Abstract
Coronaviruses infections, culminating in the recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic beginning in 2019, have highlighted the importance of effective vaccines to induce an antibody response with cross-neutralizing activity. COVID-19 vaccines have been rapidly developed to reduce the burden of SARS-CoV-2 infections and disease severity. Cross-protection from seasonal human coronaviruses (hCoVs) infections has been hypothesized but is still controversial. Here, we investigated the neutralizing activity against ancestral SARS-CoV-2 and the variants of concern (VOCs) in individuals vaccinated with two doses of either BNT162b2, mRNA-1273, or AZD1222, with or without a history of SARS-CoV-2 infection. Antibody neutralizing activity to SARS-CoV-2 and the VOCs was higher in BNT162b2-vaccinated subjects who were previously infected with SARS-CoV-2 and conferred broad-spectrum protection. The Omicron BA.1 variant was the most resistant among the VOCs. COVID-19 vaccination did not confer protection against hCoV-HKU1. Conversely, antibodies induced by mRNA-1273 vaccination displayed a boosting in their neutralizing activity against hCoV-NL63, whereas AZD1222 vaccination increased antibody neutralization against hCoV-229E, suggesting potential differences in antigenicity and immunogenicity of the different spike constructs used between various vaccination platforms. These data would suggest that there may be shared epitopes between the HCoVs and SARS-CoV-2 spike proteins.
Keywords: SARS-CoV-2, seasonal, HKU1, 229E, NL63, neutralisation
1. Introduction
In December 2019, the outbreak of a novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly spread around the world, resulting in a global pandemic [1]. Since then, international efforts to generate a suitable therapeutic have resulted in the development of multiple vaccination platforms and other antiviral pharmaceuticals. The gradual rise of variants has had a reduced impact on the efficacy of neutralising antibodies raised either by previous infection of SARS-CoV-2 or by vaccination [2,3]. The World Health Organisation (WHO) has categorised the troubling variants as variants of concern (VOC), whereas other variants that do not meet the same criteria fall under variants of interest (VOI) or variants under investigation (VUI). There has been a substantial amount of focus on variants and their characteristics, such as antibody evasion and replication rates, with many studies comparing variants and their ability to be neutralised [4,5,6,7,8], as the pandemic continues to progress.
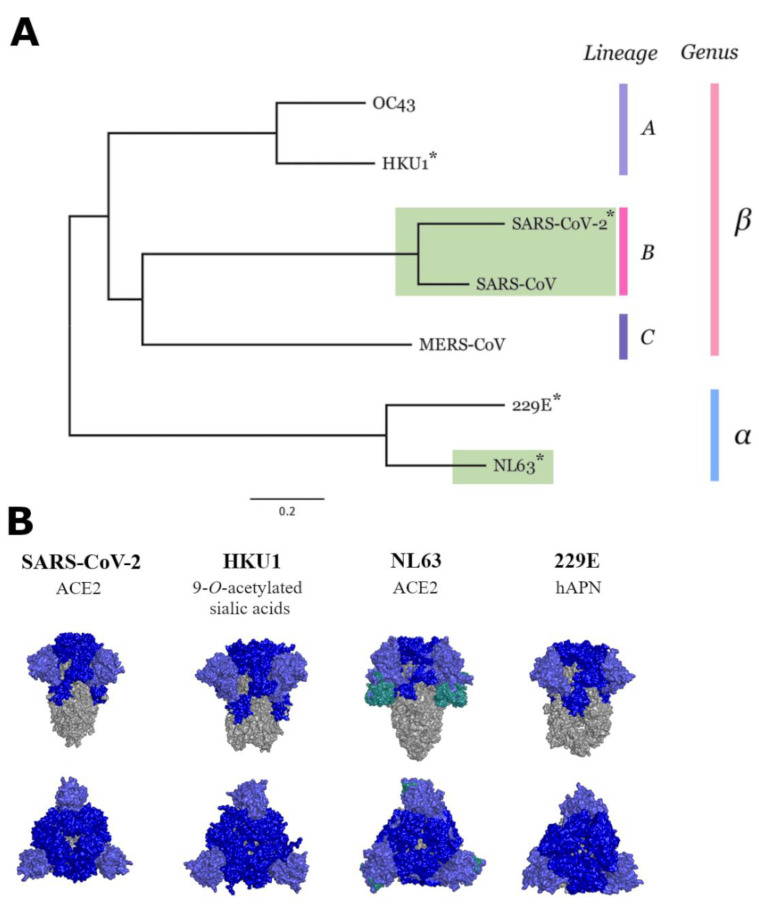
SARS-CoV-2 belongs to the Coronaviridae family that includes SARS-CoV-1 [9], middle eastern respiratory virus (MERS) [10] and four human coronaviruses 229E, HKU-1, NL63, and OC43 [11] (Figure 1A). Whilst SARS-CoV-1 and MERS have had outbreaks that caused severe disease in humans [12], the four other coronaviruses, commonly referred to as seasonal or human coronaviruses (HCoVs), typically cause mild disease similar to a common cold [11,13]. On rare occasions, however, the HCoVs may cause severe diseases [14,15,16]. SARS-CoV-2, together with NL63, use angiotensin-converting enzyme 2 (ACE2) as their major cell entry receptor [17,18]. Despite HKU1 and OC43 being more closely related to SARS-CoV-2, they bind to sialic acids as a mode of entry [19], whereas more distantly related 229E uses human aminopeptidase (hAPN) [20]. (Figure 1B).
Figure 1.
Phylogenetic tree of the members in the Coronaviridae family. * denotes spike proteins that were used in this study (A). Structures of spike proteins of SARS-CoV-2, and three of the seasonal HCoVs; HKU1, NL63 and 229E, which were used in this study (B). (PDB codes: 6VXX, 60HW, 6U7H, 5I08, and 5SZS). Grey denotes the S2 domain, whereas light blue is the N-terminal domain of S1 subunit, and dark blue represents the remaining S1 subunit. NL63 has an additional teal coloured section representing a unique region in the S1 domain not observed in other coronaviruses [21].
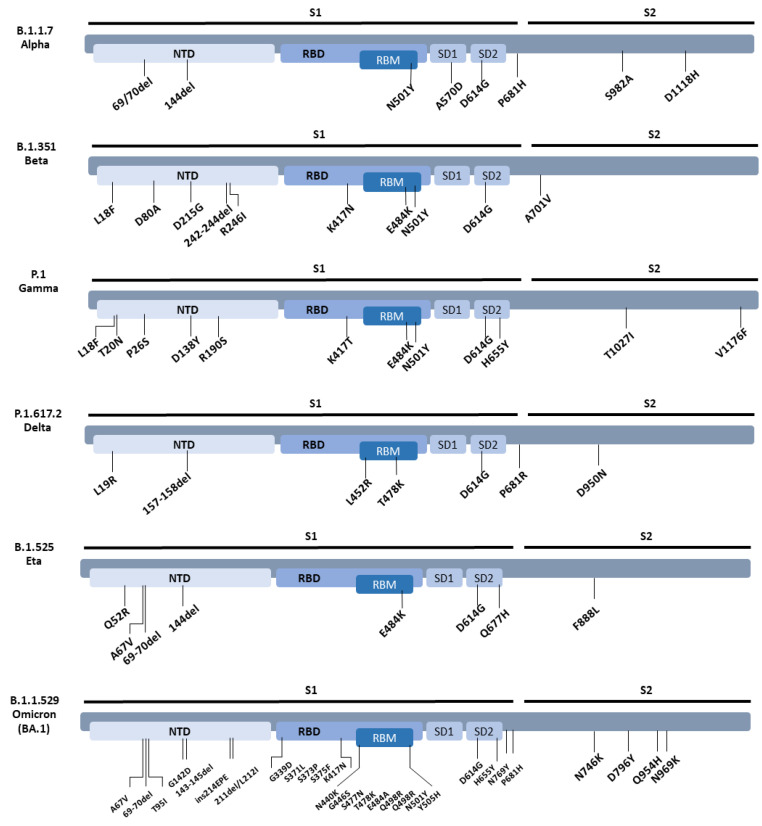
At the start of the pandemic, there was a debate as to the possibility that antibodies raised against the HCOVs had any role in protection against SARS-CoV-2 [22,23,24,25]. Since then, rising interest in HCoVs has led to an increase in understanding of the immune response they generate. Several publications that investigated the effect of HCoVs relied on the use of binding assays such as enzyme linked immunosorbent assays (ELISA) that measure antibody binding but did not elucidate their neutralising capabilities. Moreover, successful generation of various vaccine platforms have been used to protect individuals from infection and severe disease [26], though their effectiveness is diminished as newer and more immune evasive variants arise [27]. Here, we use lentiviral-based pseudotyped viruses of SARS-CoV-2, the VOCs/VOI, and HCoVs, to measure the strength of neutralising antibodies induced by two doses of either BNT162b2 (Pfizer), AZD1222 (Astrazeneca), or mRNA-1273 (Moderna) against SARS-CoV-2 and variants B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), B.1.525 (Eta), and B.1.1.529 (Omicron BA.1) (Figure 2), and whether any of these vaccines are able to augment neutralising antibodies against HCoVs 229E, HKU1, or NL63.
Figure 2.
SARS-CoV-2 variants spike mutations used in this study.
2. Materials and Methods
2.1. Patient Serum Collection/Ethics Information
Sera samples were collected from 36 healthy vaccinated subjects. The study was approved by San Raffaele Scientific Hospital Ethical Committee (protocol number 68/INT/2020). All enrolled patients gave written, informed consent.
2.2. Phylogenetic Tree and Similarity Plot
A maximum likelihood phylogenetic reconstruction based on the spike gene codon alignment, was constructed using iqtree (version 1.6.12) [28] with 10,000 ultra-fast bootstrap replicates [29] and a TVM+F+I+G4 substitution model, selected using ModelFinder [30]. The sequence similarity plot was constructed by aligning spike protein sequences of SARS-CoV-2 (QHD43416.1), HKU1 (YP_173238.1) 229E (NP_073551.1) and NL63 (YP_003767.1), using mafft (version 7.453) [31] (genafpair option) and visualised using the D3 JavaScript package implemented in observable (https://observablehq.com/@spyros-lytras/seasonal-cov-spike accessed on 3 November 2022).
2.3. Tissue Culture
Human embryonic kidney 293T/17 (HEK293T/17) cells and human hepatocytes Huh-7 cells were maintained in DMEM supplemented with 10% foetal bovine serum and 1% penicillin/streptomycin. Chinese ovarian hamster (CHO) cells were maintained in Ham’s F12 supplemented with 10% foetal bovine serum and 1% penicillin/streptomycin. Cells were routinely passaged to prevent confluency by washing with phosphate-buffered saline solution and detached with trypsin-EDTA. All cells were incubated at 37 °C and 5% CO2.
2.4. Pseudotype Virus Production
All pseudotypes (PVs) were generated as previously described [32]. Briefly, 1000 ng of pc-DNA 3.1+ plasmid bearing the spike of ancestral SARS-CoV-2, variants Alpha, Beta, Delta, Gamma, Eta, Omicron Ba.1, or HCoVs 229E, HKU1, and NL63 was mixed with 1000 ng of p8.91 plasmid encoding the HIV Gag-pol and 1500 ng of pCSFLW plasmid containing the Renilla firefly luciferase reporter gene, and co-transfected onto HEK293T cells at 50% confluency in T-75 flasks using FuGENE-HD. HKU-1 required an additional step of adding 1.5 U of exogenous neuraminidase (Sigma) in 10 mL of replenished DMEM 24 h after transfection. To harvest the pseudotyped viruses, media was aspirated 48 h after day of transfection and filtered using a 0.45 µm cellulose acetate filter. All PVs were aliquoted and stored at −80 °C for storage. After repeated attempts, we were unable to pseudotype HCoV OC43.
2.5. Pseudotype Virus Titration
All PVs were titrated as previously described [32]. Target cells for SARS-CoV-2, variants, and HCoV NL63 were prepared the day before titration by transfecting ACE-2 and TRSSMP2. CHO cells were used as target cells for HKU-1, and Huh-7 cells were used as target cells for 229E. Briefly, 50 µL of harvested PV were added in the top row of a white F-bottom 96-well plate (Nunc), and serially diluted using DMEM or Ham’s F-12 for HKU-1 PVs in half steps to the bottom row of the plate prior to addition of 10,000 target cells in each well. Plates were returned to the incubator for 48 h prior to lysis with Bright-Glo reagent and assaying luciferase reporter gene activity in relative line units (RLU) using a Glo-Max luminometer. PV titres are reported in RLU/mL.
2.6. Pseudotype Microneutralisation (pMN) Assays
The pMN assay was carried out as previously described. Briefly, convalescent sera were mixed with either DMEM or Ham’s F-12 at an initial 1:40 dilution and then serially diluted 2-fold in a white flat-bottomed 96-well plate to a final dilution of 1:5120. All samples were repeated in duplicate. PVs were then added to each well at an input of 1 × 106 RLU/mL. Plates were returned to the tissue culture incubator for 1 h, prior to addition of pre-transfected ACE-2/TRSSMP2 HEK293T target cells or CHO cells for HKU-1 and Huh-7 cells for 229E, at a density of 1 × 104 cells per well. Plates were returned to the incubator for 48 h prior to lysis with Bright-Glo reagent and assaying luciferase reporter gene activity in relative line units (RLU) using a Glo-Max luminometer. IC50s were calculated using GraphPad Prism 8 software using a non-linear regression curve as described in [33].
2.7. Statistical Analysis
Wilcoxon matched-pair ranked tests were used to assess significance in matched subjects. Kruskal–Wallis ANOVA test was used to assess significance when comparing IC50 titres between three vaccine platforms. All tests were used on Graphpad Prism 8 software.
3. Results
3.1. Cohort Characteristics
To assess the neutralizing potential of SARS-CoV-2 specific antibodies against SARS-CoV-2 VOCs and hCoVs, sera obtained from double-dosed BNT162b2-vaccinated (n = 13), AZD1222-vaccinated (n = 16) and mRNA-1273-vaccinated (n = 7) individuals with and without an history of SARS-CoV-2 infection were inspected (Table 1).
Table 1.
Demographic and clinical characteristics of the cohort.
| Total | 36 |
|---|---|
| Demographics | |
| Age (median [IQR], Range) | 49 [43.5, 55.25] (24–62) |
| Sex (Male/Female) | 8/28 |
| SARS-CoV-2 Prior Infection (Yes/No) | 11/28 |
| BNT162b2 Samples (1st dose/2nd Dose) | 13/13 |
| mRNA-1273 Samples (1st dose/2nd Dose) | 7/7 |
| AZD1222 Samples (1st dose/2nd Dose) | 16/16 |
| Time of bleed after 1st dose | 21 days (BNT162b2 and mRNA-1273) 12 weeks (AZD1222) |
| Time of bleed after 2nd dose | 15 weeks |
3.2. Neutralisation of SARS-CoV-2 Variants
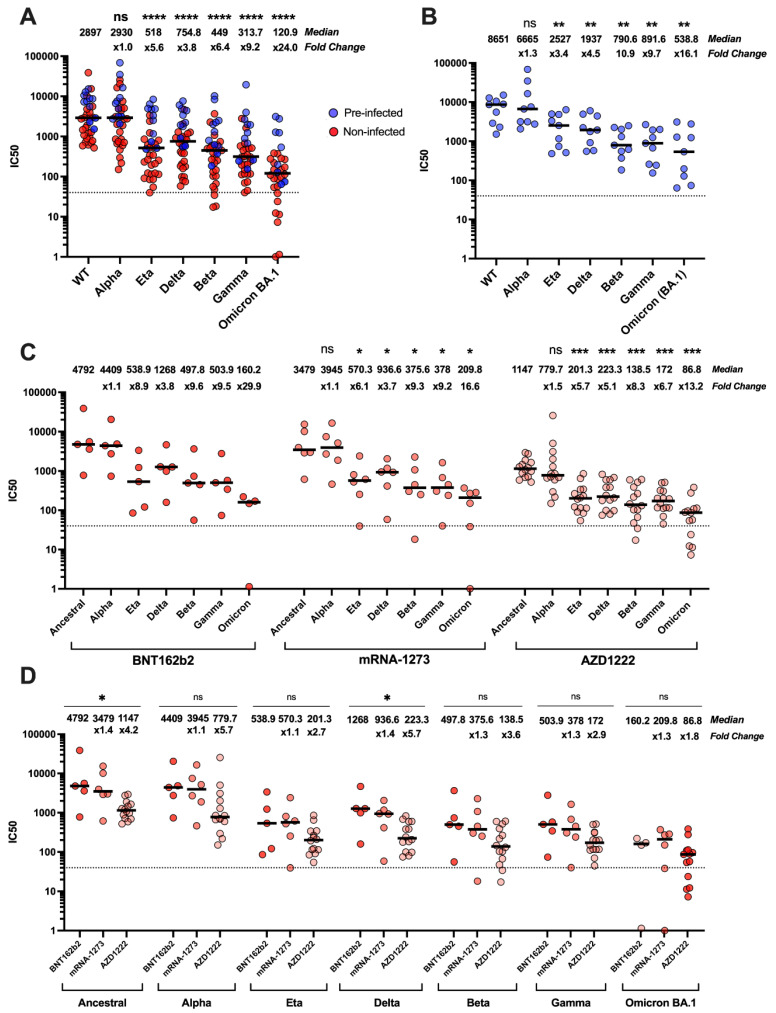
We first carried out pMN assays to analyse the magnitude of neutralising antibody responses against ancestral SARS-CoV-2 and variants, irrespective of vaccine type (Figure 3A). Our results showed that Omicron BA.1 was the least neutralised VOC (24-fold decrease, p =< 0.0001). As expected, we observed the samples from individuals with prior infection had higher neutralisation titres compared with immunological naïve subjects.
Figure 3.
Ability of serum antibodies to neutralise SARS-CoV-2 and VOCs from individuals vaccinated with two doses of either BNT162b2, AZD1222, or mRNA-1273. Neutralizing antibody response against the ancestral SARS-CoV-2 and variants, in previously infected individuals (blue) and non-infected individuals (red) receiving two doses of either BNT162b2, AZD1222, or mRNA-1273 vaccines (A). Wilcoxon matched-pairs signed rank tests statistical analysis was used to compare ancestral SARS-CoV-2 against each variant (A). Neutralisation profiles of sera from BNT162b2-vaccinated subjects with a history of prior infection. No statistical test was used for BNT162b2 in panel C due to small sample size with large variation. (B). Neutralisation profiles of the three vaccine types against variants (C) and compared between vaccine platforms. (D) Wilcoxon matched-pairs signed rank tests statistical analysis was used to compare ancestral SARS-CoV-2 against each variant in panel C. Kruskal–Wallis ANOVA was used for statistical analysis in panel D. ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p ≤ 0.0001.
The serum from previously infected individuals (Figure 3B), neutralized the Alpha variant more effectively compared with the ancestral strain, as it showed a 1.3-fold decrease in median IC50 titre, followed by Eta and Delta variants, (3.4- and 4.5-fold decrease, respectively). Beta and Gamma variants were more resistant to neutralization (10.9- and 9.7-fold decrease, respectively), and Omicron BA.1 reached a 16.1-fold decrease compared with ancestral SARS-CoV-2. Notably, the majority of these subjects had received the BNT162b2 vaccine.
Taken together, these results suggested that in vaccinated subjects the pre-existing immunity raised by natural infection with SARS-CoV-2, or a VOC is more effective in protecting against the spectrum of variants that emerged later over time, compared with immunity triggered by vaccination only. However, the recently emerged variants evolved mechanisms to evade the neutralizing antibody response.
We then analysed subjects who had not experienced SARS-CoV-2 infection before vaccine administration (Figure 3C). The efficacy of each vaccine platform was analysed with respect to the capability to neutralize both the ancestral strain and its variants. We observed that the sera from BNT162b2-vaccinated subjects had high median IC50 titres compared with those obtained from mRNA-1273- and AZD1222-vaccinated individuals. Whereas the Alpha variant did not show immune escape in any of the vaccinated subjects, all the VOCs were resistant to antibody neutralization to different degrees (Figure 3C). We were unable to extract meaningful significance scores from the BNT162b2 samples due to few samples (n = 5) with very large spread in IC50 titres.
We did not observe any statistically significant difference between the three vaccine platforms with respect to their abilities to neutralize the Alpha, Eta, Beta, Gamma, and BA.1 variants. Conversely, the biggest difference between the three vaccine types was observed with ancestral and Delta variant, as mRNA-1273 showed a 1.6- and 1.4-fold decrease in median IC50 titres, respectively, compared with BNT162b2, whereas AZD1222 showed a 3.8- and 4.1-fold decrease (Figure 3D).
3.3. Neutralisation of Seasonal HCoVs
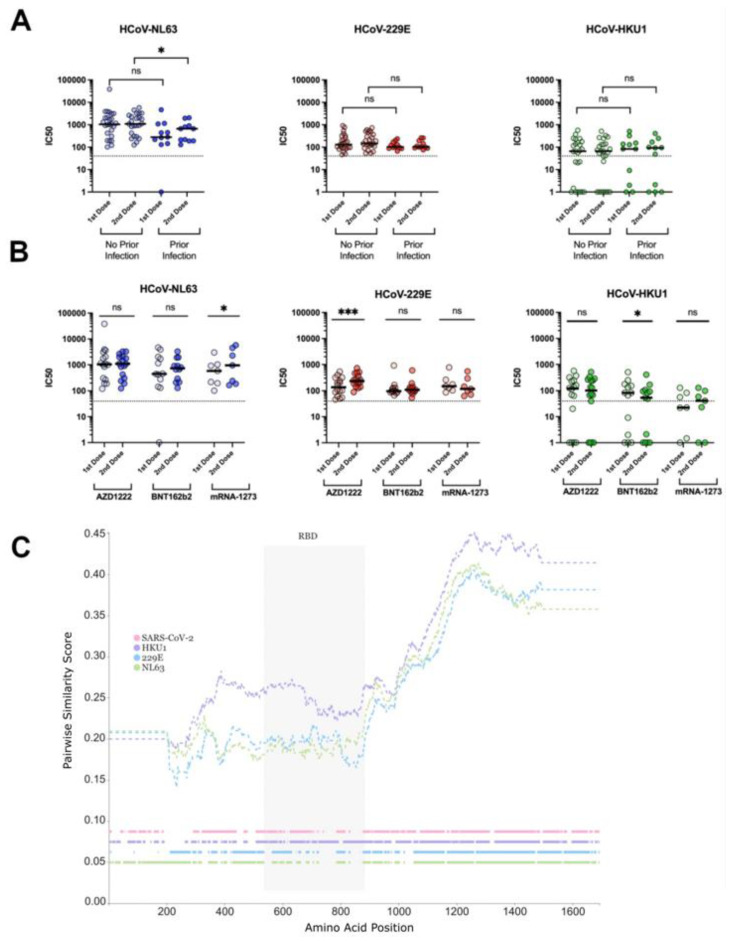
To determine whether vaccination against SARS-CoV-2 may cross-protect against seasonal HCoVs, we asked whether a prior infection with SARS-CoV-2 had any impact on antibody-mediated neutralisation of the HCoVs (Figure 4A). We observed no statistically significant increases in neutralizing titres against either 229E or HKU-1 between previously SARS-CoV-2 infected and naïve individuals. Conversely, a statistically significant decrease in neutralizing titres against NL63 after the second dose administration was found in vaccinated subjects who experienced SARS-CoV-2 infection (p = 0.033) compared with the naïve (p = 0.063).
Figure 4.
Comparing neutralising responses in HCoVs NL63, 229E, and HKU1 between first- and second-dose vaccination against SARS-CoV-2. Neutralization profile against HCoVs NL63, 229E, and HKU1 in double-dosed BNT162b2, mRNA-1273 or AZD1222 –vaccinated subjects with or without a history of SARS-CoV-2 infection. (A). Neutralizing antibody titres against the aforementioned HCoVs after the first and second dose administration of BNT162b2, mRNA-1273 or AZD1222 vaccines. (B). Wilcoxon matched-pairs signed rank tests statistical analysis was used in A and B. Similarity plots (C) show HKU-1 spike as having more similar amino acid sequence to SARS-CoV-2 compared with both NL63 and 229E in all regions of the spike protein. Dashed lines on the top show amino acid pairwise similarity between SARS-CoV-2 and the 3 HCoV Spike proteins, plotted using a 400 amino acid window size and a step of 1. Positions with gaps were excluded from the windows. Horizontal lines on the bottom indicate residue presence for each of the 4 aligned coronaviruses across the alignment length (colour presence = amino acid presence; colour absence = gap). ns = not significant, * p < 0.05, *** p < 0.001.
We then assessed whether one or more of the vaccine platforms would boost titres against the HCoVs in all subjects, irrespective of their previous infection status (Figure 4B). Overall, in vaccinated subjects, the median antibody neutralization titres against NL63 were higher compared with those against 229E and HKU1, irrespective of the vaccine platform (Figure 4B). Notably, NL63 uses ACE2 as entry receptor into the target cells, as does SARS-CoV-2.
The type of vaccine did not have an impact in boosting the neutralizing activities against the three seasonal coronaviruses we studied after the second dose administration, with the exception of NL63 and HKU1. IC50 titres against NL63 increased after the second dose using mRNA-1273 (p = 0.03), whereas 229E showed a statistically significant increase in IC50 titre in only AZD1222-vaccinated individuals (p =< 0.001). Conversely, after the second boost of BNT162b2 vaccine, neutralization titres against HCoV HKU-1 decreased, probably due to the selection of antigen-specific plasma cells with lower affinity for the HKU1 spike.
To better understand the impact of COVID-19 vaccination on the protection from seasonal HCoVs in subjects with or without a history of SARS-CoV-2 infection, we analysed the spike protein similarity of HCoVs HKU1, NL63, 229E and SARS-CoV-2 to investigate whether a particular region could explain the neutralisation differences (Figure 4C). The similarity plot generated by comparing pairwise similarity showed HKU1 had higher similarity in all spike regions to SARS-CoV-2 spike compared with 229E and NL63, consistent with the viruses’ taxonomy. However, HKU1 seems to have extra insertions at the C-terminal end of the RBD compared with the other two seasonals and SARS-CoV-2. Furthermore, the S2 region shows much higher similarity to SARS-CoV-2 in all three HCoVs compared with the S1 region (Figure 4C).
4. Discussion
In this study, we were able to directly compare the antibody neutralisation titres induced by two m-RNA-based vaccines, BNT162b2 and mRNA-1273, and an adenoviral-based vaccine, AZD1222, against SARS-CoV-2, its emerged variants, and three seasonal HCoVs.
Our data on antibody neutralization against SARS-CoV-2 and its variants in vaccinated subjects, with or without a history of previous infection, agree with what is reported in the literature [2,4,5,7,34,35,36,37,38]. We confirmed that vaccination with two doses of vaccines induced antibodies able to neutralize SARS-CoV-2 and VOCs, with BNT162b2 eliciting the highest neutralization titres, followed by mRNA-1273 and AZD1222. Despite their differences in neutralization titres, all three vaccines have been reported to have high efficacy at preventing severe COVID-19 [39,40,41]. The Omicron BA.1 variant was the most evasive of all VOCs analysed in this study (Figure 3). Indeed, the heavily mutated spike protein of BA.1 variant posed challenges to the effectiveness of the current vaccines to protect against COVID-19 and pointed out the need to monitor the protection conferred against this and the newly emerged SARS-CoV-2 variants, namely Omicron BA.4 and BA.5. Bivalent formulations of mRNA-based vaccines, containing both the mRNA of the spike of the ancestral SARS-CoV-2 and the one in common between the BA.4 and BA.5 lineages have been designed and authorized in order to counteract the evasion of the immune response elicited by the original vaccine design.
HCoVs are globally distributed and believed to induce short-lasting protective antibodies [42]. Therefore, there is a high likelihood of reinfection remaining elevated, especially during the winter periods [13,43,44,45] despite high seroprevalence [43,45,46]. It is currently debated whether prior infection with seasonal HCoVs elicits cross-reactive antibodies against SARS-CoV-2, and more importantly, if this translates into protection against SARS-CoV-2. Cross-reactive antibodies [47,48,49,50,51,52,53,54] and T-cell responses [55,56,57,58,59,60,61] were detected in pre-pandemic sera and healthy donors; however, similar experimental approaches have shown the opposite to be true by other investigators [62]. In addition, in many of the aforementioned articles that revealed cross-reactive antibodies in pre-pandemic samples, the number of cross-reactive samples was a small portion of the total sera analysed, suggesting that cross-reactivity, whilst it exists, is low.
The same question has been raised about antibodies elicited by COVID-19 vaccines, with studies showing cross-reactive antibodies to some but not all the seasonal HCoVs [63,64,65]. SARS-CoV-2 spike protein vaccination was shown to induce cross-reactive antibodies to both Alpha- and Betacoronaviruses in macaques [66]. It is important to deduce whether cross-reactive antibodies translate into protective, neutralising antibodies against SARS-CoV-2. Some reports suggested that whilst there is a small boost in antibodies towards HCoVs during SARS-CoV-2 infection, they are not associated with protection [67]. Similarly, studies showed that prior infection with HCoVs did not protect against SARS-CoV-2 infection and disease [68,69].
We did not find any boost of neutralizing antibody titres against HKU1 in our cohort of SARS-CoV-2-vaccinated subjects, irrespective of their SARS-CoV-2 pre-infectious status, with the exception of subjects administered with BNT162b2. This is in contrast with two reports that observed a boost in HKU-1 titres post vaccination against SARS-CoV-2 by BNT162b2 [63,64]. Hicks et al. showed that antibodies reacting to HCoV-OC43 and HCoV-HKU1 had minimal cross-reactivity with SARS-CoV-2, in accordance with the sequence homology of these proteins [54]. Moreover, a previous SARS-CoV-2 infection did not boost the cross-neutralization against either HKU1 or the more phylogenetically related HCoV-229E (Figure 4A). One report suggested that HKU1 may have another candidate receptor that has yet to be identified, due to the presence of a putative RBD, distant from the sialic acid binding regions [70,71]. It should also be noted that neutralising ability might not only be dependent on the pairwise similarity between amino acids in the protein, but also short insertions and deletions that can alter the protein’s structural conformation. For instance, the HKU-1-specific insertion at the C-terminal end of the RBD (Figure 4C) might partly explain our neutralisation results.
Conversely, we found that the second dose administration in naïve subjects increased the protective antibody response against NL63 compared with that obtained in previously infected subjects receiving the same dose. This was probably due to the fact that additional exposures to the spike antigen did not have an effect on antibody neutralization against NL63.
The differences in antibody neutralization between the HCoVs may be due to the differences in the spikes used by the vaccination platforms. BNT162b27 encodes full-length spike with the K986P and V987P mutation sites to stabilize the pre-fusion conformation of the protein [72]. The mRNA-1273 vaccine contains the coding sequence for a spike glycoprotein stabilized by the same proline substitutions used in the BNT162b2 vaccine, with a transmembrane anchor and an intact S1-S2 cleavage site. The pre-fusion conformation is stabilized by the consecutive proline substitutions, which are located in the S2 subunit at the top of the central helix [73]. Conversely, a native-like spike is expressed by the AZD1222 vaccine. As our naïve subjects were administered with the AZD1222 vaccine, we can speculate that the native form of the spike protein triggered the development of higher neutralizing antibodies titres compared with that induced by the pre-fusion-stabilized protein.
Conversely, the second immunogenic exposure to SARS-CoV-2 spike boosted the neutralizing response against NL63 or 229E (Figure 4B), as has been previously reported [67], depending on the vaccine platform, irrespective of the pre-infection status. Interestingly, another report observed the same cross-neutralizing activity, though this was irrespective of vaccine platform [74]. We speculate that cross reactivity can arise due to similarity in epitopes in the receptor-binding motif (RBM) of NL63 to SARS-CoV-2, since both viruses share ACE-2 as their entry receptor [75]. Similarly, an epitope overlapping the S2 fusion peptide in 229E has been reported to elicit cross-reactivity against SARS-CoV-2 [48]. Song et al. described protective neutralizing antibodies targeting the S2 subdomain [53]. Furthermore, a report during the original SARS-CoV-1 outbreak also found cross reactive antibodies against NL63 and 229E [76], strengthening the hypothesis of shared epitopes between Alphacoronaviruses and Betacoronaviruses. The S protein of NL63 does not contain the furin-recognition site and is not cleaved during biogenesis [77]. Similarly, the spike protein expressed by the mRNA1273 vaccine lacks the cleavage site; therefore, the conformation of the protein might be similar and might trigger neutralizing antibodies against shared epitopes and that are boosted after a second exposure to the same antigen.
The antigenic nature of the spike protein expressed by the different vaccines, together with multiple conformations they can acquire, might affect the development of neutralizing antibodies with different affinities towards several epitopes in the spike protein. Since the AZD-1222 spike does not contain the two proline mutations to stabilise its spike into a trimeric pre fusion structure [78,79], the presence of a post fusion spike could potentially elicit a larger immune response towards epitopes in the S2 domain. This may explain why we did not observe any boost in neutralizing titres against 229E in either mRNA-based, pre-fusion-stabilized immunogen, vaccinated samples. Ultimately, despite observing a boost in titres, it is impossible for us to state whether this translates into protective titres since correlates of protection against SARS-CoV-2 have yet to be defined.
There are several limitations in our study to consider. Our data would have benefitted from larger numbers of samples in all vaccine platform types, and control samples of non-vaccinated individuals who either have been infected with SARS-CoV-2 or not. Furthermore, we did not analyse the baseline levels of cross-reactive neutralizing antibodies against seasonal coronaviruses in our cohort of vaccinated subjects.
A pan-coronavirus vaccine would elicit antibodies that recognise and neutralise a broad range of coronaviruses. This is challenging because of the genetic nature of these RNA viruses that frequently mutate and induce an immunity that wanes over time, increasing the likelihood of reinfection. Therefore, identifying the key epitopes located at the most conserved regions of the spike protein, especially at the S2 subunit, is relevant to potentially induce neutralizing antibodies with broader affinity to the cellular receptors that mediate viral entry. Several vaccine candidates have been formulated, and some are based on dual antigens including both spike and nucleocapsid (N) components [80]. These formulations are at the pre-clinical stage as they might provide broader and more durable humoral and cellular immune responses against coronaviruses [80].
Acknowledgments
We would like to acknowledge the Ambulatorio Medico San Luca Villanuova Group members (Bettini, G.L., Bonvicini, A., Braga, A., Chappini, F., Inverardi, F., Ravera, S., Rossi, M.G., Sas, A., Tuttini, A.) and patients who enrolled in this study.
Author Contributions
Conceptualization, D.C., G.S., L.L. and N.T.; methodology, D.C, G.S, L.L. and N.T.; investigation, D.C., G.S., M.M.-N., C.P., T.F., S.L., C.D.G. and J.H.; data curation, D.C.; writing—original draft preparation, D.C and G.S.; writing—review and editing, D.C., G.S., M.M.-N., C.P., T.F., S.L., C.D.G., J.H., L.L., N.T. and Ambulatorio Medico San Luca Villanuova Group; funding acquisition, L.L. and N.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of San Raffaele Scientific Hospital Ethical Committee (protocol number 68/INT/2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All datasets are available by contacting the corresponding author upon kind request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The Temperton group (N.T., D.C., C.D.G. and M.M.N.) is funded by the Wellcome Trust (GB-CHC-210183), the MRC (MC_PC_19060) and MRC/NIHR (MC_PC_20016). S.L. and J.H. are funded by MC_UU_12014/12. L.L. was supported by the Scientific Direction of San Raffaele Scientific Institute (Immuno-COVID) and ANR France (MUCOLUNG).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmann D.M., Boyton R.J., Beale R. Immunity to SARS-CoV-2 Variants of Concern. Science. 2021;371:1103–1104. doi: 10.1126/science.abg7404. [DOI] [PubMed] [Google Scholar]
- 3.Burki T. Understanding Variants of SARS-CoV-2. Lancet. 2021;397:462. doi: 10.1016/S0140-6736(21)00298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 Variants B.1.351 and P.1 Escape from Neutralizing Antibodies. Cell. 2021;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantoni D., Mayora-Neto M., Nadesalingam A., Wells D.A., Carnell G.W., Ohlendorf L., Ferrari M., Palmer P., Chan A.C.Y., Smith P., et al. Neutralisation Hierarchy of SARS-CoV-2 Variants of Concern Using Standardised, Quantitative Neutralisation Assays Reveals a Correlation With Disease Severity; Towards Deciphering Protective Antibody Thresholds. Front. Immunol. 2022;13:773982. doi: 10.3389/fimmu.2022.773982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of Infectious SARS-CoV-2 B.1.1.7 and B.1.351 Variants to Neutralizing Antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 8.Lustig Y., Zuckerman N., Nemet I., Atari N., Kliker L., Regev-Yochay G., Sapir E., Mor O., Alroy-Preis S., Mendelson E., et al. Neutralising Capacity against Delta (B.1.617.2) and Other Variants of Concern Following Comirnaty (BNT162b2, BioNTech/Pfizer) Vaccination in Health Care Workers, Israel. Eurosurveillance. 2021;26:2100557. doi: 10.2807/1560-7917.ES.2021.26.26.2100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 10.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 11.Liu D.X., Liang J.Q., Fung T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae) In: Bamford D.H., Zuckerman M., editors. Encyclopedia of Virology. 4th ed. Academic Press; Oxford, UK: 2021. pp. 428–440. [Google Scholar]
- 12.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: Recent Insights into Emerging Coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and Clinical Presentations of the Four Human Coronaviruses 229E, HKU1, NL63, and OC43 Detected over 3 Years Using a Novel Multiplex Real-Time PCR Method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Da Veiga A.B.G., Martins L.G., Riediger I., Mazetto A., Debur M.d.C., Gregianini T.S. More than Just a Common Cold: Endemic Coronaviruses OC43, HKU1, NL63, and 229E Associated with Severe Acute Respiratory Infection and Fatality Cases among Healthy Adults. J. Med. Virol. 2021;93:1002–1007. doi: 10.1002/jmv.26362. [DOI] [PubMed] [Google Scholar]
- 15.Arden K.E., Nissen M.D., Sloots T.P., Mackay I.M. New Human Coronavirus, HCoV-NL63, Associated with Severe Lower Respiratory Tract Disease in Australia. J. Med. Virol. 2005;75:455–462. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hand J., Rose E.B., Salinas A., Lu X., Sakthivel S.K., Schneider E., Watson J.T. Severe Respiratory Illness Outbreak Associated with Human Coronavirus NL63 in a Long-Term Care Facility. Emerg. Infect. Dis. 2018;24:1964–1966. doi: 10.3201/eid2410.180862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human Coronavirus NL63 Employs the Severe Acute Respiratory Syndrome Coronavirus Receptor for Cellular Entry. Proc. Natl. Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulswit R.J.G., Lang Y., Bakkers M.J.G., Li W., Li Z., Schouten A., Ophorst B., van Kuppeveld F.J.M., Boons G.-J., Bosch B.-J., et al. Human Coronaviruses OC43 and HKU1 Bind to 9-O-Acetylated Sialic Acids via a Conserved Receptor-Binding Site in Spike Protein Domain A. Proc. Natl. Acad. Sci. USA. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human Aminopeptidase N Is a Receptor for Human Coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu K., Li W., Peng G., Li F. Crystal Structure of NL63 Respiratory Coronavirus Receptor-Binding Domain Complexed with Its Human Receptor. Proc. Natl. Acad. Sci. USA. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerholz D.K., Perlman S. Does Common Cold Coronavirus Infection Protect against Severe SARS-CoV-2 Disease? J. Clin. Investig. 2021;131 doi: 10.1172/JCI144807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringlander J., Martner A., Nilsson S., Westin J., Lindh M., Hellstrand K. Incidence and Severity of Covid-19 in Patients with and without Previously Verified Infections with Common Cold Coronaviruses. J. Infect. Dis. 2021;223:1831–1832. doi: 10.1093/infdis/jiab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnierle B.S. Reply to Ringlander et al. J. Infect. Dis. 2021;223:1833. doi: 10.1093/infdis/jiab090. [DOI] [PubMed] [Google Scholar]
- 25.Beretta A., Cranage M., Zipeto D. Is Cross-Reactive Immunity Triggering COVID-19 Immunopathogenesis? Front. Immunol. 2020;11:2695. doi: 10.3389/fimmu.2020.567710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creech C.B., Walker S.C., Samuels R.J. SARS-CoV-2 Vaccines. JAMA. 2021;325:1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 27.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The Biological and Clinical Significance of Emerging SARS-CoV-2 Variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genova C.D., Sampson A., Scott S., Cantoni D., Mayora-Neto M., Bentley E., Mattiuzzo G., Wright E., Derveni M., Auld B., et al. Production, Titration, Neutralisation, Storage and Lyophilisation of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Lentiviral Pseudotypes. Bio-Protocol. 2021;11:e4236. doi: 10.21769/BioProtoc.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrara F., Temperton N. Pseudotype Neutralization Assays: From Laboratory Bench to Data Analysis. Methods Protoc. 2018;1:8. doi: 10.3390/mps1010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced Neutralization of SARS-CoV-2 B.1.617 by Vaccine and Convalescent Serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen X., Tang H., Pajon R., Smith G., Glenn G.M., Shi W., Korber B., Montefiori D.C. Neutralization of SARS-CoV-2 Variants B.1.429 and B.1.351. N. Engl. J. Med. 2021;384:2352–2354. doi: 10.1056/NEJMc2103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., et al. SARS-CoV-2 Variant B.1.1.7 Is Susceptible to Neutralizing Antibodies Elicited by Ancestral Spike Vaccines. Cell Host Microbe. 2021;29:529–539. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willett B.J., Grove J., MacLean O.A., Wilkie C., De Lorenzo G., Furnon W., Cantoni D., Scott S., Logan N., Ashraf S., et al. SARS-CoV-2 Omicron Is an Immune Escape Variant with an Altered Cell Entry Pathway. Nat. Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siracusano G., Ruggiero A., Bisoffi Z., Piubelli C., Carbonare L.D., Valenti M.T., Mayora-Neto M., Temperton N., Lopalco L., Zipeto D. Different Decay of Antibody Response and VOC Sensitivity in Naïve and Previously Infected Subjects at 15 Weeks Following Vaccination with BNT162b2. J. Transl. Med. 2022;20:22. doi: 10.1186/s12967-021-03208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emary K.R.W., Golubchik T., Aley P.K., Ariani C.V., Angus B., Bibi S., Blane B., Bonsall D., Cicconi P., Charlton S., et al. Efficacy of ChAdOx1 NCoV-19 (AZD1222) Vaccine against SARS-CoV-2 Variant of Concern 202012/01 (B.1.1.7): An Exploratory Analysis of a Randomised Controlled Trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal Coronavirus Protective Immunity Is Short-Lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 43.Aldridge R.W., Lewer D., Beale S., Johnson A.M., Zambon M., Hayward A.C., Fragaszy E.B. Seasonality and Immunity to Laboratory-Confirmed Seasonal Coronaviruses (HCoV-NL63, HCoV-OC43, and HCoV-229E): Results from the Flu Watch Cohort Study. Wellcome Open Res. 2020;5:52. doi: 10.12688/wellcomeopenres.15812.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S.-F., Tuo J.-L., Huang X.-B., Zhu X., Zhang D.-M., Zhou K., Yuan L., Luo H.-J., Zheng B.-J., Yuen K.-Y., et al. Epidemiology Characteristics of Human Coronaviruses in Patients with Respiratory Infection Symptoms and Phylogenetic Analysis of HCoV-OC43 during 2010-2015 in Guangzhou. PLoS ONE. 2018;13:e0191789. doi: 10.1371/journal.pone.0191789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byington C.L., Ampofo K., Stockmann C., Adler F.R., Herbener A., Miller T., Sheng X., Blaschke A.J., Crisp R., Pavia A.T. Community Surveillance of Respiratory Viruses Among Families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) Study. Clin. Infect. Dis. 2015;61:1217–1224. doi: 10.1093/cid/civ486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells D.A., Cantoni D., Mayora-Neto M., Genova C.D., Sampson A., Ferrari M., Carnell G., Nadesalingam A., Smith P., Chan A., et al. Human Seasonal Coronavirus Neutralization and COVID-19 Severity. J. Med. Virol. 2022;94:4820–4829. doi: 10.1002/jmv.27937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shrwani K., Sharma R., Krishnan M., Jones T., Mayora-Neto M., Cantoni D., Temperton N.J., Dobson S.L., Subramaniam K., McNamara P.S., et al. Detection of Serum Cross-Reactive Antibodies and Memory Response to SARS-CoV-2 in Prepandemic and Post–COVID-19 Convalescent Samples. J. Infect. Dis. 2021;224:1305–1315. doi: 10.1093/infdis/jiab333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de Novo Humoral Immunity to SARS-CoV-2 in Humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woudenberg T., Pelleau S., Anna F., Attia M., Donnadieu F., Gravet A., Lohmann C., Seraphin H., Guiheneuf R., Delamare C., et al. Humoral Immunity to SARS-CoV-2 and Seasonal Coronaviruses in Children and Adults in North-Eastern France. EBioMedicine. 2021;70:103495. doi: 10.1016/j.ebiom.2021.103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tso F.Y., Lidenge S.J., Peña P.B., Clegg A.A., Ngowi J.R., Mwaiselage J., Ngalamika O., Julius P., West J.T., Wood C. High Prevalence of Pre-Existing Serological Cross-Reactivity against Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in Sub-Saharan Africa. Int. J. Infect. Dis. 2021;102:577–583. doi: 10.1016/j.ijid.2020.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laing E.D., Sterling S.L., Richard S.A., Phogat S., Samuels E.C., Epsi N.J., Yan L., Moreno N., Coles C., Mehalko J., et al. A Betacoronavirus Multiplex Microsphere Immunoassay Detects Early SARS-CoV-2 Seroconversion and Controls for Pre-Existing Seasonal Human Coronavirus Antibody Cross-Reactivity. medRxiv. 2020 doi: 10.1101/2020.10.14.20207050. [DOI] [Google Scholar]
- 52.Ladner J.T., Henson S.N., Boyle A.S., Engelbrektson A.L., Fink Z.W., Rahee F., D’ambrozio J., Schaecher K.E., Stone M., Dong W., et al. Epitope-Resolved Profiling of the SARS-CoV-2 Antibody Response Identifies Cross-Reactivity with Endemic Human Coronaviruses. Cell Rep. Med. 2021;2:100189. doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song G., He W., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-Reactive Serum and Memory B-Cell Responses to Spike Protein in SARS-CoV-2 and Endemic Coronavirus Infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hicks J., Klumpp-Thomas C., Kalish H., Shunmugavel A., Mehalko J., Denson J.-P., Snead K.R., Drew M., Corbett K.S., Graham B.S., et al. Serologic Cross-Reactivity of SARS-CoV-2 with Endemic and Seasonal Betacoronaviruses. J. Clin. Immunol. 2021;41:906–913. doi: 10.1007/s10875-021-00997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and Cross-Reactive SARS-CoV-2 T Cell Epitopes in Unexposed Humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., et al. Characterization of Pre-Existing and Induced SARS-CoV-2-Specific CD8+ T Cells. Nat Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 59.Reynolds C.J., Swadling L., Gibbons J.M., Pade C., Jensen M.P., Diniz M.O., Schmidt N.M., Butler D.K., Amin O.E., Bailey S.N.L., et al. Discordant Neutralizing Antibody and T Cell Responses in Asymptomatic and Mild SARS-CoV-2 Infection. Sci. Immunol. 2020;5:eabf3698. doi: 10.1126/sciimmunol.abf3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Low J.S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., Jarrossay D., Jovic S., Perez L., Cacciatore R., et al. Clonal Analysis of Immunodominance and Cross-Reactivity of the CD4 T Cell Response to SARS-CoV-2. Science. 2021;372:1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Echeverría G., Guevara Á., Coloma J., Ruiz A.M., Vasquez M.M., Tejera E., de Waard J.H. Pre-Existing T-Cell Immunity to SARS-CoV-2 in Unexposed Healthy Controls in Ecuador, as Detected with a COVID-19 Interferon-Gamma Release Assay. Int. J. Infect. Dis. 2021;105:21–25. doi: 10.1016/j.ijid.2021.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poston D., Weisblum Y., Wise H., Templeton K., Jenks S., Hatziioannou T., Bieniasz P. Absence of Severe Acute Respiratory Syndrome Coronavirus 2 Neutralizing Activity in Prepandemic Sera From Individuals With Recent Seasonal Coronavirus Infection. Clin. Infect. Dis. 2020;73:e1208–e1211. doi: 10.1093/cid/ciaa1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amanat F., Thapa M., Lei T., Ahmed S.M.S., Adelsberg D.C., Carreño J.M., Strohmeier S., Schmitz A.J., Zafar S., Zhou J.Q., et al. SARS-CoV-2 MRNA Vaccination Induces Functionally Diverse Antibodies to NTD, RBD, and S2. Cell. 2021;184:3936–3948.e10. doi: 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Angyal A., Longet S., Moore S.C., Payne R.P., Harding A., Tipton T., Rongkard P., Ali M., Hering L.M., Meardon N., et al. T-Cell and Antibody Responses to First BNT162b2 Vaccine Dose in Previously Infected and SARS-CoV-2-Naive UK Health-Care Workers: A Multicentre Prospective Cohort Study. Lancet Microbe. 2022;3:e21–e31. doi: 10.1016/S2666-5247(21)00275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skelly D.T., Harding A.C., Gilbert-Jaramillo J., Knight M.L., Longet S., Brown A., Adele S., Adland E., Brown H., Tipton T., et al. Two Doses of SARS-CoV-2 Vaccination Induce Robust Immune Responses to Emerging SARS-CoV-2 Variants of Concern. Nat. Commun. 2021;12:5061. doi: 10.1038/s41467-021-25167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grobben M., van der Straten K., Brouwer P.J., Brinkkemper M., Maisonnasse P., Dereuddre-Bosquet N., Appelman B., Lavell A.A., van Vught L.A., Burger J.A., et al. Cross-Reactive Antibodies after SARS-CoV-2 Infection and Vaccination. eLife. 2021;10:e70330. doi: 10.7554/eLife.70330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal Human Coronavirus Antibodies Are Boosted upon SARS-CoV-2 Infection but Not Associated with Protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sermet-Gaudelus I., Temmam S., Huon C., Behillil S., Gajdos V., Bigot T., Lurier T., Chrétien D., Backovic M., Delaunay-Moisan A., et al. Prior Infection by Seasonal Coronaviruses, as Assessed by Serology, Does Not Prevent SARS-CoV-2 Infection and Disease in Children, France, April to June 2020. Eurosurveillance. 2021;26:2001782. doi: 10.2807/1560-7917.ES.2021.26.13.2001782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gombar S., Bergquist T., Pejaver V., Hammarlund N.E., Murugesan K., Mooney S., Shah N., Pinsky B.A., Banaei N. SARS-CoV-2 Infection and COVID-19 Severity in Individuals with Prior Seasonal Coronavirus Infection. Diagn. Microbiol. Infect. Dis. 2021;100:115338. doi: 10.1016/j.diagmicrobio.2021.115338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qian Z., Ou X., Góes L.G.B., Osborne C., Castano A., Holmes K.V., Dominguez S.R. Identification of the Receptor-Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1. J. Virol. 2015;89:8816–8827. doi: 10.1128/JVI.03737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ou X., Guan H., Qin B., Mu Z., Wojdyla J.A., Wang M., Dominguez S.R., Qian Z., Cui S. Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1. Nat Commun. 2017;8:15216. doi: 10.1038/ncomms15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b Vaccines Protect Rhesus Macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 73.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An MRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawrenz J., Xie Q., Zech F., Weil T., Seidel A., Krnavek D., van der Hoek L., Münch J., Müller J.A., Kirchhoff F. Severe Acute Respiratory Syndrome Coronavirus 2 Vaccination Boosts Neutralizing Activity Against Seasonal Human Coronaviruses. Clin. Infect. Dis. 2022;75:e653–e661. doi: 10.1093/cid/ciac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simula E.R., Manca M.A., Jasemi S., Uzzau S., Rubino S., Manchia P., Bitti A., Palermo M., Sechi L.A. HCoV-NL63 and SARS-CoV-2 Share Recognized Epitopes by the Humoral Response in Sera of People Collected Pre- and during CoV-2 Pandemic. Microorganisms. 2020;8:1993. doi: 10.3390/microorganisms8121993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan K.H., Cheng V.C.C., Woo P.C.Y., Lau S.K.P., Poon L.L.M., Guan Y., Seto W.H., Yuen K.Y., Peiris J.S.M. Serological Responses in Patients with Severe Acute Respiratory Syndrome Coronavirus Infection and Cross-Reactivity with Human Coronaviruses 229E, OC43, and NL63. Clin. Vaccine Immunol. 2005;12:1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin H.-X., Feng Y., Tu X., Zhao X., Hsieh C.-H., Griffin L., Junop M., Zhang C. Characterization of the Spike Protein of Human Coronavirus NL63 in Receptor Binding and Pseudotype Virus Entry. Virus Res. 2011;160:283–293. doi: 10.1016/j.virusres.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heinz F.X., Stiasny K. Distinguishing Features of Current COVID-19 Vaccines: Knowns and Unknowns of Antigen Presentation and Modes of Action. npj Vaccines. 2021;6:1–13. doi: 10.1038/s41541-021-00369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martínez-Flores D., Zepeda-Cervantes J., Cruz-Reséndiz A., Aguirre-Sampieri S., Sampieri A., Vaca L. SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dolgin E. Pan-Coronavirus Vaccine Pipeline Takes Form. Nat. Rev. Drug Discov. 2022;21:324–326. doi: 10.1038/d41573-022-00074-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets are available by contacting the corresponding author upon kind request.