Abstract
Background:
Though germline TP53 pathogenic/likely pathogenic variants (PV) are associated with Li-Fraumeni syndrome, many detected by multigene panels represent aberrant clonal expansion (ACE), most due to clonal hematopoiesis (CH). Discerning ACE/CH from germline variants and post-zygotic mosaicism (PZM) is critically needed for risk assessment and management.
Methods:
Participants in the Li Fraumeni & TP53 Understanding & Progress (LiFT UP) study with a TP53 PV were eligible. Demographics, personal/family cancer history and clinical laboratory test reports were obtained. DNA from multiple tissues was analyzed using a custom QIAseq® assay (ACE panel) that included TP53 and other CH-associated genes; the ACE panel and eyebrow follicles were assessed in a workflow to discern TP53 PV clinical categories.
Results:
Among 134 participants there was a significant difference for the age at diagnosis (p=<0.001), component cancers (p=0.007), and clinical testing criteria (p=<0.001), comparing germline vs PZM or ACE. ACE panel analysis of DNA from 55 sets of eyebrow follicles (mean 1.4 ug) and 36 formalin-fixed paraffin imbedded tissues demonstrated low variance (standard error 3%; p = 0.993) for TP53 variant allele fraction, with no significant difference (p=0.965) between tissue types, and detected CH gene PVs. Of 55 multi-tissue cases, germline status was confirmed for 20, PZM in 7, ACE for 25 and 3 were indeterminate. Additional CH variants were detected in 6 ACE and 2 germline cases.
Conclusions:
We demonstrated an effective approach and tools for discerning germline TP53 status.
Impact:
Discernment of PZM and TP53-driven CH increases diagnostic accuracy and enables risk-appropriate care.
Keywords: Li-Fraumeni Syndrome, aberrant clonal expansion, clonal hematopoiesis, clonal hematopoiesis of indeterminate potential, germline, pathogenic variant, variant allele fraction
Introduction
Germline TP53 pathogenic/likely pathogenic variants (PV) underlie Li-Fraumeni Syndrome (LFS), a rare inherited cancer susceptibility syndrome characterized by a predisposition to a broad spectrum of cancers and risk for multiple primary cancers at early ages (1–3). Core cancers include sarcoma, brain, breast, and adrenocortical cancers (4–6).
Putative germline TP53 variants identified through hereditary cancer multi-gene panel tests (MGPT) are often found in individuals and families who share limited features with classic LFS (7), raising questions about the validity of established penetrance estimates. In addition, low (<30%) variant allele fraction (VAF) TP53 PVs identified by next generation sequencing (NGS)-based MGPT are frequently due to aberrant clonal expansion (ACE) of marrow precursors with an acquired TP53 PV (8–12), and must be distinguished from germline (constitutional) variants and true post-zygotic mosaicism (PZM), given the different clinical implications.
Clonal hematopoiesis (CH) is increasingly recognized as a somatic finding that has clinical implications regarding therapy-related hematologic malignancies and all-cause mortality (13). Skin biopsy with fibroblast culture may be used when a non-hematological source of germline DNA is needed, but is invasive, costly and time consuming, and not all diagnostic laboratories can process the samples. We developed tools for detecting CH and discerning constitutional TP53 status, including an ACE panel to confirm the prevalence and clinical impact of TP53-associated ACE, determine co-occurrence of PVs in myeloid genes, and validate eyebrow follicles as a component of multiplex tissue analyses, including skin biopsies and formalin fixed paraffin-embedded (FFPE) normal and tumor tissues.
Finally, we include a schema and clinical criteria to validate the constitutional status of TP53 PVs across tissues, and systematically characterize TP53-driven CH and PZM cases.
Materials and Methods
Participants
Eligibility criteria included all individuals (n=134) with commercial germline test results reporting TP53 PV(s), including those with: (a) putative heterozygous germline, (b) VAF of less than 30%, (c) clinical history or molecular results suspicious for ACE (e.g., active hematologic neoplasia), or (d) an atypical clinical phenotype (i.e., not meeting TP53 clinical testing guidelines). Affected cases with a TP53 PV who met Li-Fraumeni syndrome (LFS) diagnostic criteria (supplemental table 1), and/or had informative cascade testing were considered germline (n=69), and additional tissues were requested for the remaining cases (n=65); tissues were obtained for 55 of the 65 cases (85%). All participants were accrued to the Li-Fraumeni & TP53 Understanding & Progress (LiFT UP) study (https://liftupstudy.org) and provided written informed consent for City of Hope IRB-approved protocol number 96144 (NIH clinical trial ID NCT04185935) June 2002 through October 2020. Participants were referred to study from the Li-Fraumeni Exploration Consortium (LiFE) (14,15), the Li-Fraumeni Syndrome Association (https://www.lfsassociation.org/), Living LFS (https://livinglfs.org/), the Clinical Cancer Genomics Community Research Network (16,17), genetic testing laboratories, and self-referral via a RedCap survey on the LiFT UP website.
Clinical Data
We obtained demographic information, personal/family cancer history, risk factor data via questionnaire, complete blood count and genetic testing reports (for the individual [including fibroblast testing from skin biopsies if available], as well as for cascade/transmission testing for family members) (17). We determined whether an individual met TP53 testing criteria (supplemental table 1) (5,18). Quantitative VAF data was obtained from genetic testing laboratories when possible; Agilent hybrid capture sequencing library preparation methodology was used for most of the respective CLIA-certified Laboratory Designed Tests.
Biospecimens
Whole blood and/or a saliva sample, eyebrow plucks, and an archival normal and/or tumor FFPE tissue sample were collected. Blood (serial samples over time for a subset) and saliva were collected and processed as reported previously (16). Eyebrows were cleaned with an isopropanol wipe and 15–20 undyed eyebrow follicles were self-collected with a sterile tweezer into a falcon tube, dry, or in 1x PBS, or in alcohol-free mouthwash. Eyebrows were plucked close to the base of the shaft and in the direction of the growth (19). Both normal tissue (non-lymphatic) and tumor tissues were micro dissected from FFPE tissue blocks on 10-micron unstained slides for DNA extraction.
DNA Extraction
DNA was extracted from biospecimens using QIAGEN extraction kits (QIAGEN, Inc., Germantown, MD, USA); the Flexigene® DNA kit was used for whole blood; the Gentra Puregene® Cell Kit was used for mouthwash; the QIAamp® DNA Mini Kit was used for the eyebrow follicles (20) and for FFPE tissue extraction (COH Molecular Pathology Core). Genomic DNA quality was determined by optical density (OD) 260/280 value, using a NanoDrop 2000c spectrophotometer (Thermo Fisher, Chino, CA).
Genotyping
We designed a custom QIAseq® (QIAGEN, Inc., Germantown, MD, USA) amplicon-based 81-gene panel (ACE panel) for detecting cancer predisposition variants, including TP53, and genes known or thought to be involved in CH (11,13,21,22). Content includes common ACE/CH-associated variants in genes such as ASXL1, ATM, CBL, CHEK2, EZH2, ETV6, DNMT3A, GNB1, GNAS, IDH1, IDH2, JAK2, KIT, NF1, MPL, NPM1, NRAS, MYD88, PHF6, PPM1D, RUNX1, SETBP1, SF3B1, SRSF2, STAT3, TET2, TP53, U2AF1, ZRSR2, and FLT3, and the design includes the 5’ and 3’ untranslated regions, full exonal gene coverage, and extends 10 base-pairs into introns, and results in a consistent coverage of over 300–500x. 150 base-pair, paired-end sequencing was performed on the HiSEQ 2500 Genetic Analyzer (Illumina Inc., San Diego, CA).
To assess test/retest reliability we created ACE panel libraries in triplicate for 14 samples. Sequencing libraries were also created with a custom 760 cancer gene panel (23,24) based on Agilent hybrid capture technology. TP53 PV-specific Sanger resequencing was also performed for 6 samples.
Genotyping Curation and Quality Control
Variant call format (VCF) files were evaluated using Ingenuity Variant Analysis (IVA) version 4 (QIAGEN, Inc., Germantown, MD, USA) as per previous studies (21). We removed variants with a call quality less than 20, read depth less than 10x or alleles with a frequency greater than 3% in the 1000 Genomes project, National Heart, Lung and Blood Institute, Exome Sequencing Project exome or ExAC databases. According to contemporary definitions of CH, all identified PVs with >1% VAF and at least 50x locus coverage were considered (21).
Data Analysis
All cases were included in analyses of clinical characteristics, and cases with additional tissue(s) were analyzed by the ACE panel and categorized according to the schema in Figure 1. Statistical analyses were performed by SAS® (v9.4TS1M3, Cary, NC, USA) and RStudio (version 1.4.1103). χ2 test of independence or Fisher’s exact test was carried out for descriptive analysis. The two-sided significance level was set at 0.05. Performance characteristics of the ACE panel and eyebrow pluck DNAs were also analyzed. Within-participant analysis of variance (ANOVA) test was used to determine variability of VAFs for triplicate samples on the ACE panel. VAF measured by the ACE panel, the 760 gene Agilent capture panel, and original commercial laboratory testing (if available) were compared, noting sampling timeframe. Pearson correlation coefficient (r) that measures a linear association was performed between number of eyebrow follicles and extracted DNA quantity. Between-participant ANOVA test was conducted to compare VAFs among normal/tumor tissues and eyebrows.
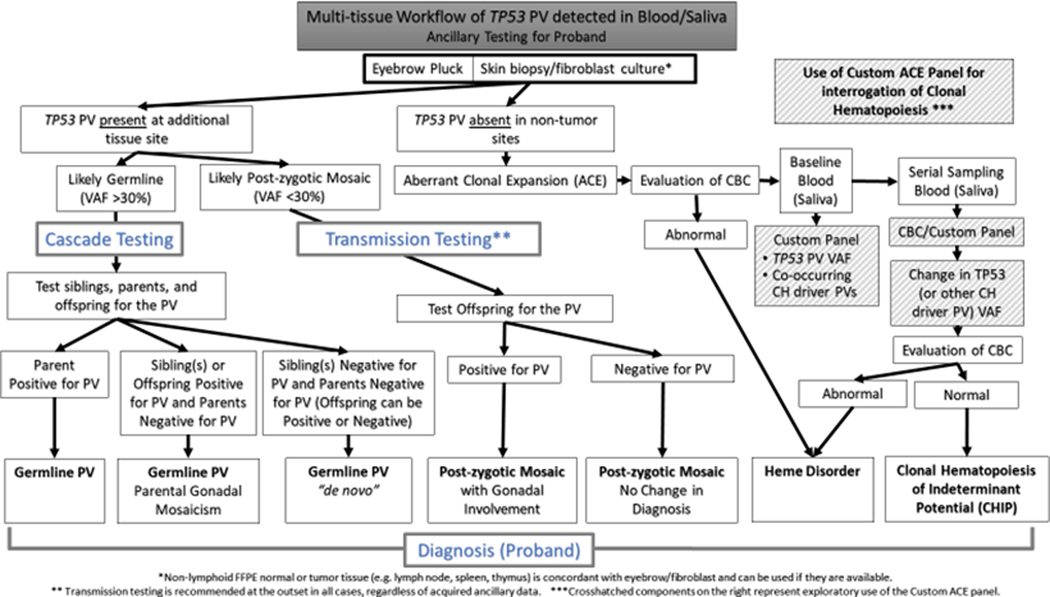
Figure 1.
Multi-tissue workup of TP53 pathogenic variants (PV) identified in germline (blood/saliva) commercial testing. This diagram depicts a diagnostic flowchart for establishing the presence/absence and variant allele fraction (VAF) of TP53 PVs in multiple tissues, often representing different embryonic germ layers, to discern and subclassify germline status including parental gonadal mosaicism, “de novo” germline status, post-zygotic mosaicism (PZM), or clonal hematopoiesis (CH) as ACE in a single tissue compartment. CH is further differentiated into clonal hematopoiesis of indeterminant potential (CHIP) if the complete blood count (CBC) is normal or extant hematologic neoplasia if not, and the approach for prospective evaluation of clonal evolution and genomic drivers of CH, and clinical state change is outlined.
Data Availability
All observed TP53 PVs have been reported previously in ClinVar by the respective commercial laboratories. All clonal hematopoiesis gene variants were included in Figure 3, and are also reported in ClinVar. All data necessary to replicate our findings are included in the manuscript and supplemental tables.
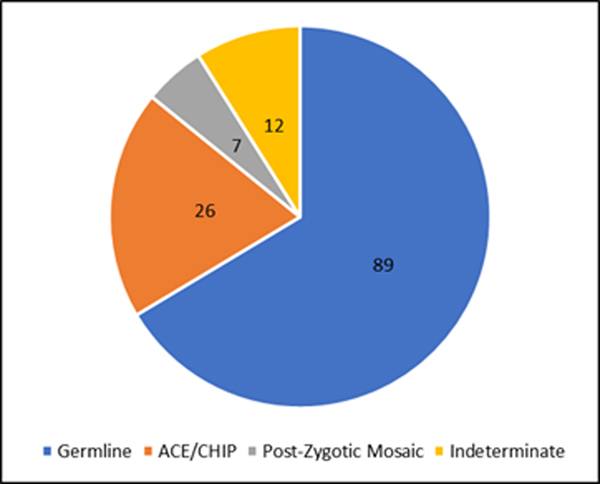
Figure 3.
Clinical category assignment for individuals with a TP53 pathogenic variants. The figure depicts the overall proportion of the clinical categories (germline, ACE/CHIP, PZM) found in this clinic based genetic testing cohort. Based on cascade/transmission testing, clinical criteria, and/or multi tissue analyses, 89/134 (66%) cases had evidence for germline, 26/134 (19%) were categorized as ACE, and 7/134 (5.2%) had evidence for PZM. Additional tissue or transmission data would be needed to discern PZM from broader ACE category for the 12/134 (9%) that were indeterminate.
Ethics Declaration
Protocol number 96144 (NIH clinical trial ID NCT04185935) was approved by the City of Hope IRB. All participants provided written informed consent.
Results
In total, 134 cases had commercial germline test results reporting TP53 PV(s), along with 125 blood samples, 31 saliva samples, 57 eyebrow samples, 17 tumor blocks, and 19 normal tissue samples that were analyzed as part of the study. Each case was assigned to a clinical category by way of pre-existing ancillary data. After an initial review, 69 cases were determined to be germline. Of the remaining 65 cases, additional tissues were obtained for 55 (85%) cases. After applying our analyses, we were able to assign clinical categories for 122/134 cases (12 cases remained indeterminate).
Participant characteristics and clinical categories are summarized in Table 1. Most participants were female (89%), white (57%) or Hispanic (26%), under the age of 50 years (70%) at accrual (average 43 years; range 1 – 82 years), had a personal history of cancer (89%) with an average age at diagnosis of 37 years (range 1 – 80 years). The most common cancer types included breast (n=100), sarcoma (n=28), lung (n=10), colorectal (n=9), thyroid (n=8), leukemia (n=7), brain (n=3), and adrenocortical (n=3). There was a significant difference for the age at entry (p=0.001), age at diagnosis (p=<0.001), component cancers (p=0.007), and presence/absence of clinical testing criteria (p=<0.001), among those discerned to be germline vs PZM or ACE (Table 1).
Table 1.
Participant characteristics.
| Characteristic | Range/Groups | N (%)1 | Clinical Categories1 | p-value7 | ||
|---|---|---|---|---|---|---|
|
|
||||||
| Germline | PZM | ACE | ||||
|
| ||||||
| Sex | Male | 15 (11) | 10 | 0 | 4 | 0.61 |
| Female | 119 (89) | 79 | 7 | 22 | ||
|
| ||||||
| Age at entry | 0–50 | 89 (70) | 68 | 6 | 10 | 0.001 |
| 51–71+ | 45 (30) | 21 | 1 | 16 | ||
|
| ||||||
| Race | White | 76 (57) | 47 | 5 | 20 | |
| Hispanic | 35 (26) | 28 | 2 | 3 | ||
| Asian American | 13 (10) | 9 | 0 | 2 | ||
| American Indian | 4 (3) | 1 | 0 | 1 | 0.559 | |
| African American | 1 (0.8) | 1 | 0 | 0 | ||
| Pacific Islander | 1 (0.8) | 1 | 0 | 0 | ||
| Unknown | 4 (3) | 2 | 0 | 0 | ||
|
| ||||||
| Cancer status | No | 15 (11) | 7 | 1 | 3 | 0.502 |
| Yes | 119 (89) | 82 | 6 | 23 | ||
|
| ||||||
| Age at cancer | 0–50 | 92 (77) | 71 | 6 | 11 | <0.001 |
| diagnosis 2 | 51–71+ | 27 (23) | 11 | 0 | 12 | |
|
| ||||||
| Cancer type 3 | Breast 4 | 100 | 63 | 5 | 26 | |
| Sarcoma | 28 | 28 | 0 | 0 | ||
| Adrenocortical carcinoma | 3 | 3 | 0 | 0 | ||
| Brain | 3 | 3 | 0 | 0 | ||
| Lung | 10 | 9 | 1 | 0 | 0.007 | |
| Colorectal | 9 | 5 | 1 | 1 | ||
| Thyroid | 8 | 4 | 1 | 3 | ||
| Leukemia | 7 | 3 | 0 | 4 | ||
| Other 5 | 31 | 18 | 2 | 9 | ||
|
| ||||||
| TP53 Testing | Meets | 71 (53) | 74 | 1 | 0 | <0.001 |
| Criteria 6 | Does Not Meet | 63 (47) | 15 | 6 | 26 | |
Total includes 12 indeterminate cases; these are not included in the statistical analyses of the clinical categories (n=122).
Age at first cancer diagnosis if multiple primaries
Percentage not provided due to some participants developing multiple primary cancers.
Including invasive breast cancer (n=90) and ductal carcinoma in situ (n=10).
Other cancers include: ovarian (n=5), melanoma (n=4), uterine (n=4), renal (n=4), gastric (n=3), pancreatic (n=2), aplastic anemia (n=1), bladder (n=1), cervical (n=1), non-Hodgkin’s lymphoma (n=1), parathyroid (n=1), phyllodes (n=1), prostate (n=1), tongue (n=1), and unknown (n=1).
TP53 Testing Criteria found in Supplemental table 3
χ2 test of independence or Fisher’s exact test on the three clinical categories.
PZM = post-zygotic mosaic; ACE = aberrant clonal expansion
Clinical categories were discerned according to a derived flowchart (Figure 1) for the 55 participants with multiple tissues analyzed with the ACE panel (Table 2 and supplemental figure 1), and additional CH-driver PVs were detected (Table 3).
Table 2.
TP53 pathogenic variant (PV) subtypes and variant allele fractions (VAFs) in different tissues
| TP53 PV SUBTYPES | Sample ID | Cancer Status (Y/N) | Cancer Diagnosis1 | Age at Diagnosis (year)2 | Blood VAF (%) | Eyebrow VAF (%) | Normal Tissue VAF (%) | Tumor Tissue VAF (%) | Skin Fibroblast VAF (%) | TP53 Testing Criteria | Transmission Testing |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GERMLINE | 001 | Y | Br | 54 | 53 | 47 | 43 | 46 | - | N | U |
| 002 | Y | Gastric | 47 | 50 | 50 | - | - | - | Y | - | |
| 003 | Y | Br | 47 | 52 | 50 | - | - | - | N | U | |
| 004 | Y | Br | 28 | 50 | 47 | 36 | - | - | Y | U | |
| 005 | Y | Br | 35 | 50 | 48 | - | - | - | Y | - | |
| 006 | Y | Br | 28 | 48 | 50 | - | - | - | Y | - | |
| 007 | Y | Br | 51 | 53 | 50 | - | - | - | N | - | |
| 008 | Y | CML | 33 | 50 | 51 | - | - | - | Y | Y | |
| 009 | Y | Mel | 43 | 46 | 48 | - | - | - | N | Y | |
| 010 | Y | Ov | 31 | 52 | 56 | - | - | - | Y | - | |
| 011 | Y | Pan | 46 | 44 | 52 | - | - | - | N | - | |
| 012 | N | - | 35 | 53 | 53 | - | - | - | N | U | |
| 013 | N | - | 24 | 54 | 47 | - | - | - | N | - | |
| 014 | Y | Ut | 42 | 49 | 51 | - | - | - | N | - | |
| 015 | Y | Br | 33 | 48 | 52 | - | - | - | Y | - | |
| 016 | Y | Ut | 26 | 50 | 50 | - | - | - | Y | - | |
| 017 | Y | Lung | 69 | 443 | 47 | - | - | - | Y | - | |
| 018 | Y | Br | 43 | 35 | 15 | 41 | 47 | >30 | N | U | |
| 019 | Y | Br | 33 | 463 | 50 | 51 | - | - | Y | - | |
| 020 | Y | Thyroid | 26 | 43 | 45 | - | - | - | N | Y | |
| PZM | 021 | Y | CRC | 27 | 183 | 47 | 7 | 6 | - | N | - |
| 022 | Y | Br | 28 | 28 | 18 | 20 | - | 20 | Y | U | |
| 023 | N | - | 30 | 28 | 14 | - | - | 29 | N | U | |
| 024 | Y | Br | 36 | 15 | 36 | 12 | 9 | - | N | - | |
| 025 | Y | DCIS | 42 | 14 | 8 | 8 | 33 | - | N | - | |
| 026 | Y | Br | 45 | 40 | - | 20 | 0 | 20 | N | - | |
| 027 | Y | Br | 43 | 383 | 17 | 30 | - | - | N | - | |
| ACE/CH | 028 | Y | Br | 37 | 20 | 0 | - | - | - | N | - |
| 029 | Y | Br | 52 | 33 | 0 | - | - | - | N | - | |
| 030 | Y | Br | 42 | 13 | 0 | - | - | - | N | - | |
| 031 | Y | Br | 57 | 17 | 0 | 0 | 0 | - | N | - | |
| 032 | N | - | 42 | 9.4 | 0 | - | - | - | N | - | |
| 033 | N | - | 38 | 203 | 0 | - | - | - | N | - | |
| 034 | Y | Br | 61 | 153 | 0 | 0 | - | - | N | - | |
| 035 | Y | Pr | 80 | 203 | 0 | 0 | - | 0 | N | U | |
| 036 | Y | Br | 28 | 33 | 0 | - | - | - | Y | - | |
| 037 | Y | Br | 56 | 20 | 0 | 0 | - | 0 | N | - | |
| 038 | Y | Ov | 65 | 23 | 0 | 0 | - | - | N | - | |
| 039 | Y | Br | 43 | 36 | 0 | - | - | - | N | - | |
| 040 | Y | Br | 63 | 52 | 0 | 0 | - | 0 | N | - | |
| 041 | Y | Br | 41 | 19 | 0 | - | - | - | N | - | |
| 042 | Y | Br | 37 | 39 | - | - | - | 0 | N | - | |
| 043 | Y | Br | 32 | 22 | - | - | - | 0 | N | - | |
| 044 | Y | Br | 72 | 33 | - | - | - | 0 | N | - | |
| 045 | Y | DCIS | 46 | 50 | - | - | - | 0 | N | U | |
| 046 | Y | Thyroid | 50 | 17 | - | - | - | 0 | N | - | |
| 047 | N | - | 68 | 22 | 0 | - | - | - | N | - | |
| 048 | Y | CRC | 20 | 0,<304 | - | - | >30 5 | - | N | - | |
| 049 | Y | Br | 67 | <306 | 06 | 0 | 0 | - | N | - | |
| 050 | N | - | 26 | 36 | 0 | - | - | - | N | - | |
| 051 | Y | Br | 61 | 16 | 0 | - | - | - | N | - | |
| 052 | Y | Renal | 71 | 30 | 0 | - | - | - | N | - | |
| INDETERMINATE | 053 | Y | Br | 31 | 20 | - | - | - | - | N | - |
| 054 | Y | Br | 56 | 23 | 0 | 5 | 0 | - | N | U | |
| 055 | Y | Ov | 63 | 16 | - | - | - | - | N | - |
ACE/CH=ABERRANT CLONAL EXPANSION/CLONAL HEMATOPOIESIS; PZM=POST-ZYGOTIC MOSAICISM; U=PERFORMED; UNINFORMATIVE; Y=PERFORMED; INFORMATIVE
FIRST CANCER, IF MULTIPLE. ABBREVIATIONS: BR=BREAST CANCER; CML=CHRONIC MYELOGENOUS LEUKEMIA; MEL=MELANOMA; OV=OVARIAN CANCER; PAN=PANCREATIC ADENOCARCINOMA; UT=UTERINE CANCER; CRC=COLORECTAL CANCER; DCIS=DUCTAL CARCINOMA IN SITU;
AGE AT ENTRY IF UNAFFECTED.
ADDITIONAL CH DRIVER VARIANTS FOUND IN BLOOD.
CASE TESTED TWICE.
DOCUMENTED TO BE CIRCULATING TUMOR DNA.
WHOLE GENE DELETION; SEMI-QUANTITATIVE FROM COMMERCIAL LABORATORY; NOT DETECTED BY ACE PANEL, THOUGH CNV ANALYSES NOT YET VALIDATED.
Table 3.
Additional CH variants found on the ACE panel.
| Gene | Variant | VAF | |
|---|---|---|---|
|
| |||
| ACE | |||
|
| |||
| ATM | c.8096C>T | 3.8, 2.7, 3.81 | |
| DNMT3A | c.*3595del | 2.2 | |
| TET2 | c.1360A>T | 23.4, 18.7, 21.31 | |
| TET2 | c.3467del | 1.1, 1.4, 1.11 | |
| TP53 | c.350G>T | 16.5, 13.2, 15.01 | |
| TP53 | c.669del | 1.0, 1.0, 1.01 | |
|
| |||
| Germline | |||
|
| |||
| DNMT3A | c.1308C>G | 2.0 | |
| DNMT3A | c.894dup | 2.8 | |
Test/retest VAF consistency
High quality genomic DNA was obtained from 55 of 57 eyebrow samples. Two failed due to insufficient quantity of follicles. DNA yield from eyebrows ranged from 0.5 μg to 4.7 μg; (average 1.4 μg) (Supplemental Table 2); OD 260/280 value ranged from 1.77 to 2.96 (average 2.10), and there was a linear correlation between number of follicles and DNA quantity (Supplemental Figure 2). Eyebrow DNA yield (range 0.5 μg to 18.4 μg; average 3.61 μg; average OD ratio 2.19).
We compared TP53 VAFs in eyebrows to normal and tumor tissues (Supplemental Table 3). Normal and tumor tissues were micro-dissected from archival tumor blocks and showed comparable TP53 VAF. TP53 VAFs detected in eyebrows were highly correlated with VAFs in normal and tumor tissues, with a between-participant ANOVA test of F ~ F (2,35) = 0.0357 and p = 0.965.
There was no significant variability (p = 0.993) of VAF for within-participant ACE panel analysis (standard error 3%) with 3 separate libraries created from the same DNA sample (Figure 2). In addition, we compared the result from our custom capture, and the VAF obtained from commercial laboratory MGPT. There was no significant difference for VAF between ACE panel and Agilent capture/commercial testing (p=0.895). Semi-quantitative Sanger sequencing was done for 6 cases, but not included in the statistical analysis.
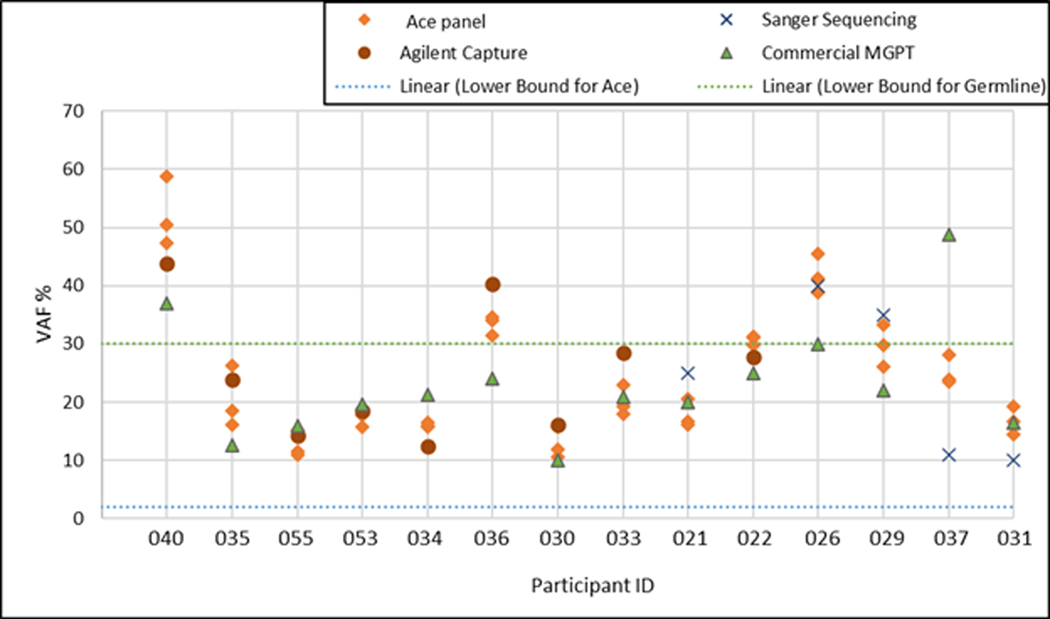
Figure 2.
Variant allele fraction (VAF) of a TP53 pathogenic variant tested on different platforms. The custom The QIAseq® amplicon-based 81-gene panel (ACE Panel) was used for assessment of VAF across blood samples from 14 participants. Test-retest variability (reproducibility) was assessed in triplicate. TP53 VAFs detected on the ACE panel were highly reproducible, as the within-participant standard error was 3%, with no statistically significant variability by ANOVA (p = 0.993). There was consistency/correlation of VAF across samples as well as across platforms (commercial laboratory multigene panel testing (MGPT); custom Agilent Hybrid capture panel) (p=0.895). Sanger sequencing values noted but not included in analysis as a semi-quantitative method. Note that Table 3 lists additional CH driver variants detected by the ACE panel, wherein, the test/retest VAF consistency is also shown for these secondary CH variants.
Beyond quantification and validation of the reported TP53 VAF, all 55 cases in the multi-tissue component cohort were also analyzed by the ACE panel for evidence of known CH driver PVs. Additional CH-associated PVs (ATM, DNMT3A [n=3], TET2 [n=2] and TP53 PVs [n=2]) were identified in addition to the known TP53 PV in 6 of 25 ACE cases and in 2 germline cases (Table 3). Note that detection of additional CH-driver PVs, while of great interest and more prevalent in the ACE cases, were not used to categorize a given case.
The VAF for these CH-PV also demonstrated consistent results on test-retest for the 4/7 cases included in the validation experiment (marked with * in Figure 2; respective VAFs noted in Table 3) and approximated that for the known TP53 PV for 2 cases; the others were at a lower VAF (range 1–3.4%). Four of seven cases with an additional CH-PV received prior cytotoxic therapy for cancer, and one is in long-term clinical remission from aplastic anemia. One case with two TET2 PVs was a 79-year-old male with a family history of pancreatic cancer and was unaffected at the time of commercial testing but was diagnosed with locally advanced prostate cancer one year later.
There were 69 affected cases with a TP53 PV who met Li-Fraumeni syndrome (LFS) diagnostic criteria, and/or had informative cascade testing and were considered germline. Additional tissues were obtained for 55 of the remaining 65 cases (85%). Multi tissue discernment according to the schema in Figure 1 was successful for 52 of 55 cases. Figure 3 depicts the proportions in context for the total cohort of 134: 89 (66%) were germline, 26 (19%) were ACE, and 7 (5.2%) were PZM; each of the PZM cases demonstrated the presence of the TP53 PV in tissues representing at least 2 germ layers. Of the 12 (9%) cases that remained indeterminate, tissues were not available for 8 cases, 3 cases the secondary biospecimen failed and 1 case (case 54) where potential infiltrating inflammatory cells confounded our categorization. Quality check indicated that the eyebrow samples for these cases failed due to collection of < 5 follicles. There is a direct relationship between the number of follicles and DNA quantity and quality, with a yield of at least 0.5 ug from ≥15 follicles (Supplemental Table 2 and Supplemental Figure 2).
Discussion
Prior to the advent of NGS based-MGPT, commercial germline testing of TP53 has been limited to individuals and families who met specific LFS testing criteria, such as classic, Chompret, Birch, Eeles, or NCCN (5,18,25). Our multi-tissue testing schema enabled discernment of patients with TP53 PVs identified on MGPT that did not have clinical characteristics (or meet TP53 testing criteria) of LFS, revealing a high proportion with ACE. These participants rarely met TP53 testing criteria (4%), had no core LFS cancers beyond breast cancer (5), and if affected were diagnosed with cancer at ages that were more typical of sporadic disease. Two recent studies noted a similar discordance between testing criteria and clinical category in the context of ACE/CH cases (12,26).
Our approach also illuminated participants with mosaicism. We observed that 59% of the cases selected for multi-tissue analysis were either ACE (46%) or PZM (13%), presumably reflecting selection bias from focus on cases that raised concerns about the phenotype. The overall categorical assignments across our clinical genetic testing cohort reflect a 19% rate of ACE plus 5% PZM (total is 24%), which is similar to the reported rates of ACE from three commercial laboratory series wherein 22–40% MGPT-detected TP53 PVs represent ACE (8,9,12,26,27). Clearly one cannot discern PZM cases, where there is an undefined but likely increased cancer risk depending on tissue distribution, without a multi-tissue approach representing the respective germ layers (9,28,29). Based on our study, we believe there is likely a similar proportion of PZM cases among the ACE cases in the other reported studies. Similar to a report from one genetic testing vendor (12), we observed apparent PZM among 13% of the 55 cases selected for multi-tissue analysis; this represents 5% of our overall genetic testing cohort (Figure 3). Importantly, TP53-associated PZM is still potentially transmissible to offspring (30) and is likely associated with increased cancer risk depending on tissue distribution of the PV. In addition, beyond potential misapplication of LFS-related care, failure to exclude ACE and PZM cases would bias TP53 penetrance estimates. VAF for a heterozygous germline PV is typically around 50% but can range from 30 – 60% (31). PZM is suspected when multiple tissues demonstrate a comparable low VAF (<30%). Our data for the PZM cases showed that the VAF varied, and was as high as 47% which overlaps the germline range. Further TP53-associated ACE may also have a VAF overlapping with the range seen in PZM. Thus, TP53 testing of ancillary tissues and transmission testing is paramount to distinguish between these two states.
In many cases, ACE reflects CH of indeterminate potential (CHIP) (32), which is observed in healthy populations at increasing frequency with increasing age (33,34). We created an amplicon-based MGPT that includes ACE/CH-specific genes for this study (LiFT UP). We demonstrated the validity of the ACE panel through test/re-test (p=0.993; 3% within-participant standard error) as well as across other NGS-based MGPT platforms (p = 0.895). The ACE panel performed equally well on all biospecimens, thus allowing comparison across tissues, and performed as designed to detect additional CH-driver PVs. TP53-associated CHIP is the 5th most common finding in previous studies, and along with other acquired myeloid neoplasia-related gene PVs, has been linked with increased risk of hematological neoplasia, cardiovascular diseases, and all-cause mortality (11,13,33).
The prevalence of additional CH variants in ACE cases is unknown, since hematopoietic genes are not included in most available clinical MGPT. We observed additional PVs in 24% of ACE cases and 10% of the germline cases that were analyzed with the ACE panel. Previous reports on CH suggest that there is an increased risk for progression to hematologic neoplasia when more than 1 driver PV is identified (13,32,35,36). Data from a large institutional tumor sequencing project provided evidence for both induction of CH-PVs in hematologic stem cell precursors by cytotoxic cancer therapies, as well as selection for CH (36). Understanding the clinical implications of TP53-driven CH is important, given the evidence suggesting that CH initiating variants often precede exposures associated with therapy-related myeloid neoplasia (tMN) (36–38), rather than the traditional thought that tMN develops from cancer therapy per se. Further, there is evidence from studies of the genomic landscape of MDS that highlight the critical effect of acquired TP53 allelic state in particular, demonstrating poorer outcome for multi-hit cases, and affirm the relevance of other CH PVs (e.g., TET2, ASXL1) in the setting of monoallelic TP53 PVs (22). Our observation of additional CH-PVs documented in 2 of 20 (10%) germline cases poses a related question about whether a patient with a germline TP53 PV is at risk for developing CH and/or myeloid neoplasms. Though leukemia was part of the original LFS phenotype (39), it is no longer considered a core cancer in the syndrome. Further studies of the prevalence of CH among germline TP53 carriers is warranted. There may be clinical utility for serial CBCs and ACE panel analyses (ideally with a CLIA certified assay) in ACE/CH cases to measure VAF change over time, subsequent acquisition of other CH-driver PVs, and monitoring for the potential development of hematological neoplasia. Given limited access to germline hematologic panels and lack of consensus in guidelines regarding workup of apparent CH/CHIP, clinicians may consider use of the ACE panel developed for this study, exploratory though we believe such a tool should be made clinically available.
Clinically, a skin biopsy with fibroblast culture is used when a non-hematological source of germline DNA is needed. Our data shows reliable and reproducible extraction of high-quality genomic DNA from eyebrows, correlation with results from skin biopsies and FFPE normal and tumor tissues. This supports the integration of eyebrows as a biospecimen type by commercial laboratories, as suggested by a recent statement of the American College of Medical Genetics and Genomics (40). Plucked hair follicles are an easily collected and less invasive alternative to skin biopsy, particularly if a small quantity of high-quality DNA will suffice for clinical diagnostics. We recently used eyebrow follicles to help characterize the distribution of a DNMT3A PV in a patient with PZM for that gene (28). Our procedures and schema (Figure 1) may be applicable to other testing scenarios where there is the possibility of ACE confounding diagnosis, such as ATM or CHEK2 PVs with low VAF (8).
Selection of cases for application of the multi-tissue strategy was biased toward inclusion of those lacking classic LFS features or evidence of transmission among family members, so it was not surprising that half of the germline cases did not meet testing criteria for LFS, and that ACE was prevalent (46%; supplemental figure 2). Cascade or transmission testing was subsequently able to independently discern germline status in 3 cases. When considering our cohort as a whole (Figure 3), the proportion of cases with ACE and PZM was 24%, supporting that the respective population was comparable to published series from commercial laboratories, where the rate of ACE ranges from 22%−40% (8,9,12,27).
For all ACE and PZM cases, more than one additional tissue was needed for discernment, and demonstrated the absence of the TP53 PV in additional tissues associated with ACE/CH or the presence of the TP53 PV in low levels (2–30%) across other tissues in the case of PZM. Somewhat surprising was the prevalence of PZM (5% overall; 13% among cases selected for analysis), which could not have been discerned without the multi-tissue approach. Transmission testing is indicated as a part of clinical care due to the possibility of transmission to offspring and should be evaluated in all suspected ACE/PZM cases, though it may only help in discernment of the clinical category if it is positive (30).
Circulating tumor DNA (ctDNA) in an advanced case may confound blood/saliva-based testing and may require ancillary testing at different timepoints to distinguish from PZM or germline. For example, case 48, the single ctDNA case in our cohort, was categorized as such as the TP53 variant was not initially detected in blood used as companion normal to his colorectal tumor testing,but was detected approximately a year later when he underwent MGPT pre-terminally with widespread metastatic disease. Had the initial blood sample not been reviewed, one may have categorized this case as germline based on our proposed schema.
In case 54, the absence of the TP53 PV in eyebrow plucks and normal tissue are the strongest indicator that this is an ACE case with likely CH. The gradient of 23% VAF in blood compared to 5% in tumor suggested contamination of the tissue with hemorrhage/inflammatory cell infiltrate. Nonetheless, to be conservative we designated the case as indeterminate. As for case 48 above, this illustrates that nuanced judgement of the multi-tissue genomic data may be warranted in some cases beyond the suggested workflow in Figure 1. Cases cited in our previous publication demonstrated low level detection of the respective PV in the tumor and/or “normal” tissues commensurate with inflammatory cell infiltrate (9). Thus, reliably “normal” tissues such as skin fibroblasts or in our case eyebrows were the most valuable for discernment. Given that 8 cases remained indeterminate due to inability to obtain additional samples such as fibroblast cultures or eyebrow plucks, and 3 cases in which the secondary biospecimen failed, FFPE tissue analysis may be valuable for discernment if preferred ancillary normal tissues are not available and for more in depth interrogation of PZM cases.
To conclude, our work has direct translational impact, as discerning the appropriate TP53 clinical category will increase the diagnostic accuracy of genetic testing and direct clinical care for individuals with TP53 PVs (40), as well as allow identification of additional CH-associated PVs and the potential for progression to myeloid neoplasia. False positive or incidental findings on surveillance can lead to diagnostic procedures and surgical misadventure and can have enormous psychological and financial impacts on individuals and families. In addition, unrecognized TP53-associated ACE will confound studies of prevalence and penetrance for individuals with LFS (8). We are currently using the tools and approach in this study to validate TP53 status and answer these questions in LiFT UP, a large prospective cohort study, that also includes a prospective natural history study of TP53-driven CH. More precise estimation of penetrance with specific TP53 PV subtypes, coupled with new insights about clinical and molecular genetic risk modifiers, and reliably discerning TP53 clinical category will enable risk-appropriate allocation of cancer surveillance and risk-reduction interventions.
Supplementary Material
Acknowledgements
We thank the Li Fraumeni Syndrome Association for their support of this and related studies, David Goldgar for his contributions to study design, and Thomas Springer for assistance during his summer internship.
Funding
The work was supported in part by the National Institutes of Health, (Grant numbers R01 CA242218 [MPI: JNW, JEG, CA], RC4 CA153828 [JNW]); National Institutes of Health Cancer Center Support Grant (P30 CA033572); Breast Cancer Research Foundation (#20-172 [JNW]), and American Society of Clinical Oncology Conquer Cancer® Research Professorship in Breast Cancer Disparities (JNW), and the Li Fraumeni Syndrome Association.
The Role of the funders:
support for subject recruitment, acquisition processing and analysis of biospecimens and data.
J.N.W. is a speaker for the Bureau for AstraZeneca, and an employee for Natera. T.S. is an employee shareholder at Myriad Genetics Laboratory. B.L.E received research funding from Celgene, Deerfield, Novartis, Calico and consulting fees from GRAIL. He is also a member and shareholder of the scientific advisory board for Neomorph Therapeutics, Skyhawk Therapeutics and Exo Therapeutics, none of which are directly related to the content of this paper. J.E.G. received commercial research support from Novartis and is a consultant/advisory board member for GTx, Helix, and Novartis.
Footnotes
Prior presentations
Portions of the work were presented as a poster at the 2020 annual meeting of the American Society of Clinical Oncology.
Conflicts of Interest
The other authors declare no conflict of interest.
References
- 1.Li FP, Fraumeni JF Jr., Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. Journal of the National Cancer Institute 1969;43(6):1365–73. [PubMed] [Google Scholar]
- 2.Malkin D, Li FP, Strong LC, Fraumeni JF Jr., Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 1990;250:1233–7. [DOI] [PubMed] [Google Scholar]
- 3.Frebourg T, Kassel J, Lam KT, Gryka MA, Barbier N, Andersen TI, et al. Germ-line mutations of the p53 tumor suppressor gene in patients with high risk for cancer inactivate the p53 protein. Proceedings of the National Academy of Science USA 1992;89:6413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBride KA, Ballinger ML, Killick E, Kirk J, Tattersall MHN, Eeles RA, et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol 2014;11(5):260–71 doi 10.1038/nrclinonc.2014.41. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, et al. Beyond Li Fraumeni Syndrome: Clinical characteristics of families with p53 germline mutations. Journal of Clinical Oncology 2009; 27( 8):1250–6 doi 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 6.Bougeard G, Renaux-Petel M, Flaman J-M, Charbonnier C, Fermey P, Belotti M, et al. Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. Journal of Clinical Oncology 2015. doi 10.1200/jco.2014.59.5728. [DOI] [PubMed] [Google Scholar]
- 7.Rana HQ, Gelman R, LaDuca H, McFarland R, Dalton E, Thompson J, et al. Differences in TP53 Mutation Carrier Phenotypes Emerge From Panel-Based Testing. J Natl Cancer Inst 2018;110(8):863–70 doi 10.1093/jnci/djy001. [DOI] [PubMed] [Google Scholar]
- 8.Slavin TP, Coffee B, Bernhisel R, Logan J, Cox HC, Marcucci G, et al. Prevalence and characteristics of likely-somatic variants in cancer susceptibility genes among individuals who had hereditary pan-cancer panel testing. Cancer Genet 2019;235–236:31–8 doi 10.1016/j.cancergen.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weitzel JN, Chao EC, Nehoray B, Van Tongeren LR, LaDuca H, Blazer KR, et al. Somatic TP53 variants frequently confound germ-line testing results. Genet Med 2018;20(8):809–16 doi 10.1038/gim.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swisher EM, Harrell MI, Norquist BM, Walsh T, Brady M, Lee M, et al. Somatic Mosaic Mutations in PPM1D and TP53 in the Blood of Women With Ovarian Carcinoma. JAMA Oncol 2016;2(3):370–2 doi 10.1001/jamaoncol.2015.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolton KL, Gillis NK, Coombs CC, Takahashi K, Zehir A, Bejar R, et al. Managing Clonal Hematopoiesis in Patients With Solid Tumors. Journal of Clinical Oncology 2019;37(1):7–11 doi 10.1200/jco.18.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mester JL, Jackson SA, Postula K, Stettner A, Solomon S, Bissonnette J, et al. Apparently Heterozygous TP53 Pathogenic Variants May Be Blood Limited in Patients Undergoing Hereditary Cancer Panel Testing. J Mol Diagn 2020;22(3):396–404 doi 10.1016/j.jmoldx.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science 2019;366(6465):eaan4673 doi 10.1126/science.aan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mai PL, Malkin D, Garber JE, Schiffman JD, Weitzel JN, Strong L, et al. Li-Fraumeni syndrome: Report of a clinical research workshop and creation of a research consortium. Cancer Genetics 2012;205(10):479–87 doi 10.1016/j.cancergen.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mai PL, Sand SR, Saha N, Oberti M, Dolafi T, DiGianni L, et al. Li-Fraumeni Exploration Consortium Data Coordinating Center: Building an Interactive Web-Based Resource for Collaborative International Cancer Epidemiology Research for a Rare Condition. Cancer Epidemiol Biomarkers Prev 2020;29(5):927–35 doi 10.1158/1055-9965.Epi-19-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slavin T, Neuhausen SL, Rybak C, Solomon I, Nehoray B, Blazer K, et al. Genetic Gastric Cancer Susceptibility in the International Clinical Cancer Genomics Community Research Network. Cancer Genet 2017;216–217:111–9 doi 10.1016/j.cancergen.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavarri-Guerra Y, Hendricks CB, Brown S, Marcum C, Hander M, Segota ZE, et al. The Burden of Breast Cancer Predisposition Variants Across The Age Spectrum Among 10 000 Patients. Journal of the American Geriatrics Society 2019;67(5):884–8 doi 10.1111/jgs.15937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daly MB, Pal T, Berry MP, Buys SS, Dickson P, Domchek SM, et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;19(1):77–102 doi 10.6004/jnccn.2021.0001. [DOI] [PubMed] [Google Scholar]
- 19.Castillo D, Herzog J, Sand S, O’Connor T, Clark C, Weitzel J. Well-groomed participants: eyebrow plucks as surrogates for biomarker samples and a viable source of constitutional DNA. 2016. October 18-22, 2016; Vancouver, Canada. The American Society of Human Genetics. [Google Scholar]
- 20.Suenaga E, Nakamura H. Evaluation of three methods for effective extraction of DNA from human hair. J Chromatogr B Analyt Technol Biomed Life Sci 2005;820(1):137–41 doi 10.1016/j.jchromb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 21.Slavin TP, Teh JB, Weitzel JN, Peng K, Wong FL, Qin H, et al. Association between Clonal Hematopoiesis and Late Non-Relapse Mortality after Autologous Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant 2019. doi 10.1016/j.bbmt.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernard E, Nannya Y, Hasserjian RP, Devlin SM, Tuechler H, Medina-Martinez JS, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nature Medicine 2020;26(10):1549–56 doi 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slavin TP, Neuhausen SL, Nehoray B, Niell-Swiller M, Solomon I, Rybak C, et al. The spectrum of genetic variants in hereditary pancreatic cancer includes Fanconi anemia genes. Fam Cancer 2018;17(2):235–45 doi 10.1007/s10689-017-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reckamp KL, Behrendt CE, Slavin TP, Gray SW, Castillo DK, Koczywas M, et al. Germline mutations and age at onset of lung adenocarcinoma. Cancer 2021;127(15):2801–6 doi 10.1002/cncr.33573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tinat J, Bougeard G, Baert-Desurmont S, Vasseur S, Martin C, Bouvignies E, et al. 2009 version of the Chompret criteria for Li Fraumeni syndrome. J Clin Oncol 2009;27(26):e108–9; reply to J. Tinat e10. [DOI] [PubMed] [Google Scholar]
- 26.Coffee B, Cox HC, Bernhisel R, Manley S, Bowles K, Roa BB, et al. A substantial proportion of apparently heterozygous TP53 pathogenic variants detected with a next-generation sequencing hereditary pan-cancer panel are acquired somatically. Hum Mutat 2020;41(1):203–11 doi 10.1002/humu.23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coffee B, Cox HC, Kidd J, Sizemore S, Brown K, Manley S, et al. Detection of somatic variants in peripheral blood lymphocytes using a next generation sequencing multigene pan cancer panel. Cancer Genetics 2017;211(Supplement C):5–8 doi 10.1016/j.cancergen.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Tovy A, Reyes JM, Gundry MC, Brunetti L, Lee-Six H, Petljak M, et al. Tissue-Biased Expansion of DNMT3A-Mutant Clones in a Mosaic Individual Is Associated with Conserved Epigenetic Erosion. Cell Stem Cell 2020;27(2):326–35.e4 doi 10.1016/j.stem.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renaux-Petel M, Charbonnier F, Thery JC, Fermey P, Lienard G, Bou J, et al. Contribution of de novo and mosaic TP53 mutations to Li-Fraumeni syndrome. J Med Genet 2018;55(3):173–80 doi 10.1136/jmedgenet-2017-104976. [DOI] [PubMed] [Google Scholar]
- 30.Wright CF, Prigmore E, Rajan D, Handsaker J, McRae J, Kaplanis J, et al. Clinically-relevant postzygotic mosaicism in parents and children with developmental disorders in trio exome sequencing data. Nature Communications 2019;10(1):2985 doi 10.1038/s41467-019-11059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batalini F, Peacock EG, Stobie L, Robertson A, Garber J, Weitzel JN, et al. Li-Fraumeni syndrome: not a straightforward diagnosis anymore-the interpretation of pathogenic variants of low allele frequency and the differences between germline PVs, mosaicism, and clonal hematopoiesis. Breast Cancer Res 2019;21(1):107 doi 10.1186/s13058-019-1193-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126(1):9–16 doi 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. New England Journal of Medicine 2017;377(2):111–21 doi 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371(26):2477–87 doi 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371(26):2488–98 doi 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 2020;52(11):1219–26 doi 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 2017;21(3):374–82.e4 doi 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwan TT, Oza AM, Tinker AV, Ray-Coquard I, Oaknin A, Aghajanian C, et al. Preexisting TP53-Variant Clonal Hematopoiesis and Risk of Secondary Myeloid Neoplasms in Patients With High-grade Ovarian Cancer Treated With Rucaparib. JAMA Oncol 2021;7(12):1772–81 doi 10.1001/jamaoncol.2021.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garber JE, Burke EM, Lavally BL, Billett AL, Sallan SE, Scott RM, et al. Choroid plexus tumors in the breast cancer-sarcoma syndrome. Cancer 1990;66(12):2658–60. [DOI] [PubMed] [Google Scholar]
- 40.Chao EC, Astbury C, Deignan JL, Pronold M, Reddi HV, Weitzel JN. Incidental detection of acquired variants in germline genetic and genomic testing: a points to consider statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021. doi 10.1038/s41436-021-01138-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All observed TP53 PVs have been reported previously in ClinVar by the respective commercial laboratories. All clonal hematopoiesis gene variants were included in Figure 3, and are also reported in ClinVar. All data necessary to replicate our findings are included in the manuscript and supplemental tables.
Figure 3.
Clinical category assignment for individuals with a TP53 pathogenic variants. The figure depicts the overall proportion of the clinical categories (germline, ACE/CHIP, PZM) found in this clinic based genetic testing cohort. Based on cascade/transmission testing, clinical criteria, and/or multi tissue analyses, 89/134 (66%) cases had evidence for germline, 26/134 (19%) were categorized as ACE, and 7/134 (5.2%) had evidence for PZM. Additional tissue or transmission data would be needed to discern PZM from broader ACE category for the 12/134 (9%) that were indeterminate.