Abstract
The ability of Actinomyces naeslundii to convert sucrose to extracellular homopolymers of fructose and to catabolize these types of polymers is suspected to be a virulence trait that contributes to the initiation and progression of dental caries and periodontal diseases. Previously, we reported on the isolation and characterization of the gene, ftf, encoding the fructosyltransferase (FTF) of A. naeslundii WVU45. Allelic exchange mutagenesis was used to inactivate ftf, revealing that FTF-deficient stains were completely devoid of the capacity to produce levan-type (β2,6-linked) polysaccharides. A polyclonal antibody was raised to a histidine-tagged, purified A. naeslundii FTF, and the antibody was used to localize the enzyme in the supernatant fluid. A sensitive technique was developed to detect levan formation by proteins that had been separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the method was used to confirm that the levan-synthesizing activity of A. naeslundii existed predominantly in a cell-free form, that a small amount of the activity was cell associated, and that the ftf mutant was unable to produce levans. By using the nucleotide sequence of the levanase gene of a genospecies 2 A. naeslundii, formerly Actinomyces viscosus, a portion of a homologue of this gene (levJ) was amplified by PCR and inserted into a suicide vector, and the resulting construct was used to inactivate the levJ gene in the genospecies 1 strain WVU45. A variety of physiologic and biochemical studies were performed on the wild-type and LevJ-deficient strains to demonstrate that (i) this enzyme was the dominant levanase and sucrase of A. naeslundii; (ii) that LevJ was inducible by growth in sucrose; (iii) that the LevJ activity was found predominantly (>90%) in a cell-associated form; and (iv) that there was a second, fructose-inducible fructan hydrolase activity produced by these strains. The data provide the first detailed molecular analysis of fructan production and catabolism in this abundant and important oral bacterium.
Actinomyces naeslundii is a gram-positive bacterium found in large numbers on mucosal and tooth surfaces. This organism is one of the first species to inhabit the oral cavity after birth and is an early colonizer of cleaned tooth surfaces. Also, by serving as a recipient for other early colonizers of the tooth in coadhesion events and by elaborating enzymatic activities that can modify enamel pellicle receptors for bacteria, such as neuraminidase, A. naeslundii appears to be a key participant in modulating the composition of the biofilms that form on teeth.
A variety of studies support a critical role for Actinomyces in oral health and in disease. Numerous microbiological studies have suggested a role for Actinomyces in coronal and root surface caries or in periodontal diseases (34, 43, 52, 55), whereas other studies have found no positive correlations between the presence of the organisms and oral diseases (27, 42, 59). These inconsistencies are probably due, at least in part, to some previous taxonomic inconsistencies and to the marked phenotypic heterogeneity demonstrated by the various species of oral Actinomyces (5, 7, 10, 66). Although these differences are not resolved and the exact role of the organisms in biofilm formation, ecology, and pathogenesis remain to be elucidated, Actinomyces spp. are consistently among the most abundant organisms in supra- and subgingival dental biofilms (8, 42), and they possess a variety of biological activities that would indicate that they are capable of playing major roles in oral biofilm ecology (62). Among the various phenotypic characteristics of the oral Actinomyces, the capacity to produce extracellular homopolysaccharides of fructose is thought to impact the composition and virulence of oral biofilms (58, 62).
Some of the most ecologically significant, gram-positive, oral bacteria produce fructosyltransferases (FTFs), or levansucrases, which are enzymes that hydrolyze sucrose and concomitantly incorporate the fructose moiety into a fructan homopolymer. The fructans produced by oral streptococci and Actinomyces are of two general types. Streptococcus mutans produces an inulin-type fructan, composed predominantly of β2,1 linkages, whereas A. naeslundii and Streptococcus salivarius make a levan-type polymer, made up mostly of β2,6 linkages (1, 24, 26, 38, 54). When human subjects are given a sucrose-containing rinse, fructans rapidly accumulate in dental plaque (28, 30), where they are thought to serve as storage carbohydrates that can be hydrolyzed when other more readily metabolized carbohydrate sources are exhausted (20). Fructan metabolism extends the depth and duration of dental plaque acidification and thus contributes to the initiation and progression of dental caries (12). In support of this idea, mutants of S. mutans with defects in fructan metabolism are less virulent in a rat caries model (13). Additionally, bacterial levans, such as those produced by A. naeslundii (1), have been postulated to contribute to periodontal diseases, because these polysaccharides can trigger inflammatory reactions and act as mitogens for B cells (19, 22, 53).
Recently, the gene encoding the FTF of a genospecies 1 A. naeslundii, strain WVU45, was cloned and characterized (3). FTFs of many bacteria can be found either in a cell-associated form or secreted into the culture fluid. The levansucrase enzymes of B. subtilis and S. salivarius are secreted in a two-step process with a cell-associated intermediate (17, 41, 48, 57). Early studies on the FTF activity of A. naeslundii suggested that the enzyme was primarily secreted but was also present in a cell-associated form (47, 58). Cell-bound FTF was postulated to provide some advantages to the cells because the fructans produced on the cell surface might act like a capsular material that could protect the organisms from inimical influences (60). Also, the fructans produced by a cell-bound enzyme would be in close contact with the organisms when the conditions were favorable for hydrolysis of the levans, giving the organism an advantage in competition for this valuable nutrient source. However, these early studies did not employ methods that could discriminate between FTF activity and other β-fructosidases that might be produced by oral Actinomyces.
Bacteria that produce fructans also produce enzymes that degrade these polymers (11). Previous studies have indicated that oral Actinomyces have the capacity to hydrolyze a variety of fructans via enzymes that specifically break down (i) only levans, (ii) only inulins, sucrose, and raffinose, but not levans; or (iii) levans, inulins, sucrose, and raffinose (40, 62). Enzymes that can attack levans, regardless of whether they attack multiple other fructosides, are often referred to as levanases. The levanase from A. naeslundii T14V, a genospecies 2 organism formerly designated as Actinomyces viscosus, is the most thoroughly studied levanase of the oral Actinomyces spp. (44, 45). The T14V levanase, encoded by the levJ gene, is a 99-kDa enzyme with significant homology to other known levanase enzymes from eubacteria. LevJ has a putative signal sequence and cell-anchoring domain, an LARTG sequence (45), which is similar to the LPXTG sorting sequence of gram-positive bacterial surface proteins (51). The levJ gene was expressed in Escherichia coli and was shown to hydrolyze levans, inulins, raffinose, and sucrose (45), similar to the S. mutans levanase, FruA (15). The levanase of A. viscosus ATCC 15987 has also been examined biochemically and shown to have substrate specificities similar to those of LevJ, and this enzyme is both cell surface associated and cell-free (40). In contrast, the A. viscosus ATCC 19246 levanase is capable of hydrolyzing only levans, similar to a levanase enzyme isolated from S. salivarius (32). Thus, levanases of Actinomyces spp. have different properties depending on the species, genospecies, and strain. The differences in substrate preference may be important in the oral cavity, where there are probably multiple types of fructans available as a result of the variable composition of oral biofilms at different sites in the mouth. The purpose of this study was to provide fundamental information on the genetics and biochemistry of the metabolism of fructans by genospecies 1 A. naeslundii.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
A. naeslundii strains WVU45 (ATCC 12104) (18), FTF1, and Lev1 (this study) were grown in either brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) or Lactobacillus-carrying medium (25). Alternatively, A. naeslundii was grown in a semidefined medium (Actinomyces defined medium [ADM]) (6) containing 1% glucose, sucrose, or fructose as the carbohydrate source. E. coli DH10B and M15 were grown in Luria broth and S. salivarius 57.1 was grown in BHI broth. Kanamycin (50 or 25 μg ml−1), streptomycin (50 μg ml−1), or ampicillin (100 μg ml−1) were added to culture media, when necessary. All chemical reagents were obtained from Sigma Chemical Co.
DNA manipulations.
Chromosomal DNA was isolated from Actinomyces strains by the method of Donkersloot et al. (23). E. coli plasmid DNA was isolated by a rapid boiling method (31) or by using the QIAprep Miniprep kit (Qiagen, Inc., Chatsworth, Calif.). Restriction and DNA modifying enzymes were obtained from Life Technologies (LTI Rockville, Md.), MBI Fermentas (Amherst, N.Y.), New England Biolabs (Beverly, Mass.), or U.S. Biochemicals (Cleveland, Ohio). Nucleotide sequence analysis was obtained using TaqTrak sequencing reagents (Promega, Madison, Wis.). Sequencing reactions were radiolabeled using [α-35S]dATP (New England Nuclear, Boston, Mass). Southern blotting experiments were performed as described by Sambrook et al. (49) under conditions of high stringency. Genetic transformation of A. naeslundii was performed as previously detailed (64, 65).
Mutants of A. naeslundii with insertions in the ftf and levJ genes were constructed as follows. Briefly, a 2-kb StuI-XhoI fragment containing the FTF open reading frame was subcloned into pGEM-7, which does not replicate in A. naeslundii (63). A kanamycin resistance (Kmr) gene was released from pJRD251 (65) by DraI-XhoI digestion, and blunt ends were created with T4 DNA polymerase (LTI) and deoxynucleoside triphosphates. The Kmr determinant was inserted into the ftf gene, leaving 1.2 kbp upstream and a 0.8 kbp downstream of the insertion site. The resulting construct, pSXKM, was electroporated into A. naeslundii WVU45, as previously described (64, 65), and potential mutants were selected on BHI supplemented with kanamycin (50 μg ml−1). Mutant strains with insertions into the ftf gene by double-crossover recombination were utilized for further analyses. To prepare a levanase-deficient mutant of A. naeslundii WVU45, a 0.5-kbp EcoRI fragment of the A. naeslundii levanase gene was amplified by PCR using primers based on the sequence of the genospecies 2 A. naeslundii strain T14V (44) (GenBank accession no. U12274). The levanase PCR product was cloned in plasmid pCR2.1, and nucleotide sequencing was done to confirm that the insert was homologous to levJ. The deduced amino acid sequences of the characterized portions of the PCR product were 92% identical to the levanase of A. naeslundii T14V. The product was cloned onto EcoRI-digested pBSK to create pLjPB. A Dral-XhoI digest of pJRD215 was used to release the Kmr gene, which was then cloned adjacent to the levanase gene fragment after digestion of pLjPB with EcoRV and XhoI. The resulting plasmid, pLjKm, was used to transform A. naeslundii WVU45 and integration of the plasmid by Campbell insertion was confirmed by Southern blotting.
Construction and purification of a six-histidine-tagged FTF.
A partial ftf gene, encoding the FTF lacking the predicted signal sequence, was amplified from plasmid pFTFLB4 (3) using PCR, with Pfu polymerase and primers (PCR 480, 5′-ACCACCGGATCCGCCGATGAGACCCCCT-3′, and PCR 2360, 5′-ACGCGCCTGCAGTCAAACGACGACGAGCGG-3′) into which BamHI and PstI sites (underlined) were incorporated to facilitate subsequent in-frame fusion of the ftf gene with the His6 tag. A 1.6-kbp PCR product containing an internal portion of the A. naeslundii ftf gene was cloned into pCR2.1 (Invitrogen), released by digestion with BamHI and PstI, and subsequently cloned into BamHI- and PstI-digested pQE-30 (Qiagen) in E. coli DH10B. Nucleotide sequence analysis confirmed that the orientation of the insert was correct and that the ftf gene was fused in the correct reading frame. The plasmid containing the partial A. naeslundii ftf open reading frame in pQE-30, designated pHis-ls, was transformed into E. coli M15. The resulting recombinant strain, E. coli His-ls M15, was grown to exponential phase, and expression of the fusion protein was induced with IPTG (isopropyl-β-D-thiogalactopyranoside) at a final concentration of 1 mM. The 68-kDa His-tagged FTF (1.4 mg) was purified by the denaturing protocol described in The QIA Expressionist kit (Qiagen) and then further purified after preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein was excised from the gel and used to elicit a polyclonal rabbit sera at Lampire Biological Laboratories (Pipersville, Pa.).
Amplification of an internal fragment of the levanase gene of A. naeslundii WVU45.
Primers prepared from the known sequence of the levanase gene, levJ, of the genospecies 2 A. naeslundii strain T14V (44) were prepared (FP1, 5′-CAACGGGCTCGTCTATTAC-3′, and BPI, 5′-GCAGCGGGAAGAAGTC-3′) and used to amplify a 0.5-kbp internal fragment of the levanase gene of A. naeslundii WVU45. PCR products were analyzed on a 1% agarose gel, a 0.5-kbp product was isolated after excision from an agarose gel, and the fragment was ligated into pCR2.1 (Invitrogen). Using primers specific for the pCR2.1 cloning vector, the 5′ and 3′ portions of the nucleotide sequence of the PCR product were determined to confirm that the correct fragment was obtained.
Detection of levans using an anti-levan monoclonal antibody.
To detect levan synthesis in an enzyme-linked immunosorbent assay (ELISA)-based assay, bacteria were grown overnight in BHI, with or without 1% sucrose, in a 24-well plate (Falcon 3047; Becton Dickinson, Lincoln Park, N.J.). Under both conditions, A. naeslundii forms biofilms very efficiently on the polystyrene. Plates were inverted to remove media and suspended cells, and the wells were gently washed with phosphate-buffered saline (PBS; 10 mM sodium phosphate [pH 7.4], 0.9% NaCl). Levans were detected by using a mouse monoclonal antibody, UPC-10 (ICN Pharmaceuticals, Aurora, Ohio), which is specific for the β2,6-linked fructans composing the majority of A. naeslundii fructan polymers, at a concentration of 1 μg ml−1 for 1.5 h. Plates were washed with PBS containing 0.05% Tween 80 and subsequently incubated with peroxidase-conjugated rabbit anti-mouse immunoglobulin G (IgG) antibody (Kirkegaard and Perry, Gaithersburg, Md.). Unbound antibody was removed by three washes with PBS containing 0.05% Tween 80 and then three washes with PBS alone. Positive reactions were detected using 3,3′,5,5′-tetramethylbenzidine in citrate buffer with hydrogen peroxide as detailed elsewhere (2). The reaction was stopped by addition of 0.5 M H2SO4, and the absorbance at 450 nm was determined. Cells grown without sucrose and wells in which the UPC-10 antibody was omitted served as negative controls. S. salivarius, which produces copious amounts of levans (4), was used as the positive control.
Protein preparations.
For measurement of enzyme activities and for preparation of proteins for blotting, strains of A. naeslundii cells were cultivated in the desired medium to mid-exponential phase, and cells were harvested by centrifugation at 5,000 × g. Culture supernatant fluid was concentrated approximately 100-fold using a Centricon Centriprep concentrator (Millipore, Bedford, Mass.) with a molecular weight cutoff of 3,000. Whole-cell extracts were prepared from 50 ml of cells. After the cells were washed in 10 mM potassium phosphate buffer (pH 6.0) and resuspended in 1 ml of the same buffer, the cells were homogenized in three 20-s intervals using a BeadBeater (Biospec, Bartlesville, Okla.) in the presence of 600 μl of glass beads (average diameter, 0.1 mm). The lysates were centrifuged at 14,000 × g for 15 min at 4°C, and the resulting supernatant fluid was used for analyses. Cleared lysates of E. coli were prepared as previously described (15).
Electrophoresis.
For Western blot analysis, or for analysis of the purity of proteins, protein preparations were boiled for 5 min in SDS sample buffer and subjected to reducing and denaturing SDS-PAGE as previously described (36). Proteins were eletrophoretically transferred to Immobilon-P membrane in a Tris-glycine-methanol buffer for analysis by Western blotting or the gels were stained with Coomasie blue (2). A novel protocol was developed to detect levansucrase activity in protein preparations separated by SDS-PAGE. In these cases, protein samples were mildly denatured in SDS sample buffer (36) for 1.5 h at 37°C prior to separation by SDS-PAGE. After electrophoresis, the proteins were transferred to Immobilon-P membranes, and the filters were incubated overnight at 37°C with 50 mM sucrose in 10 mM potassium phosphate buffer (pH 6.0). Levan production was detected using a mouse monoclonal antibody as detailed below.
Western analysis.
Western blots were performed essentially as detailed elsewhere (56). Prior to use, the polyclonal anti-FTF antisera was adsorbed against cell extracts of E. coli M15 (29). Serum containing anti-FTF antibodies was used at a 1:100 dilution, and peroxidase-conjugated goat-anti-rabbit IgG was used at a dilution of 1:1,500. Following incubation with the antibodies, filter membranes were washed three times with Tris-buffered saline (TBS; 10 mM Tris-HCl, 0.9% NaCl) containing 1% Triton X-100, followed by two more washes with TBS. Conditions for detection of levans by Western blotting were essentially as described above except that the number of TBS washes after exposure to the primary antibody, UPC-10, was reduced to two and, after incubation with the secondary antibody, the blot was washed once with TBS containing 1% Triton X-100, followed by two washes with TBS for 5 min each time.
Enzyme assays.
For measuring the hydrolysis of levans and inulins or the sucrase activity, protein preparations were incubated with 1 mg of levan or 1 mg of inulin per ml, and the reducing sugar that was released was measured as described elsewhere (37). Enzyme activities were normalized to the protein concentrations, which were determined by the method of Bradford (9) using a commercially available reagent (Bio-Rad, Hercules, Calif.). Bovine serum albumin was used as the protein standard, and standard curves were prepared for each experiment. Enzyme activity was expressed as units, which are defined as the amount of enzyme needed to liberate 1 μmol of reducing sugar in 1 h. Enzyme activities are expressed as specific activities. To calculate the total amount of levan-, inulin-, or sucrose-hydrolyzing activity present in cell or supernatant fractions from A. naeslundii growing under various conditions, the specific activities in the various fractions were multiplied by the total amount of protein present in those same fractions.
RESULTS
Construction of an FTF-defective A. naeslundii.
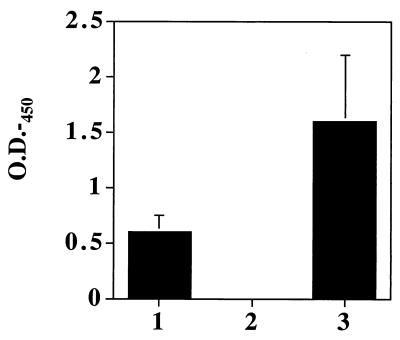
A strain of A. naeslundii in which the ftf gene was inactivated was constructed by allelic exchange as detailed in Materials and Methods. Chromosomal DNAs from Kmr bacteria were analyzed by Southern blotting by probing with the kanamycin gene and an internal fragment of the ftf gene (data not shown). The recombinant A. naeslundii strain, designated FTF1, resulted from an insertion of the kanamycin resistance cassette by double-crossover recombination into the ftf open reading frame. A. naeslundii FTF1 was assayed for formation of levans in the ELISA-based assay detailed in the methods section using a mouse monoclonal IgG antibody, UPC-10, which specifically recognizes β2,6-linkages (Fig. 1). The assay detected levan production in A. naeslundii WVU45 grown in the presence of sucrose, but not in the ftf mutant, indicating that all levansucrase activity was absent in strains lacking an intact ftf gene.
FIG. 1.
ELISA-based detection of levan production with UPC-10. Biofilms of A. naeslundii WVY45 (column 1) or FTF1 (column 2) or of S. salivarius 57.1 (column 3) were formed in microtiter wells in the presence of sucrose. Levan production by the bacteria was detected with a mouse monoclonal antibody against β2,6-linked fructans as detailed in Materials and Methods. The values presented are the means of three separate experiments performed in triplicate, and the error bars show the standard deviations. The negative controls were cells grown without sucrose.
Detection of FTF by Western blotting.
In order to obtain antibodies to assist in the characterization of the FTF from A. naeslundii, a six-histidine-tagged FTF was constructed and purified. A recombinant E. coli strain overexpressing the A. naeslundii FTF enzyme and lacking the predicted signal sequence (3) was constructed as described in Materials and Methods. The E. coli strain harboring the expression plasmid, pQE-ls, synthesized an IPTG-inducible protein with an estimated mass of about 70 kDa, consistent with the predicted mass of the protein encoded by the ftf gene, i.e., 68,215 Da (3). After induction with IPTG, the cells were visualized under the microscope, and inclusion bodies were evident. Not surprisingly then, when crude extracts of induced M15 cells harboring the pQE-ls plasmid were analyzed for the ability to hydrolyze sucrose, none was detected. Attempts to purify the protein under nondenaturing conditions were unsuccessful, so denaturing methods were then used to purify the histidine-tagged FTF.
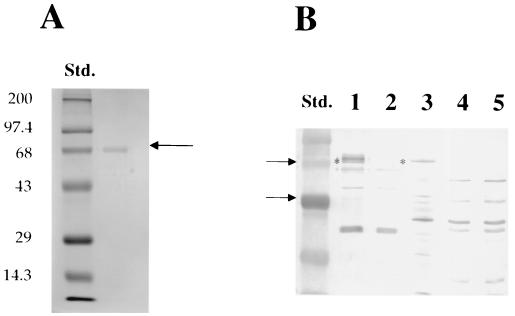
The purified protein (Fig. 2) migrated with an apparent Mr of approximately 70 kDa. The recombinant protein was further purified by isolation after SDS-PAGE and was used to elicit antibodies in a rabbit. The reactivity of anti-FTF polyclonal antibody with culture supernatants of A. naeslundii was detected by Western blot analysis. The anti-FTF antibody recognized a protein of about 70 kDa in A. naeslundii culture supernatants that was absent in the ftf mutant strain, FTF1 (Fig. 2). Some slightly higher Mr proteins were detected in the wild type, but not in the mutant, perhaps representing FTF that was aggregated, posttranslationally modified or covalently coupled to cell wall fragments. Cell-associated FTF was not detected under the same conditions, but when the gels were grossly overloaded with cell lysates, FTF could be detected by the anti-FTF antibody (data not shown), suggesting that the vast majority of FTF is secreted from the cells under the growth conditions tested. Some nonspecific reactivity was also noted with the rabbit antisera, apparently because rabbits naturally mount an immune response to bacterial antigens that cross-react with those of A. naeslundii.
FIG. 2.
(A) Coomassie blue-stained, histidine-tagged FTF. The recombinant, histidine-tagged FTF protein was purified from E. coli after induction with 1 mM IPTG using the denaturing protocol recommended by Qiagen. The purified protein (1 μg) was electrophoresed in an SDS–10% PAGE gel, and the gel was stained using Coomassie blue. Molecular weight standards were from LTI. (B) Western blot analysis with a rabbit anti-FTF antisera. Antisera was raised to the purified histidine-tagged FTF protein (1.4 mg of total protein) in rabbits at Lampire Biologicals. Following separation of the supernatant proteins or cell extracts of A. naeslundii WVU45 and FTF1 and E. coli FTFLB4 (3), which carries and expressed the ftf gene on a multicopy plasmid, the proteins were transferred to PVDF membranes. The blots were probed with the anti-FTF antisera at a dilution of 1:100 and subsequently probed with a goat anti-rabbit IgG peroxidase-conjugated antibody at a dilution of 1:1,500. Supernatant proteins from A. naeslundii WVU45 (lane 1) or A. naeslundii FTF1 (lane 2), whole-cell lysates from E. coli FTFLB4 (3) (lane 3), and whole-cell lysates of A. naeslundii WVU45 (lane 4) and FTF1 (lane 5) were tested. Std., prestained molecular weight standards from LTI. The asterisk indicates the position of the 70-kDa FTF protein in A. naeslundii WVU45 supernatant and in E. coli FTFLB4 (3).
Detection of levan synthesis following SDS-PAGE.
A protocol was developed to detect levan synthesis after separation of proteins by SGS-PAGE. Briefly, protein preparations from A. naeslundii WVU45 and from FTF1 were mildly denatured and electrophoresed by SDS–10% PAGE. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane, and the filters were incubated in sucrose overnight. Levan production by the levansucrase enzymes were detected in a Western blot analysis with mouse monoclonal antibody UPC-10, which is specific for β2,6 linkages. Levan production was detected in the culture supernatants of A. naeslundii (Fig. 3), with a prominent band migrating with an apparent molecular mass of about 70 kDa, consistent with the predicted size of the mature FTF enzyme and the protein detected in Western blots. There were two bands produced in the cell-associated lanes that migrated with apparent Mr values of 70 and 72 kDa, a result consistent with the size of the FTF before and after signal sequence removal, but there were also a number of higher-molecular-weight bands, possibly arising due to aggregation or to covalent coupling of the FTF to petidoglycan. All levan synthetic activity was absent in the ftf mutant strain FTF1 (Fig. 3). It is not at all surprising that the levan detection strategy was more sensitive than the Western analysis with the anti-FTF antibody, since each FTF molecule should produce many binding sites for UPC-10.
FIG. 3.
Detection of levan production by proteins following SDS-PAGE. Protein samples from culture supernatants of A. naeslundii WVU45 (lane 1) and FTF1 (lane 3) or from whole-cell extracts of WVU45 (lane 2) and FTF1 (lane 4) were separated in a 10% polyacrylamide gel and subsequently transferred to PVDF membranes. The filters were incubated overnight at 37°C in 50 mM sucrose. The membranes were briefly rinsed in TBS and incubated with monoclonal antibody UPC-10 as detailed in Materials and Methods. After multiple washes, the membranes were incubated with a goat anti-mouse, peroxidaase-conjugated antibody, and the immune reactivity with levans was disclosed. Std., prestained protein molecular weight standards.
Role of LevJ in fructan and sucrose utilization by A. naeslundii.
A levJ mutant was constructed as detailed in Materials and Methods. Chromosomal DNAs isolated from Kmr bacteria were analyzed by Southern blot analysis using the 0.5-kbp levJ fragment or the Kmr gene (data not shown). Insertion of pLjKm into the chromosome of A. naeslundii WVU45 in all cases was achieved by single-crossover recombination at the levJ locus. A strain of A. naeslundii, designated Lev1, that was confirmed to have an insertion of pLjKm into the chromosome at the correct locus (data not shown) was selected for further study.
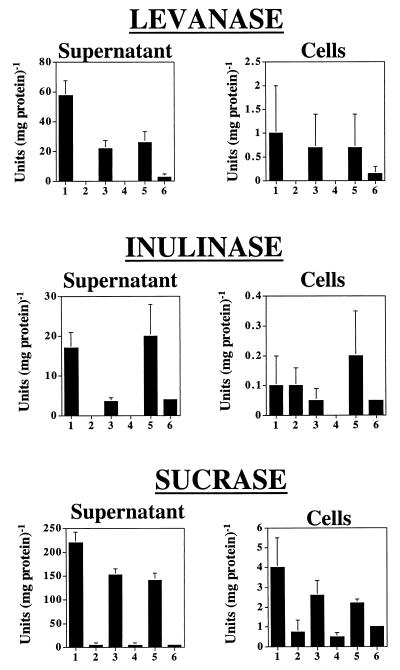
Levanase, inulinase, and sucrase activities produced by the wild-type strain and a LevJ-deficient strain were measured in cultures grown in ADM with sucrose, fructose, or glucose as the sole carbohydrate source. Overall, levanase activity of the wild-type strain (Fig. 4) was greater than inulinase activity, and inactivation of the levJ gene eliminated all levanase activity in sucrose- and glucose-grown cells. Interestingly, there was a low level of fructan hydrolase activity in supernatants from fructose-grown cells that was lacking in glucose- or sucrose-grown cells, indicating that A. naeslundii may have a fructose-inducible fructanase that is distinct from LevJ. Similarly, inulinase activity in whole-cell extracts of fructose- and sucrose-grown cells was elevated, again supporting the presence of at least one additional β-fructosidase, other than LevJ, that is inducible by fructose or fructose-containing carbohydrates. Levanase and inulinase specific activities were highest in the supernatant fractions, in large part because the total protein concentration was much lower than that in cell lysates. Importantly, though, when the total amount of fructanase activity present in cell extracts and culture supernatants was calculated, it was found that more than 90% of the total the fructanase activity was found in the cell-associated extracts, as was more than 95% of the total sucrase activity of A. naeslundii WVU45.
FIG. 4.
Levanase-, inulinase-, and sucrase-specific activities in supernatant and whole-cell lysates of A. naeslundii strains WVU45 and Lev1. Cells were grown to mid-exponential phase in ADM containing sucrose (columns 1 and 2), glucose (columns 3 and 4), or fructose (columns 5 and 6), and the levanase activity was measured in the culture fluid (supernatant) or whole-cell lysates (cells) of the wild-type strain WVU45 (columns 1, 3, and 5) or the LevJ-deficient strain (columns 2, 4, and 6). The results represent the mean of at least three separate experiments performed in a minimum of triplicate. Error bars show the standard deviation. In all cases of measurements of supernatant activities, the mutant and wild-type strains are statistically different (P < 0.05) by t test, as is the case for cell-associated levanase activity in sucrose- and glucose-grown cells (columns 1 to 4) and for the cell-associated sucrase activity in all carbohydrates tested.
The sucrase activity in culture supernates of A. naeslundii Lev1 grown with sucrose as a carbohydrate source was 7.3 U mg of protein−1 compared to the wild-type strain, which produced 224.8 U mg of protein−1. Glucose-grown Lev1 culture supernatants had a sucrase specific activity of 7.8 U mg of protein−1, compared to 153.7 U mg of protein−1 in the wild-type strain of A. naeslundii. Thus, LevJ is a major contributor to total extracellular sucrase activity. Cell-associated sucrase activity of Lev1 was four to five fold lower than that of the wild-type A. naeslundii in glucose- and sucrose-grown cells, respectively, indicating that LevJ probably can exist in a cell-associated form and contributes to total cell-associated sucrase activity. Not surprisingly, however, the data also support that there are other pathways for sucrose dissimilation possessed by these organisms besides LevJ, including FTF.
Regulation of A. naeslundii WVU45 sucrases and fructanases.
The fructanase activities of Lev1 and WVU45 were compared in cells grown in ADM on different carbohydrate sources. The carbohydrate source on which the organisms were grown had a significant impact on the expression of fructanase and sucrase activities. Levanase and inulinase activity in culture supernatants also was two- to three-fold higher when the bacteria were grown with sucrose as the sole carbohydrate source (Fig. 4). Consistent with LevJ accounting for a major component of the extracellular sucrase activity, total sucrase activity in culture supernatants increased about 50% when A. naeslundii was grown in the presence of 1% sucrose, compared to cells grown on glucose or fructose (Fig. 4). A similar pattern was observed using cell extracts prepared from WVU45 when sucrose was used as the substrate.
DISCUSSION
The functions of fructans in the oral cavity have been most thoroughly examined in S. mutans, the primary etiological agent of dental caries. S. mutans produces a secreted FTF (35), which synthesizes fructans composed predominantly of the β2,1-linked variety (4). For S. mutans, fructans are thought to serve primarily as storage polysaccharides, which can be hydrolyzed by fructanase enzymes when other carbohydrate sources are exhausted (12). Like S. mutans, A. naeslundii produces an FTF and a fructan-hydrolyzing enzyme, so it is possible that the fructans in the Actinomyces spp. play a major role in the nutrition of the organism. However, unlike S. mutans, Actinomyces spp. are typically major constituents of the subgingival flora (7, 8). In this environment, the production of levans may have adverse consequences for the host because of the ability of these polysaccharides to act as mitogens and to stimulate the inflammatory response (19, 22). Defined mutants of A. naeslundii with defects in fructan metabolism will be useful in evalvating the contribution of levans to the virulence of the organism in supra- and subgingival biofilms.
It has been suggested that levans of Actinomyces spp. may be associated with the bacterial surface and form a capsule that can be hydrolyzed by cell-associated levanases. Previous studies have indicated that levansucrase activity of Actinomyces spp. can occur both in the culture supernatant fluid and in cell-associated fractions (46, 47, 58). We have made repeated attempts to measure FTF activity in preparations from A. naeslundii by standard biochemical assays, but these efforts have not yielded repeatable results, due in large part to the fact that the enzyme may produce small polymers that are not efficiently precipitated (3) and due to rapid proteolytic digestion. In this regard, the antibody-based strategies were useful as sensitive methods to detect levans and FTF. It is now clear that levan production by A. naeslundii is attributable to a 70-kDa protein in culture supernatants and that cell-associated FTF migrates as two bands at 70 and 72 kDa, the slower-migrating species possibly representing FTF before the signal sequence was removed. The ftf mutant FTF1 lacked all ability to produce levans, both in the culture supernatants and in the cell extracts, demonstrating that the levan-synthesizing capacity of this organism may be solely due to the functional ftf gene product. However, without a more detailed analysis, we cannot exclude that another FTF activity can be produced under conditions that differ from those used in this study.
Examination of fructan hydrolysis by genospecies 1 A. naeslundii, using strain WVU45 and an otherwise-isogenic levJ mutant of this strain, revealed that this organism could efficiently hydrolyze levans and inulins and that inactivation of the levJ homologue eliminated detectable levanase and inulinase activity in cells grown on sucrose or glucose. Interestingly, there was some detectable levanase and inulinase activity in supernatant preparations of the levJ mutant when cells were grown with fructose as a sole carbohydrate source. Similarly, inulinase activity was enhanced in the levJ strain in fructose- and sucrose-grown cells. The identity of this inducible activity or activities is not currently known. However, the data can be easily explained if A. naeslundii has a fructose-inducible fructan hydrolase and a sucrose- and possibly fructose-inducible invertase with activity on inulins. Notwithstanding, it is clear that the levJ gene product is the major fructan hydrolase in A. naeslundii WVU45. Our results also indicate that LevJ of WVU45 represents a major pathway for sucrose dissimilation, a view consistent with the observation that LevJ of the genospecies 2 strain T14V has high levels of activity both on fructans and sucrose (45).
The expression of levan-, inulin-, and sucrose-hydrolyzing activities in eubacteria is generally regulated at the transcriptional level in response to carbohydrate source and availability (for example, see references 16, 39, and 57). LevJ activity of A. naeslundii WVU45 was much higher in cells grown with sucrose as the sole carbohydrate source than in cells grown on glucose or fructose. Sucrase activity displayed the same pattern of regulation, consistent with the idea that LevJ represents the primary extracellular sucrase activity of A. naeslundii. The A. viscosus ATCC 15987 levanase was also shown to be induced by growth on sucrose (40). We previously reported that the ftf gene is transcribed independently of carbohydrate source (3), which is unusual for gram-positive bacteria. Therefore, although A. naeslundii produces FTF constitutively, the organism appears to have evolved a somewhat more elaborate mechanism for regulating the catabolism of fructans.
It is noteworthy that the A. naeslundii WVU45 fructanase is not completely repressed by glucose, since many other fructanases are very sensitive to catabolite repression (14, 16, 21). One possible reason that the production of LevJ is not as highly repressed as other levanase activities may be that an important role of this enzyme is in sucrose catabolism. Many of the sucrases of oral bacteria are constitutively expressed regardless of carbohydrate source, but are upregulated two- to threefold in cells growing on sucrose (50, 61). Incomplete repression of sucrase activities of oral bacteria may reflect the fact that sucrose is present frequently, but only transiently, in the diet. Consequently, organisms in the mouth may need to have sucrase enzymes constitutively produced at a relatively high level so that sucrose can be optimally assimilated when it is introduced as a part of the diet.
Some fructanase activity was detectable in culture supernatants, but more than 90% of the inulinase and levanase activity of A. naeslundii was in a cell-associated form, and more than 95% of the total sucrase activity was found in the cell extracts. The use of the Lev1 mutant confirmed that essentially all of the cell-associated and cell-free fructanase activity was attributable to LevJ. Therefore, in terms of localization and substrate specificity, the fructanase of A. naeslundii WVU45 seems to be most similar to the levanase from A. viscosus ATCC 15987. Our studies also support that in addition to LevJ, there is at least one other fructanase capable of attacking both levans and inulins in this genospecies 1 A. naeslundii strain. This fructanase may differ from LevJ in cellular localization and substrate preference and thus may play different roles in fructan and/or sucrose metabolism than LevJ.
Actinomyces spp. have been shown to produce a levan-type polymer (1, 46, 47, 58). The reactivity of the monoclonal antibody UPC-10 with polymers synthesized by the wild-type organism, but lacking in the ftf mutant, confirmed the presence of β2,6 linkages in the fructans produced by this organism. Consistent with this information, the A. naeslundii WVU45 fructanase appears to have a marked substrate preference for levans, based on the observation that levanase activity is much greater than inulinase activity. Similarly, the A. viscosus ATCC 15987 levanase also has a higher specific activity with levan as a substrate compared to inulin (40), whereas the A. viscosus ATCC 19246 has no activity on inulins or sucrose and only attacks levans (33). It is logical that Actinomyces spp. would preferentially hydrolyze levans, since that is the type of polymer they synthesize. Still, it may be beneficial for Actinomyces spp. to hydrolyze inulin-type polymers, since they inhabit an environment colonized by inulin-producing organisms, including S. mutans. Retention of inulin hydrolase activity by genospecies 1 A. naeslundii may reflect selective pressures imposed by evolution in an environment rich in organisms that can produce multiple types of fructan polymers.
In summary, this study clarifies the molecular basis for fructan synthesis and degradation in A. naeslundii, definitively localized the enzymes responsible for fructan metabolism, and shed light on the biochemical basis for the catabolism of an abundant dietary carbohydrate, sucrose, as well as abundant, microbially produced polysaccharides, fructans, in oral biofilms. Future studies oriented toward further dissection of the fructan and sucrose catabolic pathways, coupled with the use of mutant with defects in fructan metabolism in animal models of oral diseases, will clarify the role of FTF, LevJ, and other fructosidases in the physiology, persistence, and virulence of oral Actinomyces.
ACKNOWLEDGMENTS
We thank Margaret Chen and Thomas Wen for critical review of the manuscript. We also acknowledge the late Maria Yeung for providing strains, plasmids, and encouragement and for her pioneering work in development of gene transfer technologies for the Actinomyces.
This work was supported by RO1 DE12236 and T32 DE07165 from the National Institute for Dental and Craniofacial Research.
REFERENCES
- 1.Allen P Z, Bowen W H. Immunochemical studies on levans from several strains of Actinomyces viscosus. Arch Oral Biol. 1990;35:55–62. doi: 10.1016/0003-9969(90)90115-q. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. [Google Scholar]
- 3.Bergeron L J, Morou-Bermudez E, Burne R A. Characterization of the fructosyltransferase gene of Actinomyces naeslundii WVU45. J Bacteriol. 2000;182:3649–3654. doi: 10.1128/jb.182.13.3649-3654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkhed D, Rosell K-G, Granath K. Structure of extracellular water-soluble polysaccharides synthesized from sucrose by oral strains of Streptococcus mutans, Streptococcus salivarius, Streptococcus sanguis, and Actinomyces viscosus. Arch Oral Biol. 1979;24:53–61. doi: 10.1016/0003-9969(79)90175-4. [DOI] [PubMed] [Google Scholar]
- 5.Bowden G H, Ekstrand J, McNaughton B, Challacombe S J. Association of selected bacteria with the lesions of root surface caries. Oral Microbiol Immunol. 1990;5:346–351. doi: 10.1111/j.1399-302x.1990.tb00439.x. [DOI] [PubMed] [Google Scholar]
- 6.Bowden G H, Hardie J M, Fillery E D. Antigens from Actinomyces species and their value in identification. J Dent Res. 1976;55:A192–A204. doi: 10.1177/002203457605500112011. [DOI] [PubMed] [Google Scholar]
- 7.Bowden G H, Nolette N, Ryding H, Cleghorn B M. The diversity and distribution of the predominant ribotypes of Actinomyces naeslundii genospecies 1 and 2 in samples from enamel and from healthy and carious root surfaces of teeth. J Dent Res. 1999;78:1800–1809. doi: 10.1177/00220345990780120601. [DOI] [PubMed] [Google Scholar]
- 8.Bowden G H W, Ellwood D C, Hamilton I R. Microbial ecology of the oral cavity. In: Alexander M, editor. Advances in microbial ecology. New York, N.Y: Plenum Press; 1979. pp. 135–217. [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Brailsford S R, Tregaskis R B, Leftwich H S, Beighton D. The predominant Actinomyces spp. isolated from infected dentin of active root caries lesions. J Dent Res. 1999;78:1525–1534. doi: 10.1177/00220345990780090701. [DOI] [PubMed] [Google Scholar]
- 11.Burne R A. Oral ecological disasters: the role of short-term extracellular storage polysaccharides. In: Bowen W H, Tabak L A, editors. Cariology for the nineties. Rochester, N.Y: University of Rochester Press; 1991. pp. 351–364. [Google Scholar]
- 12.Burne R A. Oral streptococci: products of their environment. J Dent Res. 1998;77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 13.Burne R A, Chen Y Y, Wexler D L, Kuramitsu H, Bowen W H. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J Dent Res. 1996;75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- 14.Burne R A, Penders J E, Wexler D L, Jayaraman G C, Clancy K A. Regulation of fructan degradation by Streptococcus mutans. Dev Biol Stand. 1995;85:323–331. [PubMed] [Google Scholar]
- 15.Burne R A, Schilling K, Bowen W H, Yasbin R E R E. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J Bacteriol. 1987;169:4507–4517. doi: 10.1128/jb.169.10.4507-4517.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burne R A, Wen Z T, Chen Y M, Penders J E C. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catbolite repression. J Bacteriol. 1999;181:2863–2871. doi: 10.1128/jb.181.9.2863-2871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambert R, Petit-Glatron M F. Secretion mechanism of Bacillus subtilis levansucrase: characterization of the second step. J Gen Microbiol. 1988;134:1205–1214. doi: 10.1099/00221287-134-5-1205. [DOI] [PubMed] [Google Scholar]
- 18.Cisar J O, Sandberg A L, Reddy G P, Abeygunawardana C, Bush C A. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral Actinomyces and streptococcal lectins. Infect Immun. 1997;65:5035–5041. doi: 10.1128/iai.65.12.5035-5041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coutinho A, Moller G. B cell mitogenic properties of thymus-independent antigens. Nat New Biol. 1973;245:12–14. doi: 10.1038/newbio245012a0. [DOI] [PubMed] [Google Scholar]
- 20.DaCosta T, Gibbons R J. Hydrolysis of levan by human plaque streptococci. Arch Oral Biol. 1968;13:609–617. doi: 10.1016/0003-9969(68)90139-8. [DOI] [PubMed] [Google Scholar]
- 21.Débarbouillé M, Martin-Verstraete I, Arnaud M, Klier A, Rapoport G. Positive and negative regulation controlling expression of the sac genes in Bacillus subtilis. Res Microbiol. 1991;142:757–764. doi: 10.1016/0923-2508(91)90052-c. [DOI] [PubMed] [Google Scholar]
- 22.Desaymard C, Ivanyi L. Comparison of in vitro immunogenicity, tolerogenicity and mitogenicity of dinitrophenyl-levan conjugates with varying epitope density. Immunology. 1976;30:647–53. [PMC free article] [PubMed] [Google Scholar]
- 23.Donkersloot J A, Cisar J O, Wax M E, Harr R J, Chassy B M. Expression of Actinomyces viscosus antigens in Escherichia coli: Cloning of a structural gene (fimA) for type 2 fimbriae. J Bacteriol. 1985;162:1075–1078. doi: 10.1128/jb.162.3.1075-1078.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebisu S, Keijiro K, Kotani S, Misaka A. Structural differences in the fructans elaborated by Streptococcus mutans and Strep. salivarius. J Biochem. 1975;78:879–887. doi: 10.1093/oxfordjournals.jbchem.a130993. [DOI] [PubMed] [Google Scholar]
- 25.Efthymiou C, Hansen C A. An antigenic analysis of Lactocbacillus acidophilus. J Infect Dis. 1962;110:258–267. doi: 10.1093/infdis/110.3.258. [DOI] [PubMed] [Google Scholar]
- 26.Ehrlich J, Stivala S S, Bahary W S, Garg S K, Long L W, Newbrun E. Levans: fractionation, solution viscosity, and chemical analysis of levans produced by Streptococcus salivarius. J Dent Res. 1975;54:290–297. [PubMed] [Google Scholar]
- 27.Ellen R P, Banting D W, Fillery E D. Longitudinal microbiological investigation of a hospitalized population of older adults with a high root surface caries risk. J Dent Res. 1985;64:1377–1381. doi: 10.1177/00220345850640121001. [DOI] [PubMed] [Google Scholar]
- 28.Gold W, Preston F B, Lache M C, Blechman H. Production of levan and dextran in plaque in vivo. J Dent Res. 1974;53:442–449. doi: 10.1177/00220345740530024401. [DOI] [PubMed] [Google Scholar]
- 29.Heinzerling H F, Olivares M, Burne R A. Genetic and transcriptional analysis of flgB flagellar operon constituents in the oral spirochete Treponema denticola and their heterologous expression in enteric bacteria. Infect Immun. 1997;65:2041–2051. doi: 10.1128/iai.65.6.2041-2051.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higuchi M, Iwani Y, Yamada T, Araya S. Levan synthesis and accumulation by human dental plaque. Arch Oral Biol. 1970;15:563–567. doi: 10.1016/0003-9969(70)90111-1. [DOI] [PubMed] [Google Scholar]
- 31.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 32.Igarashi T, Takahashi M, Yamamoto A, Etoh Y, Takamori K. Purification and characterization of levanase from Actinomyces viscosus ATCC 19246. Infect Immun. 1987;55:3001–3005. doi: 10.1128/iai.55.12.3001-3005.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igarashi T, Takajashi M, Yamamoto A, Etoh Y, Takamori K. Purification and characterization of levanase from Actinomyces viscosus ATCC 19246. Infect Immun. 1987;55:3001–3005. doi: 10.1128/iai.55.12.3001-3005.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordan H V, Hammond B F. Filamentous bacteria isolated from human root surface caries. Arch Oral Biol. 1972;17:1333–1342. doi: 10.1016/0003-9969(72)90166-5. [DOI] [PubMed] [Google Scholar]
- 35.Kuramitsu H K. Recent advances in defining the cariogenicity of mutans streptococci: molecular genetic approaches. Eur J Epidemiol. 1987;3:257–260. doi: 10.1007/BF00149733. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli U K. Cleavage of structural proteins during the assembly of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Luchsinger W W, Cornesky R A. Reducing power by the dinitrosalicylic acid method. Anal Biochem. 1962;4:346–347. doi: 10.1016/0003-2697(62)90098-2. [DOI] [PubMed] [Google Scholar]
- 38.Marshall K, Weigel H. Evidence of multiple branching in the levan elaborated by Streptococcus salivarius strain 51. Carbohydr Res. 1980;83:321–326. doi: 10.1016/s0008-6215(00)84544-9. [DOI] [PubMed] [Google Scholar]
- 39.Martin-Verstraete I, Debarbouille M, Klier A, Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990;214:657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- 40.Miller C H, Somers P J B. Degradation of levan by Actinomyces viscosus. Infect Immun. 1978;22:266–274. doi: 10.1128/iai.22.1.266-274.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milward C P, Jacques N A. Secretion of fructosyltransferase by Streptococcus salivarius involves the sucrose-dependent release of the cell-bound form. J Gen Microbiol. 1990;136:165–169. doi: 10.1099/00221287-136-1-165. [DOI] [PubMed] [Google Scholar]
- 42.Moore W E. Microbiology of periodontal disease. J Periodont Res. 1987;22:335–341. doi: 10.1111/j.1600-0765.1987.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 43.Newman M G, Socransky S S. Predominant cultivable microbiota in periodontosis. J Periodont Res. 1977;12:120–128. doi: 10.1111/j.1600-0765.1977.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 44.Norman J M, Bunny K L, Giffard P M. Characterization of levJ, a sucrase/fructanase-encoding gene from Actinomyces naeslundii T14V, and comparison of its product with other sucrose-cleaving enzymes. Gene. 1995;152:93–98. doi: 10.1016/0378-1119(94)00695-o. [DOI] [PubMed] [Google Scholar]
- 45.Norman J M, Giffard P M. Biochemical studies on LevJ, a fructanase from Actinomyces naeslundii T14V. Arch Oral Biol. 1996;41:565–570. doi: 10.1016/0003-9969(96)00017-9. [DOI] [PubMed] [Google Scholar]
- 46.Pabst M J. Levan and levansucrase of Actinomyces viscosus. Infect Immun. 1977;15:518–526. doi: 10.1128/iai.15.2.518-526.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pabst M J, Cisar J O, Trummel C L. The cell wall-associated levansucrase of Actinomyces viscosus. Biochim Biophys Acta. 1979;566:274–282. doi: 10.1016/0005-2744(79)90031-7. [DOI] [PubMed] [Google Scholar]
- 48.Petit-Glatron M F, Benyahia F, Chambert R. Secretion of Bacillus subtilis levansucrase: a possible two-step mechanism. Eur J Biochem. 1987;163:379–387. doi: 10.1111/j.1432-1033.1987.tb10810.x. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch J E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Sato Y, Yamamoto Y, Suzuki R, Kizaki H, Kuramitsu H K. Construction of a scrA::lacZ gene fusion to investigate regulation of the sucrose PTS of Streptococcus mutans. FEMS Microbiol Lett. 1991;79:339–346. doi: 10.1111/j.1574-6968.1991.tb04552.x. [DOI] [PubMed] [Google Scholar]
- 51.Schneewind O, Pancholi V, Fischetti V A. Surface proteins of gram-positive cocci have a common motif for membrane anchoring. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C.: American Society for Microbiology Press; 1991. pp. 152–153. [Google Scholar]
- 52.Schupbach P, Osterwalder V, Guggenheim B. Human root caries: microbiota in plaque covering sound, carious and arrested carious root surfaces. Caries Res. 1995;29:382–395. doi: 10.1159/000262097. [DOI] [PubMed] [Google Scholar]
- 53.Shilo M, Wolman B. Activity of bacterial levans and of lipopolysaccharides in the process of inflammation and infection. Br J Exp Pathol. 1958;39:652–661. [PMC free article] [PubMed] [Google Scholar]
- 54.Shimamura A, Kenji T, Nagase T, Ito M, Tsumori H, Mukasa H. Structural determination of d-fructans from Streptococcus mutans, serotype b, c, e, and f strains, by 13C-n.m.r. spectroscopy. Carbohydr Res. 1987;165:150–154. doi: 10.1016/0008-6215(87)80091-5. [DOI] [PubMed] [Google Scholar]
- 55.Socransky S S. Microbiology of periodontal disease – present status and future considerations. J Periodontol. 1977;48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 56.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Townsend-Lawman P, Bleiweiss A S. Multilevel control of extracellular sucrose metabolism in Streptococcus salivarius by sucrose J. Gen Microbiol. 1991;137:5–13. doi: 10.1099/00221287-137-1-5. [DOI] [PubMed] [Google Scholar]
- 58.van der Hoeven J S, Vogels G D, Bekkers M F J. A levansucrase from Actinomyces viscosus. Caries Res. 1976;10:33–48. doi: 10.1159/000260187. [DOI] [PubMed] [Google Scholar]
- 59.van Houte J, Lopman J, Kent R. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res. 1994;73:1727–1734. doi: 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 60.Warner T N, Miller C H. Cell-associated levan of Actinomyces viscosus. Infect Immun. 1978;19:711–719. doi: 10.1128/iai.19.2.711-719.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wexler D L, Hudson M C, Burne R A. Streptococcus mutans fructosyltransferase (ftf) and glucosyltransferase (gtfBC) operon fusion strains in continuous culture. Infect Immun. 1993;61:1259–1267. doi: 10.1128/iai.61.4.1259-1267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeung M K. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med. 1999;10:120–138. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
- 63.Yeung M K, Donkersloot J A, Cisar J O, Ragsdale P A. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect Immun. 1998;66:1482–1491. doi: 10.1128/iai.66.4.1482-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeung M K, Kozelsky C S. Transfection of Actinomyces spp. by genomic DNA of bacteriophages from human dental plaque. Plasmid. 1997;37:141–153. doi: 10.1006/plas.1997.1285. [DOI] [PubMed] [Google Scholar]
- 65.Yeung M K, Kozelsky C S. Transformation of Actinomyces spp. by a gram-negative broad-host-range plasmid. J Bacteriol. 1994;176:4173–4176. doi: 10.1128/jb.176.13.4173-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zambon J J, Kasprzak S A. The microbiology and histopathology of human root caries. Am J Dent. 1995;8:323–8. [PubMed] [Google Scholar]