (7.2) |
| COVID-19 pneumonia | 23 (7.2) |
| Invasive procedures | |
| Any surgical procedure | 93 (29.1) |
| Mechanical ventilation | 72 (22.5) |
| Central venous catheter | 42 (13.1) |
| Urinary catheter | 139 (43.4) |
2.1. Antimicrobial Resistance
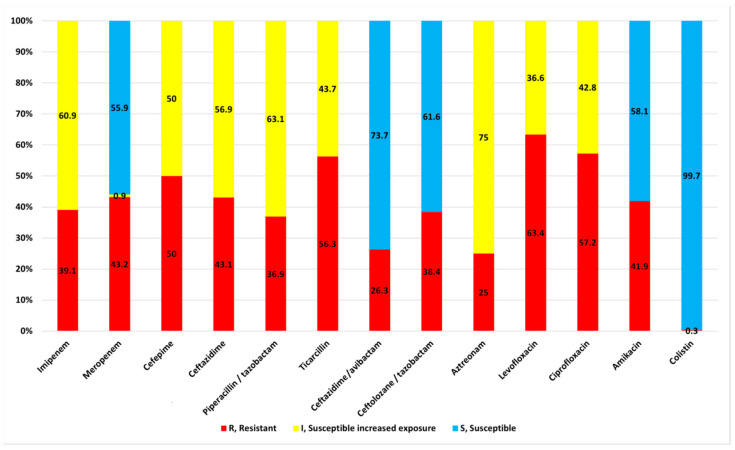
The highest resistance prevalence (>50%) was observed for fluoroquinolones, levofloxacin, and ciprofloxacin. Resistance prevalence against the tested beta-lactam antibiotics ranged from 25–56.3%, being 43.1% for carbapenems (Figure 1). The lowest resistance rates were observed for ceftazidime—avibactam, aztreonam, and colistin. Indeed, 99.7% of the isolates displayed in vitro susceptibility to colistin. All isolates had MIC values of colistin < 2 mg/L, except one isolate that had a MIC value > 16 mg/L.
Figure 1.
Antimicrobial resistance of the 320 clinical isolates of Pseudomonas aeruginosa.
Overall, 154 (48.1%) and 114 (35.6%) out of 320 P. aeruginosa isolates were MDR and XDR, respectively. The most common MDR phenotype observed was non-susceptibility to ceftazidime, piperacillin-tazobactam, ticarcillin, meropenem, levofloxacin, and ciprofloxacin (42%).
Among the 320 isolates of P. aeruginosa, 138 (43.1%) were CRPA, being resistant to meropenem and/or imipenem. The respective MICs were >8 mg/L for meropenem and/or >4 mg/L for imipenem. Of note, 23 out of 138 (16.7%) isolates of CRPA were susceptible to amikacin.
The frequency of CRPA isolates recovered from hospital wards (n = 138) was as follows: ICUs—n = 57 (41.3%), cardiovascular surgery—n = 32 (23.2%), general surgery—n = 10 (7.2%), neurosurgery—5 (3.6%), orthopedic surgery—n = 4 (2.9%), plastic surgery—n = 2 (1.4%), internal medicine—n = 10 (7.2%%), geriatric department—n = 9 (6.5%), department for COVID-19-positive patients—n = 6 (4.3%), and department of oncology—n = 3 (2.2%).
The 138 CRPA isolates were identified as adequate candidates for the detection of the MBL-encoding genes.
2.2. Molecular Detection of MBL- and ESBL-Encoding Genes
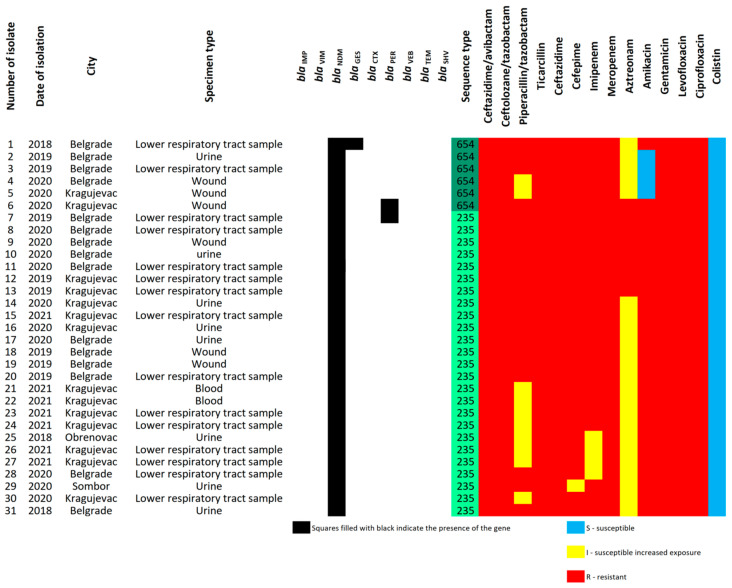
Among the 138 CRPA isolates, 31 (22.5%) harbored MBL-encoding genes, namely blaNDM-1 (Supplementary Figure S1). P. aeruginosa harboring blaNDM were obtained from four hospitals located in three cities (Supplementary Figure S2). The highest number of MBL genes was identified in isolates from patients in ICUs (n = 12; 38.7%), departments for cardiovascular surgery (n = 9; 29.7%), or corona care centers (n = 5; 16.1%). The blaVIM and blaIMP genes were not identified in any of the tested isolates.
Only three of the 31 isolates (9.7%) found to be blaNDM-positive carried ESBL-encoding genes. Two isolates harbored simultaneously blaNDM-1 and blaPER-1, whereas one isolate carried blaNDM-1 and blaGES-5. Among the tested CRPA isolates, blaCTX-M, blaTEM, blaSHV, and blaVEB genes were not detected (Figure 2).
Figure 2.
Presence/absence matrix of antimicrobial resistance genes and corresponding sequence types (STs) among 31 Pseudomonas aeruginosa isolates harboring blaNDM gene.
2.3. Multilocus Sequence Typing Analysis
MLST analysis was performed for all blaNDM-1 positive P. aeruginosa isolates (n = 31). Among them, 25 and 6 isolates were classified as ST235 and ST654, respectively (Figure 3). One isolate exhibiting resistance to all antimicrobial agents tested in the study, including colistin, belonged to ST235. In addition, two isolates co-harboring blaPER-1 and blaNDM-1 were assigned to ST235 (n = 1) and ST654 (n = 1), whereas the isolate harboring blaGES-5 gene was ST654 (n = 1). In summary, CRPA types identified in this study in Serbia were the following: ST235/blaNDM-1, ST235/blaNDM-1/blaPER-1, ST654/blaNDM-1, ST654/blaNDM-1/blaPER-1, and ST654/blaNDM-1/blaGES-5. Genotypes ST235/blaNDM-1 and ST654/blaNDM-1 were found in at least three different cities across Serbia, indicating dissemination throughout the country.
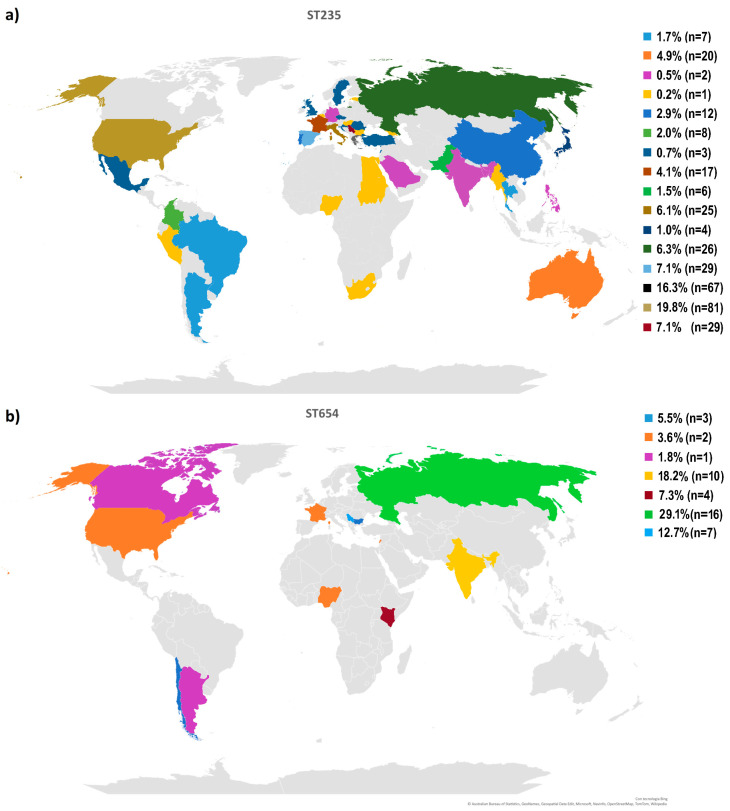
Figure 3.
Geographical distribution of the Pseudomonas aeruginosa ST235 (a) and ST654 (b) high-risk clones based on the data available in the Pseudomonas Genome Database (Date accessed 2 January 2023). Colors represent the percentage of clones ST235 and ST654 among isolates of P. aeruginosa within a country, as indicated in the geographic map.
The geographic distribution of genomes and the antibiotic resistance profiles of genomes affiliated with ST235 and ST654 were assessed based on the genomes available in the Pseudomonas Genome Database (Figure 3). From the public database were retrieved all available sequences and information of ST235 and ST654 P. aeruginosa isolates. The P. aeruginosa ST235 (n = 384) isolates presented a worldwide distribution, mostly in the United States of America (USA) (21.1% of the analyzed isolates). Serbia represented 7.1% of the ST235 isolates available in the online database (Figure 3a). Although submitted in the lower number in the online database (n = 49), most of the ST654 isolates came from Russia (29.1%), followed by India and Serbia (18.2% and 12.7%) (Figure 3b). Analyzing the antibiotic resistance genes profile in both ST235 and ST654, isolates were annotated with different aminoglycoside resistance genes and carbapenemase resistance genes of different classes. Moreover, blaOXA-396 was observed in ST654 isolates, while the blaOXA-488 gene was present in all genomes belonging to ST235 (Supplementary Figure S3).
The ST235 strains’ antibiotic resistance profile indicates the ARGs blaGES (31 isolates), blaNDM (30 isolates), and blaVIM (29 isolates) as the main carbapenem resistance genes harbored. Instead, ST654 isolates presented as the main carbapenems resistance gene blaNDM (10 isolates) followed by blaGES (five isolates) and blaVIM (two isolates).
2.4. Phylogenomic Analysis of blaNDM-Positive Strains
Details on genome sequencing and assembly quality of four randomly selected blaNDM-positive P. aeruginosa (NDM1_1, NDM1_2, NDM1_3, and NDM1_4) isolates can be found on the National Center for Biotechnology Information site, under BioProject Accession number PRJNA753000. Briefly, the genome sizes ranged from 6.9 to 7.1 Mbp, organized in 1–35 contigs, and coverages ranging from 100–300 x, as shown in Supplementary Table S2. No plasmids were detected among the four blaNDM-positive P. aeruginosa isolates.
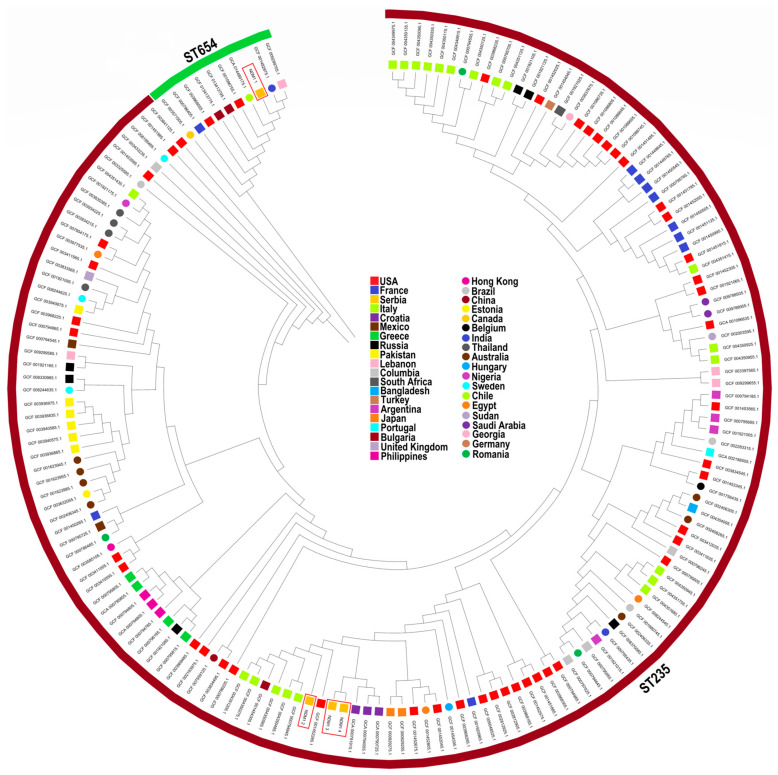
The results of phylogenomic analysis of the four genomes of P. aeruginosa harboring the blaNDM gene together with 161 previously published genomes of the same STs with a known geographical location, available in the NCBI Pathogen Detection database (Supplementary Table S3) are illustrated in Figure 4.
Figure 4.
Phylogenetic tree constructed by calling SNPs from core gene alignment of the 165 high-risk clones of Pseudomonas aeruginosa (ST235 and ST654) using the neighbor-joining method with 1000 bootstraps. The different countries can be distinguished by different shapes and color codes displayed in the figure legend; the fragments with different colors in the outer ring represent corresponding sequence types (STs), as shown.
Overall, the phylogenetic tree of the 165 P. aeruginosa isolates could be divided into two clades according to STs, as illustrated in Figure 4. The ST235 isolates NDM1_2, NDM1_3, and NDM1_4 grouped were closely related to isolates from Croatia, Italy, the USA, and Bulgaria, with detected SNP differences ranging from 59 to 1157 (Supplementary Table S4). The ST654 isolate NDM1_1 was grouped with isolates from Lebanon, India, Chile, and the USA, with detected SNP differences ranging from 447 to 4048 (Supplementary Table S4).
2.5. The Genetic Context of the Detected MBL Genes
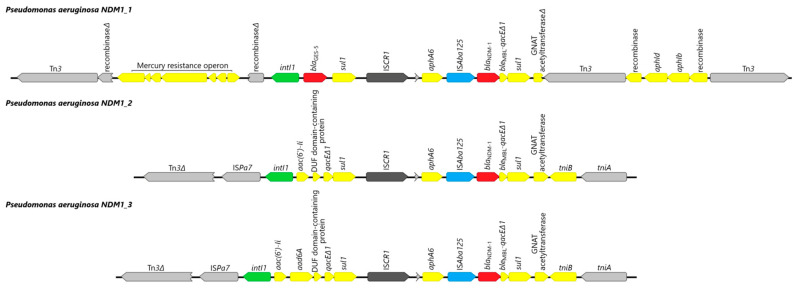
The genetic contexts of the three sequenced P. aeruginosa harboring blaNDM-1 gene have been analyzed and shown in Figure 5.
Figure 5.
Schematic representation of the blaNDM-1 region in three analyzed Pseudomonas aeruginosa isolates. Δ, an interrupted gene; genes and their transcription orientations are indicated by arrows.
Because the blaNDM gene in the NDM1_4 isolate was located at the very start of the assembled contig, its genetic context was not examined.
The blaNDM-1 gene of NDM1_1, NDM1_2, and NDM1_3 is contained within a class 1 integron-bearing insertion sequence (IS) common region 1 (ISCR1) (Figure 5). ISCR1 element is followed by the partial transposase gene ISAba14 (accession no. JQ080305.1), the aminoglycoside resistance gene aphA6, the transposase ISAba125 gene, the blaNDM-1 gene, the truncated bleomycin/qac resistance gene, and the sul1 end of a class 1 integron.
In NDM1_2 and NDM1_3 isolates, the region upstream of ISCR1 corresponds to a class 1 integron, with the intI1 gene followed by several gene cassettes: the genes aac(6′)Ii and aadA6 encoding the aminoglycoside-modifying enzymes AAC-6′(I) and AADA6, respectively, and DUF domain-containing protein. This last cassette is followed by a truncated qac/sul conserved sequence. Upstream of this complex class 1 integron, ISPa7 transposase was detected, as well as incomplete transposase belonging to the Tn3 family.
In the NDM1_1 isolate, the region upstream of ISCR1 also corresponds to a class 1 integron, with the intI1 gene, but is followed by a blaGES-5 gene and gene cassette sul1. Upstream of class 1 integron, the mercury resistance operon was detected between partial recombinase genes. In the region downstream of ISCR1, aminoglycoside resistance genes aph(6′)Id and aph(6′)Ib were identified, located between two Tn3 family transposases.
Of note, in all four genomes, the aminoglycoside resistance gene aphA6, encoding enzyme aminoglycoside 3′-phosphotransferase type VI (Aph(3′)-VI) conferring resistance to amikacin, was detected. In three analyzed genomes, it was located upstream of ISAba125 and blaNDM-1 gene. Other aminoglycoside resistance genes identified in four sequenced isolates include aadA6 (n = 3), aph(3′)-IIb (n = 3), aph(6′)Id (n = 1), aph(6′)Ib (n = 1).
3. Discussion
This study presents the first Serbian nationwide multicenter study, providing data on the molecular epidemiology of CRPA isolates recovered from different hospitals throughout the country. Moreover, we assessed the global phylogenetic analysis of the P. aeruginosa ST235 and ST654 using four genomes of strains analyzed in the current study together with those available in online genome databases.
In the current study, two-thirds of the isolated P. aeruginosa were from wounds and the lower respiratory tract. Due to its ability to grow in simple aqueous solutions, P. aeruginosa easily contaminates respiratory medical devices, anesthesia equipment, intravenous fluid, and even distilled water [18]. In the current study, ICU stay seems to be a significant risk factor for infection with MDR P. aeruginosa due to the use of invasive devices, such as mechanical ventilation and central lines. Such a finding is in accordance with the previously published report [19].
Alarmingly, in the present study, 43% of the tested isolates were CRPA. Similar results were reported by Radovanovic et al. who found that the resistance rate of hospital isolates of P. aeruginosa in Serbia during 2017/2018 was 41.7% [20]. Moreover, a higher resistance rate of 69.5% was assessed by the European Centre for Disease Prevention and Control in 2020 [21]. Based on the previously published reports, we can speculate that the overuse of carbapenems has led to the emergence of carbapenem resistance [22]. Despite aminoglycosides remaining useful antipseudomonal agents, resistance to these drugs continues to be a therapeutic concern, as these genes may be located on mobile genetic elements and thus can be transferred horizontally [23]. Genomic analysis of the present study revealed the simultaneous presence of the aminoglycoside resistance gene aphA6, conferring resistance to amikacin, upstream of blaNDM gene in the same isolates, suggesting dissemination of resistance to aminoglycosides and carbapenems in the hospital environment. An interesting observation in this study was that only 61 out of 138 (44.2%) of the CRPA isolates were resistant to aztreonam. This might be due to the fact that aztreonam is not a substrate for class B β-lactamases [24]. In addition, the retained aztreonam activity in some MDR P. aeruginosa despite resistance to other antipseudomonal β-lactams may be explained by the absence of class A ESBL in those isolates. Aztreonam is the only available monobactam that has intermediate activity against P. aeruginosa and can be a useful alternative to aminoglycosides or third-generation cephalosporin in patients with serious Gram-negative infections. The resistance rates of the tested P. aeruginosa to ceftazidime–avibactam and ceftolozane-tazobactam were comparable to those observed in Hungary in 2017, 33.6 and 32.4, respectively) [25]. According to the results obtained in this study, colistin is considered the best choice for the treatment of MBL-producing CRPA, since the overall assessed resistance rate was less than 1%. The obtained result is much lower than colistin resistance found in Qatar (3.4%) or Egypt (21.3%) [26,27]. Although known for its toxicity, colistin is being reintroduced to the treatment protocols, and the current international consensus is that the optimal use of this drug would be the option for the treatment of infections with MDR Gram-negative bacilli, particularly for P. aeruginosa and A. baumannii [28].
The blaNDM-1 was the only MBL gene detected in the present study. It was found in 9.7% of the 320 isolates of P. aeruginosa. Remarkably, the first report of NDM-positive P. aeruginosa was detected in Serbia [14]. However, in most countries, the most frequently reported MBLs globally are VIM and IMP types, whereas NDM has been detected only occasionally [29,30].
In the present study, ESBL-encoding genes detected were blaPER-1 and blaGES-5, found in 6.5% and 3.2% of the tested blaNDM-positive isolates, respectively. It is noteworthy that the PER-1-type ESBLs are the first reported ESBLs in P. aeruginosa and like most other ESBLs can hydrolyze different types of beta-lactam antibiotics except for carbapenem and cephamycin [31]. To the best of our knowledge, the first occurrence of blaPER-1-positive infections in patients from Serbia was reported in 2008 among a small cluster of patients (n = 4) admitted to one hospital in Belgrade [32]. The identification of PER-1 producers in several European countries, such as Italy and Turkey, also in the Far East, suggests their proceeding dissemination [33,34,35,36]. Globally, GES-type-producing P. aeruginosa have already been identified [37]. However, this is the first study that identified blaGES-5-producing P. aeruginosa in Serbia.
The results of our study cast a new light on the genetic characteristics of the P. aeruginosa harboring beta-lactamases in Serbia. They all belonged to the dominant clone ST235 and the less prevalent ST654. Overall, these findings are in accordance with findings reporting the global emergence of the widespread P. aeruginosa ST235 clone [15,38,39]. Curiously, an NDM-1-producing P. aeruginosa ST235 strain was isolated in France from the urine culture of a patient hospitalized in Serbia 3 months earlier (2012) and in Italy from a patient with sepsis who had been hospitalized previously in Serbia (2013) [40,41]. Of note, the blaNDM-1 gene in ST235 isolates NDM1_2 and NDM1_3 was detected in the same genetic environment as previously described in P. aeruginosa isolates from France and Italy [40,41]. To the best of our knowledge, this is the first detection of the P. aeruginosa strain ST654/blaNDM-1 and ST654/blaNDM-1/blaGES-5 in Serbia. According to the genomic data that are currently available on ST654, it seems that it is mostly confined to Russia, India, and Serbia, possibly representing the hotspots for this clone. Similarly, strain ST654, co-harboring blaNDM-1, and blaGES-5 was also isolated in Bulgaria [42], and the blaNDM gene was also located in a class I integron In1884.
A few limitations of this study should be mentioned. First, the lack of comprehensive clinical data, which could be used to make a difference between colonization and infection. Second, it was impossible to distinguish nosocomial from community-acquired infections, although the genetic relatedness of the isolates indicates that the source of the infection might be the hospital environment. When interpreting the data, these limitations should be considered, and further studies are warranted to clarify these issues.
4. Materials and Methods
4.1. Bacterial Isolates
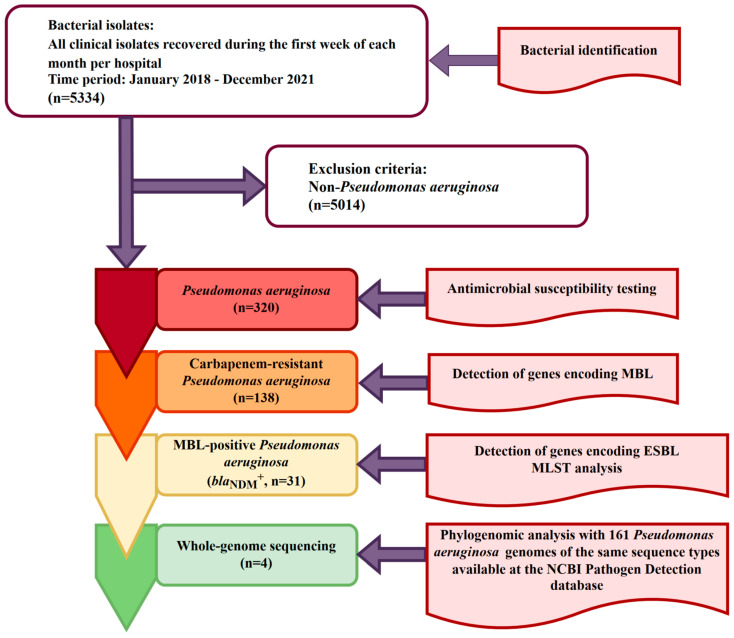
A total of 320 non-redundant isolates of P. aeruginosa obtained from patients with bacterial infections, admitted to 11 hospitals throughout Serbia and recovered during the first week of each month from 2018 to 2021, were included in the study. The ethical committee of the Medical Faculty, University of Belgrade (1550/IX-16) approved the study. A flowchart showing inclusion and exclusion criteria for the selection of bacterial isolates is shown in Figure 6.
Figure 6.
Flowchart of the process of bacterial isolates selection for phenotypic and genotypic characterization; inclusion criteria was the isolation of the Pseudomonas aeruginosa from non-redundant clinical samples (one per infected patient) obtained during the routine laboratory work. MBL—Metallo-β-lactamase; ESBL—Extended-spectrum beta-lactamase; MLST—Multilocus sequence typing.
The bacteria were isolated from various clinical samples using appropriate differential and selective media for Pseudomonas and classical microbiological culturing techniques. P. aeruginosa were identified by the VITEK2 system (bioMérieux, Marcy-l’Étoile, France), analytical profile index procedure (API 20NE test; bioMérieux, Brussels, Belgium), and MALDI-TOF (Matrix assisted laser desorption-ionisation time of flight) mass spectrometry, according to the manufacturer’s instructions. Pure bacterial cultures identified as P. aeruginosa (n = 320) were shipped on Amies transport medium to the coordinating laboratory at the Institute of Microbiology and Immunology of the Medical Faculty in Belgrade for further phenotypic and molecular testing. All isolates were stored at −70 °C in skim milk broth (Merck, Germany) until used in subsequent experiments.
4.2. Antibiotic Susceptibility Testing
Antimicrobial susceptibility was tested by employing disk diffusion, gradient test, and broth microdilution test according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations, 2021 [43]. The disks impregnated with the following antimicrobial agents were tested: imipenem (10 μg), meropenem (10 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), ceftazidime (10 µg), cefepime (30 µg), amikacin (30 µg), piperacillin/tazobactam (30/6 µg), aztreonam (30 µg), ticarcillin (75 µg), ceftazidime-avibactam (10–4 µg), and ceftolozane/tazobactam (38–10 µg) (BioRad, Watford, UK). Minimum inhibitory concentrations (MICs) for colistin, imipenem, and meropenem were evaluated by ComASP Colistin (Liofilchem, Roseto, Italy) and Gradient strip test (Liofilchem, Roseto, Italy), respectively. P. aeruginosa ATCC 27853 was used as the control strain in the antibiotic susceptibility testing. MDR P. aeruginosa were defined as isolates that tested resistant to at least one antimicrobial agent of three or more different classes. Extensively drug-resistant (XDR) P. aeruginosa were defined as a subset of MDR isolates that tested non-susceptible to at least one antimicrobial agent of five different classes. P. aeruginosa was defined as pandrug-resistant (PDR) when the organism was resistant to all tested agents.
4.3. PCR Amplification of Resistance Genes
All carbapenem-resistant P. aeruginosa isolates were screened by PCR following Sanger sequencing in case of positive PCR, for the presence of the three genes encoding MBL, blaVIM, blaIMP, and blaNDM [44]. Thereafter, all P. aeruginosa harboring MBL genes were subjected to the PCR for the detection of the genes encoding the most common ESBL-type enzymes in P. aeruginosa, namely blaCTX-M, blaPER, blaTEM, blaSHV, blaVEB, and blaGES [45,46,47,48]. The primer pairs used in the study are shown in Supplementary Table S1.
4.4. Multilocus Sequence Typing
The MLST analysis for P. aeruginosa harboring beta-lactamase genes was performed as described previously [49]. The primer pairs used for MLST analysis are shown in Supplementary Table S1. The obtained sequences of each housekeeping gene were compared with sequences in the MLST database (https://pubmlst.org/paeruginosa/, accessed on 1 July 2022) for the assignment of allelic numbers and STs.
A Pseudomonas Genome database search (Available at: www.pseudomonas.com, accessed on 1 July 2022 [50]) for P. aeruginosa isolates belonging to the same STs was carried out to create a database to use for phylogenetic analysis and to determine the clones’ geographical distribution. For these strains, information relative to the antibiotic resistance profile emphasizing the presence of carbapenems resistance genes was retrieved.
4.5. Phylogenomic and Genomic Analysis of blaNDM-Positive Strains
Four blaNDM-positive P. aeruginosa isolates were randomly selected for whole-genome sequencing (WGS) and further analysis (Illumina HiSeq paired-end and Oxford Nanopore MinION). De novo assemblies of the four P. aeruginosa genomes were generated using Unicycler v.0.4.8 [51]. The quality and completeness of the genome assemblies were assessed by testing the contamination with ContEst16S [52], and visualization with Bandage [53]. The genomes were then annotated using Prokka v1.12 [54], and rRNAs were identified by RNAmmer v1.2 [55]. PlasmidFinder 2.1, available on the Center for Genomic Epidemiology database was used to identify plasmids in whole genome sequencing data [56]. The core genes of the four P. aeruginosa genomes and of 161 (152 ST235 + nine ST654) previously published genomes of the same STs available in the NCBI Pathogen Detection database with a known geographical location were analyzed using Roary v3.13.0 [57], with a 95% minimum blastp identity and a 99% core definition threshold. Then, the SNPs of core genes were called to reduce the computational complexity for phylogenetic tree construction using SNP-sites v2.4.1 [58]. Afterward, a phylogenetic tree was built by raxmlHPC-PTHREADS v8.2.12 with the neighbor-joining method using 1000 bootstraps [59]. Visualization of the phylogenetic tree was performed using MEGA v.11 [60]. Genetic environment analysis of the flanking regions of the detected blaNDM genes was manually inspected using Geneious Prime software and ISfinder database [61].
4.6. Statistical Analysis
Statistical analysis was performed using SPSS version 14.0 (Chicago, IL, USA). The χ2 test and Fisher’s exact test were used to compare categorical data. A p ≤ 0.05 value indicated significantly different prevalence values.
5. Conclusions
In summary, this first Serbia nationwide study highlights the high frequency of several clones of MDR and XDR P. aeruginosa, the majority harboring β-lactamase encoding genes belonging to class B (MBL: blaNDM-1) and A (ESBL: blaPER-1, blaGES-5) beta-lactamases. Based on our findings, we conclude that globally disseminated ST235 also predominated throughout the country. The obtained results go beyond previous reports, showing the emergence of P. aeruginosa ST654 in Serbia. The association of the major genetic lineages and multi-resistant phenotypes is a cause for considerable concern.
Acknowledgments
We gratefully acknowledge the following persons for supplying bacterial isolates used in this study: Sanja Zornic, Snezana Jovanovic, Bojan Rakonjac, Momir Djuric, Lidija Boskovic, Teodora Vitorovic, Snezana Delic, Dragana Andjelkovic, and Vesna Karanovic. We would like to thank Dusanka Savic-Pavicevic for enabling sequencing analysis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24021519/s1.
Author Contributions
Conceptualization, I.G. and C.M.M.; methodology, I.V.-M., D.K., M.J., J.P., L.R., N.O., C.M.M. and I.G.; formal analysis, J.K., G.F., I.V.-M., D.K., M.J., J.P., L.R., N.O., C.M.M. and I.G.; investigation, J.K. and G.F.; resources, J.P., C.M.M. and I.G.; data curation, I.V.-M., D.K. and M.J.; writing—original draft preparation, J.K., G.F., I.V.-M., D.K., M.J., J.P., L.R., N.O., C.M.M. and I.G.; writing—review and editing, L.R., N.O., C.M.M. and I.G.; visualization, J.K. and G.F.; supervision, I.G. and C.M.M.; project administration, I.G. and C.M.M.; funding acquisition, I.G. and C.M.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was approved by the ethical committee of the Medical Faculty, University of Belgrade (permission number No. 1550/IX-16). Informed consent was not required for this research according to the research regulations.
Data Availability Statement
The genome sequences of four randomly selected blaNDM-positive P. aeruginosa (NDM1_1, NDM1_2, NDM1_3, and NDM1_4) isolates associated with this study have been submitted to the National Center for Biotechnology Information (NCBI) site, under BioProject Accession number PRJNA753000.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia [Grant No. 175039], the Science Fund of the Republic of Serbia [IN-DEPTH, Grant No. 6059147], the European Union’s Horizon 2020, under the Innovative Training Networks (ITN-ETN) programme Marie Skłodowska-Curie grant (ANtibioticS and mobile resistance elements in WastEwater Reuse applications: risks and innovative solutions) agreement No 675530, and by FEDER through project RISK.AR “Assessing the risks associated with environmental antibiotic resistant bacteria: propagation and transmission to humans” (PTDC/CTA-AMB/28196/2017)—Programa Operacional Competitivida de e Internacionalização. The authors thank the scientific collaboration under the FCT project UID/Multi/50016/2019.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Thaden J.T., Park L.P., Maskarinec S.A., Ruffin F., Fowler V.G., van Duin D. Results from a 13-Year Prospective Cohort Study Show Increased Mortality Associated with Bloodstream Infections Caused by Pseudomonas aeruginosa Compared to Other Bacteria. Antimicrob. Agents Chemother. 2017;61:e02671-16. doi: 10.1128/AAC.02671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pachori P., Gothalwal R., Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6:109–119. doi: 10.1016/j.gendis.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Y.-Y., Cao J.-M., Yang Q., Chen S., Lv H.-Y., Zhou H.-W., Wu Z., Zhang R. Risk Factors for Carbapenem-Resistant Pseudomonas aeruginosa, Zhejiang Province, China. Emerg. Infect. Dis. 2019;25:1861–1867. doi: 10.3201/eid2510.181699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray C.J., Ikuta K.S., Sharara F., Swetschinski L., Aguilar G.R., Gray A., Han C., Bisignano C., Rao P., Wool E., et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malkoçoğlu G., Aktaş E., Bayraktar B., Otlu B., Bulut M.E. VIM-1, VIM-2, and GES-5 Carbapenemases Among Pseudomonas aeruginosa Isolates at a Tertiary Hospital in Istanbul, Turkey. Microb. Drug Resist. 2017;23:328–334. doi: 10.1089/mdr.2016.0012. [DOI] [PubMed] [Google Scholar]
- 6.Tacconelli E., Sifakis F., Harbarth S., Schrijver R., van Mourik M., Voss A., Sharland M., Rajendran N.B., Rodríguez-Baño J., Bielicki J., et al. Surveillance for control of antimicrobial resistance. Lancet Infect. Dis. 2018;18:e99–e106. doi: 10.1016/S1473-3099(17)30485-1. [DOI] [PubMed] [Google Scholar]
- 7.Rice L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 8.Central Asian and Eastern European Surveillance of Antimicrobial Resistance. Annual Report. 2020. [(accessed on 15 July 2022)]. Available online: https://www.euro.who.int/__data/assets/pdf_file/0003/469200/Central-Asian-and-European-Surveillance-of-Antimicrobial-Resistance.-Annual-report-2020-eng.pdf.
- 9.Potron A., Poirel L., Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents. 2015;45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Davies T.A., Queenan A.M., Morrow B.J., Shang W., Amsler K., He W., Lynch A.S., Pillar C., Flamm R.K. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007–09. J. Antimicrob. Chemother. 2011;66:2298–2307. doi: 10.1093/jac/dkr290. [DOI] [PubMed] [Google Scholar]
- 11.Choudhury D., Das Talukdar A., Choudhury M.D., Maurya A.P., Paul D., Chanda D.D., Chakravorty A., Bhattacharjee A. Transcriptional Analysis of MexAB-OprM Efflux Pumps System of Pseudomonas aeruginosa and Its Role in Carbapenem Resistance in a Tertiary Referral Hospital in India. PLoS ONE. 2015;10:e0133842. doi: 10.1371/journal.pone.0133842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirsalehian A., Kalantar-Neyestanaki D., Nourijelyani K., Asadollahi K., Taherikalani M., Emaneini M., Jabalameli F. Detection of AmpC-β-lactamases producing isolates among carbapenem resistant P. aeruginosa isolated from burn patient. Iran. J. Microbiol. 2014;6:306–310. [PMC free article] [PubMed] [Google Scholar]
- 13.Livermore D.M. Multiple Mechanisms of Antimicrobial Resistance in Pseudomonas aeruginosa: Our Worst Nightmare? Clin. Infect. Dis. 2002;34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 14.Jovcic B., Lepsanovic Z., Suljagic V., Rackov G., Begovic J., Topisirovic L., Kojic M. Emergence of NDM-1 Metallo-β-Lactamase in Pseudomonas aeruginosa Clinical Isolates from Serbia. Antimicrob. Agents Chemother. 2011;55:3929–3931. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Treepong P., Kos V., Guyeux C., Blanc D., Bertrand X., Valot B., Hocquet D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018;24:258–266. doi: 10.1016/j.cmi.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 16.Woodford N., Turton J., Livermore D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 17.Bocharova Y., Savinova T., Lazareva A., Polikarpova S., Gordinskaya N., Mayanskiy N., Chebotar I. Genotypes, carbapenemase carriage, integron diversity and oprD alterations among carbapenem-resistant Pseudomonas aeruginosa from Russia. Int. J. Antimicrob. Agents. 2020;55:105899. doi: 10.1016/j.ijantimicag.2020.105899. [DOI] [PubMed] [Google Scholar]
- 18.Bush L.M., Perez M.T. Typhoid Fever. Pseudomonas and Related Infections—Infectious Diseases—MSD Manual Professional Edition. 2020. [(accessed on 19 July 2022)]. Available online: https://www.merckmanuals.com/professional/infectious-diseases/gram-negative-bacilli/pseudomonas-and-related-infections.
- 19.Aloush V., Navon-Venezia S., Seigman-Igra Y., Cabili S., Carmeli Y. Multidrug-Resistant Pseudomonas aeruginosa: Risk Factors and Clinical Impact. Antimicrob. Agents Chemother. 2006;50:43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radovanovic R.S., Savic N.R., Ranin L., Smitran A., Opavski N.V., Tepavcevic A.M., Ranin J., Gajic I. Biofilm Production and Antimicrobial Resistance of Clinical and Food Isolates of Pseudomonas spp. Curr. Microbiol. 2020;77:4045–4052. doi: 10.1007/s00284-020-02236-4. [DOI] [PubMed] [Google Scholar]
- 21.European Centre for Disease Prevention and Control Antimicrobial Resistance Surveillance in Europe 2022–2020 Data. 2022. [(accessed on 1 July 2022)]. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2022-2020-data.
- 22.Durante-Mangoni E., Andini R., Zampino R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 2019;25:943–950. doi: 10.1016/j.cmi.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Karlowsky J.A., Kazmierczak K.M., de Jonge B.L.M., Hackel M.A., Sahm D.F., Bradford P.A. In Vitro Activity of Aztreonam-Avibactam against Enterobacteriaceae and Pseudomonas aeruginosa Isolated by Clinical Laboratories in 40 Countries from 2012 to 2015. Antimicrob. Agents Chemother. 2017;61:e00472-17. doi: 10.1128/AAC.00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Neall D., Juhász E., Tóth A., Urbán E., Szabó J., Melegh S., Katona K., Kristóf K. Ceftazidime–avibactam and ceftolozane–tazobactam susceptibility of multidrug resistant Pseudomonas aeruginosa strains in Hungary. Acta Microbiol. Immunol. Hung. 2020;67:61–65. doi: 10.1556/030.2020.01152. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed M.A.S., Hassan A., Abu Jarir S., Hadi H.A., Bansal D., Wahab A.A., Muneer M., Mohamed S., Zahraldin K., Hamid J., et al. Emergence of Multidrug- and Pandrug- Resistant Pseudomonas aeruginosa from Five Hospitals in Qatar. Infect. Prev. Pract. 2019;1:100027. doi: 10.1016/j.infpip.2019.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Baky R.M.A., Masoud S.M., Mohamed D.S., Waly N.G., A Shafik E., A Mohareb D., Elkady A., Elbadr M.M., Hetta H.F. Prevalence and Some Possible Mechanisms of Colistin Resistance Among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa. Infect. Drug Resist. 2020;13:323–332. doi: 10.2147/IDR.S238811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuji B.T., Pogue J.M., Zavascki A.P., Paul M., Daikos G.L., Forrest A., Giacobbe D.R., Viscoli C., Giamarellou H., Karaiskos I., et al. International Consensus Guidelines for the Optimal Use of the Polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP) Pharmacother. J. Hum. Pharmacol. Drug Ther. 2019;39:10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urbanowicz P., Izdebski R., Baraniak A., Żabicka D., Hryniewicz W., Gniadkowski M. Molecular and genomic epidemiology of VIM/IMP-like metallo-β-lactamase-producing Pseudomonas aeruginosa genotypes in Poland. J. Antimicrob. Chemother. 2021;76:2273–2284. doi: 10.1093/jac/dkab188. [DOI] [PubMed] [Google Scholar]
- 30.Papagiannitsis C.C., Medvecky M., Chudejova K., Skalova A., Rotova V., Spanelova P., Jakubu V., Zemlickova H., Hrabak J. Molecular Characterization of Carbapenemase-Producing Pseudomonas aeruginosa of Czech Origin and Evidence for Clonal Spread of Extensively Resistant Sequence Type 357 Expressing IMP-7 Metallo-β-Lactamase. Antimicrob. Agents Chemother. 2017;61:e01811-17. doi: 10.1128/AAC.01811-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordmann P., Ronco E., Naas T., Duport C., Michel-Briand Y., Labia R. Characterization of a novel extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1993;37:962–969. doi: 10.1128/AAC.37.5.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libisch B., Poirel L., Lepsanovic Z., Mirovic V., Balogh B., Pászti J., Hunyadi Z., Dobák A., Füzi M., Nordmann P. Identification of PER-1 extended-spectrum β-lactamase producing Pseudomonas aeruginosa clinical isolates of the international clonal complex CC11 from Hungary and Serbia. FEMS Immunol. Med. Microbiol. 2008;54:330–338. doi: 10.1111/j.1574-695X.2008.00483.x. [DOI] [PubMed] [Google Scholar]
- 33.Weldhagen G.F., Poirel L., Nordmann P. Ambler Class A Extended-Spectrum β-Lactamases in Pseudomonas aeruginosa: Novel Developments and Clinical Impact. Antimicrob. Agents Chemother. 2003;47:2385–2392. doi: 10.1128/AAC.47.8.2385-2392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagani L., Mantengoli E., Migliavacca R., Nucleo E., Pollini S., Spalla M., Daturi R., Romero E., Rossolini G.M. Multifocal Detection of Multidrug-Resistant Pseudomonas aeruginosa Producing the PER-1 Extended-Spectrum β-Lactamase in Northern Italy. J. Clin. Microbiol. 2004;42:2523–2529. doi: 10.1128/JCM.42.6.2523-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Çiçek A., Ertürk A., Ejder N., Rakici E., Kostakoğlu U., Yıldız I.E., Özyurt S., Sönmez E. Screening of Antimicrobial Resistance Genes and Epidemiological Features in Hospital and Community-Associated Carbapenem-Resistant Pseudomonas aeruginosa Infections. Infect. Drug Resist. 2021;14:1517–1526. doi: 10.2147/IDR.S299742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhi M.T., Khalili Y., Ghottaslou R., Aghazadeh M., Seroush Bar Hagh M.H., Yousefi S. Prevalence of PER-1- type ex-tended-spectrum beta-lactamases in clinical strains of Pseudomonas aeruginosa isolated from Tabriz, Iran. Iran. J. Basic. Med. Sci. 2012;15:678–682. [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon E.-J., Jeong S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021;12:614058. doi: 10.3389/fmicb.2021.614058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abril D., Marquez-Ortiz R.A., Castro-Cardozo B., Moncayo I., Escobar N.M.O., Rozo Z.L.C., Reyes N., Tovar C., Sánchez H.F., Castellanos J., et al. Genome plasticity favours double chromosomal Tn4401b-blaKPC-2 transposon insertion in the Pseudomonas aeruginosa ST235 clone. BMC Microbiol. 2019;19:45. doi: 10.1186/s12866-019-1418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Barrio-Tofiño E., López-Causapé C., Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents. 2020;56:106196. doi: 10.1016/j.ijantimicag.2020.106196. [DOI] [PubMed] [Google Scholar]
- 40.Janvier F., Jeannot K., Tessé S., Robert-Nicoud M., Delacour H., Rapp C., Mérens A. Molecular Characterization of bla NDM-1 in a Sequence Type 235 Pseudomonas aeruginosa Isolate from France. Antimicrob. Agents Chemother. 2013;57:3408–3411. doi: 10.1128/AAC.02334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carattoli A., Fortini D., Galetti R., Garcia-Fernandez A., Nardi G., Orazi D., Capone A., Majolino I., Proia A., Mariani B., et al. Isolation of NDM-1-producing Pseudomonas aeruginosa sequence type ST235 from a stem cell transplant patient in Italy, May 2013. Eurosurveillance. 2013;18:20633. doi: 10.2807/1560-7917.ES2013.18.46.20633. [DOI] [PubMed] [Google Scholar]
- 42.Kostyanev T., Nguyen M., Markovska R., Stankova P., Xavier B., Lammens C., Marteva-Proevska Y., Velinov T., Cantón R., Goossens H., et al. Emergence of ST654 Pseudomonas aeruginosa co-harbouring blaNDM-1 and blaGES-5 in novel class I integron In1884 from Bulgaria. J. Glob. Antimicrob. Resist. 2020;22:672–673. doi: 10.1016/j.jgar.2020.06.008. [DOI] [PubMed] [Google Scholar]
- 43.European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2021. [(accessed on 1 July 2022)]. Version 11.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf.
- 44.Poirel L., Walsh T.R., Cuvillier V., Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Opazo A., Sonnevend A., Lopes B., Hamouda A., Ghazawi A., Pal T., Amyes S.G.B. Plasmid-encoded PER-7 -lactamase responsible for ceftazidime resistance in Acinetobacter baumannii isolated in the United Arab Emirates. J. Antimicrob. Chemother. 2012;67:1619–1622. doi: 10.1093/jac/dks087. [DOI] [PubMed] [Google Scholar]
- 46.Celenza G., Pellegrini C., Caccamo M., Segatore B., Amicosante G., Perilli M. Spread of blaCTX-M-type and blaPER-2 β-lactamase genes in clinical isolates from Bolivian hospitals. J. Antimicrob. Chemother. 2006;57:975–978. doi: 10.1093/jac/dkl055. [DOI] [PubMed] [Google Scholar]
- 47.Yoshizumi A., Ishii Y., Aoki K., Testa R., Nichols W.W., Tateda K. In vitro susceptibility of characterized β-lactamase-producing Gram-negative bacteria isolated in Japan to ceftazidime-, ceftaroline-, and aztreonam-avibactam combinations. J. Infect. Chemother. 2015;21:148–151. doi: 10.1016/j.jiac.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 48.Bae I.K., Jang S.J., Kim J., Jeong S.H., Cho B., Lee K. Interspecies Dissemination of the bla Gene Encoding PER-1 Extended-Spectrum β-Lactamase. Antimicrob. Agents Chemother. 2011;55:1305–1307. doi: 10.1128/AAC.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curran B., Jonas D., Grundmann H., Pitt T., Dowson C.G. Development of a Multilocus Sequence Typing Scheme for the Opportunistic Pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 2004;42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2015;44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee I., Chalita M., Ha S.-M., Na S.-I., Yoon S.-H., Chun J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017;67:2053–2057. doi: 10.1099/ijsem.0.001872. [DOI] [PubMed] [Google Scholar]
- 53.Wick R.R., Schultz M.B., Zobel J., Holt K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 55.Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.-H., Rognes T., Ussery D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carattoli A., Zankari E., Garcìa-Fernandez A., Larsen M., Lund O., Voldby Villa L., Møller Aarestrup F., Hasman H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Page A.J., Taylor B., Delaney A.J., Soares J., Seemann T., Keane J.A., Harris S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences of four randomly selected blaNDM-positive P. aeruginosa (NDM1_1, NDM1_2, NDM1_3, and NDM1_4) isolates associated with this study have been submitted to the National Center for Biotechnology Information (NCBI) site, under BioProject Accession number PRJNA753000.