Abstract
The oral bacterium Actinobacillus actinomycetemcomitans is implicated as a causative agent of localized juvenile periodontitis (LJP). A. actinomycetemcomitans is classified into five serotypes (a to e) corresponding to five structurally and antigenically distinct O polysaccharide (O-PS) components of their respective lipopolysaccharide molecules. Serotype b has been reported to be the dominant serotype isolated from LJP patients. We determined the lipopolysaccharide O-PS structure from A. actinomycetemcomitans CU1000, a strain isolated from a 13-year-old African-American female with LJP which had previously been classified as serotype b. The O-PS of strain CU1000 consisted of a trisaccharide repeating unit composed of l-rhamnose and 2-acetamido-2-deoxy-d-galactose (molar ratio, 2:1) with the structure →2)-α-l-Rhap-(1–3)-2-O-(β-d-GalpNAc)-α-l-Rhap-(1→. O-PS from strain CU1000 was structurally and antigenically distinct from the O-PS molecules of the five known A. actinomycetemcomitans serotypes. Strain CU1000 was mutagenized with transposon IS903φkan, and three mutants that were deficient in O-PS synthesis were isolated. All three transposon insertions mapped to a single 1-kb region on the chromosome. The DNA sequence of a 13.1-kb region surrounding these transposon insertions contained a cluster of 14 open reading frames that was homologous to gene clusters responsible for the synthesis of A. actinomycetemcomitans serotype b, c, and e O-PS antigens. The CU1000 gene cluster contained two genes that were not present in serotype-specific O-PS antigen clusters of the other five known A. actinomycetemcomitans serotypes. These data indicate that strain CU1000 should be assigned to a new A. actinomycetemcomitans serotype, designated serotype f. A PCR assay using serotype-specific PCR primers showed that 3 out of 20 LJP patients surveyed (15%) harbored A. actinomycetemcomitans strains carrying the serotype f gene cluster. The finding of an A. actinomycetemcomitans serotype showing serological cross-reactivity with anti-serotype b-specific antiserum suggests that a reevaluation of strains previously classified as serotype b may be warranted.
Actinobacillus actinomycetemcomitans is a gram-negative, capnophilic coccobacillus that colonizes the human oral cavity (25). A. actinomycetemcomitans is implicated in the etiology of localized juvenile periodontitis (LJP), a severe and rapid form of periodontal disease that affects more than 70,000 predominantly African-Americans in the United States annually (63). A. actinomycetemcomitans is also a member of the clinically important HACEK group of bacteria (Haemophilus parainfluenzae, Haemophilus aphrophilus, and Haemophilus paraphrophilus, A. actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella kingae) (4). These fastidious, gram-negative organisms are considered normal flora of the human oral cavity but can infrequently enter the submucosa and cause extraoral infections including endocarditis, bacteremias, and abscesses (16, 23, 29, 33, 55).
Serological investigations with polyclonal rabbit antisera have led to the recognition of five A. actinomycetemcomitans serotypes, designated a to e (1, 15, 39, 44, 47, 56, 64) Approximately 3 to 9% of A. actinomycetemcomitans isolates do not react with any of the five serotype-specific antisera (40). Different serotypes have been shown to be associated with periodontal health, periodontitis, and nonoral infections (1, 2, 7, 17, 65) and with different racial, ethnic, and geographic populations (18, 19, 20, 30, 31, 32), suggesting that serotype-specific strain differences may be responsible for host specificity and virulence. The immunodominant outer membrane antigen of A. actinomycetemcomitans is located in the high-molecular-mass O polysaccharide (O-PS) component of lipopolysaccharide (LPS) (6, 38, 49, 58). A. actinomycetemcomitans LPS may be an important virulence factor (9, 12, 24, 64) and vaccine candidate (51).
The chemical structures of the A. actinomycetemcomitans serotype a to e antigenic O-PS molecules are known (42, 43), and the DNA sequences of the genes involved in their synthesis have been described (35, 36, 50, 61, 62). In this report we analyze the O-PS antigen from A. actinomycetemcomitans CU1000, a strain isolated from a 13-year-old African-American female with classical symptoms of LJP (12). We show that strain CU1000 contains an O-PS component that is structurally, antigenically, and genetically distinct from those of the five known A. actinomycetemcomitans serotypes. We propose that strain CU1000 be assigned to a new A. actinomycetemcomitans serotype, designated serotype f.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
A. actinomycetemcomitans strain CU1000 was isolated in 1992 in New York City from the first-molar site of a 13-year-old, medically healthy, African-American female with classical symptoms of LJP (12). Strain CU1000 was the source of LPS and O-PS for structural analysis. Strain CU1000N, an isogenic nalidixic acid-resistant derivative of CU1000 (22), was used for transposon mutagenesis. A. actinomycetemcomitans strains SUNYab75 (ATCC 43717 [American Type Culture Collection]), Y4 (ATCC 43718), NJ2700 (D. H. Fine laboratory collection), IDH781 (provided by H. Reynolds, State University of New York at Buffalo), and NJ9500 (D. H. Fine laboratory collection) are serotypes a, b, c, d, and e, respectively. A total of 20 A. actinomycetemcomitans clinical strains isolated from 20 different LJP patients (average age, 18.4 years; range, 6 to 40 years) were from the laboratory collection of D. H. Fine. All A. actinomycetemcomitans strains were preserved as −70°C frozen stocks in 10% dimethyl sulfoxide. Bacteria were grown on Trypticase soy agar (TSA; BD Biosystems) supplemented with 6 g of yeast extract and 8 g of glucose/liter. Broth cultures were in Trypticase soy broth (TSB) supplemented as described above. For large-scale preparation of LPS, strain CU1000 was grown in brain heart infusion broth (Difco) incubated with constant agitation (200 rpm). All A. actinomycetemcomitans strains were grown at 37°C in 10% CO2. When appropriate, media were supplemented with 3 μg of chloramphenicol, 20 μg of nalidixic acid, or 20 μg of kanamycin/ml or 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Escherichia coli strain SK338 (22) was used for transposon mutagenesis of A. actinomycetemcomitans. E. coli SK338 harbors plasmids pVJT128 (carrying a chloramphenicol resistance marker and the cryptic IS903φkan transposon) and pRK21761 (an oriT-defective derivative of RK2) and was grown at 37°C in Luria-Bertani medium (48) supplemented with chloramphenicol and kanamycin (50 μg/ml each).
Large-scale preparation of LPS and O-PS for structural analysis.
Bacterial cultures were chilled to 4°C, killed by the addition of phenol to a final concentration of 2%, and harvested by Sharples continuous centrifugation. The collected bacteria (41 g wet weight) were washed in 0.9% NaCl and extracted with 50% aqueous phenol for 12 min at 60 to 70°C. Following the addition of 2 volumes of water, the cooled mixture (4°C) was subjected to low-speed centrifugation to remove solid material and the clear extract was dialyzed against running tap water until it was free of phenol. The lyophilized dialysate was dissolved in 10 mM Tris (pH 8.0, 35 ml) and treated sequentially with 200 μg of DNase and 60 μg of RNase/ml for 3 h at 37°C, followed by proteinase K (Sigma) at a concentration of 200 μg/ml for a further 10 h. The digest was dialyzed against tap water, and the dialysate was subjected to ultracentrifugation at 105,000 × g at 4°C for 12 h to yield LPS as a gel pellet, which was dissolved in distilled water and lyophilized (402 mg). Solutions of the LPS (2% [wt/vol]) in 2% acetic acid were hydrolyzed at 100°C for 2 h, and, following removal of precipitated lipid A, the lyophilized water-soluble products were dissolved in 0.05 M pyridinium acetate (pH 4.6, 4 ml) and chromatographed on a Sephadex G-50 column (3 by 92 cm) equilibrated with the same buffer. Collected samples (10 ml) were analyzed for neutral glycose by the phenol-sulfuric acid method (8) and for 2-amino-2-deoxyhexose (14).
NMR spectroscopy.
Nuclear magnetic resonance (NMR) spectra were obtained with a Varian Inova 500 spectrometer equipped with standard Varian software. Carbohydrate samples were exchanged twice with D2O and redissolved in 99.9% D2O. Measurements were made at 30°C. Proton spectra were obtained using a spectral width of 2.5 kHz and a 90° pulse. Chemical shifts are reported relative to those for internal acetone (2.225 ppm for 1H and 31.07 ppm for 13C). The two-dimensional (2D) homonuclear and heteronuclear experiments correlated spectroscopy (COSY [3]), nuclear Overhauser effect spectroscopy (NOESY [27]), and heteronuclear single quantum coherence (HSQC [26]) were performed under standard conditions.
GC.
Gas chromatography (GC) was performed with a Hewlett-Packard 5890A gas chromatograph fitted with a flame ionization detector by means of a capillary column (0.25 mm by 30 m; fused silica DB-17) and a temperature program (either 180 [delay of 2 min] to 240°C at 2°C/min for alditol acetate derivatives or 200 to 240°C at 1°C/min for methylated alditol acetate derivatives]. GC-mass spectrometry (GC-MS) was performed under the same conditions using a Hewlett-Packard 5985B GC-MS system and an ionization potential of 70 eV. Retention times of derivatives were recorded relative to those for hexa-O-acetyl-d-glucitol (tG) or 1,5-di-O-acetyl-2,3,4,6-tetra-O-methyl-d-glucitol (tGM).
PC.
Paper chromatography (PC) was performed on water-washed Whatman 3MM paper using butan-1-ol–pyridine–water (10:3:3 by volume) for neutral sugars and butan-1-ol–ethanol–water (4:1:5; top layer) for amino sugars. Detection was made with periodate-alkaline AgNO3 spray reagent, and mobilities are quoted relative to those for d-galactose (RGal).
Gel electrophoresis.
Deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE) was performed on separating gels of 14% acrylamide and 9% sodium deoxycholate. Gels were silver stained after oxidation with periodate (54).
O-PS deamination.
O-PS (60 mg) was N deacetylated by treatment with 4 M NaOH (5 ml) containing NaBH4 (3 mg) at 100°C for 3 h. The diluted solution was neutralized in the cold with acetic acid, dialyzed against water, and lyophilized. The product (42 mg) in 30% acetic acid (4 ml) was treated with 5% aqueous sodium nitrite (4 ml) and kept at 20°C for 45 min. The reaction mixture was passed through a column of Rexyn 101(H+) ion-exchange resin (6 ml), and the concentrated eluate was collected as the void volume fraction eluting from a Sephadex G-50 column (20 mg).
Small-scale preparation of LPS for enzyme-linked immunosorbent assays (ELISAs).
LPS was prepared from test strains using the hot aqueous phenol method (57) with slight modifications. Briefly, cells from TSB cultures were washed two times with phosphate-buffered saline and pelleted in a 1.5-ml microcentrifuge tube (5,000 × g for 1 min). One hundred microliters of water (68°C) per 10 mg of cells (wet weight) was added, and the cell pellet was homogenized with a disposable polypropylene pellet pestle (Kontes). An equal volume of phenol (68°C) was added, and the tube was then vortexed for 5 s and incubated at 65°C for 15 min and at 10°C for 10 min. After centrifugation (5,000 × g for 5 min), the upper aqueous phase containing crude LPS was transferred to a new tube and diluted with 3 volumes of water.
ELISAs.
All of the following steps were carried out at room temperature. The wells of a flat-bottom Nunc-Immuno microtiter plate were coated (in triplicate) with 50 μl of LPS solution overnight, washed three times with water, and blocked for 30 min with blocking buffer (phosphate-buffered saline containing 0.05% Tween 20, 1 mM EDTA, and 0.25% bovine serum albumin). The wells were washed two times with water, and 50 μl of primary anti-Y4 (serotype b) rabbit antiserum (provided by J. Zambon, State University of New York at Buffalo) or anti-CU1000 (serotype f) rabbit antiserum (12) (both diluted 1:1,000 in blocking buffer) was added. The plate was incubated for 2 h, and the wells were then washed two times with water, blocked for an additional 10 min, and rewashed. Fifty microliters of a solution of goat anti-rabbit immunoglobulin M (IgM) plus IgG alkaline phosphatase-conjugated antibody (Fisher; diluted 1:2,000 in blocking buffer) was added to each well, and the plate was incubated an additional 2 h. The wells were washed two times with water, blocked for an additional 10 min, and rewashed. Seventy-five microliters of a 1-mg/ml solution of p-nitrophenyl phosphate (in 10% diethanolamine–0.05 mM MgCl2, pH 9.8) was added to each well, and the plate was incubated for 45 min. The end product was measured in a Bio-Rad model 3550 microplate reader set at 405 nm.
IS903φkan mutagenesis.
Donor strain (E. coli SK338; washed three times with phosphate-buffered saline) and recipient strain (A. actinomycetemcomitans CU1000N) cells were mixed in a ratio of 1:10, spotted on TSA plates, and incubated at 37°C in 10% CO2 for 16 h. The mating mixture was scraped from the plate with a sterile loop, resuspended in 1 ml of TSB, and plated on TSA supplemented with chloramphenicol and nalidixic acid to select for transconjugants. Approximately 40 colonies of A. actinomycetemcomitans transconjugants carrying plasmid pVJT128 were resuspended in 1 ml of TSB, plated on TSA containing chloramphenicol and IPTG, and grown for 24 h to allow transposition. Colonies were pooled and plated on TSA supplemented with kanamycin to select for transposon mutants. Colony morphology of the transposon mutants was observed after 2 days of growth using an Olympus IMT-2 inverted microscope at ×40. Three mutants (JK1002, JK1005, and JK1022) that displayed a colony morphology that was rougher than the wild-type CU1000 rough-colony phenotype (12) were streaked to purity and frozen.
Cloning and sequencing the IS903φkan transposon insertion sites.
Genomic DNA was prepared from mutants JK1002, JK1005, and JK1022 using a DNeasy tissue kit (Qiagen) according to the instructions provided by the manufacturer. Inverse PCR (53) was performed in 0.5-ml thin-wall tubes using primers kanStart (5′-GTTTCCCGTTGAATATGGCTGGG-3′) and kanStop (5′-GCAGTTTCATTTGATGCTCGA-3′), which hybridize within the transposon and which are oriented upstream and downstream, respectively (52). The reaction mixture contained 50 pmol of each primer, 10 μl of 10× PCR buffer, 1 mM deoxynucleoside triphosphates, 2.5 U of Taq DNA polymerase (PE Biosystems), and 100 ng of HindIII-digested and ligated genomic DNA as a target in a 100-μl reaction volume overlaid with 100 μl of light mineral oil. Thirty cycles of PCR were performed under the following conditions: denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. In this experiment, DNAs from mutants JK1005 and JK1022 both yielded a 1.2-kb PCR product. The ends of the JK1005 and JK1022 PCR products were made flush using E. coli DNA polymerase (Klenow fragment) and ligated into the EcoRV site of plasmid LITMUS28 (New England BioLabs) as described previously (48). Plasmid DNAs were prepared using a QIAprep spin miniprep kit (Qiagen) and subjected to DNA sequence analysis on an ABI model 377 PRISM automated sequencer. The DNA sequences of these two fragments indicated that the transposons in mutants JK1005 and JK1022 inserted 100 bp apart into the same 1.2-kb HindIII fragment and in the same orientation (see Fig. 6). PCR analysis of genomic DNA from mutant JK1002 using primers kanStop (see above) and P1 (5′-TTAAACATAATGAGTAACCC-3′), which hybridizes within the HindIII fragment and which is oriented upstream (see Fig. 6), showed that the transposon in JK1002 inserted 900 bp upstream from the transposon in JK1005 and in the same orientation as the transposons in JK1005 and JK1022.
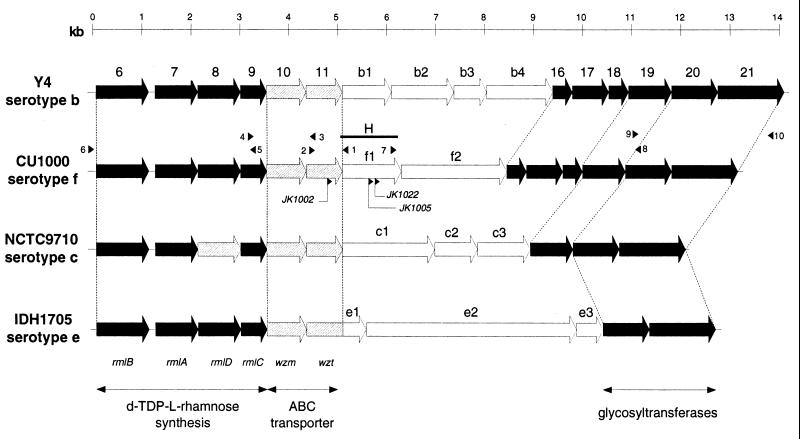
FIG. 6.
Comparison of the serotype-specific O-PS gene clusters from A. actinomycetemcomitans strains Y4, CU1000, NCTC9710, and IDH1705. Top line, scale in kilobases (kb). Arrows, ORFs and direction of transcription. Black arrows, ORFs with greater than 90% identity among strains; hatched arrows, ORFs with 40 to 60% identity; open arrows, ORFs with no corresponding gene in the other strains. Dashed lines demarcate homologous regions. ORFs numbered 6 to 11 and 16 to 21 correspond to ORFs 6 to 11 and 16 to 21 in reference 62, respectively. Bar H, position of the HindIII fragment amplified by inverse PCR. Numbered arrowheads above the CU1000 sequence, position and orientation of primers P1 to P10; arrowheads below CU1000 ORF11 and ORFf1, insertion sites and direction of transcription of transposon IS903φkan insertion mutations in JK1002, JK1005, and JK1022. The names and functions of genes shared by all four clusters are indicated at the bottom.
Cloning and sequencing the CU1000 O-PS gene cluster.
Primer pairs P1 (see above) and P2 (5′-GGATGATATTGTAGTTAATG-3′), P3 (5′-CATTAACTACAATATCATCC-3′) and P4 (5′-GTAAACCGAGTTTATCGGC-3′), and P5 (5′-CGATAAACTCGGTTTAGGATA-3′) and P6 (5′-CAAGAYTTAYGGCTGGTAAG-3′) were used to amplify by PCR three overlapping fragments (755 bp, 1.3 kb, and 3.4 kb, respectively) upstream from the transposon insertion site in JK1002 (Fig. 6). Primer pairs P7 (5′-GGATTAGAAGAAGATATTGG-3′) and P8 (5′-CTTGCCATTGCTGGTCTGCG-3′), and P9 (5′-ACAACGCAGACCAGCAATGG-3′) and P10 (5′-AAACTCCGCCCAAATCGGTGC-3′) were used to amplify two overlapping fragments (5.2 and 2.9 kb, respectively) downstream from the JK1022 transposon insertion site. PCR conditions were as described above, except that 5 ng of CU1000 genomic DNA was used as a target. All five of these PCR products were cloned into the EcoRV site of LITMUS28 and subjected to DNA sequence analysis as described above. Universal and internal primers were used to sequence both strands of all plasmids.
Serotype-specific PCR assay.
Four PCRs were performed on each clinical isolate (Table 1). All reaction mixtures contained 50 pmol of each of the indicated primers and 3 ng of genomic DNA in a 100-μl volume as described above. PCR conditions were 30 cycles at 94°C for 30 s and 55°C for 30 s. Reaction 1 was used to detect serotype b, c, and f sequences in a single multiplex reaction mixture containing primers P11, P12, P13, and P14. Primer P14 is a universal forward primer that hybridizes to ORF11 of serotypes b, c, and f (Fig. 6). Primers P11, P12, and P13 are serotype b-, c-, and f-specific reverse primers, respectively, that hybridize to serotype-specific sequences at the 3′ end of ORF11. When combined with P14, primers P11, P12, and P13 produce PCR products of 333, 268, and 232 bp, respectively. Reaction 2 was used to detect serotype a with primers that hybridize to ORF8 of the serotype a gene cluster (50) and produce a 293-bp PCR product. Reaction 3 detected serotype d with primers that hybridize to ORF9 of the serotype d gene cluster (35) and produce a 411-bp PCR product. Reaction 4 detected serotype e with primers that hybridize to ORFe3 of the serotype e gene cluster (see Fig. 6) (61) and produce a 311-bp PCR product. A 10-μl aliquot of each reaction mixture was electrophoresed through a 5% acrylamide–0.17% bisacrylamide gel in 1× Tris-borate-EDTA buffer, and the PCR products were visualized by staining with ethidium bromide (48). Assays were validated with DNAs from strains SUNYab75, Y4, NJ2700, IDH781, NJ9500, and CU1000 (serotypes a, b, c, d, e, and f, respectively). DNA from each of these strains produced a single PCR product of the predicted size in the appropriate reaction, and no DNA produced a PCR product in more than one reaction. Results of the serotype-specific PCR assay were consistent with those of conventional seroclassification using serotype-specific rabbit antisera (data not shown).
TABLE 1.
A. actinomycetemcomitans serotype-specific PCR assay
| Reaction | Primer | Sequencea (5′–3′) | Serotype | PCR product size (bp) | Region amplifiedb | GenBank accession no. |
|---|---|---|---|---|---|---|
| 1 | P11 | TCTCCACCATTTTTGAGTGG | b | 333 | 12780–13112 | AB002668 |
| P12 | GAAACCACTTCTATTTCTCC | c | 268 | 12808–13075 | AB010415 | |
| P13 | CCTTTATCAATCCAGACAGC | f | 232 | 4813–5044 | AF213680 | |
| P14 | ARAAYTTYTCWTCGGGAATG | |||||
| 2 | P15 | TGGGTCATGGGAGGTACTCC | a | 293 | 8021–8313 | AB046360 |
| P16 | GCTAGGAACAAAGCAGCATC | |||||
| 3 | P17 | TGGAACGGGTATGGGAACGG | d | 411 | 11462–12034 | AB041226 |
| P18 | GGATGCTCATCTAGCCATGC | |||||
| 4 | P19 | ATTCCAGCCTTTTGGTTCTC | e | 311 | 17118–17427 | AB030032 |
| P20 | TGGTCTGCGTTGTAGGTTGG |
R = A or G; Y = C or T; W = A or T.
GenBank nucleotide sequence coordinates.
DNA sequence accession number.
The DNA sequence of the CU1000 O-PS gene cluster was deposited in GenBank under accession no. AF213680.
RESULTS
Chemical analysis of O-PS from A. actinomycetemcomitans strain CU1000.
A. actinomycetemcomitans strain CU1000 cells were extracted by a modified hot aqueous phenol method (21) and, following sequential treatment with RNase, DNase, and protease K and ultracentrifugation, yielded a precipitated gel, which was dissolved in water and lyophilized to yield LPS (10% yield based on dry cell weight). The LPS on DOC-PAGE analysis gave a typical smooth-type LPS ladder pattern with a band spacing indicative of an O-PS composed of a repeating trisaccharide unit (41).
The LPS was hydrolyzed by hot dilute acetic acid to give an insoluble lipid A (21%) and a water-soluble product, which on Sephadex G-50 gel filtration chromatography yielded a high-molecular-weight O-PS fraction (40%; Kav, 0.02), a core oligosaccharide (14%; Kav, 0.14), and a low-molecular-weight fraction containing 3-deoxy-d-manno-oct-2-ulosonic acid and phosphate (12%; Kav, 0.95).
The O-PS had the following characteristics: [α]D, −36.8o (c 0.3, water); analytical composition results for C, 41.40%; H, 6.04%; N, 1.84%; ash, nil. The O-PS on hydrolysis and GC-MS of derived alditol acetate derivatives was found to be composed of rhamnose and 2-amino-2-deoxy-galactose in the molar ratio of 2:1. The two component glycoses were separated by preparative PC, and the collected glycoses were identified as l-rhamnose and 2-amino-2-deoxy-d-galactose. The l-rhamnose fraction (RGal, 2.90) had an [α]D of +7o (c 0.2, water) on reduction (sodium borodeuteride [NaBD4]), and acetylation afforded penta-O-acetyl-l-rhamnitol-1-d, which gave a single peak on GC (tG, 0.60) corresponding in retention time and MS analysis results to that of a reference sample; on methanolysis (2.5% methanol–HCl, 1 h, 100°C) the fraction gave methyl α-l-rhamnopyranoside having an [α]D of −60.2o (c 0.1, water). The aminoglycose fraction was characterized as its N-acetyl derivative 2-acetamido-2-deoxy-d-galactose, having an [α]D of +82o (c 0.3, water), and on reduction (NaBD4) and acetylation afforded 1,3,4,5,6-penta-O-acetyl-2-acetamido-2-deoxy-d-galactitol-1-d with a GC retention time (tG, 1.33) and MS spectrum identical with those of a reference sample. The 2-acetamido-2-deoxy-d-galactose was also characterized by GC-MS as its trimethylsilyl 2-acetamido-2-deoxy-3,4,6-tri-O-trimethylsilyl-α-d-galactopyranoside and -β-d-galactopyranoside derivatives (41).
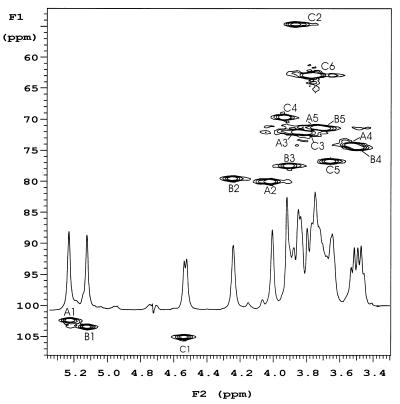
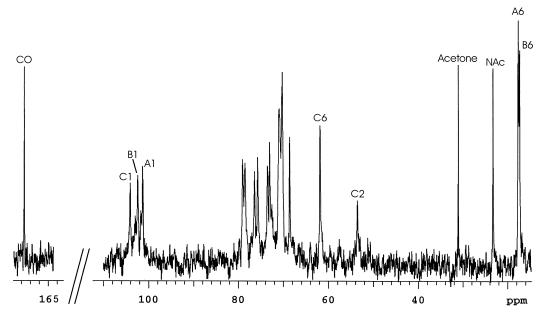
The 1H NMR spectrum of the O-PS (Fig. 1) showed inter alia signals for three anomeric protons at 5.220 (J1,2, ∼1 Hz), 5.123 (J1,2, 2.2 Hz), and 4.550 ppm (J1,2, 7.3 Hz), a signal at 2.073 ppm (3H) from an acetamido substituent, and signals at 1.332 (3H) and 1.640 ppm (3H) arising from the 6-deoxy functions of the identified l-Rha constituents. The 13C NMR spectrum of the O-PS (Fig. 2) showed inter alia anomeric carbon signals at 103.99 (JC1-H1, 161 Hz), 102.34 (JC1-H1, 175 Hz), and 101.28 ppm (JC1-H1, 174 Hz), carbon signals (underlined) at 23.27 (NHCOCH3), 175.25 (NHCOCH3), and 53.54 ppm (C-2) arising from the 2-acetamido-2-deoxy function of the identified d-GalNAc component residue and two methyl signals at 17.66 and 17.33 ppm arising from the two l-Rha residues. The composition and NMR data were consistent with the O-PS being a polymer of a regular repeating trisaccharide unit.
FIG. 1.
Partial HSQC and one-dimensional proton NMR spectra of the LPS O-PS from A. actinomycetemcomitans strain CU1000 showing H/C cross-peak assignments for glycose residues A [-2)-α-l-Rhap-(1-], B [-2,3)-α-l-Rhap-(1-], and C [β-d-GalpNAc-(1-].
FIG. 2.
13C NMR spectrum of the LPS O-PS from A. actinomycetemcomitans strain CU1000 showing carbon atom resonances for glycose residues A [-2)-α-l-Rhap-(1-], B [-2,3)-α-l-Rhap-(1-], and C [β-d-GalpNAc-(1-]. NAc, N-acetyl.
GC-MS analysis of the reduced (NaBD4) and acetylated hydrolysis products of methylated O-PS identified 1,2,5- tri-O-acetyl-3,4-di-O-methyl-l-rhamnitol-1-d (tGM, 0.87), 1,2,3,5-tetra-O-acetyl-4-O-methyl-l-rhamnitol-1-d (tGM,1.13), and 1,5-di-O-acetyl-2-deoxy-3,4,6-tri-O-methyl-2-(N-methyl-acetamido)-d-galactitol-1-d (tGM, 2.69) (1:1:1), showing the repeating trisaccharide unit to contain a 2-substituted l-Rhap residue, a 2,3-disubstituted l-Rhap residue, and a terminal d-GalpNAc residue glycosidically linked to the branched l-Rhap residue.
The complete structural characterization of the O-PS, involving linkage position and anomeric configuration assignments, was obtained by the application of (2D) 1H and 13C NMR spectroscopy. The anomeric proton signals of the glycosyl residue in the NMR spectrum of the O-PS were arbitrarily assigned A to C in order of their decreasing chemical shifts, and their subspectra were identified from H-1 connectivities to corresponding ring proton resonances and the magnitude of vicinal coupling constants (Table 1). The proton subspectra of residues A and B were consistent with a manno configuration having small J1,2 (∼1 Hz) and J2,3 (∼4 Hz) coupling constants and a large J3,4 (9.3 Hz) coupling constant. Further consideration of the corresponding JC1-H1 values for residues A (174 Hz) and B (175 Hz) led to them both being identified as α-l-Rhap residues. Glycose C was identified as a β-d-GalpNAc residue from an HSQC spectrum (Fig. 1), in which H-2C correlated with the carbon resonance at 53.54 ppm (C-2C), and from its characteristic anomeric coupling constants JC1-H1 (160 Hz) and J1,2 (7.3 Hz).
The sequence and glycosidic linkage positions within the O-PS were established from observed transglycosidic nuclear Overhauser effects (NOEs), which were seen between H-1A (α-l-Rhap) and H-3B (α-l-Rhap) and its own H-2A, which, together with NOEs observed between H-1B and H-2A and its own H-2B, indicated that the O-PS had a linear backbone structure composed of a repeating disaccharide having the structure →2)-α-l-Rhap-(1→3)-α-l-Rhap-(1→.
An observed NOE between H-lC (β-d-GalpNAc) and H-2B, along with interresidue NOEs to its own H-2C, H-3C, and H-5C confirmed that residue C was a nonreducing β-d-GalpNAc end group glycosidically linked to residue B and that the repeating trisaccharide of the O-PS had the structure (I)
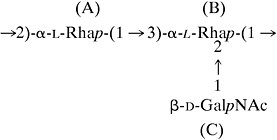 |
I |
Confirmatory proof of the above structure was obtained from nitrous acid degradation of the N-deacetylated O-PS to yield a high-molecular-weight homopolymer having an [α]D of −75o (c 1.0, water) and composed of only l-rhamnose. GC-MS analysis of the methylated l-rhamnan gave two products identified as 1,2,5-tri-O-acetyl-3,4-di-O-methyl-l-rhamnitol-1-d (tGM, 0.87) and 1,3,5-tri-O-acetyl-2,4-di-O-methyl-l-rhamnitol-1-d (tGM, 0.94) in the molar ratio 1:1, confirming the 2- and 3-monosubstitutions of the α-l-Rhap residues and also the substitution of the β-d-GalpNAc end group at the 2 position of the backbone 3-substituted α-l-Rhap residue (B) in the native O-PS. 2D NMR experiments (Table 2) on the N-deacetylated O-PS confirmed the repeating disaccharide nature of the l-rhamnan backbone chain and thus the proposed structure of the antigenic O chain of A. actinomycetemcomitans strain CU1000 LPS.
TABLE 2.
1H and 13C NMR chemical shifts for the LPS O-PS of A. actinomycetemcomitans CU1000a
| Glycose unit | Chemical shift (ppm) (coupling constant [Hz]) for:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H-1 (J1,2) | H-2 (J2,3) | H-3 (J3,4) | H-4 (J4,5) | H-5 | H-6 (J5,6) | C-1 (JC1-H1) | C-2 | C-3 | C-4 | C-5 | C-6 | |
| -2)-α-l-Rhap-(1- (A) | 5.222 (∼1) | 4.021 (∼4) | 3.842 (9.4) | 3.527 (9.3) | 3.738 | 1.342 (3.9) | 101.28 (174) | 78.93 | 70.88 | 73.24 | 70.16 | 17.66 |
| 2,3-)-α-l-Rhap-(1- (B) | 5.123 (2.2) | 4.249 (4.5) | 3.904 (9.4) | 3.513 (9.3) | 3.692 | 1.256 (4.3) | 102.34 (175) | 78.44 | 76.36 | 73.45 | 70.37 | 17.33 |
| β-d-GalpNAc-(1- (C) | 4.550 (7.3) | 3.863 (9.5) | 3.805 (∼4) | 3.933 (∼1) | 3.666 | 3.760 | 103.99 (161) | 53.44 | 71.03 | 68.57 | 75.64 | 61.80 |
Chemical shifts were recorded at 30°C.
Reactivity of CU000 LPS with anti-Y4 and anti-CU1000 rabbit sera.
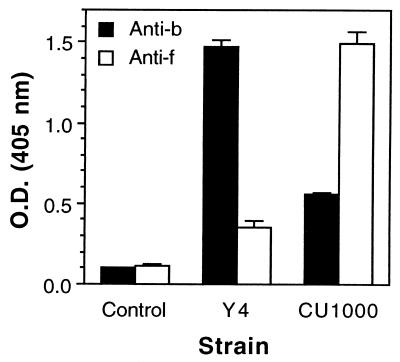
A. actinomycetemcomitans CU1000 was initially identified serologically as a serotype b strain (12). To demonstrate that LPS from strain CU1000 is antigenically distinct from that of a serotype b strain, LPS from strains CU1000 and Y4 (serotype b) were tested in an ELISA using anti-CU1000 and anti-Y4 rabbit antisera (Fig. 3). Binding of rabbit antisera to CU1000 and Y4 LPS was serotype specific, although a small amount of cross-reactivity was observed. This cross-reactivity could be due to epitopes residing in structures common to both CU1000 and serotype b O-PS antigens (see below).
FIG. 3.
Reactivities of anti-serotype b and anti-serotype f rabbit antisera with LPS prepared from A. actinomycetemcomitans strains Y4 (serotype b) and CU1000 (serotype f) as measured by ELISA. Control wells contained no LPS. The mean values (± standard errors) for triplicate wells are shown. O.D., optical density.
Construction of O-PS mutants in strain CU1000N.
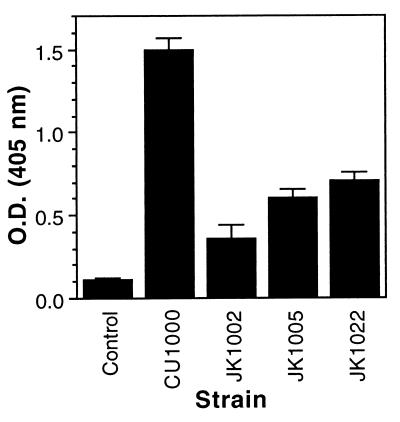
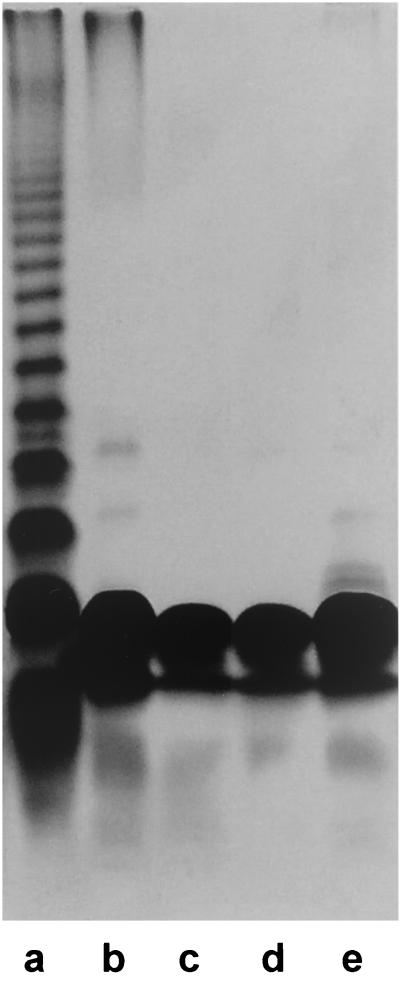
Gram-negative bacteria that are unable to synthesize the O-antigenic side chains of LPS generally exhibit a rough colony morphology (5, 45). A. actinomycetemcomitans strain CU1000N was mutagenized with transposon IS903φkan, which carries a cryptic kanamycin resistance gene that is expressed only after successful transposition into an actively transcribed gene (52), and three kanamycin-resistant mutants (out of ∼3,000) that displayed a dry, rough colony morphology on TSA plates were selected. All three mutants (designated JK1002, JK1005, and JK1022) exhibited wild-type growth rates and adherence properties in TSB (11) (data not shown). LPS preparations from all three mutants displayed reduced reactivity with anti-CU1000 rabbit antisera as measured by ELISA (Fig. 4). The residual antigenic cross-reactivity displayed by LPS prepared from the mutants may be due to reactivity of antibodies with partially assembled O-PS components or the presence of minor amounts of immunoreactive molecules that copurified with the LPS (49). LPS preparations from strain CU1000 and the three mutants were analyzed by DOC-PAGE (Fig. 5). Strain CU1000 produced a smooth-type LPS with extended O-antigen side chains, whereas LPS from the three mutants consisted of a core oligosaccharide with O-antigen side chains short or absent. These data were confirmed by analyzing hydrolyzed LPS preparations from CU1000 and the three mutants using Sephadex G-50 chromatography (data not shown). Strain CU1000 yielded an O-antigen fraction which was absent in the corresponding products of the mutants. Proton NMR spectra of core oligosaccharides from all three mutants were identical to those of strain CU1000 (data not shown). These data indicate that mutant strains JK1002, JK1005, and JK1022 are deficient in LPS O-antigen side chain biosynthesis.
FIG. 4.
Reactivities of anti-serotype f rabbit antisera with LPS prepared from A. actinomycetemcomitans strain CU1000 and three LPS mutants (JK1002, JK1005, and JK1022) as measured by ELISA. Control wells contained no LPS. The mean values (± standard errors) for triplicate wells are shown. O.D., optical density.
FIG. 5.
DOC-PAGE profiles of LPS from Salmonella enterica serovar Milwaukee (lane a) and A. actinomycetemcomitans strains CU1000 (lane b), JK1002 (lane c), JK1005 (lane d), and JK1022 (lane e). Each lane contained 10 μg of LPS in saline plus 0.002% bromphenol tracking dye. LPS was visualized by silver staining.
DNA sequence of the CU1000 O-PS gene cluster.
DNA sequence analysis indicated that the transposons in mutants JK1002, JK1005, and JK1022 inserted into a region that was homologous to the A. actinomycetemcomitans serotype b, c, and e O-PS gene clusters (36, 61, 62). Primers that hybridize to conserved sequences in the serotype b, c, and e O-PS gene clusters and flanking regions were used to amplify by PCR the region surrounding the transposon insertion sites from strain CU1000 as described in Materials and Methods. DNA sequence analysis of a 13.1-kb region surrounding the transposon insertion sites revealed a cluster of 14 closely spaced or overlapping open reading frames (ORFs) that were all transcribed in the same orientation and in the same direction as the transposon insertions in mutants JK1002, JK1005, and JK1022. The DNA sequences of the CU1000 gene cluster and predicted amino acid sequences were clearly homologous to those of the O-PS gene clusters from strains Y4, NCTC9710, and IDH1705 (Fig. 6), indicating that this region contained the serotype f O-PS gene cluster.
Comparison of the serotype b, c, e, and f O-PS gene clusters.
The serotype b, c, e, and f O-PS gene clusters each contained a set of four genes (rmlBADC) at the beginning of the cluster (ORF6, ORF7, ORF8, and ORF9 in that order) that encode the four enzymes responsible for the four-step biosynthesis of dTDP-l-rhamnose from glucose-1-phosphate. dTDP-l-rhamnose is the activated nucleotide sugar form of l-rhamnose, which is widely distributed in O antigens of gram-negative bacteria (28, 60). Homologous rmlBADC genes have been detected in a wide range of bacteria, although the gene order may vary from species to species (28). The rmlBADC genes were highly conserved (>90% amino acid identity) among A. actinomycetemcomitans serotypes b, c, e, and f, except for rmlD from serotype c, whose product showed only 60% amino acid identity to the rmlD gene products of the other three serotypes (35, 36). Although significantly divergent, both Y4 and NCTC9710 RmlD gene products are capable of converting dTDP-6-deoxy-l-lyxo-4-hexulose to dTDP-l-rhamnose, the last step in the dTDP-l-rhamnose pathway (34).
Two genes (wzm and wzt) that encode a hydrophobic integral-membrane protein component and a hydrophilic ATPase component, respectively, of an ATP-binding cassette (ABC) membrane transporter (10) immediately followed rmlBADC in the serotype b, c, e, and f O-PS gene clusters (ORF10 and ORF11 in that order; Fig. 6). These two genes showed 40 to 60% amino acid identity among the four serotypes. ABC transporters have been shown to be involved in the translocation of exopolysaccharides in a variety of gram-negative bacteria (46), and wzm and wzt have been shown to be necessary for the expression of A. actinomycetemcomitans serotype b, c, and e O-PS antigens in E. coli (36, 61, 62). One of the transposon mutations in A. actinomycetemcomitans strain CU1000 that resulted in reduced levels of LPS synthesis (mutant JK1002) was located in wzt.
The wzm and wzt genes were followed by a group of two to four genes that are unique to each serotype (Fig. 6). The O-PS gene cluster from strain CU1000 contained two unique genes, ORFf1 and ORFf2. ORFf1 encodes a putative glycosyltransferase that is homologous to the N terminus of the serotype c ORFc1 gene product (27% amino acid identity in a region of >300 residues). ORFc1 has been shown to be necessary for the expression of serotype c O-PS in E. coli (36). ORFf1 was also homologous (26 to 27% amino acid identity) to Streptococcus mutans rgpFc (59), Vibrio anguillarum virA (37), and gene y4gN from the Rhizobium sp. plasmid pNGR234a (13), all of which are involved in the expression of exopolysaccharide antigens. Two of the transposon mutations in A. actinomycetemcomitans strain CU1000 that resulted in reduced levels of LPS synthesis (those of JK1005 and JK1022) were located in ORFf1. ORFf2 has no known function and no homologues in the GenBank database, although homologues of ORFf2 were found in the unfinished genome sequences of Enterococcus faecalis and Clostridium acetobutylicum (http://www.ncbi.nlm.nih.gov /Microb_blast/unfinishedgenome.html). The function of these genes in not known. Among the genes unique to the serotype b, c, and e O-PS gene clusters are the following: ORFb3 (fcd), encoding dTDP-4-keto-6-deoxy-d-glucose reductase, which catalyzes the conversion of dTDP-4-keto-6-deoxy-d-glucose to dTDP-d-fucose, the activated nucleotide sugar form of the d-fucose present in serotype b O-PS (60); ORFc2 (tll), encoding dTDP-6-deoxy-l-lyxo-4-hexulose reductase, which catalyzes the conversion of dTDP-4-keto-l-rhamnose to dTDP-6-deoxy-l-talose, the activated nucleotide sugar form of the 6-deoxy-l-talose present in serotype c O-PS (34); ORFb1 and ORFe2, encoding putative glycosyltransferases (61, 62); ORFc3 and ORFe3, encoding putative acetyltransferases (36, 61); ORFb2 and ORFe1 (which is fused to ORF11), which are homologous to genes of unknown function present in O-antigen operons of other bacteria; ORFb4, which has no known function and no homologues in the GenBank database.
A set of three genes (ORF16, ORF17, and ORF18) was present in the serotype b and f O-PS gene clusters but not in the serotype c and e gene clusters (Fig. 6). The products of these three genes displayed >90% amino acid identity between serotypes b and f. ORF17 encodes a putative glycosyltransferase (62), ORF16 is homologous to genes of unknown function in other bacterial O-antigen operons, and ORF18 has no known function and no homologues in GenBank. Two or three highly conserved (>90% amino acid identity) putative glycosyltransferases (encoded by ORF19, ORF20, and ORF21) were located at the distal end of all four O-PS gene clusters (36, 61, 62).
The serotype b, c, and e O-PS gene clusters contain a region of low G+C content (ORF10 to ORF19 in serotypes b and c, and ORF10 to ORFe3 in serotype e) (36, 61, 62). These regions display <30% G+C, compared to 37 to 42% G+C for rmlBADC, 42 to 43% G+C for ORF20 and ORF21, and 46% for the entire A. actinomycetemcomitans genome (35). The corresponding region of the CU1000 O-PS gene cluster (ORF10 to ORF19) also displayed a low G+C content (29%), compared to 41% G+C for rmlBADC and 42% G+C for ORF20 and ORF21.
The DNA sequences of the regions flanking the CU1000 O-PS gene cluster were >90% identical to the corresponding regions flanking the serotype b, c, and e gene clusters (data not shown), indicating that the CU1000 cluster is located in the same site on the chromosome as the serotype b, c, and e O-PS clusters.
Frequency of serotype f strains among A. actinomycetemcomitans clinical isolates.
We measured the frequency of serotype a to f strains among a collection of 20 A. actinomycetemcomitans clinical strains isolated from LJP patients using a serotype-specific PCR assay (see Materials and Methods). This assay utilizes PCR primers that hybridize to unique sequences in the serotype a to f O-PS gene clusters. All 20 strains yielded a product with one of the serotype-specific primer pairs, and no strain yielded more than one product. The frequencies of the different serotypes among the 20 clinical isolates were as follows: a, 4 strains, 20%; b, 5 strains, 25%; c, 8 strains, 40%; f, 3 strains, 15%. No serotype d or e strains were detected. PCR analysis using primers that hybridize to serotype f-specific DNA sequences in the region between ORF9 and ORF20 revealed that the organization of the O-PS gene cluster in the three serotype f strains was identical to that of the CU1000 O-PS cluster (data not shown). These data indicate that the serotype f-specific O-PS cluster is a common and stable genetic locus in strains isolated from LJP patients.
DISCUSSION
Previous investigations with polyclonal rabbit sera have led to the recognition of five serotypes of A. actinomycetemcomitans, designated a to e. In this study we identified a sixth A. actinomycetemcomitans serotype, designated serotype f. We showed that serotype f O-PS was structurally, antigenically, and genetically distinct from those of the five known A. actinomycetemcomitans serotypes. We also showed that the gene cluster responsible for the synthesis of serotype f O-PS was a stable genetic locus that was evolutionarily related to the gene clusters responsible for the syntheses of A. actinomycetemcomitans serotype b, c, and e O-PS and that the serotype f gene cluster was present in 15% (3 of 20) of A. actinomycetemcomitans strains isolated from LJP patients.
Serotype f O-PS consisted of a trisaccharide repeating unit composed of l-rhamnose and 2-acetamido-2-deoxy-d-galactose (molar ratio, 2:1) having structure I. Although serotype f O-PS was structurally distinct from the O-PS molecules of the other five known A. actinomycetemcomitans serotypes (42, 43), the structure of serotype f O-PS was similar to the structure of serotype b O-PS, which consists of a repeating trisaccharide unit composed of d-Fuc, l-Rha, and d-GalNAc residues (molar ratio, 1:1:1) having the structure (II)
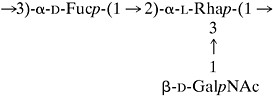 |
II |
Both serotype b and serotype f O-PS molecules contain a β-d-GalpNAc single nonreducing end group linked to a main linear polysaccharide backbone consisting of a disaccharide repeating unit (structures I and II). These common β-d-GalpNAc epitopes may account for the previously observed serological cross-reactivity between strain CU1000 and anti-serotype b-specific rabbit antiserum (12).
The gene cluster responsible for A. actinomycetemcomitans serotype f O-PS synthesis was located on a 13.1-kb segment of DNA containing 14 closely spaced or overlapping ORFs that was homologous to the gene clusters responsible for serotype b, c, and e O-PS synthesis (Fig. 6). All four of these gene clusters contain highly conserved sets of genes at the proximal and distal ends of the cluster (ORF6 to ORF9 and ORF20 and ORF21; Fig. 6) and a central region of lower G+C content, which includes two to four genes that are unique to each serotype. These observations suggest that these four serotypes evolved from a common ancestor and are consistent with a model of evolution of the A. actinomycetemcomitans serotype b, c, e, and f O-PS gene clusters which involves interspecific transfer of genes from species with low G+C content to A. actinomycetemcomitans (36, 62). The gene clusters responsible for the synthesis of A. actinomycetemcomitans serotype a and d O-PS are structurally unrelated to the serotype b, c, e, and f gene clusters (35, 50). The 13.9-kb segment of DNA containing the A. actinomycetemcomitans serotype d gene cluster is located 2 kb downstream from the site of the serotype b, c, e, and f gene clusters (35). The serotype d-specific gene cluster contains homologues of the rmlBADC genes from serotypes b, c, e, and f (>90% amino acid identity) and eight ORFs that are unique to serotype d. The A. actinomycetemcomitans serotype a gene cluster consists of a 12.9-kb segment of DNA, which is located in a different region of the chromosome from the serotype b to f gene clusters (50). The serotype a-specific gene cluster contains two genes that are homologues of wzm and wzt from serotypes b, c, e, and f (40 to 60% amino acid identity) and nine ORFs that are unique to serotype a.
Studies on the prevalence of the five previously identified A. actinomycetemcomitans serotypes have suggested that serotypes a to c occur much more frequently among oral isolates than serotypes d and e (44, 47) and that serotype b is the dominant A. actinomycetemcomitans serotype in subjects with LJP (1, 17, 18, 19, 64). Our results using a serotype-specific PCR assay to measure the frequency of the six A. actinomycetemcomitans serotypes among a collection of 20 strains isolated from LJP patients indicated that serotype a, b, c, and f isolates constitute the majority of A. actinomycetemcomitans oral isolates from LJP patients, with serotype f accounting for 15% of the strains. The identification of A. actinomycetemcomitans serotype f, which shows serological cross-reactivity with anti-serotype b-specific antiserum, suggests that a reevaluation of strains previously classified as serotype b may be warranted. Our findings indicate that serotype f may account for some strains that have been previously classified as serotype b and that serotype f may constitute a significant portion of strains isolated from LJP patients. Because of possible cross-reactivity between serotypes b and f, a serotype-specific PCR assay such as the one described in this report may be more accurate than conventional serotyping with polyclonal rabbit antisera for seroclassification of A. actinomycetemcomitans serotype b isolates.
TABLE 3.
1H and 13C NMR chemical shifts for the l-rhamnan derived from the LPS O-PS of A. actinomycetemcomitans CU1000a
| Glycose unit | Chemical shift (ppm) (coupling constant [Hz]) for:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H-1 (J1,2) | H-2 | H-3 | H-4 | H-5 | H-6 | C-1 (JC1-H1) | C-2 | C-3 | C-4 | C-5 | C-6 | |
| -2)-α-l-Rhap-(1- | 5.199 (∼1) | 4.087 | 3.960 | 3.532 | 3.850 | 1.350 | 101.82 (172) | 78.79 | 70.64 | 72.97 | 70.09 | 17.57 |
| -3)-α-l-Rhap-(1- | 4.963 (∼1) | 4.170 | 3.853 | 3.566 | 3.770 | 1.322 | 102.82 (167) | 70.64 | 78.33 | 72.44 | 70.09 | 17.47 |
Chemical shifts were recorded at 30°C.
ACKNOWLEDGMENTS
We thank Douglas Griffith for the fermenter production of bacterial cells, Ken Chan for GC-MS analyses, Aseel Toni for help with ELISAs, Paul Goncharoff and Helen Schreiner for helpful discussions, and Robert Donnelly, Kishore Kuppasani, and Anindita Sarangi of the Molecular Resource Facility, New Jersey Medical School, for assistance with DNA sequencing.
REFERENCES
- 1.Asikainen S, Chen C, Slots J. Actinobacillus actinomycetemcomitans genotypes in relation to serotypes and periodontal status. Oral Microbiol Immunol. 1995;10:65–68. doi: 10.1111/j.1399-302x.1995.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 2.Asikainen S, Lai C-H, Alaluusua S, Slots J. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral Microbiol Immunol. 1991;6:115–118. doi: 10.1111/j.1399-302x.1991.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 3.Bax A, Freeman R, Morris G. Correlation of proton chemical shifts by two-dimensional Fourier transform NMR. J Magn Reson. 1981;42:164–168. [Google Scholar]
- 4.Berbari E F, Cockerill F R, Stickelberg J M. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin Proc. 1997;72:532–542. doi: 10.4065/72.6.532. [DOI] [PubMed] [Google Scholar]
- 5.Brink B A, Miller J, Carlson R W, Noel K D. Expression of Rhizobium leguminosarum CFN42 genes for lipopolysaccharide in strains derived from different R. leguminosarum soil isolates. J Bacteriol. 1990;172:548–555. doi: 10.1128/jb.172.2.548-555.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Califano J V, Schenkein H A, Tew J G. Immunodominant antigens of Actinobacillus actinomycetemcomitans serotype b in early-onset periodontitis patients. Oral Microbiol Immunol. 1992;7:65–70. doi: 10.1111/j.1399-302x.1992.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 7.Dogan B, Saarela M H, Jousimies-Somer H, Alaluusua S, Asikainen S. Actinobacillus actinomycetemcomitans serotype e—biotypes, genetic diversity and distribution in relation to periodontal status. Oral Microbiol Immunol. 1999;14:98–103. doi: 10.1034/j.1399-302x.1999.140204.x. [DOI] [PubMed] [Google Scholar]
- 8.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric methods for the determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 9.Ebersole J L, Cappelli D. Gingival crevicular fluid antibody to Actinobacillus actinomycetemcomitans in periodontal disease. Oral Microbiol Immunol. 1994;9:335–344. doi: 10.1111/j.1399-302x.1994.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 10.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fine D H, Furgang D, Kaplan J, Charlesworth J, Figurski D H. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol. 1999;44:1063–1076. doi: 10.1016/s0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 12.Fine D H, Furgang D, Schreiner H C, Goncharoff P, Charlesworth J, Ghazwan G, Fitzgerald-Bocarsly P, Figurski D H. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999;145:1335–1347. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- 13.Freiberg C, Fellay R, Bairoch A, Broughton W T, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 14.Gatt R, Berman E R. A rapid method for the estimation of aminosugars on a micro scale. Anal Biochem. 1965;15:167–171. doi: 10.1016/0003-2697(66)90262-4. [DOI] [PubMed] [Google Scholar]
- 15.Gmür R, McNabb H, van Steenbergen T J M, Baehni P, Mombelli A, van Winkelhoff A J, Guggenheim B. Seroclassification of hitherto nontypeable Actinobacillus actinomycetemcomitans strains: evidence for a new serotype e. Oral Microbiol Immunol. 1993;8:116–120. doi: 10.1111/j.1399-302x.1993.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 16.Grance C J, Levitz R E, Katz-Pollak H, Brettman L R. Actinobacillus actinomycetemcomitans prosthetic valve endocarditis. Rev Infect Dis. 1988;10:922–929. doi: 10.1093/clinids/10.5.922. [DOI] [PubMed] [Google Scholar]
- 17.Haraszthy V I, Hariharan G, Tinoco E M B, Cortelli J R, Lally E T, Davis E, Zambon J J. Evidence for the role of highly leukotoxic Actinobacillus actinomycetemcomitans in the pathogenesis of localized juvenile and other forms of early-onset periodontitis. J Periodontol. 2000;71:912–922. doi: 10.1902/jop.2000.71.6.912. [DOI] [PubMed] [Google Scholar]
- 18.Haubek D, DiRienzo J M, Tinoco E M B, Westergaard J, Lopez N J, Chung C-P, Poulsen K, Kilian M. Racial tropism in a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J Clin Microbiol. 1997;35:3037–3042. doi: 10.1128/jcm.35.12.3037-3042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haubek D, Poulsen K, Westergaard J, Dahlèn G, Kilian M. Highly toxic clone of Actinobacillus actinomycetemcomitans in geographically widespread cases of juvenile periodontitis in adolescents of African origin. J Clin Microbiol. 1996;34:1576–1578. doi: 10.1128/jcm.34.6.1576-1578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hölttä P, Alahuusa S, Saarela M, Asikainen S. Isolation frequency and serotype distribution of mutans streptococci and Actinobacillus actinomycetemcomitans, and clinical periodontal status in Finnish and Vietnamese children. Scand J Dent Res. 1994;102:113–119. doi: 10.1111/j.1600-0722.1994.tb01165.x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson K G, Perry M B. Improved techniques for the preparation of bacterial lipopolysaccharides. Can J Microbiol. 1975;22:29–34. doi: 10.1139/m76-004. [DOI] [PubMed] [Google Scholar]
- 22.Kachlany S C, Planet P J, Bhattacharjee M K, Kollia E, DeSalle R, Fine D H, Figurski D H. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan A H, Weber D J, Oddone E Z, Perfect J R. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev Infect Dis. 1989;11:46–63. doi: 10.1093/clinids/11.1.46. [DOI] [PubMed] [Google Scholar]
- 24.Kiley P, Holt S C. Characterization of the lipopolysaccharide from Actinobacillus actinomycetemcomitans Y4 and N27. Infect Immun. 1980;30:862–873. doi: 10.1128/iai.30.3.862-873.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King E O, Tatum H W. Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. J Infect Dis. 1962;111:85–94. doi: 10.1093/infdis/111.2.85. [DOI] [PubMed] [Google Scholar]
- 26.Kogan G, Uhrin D. Current NMR methods in the structural elucidation of polysaccharides. In: Atta-w-Rahman, editor. New advances in chemistry. Amsterdam, The Netherlands: Harwood Academic Publishers; 2000. pp. 73–131. [Google Scholar]
- 27.Kumar A, Ernst R R, Wurthrich K. A 2-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complex proton-proton cross-relaxation in networks of biological molecules. Biochem Biophys Res Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Reeves P R. Genetic variation of dTDP-L-rhamnose pathway genes in Salmonella enterica. Microbiology. 2000;146:2291–2307. doi: 10.1099/00221287-146-9-2291. [DOI] [PubMed] [Google Scholar]
- 29.Martine B F, Derby B M, Budzilovich G N, Ransohoff J. Brain abscesses due to Actinobacillus actinomycetemcomitans. Neurology. 1967;17:833–837. doi: 10.1212/wnl.17.9.833. [DOI] [PubMed] [Google Scholar]
- 30.McNabb H, Mombelli A, Gmür R, Mathey-Dinç S, Lang N P. Periodontal pathogens in the shallow pockets of immigrants from developing countries. Oral Microbiol Immunol. 1992;7:267–272. doi: 10.1111/j.1399-302x.1992.tb00586.x. [DOI] [PubMed] [Google Scholar]
- 31.Mombelli A, Gmür R, Frey J, Meyer J, Zee K Y, Tam J O W, Lo E C M, DiRienzo J, Lang N P, Corbet E F. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in young Chinese adults. Oral Microbiol Immunol. 1998;13:231–237. doi: 10.1111/j.1399-302x.1998.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 32.Mombelli A, Gmür R, Lang N P, Corbet E, Frey J. Actinobacillus actinomycetemcomitans in Chinese adults. J Clin Periodontol. 1999;26:505–510. doi: 10.1034/j.1600-051x.1999.260803.x. [DOI] [PubMed] [Google Scholar]
- 33.Muhle L, Rau J, Ruskin J. Vertebral osteomyelitis due to Actinobacillus actinomycetemcomitans. JAMA. 1979;142:1824–1825. [PubMed] [Google Scholar]
- 34.Nakano Y, Suzuki N, Yoshida Y, Nezu T, Yamashita Y, Koga T. Thymidine diphosphate-6-deoxy-l-lyxo-4-hexulose reductase synthesizing dTDP-6-deoxy-l-talose from Actinobacillus actinomycetemcomitans. J Biol Chem. 2000;275:6806–6812. doi: 10.1074/jbc.275.10.6806. [DOI] [PubMed] [Google Scholar]
- 35.Nakano Y, Yoshida Y, Suzuki N, Yamashita Y, Koga T. A gene cluster for the synthesis of serotype d-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 2000;1493:259–263. doi: 10.1016/s0167-4781(00)00179-2. [DOI] [PubMed] [Google Scholar]
- 36.Nakano Y, Yoshida Y, Yamashita Y, Koga T. A gene cluster for 6-deoxy-l-talan synthesis in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 1998;1442:409–414. doi: 10.1016/s0167-4781(98)00174-2. [DOI] [PubMed] [Google Scholar]
- 37.Norqvist A, Wolf-Watz H. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect Immun. 1993;61:2434–2444. doi: 10.1128/iai.61.6.2434-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page R C, Sims T J, Engel L D, Moncla B J, Bainbridge B, Stray J, Darveau R P. The immunodominant outer membrane antigen of Actinobacillus actinomycetemcomitans is located in the serotype-specific high-molecular-mass carbohydrate moiety of lipopolysaccharide. Infect Immun. 1991;59:3451–3462. doi: 10.1128/iai.59.10.3451-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paju S, Carlson P, Jousimies-Somer H, Asikainen S. Heterogeneity of Actinobacillus actinomycetemcomitans strains in various human infections and relationships between serotypes, genotype, and antimicrobial susceptibility. J Clin Microbiol. 2000;38:79–84. doi: 10.1128/jcm.38.1.79-84.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paju S, Saarela M, Alaluusua S, Fives-Taylor P, Asikainen S. Characterization of serologically nontypeable Actinobacillus actinomycetemcomitans isolates. J Clin Microbiol. 1998;36:2019–2022. doi: 10.1128/jcm.36.7.2019-2022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry M B. The separation, determination and characterization of 2-amino-2-deoxy-D-glucose and 2-amino-deoxy-d-galactose in biological materials by gas-liquid chromatography. Can J Biochem. 1964;42:451–460. doi: 10.1139/o64-053. [DOI] [PubMed] [Google Scholar]
- 42.Perry M B, MacLean L L, Brisson J-R, Wilson M E. Structures of the antigenic O-polysaccharides of lipopolysaccharides produced by Actinobacillus actinomycetemcomitans serotypes a, c, d and e. Eur J Biochem. 1996;242:682–688. doi: 10.1111/j.1432-1033.1996.0682r.x. [DOI] [PubMed] [Google Scholar]
- 43.Perry M B, MacLean L L, Gmür R, Wilson M E. Characterization of the O-polysaccharide structure of lipopolysaccharide from Actinobacillus actinomycetemcomitans serotype b. Infect Immun. 1996;64:1215–1219. doi: 10.1128/iai.64.4.1215-1219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulsen K, Theilade E, Lally E T, Demuth D R, Kilian M. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology. 1994;140:2049–2060. doi: 10.1099/13500872-140-8-2049. [DOI] [PubMed] [Google Scholar]
- 45.Rick P D. Lipopolysaccharide biosynthesis. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1987. pp. 648–662. [Google Scholar]
- 46.Rocchetta H L, Lam J S. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saarela M, Asikainen S, Alaluusua S, Pyhälä L, Lai C-H, Jousimies-Somer H. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol Immunol. 1992;7:277–279. doi: 10.1111/j.1399-302x.1992.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Sims T J, Moncla B J, Darveau R P, Page R C. Antigens of Actinobacillus actinomycetemcomitans recognized by patients with juvenile periodontitis and periodontally normal subjects. Infect Immun. 1991;59:913–924. doi: 10.1128/iai.59.3.913-924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki N, Nakano Y, Yoshida Y, Nakao H, Yamishita Y, Koga T. Genetic analysis of the gene cluster for the synthesis of serotype a-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 2000;1517:135–138. doi: 10.1016/s0167-4781(00)00229-3. [DOI] [PubMed] [Google Scholar]
- 51.Takamatsu-Matsushita N, Yamaguchi N, Kawasaki M, Yamashita Y, Takehara T, Koga T. Immunogenicity of Actinobacillus actinomycetemcomitans b-specific polysaccharide-protein conjugate. Oral Microbiol Immunol. 1996;11:220–225. doi: 10.1111/j.1399-302x.1996.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 52.Thomson V J, Bhattacharjee M K, Fine D H, Derbyshire K M, Figurski D H. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J Bacteriol. 1999;181:7298–7307. doi: 10.1128/jb.181.23.7298-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Triglia T, Peterson M, Kemp D. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 55.Vandepitte J, De Geest H, Jousten P. Subacute bacterial endocarditis due to Actinobacillus actinomycetemcomitans. J Clin Pathol. 1977;30:842–846. doi: 10.1136/jcp.30.9.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Steenbergen T J M, Bosch-Tijhoh C J, van Winkelhoff A J, Gmür R, de Graaff J. Comparison of six typing methods for Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1994;32:2769–2774. doi: 10.1128/jcm.32.11.2769-2774.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. In: Whistler R L, editor. Methods in carbohydrate chemistry. Vol. 5. New York, N.Y: Academic Press; 1965. pp. 83–91. [Google Scholar]
- 58.Wilson M E, Schifferle R E. Evidence that the serotype b antigenic determinant of Actinobacillus actinomycetemcomitans Y4 resides in the polysaccharide moiety of lipopolysaccharide. Infect Immun. 1991;59:1544–1551. doi: 10.1128/iai.59.4.1544-1551.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamashita Y, Tsukioka Y, Tomihisa K, Nakano Y, Koga T. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J Bacteriol. 1998;180:5803–5807. doi: 10.1128/jb.180.21.5803-5807.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida Y, Nakano Y, Nezu T, Yamashita Y, Koga T. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-D-fucose. J Biol Chem. 1999;274:16933–16939. doi: 10.1074/jbc.274.24.16933. [DOI] [PubMed] [Google Scholar]
- 61.Yoshida Y, Nakano Y, Suzuki N, Nakao H, Yamashita Y, Koga T. Genetic analysis of the gene cluster responsible for synthesis of serotype e-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim Biophys Acta. 1999;1489:457–461. doi: 10.1016/s0167-4781(99)00192-x. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida Y, Nakano Y, Yamashita Y, Koga T. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect Immun. 1998;66:107–114. doi: 10.1128/iai.66.1.107-114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zambon J J. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 64.Zambon J J, Slots J, Genco R J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41:19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zambon J J, Umemoto T, DeNardin E, Nakazawa F, Christersson L A, Genco R J. Actinobacillus actinomycetemcomitans in the pathogenesis of human periodontal disease. Adv Dent Res. 1988;2:269–274. doi: 10.1177/08959374880020021101. [DOI] [PubMed] [Google Scholar]