Abstract
The DegP protease, a multifunctional chaperone and protease, has been shown to be essential for virulence in gram-negative pathogens such as Salmonella enterica serovar Typhimurium, Brucella abortus, Yersinia enterocolitica, and Pseudomonas aeruginosa. The function of DegP in pathogenesis appears to be the degradation of damaged proteins that accumulate as a result of the initial host response to infection, which includes the release of reactive oxygen intermediates. Additionally, the DegP protease plays a major role in monitoring and maintaining the Escherichia coli periplasm and influences E. coli pilus biogenesis. We report here the identification of highly homologous enzymes in Streptococcus pyogenes, Streptococcus gordonii, Streptococcus mutans, Staphylococcus aureus, and Enterococcus faecalis. Moreover, the phenotype of an insertionally inactivated degP allele in S. pyogenes is similar to that reported for E. coli, with temperature sensitivity for growth and enhanced sensitivity to reactive oxygen intermediates. Virulence studies in a mouse model of streptococcal infection indicate that a functional DegP protease is required for full virulence. These results suggest DegP as an attractive broad-spectrum target for future anti-infective drug development.
DegP (HtrA) is a highly conserved periplasmic protease found in most gram-negative bacteria (25, 30, 44). DegP homologues have also been identified in chlamydia, mycobacteria, yeast, and humans (19, 30). DegP was initially characterized as a serine protease due to cleavage activity on casein and inhibition of proteolytic activity by diisopropylfluorophosphate (25). Point mutations in residues presumed to represent two members of the catalytic triad (serine 210 and histidine 105) inactivated enzymatic activity, confirming the classification of DegP as a serine protease (38).
The DegP protease performs an essential function during growth in monitoring and degrading misfolded or aggregated proteins in the periplasm (16, 30). This is most apparent when considering that null mutations in degP (htrA) are unable to grow at high temperature (25, 37). Moreover, fusion proteins and experimentally unstable proteins have been found to be stabilized in degP knockout strains (16, 20, 30, 44). Recent studies revealed that degP is a member of a regulon that responds to extracytoplasmic stress and has a promoter sequence utilized by RNA polymerase containing ςE, the stress response sigma factor in Escherichia coli (8, 30). It has also been demonstrated that DegP is regulated by the CpxAR two-component regulatory system (8). This system “senses” and responds to periplasmic stress such as protein misfolding and aggregation. Jones et al. demonstrated that Pap pilin expression in the absence of the periplasmic chaperone, PapD, resulted in the induction of expression of the degP promoter (16). Furthermore, high-level expression of Pap pilin proteins in a degP null mutant was found to be highly toxic to the cell, resulting in stasis of growth (16).
DegP's cleavage and recognition site in a native substrate has not yet been identified, although a number of proteins have been described as DegP substrates in vitro and in vivo: colicin A lysis protein (5), K88 and K99 pilin subunits (2), HMW1 and HMW2 of Haemophilus influenzae (43), MalS (42), and the PapA pilin (C. H. Jones et al., unpublished data). The pilin subunit proteins represent a relevant in vivo substrate for DegP, and recent in vitro studies have shown that DegP cleaves PapA in both monomeric and polymeric or aggregate forms (Jones et al., unpublished). Earlier studies demonstrated that DegP shows a preference for misfolded substrate proteins, such as those that might arise from thermal or oxidative denaturation (18, 20). Two recent studies have provided evidence that under specific conditions DegP displays chaperone activity (27, 42).
In the last several years a significant body of data has accumulated demonstrating that DegP is a virulence factor for several pathogenic organisms. In Salmonella enterica serovar Typhimurium, htrA null mutants were found to be avirulent and more susceptible to oxidative stress (15). The authors of this study suggested that htrA mutants are less able to withstand oxidative killing within the macrophage. Furthermore, an htrA lesion was found to be useful in attenuating Salmonella enterica serovar Typhi for implementation as a vaccine strain (45). Similarly, Brucella abortus and Brucella melitensis htrA null mutants were attenuated for virulence in goats and found to be significantly more sensitive to oxidative killing by cultured neutrophils in vitro (10, 11, 31). Likewise, in Yersinia enterocolitica HtrA was found to be essential for virulence and an htrA mutant strain was more sensitive to oxidative stress (23). Finally, Boucher et al. (4) demonstrated recently that in Pseudomonas aeruginosa conversion to mucoidy, the so-called cystic fibrosis phenotype, involves two htrA homologues.
Thus, DegP plays a number of important and essential roles within the periplasmic compartment. Since gram-positive bacteria lack this organelle, it was quite surprising to discover that like their gram-negative brethren, gram-positive bacteria also encode and express essential DegP proteases (14, 29, 30, 33). The studies described here report the identification of DegP homologues in Streptococcus pyogenes, Staphylococcus aureus, Enterococcus faecalis, and several other gram-positive bacteria. We demonstrate that DegP is required for thermal stability and resistance to oxidative stress in S. pyogenes. Moreover, virulence of the degP knockout strain is reduced in a mouse model. As such, DegP represents a novel broad-spectrum anti-infective target for development.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. pyogenes S43 (type 6, ATCC 12348), Streptococcus gordonii (13), Streptococcus mutans (UAB159), Lactococcus lactis (ATCC 27861), and S. aureus (RN4220 and Newman strains, gift of O. Schneewind, UCLA; ATCC 25923, ATCC 35556) were obtained from laboratory stocks or the American Type Culture Collection, as indicated. INVα (Invitrogen, Inc., Carlsbad, Calif.) was used for routine cloning steps. pVA891 was a gift from F. Macrina, Medical College of Virginia. Gram-positive organisms were propagated in brain heart infusion media (BHI; Difco-Becton Dickinson, Sparks, Md.). E. coli was propagated in Luria Bertani broth (Difco). For assay of temperature sensitivity, overnight cultures, grown at 30°C, were diluted 1:50 into fresh media containing appropriate antibiotics (erythromycin, 5 μg/ml for SP10). Cultures were split and incubated at the indicated temperatures and growth was monitored by absorbance (A600) every half hour for an 8-h period. For paraquat (methyl viologen) and hydrogen peroxide sensitivity, overnight cultures grown at 30°C were streaked onto fresh BHI plates containing the appropriate antibiotics. A sterile 3M disk (Whatman filter paper, approximately 7 mm in diameter) was applied to the plate into which paraquat (50 μl, 300 mM) or H2O2 (50 μl, 300 mM) was added. The growth inhibition zone around the filter disk was measured after 12 h of incubation at 30°C.
Database search methodology and sequence alignment.
DegP sequences were collected from the following sources: S. pyogenes and S. aureus (University of Oklahoma Advanced Center for Genome Technology [OU-ACGT], The Sanger Center), E. faecalis (The Institute for Genomic Research), Streptococcus pneumoniae (14), Lactobacillus helveticus (40), Bacillus subtilis (12) and E. coli (24). The S. gordonii, S. mutans, and L. lactis sequences were identified by screening genomic preparations with PCR primers (TB273, TB274; see below) that were based on highly conserved regions of degP. The sequences were aligned using the Clustal W 1.7 alignment tool at the Baylor College of Medicine Search Launcher-Multiple Sequence Alignment website (http://dot.imgen.bcm.tmc.edu). The alignment was formatted for publication using the Boxshade 3.21 tool (http://www.ch.embnet.org).
Construction of SP10 (S43 degP::ermAM).
A 630 base-pair fragment of degP (encoding residues 28 to 237) was amplified from whole chromosomal DNA prepared from S43 by standard “boil-prep” method (3). The following primers were used: TB273 (5′CCA TCG ATG GGT TAA TAG CAG CAT C) and TB274 (5′ GCT CTA GAC ATT CAA TAA TCT CTA CCC). TB273 encodes a ClaI site and TB274 encodes an XbaI site. One hundred nanograms of DNA and 0.4 μM concentrations of the primers (TB273 and TB274) were used in the PCRs under standard conditions (3). The resultant PCR product was subcloned into pVA891 using ClaI and XbaI, creating the insertional plasmid pVA273-274. The primers TB273 and TB274 were also used to amplify degP sequences from S. mutans, S. gordonii, and L. lactis. The insertional plasmid pVA273-274 was electroporated into electrocompetent S43 and recombinants were selected on BHI plates supplemented with erythromycin (5 μg/ml) at 30°C. Recombinants were verified to have inserted the plasmid into the degP locus by PCR analysis using diagnostic primers and by Southern blot analysis (data not shown).
Mouse virulence studies.
Swiss CD-1 mice (10 weeks old) were used for 50% lethal dose (LD50) determinations with S43 and SP10. Bacteria were grown in Todd-Hewitt medium (Difco) supplemented with 2% yeast extract until late exponential growth phase (A650 = 0.75) in the presence of appropriate antibiotics (5 μg of erythromycin/ml for SP10). The bacteria were washed with and then resuspended in 1× phosphate-buffered saline (0.14 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) at 1/10 (experiment 1) or 1/100 (experiment 2) the original culture volume. Serial 10-fold dilutions were then delivered intraperitoneally in 100-μl volumes to the mice (five mice/group in experiment 1; 6 mice/group in experiment 2). The dilutions were also plated on 5% blood agar plates to determine actual CFU delivered. The mice were monitored daily for mortality through day 5 and the LD50 was calculated by the method of Reed and Muench (35).
RESULTS
degP is conserved in gram-positive pathogens.
Database searches were conducted to identify degP homologues in S. pyogenes (M1; group A streptococcus), S. aureus (NCTC 8325, methicillin-resistant strain 252, and methicillin-sensitive strain 476), and E. faecalis (V583 strain). PCR was carried out to screen S. mutans, S. gordonii, and L. lactis as well as to clone the homologues from S. pyogenes and S. aureus. In each organism screened, in silico or in vitro, at least one degP homologue was identified (Fig. 1). Interestingly, in all S. aureus strains examined two degP genes were identified (Fig. 1). Recently degP homologues have been described in L. lactis (33). S. pneumoniae (14), and L. helveticus (40), and several homologues have been identified in B. subtilis (29).
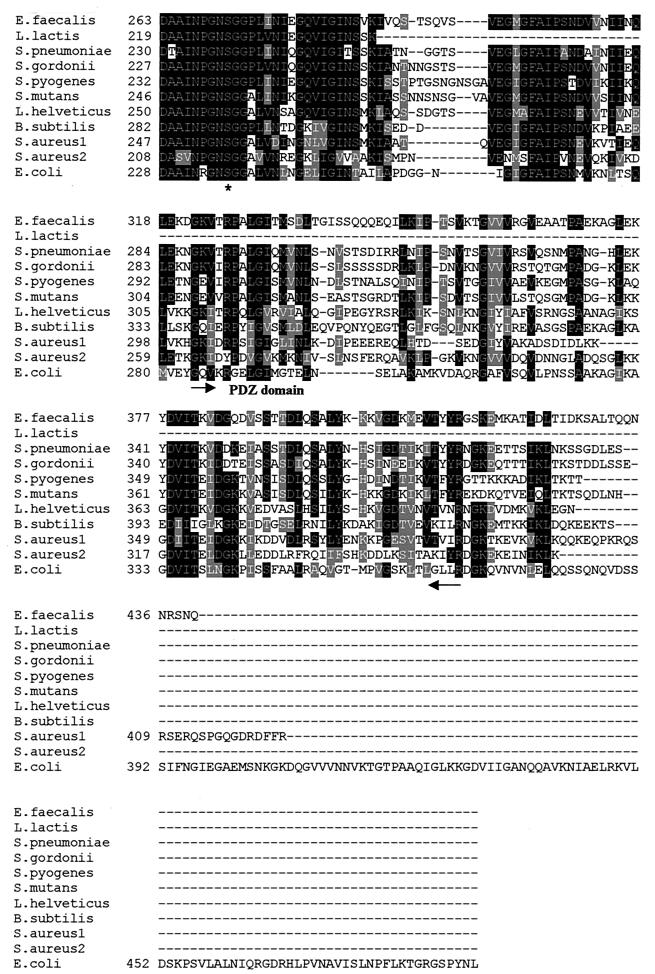
FIG. 1.
Alignment of DegP homologues from gram-positive bacteria and E. coli. Note the high degree of sequence conservation around the invariant residues of the serine-protease catalytic triad (histidine 129, aspartic acid 158, and serine 240; S. pyogenes amino acid numbering). The PDZ domain (see text) initiates with glycine 296 and terminates with arginine 385 (S. pyogenes residue numbering) in the gram-positive bacterial proteins. The putative membrane anchor region in S. pyogenes DegP lies between isoleucine 13 and isoleucine 26. The three residues of the catalytic triad are indicated by asterisks; the putative transmembrane region and the PDZ domain are also noted. The DegP sequence from L. lactis is not complete.
Figure 1 is a sequence alignment of degP homologues identified in E. faecalis, S. gordonii, S. pyogenes, S. mutans, and S. aureus, along with several other previously described homologues (14, 29, 33, 40). The central core of the protein, approximately 150 residues, is very highly conserved among the gram-positive enzymes as well as with the canonical E. coli protein. The three residues of the serine protease catalytic triad (histidine 129, aspartic acid 158, and serine 240; based on S. pyogenes numbering) are invariant. The amino acids immediately surrounding the catalytic triad residues are strongly conserved as well. The homologues share 66.8% similarity and 19.25% identity as a group. In line with the gram-negative DegP proteins and homologues, all of the gram-positive homologues possess a PDZ (postsynaptic density, discs-large, ZO-1) (22, 32, 41) domain that initiates approximately 50 residues downstream (glycine 296) of the catalytic-site serine residue. Interestingly, the gram-positive homologues have a single PDZ domain, whereas the gram-negative proteins, such as E. coli DegP (HtrA), have a second PDZ domain immediately following the first. PDZ domains have been implicated in protein-protein interactions as well as in protein-substrate interactions (22, 36, 41).
The gram-positive DegP homologues have an amino-terminal sequence (S. pyogenes DegP residues 13 to 26; S. aureus DegP1 residues 30 to 49) that is predicted to be membrane spanning, based on both the Argos transmembrane and Von Heijne transmembrane prediction algorithms (1, 40). Although the precise localization of the gram-positive proteins has not yet been determined, the amino-terminal sequence may play a role in anchoring the protease in the membrane as proposed by Poquet et al. for the L. lactis homologue, HtrALI (38), and as suggested by Noone et al. for the B. subtilis homologue, YdkA (29). Some support for this mode of anchoring gram-positive surface proteins comes from recent work on the SrtA surface protease in S. aureus (47). Schneewind's group demonstrated that SrtA contains a noncleaved amino-terminal hydrophobic region that serves to anchor the enzyme in the membrane (47). The gram-positive DegP homologues do not possess the canonical LPXTG sequence at the carboxyl terminus, which is essential for cell wall linkage, that has been described for many surface proteins in gram-positive bacteria (28).
DegP is essential for survival at high temperature and for resistance to oxidative stress in S. pyogenes.
An internal fragment of 630 nucleotides from the S. pyogenes degP gene was cloned from the S43 (type 6) strain by PCR using primers (TB273 and TB274) based on the sequence acquired from the OU-ACGT database (www.genome.ou.edu) (Fig. 2A). The degP sequence was cloned into pVA891 (E. coli-S. pyogenes shuttle vector; gift of F. Macrina, Medical College of Virginia) to create pVA273-274 (26). Primers TB273 and TB274 amplify the sequence encoding amino terminal residues 28 to 237 of DegP. pVA273-274 also carries the ermAM selectable marker (26). Insertion of this plasmid into the degP locus results in two truncated copies of the gene, since the 5′ end of degP is not included in pVA273-274. S. pyogenes S43 was transformed by electroporation with pVA273-274, and the transformants were selected on erythromycin (5 μg/ml) at 30°C. A transformant was selected and plasmid insertion by single crossover into the degP locus was confirmed by PCR (Fig. 2B) and Southern blot analysis (data not shown). The S43 degP::ermAM strain is called SP10.
FIG. 2.
Construction of a degP::ermAM knockout mutation in S. pyogenes S43. (A) Strategy for insertional inactivation of degP in S. pyogenes. Primers TB273 and TB274 were used to amplify 630 nucleotides from the 5′ end (encoding residues 28 to 237) of degP. The 273 to 274 fragment was cloned into pVA891 and the resultant plasmid, pVA273-274, was used to electroporate S. pyogenes S43. The primers used for PCR are indicated with an arrow. (B) Verification of the degP knockout in SP10 (S43 degP::ermAM). Chromosomal DNA was prepared from S43 and SP10 and diagnostic primers were used to demonstrate the insertion of sequences from pVA273-274 into the degP locus in SP10. Primer pairs TB250 and TB273 were used in lanes 2 and 3, TB332 and TB333 were used in lanes 4 and 5, and TB170 and TB173 were used in lanes 6 and 7. S43 chromosomal DNA was amplified in lanes 2, 4, and 6, while SP10 chromosomal DNA was amplified in lanes 3, 5, and 7. The presence of the amplified product in lane 3 (SP10 DNA) indicates the presence of the pVA891 DNA, while the product in lane 5 (SP10 DNA) indicates the presence of the ermAM gene. Note also that the absence of a product in lane 7 (SP10 DNA) is due to the large size of the TB173-TB170 (see panel A) product (>7 kb) that is not efficiently amplified by Taq polymerase.
DegP (htrA) knockouts in gram-negative organisms are, in all cases except one, temperature sensitive for growth (30). We tested SP10 for growth at 30, 37, and 44°C to assay temperature sensitivity. SP10 grows similar to S43 at 30°C, grows markedly slower at 37°C, and is unable to grow at 44°C (Fig. 3 and data not shown). Therefore, DegP appears to carry out a role in S. pyogenes similar to that in gram-negative organisms: providing thermoprotection by clearing heat-damaged or otherwise denatured proteins from the bacterial membrane. We next tested the resistance of S43 (wild type [wt]) and SP10 (S43 degP::ermAM) to oxidative stress using paraquat in disk diffusion assays on BHI plates. As shown by the failure of SP10 to grow near the filter disk, the degP mutant strain is more sensitive to oxidative stress than the parent strain (Fig. 3C).
FIG. 3.
DegP is essential for thermal and oxidative tolerance in S. pyogenes. (A and B) Growth curves. S43 and SP10 were grown overnight at 30°C in stationary flasks in BHI with the appropriate antibiotic, diluted 1:50 into fresh BHI, and grown at the indicated temperature. Growth was monitored by absorbance (A600) at 30-min intervals. SP10 grew slower than S43 at 30°C, although at 20 h the strains reached an A600 of 0.84 and 0.83, respectively (data not shown). SP10 grew slower than S43 at 37°C (A) and failed to grow at 44°C (B). (C) Sensitivity to paraquat. Overnight cultures (30°C) of S43 and SP10 were brought to the same optical density (A600) in BHI and spread on fresh BHI agar plates containing the appropriate antibiotics. After the plates were dry (30 min), a sterile filter disk containing 50 μl of 200 mM paraquat was placed in the center of the plate. Inhibition zones (including the 7-mm disc) were measured after 20 h of growth at 30°C. The inhibition zones indicated are the means from six identical plates. The data clearly indicate mutation of degP results in enhanced sensitivity to oxidative stress generated by exposure to paraquat (P = 0.001; paired t test).
To ensure that the observed phenotype of SP10 was linked to the plasmid insertion, chromosomal DNA was prepared from SP10 and electroporated into an erythromycin-sensitive strain to construct SP10b. The erythromycin-resistant transformants were found to be highly sensitive to paraquat (data not shown).
DegP is required for full virulence in a mouse systemic infection model.
SP10 and S43 were tested in a mouse pathogenesis model to ascertain the effect of the degP lesion on virulence. In the first experiment at the highest dose administered (1.5 × 109CFU), all of the mice receiving SP10 survived whereas all five mice that were inoculated with S43 died (Table 1). In the second experiment, an accurate LD50 was calculated for both strains, as higher inocula of bacteria were delivered. Under the conditions of the assay, the LD50 for SP10 (4.74 × 108 CFU) was found to be 35-fold higher than that for S43 (1.37 × 107 CFU), suggesting that DegP does indeed play a role in virulence of S. pyogenes.
TABLE 1.
S. pyogenes DegP is required for full virulence in mice
| S. pyogenes strain | CFU injected | Mortality (no. dead/total no. treated) |
|---|---|---|
| Experiment 1 | ||
| S43 (wt) | 1.5 × 109 | 5/5 |
| 1.5 × 107 | 1/5 | |
| SP10 (degP::ermAM) | 1.5 × 109 | 0/5 |
| 1.5 × 107 | 0/5 | |
| Experiment 2 | ||
| S43 (wt) | 2.2 × 109 | 6/6 |
| 2.2 × 108 | 4/6 | |
| 2.2 × 107 | 1/6 | |
| SP10 (degP::ermAM) | 2.2 × 109 | 5/6 |
| 2.2 × 108 | 2/6 | |
| 2.2 × 107 | 0/6 |
DISCUSSION
A mechanism to deal with thermally, oxidatively, or otherwise unfolded proteins from the periplasm and inner membrane of gram-negative bacteria has been described and shown to depend on the conserved periplasmic protease DegP (7, 8, 16, 30, 34). Somewhat surprisingly, this system is at least in part conserved in gram-positive bacteria, including several important human pathogens. We present evidence that the protease component of this system, DegP, is highly conserved in sequence in gram-positive bacteria and functions in a manner similar to the E. coli counterpart.
The gram-positive DegP homologues are well conserved, especially through the central portion of the protein, which contains the residues of the serine protease catalytic triad (Fig. 1). The proteins are 66.8% similar and 19.25% identical through the central 185 residues (extending from residue 107 to residue 294; S. pyogenes numbering). One interesting difference between the gram-negative and gram-positive enzymes is the presence of a single PDZ domain at the carboxyl terminus of the gram-positive proteins, whereas most gram-negative enzymes possess two PDZ domains (30). PDZ domains, originally described in membrane receptors from eukaryotes, have been implicated in substrate recognition as well as in protein multimerization (22, 32, 41; M. J. Pallen and C. P. Ponting, Letter, Mol. Microbiol. 26:411–413, 1997). It was demonstrated recently that deletion of both PDZ domains in E. coli DegP resulted in the failure of the enzyme to form a multimer, in this case a hexamer (36). Deletion of only one of the two PDZ domains of the E. coli enzyme resulted in an unstable protein, suggesting that both PDZ domains are required for correct folding of the E. coli enzyme (36). Moreover, the failure to form the hexamer also correlated with the lack of enzymatic activity of the mutant enzyme (36). PDZ domains have also been associated with substrate binding for several proteins (22, 41). Additional studies will be necessary to establish the multimeric state of the S. pyogenes DegP and to clarify the role of the single PDZ domain for the gram-positive DegP homologues.
There is still debate as to the localization of the DegP protease in E. coli, whether it is soluble in the periplasm or membrane associated (30, 36). However, recent work on the DegP homologue from L. lactis, referred to as HtrALI, suggests that HtrALI is the major membrane-associated surface protease in lactococci (33). Poquet et al. (33) found that in an htrALI null mutant several surface proteins failed to be appropriately processed in lactococcus. Our preliminary studies suggest that S. pyogenes DegP is membrane associated (unpublished data). However, we have not detected a difference in major surface protein expression in SP10 (S43 degP::ermAM), contrary to the results in lactococci (unpublished data). A number of proteins in gram-positive bacteria influence surface protein expression, such as the recently described SrtA protease-transpeptidase in S. aureus (28) and S. gordonii (3).
Surface proteins in gram-positive bacteria are often linked to the cell wall through a conserved surface protein expression system in which the canonical LPXTG sequence is recognized by the SrtA protease-transpeptidase and linked to the cell wall peptidoglycan (28). The LPXTG sequence is not found in the gram-positive DegP homologues. However, the DegP homologues do possess an amino-terminal sequence that is predicted to be a transmembrane anchoring sequence (Fig. 1). The surface anchoring of DegP may be similar to that described for the SrtA protease of S. aureus. SrtA is proposed to be surface exposed on the bacterial membrane by a noncleavable transmembrane leader sequence (46, 47). It may be that all membrane-associated surface proteases are anchored to the membrane in a similar manner; however, we have yet to determine if the putative leader sequence is cleaved in DegP.
The phenotype of SP10 is strikingly similar to that of many gram-negative degP null mutants, with thermal and oxidative sensitivity (30). We suggest that this is due to a similar role played by DegP in gram-positive and gram-negative organisms. Heat or oxidative stress results in an accumulation of misfolded or aggregated proteins in the bacterial membrane. A signal transduction system is triggered by the presence of the aberrant protein species that results in synthesis of chaperones and proteases. Preliminary studies reveal that in SP10 (S43 degP::ermAM) a number of novel proteins accumulate following temperature shift or exposure to paraquat, supporting the contention that in the wt strain these aberrant proteins are degraded by the DegP protease (unpublished data). In gram-negative bacteria, there are at least two systems that respond to misfolded or aggregated proteins in the periplasm and membrane: the Cpx system and the ςE system (34). Both of these signal transduction systems activate transcription of degP (34). Recent data suggest that DegP in E. coli selectively degrades denatured target proteins, such as those arising from thermal stress (18, 20). Interestingly, two recent papers presented evidence that E. coli DegP also has an intrinsic chaperone function as well as proteolytic activity (27, 42). In one case the proteolytic activity is triggered by temperature shift (to high temperature), whereas the chaperone activity predominates at low temperatures (42).
The studies presented herein demonstrate that the DegP protease from S. pyogenes is essential for survival following thermal and oxidative stress. We suggest that a mechanism, similar to that described in E. coli, senses and triggers a response to membrane damage due to the presence of denatured or aggregated proteins. A major player in the proposed pathway is the DegP protease. We are currently studying the regulation of the degP gene and investigating the possibility that a Cpx-like signal transduction system is involved in activation of DegP synthesis.
An interesting finding of these studies is the presence of two DegP homologues in S. aureus (Fig. 1). We are currently studying the regulation of the two degP genes in response to various environmental stresses. One possibility is that, similar to the E. coli DegP homologues DegQ and DegS, the two S. aureus DegP homologues have distinct as well as overlapping functions (30) relating to either substrate specificity, regulatory signals, or temporal expression.
For a number of gram-negative pathogens, a degP null mutation resulted in reduced virulence that correlated with an enhanced sensitivity to oxidative stress (10, 11, 15, 23, 31, 39). Therefore, we tested SP10 in a virulence model for S. pyogenes in mice. The results clearly indicate that the degP::ermAM lesion reduces virulence in the mouse (Table 1). Whether the reduction in virulence is due to a growth defect in the mouse (body temperature 36.5 to 38°C) or due to oxidative stress is unclear at present. Previous investigations indicate that in vitro passage of S. pyogenes may result in the loss of certain virulence factors, including M protein, and that in vivo mouse passage of these strains results in the restoration of these factors (21). Therefore, under the current nonpassaged conditions, it is surmised that the 35-fold increase in the LD50 for the degP null mutant can be considered a conservative estimate of this mutation's effect. An additional consideration is that since the insertion event results in a promoterless copy of the intact degP gene lacking only the first 28 residues, readthrough could result in a partially active DegP protein in SP10. Moreover, in the paraquat sensitivity experiment (Fig. 3C) a degree of reversion was seen. Nevertheless, these data support the role of DegP in full virulence of S. pyogenes. A number of recent studies have demonstrated that gram-positive pathogens efficiently invade host cells (6, 9, 17). These findings indicate that a mechanism to protect S. pyogenes and other gram-positive pathogens from oxidative damage would benefit the pathogen. Therefore, due to the conservation of the DegP protease, these studies indicate that DegP inhibitors may represent broad-spectrum anti-infective agents.
ACKNOWLEDGMENTS
We acknowledge technical assistance from Jenny McDonald, Travis Warren, David King, and Zachariah Blackwood and thank Emma Dutton for critical reading of the manuscript.
The research is supported by a Small Business Innovation Research grant (R43 AI 46828-01; C.H.J.).
REFERENCES
- 1.Argos P, Rao J K, Hargrave P A. Structural prediction of membrane-bound proteins. Eur J Biochem. 1982;128:565–575. doi: 10.1111/j.1432-1033.1982.tb07002.x. [DOI] [PubMed] [Google Scholar]
- 2.Bakker D, Vader C E, Roosendaal B, Mooi F R, Oudega B, de Graaf F K. Structure and function of periplasmic chaperone-like proteins involved in the biosynthesis of K88 and K99 fimbriae in enterotoxigenic Escherichia coli. Mol Microbiol. 1991;5:875–886. doi: 10.1111/j.1365-2958.1991.tb00761.x. [DOI] [PubMed] [Google Scholar]
- 3.Bolken T C, Franke C A, Jones K F, Zeller G O, Jones C H, Dutton E K, Hruby D E. Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion. Infect Immun. 2001;69:75–80. doi: 10.1128/IAI.69.1.75-80.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher J C, Martinez-Salazar J, Schurr M J, Mudd M H, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavard D, Lazdunski C, Howard S P. The acylated precursor form of the colicin A lysis protein is a natural substrate of the DegP protease. J Bacteriol. 1989;171:6316–6322. doi: 10.1128/jb.171.11.6316-6322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary P P, Cue D. High frequency invasion of mammalian cells by beta hemolytic streptococci. Subcell Biochem. 2000;33:137–166. doi: 10.1007/978-1-4757-4580-1_7. [DOI] [PubMed] [Google Scholar]
- 7.Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding systems in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 8.Danese P N, Snyder W B, Cosma C L, Davis L J B, Silhavy T J. The Cpx two-component signal transduction pathway of Escherichia coli regulates degP transcription. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 9.Dombek P E, Cue D, Sedgewick J, Lam H, Ruschkowski S, Finlay B B, Cleary P P. High-frequency intracellular invasion of epithelial cells by serotype M1 group A streptococci: M1 protein-mediated invasion and cytoskeletal rearrangements. Mol Microbiol. 1999;31:859–870. doi: 10.1046/j.1365-2958.1999.01223.x. [DOI] [PubMed] [Google Scholar]
- 10.Elzer P H, Hagius S D, Robertson G T, Phillips R W, Walker J V, Fatemi M B, Enright F M, Roop R M., II Behaviour of a high-temperature-requirement A (HtrA) deletion mutant of Brucella abortus in goats. Res Vet Sci. 1996;60:48–50. doi: 10.1016/s0034-5288(96)90130-7. [DOI] [PubMed] [Google Scholar]
- 11.Elzer P H, Phillips R W, Robertson G T, Roop R M., II The HtrA stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect Immun. 1996;64:4838–4841. doi: 10.1128/iai.64.11.4838-4841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabret C, Hoch J. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke C A, Bolman T M, Ottum S A, Jones K F, Hruby D E. Streptococcus gordonii strains resistant to fluorodeoxyuridine contain mutations in the thymidine kinase gene and are deficient in thymidine kinase activity. Antimicrob Agents Chemother. 2000;44:787–789. doi: 10.1128/aac.44.3.787-789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasc A M, Giammarinaro P, Richter S, Sicard M. Organization around the dnaA gene of Streptococcus pneumoniae. Microbiology. 1998;144:433–439. doi: 10.1099/00221287-144-2-433. [DOI] [PubMed] [Google Scholar]
- 15.Johnson K, Charles I, Dougan G, Pickard D, O'Gaora P, Costa G, Ali T, Miller I, Hormaeche C. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahl B C, Goulian M, van Wamel W, Herrmann M, Simon S M, Kaplan G, Peters G, Cheung A L. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect Immun. 2000;68:5385–5392. doi: 10.1128/iai.68.9.5385-5392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K I, Park S C, Kang S H, Cheong G W, Chung C H. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J Mol Biol. 1999;294:1363–1374. doi: 10.1006/jmbi.1999.3320. [DOI] [PubMed] [Google Scholar]
- 19.Kolmar H. DegP or protease DO CLAN SA. In: Barrett A J, Rawlings N D, Woessner J F, editors. Handbook of proteolytic enzymes (CD-ROM). London, Great Britain: Academic Press; 1998. [Google Scholar]
- 20.Kolmar H, Waller P R H, Sauer R T. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J Bacteriol. 1996;178:5925–5929. doi: 10.1128/jb.178.20.5925-5929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lancefield R C. Occurrence of R antigen specific for group A type 3 streptococci. J Exp Med. 1958;108:329–341. doi: 10.1084/jem.108.3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. PDZ-like domains mediate binding specificity in the CLP/HSP100 family of chaperones and protease regulatory subunits. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 23.Li S-R, Dorrell N, Everest P H, Dougan G, Wren B W. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect Immun. 1996;64:2088–2094. doi: 10.1128/iai.64.6.2088-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: a ς32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–10066. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macrina F L, Evans R P, Tobian J A, Hartley D L, Clewell D B, Jones K R. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983;25:145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- 27.Misra R, CastilloKeller M, Deng M. Overexpression of protease-deficient DegP(S210A) rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J Bacteriol. 2000;182:4882–4888. doi: 10.1128/jb.182.17.4882-4888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarre W W, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noone D, Howell A, Devine K M. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J Bacteriol. 2000;182:1592–1599. doi: 10.1128/jb.182.6.1592-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pallen M J, Wren B W. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 31.Phillips R W, Elzer P H, Robertson G T, Hagius S D, Walker J V, Fatemi M B, Enright F M, Roop R M., II A Brucella melitensis high-temperature-requirement A (htrA) deletion mutant is attenuated in goats and protects against abortion. Res Vet Sci. 1997;63:165–167. doi: 10.1016/s0034-5288(97)90012-6. [DOI] [PubMed] [Google Scholar]
- 32.Ponting C P. Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 1997;6:464–468. doi: 10.1002/pro.5560060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poquet I, Saint V, Seznec E, Simones N, Bolotin A, Gruss A. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol Microbiol. 2000;35:1042–1051. doi: 10.1046/j.1365-2958.2000.01757.x. [DOI] [PubMed] [Google Scholar]
- 34.Raivio T L, Silhavy T J. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- 35.Reed L J, Muench H. A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 36.Sassoon N, Arie J P, Betton J M. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol Microbiol. 1999;33:583–589. doi: 10.1046/j.1365-2958.1999.01505.x. [DOI] [PubMed] [Google Scholar]
- 37.Seol J H, Woo S K, Jung E M, Yoo S J, Lee C S, Kim K J, Tanaka K, Ichihara A, Ha D B, Chung C H. Protease Do is essential for survival of Escherichia coli at high temperatures: its identity with the htrA gene product. Biochem Biophys Res Commun. 1991;176:730–736. doi: 10.1016/s0006-291x(05)80245-1. [DOI] [PubMed] [Google Scholar]
- 38.Skorko-Glonek J, Wawrzynow A, Krzewski K, Kurpierz K, Lipinska B. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene. 1995;163:47–52. doi: 10.1016/0378-1119(95)00406-v. [DOI] [PubMed] [Google Scholar]
- 39.Skorko-Glonek J, Zurawa D, Kuczwara E, Wozniak M, Wypych Z, Lipinska B. The Escherichia coli heat shock protease HtrA participates in defense against oxidative stress. Mol Gen Genet. 1999;262:342–350. doi: 10.1007/s004380051092. [DOI] [PubMed] [Google Scholar]
- 40.Smeds A, Varmanen P, Palva A. Molecular characterization of a stress-inducible gene from Lactobacillus helveticus. J Bacteriol. 1998;180:6148–6153. doi: 10.1128/jb.180.23.6148-6153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 42.Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 43.St. Geme J W, III, Grass S. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol Microbiol. 1998;27:617–630. doi: 10.1046/j.1365-2958.1998.00711.x. [DOI] [PubMed] [Google Scholar]
- 44.Strauch K L, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci. 1988;85:1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tacket C O, Sztein M B, Losonsky G A, Wasserman S S, Nataro J P, Edelman R, Pickard D, Dougan G, Chaatfield S N, Levine M M. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect Immun. 1997;65:452–456. doi: 10.1128/iai.65.2.452-456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ton-That H, Liu G, Mazmanian S K, Faull K F, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ton-That H, Mazmanian S K, Faull K F, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 48.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]