Abstract
Borrelia burgdorferi, the causative agent of Lyme disease, produces RevA protein during the early stages of mammalian infection. B. burgdorferi apparently uses temperature as a cue to its location, producing proteins required for infection of warm-blooded animals at temperatures corresponding to host body temperature, but does not produce such virulence factors at cooler, ambient temperatures. We have observed that B. burgdorferi regulates expression of RevA in response to temperature, with the protein being synthesized by bacteria cultivated at 34°C but not by those grown at 23°C. Tissues encountered by B. burgdorferi during its infectious cycle vary in their pH values, and the level of RevA expression was also found to be dependent upon pH of the culture medium. The cellular localization of RevA was also analyzed. Borrelial inner and outer membranes were purified by isopycnic centrifugation, and membrane fractions were conclusively identified by immunoblot analysis using antibodies raised against the integral inner membrane protein MotB and outer membrane-associated Erp lipoproteins. Immunoblot analyses indicated that RevA is located in the B. burgdorferi outer membrane. These analyses also demonstrated that an earlier report (H. A. Bledsoe et al., Infect. Immun. 176:7447–7455, 1994) had misidentified such B. burgdorferi membrane fractions. RevA was further demonstrated to be exposed to the external environment, where it could facilitate interactions with host tissues.
The spirochete Borrelia burgdorferi has evolved efficient mechanisms by which it can persistently infect both warm-blooded and arthropod hosts and be efficiently transmitted between these two host types (49). Such a complex lifestyle requires that the bacteria produce proteins appropriate for each stage of the infectious cycle. These might include surface proteins that facilitate interactions with host cells or extracellular components, function in nutrient acquisition, or help protect the bacteria against host immune system responses. Throughout its infectious cycle, B. burgdorferi apparently senses its location in order to produce proteins and other factors required for each stage of the cycle. We and others have demonstrated that B. burgdorferi recognizes various environmental cues, including temperature, pH, and soluble chemicals and, as a consequence, regulates surface protein expression (1–3, 12, 13, 30, 40, 45, 46, 50, 51, 54, 55, 62).
Among the B. burgdorferi proteins known to be synthesized during mammalian infection is an approximately 17-kDa protein previously designated Rev (25, 43, 53) and herein renamed RevA (see below for the rationale behind this name modification). All analyzed Lyme disease spirochetes contain numerous different, but largely homologous, plasmids of the cp32 family (15, 16, 56). RevA proteins are encoded by some, but not all, cp32s: B. burgdorferi type strain B31 contains two revA alleles, one each on cp32-1 and cp32-6 (15). To date, only three additional strains of B. burgdorferi have been examined for this gene, but all were found to contain at least one revA gene (25, 43, 63). A recent report indicated that serum samples from many human Lyme disease patients contained antibodies that recognized a recombinant RevA protein (25). Additionally, these same researchers produced hybridomas from the spleens of mice infected with B. burgdorferi strain B31 via tick bite, some of which produced antibodies directed against the RevA protein (25). However, reverse transcriptase-PCR analyses of gene expression by B. burgdorferi during tick infection indicated that expression of revA ceased shortly after ticks became infected and then increased again during feeding of those infected ticks on a mammalian host (26).
These earlier studies indicate that B. burgdorferi expresses RevA during mammalian infection and regulates synthesis of this protein during the bacterium's natural transmission cycle. We therefore sought to determine signals responsible for the controlled expression of RevA. Since the cellular localization of a protein can be suggestive of its function, this aspect of the RevA protein was also addressed in our studies.
MATERIALS AND METHODS
Bacteria.
B. burgdorferi B31 is a wild-type strain, isolated from a tick collected on Shelter Island, N.Y. (11). Bacteria used in these studies are infectious to mice (14, 16). B. burgdorferi were grown in either Barbour-Stoenner-Kelly II (BSK-II) medium (4) prepared in our laboratories or a commercially prepared modification of that medium (BSK-H; Sigma, St. Louis, Mo.). All media contained 6% (vol/vol) rabbit serum (Sigma). Bacteria were cultivated at 34°C, unless otherwise noted.
Cloning, overexpression, and purification of N-terminally truncated MotB.
To aid in the identification of B. burgdorferi membrane fractions, a recombinant form of an integral inner membrane protein was synthesized for use in producing antibodies. The motB gene encodes part of the flagellar motor, which is embedded in the bacterial inner membrane (32, 33). The B. burgdorferi motB gene, lacking the first 105 nucleotides, was PCR amplified from strain B31 DNA and cloned into pET30 LIC (Invitrogen, Carlsbad, Calif.). The resultant construct expressed a polyhistidine-tagged MotB that was missing the N terminal signal sequence and transmembrane region (tMotB). Recombinant tMotB could not be purified by nickel column due to its aggregation in inclusion bodies. Therefore, inclusion bodies were purified from cell lysates as follows. Cell lysates were cleared by centrifugation (22,000 × g, 1 h, 4°C). The inclusion body pellet was washed in 2% (vol/vol) Triton X-100 and 2 M urea in phosphate-buffered saline (PBS; pH 8.0) and then centrifuged (22,000 × g, 30 min, 4°C). This process was repeated seven times. The inclusion body pellet was then washed once in PBS and centrifuged (22,000 × g, 30 min, 4°C).
Antibodies.
Gilmore and Mbow earlier reported the production of a panel of hybridomas derived from mice infected with B. burgdorferi B31 via tick bite (25). One of those hybridomas, designated YM.17 produced monoclonal antibodies (MAbs) that specifically recognized the strain B31 RevA protein (25). A second hybridoma, YM.26, was subsequently found to also recognize a recombinant RevA protein (R. Gilmore, Jr., unpublished results). Both YM.17 and YM.26, were provided by Lamine Mbow (Colorado State University, Ft. Collins, Colo.) and Robert Gilmore, Jr. (Centers for Disease Control and Prevention, Ft. Collins, Colo.), as were the hybridomas B5, which is directed against the strain B31 OspC protein (35), and B11, which is directed against the strain B31 ErpA/I/N protein (22). MAb H9724, which is specific for the FlaB proteins of Borrelia species (5), and MAb H5332, which recognizes the B. burgdorferi strain B31 OspA protein (6), were provided by Tom Schwan (Rocky Mountain Laboratories, National Institutes of Health [NIH], Hamilton, Mont.). Hybridomas were maintained at 37°C with a 5% CO2 environment, in RPMI supplemented with 10% (vol/vol) heat-inactivated calf serum, 10 mM HEPES, 1 mM sodium pyruvate, 2 mM l-glutamine, 1% (vol/vol) nonessential amino acid solution, 0.2% (wt/vol) sodium bicarbonate, and 0.09% (vol/vol) β-mercaptoethanol (all from Gibco, Gaithersburg, Md.).
Antibodies raised against recombinant tMotB were used to identify the inner membrane fraction of purified B. burgdorferi membranes. Antiserum was produced by immunizing a New Zealand White rabbit with approximately 500 μg of gel-purified tMotB in complete Freund adjuvant. The rabbit was boosted 3 weeks postimmunization with 500 μg of purified tMotB in incomplete Freund adjuvant. The rabbit was exsanguinated via cardiac puncture and blood was processed to serum.
RNA analyses.
Total RNA was extracted from B. burgdorferi B31 cultures incubated at various pHs (pH 7.0 and 8.0) (12) or temperatures (23 and 33°C) (55) using the Ultraspec-II RNA isolation system (Biotecx, Houston, Tex.) (9). RNA was denatured with glyoxal and dimethyl sulfoxide for 1 h at 50°C, and 10 μg per lane of total RNA was resolved by a 1.0% (wt/vol) agarose gel in 10 mM NaH2PO4, pH 7.0 (80 V 3 h). Separated RNA was transferred to Hybond N+ nylon membrane using a vacuum blotter system (60 mbar, 1 h, 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]), air dried, auto-cross-linked, and stained with methylene blue (0.03% methylene blue in 1.0% acetic acid). Millennium RNA markers (Ambion, Inc., Austin, Tex.) served as standards. RNA blots were stored dry in the dark at 24°C until probed.
To produce a revA-specific probe, a plasmid clone containing revA1 (25) (obtained from R. Gilmore, Jr.), was digested with XbaI and XhoI. The 1.5-kb fragment containing revA was purified by agarose gel electrophoresis and extracted from the gel using a GenElute agarose spin column (Sigma). The purified revA fragment served as the template for radioactive labeling a Northern blot probe, using a RadPrime labeling kit (Life Technologies, Grand Island, N.Y.) and [α-32P]dATP (3,000 Ci/mmol) (NEN Life Science Products, Inc., Boston, Mass.). RNA blots were hybridized and washed as previously described (12). Hybridizing mRNAs were detected by autoradiography of X-ray film. Signal intensities and integrated density values were measured with an AlphaImager 2000 digital imaging system (Alpha Innotech Corp., San Leandro, Calif.). All Northern blot studies were performed independently at least twice.
Protein electrophoresis and immunoblot analyses.
For one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), cultured B. burgdorferi were harvested by centrifugation and, unless noted otherwise, washed with PBS and lysed by resuspension in distilled water and incubation in a boiling water bath for 5 min. Equivalent amounts of total protein were separated by SDS-PAGE and transferred to nitrocellulose membranes as described previously (37). Membranes were blocked by incubation for <1 h in 5% (wt/vol) nonfat dried milk in Tris-buffered saline–Tween 20 (TBS-T) (37), rinsed with TBS-T, and incubated for 1 h at room temperature in appropriate primary antibody. Membranes were then washed with TBS-T, incubated for 1 h with conjugated protein A-horseradish peroxidase (Amersham, Piscataway, N.J.) in TBS-T, according to the manufacturer's instructions. Bound primary antibodies were detected by enhanced chemiluminescence (Amersham) and XAR-5 film (Kodak, Rochester, N.Y.).
Two-dimensional nonequilibrium pH gradient gel electrophoresis (2D-NEPHGE) was performed as described by O'Farrell (39) with modifications described by Carroll et al. (13). Briefly, 80 mg of solubilized membrane proteins were separated in the first dimension on a 1-mm analytical tube gel for 3000 V · h (500 V, 6 h, 14°C) using a vertical 1-D Running System (Genomic Solutions, Ann Arbor, Mich.). Membrane proteins were then separated in the second dimension by SDS–12% PAGE and were either stained with silver (Silver Stain Plus; Bio-Rad) or transferred to nitrocellulose for immunoblot analysis.
Effects of temperature and pH on protein levels.
For temperature effect studies, bacteria were grown to mid-exponential phase (approximately 107 bacteria per ml) at 23°C, diluted 1:100 into fresh medium, and grown to mid-exponential phase at 34°C (55). For pH effect studies, bacteria were grown to mid-exponential phase at 34°C in BSK-II or BSK-H medium supplemented with 25 mM HEPES and buffered to a pH of either 8.0 or 7.0 (13). The pH values of the media were measured following cell harvesting, and no detectable changes were observed.
B. burgdorferi membrane purification and analysis.
Cell lysates were separated into total membrane and soluble fractions by ultracentrifugation (14). Inner and outer membranes of B. burgdorferi B31 were separated by isopycnic centrifugation according to the method of Bledsoe et al. (8). Evidence presented in the Results section below indicate that the original designations of inner and outer membrane by Bledsoe et al. were actually reversed. To avoid confusion, we refer to our membrane fractions as “true” inner and outer membranes.
In situ protease analyses.
B. burgdorferi were grown to mid-exponential phase in BSK-H, pelleted by centrifugation, washed once with PBS, and resuspended in PBS to a final concentration of approximately 2 × 109 bacteria/ml. Examination of bacterial suspensions by phase-contrast light microscopy did not indicate detectable lysis of the bacteria. Bacteria were then incubated at room temperature in PBS containing a protease for 30 min, 1 h, or 2 h, whereupon digestion was terminated by addition of an appropriate inhibitor followed by sample boiling. One of three different proteases was used in each experiment at the following final concentrations: 40 μg of proteinase K (Sigma), 40 μg of trypsin (Sigma), or 0.05 μg of pronase (Boehringer-Mannheim, Indianapolis, Ind.) per ml. Proteinase K was inhibited by addition of paramethylsulfonyl fluoride (PMSF) to a final concentration of 1.6 mg/ml. Trypsin was inhibited by the addition of PMSF and pefabloc SC (Boehringer-Mannheim) to final concentrations of 1.6 and 0.3 mg/ml, respectively. Pronase was inhibited by addition of PMSF, pefabloc SC, and EDTA to final concentrations of 0.06, 0.3, and 0.5 mg/ml, respectively. Control aliquots of bacteria were incubated in buffer for 2 h at room temperature without added protease, followed by the addition of inhibitor and boiling as with the protease-treated bacteria. Equal volumes of each bacterial lysate were subjected to SDS-PAGE and transferred to nitrocellulose membranes, and the susceptibility of RevA to protease digestion was assessed by immunoblot analysis with MAb YM.26. As experimental controls, lysates were also immunoblotted with MAbs directed against OspC (located on the bacterial outer surface and thus susceptible to proteolysis [22, 24]) and FlaB (located in the periplasmic space and thus protected against protease digestion in intact bacteria [29]).
RESULTS
Regulation of RevA synthesis.
Three genes of strain B31 have been referred to as “rev”: two on cp32 plasmids that encode 17.9-kDa proteins and are very similar to the original rev gene of strain 297 identified by Porcella et al. (43), and a third gene encoding a 20.1-kDa protein that is located on plasmid cp9-1 (15, 23, 36). Due to the differences described below, it is apparent that the cp32 genes and the cp9-1 gene are not allelic. Thus, we have designated the cp32 genes revA and the cp9-1 gene revB. The two revA genes of strain B31 are given allele designations based upon the plasmid on which each gene is located: allele revA1 is located on cp32-1, and cp32-6 contains revA6 (15). The predicted sequences of the mature RevA1 and RevA6 proteins are identical (Fig. 1A) and so are referred to as simply “RevA” throughout this report. The revB gene has 47.5% nucleic acid identity with the revA genes and encodes a protein that shares just 28% amino acid sequence identity with the two cp32-encoded proteins (Fig. 1A). Additionally, the cp9-1 gene promoter region is very different from that of the cp32 genes (data not shown), suggesting that different mechanisms might control transcription for each locus type.
FIG. 1.
(A) Alignments of the predicted protein sequences of the two RevA alleles and one RevB gene of strain B31. Note that RevA1 and RevA2 differ only at the 13th residue of each protein, while RevB shares only 28% identical amino acids with the RevA proteins. Each protein is predicted to be lipidated at the cysteine residue marked by an asterisk (28, 57, 60). (B) Alignment of the promoter regions of the strain B31 revA and ospC genes. Identical nucleotides found in both a revA and the ospC promoter are boxed and shaded. The ospC promoter contains an inverted repeat, indicated by opposing arrows above the DNA sequences (3, 34, 59). All other analyzed ospC promoters contain identical sequences at this location (59). A similar sequence is found 5′ of all known alleles of revA.
Both revA genes are flanked on the 5′ end by a divergently transcribed gene of the mlp multigene family (mlpA and mlpF upstream of revA1 and revA6, respectively) (15, 42). These two intergenic regions, which presumably contain the promoters of both the revA and mlp genes, contain nearly identical nucleotide sequences (Fig. 1B), making it likely that both revA1 and revA6 are under similar transcriptional controls.
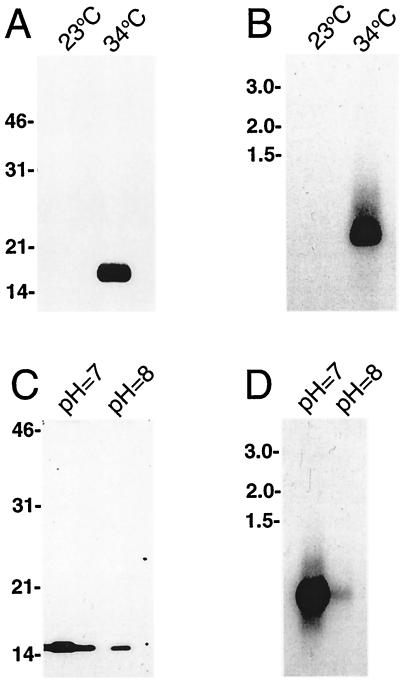
We and others have previously observed that some spirochete proteins involved in mammalian infection are regulated by temperature, with greater amounts of protein synthesized by bacteria cultivated at temperatures similar to the mammalian body temperature than by bacteria grown at ambient temperature (17, 31, 38, 46, 51, 54, 55, 62). For these reasons, we examined the levels of RevA protein synthesized by bacteria cultivated at either 23 or 34°C. Immunoblot analysis indicated that RevA was made by bacteria in the 34°C culture, while the protein was undetectable in the 23°C cultivated spirochetes (Fig. 2A). Northern blot analysis of RNA purified from bacteria that had undergone this culture temperature shift indicated that revA mRNA was present in the bacteria grown at 34°C but was undetectable in those maintained at 23°C (Fig. 2B).
FIG. 2.
Regulated expression of RevA. (A) Immunoblot of B. burgdorferi cultured at either a constant 23°C or shifted from 23 to 34°C and probed with anti-RevA MAb YM.26. (B) Northern blot of such bacteria using a revA-specific probe. (C) RevA immunoblot of bacteria grown in medium buffered to remain at either pH 7 or 8. (D) Northern blot of bacteria grown at pH 7 or 8. To the left of each panel are indicated the locations of either protein molecular mass markers (panels A and C) or RNA standards (panels B and D).
Since the production of some B. burgdorferi proteins preferentially synthesized during infection of warm-blooded animals are also regulated by pH (12, 13, 46, 62), we examined the effect of culture medium pH on RevA expression. Significantly higher levels of RevA were produced by bacteria cultivated in medium buffered to remain at pH 7.0 than by those grown in medium having a pH of 8.0 (Fig. 2C). Again, revA mRNA levels corresponded with protein levels (Fig. 2D).
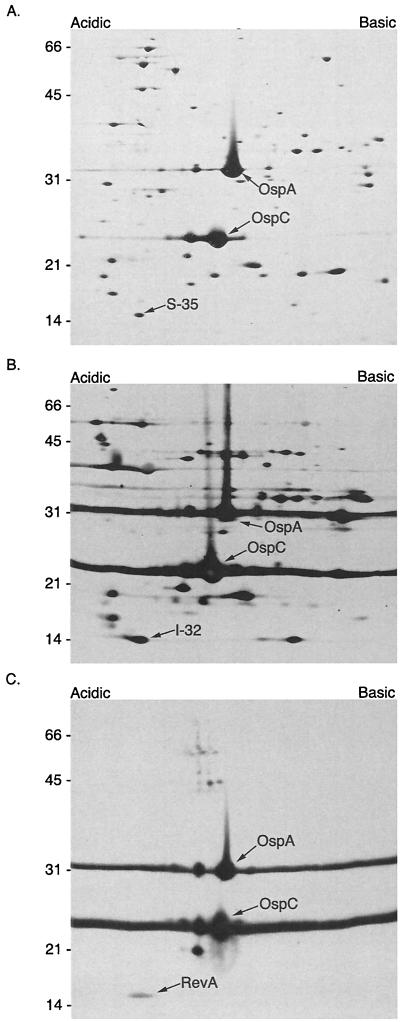
One of us (J. Carroll) had previously observed that expression of at least 37 B. burgdorferi proteins was affected by pH of the culture medium, which was most clearly seen by two-dimensional PAGE (12, 13). To determine whether RevA was among those proteins, B. burgdorferi membrane proteins were separated as in those earlier studies and then immunoblotted with RevA-specific MAb YM.26 (Fig. 3). The result was then compared with a silver-stained two-dimensional gel, and an immunoblot was performed using B. burgdorferi-infected animal serum. These comparisons revealed that the RevA spot aligned with the previously identified spots I-32 and S-35 (13) (Fig. 3).
FIG. 3.
2D-NEPHGE of membrane protein preparations from B. burgdorferi B31 were stained with silver (A), transferred and probed with hyperimmune serum (B), or transferred and probed with monoclonal antibodies to RevA, OspC, and OspA (C). The acidic protein spots S-35 and I-32 identified by Carroll et al. (13) were determined to be RevA by immunoblot. OspA and OspC are indicated for orientation. The protein spot just below OspC has been determined by MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) analysis to be an OspC breakdown product (spot S-25/I-25 from Carroll et al. [13]) (J. Carroll, unpublished results) Molecular mass standards in kilodaltons are indicated on the left of each panel.
Several other B. burgdorferi genes, including the gene encoding the infection-associated outer surface protein OspC, are known to be regulated in response to temperature in manners similar to that of revA (1, 3, 12, 13, 17, 46, 51, 54, 55, 62). These similarities led us to compare the 5′ noncoding regions adjacent to the revA genes with those of other known regulated loci. Significant similarity was found between the ospC and revA promoter regions, including an inverted repeat that may indicate a protein-binding site (Fig. 1B). No homology was evident when comparing revA promoters with those of other regulated genes, including the erp and dbpAB genes (data not shown).
RevA is an outer membrane protein.
The predicted amino acid sequences of all known RevA proteins contain a charged amino terminus followed by a hydrophobic region and a type II secretion-lipidation consensus sequence (Fig. 1A), suggesting that they are membrane-bound lipoproteins (25, 28, 43, 57, 60). Studies were thus undertaken to determine the cellular localization of RevA.
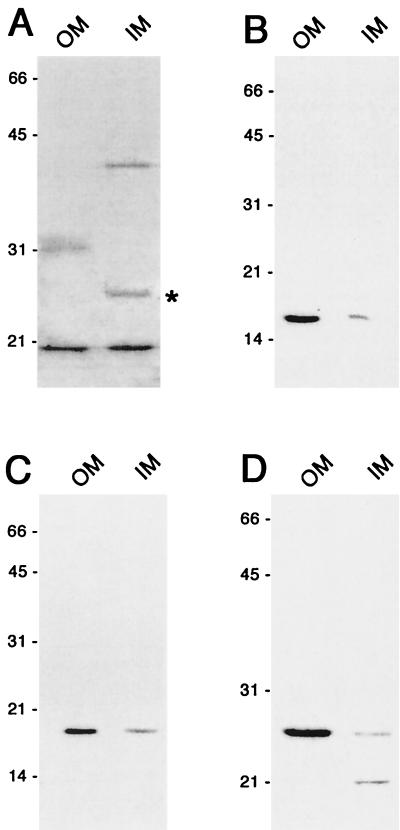
B. burgdorferi membranes were separated by isopycnic centrifugation according to a previously described procedure (8). B. burgdorferi does not contain lipopolysaccharide as do enteric gram-negative bacteria (58), raising concern that borrelial inner and outer membranes may have relative buoyancies different from those of previously characterized bacteria. For this reason, antibodies were generated against the B. burgdorferi MotB protein, a subunit of the flagellar motor and, therefore, an integral inner membrane protein. Equivalent amounts of total protein from membrane vesicle preparations were subjected to immunoblot analysis, identifying an approximately 24-kDa protein, which is the anticipated size of MotB (Fig. 4A). Immunoblot analysis of a B. burgdorferi mutant disrupted in motA, which is directly upstream of motB, makes less of the 24-kDa band, consistent with the 24-kDa band being MotB (N. W. Charon and M. A. Motaleb, unpublished data). Densitometric analysis indicated that 86% of MotB was contained in one of the fractions, indicating that this fraction consisted primarily of inner membrane proteins. Similar analyses with antibodies directed against the known outer membrane proteins ErpA/I/N and ErpL (22) indicated that these two proteins were contained in the second membrane fraction (Fig. 4C and D), indicating that this fraction consisted primarily of outer membrane proteins. An earlier study (8) used the same membrane purification technique as in the present study, yet lacked an appropriate marker to differentiate inner membranes from outer membranes and, as a result, misidentified the membrane fractions. To avoid confusion with the older terminology, we refer to the fractions as “true” inner and outer membranes according to the localization of MotB.
FIG. 4.
RevA is an outer membrane protein. Immunoblots of purified B. burgdorferi “true” outer and inner membranes (OM and IM, respectively, about each panel). Analyses were done with various antibodies. (A) polyclonal rabbit antiserum raised against B. burgdorferi MotB. The immunoblot band corresponding with MotB is marked by an asterisk, while the identities of the additional proteins are as yet unknown. Analysis of a mutant with a disrupted motAB operon indicated that the 24-kDa band corresponds with MotB (see the text). (B) MAb YM.26 directed against RevA. (C) MAb B11 directed against known outer membrane protein ErpA/I/N (22). (D) Polyclonal rabbit antiserum raised against the known outer membrane protein ErpL (22).
Purified membranes contained RevA, consistent with predictions that it is a membrane-bound lipoprotein. We determined by immunoblot and densitometry that greater than 85% of RevA localized to the true outer membrane fraction (Fig. 4B). We conclude from these studies that RevA is an outer membrane protein.
Localization of RevA on the B. burgdorferi outer surface.
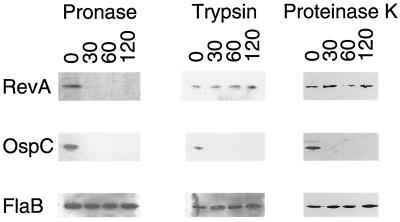
The cellular localization of RevA was addressed by examining the susceptibility of the in situ protein to proteolysis. Surface-exposed proteins of intact bacteria can be digested by proteases, while those below the surface cannot be degraded. Cultured bacteria were incubated with a protease, and the proteins were separated by SDS-PAGE and immunoblotted with MAb YM.26. Since a number of borrelial surface-exposed outer membrane proteins are resistant to proteolysis by some enzymes (10, 20, 22, 64), three different proteases were used in this study. Incubation of bacteria with pronase completely digested RevA within 30 min, a result indicative of surface exposure (Fig. 5). Control immunoblot analyses indicated that the bacterial outer membranes were intact, since there was no detectable proteolysis of FlaB, a component of the periplasmic flagella (29). However, no degradation of RevA was detected following 2 h of incubation with either proteinase K or trypsin (Fig. 5). Proteolysis of the known outer surface protein OspC (22, 24) was detected in these bacteria, indicating that both enzymes were functionally active and that RevA in situ is insensitive to these proteases. These studies demonstrated that RevA is exposed to the external environment on the B. burgdorferi outer membrane and that it is resistant to proteolysis by certain enzymes.
FIG. 5.
Demonstration of outer surface exposure of RevA by in situ protease degradation. Whole B. burgdorferi were incubated with a protease for 30, 60, or 120 min or for 120 min in buffer without protease. Proteases were then inactivated, bacteria were lysed, proteins were separated by SDS-PAGE, and the integrity of RevA, OspC, and FlaB were analyzed by immunoblot.
DISCUSSION
B. burgdorferi interacts with a wide variety of tissues during its natural infectious cycle between warm-blooded and arthropod hosts. It is not surprising, therefore, that these bacteria regulate the synthesis of surface proteins. Transcription of revA genes ceases shortly after the bacteria infect ticks and then resumes when the vector feeds on a new host (26). The RevA protein is synthesized during mammalian infection (25, 53), suggesting that this protein facilitates the infection of warm-blooded animals. The results presented here indicate that B. burgdorferi controls RevA expression in response to temperature, producing both the mRNA and the protein when cultivated at 34°C, while neither was detectable in bacteria grown at 23°C. These temperatures are comparable to the body temperature of a warm-blooded animal and the ambient temperature experienced within an unfed tick, respectively. Synthesis of RevA protein and mRNA was also affected by pH, with significantly greater quantities produced by bacteria grown at pH 7 as opposed to those grown at pH 8. The pH of a tick's midgut acidifies during ingestion of a blood meal (62), so a drop in pH could signal the bacteria that the vector is feeding and of the necessity to produce factors required for infection of the new host. The observation that mRNA levels paralleled protein levels suggests that RevA synthesis is controlled by the level of revA gene transcription.
The in vitro pattern of RevA synthesis was similar to that of OspC, another protein produced by B. burgdorferi during mammalian infection (13, 51, 55). Levels of both proteins appear to be regulated at the level of transcription, since the amount of each mRNA corresponded directly with protein levels (this work and reference 59). Comparison of the revA and ospC 5′ noncoding regions indicated numerous regions of extended identity. All sequenced ospC promoter regions contain a well-conserved 16-bp inverted repeat (3, 34, 59), suggestive of a DNA-binding protein recognition site. Since a similar sequence is found 5′ in both the revA1 and the revA6 genes of strain B31, it is possible that transcription of both revA and ospC is regulated through the binding of the same protein to DNA near the promoters of both locus types.
Analysis of purified B. burgdorferi inner and outer membranes revealed that RevA is located in the outer membrane. The technique utilized to separate the membranes was devised several years ago by Bledsoe et al. (8). However, at that time, membrane vesicle fractions were designated as being derived from either the inner or outer membrane based on their densities as compared with those of E. coli, the number of particles embedded in the membrane vesicles as determined by freeze fracture, and the presence or absence of immunoreactivity to antibody raised against the C subunit of the F0/F1 ATPase of Escherichia coli (8). Several lines of evidence suggested to us that those earlier designations were incorrect. First, the genome sequence of B. burgdorferi B31 (23) indicates that this bacterium does not encode a homolog of the E. coli F0/F1 ATPase. Second, a similar procedure was used to isolate membrane vesicles from the spirochete Serpulina (now Brachyspira) hyodysenteriae, where it was determined that the relative densities of the inner and outer membranes were opposite to those of other gram-negative bacteria (41). Third, numerous proteins known to be abundant on the B. burgdorferi cell surface (such as OspC and OspA) were seen to localize primarily to what Bledsoe et al. called the inner membrane fraction. We conclude that while the method developed by Bledsoe et al. (8) for the purification of borrelial membranes can accurately separate inner and outer membranes, the relative buoyant densities of B. burgdorferi inner and outer membranes are opposite to those of more typical gram negative organisms such as E. coli. This is likely due to the lack of classical lipopolysaccharide in the outer membranes of borreliae (58). Additionally, these and other spirochetes contain unusual types of lipids and liposaccharides that might affect membrane buoyancies (7, 18, 21, 41, 48, 61). The results of our studies also indicate a need for the reexamination of other procedures used for separation of the inner and outer membranes of borreliae (19, 27, 44, 52).
RevA was sensitive to in situ digestion with pronase, a combination of several different proteases having various endo- and exoproteolytic activities (Boehringer-Mannheim catalog). No digestion of RevA was detected when using either of the endoproteases trypsin or proteinase K. Several other B. burgdorferi surface proteins, including OspA and some members of the Erp (OspE and -F-related) protein family, are also insensitive to certain proteases in situ (10, 20, 22, 47). The resistance of RevA to proteolysis may be a consequence of its folding, as is apparently the case with some surface proteins of the related spirochete B. turicatae (64). Alternatively, RevA may interact with other proteinaceous or nonproteinaceous outer membrane components, which could have prevented the enzymes from reaching RevA. Such interactions have been observed with another B. burgdorferi outer surface protein, known as P66 or Oms66, which is protected from both proteases and antibodies by interactions between it and the OspA surface protein (10).
RevA is produced during mammalian infection and presumably performs a function for B. burgdorferi during that stage of the bacterium's life cycle. Since the control of RevA synthesis can be observed in vitro, it will be possible to define the mechanisms by which B. burgdorferi senses its environment and controls the synthesis of this antigenic protein in vivo. The function of RevA is as yet unknown, but its location on the bacterial outer surface would allow interactions with host cell surfaces or other host tissue constituents. Through a variety of techniques, some B. burgdorferi surface proteins have been found to bind specific host tissue components, and studies are under way in our laboratories to identify substances that bind RevA.
ACKNOWLEDGMENTS
Jay Carroll and Nazira El-Hage contributed equally to this work.
This study was funded by an NIH Institutional Training Award to Jay Carroll, and U.S. Public Health Service grant AI44254 to Brian Stevenson.
We thank Julie Stewart for technical assistance; Lamine Mbow, Robert Gilmore, Jr., and Tom Schwan for providing MAbs, hybridomas, and recombinant plasmids; Jerold Woodward for hybridoma advice; Nyles Charon and Mohammed Motaleb for sharing unpublished data; James Bono for providing B. burgdorferi RNA samples; Gary Hettrick and Anita Mora for graphics assistance; and Anthony Sinai, Patricia Rosa, and Philip Stewart for constructive comments on the manuscript.
REFERENCES
- 1.Akins D R, Bourell K W, Caimano M J, Norgard M V, Radolf J D. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Investig. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alban P S, Johnson P W, Nelson D R. Serum-starvation-induced changes in protein synthesis and morphology on Borrelia burgdorferi. Microbiology. 2000;146:119–127. doi: 10.1099/00221287-146-1-119. [DOI] [PubMed] [Google Scholar]
- 3.Babb K, El-Hage N, Miller J C, Carroll J A, Stevenson B. Distinct regulatory pathways control the synthesis of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect Immun. 2001;69:4146–4153. doi: 10.1128/IAI.69.6.4146-4153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour A G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour A G, Hayes S F, Heiland R A, Schrumpf M E, Tessier S L. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect Immun. 1986;52:549–554. doi: 10.1128/iai.52.2.549-554.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour A G, Tessier S L, Todd W J. Lyme disease spirochetes and ixodid tick spirochetes share a common surface antigenic determinant defined by a monoclonal antibody. Infect Immun. 1983;41:795–804. doi: 10.1128/iai.41.2.795-804.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck G, Habicht G S, Benach J L, Coleman J L. Chemical and biologic characterization of a lipopolysaccharide extracted from the Lyme disease spirochete (Borrelia burgdorferi) J Infect Dis. 1985;152:108–117. doi: 10.1093/infdis/152.1.108. [DOI] [PubMed] [Google Scholar]
- 8.Bledsoe H A, Carroll J A, Whelchel T R, Farmer M A, Dorward D W, Gherardini F C. Isolation and partial characterization of Borrelia burgdorferi inner and outer membranes by using isopycnic centrifugation. J Bacteriol. 1994;176:7447–7455. doi: 10.1128/jb.176.24.7447-7455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bono J L, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 1998;144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 10.Bunikis J, Barbour A G. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect Immun. 1999;67:2874–2883. doi: 10.1128/iai.67.6.2874-2883.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 12.Carroll J A, Cordova R M, Garon C F. Identification of eleven pH-regulated genes in Borrelia burgdorferi localized to linear plasmids. Infect Immun. 2000;68:6677–6684. doi: 10.1128/iai.68.12.6677-6684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll J A, Garon C F, Schwan T G. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll J A, Gherardini F C. Membrane protein variations associated with in vitro passage of Borrelia burgdorferi. Infect Immun. 1996;64:392–398. doi: 10.1128/iai.64.2.392-398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 16.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cassatt D R, Patel N K, Ulbrandt N D, Hanson M S. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect Immun. 1998;66:5379–5387. doi: 10.1128/iai.66.11.5379-5387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cinco M, Banfi E, Balanzin D, Godeas C, Panfili E. Evidence for (lipo)oligosaccharides in Borrelia burgdorferi and their serological specificity. FEMS Microbiol Lett. 1991;76:33–38. doi: 10.1111/j.1574-6968.1991.tb04160.x. [DOI] [PubMed] [Google Scholar]
- 19.Coleman J L, Benach J L, Beck G, Habicht G S. Isolation of the outer envelope from Borrelia burgdorferi. Zentbl Bakteriol Hyg A. 1986;263:123–126. doi: 10.1016/s0176-6724(86)80112-2. [DOI] [PubMed] [Google Scholar]
- 20.Dunn J J, Lade B N, Barbour A G. Outer surface protein A (OspA) from the Lyme disease spirochete, Borrelia burgdorferi: high-level expression and purification of a soluble recombinant form of OspA. Protein Expr Purif. 1990;1:159–168. doi: 10.1016/1046-5928(90)90011-m. [DOI] [PubMed] [Google Scholar]
- 21.Eiffert H, Lotter H, Jarecki-Khan K, Thomssen R. Identification of an immunreactive non-proteinaceous component in Borrelia burgdorferi. Med Microbiol Immunol. 1991;180:229–237. doi: 10.1007/BF00202557. [DOI] [PubMed] [Google Scholar]
- 22.El-Hage N, Babb K, Carroll J A, Lindstrom N, Fischer E R, Miller J C, Gilmore R D, Jr, Mbow M L, Stevenson B. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology. 2001;147:821–830. doi: 10.1099/00221287-147-4-821. [DOI] [PubMed] [Google Scholar]
- 23.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidmann J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs R, Jauris S, Lottspeich F, Preac-Mursic V, Wilske B, Soutschek E. Molecular analysis and expression of a Borrelia burgdorferi gene encoding a 22kDa protein (pC) in Escherichia coli. Mol Microbiol. 1992;6:503–509. doi: 10.1111/j.1365-2958.1992.tb01495.x. [DOI] [PubMed] [Google Scholar]
- 25.Gilmore R D, Jr, Mbow M L. A monoclonal antibody generated by antigen inoculation via tick bite is reactive to the Borrelia burgdorferi Rev protein, a member of the 2.9 gene family locus. Infect Immun. 1998;66:980–986. doi: 10.1128/iai.66.3.980-986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect., in press. [DOI] [PubMed]
- 27.Gondolf K B, Batsford S R, Vogt A. Isolation of an outer membrane protein complex from Borrelia burgdorferi by n-butanol extraction and high-performance ion-exchange chromatography. J Chromatogr. 1990;521:325–334. doi: 10.1016/0021-9673(90)85056-2. [DOI] [PubMed] [Google Scholar]
- 28.Haake D A. Spirochetal lipoproteins and pathogenesis. Microbiology. 2000;146:1491–1504. doi: 10.1099/00221287-146-7-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt S C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Indest K J, Ramamoorthy R, Sole M, Gilmore R D, Jr, Johnson B J B, Philipp M T. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect Immun. 1997;65:1165–1171. doi: 10.1128/iai.65.4.1165-1171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konkel M E, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2000;2:157–166. doi: 10.1016/s1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Motaleb M A, Sal M, Goldstein S F, Charon N W. Spirochete periplasmic flagella and motility. J Mol Microbiol Biotechnol. 2000;2:345–354. [PubMed] [Google Scholar]
- 33.MacNab R M. Flagella and motility. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 34.Margolis N, Hogan D, Cieplak W, Jr, Schwan T G, Rosa P A. Homology between Borrelia burgdorferi OspC and members of the family of Borrelia hermsii variable major proteins. Gene. 1994;143:105–110. doi: 10.1016/0378-1119(94)90613-0. [DOI] [PubMed] [Google Scholar]
- 35.Mbow M L, Gilmore R D, Jr, Titus R G. An OspC-specific monoclonal antibody passively protects mice from tick-transmitted infection by Borrelia burgdorferi B31. Infect Immun. 1999;67:5470–5472. doi: 10.1128/iai.67.10.5470-5472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J C, Bono J L, Babb K, El-Hage N, Casjens S, Stevenson B. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9–2. J Bacteriol. 2000;182:6254–6258. doi: 10.1128/jb.182.21.6254-6258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller J C, El-Hage N, Babb K, Stevenson B. Borrelia burgdorferi B31 Erp proteins that are dominant immunoblot antigens of animals infected with isolate B31 are recognized by only a subset of human Lyme disease patient sera. J Clin Microbiol. 2000;38:1569–1574. doi: 10.1128/jcm.38.4.1569-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nally J E, Timoney J F, Stevenson B. Temperature-regulated protein synthesis by Leptospira interrogans. Infect Immun. 2001;69:400–404. doi: 10.1128/IAI.69.1.400-404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 40.Obonyo M, Munderloh U G, Fingerle V, Wilske B, Kurtti T J. Borrelia burgdorferi in tick cell culture modulates expression of outer surface proteins A and C in response to temperature. J Clin Microbiol. 1999;37:2137–2141. doi: 10.1128/jcm.37.7.2137-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plaza H, Whelchel T R, Garczynski S F, Howerth E W, Gherardini F C. Purified outer membranes of Serpulina hyodysenteriae contain cholesterol. J Bacteriol. 1997;179:5414–5421. doi: 10.1128/jb.179.17.5414-5421.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porcella S F, Fitzpatrick C A, Bono J L. Expression and immunological analysis of the plasmid-borne mlp genes of Borrelia burgdorferi strain B31. Infect Immun. 2000;68:4992–5001. doi: 10.1128/iai.68.9.4992-5001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porcella S F, Popova T G, Akins D R, Li M, Radolf J D, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multi-copy tandem open reading frames and a lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radolf J D, Goldberg M S, Bourell K, Baker S I, Jones J D, Norgard M V. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect Immun. 1995;63:2154–2163. doi: 10.1128/iai.63.6.2154-2163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramamoorthy R, Philipp M T. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramamoorthy R, Scholl-Meeker D. Borrelia burgdorferi proteins whose expression is similarly affected by culture temperature and pH. Infect Immun. 2001;69:2739–2742. doi: 10.1128/IAI.69.4.2739-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadziene A, Thomas D D, Barbour A G. Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect Immun. 1995;63:1573–1580. doi: 10.1128/iai.63.4.1573-1580.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz C P, Wolf V, Lange R, Mertens E, Wecke J, Naumann D, Zähringer U. Evidence for a new type of outer membrane lipid in oral spirochete Treponema denticola. J Biol Chem. 1998;273:15661–15666. doi: 10.1074/jbc.273.25.15661. [DOI] [PubMed] [Google Scholar]
- 49.Schwan T G, Burgdorfer W, Rosa P A. Borrelia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 746–758. [Google Scholar]
- 50.Schwan T G, Piesman J. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shang E S, Skare J T, Exner M M, Blanco D R, Kagan B L, Miller J N, Lovett M A. Isolation and characterization of the outer membrane of Borrelia hermsii. Infect Immun. 1998;66:1082–1091. doi: 10.1128/iai.66.3.1082-1091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skare J T, Foley D M, Hernandez S R, Moore D C, Blanco D R, Miller J N, Lovett M A. Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect Immun. 1999;67:4407–4417. doi: 10.1128/iai.67.9.4407-4417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevenson B, Bono J L, Schwan T G, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevenson B, Zückert W R, Akins D R. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J Mol Microbiol Biotechnol. 2000;2:411–422. [PubMed] [Google Scholar]
- 57.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takayama K, Rothenberg R J, Barbour A G. Absence of lipopolysaccharide in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1987;55:2311–2313. doi: 10.1128/iai.55.9.2311-2313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tilly K, Casjens S, Stevenson B, Bono J L, Samuels D S, Hogan D, Rosa P. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 60.von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 61.Wheeler C M, Monco J C G, Benach J L, Golightly M G, Habicht G S, Steere A C. Nonprotein antigens of Borrelia burgdorferi. J Infect Dis. 1993;167:665–674. doi: 10.1093/infdis/167.3.665. [DOI] [PubMed] [Google Scholar]
- 62.Yang X, Goldberg M S, Popova T G, Schoeler G B, Wikel S K, Hagman K E, Norgard M V. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- 63.Yang X, Popova T G, Hagman K E, Wikel S K, Schoeler G B, Caimano M J, Radolf J D, Norgard M V. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zückert W R, Kerentseva T A, Lawson C L, Barbour A G. Structure analysis of the neurotropism-associated Borrelia turicatae VspA lipoprotein. J Biol Chem. 2001;276:457–463. doi: 10.1074/jbc.M008449200. [DOI] [PubMed] [Google Scholar]