Abstract
Giardia lamblia infections are associated with antigenic variation of the parasite, which is generated by a continuous change of the variant-specific surface proteins (VSPs). Many investigations on the process of antigenic variation were based on the use of G. lamblia clone GS/M-83-H7, which expresses VSP H7 as its major surface antigen. In the present study, mice were infected with the aforementioned clonal line to investigate vsp gene expression during the complex process of antigenic variation of the parasite. Trophozoites collected from the intestines of individual animals at different time points postinfection (p.i.) were analyzed directly for their vsp gene expression patterns, i.e., without cultivating the recovered parasites in vitro. Because few trophozoites were recovered at late time points p.i., a combined 5′ rapid amplification of cDNA ends–reverse transcription-PCR approach was utilized. This allowed detection and subsequent sequence analysis of vsp gene transcripts upon generation of amplified cDNA analogues. The same PCR approach was applied for analysis of vsp gene expression in variants obtained after negative selection of axenic GS/M-83-H7 trophozoites by treatment with a cytotoxic, VSP H7-specific monoclonal antibody. In an overall view of the entire panel of in vivo- and in vitro-derived parasite populations, expression of 29 different vsp gene sequences could be demonstrated. In vivo antigenic variation of G. lamblia clone GS/M-83-H7 was shown to be a continuous process involving the consecutive appearance of relatively distinct sets of vsp transcripts. During the 42-day infection period investigated, this process activated at least 22 different vsp genes. Comparative molecular analyses of the amino acid level demonstrated that all cDNA segments identified encode structural elements typical of the terminal segment of Giardia VSP. The similarity of most of the GS/M-83-H7 VSP sequences identified in the present study supports previous suggestions that vsp gene diversification in G. lamblia is the result of ancestral gene duplication, mutation, and/or recombination events.
The protozoan parasite Giardia lamblia is an important causative agent of acute or chronic diarrhea in humans and various animals. G. lamblia is characterized by a considerable potential to alter its surface antigens, a phenomenon that is mediated by a unique protein family, the variant-specific surface proteins (VSPs) (19). Individual trophozoites basically produce a single VSP, which constitutes the major surface antigen and which covers the entire cell including the flagella (27). The proteins are cysteine rich, exhibiting a common four-amino-acid CXXC repeat motif and remarkable similarity across the hydrophobic, membrane-spanning region at the extreme C terminus (15, 26).
The antigenic surfaces of different isolates of G. lamblia are basically variable (23, 25, 34), and surface antigenic variation may occur within an isolate (1–3, 20, 21). Surface antigen alterations have been observed within proliferating trophozoite populations inside the host (3, 7, 8, 32), as well as among trophozoites upon release from nonproliferative cysts (13, 33). This change of the surface protein coat is supposed to allow the parasite to persist within the hostile immunological and physiological environment of the gut (reviewed in references 16 and 18). Since a maturating humoral immune response in both experimental and natural Giardia-infected hosts coincides with the elimination of the original variant phenotype, a functional role of antibodies in the selection of variants was proposed (11, 21). Support for this conclusion was provided by the finding of a parasiticidal effect of anti-Giardia antibodies on in vitro-cultivated trophozoites. Thus, incubation of G. lamblia trophozoites with sera from infected individuals (11, 21) and with monoclonal antibodies (MAbs) to different VSPs (21) resulted in a complement-dependent lysis of the parasites. Furthermore, a MAb (G10/4) to VSP H7 (2) produced by G. lamblia clone GS/M-83-H7 and a MAb (6E7) to VSPA6 produced by G. lamblia isolate WB (20) induced complement-independent lysis of the trophozoites with respective variant antigen types (9, 20, 31). High concentrations of MAbs G10/4 (31) and 6E7 (20) caused immediate immobilization as well as detachment and aggregation of the trophozoites. The direct cytotoxic effect of MAb G10/4 was associated with shedding VSP-containing membrane vesicles from the parasite surface and a concomitant partial disruption of the cellular membrane (9).
So far, the process of antigenic variation of G. lamblia in vivo and the host's immune response against the parasite have best been studied in experimentally infected mice (reviewed in references 5 and 16). Investigations have been mostly based on the use of G. lamblia clone GS/M-83-H7, which expresses major surface antigen VSP H7 (2). Experimental G. lamblia GS/M-83-H7 infections in a combined mother-offspring mouse model involving simultaneous infection of mother and offspring animals indicated that ingestion of anti-VSP H7 secretory immunoglobulin A antibodies by neonatal mice is associated with an immediate increase in the prevalence of new variant antigen types within the intestinal parasite population of these animals (32). These antibodies display a direct cytotoxic effect against VSP H7-type trophozoites (32). We have now investigated in detail at the transcriptional level the complexity of the antigenic diversification process during an experimental infection with G. lamblia clone GS/M-83-H7 in the murine host. Antigenic variation in vivo was found to be associated with activation of a large portion of the vsp gene repertoire of the parasite. A remarkable diversification in vsp gene expression was also observed in vitro upon antibody (MAb G10/4)-mediated growth selection for VSP H7-negative trophozoites within a culture of G. lamblia clone GS/M-83-H7 trophozoites.
MATERIALS AND METHODS
Mice.
Gravid 10- to 12-week-old outbred CD-1(ICR)BR mice were obtained from Charles River GmbH, Germany. Animals were kept according to the Swiss regulations of animal experiments with free access to germ-free food and sterile water.
Parasite.
The origin, axenization, and cloning of G. lamblia clone GS/M-83-H7 have been described by Aggarwal and coworkers (2). This clone expresses on its surface a major 72-kDa antigen (VSP H7), which is recognized by MAb G10/4. Trophozoites from a clonal GS/M-83-H7 line of G. lamblia were cultivated in modified TYI-S-33 medium with antibiotics as previously described (12).
Immunofluorescence assays.
The kinetics of expression of the major surface antigen on trophozoites isolated from the duodena of experimentally infected mice (see below) were assessed by immunofluorescence using MAb G10/4 as described previously (7).
Experimental parasite infection, sample collection, and total RNA extraction.
Three-day-old offspring and the respective mothers were infected with G. lamblia trophozoites (clone GS/M-83-H7) as described previously (32).
The course of the G. lamblia infection within offspring was determined as described by Gottstein and coworkers (6) by quantifying the parasite burden through microscopical examination of adherent trophozoites from intestinal washes.
For total RNA extraction (see below), intestinal parasites were isolated as follows. Sections of about 1 cm from the upper part of the duodenum were slit longitudinally and subsequently incubated for 20 min in 1 ml of phosphate-buffered saline (PBS; containing 0.15 M NaCl), pH 7.2, on ice to detach trophozoites from the intestinal surface. Then, 0.5 ml of PBS supernatant containing detached trophozoites was transferred into a well of a microtiter plate (Cellstar TC plate; 24 wells; Greiner Labortechnik GmbH, Frickenhausen, Switzerland), and viable parasites were allowed to adhere to the bottom of the well by incubation for 20 min at 37°C. After the parasites were washed three times with 1 ml of prewarmed (37°C) PBS, residual, adherent trophozoites were resuspended in 100 μl of lysis buffer–β-mercaptoethanol (β-ME) mixture from the StrataPrep Total RNA Microprep kit (Stratagene, La Jolla, Calif.). The lysates were processed for extraction of total RNA as instructed including treatment with RNase-free DNase I. Finally, total RNA preparations (solubilized in 30 μl of elution buffer) were stored at −80°C until further used.
Antigen switching by GS/M-83-H7 trophozoites in vitro, sample collection, and total-RNA extraction.
The procedure used to detect antigen switching of in vitro-cultivated G. lamblia clone GS/M-83-H7 trophozoites from a VSP H7-positive to a VSP H7-negative parasite population was adapted from a method of Nash et al. (21). Adherent GS/M-83-H7 trophozoites from late-log-phase cultures were washed twice in prewarmed (37°C) PBS and then detached from the bottom of the culture tube by a 15-min incubation in ice-cold PBS. About 105 cells (corresponding to 200 μl from the resulting parasite suspension) were transferred into a well from a culture plate and subsequently allowed to readhere for 15 min at 37°C. To select against VSP H7-positive trophozoites within this parasite population, incubation was continued for 15 min after adding 2 μl of ascites fluid containing cytotoxic, VSP H7-specific MAb G10/4. Nonadherent, dead trophozoites were removed by washing three times with 1 ml of prewarmed (37°C) PBS, after which the residual adherent (MAb G10/4-resistant) variant trophozoites were detached in 500 μl of ice-cold PBS. Released parasites were grown in vitro for 3 days in 15 ml of TYI-S-33 medium and then re-treated with MAb G10/4 and recultivated as described above. After confirmation by immunofluorescence that the resulting MAb G10/4-resistant parasite population was essentially (>99%) VSP H7 negative, about 104 of these negatively selected variant trophozoites (<1% VSP H7 positive) treated with MAb G10/4 were resuspended in 100 μl of a lysis buffer–β-ME mixture from the StrataPrep Total RNA Microprep kit (Stratagene). In parallel, an analogous cellular lysate was prepared from about 104 GS/M-83-H7 trophozoites that had not been treated with MAb G10/4. Both cellular lysates were then further processed for total-RNA extraction as described above. Finally, total-RNA preparations were solubilized in 30 μl of elution buffer and stored at −80°C until further used.
Analysis of vsp gene transcripts by reverse transcription-PCR.
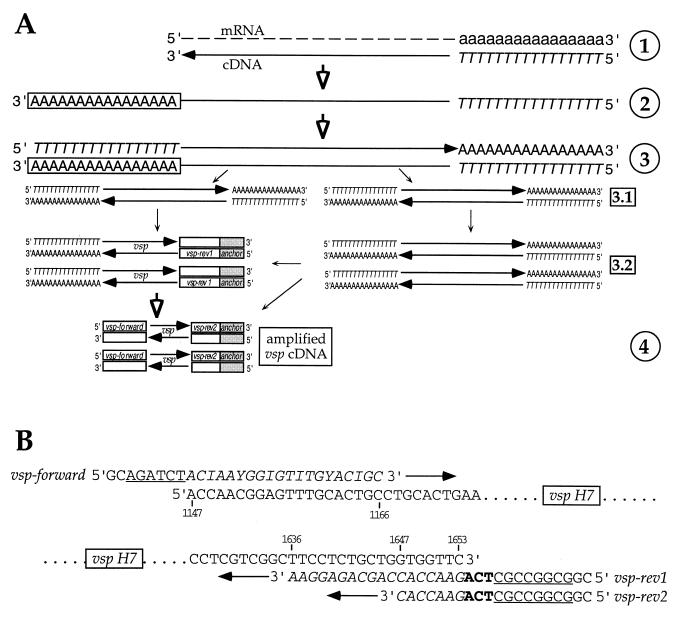
cDNA was synthesized by reverse transcription from total RNA, prepared from in vitro-cultivated and intestinal trophozoite populations, by using an oligo(dT) primer, avian myeloblastosis virus reverse transcriptase, and other components of the 5′-to-3′ rapid amplification of cDNA ends (RACE) kit (Roche Diagnostics GmbH, Rotkreuz, Switzerland) (Fig. 1A, step 1) as instructed. The same kit system was used for 5′ RACE (Fig. 1A, steps 2 and 3) by the addition of a 3′ poly(dA) tail to the 3′ end of the cDNA and a subsequent amplification (PCR-1; 40 cycles of 94°C for 30 s, 38°C for 30 s, and 72°C for 150 s, ending with a 15-min extension at 72°C) of the tailed cDNA using the kit's oligo(dT) as the forward primer (complementary to the artificially introduced poly(dA) tail; see above) and vsp gene-specific reverse primer vsp-rev1 (5′-CGGCGGCCGCTCAGAACCACCAGCAGAGGAA-3′) (Fig. 1B). The latter contained a NotI restriction site (underlined) as an anchor sequence, including the stop codon (in boldface), and a sequence complementary to nucleotides (nt) 1636 to 1653 (in italics) of the vsp H7 gene. This vsp-specific PCR occurred as a seminested reaction concomitant with general “nonspecific” single-primer amplification of all cDNA by oligo(dT) priming (Fig. 1A, steps 3.1 and 3.2). Finally, selective amplification of vsp cDNA molecules (PCR-2; 5 cycles of 94°C for 30 s, 38°C for 30 s, and 72°C for 150 s and then 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 150 s, with a final 15-min extension at 72°C) was achieved using degenerate vsp gene-specific forward primer vsp-forward (5′-GCAGATCTCACIAAYGGIGTITGYACIGC-3, where I = deoxyinosine and Y = C or T) containing a Bgl2 restriction site (underlined) followed by nt 1147 to 1166 (in italics) of the vsp H7 gene and reverse anchor primer vsp-rev2 (5′-CGGCGGCCGCTCAGAACCAC-3′), representing the first 20 nt from the vsp-rev1 primer, used for PCR-1 (Fig. 1A, step 4, and B). Introduction of artificial mutations during amplification was minimized by using the Expand High Fidelity PCR system (Roche Diagnostics) for all PCRs. Controls were included for both PCR steps using total RNA equivalents that had not been reverse transcribed into cDNA (not shown) to check that no DNA was amplified from any residual genomic DNA that might have resisted DNase I digestion. PCR products were analyzed by electrophoresis in 2% agarose gels. The amplification products from the predominant band (migrating with a size of around 550 bp) were tested for restriction fragment length polymorphism (RFLP) by cleavage with AluI. Additionally, the amplification products were cloned into the pGEMeasy vector (Promega, Madison, Wis.), and cloned inserts were sequenced by a commercial sequencing service (Solvias AG, Basel, Switzerland). All recombinant DNA methods, unless otherwise stated, were those of Sambrook et al. (29). For DNA cloning, Escherichia coli XL1-Blue (Stratagene) was used as the bacterial host.
FIG. 1.
Strategy used for PCR-based amplification and analysis of vsp cDNA molecules. (A) In step 1, polyadenylated (lowercase) mRNA present within total trophozoite RNA was reverse transcribed into cDNA using an oligo(dT) primer (italic uppercase). In step 2, the first-strand cDNA molecules were polyadenylated (boxed uppercase) at the 3′ end using terminal transferase. In step 3, 3′-polyadenylated cDNA was amplified by PCR using oligo(dT) as the forward and reverse primers (step 3.1). This increased the amount of cDNA template available for amplification of vsp cDNA mediated by oligo(dT) (as the forward primer) and reverse vsp-specific (open box) primer vsp-rev1 (step 3.2; see also panel B), containing a short anchor sequence (grey box). In step 4, the heterogeneous cDNA amplified in step 3 was used as a template for subsequent nested PCR, which allowed more-specific amplification using degenerate forward vsp primer vsp-forward (open box) and reverse primer vsp-rev2, modified from vsp-rev1 (see also panel B). (B) Nucleotide positions of the vsp H7 target sequences from primers vsp-forward, vsp-rev1, and vsp-rev2. Primer vsp-forward contains a 5′ GC, followed by a Bgl2 restriction site (underlined) as the anchor sequence and a degenerate sequence targeting between nt 1147 and 1166 (italics) of the vsp H7 gene (I = deoxyinosine; Y = C or T). Primer vsp-rev1 contains a 5′ GC, a NotI restriction site (underlined), and the stop codon (boldface) as the anchor sequence and a sequence complementary to nt 1636 to 1653 (italics) of the vsp H7 gene. Reverse primer vsp-rev2 represents the first 20 nt from vsp-rev1.
Sequence alignments.
Alignment and comparison of the corresponding amino acid sequences derived from the studied in vivo- and in vitro-derived parasite populations, including the original GS/M-83-H7 isolate, were done using the MultAlin and the ESPript1.9 computer software, available at the ExPASy molecular biology server using the Blosum62 symbol comparison table (10). The newly derived sequences were aligned against all homologous regions of those G. lamblia VSP sequences available in GenBank using the ClustalX, version 1.8, and the Risler symbol comparison table (28). A gene tree, based on distances calculated from amino acid sequence alignments by ClustalX, version 1.8, was generated using TreeView, version 1.6.1.
RESULTS
Detection of antigen switching in G. lamblia trophozoites cultivated in vitro.
To select for new variants within clonal line GS/M-83-H7 of G. lamblia, originally consisting of more than 95% VSP H7-type (MAb G10/4-positive) trophozoites, cultivated parasites were treated twice with VSP H7-specific, cytotoxic MAb G10/4. The efficacy of this selection was assessed by immunofluorescence, which demonstrated that the treated culture contained more than 99% non-VSP H7-type (MAb G10/4-negative) trophozoites.
Course of G. lamblia infection in mice and in vivo antigen switching of the parasite.
In our study, 3-day-old suckling mice were infected by intragastric injection with trophozoites from clonal line GS/M-83-H7. At different time points during (days 7 and 14 postinfection [p.i.]) and after (days 21 and 42 p.i.) the lactation phase, offspring individuals were sacrificed by CO2 euthanasia. The follow-up study of the parasite burden confirmed previously published results (17) in that it revealed high infection intensities at days 7 and 14 p.i. but a significant regression of infection by day 21 p.i. (data not shown). At day 42 p.i., intestinal trophozoites had nearly disappeared and only one of the three animals tested contained a parasite burden sufficient for further investigations (see below). Immunofluorescence analysis demonstrated that trophozoites from both the inoculum (representing day 0 p.i.) and the parasites isolated at day 7 p.i. were mostly (>95%) VSP H7 positive. In contrast, the intestinal parasite populations recovered after day 7 p.i., i.e., on days 14, 21, and 42 p.i., did not possess detectable numbers of VSP H7-type trophozoites in any of the respective animals tested.
Characterization of cloned vsp cDNA derived from variants emerging during antigen switching of the parasites in vitro and in vivo.
For molecular analysis of vsp gene expression in different G. lamblia clone GS/M-83-H7 populations during antigen switching in vitro or in vivo, trophozoites were sampled either from (i) an original VSP H7-type GS/M-83-H7 culture (which represents both the nontreated culture prior to detectable antigen switching and the parasite inoculum corresponding to day 0 of the experimental infection; see below), (ii) a culture treated with cytotoxic MAb G10/4, or (iii) the intestines of individual animals at days 7, 14, 21, and 42 p.i. From these different sample groups, total RNA was prepared and then used as a template for cDNA synthesis (Fig. 1A, step 1). To avoid ex vivo antigenic drift of the intestinal parasite populations, sampled trophozoites were further processed without previous proliferation of the parasites through in vitro cultivation. Since sampling of parasites from late-stage infected animals (at days 21 and 42 p.i.) provided only trace amounts of parasite material, cDNA containing vsp gene segments had to be amplified in several PCR steps prior to cloning and subsequent analysis (Fig. 1A, steps 2 to 4). The final PCR, specifically amplifying vsp cDNA, was performed using a degenerate forward primer corresponding to nt 1147 to 1166 of vsp H7 and covering a relatively conserved vsp region (see Discussion) and a reverse primer that was complementary to nt 1636 to 1653 of the vsp H7 coding sequence (Fig. 1A, step 4, and B). The latter encodes part of the highly conserved transmembrane domain of the surface protein. A preevaluation of the PCR on serial dilutions of genomic DNA from G. lamblia clone GS/M-83-H7 demonstrated that use of the degenerate forward primer generated significant amounts of PCR amplicons only in the presence of relatively high template DNA concentrations (data not shown). Accordingly, we had to introduce a preceding 5′ RACE-PCR step (Fig. 1, step 3.1), which nonspecifically increased the amount of template cDNA and thus provided the basis for successful PCR amplification of vsp gene sequences (Fig. 1A, steps 3.2 and 4).
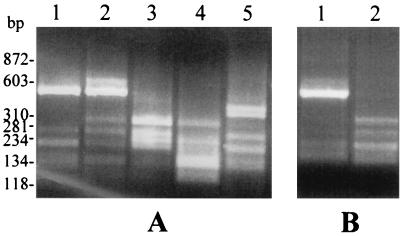
In all samples investigated, the multistep PCR provided final cDNA amplification products which migrated in agarose gels as diffuse bands of around 550 bp (data not shown). RFLP analysis using AluI for digestion of cDNA amplicons revealed alterations of the banding patterns which correlated with the in vivo (Fig. 2A) and in vitro antigen switching (Fig. 2B) of the parasite. This indicated that the composition of the vsp gene sequences within the amplification products had changed as a consequence of the antigenic-diversification process.
FIG. 2.
RFLP analysis of PCR-amplified vsp cDNAs. Banding patterns (2% agarose gel) of AluI-digested cDNA amplicons derived from RNA extracted from trophozoites recovered from infected mice on days 0 (lane 1), 7 (lane 2), 14 (lane 3), 21 (lane 4), and 42 (lane 5) p.i. (A) or from untreated (lane 1) or MAb G10/4-treated (lane 2) trophozoites grown in axenic culture (B) are shown. Size markers are shown at left.
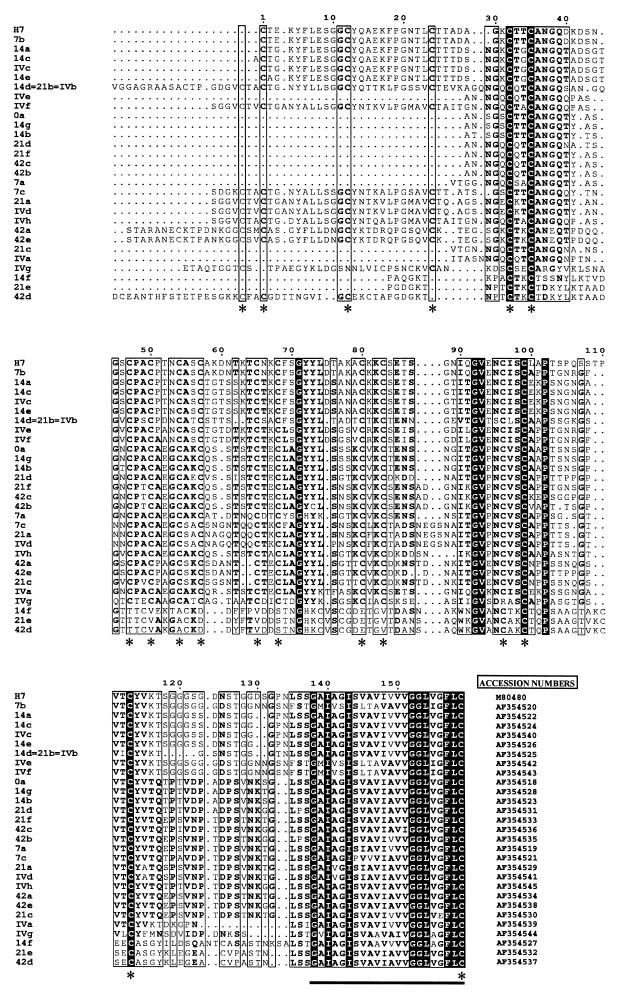
Amplified cDNA molecules were cloned into plasmid vector pGEMeasy, and 12 clones from each sample group were further investigated. DNA sequence analysis indicated that all of the cloned cDNA inserts represented vsp gene segments. The open reading frames from the cloned cDNA molecules encoded 29 peptide sequences, which all included a combination of typical structural elements of the C-terminal regions of the G. lamblia VSPs: (i) a high content of cysteine residues, (ii) the presence of at least three four-amino-acid CXXC motifs, and (iii) a hydrophobic segment corresponding to the highly conserved transmembrane portion located at the very C terminus of these surface proteins. The amino acid sequences encoded by cDNA clones H7, 7b, 14a, 14c, IVc, 14e, 14d/21b/IVb, IVf, 21a, IVd, and IVh (Fig. 3) also contained a four-amino-acid GGCY motif, which may be taken as an additional marker sequence of the VSP family (22).
FIG. 3.
Aligned amino acid sequences representing the C-terminal segments of VSP H7 (H7) and subvariant VSPs derived from G. lamblia clone GS/M-83-H7. Sequences were deduced from cloned vsp cDNA derived from parasite populations recovered on days 0 (cDNA clones H7 and 0a), 7 (clones H7 and 7a to -c), 14 (clones 14a to -g), 21 (clones 21a to -f), and 42 (clones 42 a to -e) p.i. from infected mice or clones IVa to -h from negatively selected axenic trophozoites. Similarities between VSP H7 and the corresponding segments of the subvariant VSPs identified ranged between 83 (clone 7b) and 31% (clone 42d). Black, invariant positions; boxes, regions of high sequence similarities (conserved residues are in boldface); dots, alignment gaps; asterisks, positions with frequent occurrence of cysteines (C); bar, highly conserved transmembrane segment (positions 139 to 159 of VSP H7). GenBank accession numbers are at the right.
Molecular analysis of vsp gene expression during in vitro antigen switching of the parasites.
A sequence comparison that focused on those cloned vsp cDNA molecules that originated from parasite populations before and after in vitro antigen switching provided the following results (Fig. 3). For the original G. lamblia clone GS/M-83-H7 culture, 11 of the 12 cDNA clones encoded a 159-residue C-terminal sequence identical to that of VSP H7. This result indicated predominant expression of gene vsp H7 in clone GS/M-83-H7 prior to antigen switching. In contrast, analogous analysis of the axenic parasite population that had been selected against MAb G10/4-reactive trophozoites (see above) yielded a set of eight distinct vsp cDNAs, which all encoded VSP amino acid sequences different from that of VSP H7 (Table 1 and Fig. 3). These findings demonstrated that elimination of MAb G10/4-reactive trophozoites left variants whose progeny exhibited a strong diversification in vsp gene expression.
TABLE 1.
Prevalence of different vsp gene sequences within sets of 12 cloned cDNA inserts derived from different tested Giardia populations
| Source of cDNA | Time sampled (day p.i.) or culture type | Prevalence of vsp transcriptsa |
|---|---|---|
| Infecting parasites | 0b | H7, 11c; 0a, 1 |
| 7 | H7, 9c; 7a, 1; 7b, 1; 7c, 1 | |
| 14 | 14a, 2; 14b, 2; 14c, 2; 14d, 2; 14e, 1; 14f, 1; 14g, 2 | |
| 21 | 21a, 1; 21b, 7c; 21c, 1; 21d, 1; 21e, 1; 21f, 1 | |
| 42 | 42a, 2; 42b, 5c; 42c, 1; 42d, 2; 42e, 2 | |
| Axenic culture | Unselected | H7, 11c; 0a, 1 |
| Negatively selected | IVa, 2; IVb, 2; IVc, 1; IVd, 1; IVe, 1; IVf, 1; IVg, 2; IVh, 2 |
Within sets of 12 sequenced cDNA inserts.
Original inoculum; same as for parasites of the unselected axenic culture.
Clones frequently occurring within a sample group.
Molecular analysis of vsp gene expression during in vivo antigen switching of the parasites.
Examination of the cloned cDNA that was derived from the different parasite populations consecutively emerging during a G. lamblia GS/M-83-H7 infection in mice provided the following results (Table 1 and Fig. 3). G. lamblia GS/M-83-H7 trophozoites sampled at day 7 p.i. yielded transcripts that were predominantly from vsp H7, as evidenced by detection of 9 vsp H7 versus 3 non-vsp H7 cDNA clones among 12 clones examined. Trophozoite populations consecutively sampled later in the infection exhibited concomitant expression of at least 7 (day 14 p.i.), 6 (day 21 p.i.), or 5 (day 42 p.i.) individual vsp genes that were different from vsp H7. The sets of vsp genes expressed at the different stages of infection were rather distinct from each other and contained only one common vsp cDNA (14d and 21b), which appeared within sample groups from days 14 and 21 p.i. The same cDNA was also found within the repertoire of cloned vsp amplification products from axenic trophozoites that had been negatively selected for antigenic variants (clone IVb).
Taken together, these data indicate that antigenic variation of G. lamblia clone GS/M-83-H7 during an infection in mice leads to the consecutive expression of relatively distinct and rather heterogeneous sets of vsp genes. Both diversification of vsp gene expression after day 7 p.i. and the relative individual patterns of vsp gene expression at the different experimental time points of infection were confirmed by RFLP analysis using AluI for digestion of corresponding cDNA amplicons (Fig. 2A). In this analysis, the DNA banding pattern significantly changed after day 7 p.i. and showed distinct features at the different time points after antigen switching had occurred. Analogous results were achieved by digestion of amplicons with another set of frequently cutting DNA restriction enzymes (data not shown).
Determination of the genetic relationship between the different subvariant-type VSP sequences.
Alignment of the amino acid sequences indicated that the similarities between VSP H7 (expressed by the parent G. lamblia GS/M-83-H7 line) and the corresponding regions from the subvariant VSPs ranged between 83 (clone 7b) and 31% (clone 42d) (Fig. 3). Loss of similarity of the subvariant VSP sequences related to VSP H7 did not quantitatively correlate with the time points at which the variants appeared during infection. Furthermore, subvariant VSP sequences identified within the negatively selected axenic trophozoites were heterogeneously distributed within the entire panel of VSP sequences analyzed.
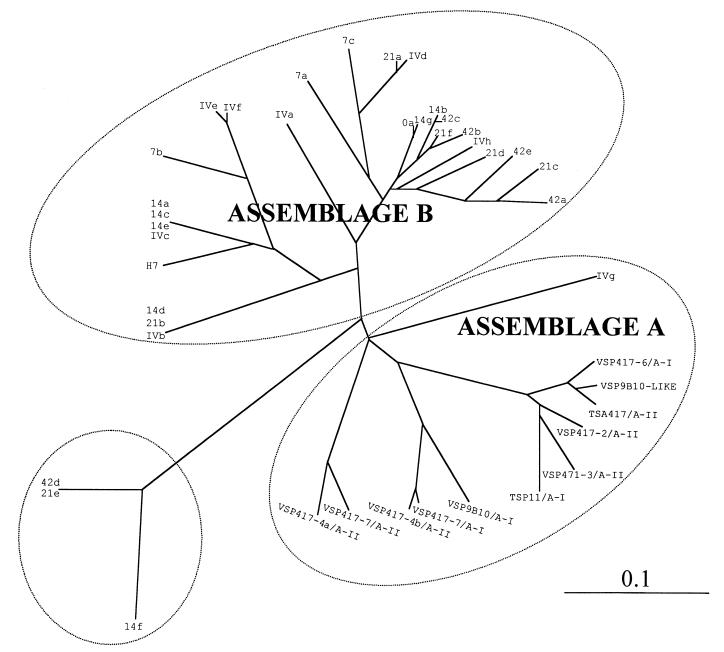
A comparison of the identified VSP sequences with corresponding regions from other VSP sequences available in GenBank also demonstrated this heterogeneous distribution (Fig. 4). This comparison included all representative VSP sequences which were available from GenBank and which had previously been used for the definition of VSP-based phylogenetic assemblage groups A and B (14). From the other assemblages (C and D) previously characterized (13), no comparable VSP sequences could be found in the database. In the comparative analysis, the majority of GS/M-83-H7 subvariant VSP sequences were clustered in the same group (assemblage B) as VSP H7 (Fig. 4). However, four VSP sequences identified in the present study were as distinct from the majority of the assemblage B-derived sequences as were vsp sequences derived from assemblage A isolates, e.g., in vitro-derived cDNA clone IVg and in vivo-derived clones 14f, 21e, and 42d.
FIG. 4.
Phylogenetic relationship inferred from the C-terminal amino acid sequences of VSP H7, the subvariant VSPs identified in this study from cDNA, and corresponding VSP sequences available from GenBank. Segments corresponding to residues 29 to 159 (the C-terminal segment) of VSP H7 (Fig. 3) were aligned using ClustalX, version 1.8, and the resulting distance data were viewed using TreeView, version 1.6.1. Accession numbers of VSP sequences obtained for GS/M-83-H7-derived G. lamblia populations are given in Fig. 3. Those of the heterologous VSPs are as follows: AAD28789 (VSP417-4a/A-II), AAF31772 (VSP417-7/A-II), AAD04339 (VSP417-4b/A-II), AAF04387 (VSP417-7/A-I), AAG16629 (VSP9B10/A-I), A48579 (TSP11/A-I), AAD03497 (VSP417-3/A-II), AAD28790 (VSP417-2/A-II), AAD05040 (TSA417/A-II), A35502 (VSP9B10-LIKE), and AAF02907 (VSP417-6/A-I). Bar, index of dissimilarity (0.1 units) among the different sequences. Ellipses enclose sequences derived from the GS/M-83-H7 isolate (assemblage B) or from isolates belonging to genetic assemblage A and a third group of sequences apparently lying outside these assemblages.
DISCUSSION
In the past, multiple studies addressed the ability of G. lamblia to alter its VSP surface coat. Experimental infections in rodents as well as in vitro studies revealed that exposure of G. lamblia trophozoites to VSP-specific antibodies can result in a change of the variant-type composition (reviewed in references 16 and 18). These studies were mostly based on biochemical or immunobiochemical methods and did not evaluate in detail molecular aspects of the antigenic diversification process. In the present study, we have for the first time extensively explored at the transcriptional level the nature of in vitro and in vivo antigenic variation of G. lamblia using well-characterized clone GS/M-83-H7 as the model parasite.
The most important goal of our study was to generate data which characterized vsp gene expression during an experimental G. lamblia GS/M-83-H7 infection in mice. Our previous investigations performed with the mother-offspring mouse model had shown that antigenic variation of the parasite is initiated by selection of variant trophozoites mediated through a direct parasiticidal effect of lactogenic immunoglobulin A against the parental VSP H7-type trophozoites (32). Furthermore, a strong fluctuation of the parasite burden during the initial stages of antigen switching had indicated that the original VSP H7 trophozoites were eliminated and replaced by a second variant-type population. The variant-type formation during this process of antigenic variation was investigated in various studies (reviewed in references 16 and 18). However, detailed biochemical and/or immunobiochemical analysis monitoring surface antigen alterations of G. lamblia variants emerging within clone GS/M-83-H7 mostly relied on procedures which included a step of in vitro cultivation of the parasite (2, 7, 24). Since cultivation of subvariant types from G. lamblia clone GS/M-83-H7 had been shown to gradually change the variant-type pattern within the parasite population (24, 30), results from experimental infections involving a subsequent in vitro proliferation step may distort the real in vivo situation regarding variant-type diversification.
In the present study, we chose a PCR approach (Fig. 1) which avoided the possibility of those experimental constraints listed above. Our aim was to isolate different vsp cDNA molecules that were derived from G. lamblia clone GS/M-83-H7 before and after antigenic diversification. The PCR strategy relied on the use of primers which targeted conserved regions in vsp genes and thus might be expected to simultaneously amplify most of the sequences from the vsp gene repertoire. The pair of primers finally used was deduced from the highly conserved 3′-terminal region of vsp H7, which encodes the C-terminal transmembrane domain (primer vsp-rev1; Fig. 1B), and an upstream region of vsp H7 (target of primer vsp-forward; Fig. 1B), which in a sequence comparison study using data from GenBank (M. Bienz, unpublished data) had been assessed to be relatively similar to those of vsp genes from heterologous G. lamblia isolates. However, in applying this experimental strategy, we had to take into account that selective processes during both the first-strand cDNA synthesis and the different cDNA amplification steps (e.g., caused by inefficient priming, or lack of priming, of vsp-rev1 and/or degenerate vsp-forward to a subset of vsp genes) provided results which did not quantitatively reflect the vsp gene expression within the different parasite populations. Accordingly, and also because of statistical limitations due to only having sequenced 12 cloned cDNAs per experimental group, our investigation represents a semiquantitative approach, which could not assess the entire complexity related to antigenic variation of G. lamblia clone GS/M-83-H7.
By applying this PCR-based strategy, we were able to demonstrate that (i) GS/M-83-H7 trophozoites, prior to in vitro and in vivo antigen switching, preferentially express vsp H7 and predominantly synthesize VSP H7 (as shown by the immunofluorescence assay), (ii) in vitro and in vivo antigen switching of the parasite is associated with a strong diversification in vsp gene expression, (iii) in vivo antigenic variation involves at least 22 vsp genes, and (iv) in vivo antigenic variation represents a continuous process which generates consecutively emerging and rather distinct variants that exhibit, at the population level, a highly heterogeneous vsp gene expression.
In the present study, comparative sequence analysis demonstrated high similarities between VSP H7 and the different subvariant VSPs identified and related most of these sequences to the same phylogenetic assemblage (assemblage B). This close relationship of most GS/M-83-H7-derived VSP sequences further supported a previous hypothesis suggesting that the formation of the different Giardia vsp gene repertoires is the consequence of ancestral combinatorial processes that included serial gene duplication, mutation and/or recombination events within the individual isolates (4, 35, 36). However, since four vsp cDNA clones identified turned out to be rather distinct from the assemblage B-type vsp gene sequences, this ancestral gene duplication model may be insufficient to explain the entire complexity of the evolutionary vsp gene diversification within G. lamblia.
ACKNOWLEDGMENTS
We acknowledge A. Hemphill for carefully reading the manuscript and T. E. Nash (NIH, Bethesda, Md.) for his gift of MAb G10/4 and G. lamblia clone GS/M-83-H7.
This work was supported by grants obtained from the Swiss National Science Foundation (no. 31-49439.96 and 31-58973.99).
REFERENCES
- 1.Adam R D, Aggarwal A, Lal A A, De La Cruz V F, McCutchan T, Nash T E. Antigenic variation of a cysteine-rich protein in Giardia lamblia. J Exp Med. 1988;167:109–118. doi: 10.1084/jem.167.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal A, Merritt J W, Nash T E. Cysteine-rich variant surface proteins of Giardia lamblia. Mol Biochem Parasitol. 1989;32:39–48. doi: 10.1016/0166-6851(89)90127-8. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal A, Nash T E. Antigenic variation of Giardia lambliain vivo. Infect Immun. 1988;56:1420–1423. doi: 10.1128/iai.56.6.1420-1423.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ey P L, Darby J M, Mayrhofer G. A new locus (vsp417-4) belonging to the tsa417-like subfamily of variant-specific surface protein genes in Giardia intestinalis. Mol Biol Biochem. 1999;99:55–68. doi: 10.1016/s0166-6851(98)00183-2. [DOI] [PubMed] [Google Scholar]
- 5.Faubert G. Immune response to Giardia duodenalis. Clin Microbiol Rev. 2000;13:35–54. doi: 10.1128/cmr.13.1.35-54.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottstein B, Deplazes P, Tanner I. In-vitro synthesized immunoglobulin A from nu/+ and reconstituted nu/nu mice against a dominant surface antigen of Giardia lamblia. Parasitol Res. 1993;79:644–648. doi: 10.1007/BF00932506. [DOI] [PubMed] [Google Scholar]
- 7.Gottstein B, Harriman G R, Conrad J T, Nash T E. Antigenic variation in Giardia lamblia: cellular and humoral immune response in a mouse model. Parasite Immunol. 1990;12:659–673. doi: 10.1111/j.1365-3024.1990.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 8.Gottstein B, Nash T E. Antigenic variation in Giardia lamblia: infection of congenitally athymic nude and scidmice. Parasite Immunol. 1991;13:649–659. doi: 10.1111/j.1365-3024.1991.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 9.Hemphill A, Stäger S, Gottstein B, Müller N. Electron microscopical investigation of surface alterations on Giardia lambliatrophozoites after exposure to a cytotoxic monoclonal antibody. Parasitol Res. 1996;82:206–210. doi: 10.1007/s004360050096. [DOI] [PubMed] [Google Scholar]
- 10.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill D R, Burge J J, Pearson R D. Susceptibility of Giardia lambliatrophozoites to the lethal effect of human serum. J Immunol. 1984;132:2046–2052. [PubMed] [Google Scholar]
- 12.Keister D B. Axenic culture of Giardia lambliain TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- 13.Meng T C, Hetsko M L, Gillin F D. Antigenic switching of TSA 417, a trophozoite variable surface protein, following completion of the life cycle of Giardia lamblia. Infect Immun. 1993;61:5394–5397. doi: 10.1128/iai.61.12.5394-5397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monis P T, Andrews R H, Mayrhofer G, Mackrill J, Kulda J, Isaac-Renton J L, Ey P L. Novel lineages of Giardia intestinalisidentified by genetic analysis of organisms isolated from dogs in Australia. Parasitology. 1998;116:7–19. doi: 10.1017/s0031182097002011. [DOI] [PubMed] [Google Scholar]
- 15.Mowatt M R, Aggarwal A, Nash T E. Carboxy-terminal sequence conservation among variant-specific surface proteins of Giardia lamblia. Mol Biochem Parasitol. 1991;49:215–228. doi: 10.1016/0166-6851(91)90065-e. [DOI] [PubMed] [Google Scholar]
- 16.Müller N, Gottstein B. Antigenic variation and the murine immune response to Giardia lamblia. Int J Parasitol. 1998;28:1829–1839. doi: 10.1016/s0020-7519(98)00137-4. [DOI] [PubMed] [Google Scholar]
- 17.Müller N, Stäger S. Periodic appearance of a predominant variant antigen type during a chronic Giardia lambliainfection in a mouse model. Int J Parasitol. 1999;29:1917–1923. doi: 10.1016/s0020-7519(99)00147-2. [DOI] [PubMed] [Google Scholar]
- 18.Nash T E. Antigenic variation in Giardia lambliaand the host's immune response. Philos Trans R Soc Lond B. 1997;352:1369–1375. doi: 10.1098/rstb.1997.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash T E. Surface antigen variability and variation in Giardia lamblia. Parasitol Today. 1992;8:229–234. doi: 10.1016/0169-4758(92)90119-m. [DOI] [PubMed] [Google Scholar]
- 20.Nash T E, Aggarwal A. Cytotoxicity of monoclonal antibodies to a subset of Giardiaisolates. J Immunol. 1986;136:2628–2632. [PubMed] [Google Scholar]
- 21.Nash T E, Aggarwal A, Adam R D, Conrad J T, Merritt J W. Antigenic variation in Giardia lamblia. J Immunol. 1988;141:636–641. [PubMed] [Google Scholar]
- 22.Nash T E, Conrad J T, Mowatt M R. Giardia lamblia: identification and characterization of a variant-specific surface protein gene family. J Eukaryot Microbiol. 1995;42:604–609. doi: 10.1111/j.1550-7408.1995.tb05914.x. [DOI] [PubMed] [Google Scholar]
- 23.Nash T E, Gillin F D, Smith P D. Excretory-secretory products of Giardia lamblia. J Immunol. 1983;131:2004–2010. [PubMed] [Google Scholar]
- 24.Nash T E, Herrington D A, Levine M M, Conrad J T, Merritt J W., Jr Antigenic variation of Giardia lambliain experimental human infections. J Immunol. 1990;144:4362–4369. [PubMed] [Google Scholar]
- 25.Nash T E, Keister D B. Differences in excretory-secretory products and surface antigens among 19 isolates of Giardia. J Infect Dis. 1985;152:1166–1171. doi: 10.1093/infdis/152.6.1166. [DOI] [PubMed] [Google Scholar]
- 26.Nash T E, Mowatt M R. Characterization of a Giardia lambliavariant-specific surface protein (VSP) gene from isolate GS/M and estimation of the VSP gene repertoire size. Mol Biochem Parasitol. 1992;51:219–228. doi: 10.1016/0166-6851(92)90072-r. [DOI] [PubMed] [Google Scholar]
- 27.Pimenta P F P, da Silva P P, Nash T E. Variant surface antigens of Giardia lambliaare associated with the presence of a thick cell coat: thin section and label fracture immunocytochemistry survey. Infect Immun. 1991;59:3989–3996. doi: 10.1128/iai.59.11.3989-3996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risler J L, Delorme M D, Delacroix H, Henant A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J Mol Biol. 1988;204:1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Singer S M, Elmendorf H G, Conrad J T, Nash T E. Biological selection of variant-specific surface proteins in Giardia lamblia. J Infect Dis. 2001;183:119–124. doi: 10.1086/317659. [DOI] [PubMed] [Google Scholar]
- 31.Stäger S, Felleisen R, Gottstein B, Müller N. Giardia lambliavariant surface protein H7 stimulates a heterogeneous repertoire of antibodies displaying differential cytological effects on the parasite. Mol Biochem Parasitol. 1997;85:113–124. doi: 10.1016/s0166-6851(96)02818-6. [DOI] [PubMed] [Google Scholar]
- 32.Stäger S, Gottstein B, Sager H, Jungi T W, Müller N. Influence of antibodies in mother's milk on antigenic variation of Giardia lambliain the murine mother-offspring model of infection. Infect Immun. 1998;66:1287–1292. doi: 10.1128/iai.66.4.1287-1292.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svärd S G, Meng T C, Hetsko M L, McCaffery J M, Gillin F D. Differentiation-associated surface antigen variation in the ancient eukaryote Giardia lamblia. Mol Microbiol. 1998;39:979–989. doi: 10.1046/j.1365-2958.1998.01125.x. [DOI] [PubMed] [Google Scholar]
- 34.Ungar B L P, Nash T E. Cross-reactivity among different Giardia lambliaisolates using immunofluorescent antibody and enzyme immunoassay techniques. Am J Trop Med Hyg. 1987;37:283–289. doi: 10.4269/ajtmh.1987.37.283. [DOI] [PubMed] [Google Scholar]
- 35.Yang J M, Adam R D. A group of Giardia lambliavariant-specific surface protein (VSP) genes with near identical 5′ regions. Mol Biochem Parasitol. 1995;75:69–74. doi: 10.1016/0166-6851(95)02514-6. [DOI] [PubMed] [Google Scholar]
- 36.Yang J M, Adam R D. Analysis of a repeat-containing family of Giardia lambliavariant-specific surface protein (VSP) genes: diversity through gene duplication and divergence. J Eukaryot Microbiol. 1995;42:439–444. doi: 10.1111/j.1550-7408.1995.tb05888.x. [DOI] [PubMed] [Google Scholar]