Abstract
The natural resistance-associated macrophage protein (NRAMP) family plays crucial roles in metal uptake and transport in plants. However, little is known about their functions in peanut. To understand the roles of AhNRAMP genes in iron/cadmium interactions in peanut, genome-wide identification and bioinformatics analysis was performed. A total of 15 AhNRAMP genes were identified from the peanut genome, including seven gene pairs derived from whole-genome duplication and a segmental duplicated gene. AhNRAMP proteins were divided into two distinct subfamilies. Subfamily I contains eight acid proteins with a specific conserved motif 7, which were predicted to localize in the vacuole membrane, while subfamily II includes seven basic proteins sharing specific conserved motif 10, which were localized to the plasma membrane. Subfamily I genes contained four exons, while subfamily II had 13 exons. AhNRAMP proteins are perfectly modeled on the 5m94.1.A template, suggesting a role in metal transport. Most AhNRAMP genes are preferentially expressed in roots, stamens, or developing seeds. In roots, the expression of most AhNRAMPs is induced by iron deficiency and positively correlated with cadmium accumulation, indicating crucial roles in iron/cadmium interactions. The findings provide essential information to understand the functions of AhNRAMPs in the iron/cadmium interactions in peanuts.
Keywords: Arachis hypogaea, NRAMP, iron deficiency, cadmium uptake and transport, cultivar difference
1. Introduction
Iron (Fe) is one of the essential micronutrients for virtually all organisms. Due to its ability to alternate between ferric (Fe3+) and ferrous (Fe2+) ions, Fe acts as a structural cofactor in many enzymes and plays important roles in many metabolic processes such as photosynthesis, respiration, sulfur assimilation, and nitrogen fixation [1]. The shortage of Fe can inhibit chlorophyll synthesis, reduce photosynthesis, and interrupt the respiratory electron transport and tricarboxylic acid cycle [2]. Although Fe is abundant in soils, its bioavailability is limited because it mainly exists in insoluble ferric hydroxides [3]. Fe deficiency in the edible tissues of plants is a critical issue for human health as most people rely on plant-based diets for their primary Fe source. Therefore, understanding the physiological and molecular mechanisms of Fe uptake and translocation in plants is critical to improving their capacity for Fe acquirement.
Cadmium (Cd) is a non-essential trace metal with high toxicity to nearly all living organisms. Since there are no specific transporters for Cd in plant cells, it enters into roots and transfers to the aerial parts by hijacking the transport pathways of micronutrients, including Fe [4]. Therefore, there is an interaction between Cd and Fe in the process of uptake and transport. Cd exposure significantly reduces Fe concentrations in plants [5]. Fe deficiency increased the uptake and accumulation of Cd [6,7,8], while excess Fe reduced Cd uptake in plants [5,9].
The NRAMPs (natural resistance-associated macrophage proteins) are the integral membrane transporters that play an essential role in divalent metal transport across the cellular membrane. NRAMP proteins are highly conserved from bacteria to mammals, with 10–12 transmembrane domains (TMD) and a consensus transport signature [10]. In Arabidopsis, six members (NRAMP1-6) were identified, which are classified into two subfamilies based on phylogenetic analysis [11]. AtNRAMP1 is a plasma membrane-localized transporter that acts as a high-affinity manganese (Mn) transporter in roots under low Mn conditions [12] and plays a pivotal role in Fe transport by cooperating with IRT1 [13]. AtNRAMP2 is a resident protein of the trans-Golgi network and likely exports Mn from the trans-Golgi network lumen to the cytosol under Mn-deficient conditions [14,15]. Both AtNRAMP3 and AtNRAMP4 are vacuolar metal transporter and redundantly mediate the vacuolar export of Fe and Mn under limiting conditions [16,17,18]. AtNRAMP6 functions as an intracellular metal transporter and are involved in cellular Cd distribution, contributing to Cd toxicity [19]. To date, functions of AtNRAMP5 have not been characterized in detail.
Rice (Oryza sativa) has seven NRAMP members, and most of them have been functionally characterized. OsNRAMP5 is a plasma membrane-localized transporter that contributes to the uptake of Fe, Mn, Cd, and lead (Pb) [20,21,22,23,24,25]. Knockout of OsNRAMP5 significantly reduces concentrations of Mn and Cd in the roots, shoots, and grains [22,24] but simultaneously facilitates Cd translocation from roots to shoots [21]. Overexpression of OsNRAMP5 markedly decreased root-to-shoot Cd translocation by disrupting its radial transport into the stele for xylem loading [20]. OsNRAMP1, a close homolog of OsNRAMP5 (72.8% similarity in the amino acid sequences), is localized to the plasma membrane of root and leaf cells, having similar functions with OsNRAMP5 but not redundant [26,27,28]. OsNRAMP2 is a tonoplast-localized transporter that transports Fe from the vacuole to the cytosol [29]. OsNRAMP3 is a vascular bundles-specific Mn transporter that is highly expressed in the node and preferentially transports Mn to young leaves under Mn-limiting conditions [30,31]. OsNRAMP4 (also known as Nrat1) is a plasma membrane-localized transporter for aluminum (Al) but not for divalent cations in yeast [32,33]. OsNRAMP6 is a plasma membrane-localized transporter that functions as iron and Mn transporters and contributes to disease resistance [34].
Peanut (Arachis hypogaea L.) is an allotetraploid (2n = 4x = 40) legume widely cultivated in temperate and tropical regions of the world for edible oil production and digestible protein sources. However, peanut is sensitive to iron deficiency and frequently suffers from Fe deficiency chlorosis in calcareous soil, which greatly limits the yield and quality [35]. More seriously, peanut has a high ability to accumulate Cd in seeds and vegetative tissues [36,37], and iron deficiency significantly increases the absorption and accumulation of Cd in peanut plants [6,7,38]. Fe deficiency chlorosis and Cd pollution have become important constraints in peanut cultivation. In order to avoid these adverse effects, it is necessary to understand the mechanism of Fe/Cd interaction during their uptake and translocation.
Although AhNRAMP1 has been confirmed to be involved in Fe accumulation in young leaves and tolerance to Fe deprivation [39], little data is available regarding the NRAMP gene family in peanut. Here, 15 NRAMP genes were identified from the peanut genome, and their structure, function, and evolution were analyzed. Additionally, the expression of NRAMP genes in response to Fe deficiency and Cd exposure was evaluated. These findings would provide clues to further characterize the functions of AhNRAMP genes and to understand the mechanisms of Fe/Cd interaction in peanut plants.
2. Results
2.1. Identification and Phylogenetic Analysis of the AhNRAMP Family in Peanut
A total of 19 candidate sequences were identified by a BLASTP search against the peanut genome. Although all of them contain the NRAMP domain, four sequences were excluded because they are homologous with ethylene-insensitive proteins (EINs, Figure S1). Among the remaining 15 sequences, a short sequence (NCBI gene ID: 112717764) only contains the last five residues (MQGFL) of the typically conserved amino acid residues GQSSTITGTYAGQY(/F)V(/I)MQGFL [40]. However, we still retained it (named AhNRAMP1.2) because synteny analysis revealed that it is the segmental duplicated gene of AhNRAMP1.1. Thus, a total of 15 putative AhNRAMP genes were identified in peanut, and were named based on the phylogenetic relationships with AtNRAMPs from Arabidopsis (Table 1).
Table 1.
Molecular characterization of AhNRAMP genes and corresponding proteins in peanut.
| Gene Name | Gene ID | Gene Length (bp) |
CDS (bp) |
Aa a | MW b (kDa) |
Instability | Aliphatic Index |
GRAVY c | pI d | No. of TMD e |
Location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AhNRAMP1.1 | 112706038 | 2062 | 1638 | 546 | 58.78 | 36.06 | 120.26 | 0.564 | 8.93 | 12 | PM f |
| AhNRAMP1.2 | 112717764 | 742 | 570 | 190 | 20.83 | 31.17 | 123.76 | 0.584 | 8.74 | 4 | PM |
| AhNRAMP1.3 | 112766867 | 2393 | 1638 | 546 | 58.77 | 37.32 | 120.79 | 0.566 | 8.93 | 12 | PM |
| AhNRAMP2.1 | 112717946 | 1638 | 1638 | 546 | 60.18 | 41.85 | 111.45 | 0.494 | 5.21 | 12 | VM g |
| AhNRAMP2.2 | 112706457 | 2334 | 1680 | 560 | 61.35 | 35.46 | 108.53 | 0.355 | 5.35 | 12 | VM |
| AhNRAMP2.3 | 112729510 | 1756 | 1638 | 546 | 60.24 | 42.40 | 110.55 | 0.486 | 5.29 | 12 | VM |
| AhNRAMP2.4 | 112764447 | 2341 | 1680 | 560 | 61.39 | 34.44 | 108.87 | 0.363 | 5.27 | 12 | VM |
| AhNRAMP3.1 | 112799830 | 1868 | 1563 | 521 | 57.34 | 38.79 | 119.81 | 0.530 | 5.23 | 12 | VM |
| AhNRAMP3.2 | 112800530 | 2085 | 1515 | 505 | 55.15 | 39.60 | 122.66 | 0.667 | 4.94 | 12 | VM |
| AhNRAMP3.3 | 112720549 | 1984 | 1563 | 521 | 57.37 | 37.77 | 119.25 | 0.532 | 5.42 | 12 | VM |
| AhNRAMP3.4 | 112748442 | 2056 | 1494 | 498 | 54.41 | 41.17 | 124.19 | 0.693 | 4.92 | 12 | VM |
| AhNRAMP6.1 | 112703806 | 2277 | 1674 | 558 | 59.78 | 31.48 | 118.01 | 0.580 | 8.62 | 12 | PM |
| AhNRAMP6.2 | 112709742 | 2141 | 1608 | 536 | 58.14 | 38.23 | 118.52 | 0.598 | 8.69 | 12 | PM |
| AhNRAMP6.3 | 112773194 | 2505 | 1674 | 558 | 59.78 | 31.48 | 118.01 | 0.580 | 8.62 | 12 | PM |
| AhNRAMP6.4 | 112776188 | 1869 | 1605 | 535 | 58.04 | 38.65 | 118.56 | 0.598 | 8.57 | 12 | PM |
a amino acid number, b molecular weight, c grand average of hydropathicity, d isoelectric points, e transmembrane domain, f plasma membrane, g vacuole membrane.
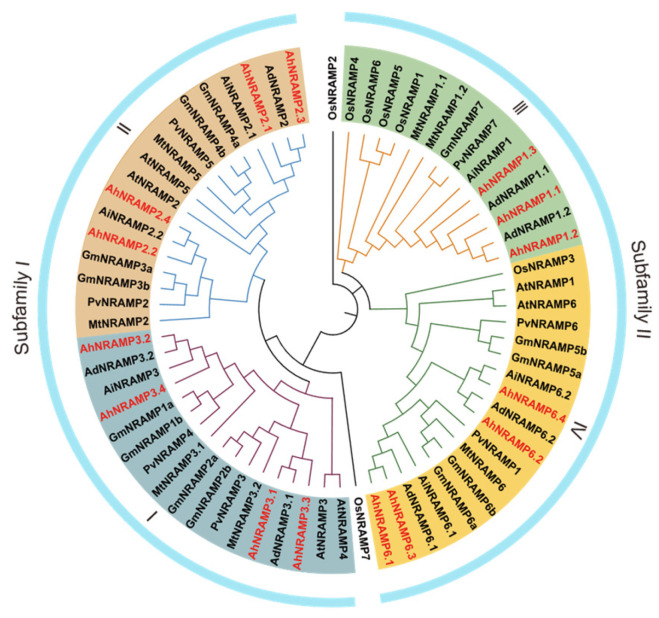
To elucidate the phylogenetic distribution of AhNRAMPs, a tree was constructed based on 58 NRAMP protein sequences from peanut, A. duranensis, A. ipaensis, Medicago truncatula, soybean, common bean, Arabidopsis, and rice. The tree topology showed that the 58 NRAMP members were divided into two distinct subfamilies: subfamily I and II (Figure 1). Each subfamily contained members from different species, suggesting close genetic conservation among NRAMPs. Subfamily I, typified by the AtNRAMP2/3/4/5, contained eight AhNRAMP members. Subfamily II is typified by the AtNRAMP1/6 and consists of seven AhNRAMP members (Figure 1). Notably, NRAMP members from the six legumes are located on adjacent branches with good genetic relationships.
Figure 1.
Phylogenetic relationships of NRAMP proteins from peanut and other plant species. The 15 AhNRAMP proteins of peanut were marked in red color. The species involved in the evolutionary tree include Arabidopsis thaliana (AtNRAMPs), Oryza sativa (OsNRAMPs), Glycine max (GmNRAMPs), A. duranensis (AdNRAMPs), A. ipaensis (AiNRAMPs), Medicago truncatula (MtNRAMPs), and Phaseolus vulgaris (PvNRAMPs).
The length of AhNRAMP genes varied from 742 bp (AhNRAMP1.2) to 2505 bp (AhNRAMP6.3), with CDS lengths from 570 bp (AhNRAMP1.2) to 1680 bp (AhNRAMP2.2/2.4). The amino acid number of AhNRAMP proteins ranged from 190 (AhNRAMP1.2) to 560 (AhNRAMP2.2/2.4), and the molecular weight varied from 20.83 kDa (AhNRAMP1.2) to 61.39 kDa (AhNRAMP2.4). The instability, aliphatic index, and GRAVY of AhNRAMP proteins ranged from 31.17 (AhNRAMP1.2) to 42.40 (AhNRAMP2.3), from 108.53 (AhNRAMP2.2) to 124.19 (AhNRAMP3.4), and from 0.355 (AhNRAMP2.2) to 0.693 (AhNRAMP3.4), respectively. The isoelectric point (pI) of subfamily I members less than 7, while that of subfamily II members is larger than 7 (Table 1). All AhNRAMP proteins contained 12 TMDs except AhNRAMP1.2, which has only four TMDs (Table 1). The eight subfamily I proteins were predicted to localize in the vacuole membrane, whereas the seven subfamily II proteins localized to the plasma membrane (Table 1).
2.2. Conserved Motifs, Domains, and Models of AhNRAMP Proteins
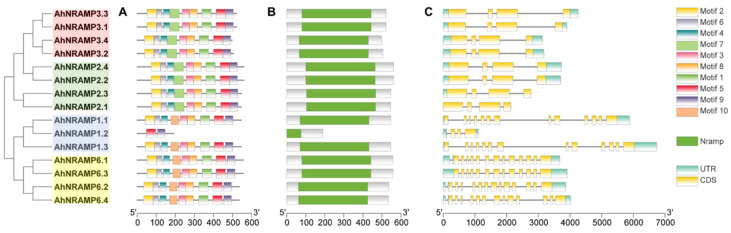
A total of ten conserved motifs were identified in the sequences of AhNRAMP proteins, and all of them were annotated to be NRAMP according to the Pfam tool (Figure 2A and Table S1). As shown in Figure 2A, all AhNRAMP proteins contain nine motifs except AhNRAMP1.2. The members of the same subfamily shared the same motif composition; however, considerable differences were also observed between the two subfamilies. Subfamily I members specifically contained motif 7, while subfamily II specifically contained motif 10. All AhNRAMP proteins shared motifs 1, 2, 3, 4, 5, 6, 8, and 9 except AhNRAMP1.2, which only contains the last two motifs of AhNRAMPs, indicating a distinct evolutionary process and physiological function. All AhNRAMP proteins contained one domain named NRAMP, which is the typical domain of the NRAMP family (Figure 2B).
Figure 2.
Conserved motifs (A) and domains (B) in AhNRAMP proteins as well as exon-intron structure (C) of AhNRAMP genes from peanut. UTR and CDS represent untranslated regions and coding sequences, respectively.
2.3. Exon-Intron Structure, Duplication, and Ka/Ks of the AhNRAMP Family
The two subfamilies differed from each other in the exon-intron structure (Figure 2C). AhNRAMP genes belonging to subfamily I contained four exons with three introns, while those of subfamily II had 13 exons except AhNRAMP1.2, which had three exons and three introns (Figure 2C). However, AhNRAMP genes that clustered in the same branch of the evolutionary tree are similar in the exon-intron organization.
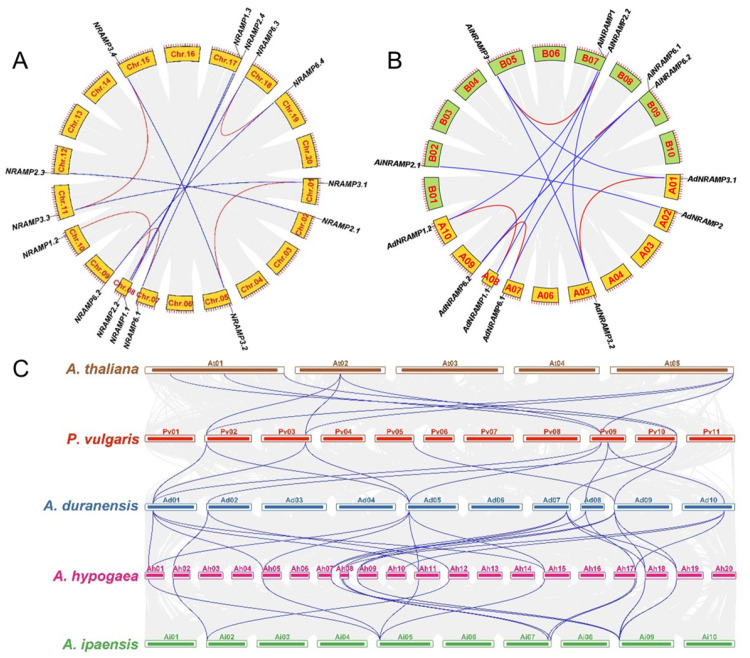
Fifteen AhNRAMP genes are distributed unevenly on 13 chromosomes of the two subgenomes, and the subgenome A (Chr.01–10) and B (Chr.11–20) contained eight and seven AhNRAMP genes, respectively (Figure 3A and Figure S2). Chromosomes 08 and 17 contained two AhNRAMP genes, respectively, while no AhNRAMP gene was distributed on chromosomes 03, 04, 06, 13, 14, 16, and 20, and each of the remaining 11 chromosomes contained only one gene. All AhNRAMP genes experienced duplication events, including seven pairs of whole-genome duplication (WGD) genes (AhNRAMP1.1/1.3, AhNRAMP2.1/2.3, AhNRAMP2.2/2.4, AhNRAMP3.1/3.3, AhNRAMP3.2/3.4, AhNRAMP6.1/6.3, and AhNRAMP6.2/6.4) and five pairs of the segmental duplicated genes (AhNRAMP1.1/1.2, AhNRAMP3.1/3.2, AhNRAMP3.3/3.4, AhNRAMP6.1/6.2, and AhNRAMP6.3/6.4) (Figure 3A). No tandem duplication was identified in AhNRAMP genes.
Figure 3.
Synteny relationship of NRAMP gene pairs in peanut and other four species. (A) Synteny relationship of AhNRAMP gene pairs in peanut. (B) Synteny relationship of NRAMP gene pairs between A. duranensis and A. ipaensis. (C) Synteny relationship of NRAMP gene pairs among five plant species. The red and blue lines represent segmental duplicated genes and synteny genes, respectively. The gray lines show the collinear blocks of the plant genomes.
To better understand the evolutionary process of the AhNRAMP family, a collinear map was constructed between the two ancestral species, A. duranensis and A. ipaensis (Figure 3B and Table S2). As presented in Figure 3B, there were five orthologous gene pairs between A. duranensis and A. ipaensis, which are less than that of WGD-derived genes in peanut (seven gene pairs). Compared with the peanut genome, a homologous gene of AdNRAMP2 was lost on chromosome A08 of the A. duranensis genome, and a homologous gene of AiNRAMP1 was lost on chromosome B01 of the A. ipaensis genome. Moreover, AiNRAMP6.1, which was originally located on chromosome B08, was translocated to chromosome B09 in the A. ipaensis genome (Figure 3B).
To further reveal the evolution of the NRAMP family, a comparative syntenic analysis was carried out on peanut, A. duranensis, A. ipaensis, common beans, and Arabidopsis. A total of 23 and 18 collinear blocks were detected between peanut (15 genes) and A. duranensis (7 genes) and between peanut (14 genes) and A. ipaensis (6 genes), respectively (Figure 3C). All AhNRAMP genes were collinear with the seven PvNRAMPs of the common bean through the seven AdNRAMP, suggesting a close evolutionary relationship among the three species. Four PvNRAMPs of common bean showed a collinear relationship with five AtNRAMP genes of Arabidopsis, while the other three were not collinear with AtNRAMP genes (Figure 3C).
The Ka/Ks ratios of all duplicated pairs (0.01–0.90) were less than 1 (Table 2), indicating a purifying selection in the evolutionary process of AhNRAMP genes [41]. The divergence time for the ten whole-genome duplicated gene pairs ranged from 0.000 Mya to 3.697 Mya, which was dramatically less than most of the segmental duplicated genes (51.236–58.221 Mya) (Table 2).
Table 2.
Ka/Ks analysis of all gene duplication pairs for AhNRAMP genes.
| Gene Pairs | Duplicate Type | Ka a | Ks b | Ka/Ks c | Positive Selection | Divergence Time (Mya) |
|---|---|---|---|---|---|---|
| AhNRAMP1.1/1.2 | Segmental | 0.0069 | 0.0077 | 0.9017 | No | 0.472 |
| AhNRAMP3.1/3.2 | Segmental | 0.0966 | 0.9455 | 0.1022 | No | 58.221 |
| AhNRAMP3.3/3.4 | Segmental | 0.0865 | 0.8961 | 0.0966 | No | 55.178 |
| AhNRAMP6.1/6.2 | Segmental | 0.1076 | 0.8321 | 0.1294 | No | 51.236 |
| AhNRAMP6.3/6.4 | Segmental | 0.1074 | 0.8417 | 0.1276 | No | 51.826 |
| AhNRAMP1.1/1.3 | Whole-genome | 0.0065 | 0.0201 | 0.3237 | No | 1.240 |
| AhNRAMP2.1/2.3 | Whole-genome | 0.0040 | 0.0481 | 0.0834 | No | 2.963 |
| AhNRAMP2.2/2.4 | Whole-genome | 0.0047 | 0.0329 | 0.1440 | No | 2.023 |
| AhNRAMP3.1/3.3 | Whole-genome | 0.0051 | 0.0368 | 0.1399 | No | 2.265 |
| AhNRAMP3.2/3.4 | Whole-genome | 0.0009 | 0.0219 | 0.0407 | No | 1.351 |
| AhNRAMP6.1/6.3 | Whole-genome | 0.0000 | 0.0000 | - | No | 0.000 |
| AhNRAMP6.2/6.4 | Whole-genome | 0.0008 | 0.0600 | 0.0136 | No | 3.697 |
a The number of nonsynonymous substitutions per nonsynonymous site, b the number of synonymous substitutions per synonymous site, c Ka/Ks ratios.
2.4. 3D Model Predictions and Multiple Sequence Alignment of AhNRAMP Proteins
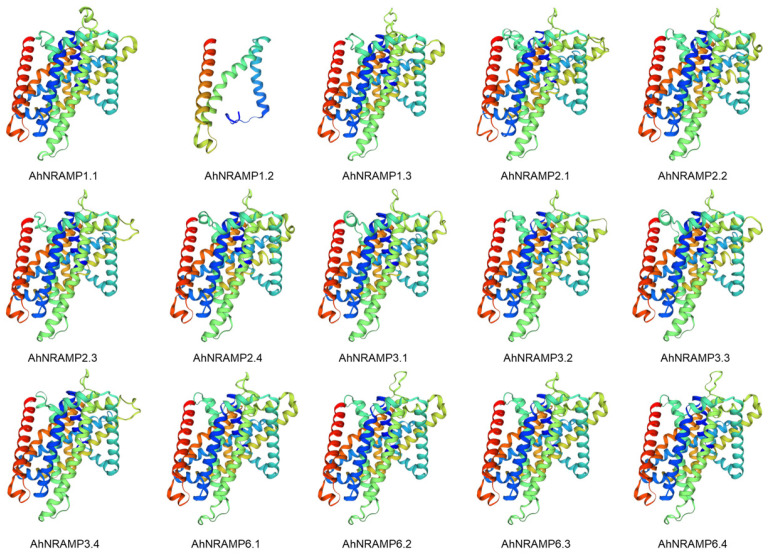
All AhNRAMP proteins were well modeled with the template, 5m94.1.A (Figure 4 and Table S3). Sequence identity ranged from 36.52% to 42.19%, the value of GMQE ranged from 0.32 to 0.59, and the QMEANDisCo global score ranged from 0.63 to 0.70 (Table S3). These data suggest a high quality of 3D model predictions for AhNRAMP proteins.
Figure 4.
Predicted 3D structure of peanut AhNRAMP proteins by SwissModel. Models were visualized by rainbow color from N to C terminus.
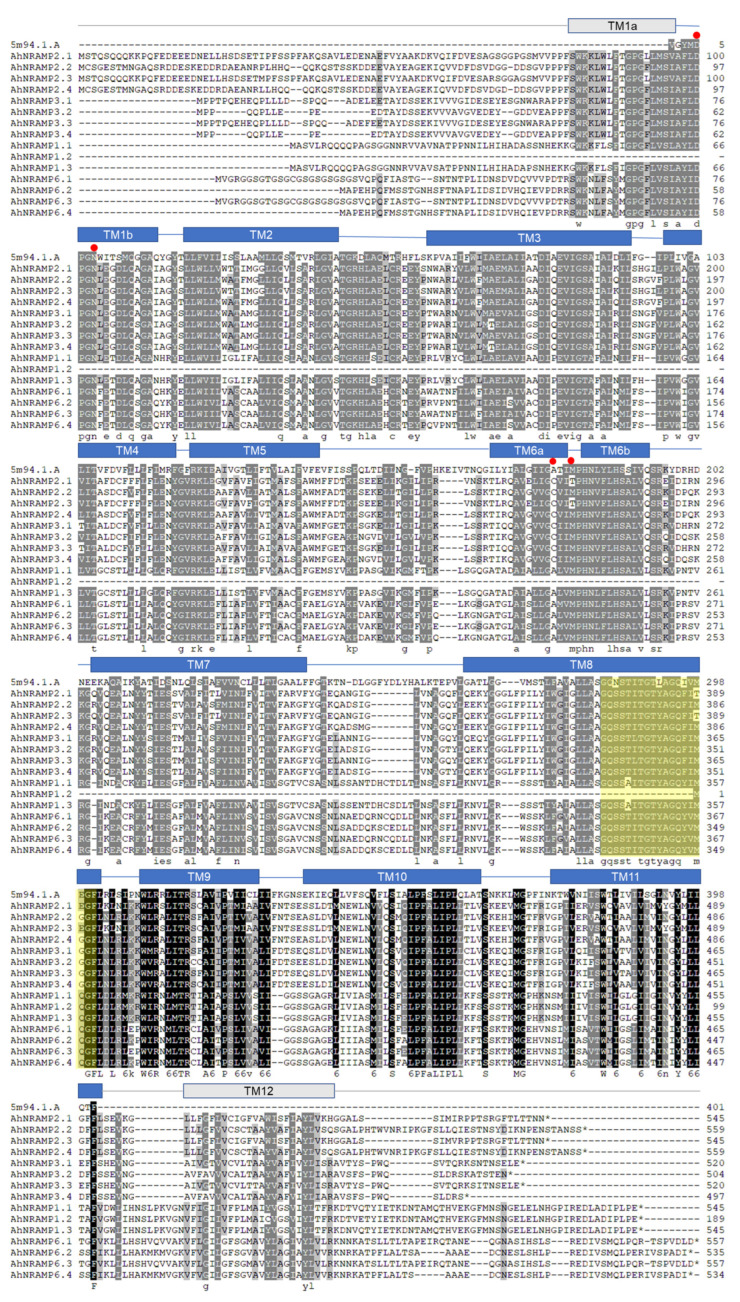
Multiple sequence alignment showed considerable homology between the ScaDMT and AhNRAMP proteins throughout the TMDs (Figure 5). Compared with ScaDMT, which has 11 TMDs, an additional TMD (TM12) was harbored in the C terminus of AhNRAMP proteins. All AhNRAMPs contained typical conserved amino acid residues GQSSTITGTYAGQY(/F)V(/I)MQ(/G/E)GFL except AhNRAMP1.2, in which the last five residues (MQGFL) were reserved. Besides, conserved amino acid residues were also found in different TMDs, e.g., DPGN in TM1, EVIGS(/T)AI(/L)A in TM3, M(/T)PHNV(/L) FLHSS(/A)LVQ(/L)SF in TM6, A(/G)LLAS(/A) in TM8 (Figure 5).
Figure 5.
Sequence alignment of ScaDMT and AhNRAMP with ClustalW. Identical residues are highlighted in black and homologous residues in gray, while the typical conserved amino acid residues GQSSTITGTYAGQY(/F)V(/I)MQ(/G/E)GFL are highlighted in yellow. Secondary structure elements are shown above the sequences. The red circles indicate residues of the transition metal binding site in ScaDMT.
2.5. cis-Acting Elements of AhNRAMP Genes
To better understand the potential regulation of AhNRAMP genes, the cis-acting elements were predicted and shown in Table S4. The promoter regions of all AhNRAMP genes harbored CAAT-box, TATA-box, and Box 4 (Table S4). The number ranged from 23 to 43 for CAAT-box, from 45 to 137 for TATA-box, and from 1 to 12 for Box 4, respectively. Besides, a number of cis-acting elements such as ABRE, G-box, ARE, TCT-motif, TC-rich repeats, CGTCA-motif, TGACG-motif, TCA-element, AT1-motif, GATA-motif, GT1-motif, circadian, MBS, and LTR, were frequently identified in promoter regions of AhNRAMP genes. Most of them are involved in light, abiotic stress, and hormone responsiveness (Table S4).
2.6. Tissue-Specific Expression Profiles of AhNRAMP Genes
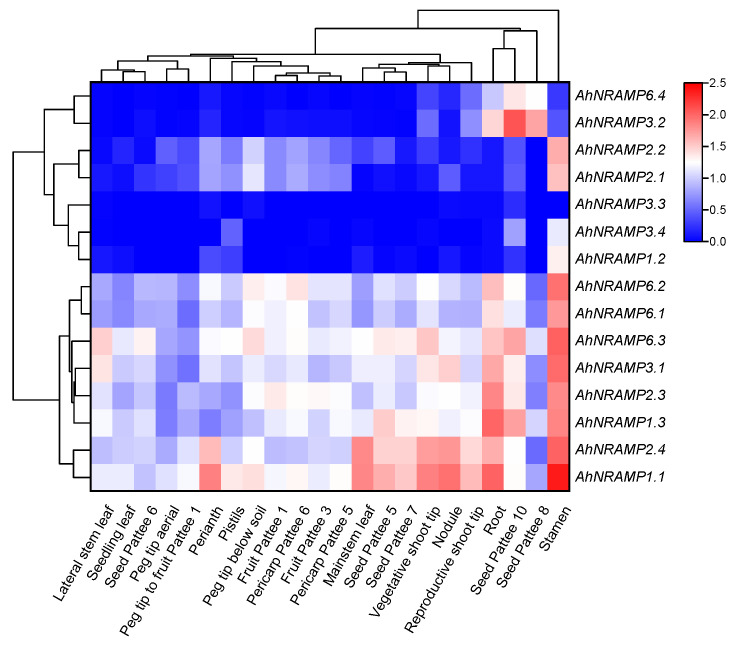
RNA-seq data showed that all AhNRAMP genes exhibited a tissue-specific expression in peanut plants, and their expression profiles were independent of phylogeny (Table S5 and Figure 6). Fifteen AhNRAMP genes could be classified into two distinct groups. Group 1 included AhNRAMP1.2, AhNRAMP2.1/2.2, AhNRAMP3.2/3.3/3.4, and AhNRAMP6.4, which show a low expression in most of the peanut tissues. Among them, AhNRAMP3.3 was not or lowly expressed in all peanut tissues, AhNRAMP3.2 and AhNRAMP6.4 were mainly expressed in roots and seeds (Pattee 8/10), while the other four genes were preferentially expressed in the stamens. Group 2 is composed of the remaining eight genes representing high gene expression, and all of them were preferentially expressed in the roots and stamens. Besides, AhNRAMP1.1 and AhNRAMP2.4 also showed high expression in nodules, shoot tips, mainstem leaves, perianths, and developing seeds.
Figure 6.
Tissue-specific expression profiles of AhNRAMP genes in peanut. Gene expression is expressed in lg(TPM + 1). Pattee 1, 3, 5, 6, 7, 8, and 10 represent different developmental stages of peanut pods based on Pattee’s classification.
2.7. Gene Expression of AhNRAMPs in Response to Fe-Deficiency and Cd Exposure
To elucidate the responses of AhNRAMP genes to Fe-deficiency and Cd exposure, two contrasting peanut cultivars (Fenghua 1 and Silihong) were used for qRT-PCR analysis. As presented in Figure S3, compared with Silihong, the reduction of shoot dry weight and leaf chlorophyll contents (SPAD value) caused by Fe deficiency was more pronounced in Fenghua 1. Under Fe deficient conditions, Cd exposure resulted in larger decreases in shoot dry weight and leaf chlorophyll contents in Silihong compared with Fenghua 1 (Figure S3). The results, in agreement with the previous study [37], suggested that Silihong is more tolerant to Fe deficiency but more sensitive to Cd stress than Fenghua 1.
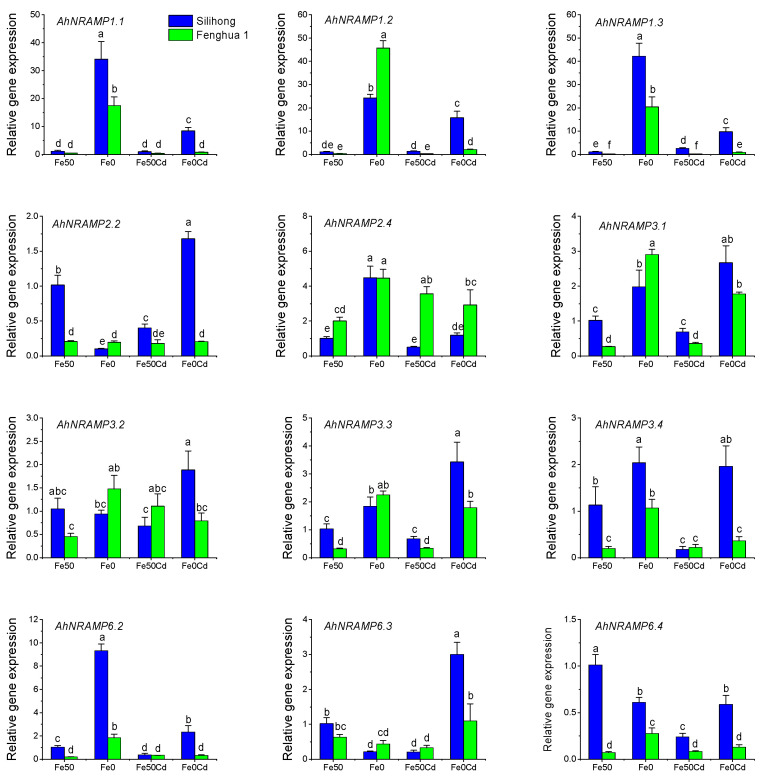
The expression of AhNRAMP genes differed between Silihong and Fenghua 1, and was significantly influenced by Fe-deficiency and/or Cd exposure (Figure 7). Except for AhNRAMP2.4, the expression of all genes in Silihong was higher than that of Fenghua 1 under normal nutrition. Fe deficiency up-regulated the expression of AhNRAMP1.1/1.2/1.3, AhNRAMP2.4, AhNRAMP3.1/3.3/3.4, and AhNRAMP6.2 for both cultivars (Figure 7).
Figure 7.
Expression levels of 12 AhNRAMP genes in peanut roots exposed to 0 or 2 μM Cd under Fe-sufficient (Fe50) and Fe-deficient (Fe0) conditions for 14 days. Data (means ± SE, n = 3) sharing the same letter(s) above the error bars are not significantly different at the 0.05 level based on the Duncan multiple range test.
Under normal Fe supply, Cd exposure repressed the expression of AhNRAMP2.2, AhNRAMP3.4, and AhNRAMP6.2/6.3/6.4 in Fenghua 1 but increased the expression of AhNRAMP2.4 in Silihong. Under Fe-deficiency, Cd exposure repressed the expression of AhNRAMP1.1/1.2/1.3, AhNRAMP2.4, and AhNRAMP6.2 but induced the expression of AhNRAMP6.3 for both cultivars (Figure 7). The remaining genes showed a cultivar-specific response to Cd exposure under Fe-deficient conditions. Specifically, Cd-induced the expression of AhNRAMP2.2, AhNRAMP3.2/3.3 in Silihong but reduced the expression of AhNRAMP3.1/3.4 and AhNRAMP6.2/6.4 in Fenghua 1 (Figure 7).
2.8. Fe/Cd Accumulation and Translocation in Two Peanut Cultivars
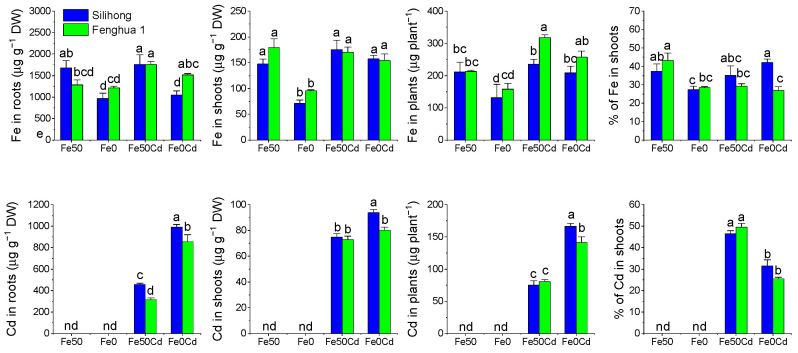
The two peanut cultivars showed a differential response in Fe accumulation to Fe deficiency (Figure 8). In Silihong, Fe deficiency significantly reduced Fe concentrations in roots and shoots, the total amount of Fe in plants, and the percentage of Fe in shoots. In Fenghua 1, Fe deficiency significantly reduced Fe concentrations in shoots and the percentage of Fe in shoots, while root Fe concentrations and the total amount of Fe in plants were not affected. Cd exposure increased Fe accumulation in peanut plants, which was more pronounced in the Fe-deficient treatment (Figure 8). In contrast, the percentage of Fe in shoots of Fenghua 1 was decreased by Cd exposure under normal Fe supply conditions.
Figure 8.
Fe/Cd accumulation and translocation in two peanut cultivars exposed to 0 or 2 μM Cd under Fe-sufficient (Fe50) and Fe-deficient (Fe0) conditions for 14 days. Data (means ± SE, n = 3) sharing the same letter(s) above the error bars are not significantly different at the 0.05 level based on the Duncan multiple range test. nd, not determinable.
Fe deficiency significantly enhanced root Cd concentrations and the total amount of Cd in plants for both cultivars, while the percentage of Cd in shoots was reduced (Figure 8). Shoot Cd concentrations in Silihong were also increased by Fe deficiency. Cd concentrations in roots were higher in Silihong than in Fenghua 1, regardless of Fe supply. Compared with Fenghua 1, higher shoot Cd concentrations and total amount of Cd in plants was also observed in Silihong under Fe-deficient conditions (Figure 8).
2.9. Relationship of Gene Expression of AhNRAMPs and Fe/Cd Accumulation
To identify the AhNRAMP genes involved in Fe/Cd accumulation, Pearson’s correlation analysis was performed. As shown in Table 3, the expression of AhNRAMP1.1/1.2/1.3, AhNRAMP3.1/3.3/3.4, and AhNRAMP6.2 was significantly and negatively correlated with Fe concentrations in roots and shoots, as well as the total amount of Fe in plants. The expression of AhNRAMP2.2/2.4 and AhNRAMP6.3 was significantly correlated with the percentage of Fe in shoots. The expression of all AhNRAMP genes except AhNRAMP2.4 and/or AhNRAMP3.2 were significantly and positively correlated with Cd concentrations in roots and shoots, as well as the total amount of Cd in plants (Table 3). Negative correlations were observed between the expression of AhNRAMP3.1/3.3 and the percentage of Cd in shoots (Table 3).
Table 3.
Correlations Fe/Cd accumulation and the expression of AhNRAMP genes in the roots of two peanut cultivars (n = 24 and 12 for Fe and Cd accumulation, respectively).
| Gene Expression |
Fe in Roots |
Fe in Shoots |
Total Fe in Plants |
% of Fe in Shoots |
Cd in Roots |
Cd in Shoots |
Total Cd in Plants |
% of Cd in Shoots |
|---|---|---|---|---|---|---|---|---|
| AHNRAMP1.1 | −0.646 ** | −0.820 ** | −0.681 ** | −0.294 | 0.699 * | 0.785 ** | 0.700 * | −0.432 |
| AHNRAMP1.2 | −0.541 ** | −0.719 ** | −0.588 ** | −0.278 | 0.699 * | 0.876 ** | 0.738 ** | −0.380 |
| AHNRAMP1.3 | −0.610 ** | −0.828 ** | −0.673 ** | −0.352 | 0.650 * | 0.754 ** | 0.616 * | −0.367 |
| AHNRAMP2.2 | −0.142 | 0.223 | −0.015 | 0.529 ** | 0.657 * | 0.830 ** | 0.656 * | −0.326 |
| AHNRAMP2.4 | −0.349 | −0.544 ** | −0.230 | −0.498 * | −0.183 | −0.261 | −0.001 | −0.026 |
| AHNRAMP3.1 | −0.570 ** | −0.555 ** | −0.501 * | −0.123 | 0.888 ** | 0.802 ** | 0.871 ** | −0.721 ** |
| AHNRAMP3.2 | −0.192 | −0.192 | −0.099 | 0.069 | 0.423 | 0.718 ** | 0.505 | −0.093 |
| AHNRAMP3.3 | −0.594 ** | −0.400 | −0.438 * | 0.010 | 0.847 ** | 0.752 ** | 0.817 ** | −0.705 * |
| AHNRAMP3.4 | −0.561 ** | −0.568 ** | −0.546 ** | 0.051 | 0.670 * | 0.809 ** | 0.678 * | −0.364 |
| AHNRAMP6.2 | −0.596 ** | −0.740 ** | −0.610 ** | −0.235 | 0.603 * | 0.748 ** | 0.622 * | −0.285 |
| AHNRAMP6.3 | −0.261 | 0.246 | 0.055 | 0.464 * | 0.808 ** | 0.908 ** | 0.872 ** | −0.550 |
| AHNRAMP6.4 | −0.178 | −0.331 | −0.445 * | 0.158 | 0.640 * | 0.761 ** | 0.596 * | −0.367 |
* p < 0.05, ** p < 0.01.
3. Discussion
Due to the important role in the uptake and transport of metal ions, a large number of NRAMP genes have been identified and functionally characterized [12,16,17,18,19,20,21,22,23,24,26,27,28,29,30,31,32,33,34]. However, there is a lack of detailed information about this family in peanuts. In this study, 15 NRAMP genes were identified from the peanut genome, which is the largest among reported plant species, including Arabidopsis (6) [11], rice (7) [42], soybean (13) [43], common bean (7) [44], potato (Solanum tuberosum, 5) [45], tea plant (Camellia sinensis, 11) [46], cacao (Theobroma cacao, 5) [47], and Spirodela polyrhiza (3) [48]. A large number of genes in the peanut genome have been reported in the gene families of metal tolerance proteins [49], zinc/iron-regulated transporter-like proteins [50], and oligopeptide transporters [51].
The expansion and functional diversification of gene families depend on gene duplication events, including WGD, segmental, and tandem duplication [48]. As an allotetraploid species, peanut contains two sets of subgenomes (A and B) from two ancestral species (A. duranensis and A. ipaensis) [52]. Syntenic analysis revealed that among the 15 AhNRAMP genes, seven paralogous gene pairs resulted from WGDs. WGD events are ubiquitous among angiosperms [53]. Peanut has experienced at least three rounds of such events together with the allopolyploidization [54]. Besides, three paralogous gene pairs in subgenome A (Chr. 01–10) and two paralogous gene pairs in subgenome B (Chr. 11–20) were found to be segmental duplicates. Similarly, the comparative syntenic analysis showed three and two pairs of segmental duplicated genes in the genome of A. duranensis (AA) and A. ipaensis (BB), respectively. These data suggest that segmental duplications of AhNRAMP genes have occurred before the allopolyploidization, which is illustrated by the considerably long divergence time of segmental duplicated genes (51.236–58.221 Mya). No tandem duplication was detected in peanut, A. duranensis, and A. ipaensis. Therefore, it might be WGD during allopolyploid speciation that contributes to the large number of AhNRAMP genes in peanut.
AhNRAMP genes are unevenly distributed in the two subgenomes of peanut. Subgenome A contained 8 AhNRAMP genes, while subgenome B contained seven genes (Figure 3A). Meanwhile, the number of AhNRAMP genes differed between the two diploid progenitors, A. duranensis (7) and A. ipaensis (6) (Figure 3B). These differences suggest an asymmetrical evolution between the two subgenomes of peanut. Notably, the number of AhNRAMP genes in the peanut genomes was higher than the total number of genes in A. duranensis and A. ipaensis, indicating a gene loss in the two ancestral species of peanut. Compared with peanut, a homologous gene of AdNRAMP2 in A. duranensis and a homologous gene of AiNRAMP3 in A. ipaensis have got lost since the allotetraploidization event. Interestingly, two paralogous genes of peanut AhNRAMP6 (AhNRAMP6.3/6.4) are located in different chromosomes (Chr.18 and Chr.19). However, their orthologous genes (AiNRAMP6.1/6.2) in A. ipaensis located in the same chromosome (B09). This might be a result of the translocation between chromosome B08 and B09 in A. ipaensis after the allotetraploidization event. Allotetraploid seems to have a higher capacity for avoiding gene loss than their diploid progenitors, and peanut is closer to A. duranensis than A. ipaensis in terms of the NRAMP family.
All AhNRAMP genes were collinear with the seven PvNRAMPs of the common bean through AdNRAMPs (Figure 3C), suggesting a close evolutionary relationship. Most of the AdNRAMPs were collinear with five AtNRAMP genes of Arabidopsis through PvNRAMPs, forming continuous colinear gene pairs. However, AdNRAMP1.1/1.2 and AdNRAMP2.1 did not form continuous colinear gene pairs with AtNRAMP genes (Figure 3C). These data suggest that, besides the common ancestor shared with Arabidopsis, some NRAMP genes of leguminous plants such as AdNRAMP1.1/1.2 and AdNRAMP2.1 in A. duranensis and PvNRAMP7 and PvNRAMP5 in common bean, might originate from other ancestors.
Gene duplication contributes to the evolution of novel genes with new functions [55]. However, newly duplicated genes are usually functionally redundant, which may lead to gene loss [56]. Thus, expression reduction facilitates the retention of duplicates and the conservation of their ancestral functions [56]. In the present study, seven AhNRAMP genes showed low expression in most of the 22 peanut tissues under normal conditions (Figure 6). Similar results have been reported in other gene families in peanut [49,50,51]. These results support the speculation that reduced expression might be beneficial to retain duplicate genes and their functional redundancy.
The NRAMPs typically consists of around 500 amino acid residues proteins with 10–12 TMDs in different species [10,42]. All PvNRAMPs from common beans have 12 TMDs, and the number of amino acid residues ranged from 507 to 554 [44]. The cacao NRAMP proteins are comprised of 510 to 557 amino acids with 11–13 TMDs [47]. In the current study, we found that, except that AhNRAMP1.2 contains 4 TMDs, all other AhNRAMP proteins contain 12 TMDs with amino acid numbers ranging from 498 to 560 (Table 1). According to syntenic analysis, AhNRAMP1.2 might be derived from AdNRAMP1.2 of the A. duranensis genome, which consists of 333 amino acids with 7 TMDs. The motif composition and exon/intron structure showed that AhNRAMP1.2 only contained the last two motifs (Motif 5 and 9), and the corresponding gene contained the last three exons. Multiple alignments revealed that AhNRAMP1.2 only contains the last five residues (MQGFL) of typical conserved amino acid residues GQSSTITGTYAGQY(/F)V(/I)MQ(/G/E)GFL. Thus, the gene was continuously shortened during evolution, and only the C-terminal fragment of AhNRAMPs was retained. However, the preferential expression of AhNRAMP1.2 in stamens and the induction by Fe deficiency in roots confirm its roles in stamen development and Fe nutrition. Short NRAMP protein sequences have been reported in several plant species, such as C. sinensis and Brassica napus [46,57].
All AhNRAMP proteins were well modeled with the 3D model template, 5m94.1.A, which is the crystal structure of a divalent metal transporter from Staphylococcus capitis (ScaDMT). ScaDMT is a close prokaryotic homolog of the SLC11 (NRAMP) family that transports divalent transition-metal ions such as Fe2+, Mn2+, and Cd2+ across cellular membranes [58]. It has been confirmed that D49 and N52 from TM1, M226 from the side chain, and A223 from TM6 coordinate the Mn2+ substrate [58]. Moreover, the Mn2+ binding site also bound other cations such as Fe2+, Co2+, Ni2+, Cd2+, and Pb2+, while the position of Cu2+ shifted somewhat but retained a close interaction with M226 [58]. Surprisingly, Zn2+ is bound in another site where it closely interacts with H233 in TM6b [58]. Multiple sequence alignment showed considerable homology between the ScaDMT and AhNRAMP proteins throughout the TMDs (36.52–42.19% sequence identity). All AhNRAMPs contained the typical conserved amino acid residues GQSSTITGTYAGQY(/F)V(/I)MQ(/G/E)GFL except AhNRAMP1.2, in which the last five residues (MQGFL) were reserved. The conserved amino acid residues were located between the TM8 and TM9 [40]. Besides, conserved amino acid residues were also found in different TMDs, e.g., DPGN in TM1, EVIGS(/T)AI(/L)A in TM3, M(/T)PHNV(/L) FLHSS(/A)LVQ(/L)SF in TM6, A(/G)LLAS(/A) in TM8 (Figure 5). The similar structures indicate that AhNRAMPs might have equivalent physiological functions as ScaDMT.
Phylogenetic analysis grouped the NRAMP proteins into two subfamilies, and each of them was further divided into two groups (Figure 1). The classification concurred with those reported in previous studies [11,43,44,46]. Subfamily I was formed by eight AhNRAMP proteins (AhNRAMP2.1/2.2/2.3/2.4 and AhNRAMP3.1/3.2/3.3/3.4), and subfamily II contained seven other AhNRAMP members (AhNRAMP1.1/1.2/1.3 and AhNRAMP6.1/6.2/6.3/6.4). The two subfamilies differed from each other in pI, conserved motif composition, and subcellular localization of AhNRAMP proteins, as well as exons/intron structure. These traits, however, were highly conserved within a subfamily. Subfamily I AhNRAMPs are acid proteins (pI ranged from 4.92 to 5.42) with a specific conserved motif (Motif 7), while subfamily II members are basic proteins (pI ranged from 8.62 to 8.93) sharing another specific conserved motif (Motif 10) except AhNRAMP1.2 (Table 1, Figure 2A). Subfamily I genes contained four exons, while those in subfamily II had 13 exons except AhNRAMP1.2 (Figure 2C). The subfamily I proteins were predicted to be localized to the vacuole membrane, while the subfamily II proteins were localized to the plasma membrane (Table 1). Similar results were reported by Qin et al. [43] in soybean. Indeed, experimental work confirmed that four subfamily I members (GmNRAMP1a, GmNRAMP2a, GmNRAMP2b, and GmNRAMP3a) from soybean were localized to the tonoplast, and two subfamily II members (GmNRAMP5a and GmNRAMP7) localized to the plasma membrane [43].
The cis-acting elements are important molecular switches that play a critical role in gene expression in plants. In the promoter regions of AhNRAMPs, many light-responsive elements (Box 4, AT1-motif, G-box, GT1-motif, and TCT-motif), stress-responsive elements (ARE, LTR, MBS, TC-rich repeats) and hormone-responsive elements (ABRE, CGTCAmotif, TGACG-motif, and TCA-element) were frequently identified, indicating that AhNRAMP genes might be regulated by complex regulatory networks.
Several homologous genes belonging to the subfamily I have been functionally characterized in Arabidopsis. AtNRAMP2 is localized to the trans-Golgi network and exports Mn from the trans-Golgi network lumen to the cytosol under Mn-deficient conditions [14,15]. AtNRAMP3 and AtNRAMP4 are vacuolar membrane-localized proteins that contribute to Fe and Mn nutrition by remobilizing vacuolar Fe stores [16,17,18]. Overexpression of AtNRAMP3 leads to Cd hypersensitivity and increased accumulation of Fe and Cd in Arabidopsis [59]. In contrast, more subfamily II genes have been characterized in Arabidopsis and rice. OsNRAMP1/4/5/6 has been demonstrated to be plasma membrane-localized transporters in rice [20,21,22,23,24,26,27,28,32,33,34]. OsNRAMP1 and OsNRAMP5 are responsible for the uptake of Fe, Mn, and Cd in rice [20,21,22,23,24,26,27,28]. OsNRAMP6 function as Fe and Mn transporters and contribute to disease resistance in rice [34]. OsNRAMP4 transport Al but not for divalent cations in yeast [32,33]. AtNRAMP1 is a plasma membrane-localized Mn transporter essential for Arabidopsis growth in low-Mn conditions [12]. AtNRAMP6 is an intracellular metal transporter, and its overexpression increases Cd sensitivity in Arabidopsis [19]. OsNRAMP3 is a vascular bundles-specific Mn transporter that regulates the distribution of Mn in plants [30,31]. These studies fully demonstrated that the NRAMP gene family participates in the uptake and transport of divalent metals, including Fe, Mn, and Cd, in plants.
In peanut, most of the AhNRAMP genes are predominantly expressed in stamens and roots and/or developing seeds (Figure 6). Similar results have been reported in potato [41]. The results suggest that the AhNRAMP family genes might play an essential role in metal uptake by the roots and the transport of divalent metals to stamens and/or seeds during their developing processes. The expression of almost all AhNRAMP genes is induced by Fe deficiency in peanut roots (Figure 7). The up-regulation of AhNRAMP genes implies a high capacity for Fe uptake. However, significant but negative correlations were found between the expression of AhNRAMP genes in roots and Fe accumulation in peanut plants (Table 3). Such a contradictory result is due to the Fe-deficient plants having no absorbable Fe despite their strong ability to take up Fe.
In agreement with previous studies [6,7,38], our results indicate that Fe deficiency significantly increases the uptake and accumulation of Cd in peanut plants (Figure 8). Moreover, the uptake and accumulation of Cd in peanut plants are positively correlated with the expression of all AhNRAMP genes except AhNRAMP2.4 and/or AhNRAMP3.2 (Table 3). These results indicated that the induction of AhNRAMPs by Fe deficiency in peanut roots might be, at least partially, responsible for the increased Cd accumulation in Fe-deficient plants.
Not unexpectedly, we found that Silihong showed a higher capacity for uptake and translocation of Cd from roots to shoots than Fenghua 1 under Fe-deficient conditions (Figure 8). This result is consistent with our previous studies [37,49,51]. Similarly, the expression of all AhNRAMP genes except AhNRAMP2.4 was higher in the roots of Silihong than that of Fenghua 1 in the treatments of Fe deficiency with Cd exposure (Figure 7). It seems that the higher expression of AhNRAMP genes contributes to the high capacity for Cd accumulation in Silihong compared with Fenghua 1.
4. Materials and Methods
4.1. Plant Materials and Treatments
Two peanut cultivars, Fenghua 1 (Fe deficiency sensitive/Cd tolerant cultivar) and Silihong (Fe deficiency tolerant/Cd sensitive cultivar) were used for experiments [37]. Seeds were surface sterilized with 5% sodium hypochlorite, rinsed in deionized water for 24 h in the dark, and then sown in the sand for germination. After germination, 3-d-old uniform seedlings were transplanted to polyethylene pots and cultured in hydroponics under controlled conditions for a week. The culture conditions and nutrient solutions were followed as described previously by Su et al. [38]. Ten-day-old seedlings were exposed to 0 or 2 μM CdCl2 under Fe-sufficient (50 μM Fe-EDTA) or Fe-deficient (0 μM Fe-EDTA) conditions, respectively. Each treatment had three biological replicates with three seedlings per replication. During the growing period, nutrient solutions were renewed twice a week. Plants were harvested following 14 days of treatment. Fresh root tissues were immediately frozen in liquid nitrogen and stored at −80 °C for RT-qPCR analysis.
4.2. Determination of Fe and Cd in Peanut Plants
The harvested plants were separated into roots and shoots. The root was rinsed with 20 mM Na2EDTA for 15 min to remove the surface-bound metal ions. Thereafter, roots and shoots were oven-dried, weighed, and ground into powder. After digestion with HNO3–HClO4 (3:1, v/v), Cd and Fe concentrations in the samples were determined by flame atomic absorbance spectrometry (WFX-110, Beijing Rayleigh Analytical Instrument Company, Beijing, China). The total Fe/Cd in plants and the percentage of Fe/Cd in shoots were calculated using the equations reported by Liu et al. [37].
4.3. Identification and Bioinformatics Analyses of NRAMP Family in Peanut
The sequences of six AtNRAMP (AtNRAMP1-6) and seven OsNRAMP proteins (OsNRAMP1-7) were used as queries for TBLASTP against the genome of peanut (cv. Tifrunner) using TBtools software v. 1. 108 [60]. After examining for the presence of the conserved NRAMP domain (PF01566) using the hmmscan tool (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan, accessed on 6 July 2022), candidates were further aligned by ClustalW, along with AtNRAMPs and OsNRAMPs. An NJ (neighbor-joining) phylogenetic tree was constructed with the Poisson model and 1000 bootstraps using the MEGA-X program (v. 10.2.6). Sequences clustered together with AtNRAMPs and OsNRAMPs could be recognized as AhNRAMPs. To investigate the phylogenetic relationships of AhNRAMPs, full-length sequences of NRAMP proteins from peanut, Arabidopsis, rice, soybean, and common beans were used to construct a phylogenetic tree as the method mentioned above. The phylogenetic tree was displayed and modified using iTOL (https://itol.embl.de/itol.cgi, accessed on 20 July 2022).
The physical and chemical properties of AhNRAMP proteins were analyzed using the ProtParam tool (https://web.expasy.org/protparam/, accessed on 6 July 2022) [61]. The number of TMDs in proteins was predicted using TOPCONS (http://topcons.net/, accessed on 8 June 2022) [62]. Subcellular localization of AhNRAMP proteins was predicted using ProtComp 9.0 (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc, accessed on 30 June 2022). The conserved domains and motifs were analyzed using the Pfam tool (http://pfam.xfam.org/search#tabview=tab1, accessed on 15 July 2022) and the MEME v. 5.3.3 (https://meme-suite.org/meme/tools/meme, accessed on 20 July 2022), respectively [63,64]. Homology-modeled 3D structures of AhNRAMP proteins were predicted using the SwissModel (https://swissmodel.expasy.org/, accessed on 20 September 2022) [65].
The exon/intron structures were detected with genomic and coding sequences using GSDS v. 2.0 (http://gsds.gao-lab.org/, accessed on 10 June 2022) [66]. The synteny relationship of NRAMP genes within peanut genome and among genomes of multiple species, including peanut, A. duranensis, A. ipaënsis, Arabidopsis, and common bean, was analyzed using one-step MCScanX integrated into the TBtools software v. 1. 108 [60]. Gene collinearity and Ka/Ks (ratio of nonsynonymous substitution rate to synonymous substitution rate) were analyzed by one-step MCScanX and simple Ka/Ks calculator (NJ) in TBtools software v. 1. 108, respectively [60]. The cis-acting elements in the promoter sequences (upstream 2.0 kb) were predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 1 January 2023) [67].
4.4. Gene Expression Analysis Based on RNA-Seq Data
Expression profiles of AhNRAMP genes in different tissues were identified using RNA-seq data of cv. Tifrunner obtained from PeanutBase (https://www.peanutbase.org/, accessed on 15 June 2022) [68]. The expression of AhNRAMP genes was normalized and represented in TPM (transcripts per kilobase of exon model per million mapped reads), and lg(TPM + 1) was used to construct the heatmap diagram.
4.5. RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from the roots using a plant RNA rapid extract Kit (Coolaber, Beijing, China). First-strand cDNA was synthesized using MonScript™ RTIII Super Mix with dsDNase (Monad, Suzhou, China) according to the manufacturer’s protocol. For qRT-PCR analysis, specific primers of AhNRAMP genes were designed, and 60S ribosomal protein L7-2 (NCBI_ID: 112697914) was used as an internal control (Table S6). qRT-PCR was performed using 10× diluted cDNAs and MonAmp™ ChemoHS Plus qPCR Mix (Monad, Suzhou, China) on the LightCycler®96 Instrument (Roche Applied Science, Indianapolis, IN, USA). Each sample was repeated with three technical replicates. The relative gene expression was calculated using the delta-delta CT method (2−ΔΔCT) [69].
4.6. Statistical Analysis
Data were subjected to a one-way analysis of variance, and significant variations among means were determined by Duncan’s Multiple Range Test (p < 0.05). Pearson’s correlation analysis was performed to determine the correlations between gene expression and Fe/Cd accumulation. All statistical analysis was conducted using IBM SPSS Statistics v.22 (IBM, New York, NY, USA).
5. Conclusions
A total of 15 AhNRAMP genes were identified from the peanut genome, including seven WGD-derived gene pairs and a segmental duplicated gene. Phylogenetic analysis grouped the NRAMP proteins into two distinct subfamilies. Subfamily I contains eight acid proteins with the specific motif 7, while subfamily II includes seven basic proteins sharing the specific motif 10. Subfamily I genes contained four exons, while those in subfamily II had 13 exons. The subfamily I proteins were predicted to localize in the vacuole membrane, while the subfamily II proteins localized to the plasma membrane. All AhNRAMP proteins are perfectly modeled on the 5m94.1.A template, suggesting that AhNRAMPs might have equivalent physiological functions as ScaDMT. Most AhNRAMP genes are preferentially expressed in roots, stamens, and developing seeds, suggesting an essential role in metal uptake and their transport to stamens and developing seeds. The expression of most AhNRAMPs is induced by Fe deficiency in peanut roots and positively correlated with Cd accumulation, indicating a crucial role in Fe/Cd interactions in peanuts. The findings provide essential information to understand the functions of the AhNRAMP genes in the Fe/Cd interactions in peanut plants, which are of great importance for screening or breeding cultivars for improving Fe nutrition while reducing Cd accumulation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24021713/s1.
Author Contributions
Conceptualization, Z.T. and G.S.; methodology, G.S.; validation, Z.T. and G.S.; formal analysis, Z.T., J.L., J.G., C.W. and G.S.; investigation, Z.T., J.L., J.G., C.W., Z.Z. and G.S.; resources, G.S.; data curation, Z.T. and G.S.; writing—original draft preparation, Z.T., Z.Z. and G.S.; writing—review and editing, Z.T. and G.S.; supervision, G.S.; project administration, G.S.; funding acquisition, Z.Z. and G.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by grants from the Natural Science Foundation of Anhui Province (2108085MC83), the Natural Science Foundation for Colleges and Universities of Anhui Province (KJ2020ZD83 and KJ2019A0587), and the Innovation Team of Scientific Research Platform of Anhui Province (KJ2015TD001).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Marschner H. Mineral Nutrition of Higher Plants. 2nd ed. Academic Press; Boston, MA, USA: 1995. [Google Scholar]
- 2.Liu F., Zhang Y., Pu X., Cai N., Sui X., Rengel Z., Chen Q., Song Z. Physiological and molecular changes in cherry red tobacco in response to iron deficiency stress. Front. Plant Sci. 2022;13:861081. doi: 10.3389/fpls.2022.861081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo C., Palumbo G., He J.-Z., Pinton R., Cesco S. Review on iron availability in soil: Interaction of Fe minerals, plants, and microbes. J. Soil Sediment. 2014;14:538–548. doi: 10.1007/s11368-013-0814-z. [DOI] [Google Scholar]
- 4.Sterckeman T., Thomine S. Mechanisms of cadmium accumulation in plants. Crit. Rev. Plant Sci. 2020;39:322–359. doi: 10.1080/07352689.2020.1792179. [DOI] [Google Scholar]
- 5.Shao G., Chen M., Wang W., Mou R., Zhang G. Iron nutrition affects cadmium accumulation and toxicity in rice plants. Plant Growth Regul. 2007;53:33–42. doi: 10.1007/s10725-007-9201-3. [DOI] [Google Scholar]
- 6.Chen C., Cao Q., Jiang Q., Li J., Yu R., Shi G. Comparative transcriptome analysis reveals gene network regulating cadmium uptake and translocation in peanut roots under iron deficiency. BMC Plant Biol. 2019;19:35. doi: 10.1186/s12870-019-1654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su Y., Wang X., Liu C., Shi G. Variation in cadmium accumulation and translocation among peanut cultivars as affected by iron deficiency. Plant Soil. 2013;363:201–213. doi: 10.1007/s11104-012-1310-8. [DOI] [Google Scholar]
- 8.Nakanishi H., Ogawa I., Ishimaru Y., Mori S., Nishizawa N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006;52:464–469. doi: 10.1111/j.1747-0765.2006.00055.x. [DOI] [Google Scholar]
- 9.Shi G., Sun L., Wang X., Liu C. Leaf responses to iron nutrition and low cadmium in peanut: Anatomical properties in relation to gas exchange. Plant Soil. 2014;375:99–111. doi: 10.1007/s11104-013-1953-0. [DOI] [Google Scholar]
- 10.Cellier M., Privé G., Belouchi A., Kwan T., Rodrigues V., Chia W., Gros P. Nramp defines a family of membrane proteins. Proc. Natl. Acad. Sci. USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mäser P., Thomine S., Schroeder J.I., Ward J.M., Hirschi K., Sze H., Talke I.N., Amtmann A., Maathuis F.J., Sanders D., et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cailliatte R., Schikora A., Briat J.F., Mari S., Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell. 2010;22:904–917. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castaings L., Caquot A., Loubet S., Curie C. The high-affinity metal transporters NRAMP1 and IRT1 team up to take up iron under sufficient metal provision. Sci. Rep. 2016;6:37222. doi: 10.1038/srep37222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alejandro S., Cailliatte R., Alcon C., Dirick L., Domergue F., Correia D., Castaings L., Briat J.-F., Mari S., Curie C. Intracellular distribution of manganese by the trans-golgi network transporter NRAMP2 is critical for photosynthesis and cellular redox homeostasis. Plant Cell. 2017;29:3068–3084. doi: 10.1105/tpc.17.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao H., Xie W., Yang C., Xu J., Li J., Wang H., Chen X., Huang C.-F. NRAMP2, a trans-Golgi network-localized manganese transporter, is required for Arabidopsis root growth under manganese deficiency. New Phytol. 2018;217:179–193. doi: 10.1111/nph.14783. [DOI] [PubMed] [Google Scholar]
- 16.Thomine S., Lelièvre F., Debarbieux E., Schroeder J.I., Barbier-Brygoo H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J. 2003;34:685–695. doi: 10.1046/j.1365-313X.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- 17.Lanquar V., Ramos M.S., Lelièvre F., Barbier-Brygoo H., Krieger-Liszkay A., Krämer U., Thomine S. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 2010;152:1986–1999. doi: 10.1104/pp.109.150946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanquar V., Lelièvre F., Bolte S., Hamès C., Alcon C., Neumann D., Vansuyt G., Curie C., Schröder A., Krämer U., et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cailliatte R., Lapeyre B., Briat J.F., Mari S., Curie C. The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem. J. 2009;422:217–228. doi: 10.1042/BJ20090655. [DOI] [PubMed] [Google Scholar]
- 20.Chang J.D., Huang S., Konishi N., Wang P., Chen J., Huang X.Y., Ma J.F., Zhao F.J. Overexpression of the manganese/cadmium transporter OsNRAMP5 reduces cadmium accumulation in rice grain. J. Exp. Bot. 2020;71:5705–5715. doi: 10.1093/jxb/eraa287. [DOI] [PubMed] [Google Scholar]
- 21.Tang L., Dong J., Qu M., Lv Q., Zhang L., Peng C., Hu Y., Li Y., Ji Z., Mao B., et al. Knockout of OsNRAMP5 enhances rice tolerance to cadmium toxicity in response to varying external cadmium concentrations via distinct mechanisms. Sci. Total Environ. 2022;832:155006. doi: 10.1016/j.scitotenv.2022.155006. [DOI] [PubMed] [Google Scholar]
- 22.Yang M., Zhang Y., Zhang L., Hu J., Zhang X., Lu K., Dong H., Wang D., Zhao F.J., Huang C.F., et al. OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. J. Exp. Bot. 2014;65:4849–4861. doi: 10.1093/jxb/eru259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishimaru Y., Bashir K., Nakanishi H., Nishizawa N.K. OsNRAMP5, a major player for constitutive iron and manganese uptake in rice. Plant Signal. Behav. 2012;7:763–766. doi: 10.4161/psb.20510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki A., Yamaji N., Yokosho K., Ma J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell. 2012;24:2155–2167. doi: 10.1105/tpc.112.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang J.-D., Gao W., Wang P., Zhao F.-J. OsNRAMP5 is a major transporter for lead uptake in rice. Environ. Sci. Technol. 2022;56:17481–17490. doi: 10.1021/acs.est.2c06384. [DOI] [PubMed] [Google Scholar]
- 26.Chang J.D., Huang S., Yamaji N., Zhang W., Ma J.F., Zhao F.J. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020;43:2476–2491. doi: 10.1111/pce.13843. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi R., Ishimaru Y., Senoura T., Shimo H., Ishikawa S., Arao T., Nakanishi H., Nishizawa N.K. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011;62:4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi R., Ishimaru Y., Nakanishi H., Nishizawa N.K. Role of the iron transporter OsNRAMP1 in cadmium uptake and accumulation in rice. Plant Signal. Behav. 2011;6:1813–1816. doi: 10.4161/psb.6.11.17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Li J., Yu Y., Dai X., Gong C., Gu D., Xu E., Liu Y., Zou Y., Zhang P., et al. The tonoplast-localized transporter OsNRAMP2 is involved in iron homeostasis and affects seed germination in rice. J. Exp. Bot. 2021;72:4839–4852. doi: 10.1093/jxb/erab159. [DOI] [PubMed] [Google Scholar]
- 30.Yamaji N., Sasaki A., Xia J.X., Yokosho K., Ma J.F. A node-based switch for preferential distribution of manganese in rice. Nat. Commun. 2013;4:2442. doi: 10.1038/ncomms3442. [DOI] [PubMed] [Google Scholar]
- 31.Yang M., Zhang W., Dong H., Zhang Y., Lv K., Wang D., Lian X. OsNRAMP3 is a vascular bundles-specific manganese transporter that is responsible for manganese distribution in rice. PLoS ONE. 2013;8:e83990. doi: 10.1371/journal.pone.0083990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J.Y., Liu J., Dong D., Jia X., McCouch S.R., Kochian L.V. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA. 2014;111:6503–6508. doi: 10.1073/pnas.1318975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J., Yamaji N., Kasai T., Ma J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA. 2010;107:18381–18385. doi: 10.1073/pnas.1004949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peris-Peris C., Serra-Cardona A., Sánchez-Sanuy F., Campo S., Ariño J., San Segundo B. Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice. Mol. Plant. Microbe Interact. 2017;30:385–398. doi: 10.1094/MPMI-01-17-0005-R. [DOI] [PubMed] [Google Scholar]
- 35.Su Y., Zhang Z., Su G., Liu J., Liu C., Shi G. Genotypic differences in spectral and photosynthetic response of peanut to iron deficiency. J. Plant Nutr. 2015;38:145–160. doi: 10.1080/01904167.2014.920392. [DOI] [Google Scholar]
- 36.Shi G., Su G., Lu Z., Liu C., Wang X. Relationship between biomass, seed components and seed Cd concentration in various peanut (Arachis hypogaea L.) cultivars grown on Cd-contaminated soils. Ecotoxicol. Environ. Saf. 2014;110:174–181. doi: 10.1016/j.ecoenv.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Liu C., Yu R., Shi G. Effects of drought on the accumulation and redistribution of cadmium in peanuts at different developmental stages. Arch. Agron. Soil Sci. 2017;63:1049–1057. doi: 10.1080/03650340.2016.1271120. [DOI] [Google Scholar]
- 38.Su Y., Liu J., Lu Z., Wang X., Zhang Z., Shi G. Effects of iron deficiency on subcellular distribution and chemical forms of cadmium in peanut roots in relation to its translocation. Environ. Exp. Bot. 2014;97:40–48. doi: 10.1016/j.envexpbot.2013.10.001. [DOI] [Google Scholar]
- 39.Xiong H., Kobayashi T., Kakei Y., Senoura T., Nakazono M., Takahashi H., Nakanishi H., Shen H., Duan P., Guo X., et al. AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J. Exp. Bot. 2012;63:4437–4446. doi: 10.1093/jxb/ers117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams L.E., Pittman J.K., Hall J.L. Emerging mechanisms for heavy metal transport in plants. Biochim. Biophys. Acta. 2000;1465:104–126. doi: 10.1016/S0005-2736(00)00133-4. [DOI] [PubMed] [Google Scholar]
- 41.Hurst L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002;18:486–487. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 42.Mani A., Sankaranarayanan K. In silico analysis of natural resistance-associated macrophage protein (NRAMP) family of transporters in rice. Protein J. 2018;37:237–247. doi: 10.1007/s10930-018-9773-y. [DOI] [PubMed] [Google Scholar]
- 43.Qin L., Han P., Chen L., Walk T.C., Li Y., Hu X., Xie L., Liao H., Liao X. Genome-wide identification and expression analysis of NRAMP family genes in soybean (Glycine max L.) Front. Plant Sci. 2017;8:1436. doi: 10.3389/fpls.2017.01436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishida J.K., Caldas D.G.G., Oliveira L.R., Frederici G.C., Leite L.M.P., Mui T.S. Genome-wide characterization of the NRAMP gene family in Phaseolus vulgaris provides insights into functional implications during common bean development. Genet. Mol. Biol. 2018;41:820–833. doi: 10.1590/1678-4685-gmb-2017-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian W., He G., Qin L., Li D., Meng L., Huang Y., He T. Genome-wide analysis of the NRAMP gene family in potato (Solanum tuberosum): Identification, expression analysis and response to five heavy metals stress. Ecotoxicol. Environ. Saf. 2021;208:111661. doi: 10.1016/j.ecoenv.2020.111661. [DOI] [PubMed] [Google Scholar]
- 46.Li J., Duan Y., Han Z., Shang X., Zhang K., Zou Z., Ma Y., Li F., Fang W., Zhu X. Genome-wide identification and expression analysis of the NRAMP family genes in tea plant (Camellia sinensis) Plants. 2021;10:1055. doi: 10.3390/plants10061055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ullah I., Wang Y., Eide D.J., Dunwell J.M. Evolution, and functional analysis of Natural Resistance-Associated Macrophage Proteins (NRAMPs) from Theobroma cacao and their role in cadmium accumulation. Sci. Rep. 2018;8:14412. doi: 10.1038/s41598-018-32819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y., Zhao X., Li G., Kumar S., Sun Z., Li Y., Guo W., Yang J., Hou H. Genome-wide identification of the Nramp gene family in Spirodela polyrhiza and expression analysis under cadmium stress. Int. J. Mol. Sci. 2021;22:6414. doi: 10.3390/ijms22126414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Wang C., Zhang Z., Shi G. Genome-wide identification of metal tolerance protein genes in peanut: Differential expression in the root of two contrasting cultivars under metal stresses. Front. Plant Sci. 2022;13:791200. doi: 10.3389/fpls.2022.791200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z., Chen N., Zhang Z., Shi G. Genome-wide identification and expression profile reveal potential roles of peanut ZIP family genes in zinc/iron-deficiency tolerance. Plants. 2022;11:786. doi: 10.3390/plants11060786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C., Wang X., Li J., Guan J., Tan Z., Zhang Z., Shi G. Genome-wide identification and transcript analysis reveal potential roles of oligopeptide transporter genes in iron deficiency induced cadmium accumulation in peanut. Front. Plant Sci. 2022;13:894848. doi: 10.3389/fpls.2022.894848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertioli D.J., Jenkins J., Clevenger J., Dudchenko O., Gao D., Seijo G., Leal-Bertioli S.C.M., Ren L., Farmer A.D., Pandey M.K., et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019;51:877–884. doi: 10.1038/s41588-019-0405-z. [DOI] [PubMed] [Google Scholar]
- 53.Soltis D.E., Visger C.J., Soltis P.S. The polyploidy revolution then…and now: Stebbins revisited. Am. J. Bot. 2014;101:1057–1078. doi: 10.3732/ajb.1400178. [DOI] [PubMed] [Google Scholar]
- 54.Chen X., Lu Q., Liu H., Zhang J., Hong Y., Lan H., Li H., Wang J., Liu H., Li S., et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant. 2019;12:920–934. doi: 10.1016/j.molp.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 55.Panchy N., Lehti-Shiu M., Shiu S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian W., Liao B.-Y., Chang A.Y.F., Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meng J.G., Zhang X.D., Tan S.K., Zhao K.X., Yang Z.M. Genome-wide identification of Cd-responsive NRAMP transporter genes and analyzing expression of NRAMP 1 mediated by miR167 in Brassica napus. Biometals. 2017;30:917–931. doi: 10.1007/s10534-017-0057-3. [DOI] [PubMed] [Google Scholar]
- 58.Ehrnstorfer I.A., Geertsma E.R., Pardon E., Steyaert J., Dutzler R. Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol. 2014;21:990–996. doi: 10.1038/nsmb.2904. [DOI] [PubMed] [Google Scholar]
- 59.Thomine S., Wang R., Ward J.M., Crawford N.M., Schroeder J.I. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 61.Duvaud S., Gabella C., Lisacek F., Stockinger H., Ioannidis V., Durinx C. Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021;49:W216–W227. doi: 10.1093/nar/gkab225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsirigos K.D., Peters C., Shu N., Käll L., Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clevenger J., Chu Y., Scheffler B., Ozias-Akins P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 2016;7:1446. doi: 10.3389/fpls.2016.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.