Abstract
The reemergence of pertussis has been reported in several countries despite high vaccination coverage. Studies in The Netherlands and Finland have investigated polymorphism in the genes coding for two important virulence factors of Bordetella pertussis, pertactin and pertussis toxin, and identified the emergence and subsequent dominance in circulating strains of pertactin and toxin variants not found in the whole-cell vaccine (WCV). The study described here investigated whether such variation had occurred in the United Kingdom, which presently has low levels of pertussis. Sequence analysis of the genes for pertactin (prnA) and the pertussis toxin S1 subunit (ptxA) among isolates of B. pertussis from 285 United Kingdom patients, from 1920 to 1999, revealed three prnA variants, prnA(1), prnA(2), and prnA(3), and two ptxA variants, ptxA(1) and ptxA(2), showing differences in nucleic acid sequence. The proportion of pertactin gene types not included in the United Kingdom WCV, i.e., prnA(2) and prnA(3), has increased in recent years and was found in 21 of 86 (24%) strains from the 1980s and 56 of 105 (53%) strains from the 1990s. To date, the presence of these nonvaccine prnA types has not been associated with a resurgence of pertussis in the United Kingdom. The distribution of prnA and ptxA types in The Netherlands, Finland, and the United Kingdom in the 1990s is distinct. The most striking difference in the United Kingdom isolates is that all 105 of the most recent circulating strains (from 1998 to 1999) are of a pertussis toxin type found in the United Kingdom WCV, i.e., ptxA(1).
Within the last decade, several countries have reported an increased incidence of infection caused by Bordetella pertussis, despite high vaccination coverage (1, 4, 10). One possible explanation for this resurgence, proposed by Mooi et al. (17), is the expansion of strains of B. pertussis antigenically distinct from those in the vaccine. Whole-cell vaccines (WCVs) against pertussis are the most widely used; however, vaccines from different manufacturers can be quite distinct. Differences can occur both in the method of strain preparation and in the choice of strain(s), which may be random or a deliberate selection of a known serotype(s). As many as eight agglutinogens (mostly fimbrial) may be present on the cell envelope of B. pertussis, six of which are reported to be unique for this species (11). Of these six agglutinogens, types 1, 2, and 3 have been considered important in the pathogenesis and immunity of pertussis, and antibodies to these agglutinogens have been useful in serological studies (11, 18). Most early vaccines included only agglutinogens 1 and 2 due to the predominance of strains with this serotype. Some later vaccines also included agglutinogen 3, and in 1979, the World Health Organization recommended the inclusion of all three agglutinogens in the final vaccine (25). The lack of any recommendations regarding the inclusion of other specific types of antigen (e.g., pertactin and pertussis toxin) has led to the use of different strains by different manufacturers, such that the vaccination history in each country where pertussis WCV has been introduced appears unique.
Pertactin (P.69) and pertussis toxin (PT) are important virulence factors of B. pertussis, and previous studies have suggested that antibodies to pertactin and PT are significant in protection against pertussis infection (8, 21, 24). It may therefore be expected that these two proteins will be under selective evolutionary pressure, and polymorphism in nucleic acid sequence has been demonstrated in the genes coding for pertactin (prnA) and the S1 subunit of pertussis toxin (ptxA) (3, 17).
Following the pertussis epidemic in The Netherlands in 1996, polymorphism in the prnA and ptxA genes of isolates of B. pertussis was examined by Mooi et al. (17). This study showed a clear shift in the genetic composition of prnA and ptxA types in the circulating strains of B. pertussis over time. From 1949 to 1980, all 35 isolates (100%) had the same pertactin genotype, prnA(1), as that found in the Dutch WCV, but by 1996, the most recent year studied, the frequency of prnA(1) had decreased to only 5 of 49 (10%). Similarly, the two pertussis toxin genotypes found in isolates from 1949 to 1954 were the same as those in the Dutch WCV: 8 of 14 (57%) were type ptxA(2), and 6 of 14 (43%) were type ptxA(3) (17), while in the period 1990 to 1996, only 2 of 15 (13%) isolates were of a Dutch WCV type, with the remaining 13 of 15 (87%) being ptxA(1) (17). A similar study on a small number of strains was also performed using isolates from Finland, where a resurgence of pertussis has not been seen (16). The Finnish WCV, containing a single strain of B. pertussis [prnA(1) ptxA(3)], was introduced in 1962 and combined with an additional strain [prnA(1) ptxA(2)] in 1976. Analysis of the Finnish isolates revealed a similar shift from all 5 isolates being of type prnA(1) in 1953 to 1964 (i.e., the same prnA type as found in the Finnish WCV) to only 3 of 43 (7%) isolates of the vaccine type [prnA(1)] from 1990 to 1996. There also appeared to have been a shift in circulating strains containing the toxin type found in the Finnish WCV, ptxA(2), from all 5 isolates in strains from 1953 to 1964 to 40 of 40 (100%) of type ptxA(1) (i.e., a non-Finnish WCV type) from 1990 to 1996 (16). Significant differences were seen between the two countries in the proportion of isolates of pertactin gene types prnA(2) and prnA(3). In Dutch isolates from 1996, 13 of 49 (27%) were prnA(2) and 31 of 49 (63%) were prnA(3). This contrasts with the Finnish isolates from 1990 to 1996, of which 31 of 43 (72%) were prnA(2), and 5 of 43 (12%) were prnA(3). In addition, one pertactin type [prnA(4)] which had not been previously reported was seen at a frequency of 4 of 43 (9%) Finnish isolates from the 1990s (16, 17). To explain the difference in the incidence of pertussis in The Netherlands and Finland, the authors of these studies postulated that pertussis WCVs containing only the pertactin type prnA(1) may confer differing levels of protection against strains of different prnA types [i.e., greater protection against prnA(2) strains predominating in Finland than against the prnA(3) strains predominating in The Netherlands] (16).
There are now a number of effective acellular vaccines (ACVs) derived from purified components of B. pertussis, including inactivated pertussis toxin, filamentous hemagglutinin, pertactin, and/or fimbrial protein(s) (9, 11). These vaccines have been introduced either as replacements for or in addition to B. pertussis WCVs in several countries (11, 24). As both pertussis toxin and pertactin are important components of these ACVs, it is prudent to investigate the genotypic characteristics of the circulating strains in countries where there is an active vaccination program.
In the United Kingdom, pertussis vaccination was introduced in 1953 and integrated into the whole-cell diphtheria-tetanus-pertussis vaccine in 1961 (22). Since 1995 the vaccination coverage has been ca. 95% (2). The number of notified cases of pertussis in the United Kingdom has reached historically low levels (23), and although the true incidence is probably much greater than notification suggests (14), the United Kingdom has not seen the resurgence of pertussis reported in The Netherlands. The aims of this study were (i) to examine the genotypic variation in the pertactin and pertussis toxin genes in both historical and current circulating strains of B. pertussis in the United Kingdom and (ii) to look for evidence of genetic variation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 285 United Kingdom clinical isolates of B. pertussis from 1920 to 1999 were analyzed. The years of isolation and numbers of strains isolated were as follows: 1920, 1; 1941, 10; 1942, 9; 1943, 3; 1944, 7; 1946, 1; 1947, 1; 1948, 6; 1949, 13; 1950, 8; 1954, 3; 1956, 6; 1963, 8; 1964, 2; 1966, 2; 1967, 1; 1977, 8; 1978, 3; 1979, 2; 1982, 32; 1983, 36; 1984, 5; 1985, 13; 1998, 37; and 1999, 68. Strains were obtained from the Wellcome Bacterial Collection, held by the National Collection of Type Cultures; the National Collection of Type Cultures; the Respiratory and Systemic Infection Laboratory, Public Health Laboratory Service Central Public Health Laboratory, London, United Kingdom; the Pertussis Reference Laboratory, Manchester, United Kingdom; the Centre for Applied Microbiology and Research, Salisbury, United Kingdom; and the University of Glasgow, Glasgow, Scotland. The year of isolation or submission was recorded for all strains, but clinical data were available for the strains only from 1998 and 1999. These isolates comprise all those submitted to the Pertussis Reference Laboratory in a 6-month period from November 1998 to April 1999. The numbers of above strains, by year of isolation, for which serotyping data were available were 1 from 1964, 3 from 1978, 2 from 1979, 37 from 1998, and 61 from 1999. Serotyping was performed by the Pertussis Reference Laboratory as described by Preston (18), and serotyping data were available on all strains from 1978 to 1999. The serotype data of the single isolate from 1964 and of the three vaccine strains were historical data from the Wellcome Collection and the National Institute for Biological Standards and Control, Potters Bar, United Kingdom. The three strains used in the United Kingdom WCV, i.e., CN2992 (serotype 1,2,3), CN5476 (serotype 1,3), and CN3099 (serotype 1,2) were obtained from the Wellcome Bacterial Collection and the National Institute for Biological Standards and Control. All isolates of B. pertussis were grown on blood charcoal agar (Media Services, Central Public Health Laboratory) at 37°C for up to 5 days with 5% CO2 and were archived by preservation in glycerol broth on glass beads at −80°C (12).
Extraction of DNA.
Genomic DNA suitable as template in the PCR was extracted with the Nucleon genomic DNA extraction kit (GeneSys Biotech Ltd., Coatbridge, Strathclyde, United Kingdom). DNA concentration was determined spectrophotometrically by measuring the absorbance at 260 nm.
Gene nomenclature.
According to the Nomenclature Committee of the ASM Publications Board, the genetic nomenclature used by Mooi et al. (15–17) to describe the genes coding for pertactin and pertussis toxin is nonstandard. We therefore propose the following: prn1, prn2, prn3, etc. (15), become prnA(1), prnA(2), prnA(3), etc., and ptxS1, ptxS2, ptxS3, ptxS4, and ptxS5 become ptxA, ptxB, ptxC, ptxD, and ptxE. Finally, we propose that variants of the pertussis toxin gene coding for the S1 subunit, ptxS1A, ptxS1B, ptxS1D, and ptxS1E (15), become ptxA(1), ptxA(2), ptxA(3), and ptxA(4). This proposed nomenclature is used throughout the text.
Gene typing.
Analysis of the genes coding for pertactin (prnA) and the S1 subunit of pertussis toxin (ptxA) was essentially as described by Mooi and colleagues (15, 17), with minor modifications. More than 95% of the prnA gene coding for the P.69 precursor, including the entire region encoding P.69 (Fig. 1), was determined for six clinical isolates, one each from 1942, 1982, 1983, and 1999, two from 1998, and the three United Kingdom WCV strains. The two polymorphic regions of the prnA gene, designated region 1 and region 2 (Fig. 1), were sequenced for a much larger number of strains. Region 1 was sequenced for all 285 clinical strains, and region 2 was sequenced for 73 of 285 clinical strains. This was done primarily in order to determine the existence of a single point mutation ca. 150 nucleotides upstream of region 2. Mooi et al. (15) proposed that strains with pertactin gene sequences identical to that of prnA(1) at region 1 but possessing this point mutation should be designated a distinct prnA type, i.e., prnA(7). Polymorphism in the gene coding for the S1 subunit of pertussis toxin (ptxA) was studied by analyzing 240 of the 285 strains comprising all available strains from each year of isolation, with the exception of the years 1982 to 1984, for which 28 of 73 available strains were analyzed (1982, 13 strains; 1983, 14 strains; and 1985, 1 strain). The ptxA genes of the three United Kingdom vaccine strains were also examined.
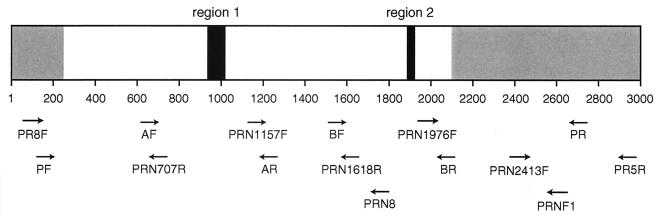
FIG. 1.
Schematic of the B. pertussis pertactin gene (prnA) coding for the P.69 precursor showing regions of polymorphism. Regions 1 and 2, which code for the repeats GGxxP and PQP, respectively, are shown in black. The region including the mature protein is shown in white. The regions removed from the precursor protein are shown in gray. The arrows show the approximate positions of primers used for PCR and sequencing. The figure was adapted with permission from the work of Mooi et al. (17). Details of primers can be found in Table 1.
PCR amplification and sequencing.
Oligonucleotide primers targeting the S1 subunit of the pertussis toxin gene (ptxA) of B. pertussis (S1F and S1R) were used to amplify a ca. 800-bp product encompassing three regions of variation (Table 1; Fig. 2). Reaction mixtures were in a total volume of 50 μl and contained 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 200 μM (each) deoxynucleotide, 10 pmol of each primer (MWG Biotech United Kingdom Ltd., Milton Keynes, United Kingdom), and 2.5 U of Taq polymerase (Life Technologies, Paisley, United Kingdom). Template DNA (ca. 10 to 100 ng) was added and reaction mixtures with no added DNA served as negative controls. Amplification was performed in a Hybaid Thermocycler (Hybaid) under the following conditions: predenaturation for 3 min at 95°C and then 30 cycles consisting of denaturation for 15 s at 95°C, annealing for 15 s at 59°C, extension for 1 min at 72°C, followed by a final extension for 10 min at 72°C. The ptxA gene was sequenced using primers S1F, S1M, and S1R (Table 1; Fig. 2).
TABLE 1.
Oligonucleotide primers used in this studya
| Primer name | Sequence (5′-3′) | Gene | Positionsb | Reference |
|---|---|---|---|---|
| PR8F | CCCATTCTTCCCTGTTCCAT | prnA | 81–100 | 15 |
| PF | TGTCTCTGTCACGCATTGTC | prnA | 152–171 | 17 |
| AF | GCCAATGTCACGGTCCAA | prnA | 649–666 | 17 |
| PRN707R | AGGGCGCCGATATGCAAG | prnA | 707–690 | This study |
| PRN1157F | CACCGCACGGCAATGTCAT | prnA | 1157–1175 | This study |
| AR | GCAAGGTGATCGACAGGG | prnA | 1234–1217 | 17 |
| BF | AGCTGGGCGGTTCAAGGT | prnA | 1542–1559 | 17 |
| PRN1618R | GGTCCGCGAAGACATTCAT | prnA | 1618–1600 | This study |
| PRN8 | AGGGTAAAGGTCGCCGCGCT | prnA | 1763–1744 | 5 |
| PRN1976F | ACGCGGCGGTCAACACG | prnA | 1976–1992 | This study |
| BR | CGGATTCAGGCGCAACTC | prnA | 2076–2059 | 17 |
| PRN2413F | GGCAAGTACCGCACCCAT | prnA | 2413–2430 | This study |
| PRNF1 | CAGTTCGATGCGCTTGCC | prnA | 2628–2611 | 5 |
| PR | ATGCCGTTGGTGTGTACCGT | prnA | 2714–2695 | 17 |
| PR5R | GCCTGAGCCTGGAGACTGG | prnA | 2949–2931 | 15 |
| S1F | TAGGCACCATCAAAACGCAG | ptxA | 474–493 | 17 |
| S1FM | ACAATGCCGGCCGTATCCTC | ptxA | 946–965 | 17 |
| S1R | TCAATTACCGGAGTTGGGCG | ptxA | 1350–1331 | 17 |
Oligonucleotide primers used for amplification and sequencing of the pertactin and pertussis toxin genes of B. pertussis.
FIG. 2.
Primary structure of the B. pertussis S1 pertussis toxin gene showing regions of polymorphism. To date, four ptxA variants have been described and designated ptxA(1), ptxA(2), ptxA(3), and ptxA(4). Dots indicate sequence identity with ptxA(3). Numbers indicate the positions of nucleotides relative to the start codon of AJ245366. Nonsilent mutations are shown in bold and associated amino acid changes are indicated beneath the relevant codons. The figure was adapted with permission from the work of Mooi et al. (15).
Oligonucleotide primers (AF and BR) targeting the pertactin gene (prnA) of B. pertussis were used to amplify a ca. 1,400-bp product encompassing region 1 and region 2 (Table 1; Fig. 1) (17). Reaction mixtures were as described above but contained 2.5 mM MgCl2 and 10% dimethyl sulfoxide. Thermal cycling was performed as follows: predenaturation for 5 min at 95°C and then 30 cycles consisting of denaturation for 20 s at 95°C, annealing for 30 s at 57°C, extension for 1 min at 72°C, followed by a final extension for 7 min at 72°C. The primary structure of >95% of the prnA gene (in the six strains described above) was determined following amplification using the primers PR8F and PR5R (Table 1). Region 1 was sequenced with primers AF and AR, while region 2 was sequenced with primers BF and BR (Table 1, Fig. 1).
The amplified products were purified using the Wizard PCR Preps purification system (Promega Corp., Madison, Wis.) and sequenced with the primers used for amplification in combination with internal primers (Table 1). The nucleotide sequence was determined by the dideoxynucleotide method (20) with purified PCR products using (i) the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase FS (PE Applied Biosystems), analyzing the products on a model 377 ABI DNA sequencer (PE Applied Biosystems), or (ii) the Dye Terminator Cycle Sequencing kit (Beckman Coulter), analyzing the products on a CEQ 2000 DNA Analysis System (Beckman Coulter).
Sequence analysis.
Sequence analyses were performed using the software package GeneBase (Applied Maths, Kortrijk, Belgium). For all analyses sequence data from forward and reverse sequencing primers were combined and aligned manually. New prnA sequence data were aligned with all available sequences for B. pertussis types prnA(1) to prnA(8), under the following GenBank accession numbers: prnA(1), AJ011091; prnA(2), AJ011092; prnA(3), AJ011093; prnA(4), AJ011015; prnA(5), AJ011016; prnA(6), AJ132095; prnA(7), AJ133784; and prnA(8), AJ133245 (Fig. 3). New ptxA sequence data were aligned with the available B. pertussis ptxA gene sequences for variants ptxA(1), ptxA(2), ptxA(3), and ptxA(4), under the following GenBank accession numbers: ptxA(1), AJ006155, AJ007363, AJ007364, and AJ245366; ptxA(2), AJ006157, AJ245367, and M14378; ptxA(3), A13359, AJ245368, and X16347; and ptxA(4), AJ006151 and AJ006159.
FIG. 3.
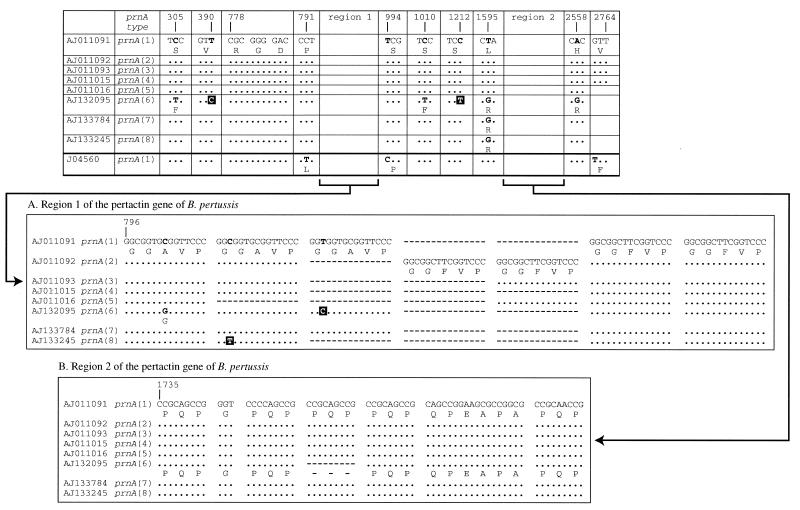
Primary structure of the B. pertussis prnA gene showing polymorphism in the eight pertactin types described to date, designated prnA(1) to prnA(8) (6, 15). Nonsilent mutations are shown in bold and associated amino acid changes are indicated beneath the relevant codons. Shaded nucleotides indicate silent mutations. Dots indicate sequence identity and dashes indicate that the sequence is not found. The two main regions of polymorphism comprising repeat units are region 1 near the tripeptide motif RGD and region 2 at the carboxyl-terminal region of the protein. (A) Region 1, comprising repeats coding for the amino acid sequence (GGxxP)4–6. (B) Region 2, comprising repeats coding for the amino acid sequence (PQP)4–5.
Statistical analysis.
Exact confidence intervals for proportions and P values were calculated using the STATA, v 6.0 (Stata Corp., College Station, Tex.), software package.
Nucleotide sequence accession numbers.
The sequences comprising >95% of the pertactin gene from the nine isolates of B. pertussis described above were deposited in GenBank, National Center for Biotechnology Information, Bethesda, Md. (http://www.ncbi.nlm.nih.gov), under the following accession numbers: AF34849 (CN351), AF348481 (DCH53), AF34842 (DCH132), AF348483 (98K300), AF348484 (98K320), AF348485 (99K45), AF348486 (CN2992), AF348487 (CN5476), and AF348488 (CN3099).
RESULTS
Polymorphism in the pertactin gene.
Results from sequencing the complete prnA coding region from the six clinical strains were as follows: CN351, prnA(1) (1942); DCH53, prnA(1) (1982); DCH132, prnA(2) (1983); 98K300, prnA(1) (1998); 98K320, prnA(2) (1998); and 99K45, prnA(3) (1999). The three strains used in the United Kingdom WCV (CN2992, CN3099, and CN5476) were all determined to belong to pertactin type prnA(1). The region sequenced comprised >95% of the prnA gene and included the entire coding region for the mature pertactin protein P.69. The nucleotide sequence of each of these nine complete sequences was identical to one of the prnA types, prnA(1) to prnA(3) previously designated by Mooi et al. (16). These three pertactin gene types vary in the number and composition of the repeat units GGxxP found in region 1 (Fig. 1 and 3).
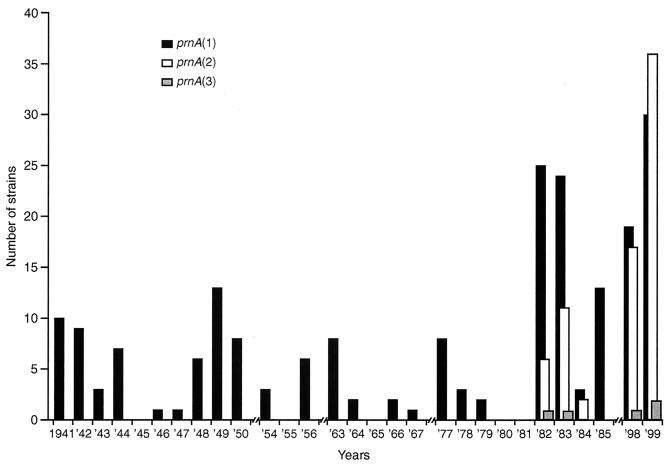
Region 1 (containing the GGxxP repeat motif) of the pertactin gene was sequenced for all of the 285 United Kingdom B. pertussis strains. Region 2 (containing the PQP repeat motif) was sequenced for 70 randomly selected strains of type prnA(1), two of type prnA(2), and one of type prnA(3). All 73 of the isolates examined for variation within region 2 contained five PQP repeat units (Fig. 3). The point mutation used to differentiate prnA(7) from prnA(1) was not found, i.e., all 70 of the 208 strains (Fig. 4) identified as prnA(1) from sequencing region 1 were confirmed as prnA(1) and not prnA(7) by sequencing region 2. Therefore, we can be 99% confident that <7.3% of the population described here as prnA(1) could actually have the single point mutation resulting in the designation prnA(7) proposed by Mooi et al. (15). In addition to the nine strains described above, all of the allelic prnA types found in the United Kingdom isolates were identical to prnA(1) to prnA(3) described by Mooi et al. (15); i.e., prnA(1) contained five GGxxP repeats, prnA(2) contained six, and prnA(3) contained five (Fig. 3). Figure 4 shows the temporal analysis of the prnA types of all the available circulating United Kingdom B. pertussis strains in the years 1941 to 1999. Cumulative figures for the frequency of prnA types between 1941 and 1979 show that all of 93 strains were type prnA(1), i.e., none were of a non-United Kingdom vaccine type (95% confidence intervals [CI], 0 to 4%). The single isolate from 1920 was also prnA(1). Pertactin types prnA(2) and prnA(3) were detected in strains isolated from 1982 onwards, and from 1982 to 1985, 21 of 86 (24%) isolates were non-prnA(1) (95% CI, 16 to 35%). No strains were available for examination from 1986 to 1997. Of the available strains analyzed from the late 1990s (1998 to 1999), 56 of 105 (53%) were non-prnA(1), i.e., of a non-United Kingdom WCV type (95% CI, 43 to 63%). While all the isolates prior to 1982 were prnA(1), the decrease in the proportion of prnA(1) strains isolated between 1982 and 1985 compared to those isolated in 1998 and 1999 is statistically significant (χ2 = 15.25, P = 0.0001).
FIG. 4.
Temporal trends in the frequency of pertactin gene (prnA) variants in the United Kingdom B. pertussis population. The number of strains containing distinct prnA variants was determined for each period shown on the x axis. Breaks in the x axis indicate periods for which no strains were available. The United Kingdom WCV contains prnA(1).
Polymorphism in the gene coding for the S1 subunit of pertussis toxin.
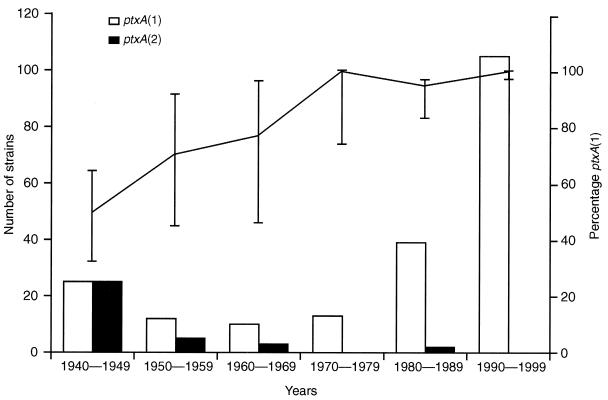
Four ptxA variants have been described to date, ptxA(1), ptxA(2), ptxA(3), and ptxA(4) (14) (Fig. 2). Only two variants, ptxA(1) and ptxA(2), were found in the United Kingdom B. pertussis population. Figure 5 shows the temporal analysis of the ptxA types of the circulating United Kingdom B. pertussis strains from 1940 to 1999. The ptxA type of the isolate from 1920 was ptxA(2). The frequency of ptxA(1) changed from 50% (25 of 50; 95% CI, 36 to 64%) in isolates from 1940 to 1949 to a predominance of ptxA(1) in the following years of 100% (105 of 105; 95% CI, 97 to 100%) in isolates from 1990 to 1999. A linear trend analysis of the change in proportion of ptxA(1) out of the total number of strains available was performed and found to be significant (χ2 = 6.394, P = 0.011). The strains used in the United Kingdom WCV comprised two ptxA types, ptxA(1) (CN3099, CN5476) and ptxA(2) (CN2992).
FIG. 5.
Temporal trends in the frequency of pertussis toxin S1 subunit gene (ptxA) variants in the United Kingdom B. pertussis population. The number of strains containing distinct ptxA variants was determined for each of the six decades shown. The proportion of strains containing ptxA(1) out of the total number of strains is shown for each period. Error bars show the 95% CI for these percentages. The United Kingdom WCV strains contain ptxA(1) and ptxA(2).
Relationship between serotype and pertactin type.
The majority of the available serotyping data were limited to the most recent years (1998 to 1999). However, to investigate any possible linkage between serotype and pertactin type, all available data are presented in Table 2. All of these isolates were pertussis toxin type ptxA(1).
TABLE 2.
Relationship between serotype and pertactin type of 61 United Kingdom isolates of B. pertussis
| Serotype | Pertactin gene (prnA) type | No. of isolates with indicated prnA type in:
|
||||
|---|---|---|---|---|---|---|
| 1964 | 1978 | 1979 | 1998 | 1999 | ||
| 1,2 | prnA(1) | 1 | 1 | 19 | 30 | |
| prnA(2) | 1 | 1 | ||||
| 1,3 | prnA(1) | 1 | 1 | |||
| prnA(2) | 16 | 28 | ||||
| prnA(3) | 1 | 2 | ||||
| 1,2,3 | prnA(1) | 1 | 1 | |||
DISCUSSION
Previous analyses of circulating strains of B. pertussis in The Netherlands and Finland (16, 17) appeared to demonstrate the emergence, over time, of pertactin and pertussis toxin gene variants distinct from those included in pertussis vaccines in each country. Concern has therefore been expressed regarding the optimal composition, and hence efficacy, of pertussis vaccines, particularly with regard to the marked increased incidence of pertussis seen in many countries, despite high vaccination coverage (17). The study presented here sought to investigate the possible genotypic change in the United Kingdom B. pertussis population following the introduction of the United Kingdom WCV. Although isolates of B. pertussis were not available for all years after 1940, a sufficiently large number of strains was examined to be considered reasonably representative of the situation in the United Kingdom both before and after the introduction of the United Kingdom WCV.
Genotypic variation of pertactin.
No variation was seen in the pertactin gene (prnA) type of any of the circulating isolates from 1941 to 1979, with 100% of the strains having the same prnA type [prnA(1)] as the three strains contained in the United Kingdom WCV: as noted previously, these three strains were chosen primarily to ensure that the final vaccine contained agglutinogens 1, 2, and 3 (25). Thus, prior to 1980, the situation in the United Kingdom would appear to be concordant with that seen in both The Netherlands and Finland (15, 16). The precise year of the emergence in the United Kingdom of prnA types distinct from those in the United Kingdom WCV cannot be determined; however, the two variants, prnA(2) and prnA(3), were found in strains from 1982 onwards. Similarly, in The Netherlands, non-prnA(1) types were first reported in isolates from 1981, and although no Finnish isolates were available between 1965 and 1981, the six isolates from 1982 were all prnA(2).
Whereas data from both The Netherlands and Finland show that by the 1990s ca. 90% of isolates were of a nonvaccine prnA type (16, 17), the United Kingdom data show that although the proportion of these nonvaccine prnA types has increased (to 24% in the 1980s and 53% in the 1990s), almost half of all circulating strains remain of the vaccine prnA type, i.e., prnA(1). If the pertactin-type content of a vaccine is a significant factor in its efficacy, then these data could explain the continued low level of pertussis in the United Kingdom compared to that in The Netherlands.
Although the study of Finnish isolates revealed a trend of strain divergence similar to that in The Netherlands, the decline in circulating prnA(1) strains was not accompanied by reemergence of pertussis (16). The authors of the above studies suggested several possible reasons for the differences between these two countries. One hypothesis was that a WCV containing prnA(1) may protect better against B. pertussis strains with certain prnA types, such as prnA(2), than against B. pertussis strains of other prnA types, such as prnA(3). In support of this, they noted that the proportion of prnA(3) in The Netherlands in isolates from 1996 was 63% compared with only 12% in Finland from 1990 to 1996. The United Kingdom data are concordant with this hypothesis, as the proportion of prnA(3) in United Kingdom isolates from 1998 to 1999 was only 3% (3 of 105).
Genotypic variation of pertussis toxin.
The hypothesis of variation driven by immune selection was also proposed to describe the emergence of pertussis toxin variants in the Dutch B. pertussis population (17). The Dutch WCV contained two strains, one of pertussis toxin type ptxA(2) and one of type ptxA(4). Both of these types were found in the 14 circulating strains analyzed from 1949 to 1954, with 8 of 14 (57%) being type ptxA(2) and 6 of 14 (43%) being type ptxA(3). In the following years no ptxA(3)-carrying strains were identified, the number of ptxA(2) strains decreased, and the variant ptxA(1) strain emerged, such that from 1990 to 1996, 87% were ptxA(1) and 13% were ptxA(2). Prior to 1976, the Finnish WCV contained only one strain (serotype 1,3) which was toxin type ptxA(3), but following an increase in the proportion of infection caused by serotype 1,2, a second strain containing agglutinogen 2 [toxin type ptxA(2)] was added to the vaccine (13, 16, 19). All of the 40 Finnish strains analyzed from the 1990s were found to comprise ptxA(1) (16).
The data for United Kingdom isolates again contrast with those reported previously. Only two pertussis toxin S1 subunit variants, ptxA(1) and ptxA(2), were found among the 240 circulating United Kingdom isolates of B. pertussis analyzed from 1920 to 1999. Both of these types are also present in the United Kingdom WCV. The proportion of ptxA(2) in circulating B. pertussis fell over time from 50% in the 1940s to 0% in the 1990s, with a corresponding increase to 100% for ptxA(1). No non-United Kingdom WCV ptxA types were found in any of the 240 isolates examined. The situation in the United Kingdom therefore appears unique in that all 105 of the most recent United Kingdom circulating isolates (from 1998 to 1999) are of a toxin type also found in the WCV used in this population. If the pertussis toxin is a major antigen in WCV, then these data might also explain why the United Kingdom WCV still appears to be highly effective.
The type ptxA(1) was not found in any of the Dutch circulating strains from 1949 to 1954 and was therefore considered a novel variant when it was identified in Dutch strains isolated between 1978 and 1985 (17). Similarly, in Finland ptxA(1) was not seen in the small number of circulating strains examined from 1953 to 1964 but was first seen in strains from 1982 (15). Clearly this toxin type was not a new variant in the 1980s but had been circulating in the United Kingdom for many years, the earliest occurrence in this series being in an isolate from 1949. These United Kingdom data, which represent by far the largest number of isolates which have been investigated for the pertussis toxin S1 (ptxA) genotype, argue against the emergence of nonvaccine ptxA types [e.g., ptxA(1)] by vaccine-driven selection.
Relationship of serotype to genotype.
Analysis of the limited serotyping data available for the United Kingdom isolates revealed a strong association between certain serotypes and pertactin types: 51 of 55 (93%) serotype 1,2 strains were prnA(1) and 44 of 47 (94%) serotype 1,3 strains were prnA(2). Strains of B. pertussis serotype 1,2 have been reported to cause more severe disease in the United Kingdom than other serotypes (23). All the clinical strains for which serotyping data were available were ptxA(1), and therefore an association between serotype and pertussis toxin type could not be investigated.
Genotypic designation of pertactin types.
To ensure that the United Kingdom data could be directly compared with those from previous studies, the protocols for both the pertactin and S1 toxin genotyping closely followed those of Mooi et al. (15, 17). For the pertactin gene this necessitated extensive sequencing of region 2 and ca. 150 nucleotides upstream of this region in order that strains could be accurately designated as either prnA(1) or prnA(7). In addition, the entire (>95%) prnA sequence was determined for a small number of isolates to investigate any additional variation outside regions 1 and 2. No point mutations outside regions 1 and 2 were found in any of the nine United Kingdom strains for which the complete prnA sequence was determined (six clinical isolates and three vaccine strains). However, on review of the original pertactin gene sequence published by Charles et al. (6), it can be seen that it differs from the prnA(1) described by Mooi et al. (16, 17) by three point mutations, two adjacent to region 1 and one downstream of region 2 (Fig. 3). Using the proposed nomenclature described by Mooi et al. (15), this strain should presumably be described as a novel prnA type. We would suggest that the designation of prnA types should be restricted to variation comprising silent or nonsilent mutations within the repeat motif (GGxxP)n of region 1. All of the following prnA types [prnA(1) to prnA(6) and prnA(8)], i.e., all except prnA(7), show such variation. Using this nomenclature, the original sequence reported by Charles et al. (6) and prnA(7) would belong to the pertactin type prnA(1). This would save the unnecessary cost of additional sequencing reactions for minimal return of information. Previous work by Charles and colleagues (7) reported that region 2 was particularly immunogenic, and thus it might be expected to be under greater selection pressure than region 1. Although no variation in region 2 was seen in this study, further work may be justified. However, for the reasons given above, variation in this second region should be described using a separate notation from that used for region 1.
Conclusions.
The resurgence of pertussis in some countries despite high vaccination coverage is certainly a cause for concern. Mooi et al. (15) have raised awareness of this, and their recommendation for a standard methodology in investigating the epidemiological typing of B. pertussis is an important step. However, the nomenclature for the description of genotypes, particularly of the pertactin gene, needs to be properly addressed to avoid confusion and unnecessary costs for any future studies of this kind.
The use of many different vaccines (both WCV and ACV) containing different strains and components, in addition to different regimens of vaccination by various countries, presents a complex picture. Further work into the phenotypic relevance of the nonsilent mutations of these and other components of pertussis vaccines, including expression, cross-protection, and transmission studies, particularly using relevant animal models, is needed to help elucidate the roles of these antigens in both pathogenesis and immunity (5, 26).
The only existing recommendations for the inclusion of specific strains in WCVs are based on the presence of agglutinogens (25). The composition of WCVs is therefore not defined with respect to individual protein variants (e.g., pertactin and pertussis toxin). Vaccine efficacy is multifactorial, and the impact of these distinct protein variants in WCVs may be relatively minor. However, acellular vaccines comprise a number of purified components of B. pertussis, such as pertactin, pertussis toxin, filamentous hemagglutinin, and fimbrial protein(s). These components are derived from individual strains of B. pertussis and therefore contain only single variants of each factor. To our knowledge the matching of such variants in ACVs with those expressed by the resident circulating strains in the population has not been considered. The impact of these distinct protein variants in ACVs may be greater than in WCVs. As the use of ACVs for pertussis increases, we would therefore recommend the vigilant surveillance of circulating B. pertussis strains, specifically monitoring any change in those factors included in the ACV, which may precede resurgence. This is particularly relevant to the situation in the United Kingdom, because since the completion of this study, ACVs for pertussis have been introduced in the United Kingdom (22). In addition to studies designed to determine more precisely the true incidence of pertussis infections, the use of comparative methods to analyze populations of B. pertussis and to determine the mechanisms responsible for their genotypic variation should allow meaningful debate on present and future vaccination policies for pertussis in the United Kingdom and other countries.
ACKNOWLEDGMENTS
We acknowledge Frits Mooi and his colleagues for providing laboratory protocols for the PCRs and for their helpful advice. We thank Ying T. Li for technical assistance with sequencing reactions and Debbie Owen, Pertussis Reference Laboratory, for serotyping data and clinical isolates. We also thank Barry Holmes, National Collection of Type Cultures; Andy Robinson, Centre for Applied Microbiology and Research; and Roger Parton, University of Glasgow, for provision of clinical isolates of B. pertussis. We gratefully acknowledge the help of Dorothy Xing, National Institute for Biological Standards and Control, in obtaining the United Kingdom whole-cell vaccine strain CN5476 and Shona Livingstone of the PHLS Statistics Unit for assistance and helpful discussion. We acknowledge the advice of John Coote, Roger Parton, Anne Moir, and the Nomenclature Committee of the ASM Publications Board regarding genetic nomenclature.
REFERENCES
- 1.Andrews R, Herceg A, Roberts C. Pertussis notifications in Australia, 1991 to 1997. Commun Dis Intell. 1997;21:145–148. doi: 10.33321/cdi.1997.21.30. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. Whooping cough notifications continue to fall: young unimmunised infants remain at highest risk. Commun Dis Rep. 1999;9:201–204. [PubMed] [Google Scholar]
- 3.Aricò B, Gross R, Smida J, Rappuoli R. Evolutionary relationships in the genus Bordetella. Mol Microbiol. 1987;1:301–308. doi: 10.1111/j.1365-2958.1987.tb01936.x. [DOI] [PubMed] [Google Scholar]
- 4.Bass J W, Wittler R R. Return of epidemic pertussis in the United States. Pediatr Infect Dis J. 1994;13:343–345. doi: 10.1097/00006454-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Boursaux-Eude C, Thiberge S, Carletti G, Guiso N. Intranasal murine model of Bordetella pertussis infection. II. Sequence variation and protection induced by a tricomponent acellular vaccine. Vaccine. 1999;17:2651–2660. doi: 10.1016/s0264-410x(99)00038-9. [DOI] [PubMed] [Google Scholar]
- 6.Charles I G, Dougan G, Pickard D, Chatfield S, Smith M, Novotny P, Morrissey P, Fairweather N F. Molecular cloning and characterization of protective outer membrane protein P.69 from Bordetella pertussis. Proc Natl Acad Sci USA. 1989;86:3554–3558. doi: 10.1073/pnas.86.10.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charles I G, Li J, Roberts M, Beesley K, Romanos M, Pickard D J, Francis M, Campbell D, Dougan G, Brennan M J, Manclark C R, Jensen M A, Heron I, Chubb A, Novotny P, Fairweather N F. Identification and characterization of a protective immunodominant B cell epitope of pertactin (P.69) from Bordetella pertussis. Eur J Immunol. 1991;21:1147–1153. doi: 10.1002/eji.1830210509. [DOI] [PubMed] [Google Scholar]
- 8.Cherry J D, Gornbein J, Heininger U, Stehr K. A search for serologic correlates of immunity to Bordetella pertussis cough illnesses. Vaccine. 1998;16:1901–1906. doi: 10.1016/s0264-410x(98)00226-6. [DOI] [PubMed] [Google Scholar]
- 9.Cherry J D, Olin P. The science and fiction of pertussis vaccines. Pediatrics. 1999;104:1381–1384. doi: 10.1542/peds.104.6.1381. [DOI] [PubMed] [Google Scholar]
- 10.De Serres G, Boulianne N, Douville-Fradet M, Duval B. Pertussis in Quebec: ongoing epidemic since the late 1980s. Can Commun Dis Rep. 1995;21:45–48. [PubMed] [Google Scholar]
- 11.Edwards K M, Decker M D, Mortimer E A. Pertussis vaccine. In: Plotkin S A, Orenstein W A, editors. Vaccines. 3rd ed. Philadelphia, Pa: W. B. Saunders Company; 1999. pp. 293–344. [Google Scholar]
- 12.Harrison T G, Taylor A G. A laboratory manual for Legionella. Chichester, United Kingdom: John Wiley and Sons, Ltd.; 1988. pp. 157–158. [Google Scholar]
- 13.Kuronen T, Huovila R. Seroresponse to pertussis vaccine. In: Hennessen W, editor. Recent advances in pertussis research. Conches, France: International Association of Biological Standardization; 1978. pp. 4–5. [Google Scholar]
- 14.Miller E, Fleming D M, Ashworth L A E, Mabbett D A, Vurdien J E, Elliot T S J. Serological evidence of pertussis in patients presenting with cough in general practice in Birmingham. Commun Dis Public Health. 2000;3:132–134. [PubMed] [Google Scholar]
- 15.Mooi F R, Hallander H, Von König C H W, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis. 2000;19:174–181. doi: 10.1007/s100960050455. [DOI] [PubMed] [Google Scholar]
- 16.Mooi F R, He Q, Van Oirschot H, Mertsola J. Variation in the Bordetella pertussis virulence factors pertussis toxin and pertactin in vaccine strains and clinical isolates in Finland. Infect Immun. 1999;67:3133–3134. doi: 10.1128/iai.67.6.3133-3134.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooi F R, Van Oirschot H, Heuvelman K, Van Der Heide H G J, Gaastra W, Willems R J L. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun. 1998;66:670–675. doi: 10.1128/iai.66.2.670-675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preston N W. Type-specific immunity against whooping cough. Br Med J. 1963;2:724–726. doi: 10.1136/bmj.2.5359.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston N W. Pertussis today. In: Wardlaw A C, Parton R, editors. Pathogenesis and immunity in pertussis. Chichester, United Kingdom: John Wiley and Sons Ltd.; 1988. pp. 1–18. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taranger J, Trollfors B, Lagergård T, Sundh V, Bryla D A, Schneerson R, Robbins J B. Correlation between pertussis toxin IgG antibodies in postvaccination sera and subsequent protection against pertussis. J Infect Dis. 2000;181:1010–1013. doi: 10.1086/315318. [DOI] [PubMed] [Google Scholar]
- 22.Therre H, Baron S. Pertussis immunisation in Europe—the situation in late 1999. Eurosurveillance. 2000;5:6–10. doi: 10.2807/esm.05.01.00001-en. [DOI] [PubMed] [Google Scholar]
- 23.Van Buynder P G, Owen D, Vurdien J E, Andrews N J, Matthews R C, Miller E. Bordetella pertussis surveillance in England and Wales: 1995–7. Epidemiol Infect. 1999;123:403–411. doi: 10.1017/s0950268899003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willems R J L, Mooi F R. From whole cell to acellular pertussis vaccines. Rev Med Microbiol. 1996;7:13–21. [Google Scholar]
- 25.World Health Organization. WHO Expert Committee on Biological Standardization, thirtieth report. Technical Report Series, no. 638. 1979. p. 61. , 65. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 26.Xing D K, Das R G, Williams L, Canthaboo C, Tremmil J, Corbel M. An aerosol challenge model of Bordetella pertussis infection as a potential bioassay for acellular pertussis vaccines. Vaccine. 1999;17:565–576. doi: 10.1016/s0264-410x(98)00235-7. [DOI] [PubMed] [Google Scholar]