Abstract
Virulence is the outcome of an interaction between the host and a microbe and is characterized by a large array of opposing reactions operating at the host-pathogen interface. Cryptococcus neoformans is an important opportunistic pathogen in immunocompromised patients, including those with human immunodeficiency virus, and expresses a virulence-associated laccase which is believed to oxidize brain catecholamines and iron as a defense against host immune cells. In the present report, we investigated the cellular location of laccase to understand more fully how it contributes to cryptococcal virulence. A monoclonal antibody to the C. neoformans laccase was generated and used to show localization in the cell walls of representative serotype A (H99) and serotype D (B-3501) strains by immunoelectron microscopy. In addition, confocal microscopy was used to show a peripheral location of green fluorescent protein-tagged laccase expressed in live H99 cells. Biochemical studies showed that laccase could be released from intact cells or cell wall fractions with glucanase enzymes but was retained in the cell wall after sequential extraction with 1 M NaCl, 6 M urea, and 1% sodium dodecyl sulfate. The presence of a hydrolyzable bond linking laccase to the cell wall was suggested by removal of laccase from cell wall preparations after they were boiled in 1% sodium dodecyl sulfate, as was the presence of a disulfide or thioester bond by removal with dithiothreitol or β-mercaptoethanol. These data show that laccase is present as a tightly associated cell wall enzyme that is readily accessible for interactions with host immune cells.
Cryptococcus neoformans is a major opportunistic pathogen in immunocompromised hosts and accounts for a significant proportion of AIDS-related infections (28). Three important virulence properties in C. neoformans are its ability to grow at 37°C, requiring the factor calcineurin (27); production of a polysaccharide capsule (4); and expression of the enzyme laccase (14, 37), which forms a melanin-like pigment when grown on substrates containing polyphenolic or polyaminobenzene compounds (5). Recently, additional virulence factors have been proposed, including urease (8), phospholipase (7), and mannitol production (6).
More than 35 years ago, Staib first described in vitro melanin pigmentation by C. neoformans and associated the phenomenon with virulence (31). In spite of considerable efforts by several investigators, many aspects of the nature of laccase-derived products in vivo remain unclear. In vitro, the yeast produces a black melanin pigment after the addition of exogenous catecholamines, a pigment which has been shown to have several immunological properties that are protective for the yeast (35). However, while laccase-derived dopamine products are clearly formed in vivo (21, 24), the exact chemical nature of the product in the host remains to be determined. Dopamine-derived laccase products formed in the brain confer acid stability to the cell wall similar to that conferred by true melanin (23) and react to antibodies made against polymerized melanin (24) but do not have the absorptive properties of a typical melanin polymer which are present in cryptococcal melanin produced in vitro (21). In addition, laccase alone has been demonstrated to confer significant protection against murine alveolar macrophages independent of dopamine by virtue of the enzyme's iron oxidase activity, which appears to diminish the host cell oxidative burst by reducing available FeII stores (20).
Likewise, the cellular localization of laccase is not fully understood. Most information on this matter is from experiments the main purpose of which was to provide soluble enzyme for purification, not to provide localization of the predominant form of the enzyme. For example, solubilization of small amounts of enzyme with detergents has suggested that laccase is a membrane-bound enzyme when cells are grown at neutral pH (29). In contrast, the finding of a minor fraction of soluble enzyme when cells are grown under acidic conditions might suggest that the enzyme has a periplasmic or cytosolic location under some conditions (14, 37). Biochemical and amino acid analysis of laccase shows a hydrophobic 20-amino-acid leader sequence which is proteolytically removed in the mature enzyme as well as four glycosylation sites which are each linked to N-acetyl glucosamine and six terminal mannose residues (37); both properties suggest transport within the secretory pathway. The implications of the cellular location of laccase are important, because its ability to act as an immune modulator at the host-parasite interface could be affected by the enzyme's proximity to the extracellular space. For example, oxidation of low levels of exogenous brain catecholamines as well as transient host cell FeII species in the phagolysosome would be most effective for an enzyme exposed directly to the reagents at a relatively superficial location and would obviate the need for additional dopamine or iron transport mechanisms.
Because of these questions, we sought to determine the cellular location of laccase in representative serotype A and D cryptococcal strains by electron microscopy and fluorescent microscopy of green fluorescent protein (GFP)-tagged laccase as well as by biochemical methods. These results show that the majority of mature laccase is strongly associated with the cell wall of the yeast, which allows the enzyme to have maximal access to host cell substrates in its role as modulator of immune response in C. neoformans.
MATERIALS AND METHODS
Strains.
C. neoformans strain ATCC 208821 (H99) was a gift of J. Perfect, and C. neoformans strain ATCC 34873 (B-3501) was a gift of K. J. Kwon-Chung. Escherichia coli strain DH10B (Life Technologies, Bethesda, Md.) was the host strain for the recovery of ligated plasmids.
Production of recombinant laccase.
Recombinant laccase was expressed in Pichia pastoris by using expression plasmid pPIC93 as previously described (20). Expressed laccase was purified on diethylaminoethyl-Sepharose (Sigma) and then subjected to gel filtration chromatography with a TosoHaas TSK-Gel G2000SW 7.8- by 300-mm column (Sulpelco, Bellefonte, Pa.). In addition, an N-terminal fragment of laccase was expressed in E. coli by using the pIH902 expression system (New England Biolabs, Beverly, Mass.). A 588-bp fragment of laccase cDNA obtained by PCR with Pfu polymerase (Stratagene, La Jolla, Calif.), plasmid p6 as a template containing laccase cDNA (37), and primers N-term-lacc-S (GCCGCCGAATTCAAGACTGATGAGTCGCCA) and N-term-lacc-A (GCCGCCTCTAGAAGTGGCTAGAGCTGCAATGAT) was endonuclease digested with EcoRI and XbaI and inserted into compatible sites of pIH902. The recombinant maltose-binding protein–laccase fusion protein (MBP-laccase) and the MBP were expressed and purified on amylose-Sepharose according to the manufacturer's directions.
Immunization.
Male BALB/c mice, 18 to 20 weeks old (National Cancer Institute, Rockville, Md.), were injected intraperitoneally with full-length recombinant laccase (15, 25, 35, 45, or 50 μg) in a 1:1 (vol/vol) emulsion of complete Freund's adjuvant (Sigma) and phosphate-buffered saline (PBS). At week 4 after immunization, mice were boosted with 25 μg of laccase in a 1:1 (vol/vol) emulsion of incomplete Freund's adjuvant (Sigma) and PBS. After the immune response was induced, the mice were bled from the retro-orbital plexus, and their sera were analyzed for antibodies to laccase by enzyme-linked immunosorbent assay (ELISA) as described below. The mouse with the highest antibody titers was boosted twice with 50 μg in PBS at a 3-h interval on week 24. After 1 day, the mouse was boosted again with 50 μg of laccase in PBS.
Production of hybridomas.
One day after receiving the last boost, the mouse was sacrificed and the spleen was removed. A single-cell suspension was fused with the myeloma P3xA63Ag8.653 at a ratio of 4:1 in the presence of 50% polyethylene glycol as previously described (10). Fused cells were suspended in hypoxanthine-aminopterin-thymidine (HAT) medium supplemented with Dulbecco's modified Eagle's medium with l-glutamine (Mediatech, Washington, D.C.) containing 20% heat-inactivated fetal bovine serum (FBS) (Harlan Bioproducts for Science, Indianapolis, Ind.), 10% NCTC-109 medium (Life Technologies, GIBCO BRL, Grand Island, N.Y.), 2% HAT (Sigma), 1% nonessential amino acids (Life Technologies, GIBCO BRL), and 1% penicillin-streptomycin (Life Technologies, GIBCO BRL); a suspension of 3 × 105 cells/ml was seeded in 20 96-well tissue culture-treated plates (Becton Dickinson) and incubated in a 10% CO2 incubator at 37°C. Hybridomas were fed with complete HAT medium and 10% Opti-Clone Hybridoma Cloning Factor (ICN Biochemicals). Supernatants were screened for the presence of monoclonal antibodies (MAbs) to laccase by ELISA. Hybridomas secreting MAbs of interest were subcloned twice in soft agarose. Single cells were selected, screened, and expanded. Complete HAT medium was substituted by complete hypoxantine-thymidine medium with 10% Opti-Clone Hybridoma Cloning Factor (ICN Biochemicals). The concentrations of hypoxantine-thymidine medium, FBS, and the Opti-Clone Hybridoma Cloning Factor were reduced gradually. Selected hybridomas were grown in medium supplemented with Dulbecco's modified Eagle's medium with l-glutamine (Mediatech) containing 10% FBS (Harlan Bioproducts for Science), 10% NCTC-109 (Life Technologies, GIBCO BRL), 1% nonessential amino acids (Life Technologies, GIBCO BRL), and 1% penicillin-streptomycin (Life Technologies, GIBCO BRL). MAbs present in the supernatants were concentrated and treated with sodium azide at a final concentration of 1 mM.
ELISA.
Polystyrene 96-well ELISA plates (Corning Glass Works, Corning, N.Y.) were incubated with laccase (1 μg/ml), MBP-laccase (1 μg/ml), 1% bovine serum albumin (BSA), or MBP (1 μg/ml) for 1 h at 37 or 4°C overnight. The plates were blocked with 200 μl of 1% BSA for 2 h. Each of the incubations was followed by three washes with 0.1% Tween 20 in Tris-buffered saline. The wells were first treated with 50 μl of hybridoma culture supernatant and incubated for 1 h at 37°C. After the wells were washed, 50 μl of a 1:1,000 alkaline phosphatase-conjugated goat anti-mouse total immunoglobulin G (IgG) (H+L) (Southern Biotechnologies, Inc., Birmingham, Ala.) was added to the wells and incubated for 1 h at 37°C. Detection of bound antibodies was determined by addition of p-nitrophenyl phosphate (Sigma). After 1 h, the absorbance was measured at 405 nm with a Ceres 900 HDi EIA Workstation (Bio-Tek Instruments, Inc., Winooski, Vt.). The isotype of each hybridoma was determined with alkaline phosphatase-conjugated antibodies specific for IgG1, IgG2a, IgG2b, and IgG3.
Electron microscopy.
C. neoformans strains H99 and B-3501 were grown on YP-glycerol agar (2% glycerol, 2% peptone, 1% yeast extract), washed twice in distilled water, then transferred to modified asparagine agar without glucose (1-g/liter asparagine, 10 mM sodium phosphate, [pH 6.5], 0.1-g/liter MgSO4, 50 μM CaCl2), and incubated for 2 days at 25°C to express laccase. Yeast cells were washed three times in sodium phosphate, pH 7.2, and then fixed in 4% (vol/vol) paraformaldehyde and 0.05% (vol/vol) glutaraldehyde in 100 mM sodium phosphate buffer, pH 7.2, for 16 h at 4°C. Cells were then washed three times for 10 min per wash in phosphate buffer and then dehydrated for 45 min in a graded series of ethanol concentrations (15, 30, 50, 75, and 100% [vol/vol]), with two changes each. Infiltration was continued with 2 parts of ethanol to 1 part of LR White resin and then 1:1 and 1:2 ratios of ethanol and LR White resin each for 2 days. Pure LR White resin infiltration was completed over 24 h with three changes, and the samples were then polymerized in 1 ml of pure LR White resin in a vacuum oven at 50°C for 3 days. Cured blocks were trimmed and thin sectioned with a diamond knife on a Riechert Ultra Cut E ultramicrotome (Leica, Inc., Deerfield, Ill.), and the sections were picked up on 200-hex-mesh Ni grids. The thin sections on the Ni grids were incubated for 45 min at room temperature in blocking solution (0.8% [wt/vol] BSA, 0.1% [wt/vol] immunogold-silver stain-quality gelatin, 5% [wt/vol] normal goat serum in PBS [pH 7.4]); washed twice in 0.8% (wt/vol) BSA, 0.1% (wt/vol) gelatin, and 0.025% (vol/vol) Tween 20 in PBS (pH 7.4); and incubated overnight with MAb clone G3P4D3 at a concentration of 2.6 μg/ml in 0.8% (wt/vol) BSA, 0.1% (wt/vol) gelatin, and 1% (vol/vol) normal goat serum in PBS. Then, the sections were washed twice in washing solution and incubated for 4 h with immunogold-labeled secondary antibody (goat anti-mouse IgG; Amersham) diluted 1:25. Grids were washed six times with the washing solution for 5 min per wash and then twice with PBS, and the samples were fixed for 10 min with 2% (vol/vol) glutaraldehyde in PBS. Grids were then washed twice in PBS and three times in distilled water, dried, stained with 2% (wt/vol) aqueous uranyl acetate for 5 min, dried, stained with lead citrate for 2 min, and photographed with a JEOL 1200 EX transmission electron microscope. Seven randomly selected cells were selected for each of the four incubation conditions; gold particles were localized by morphological criteria and counted. Cross-sectional areas of cell walls and cell interiors were assessed by weighing traced-out photographs and comparing them to equivalently traced standard areas. Values were expressed as the mean of seven values ± standard deviation.
Fluorescent microscopy of GFP-tagged laccase.
A CNLAC1-GFP fusion protein expression plasmid was constructed as follows: an HgR-actin fusion gene (a gift of J. Perfect) was inserted into the XhoI site of plasmid p5.1, which contains a 6-kb genomic fragment of CNLAC1 (37), and an AvrII restriction site was introduced into CNLAC1 within the open reading frame (ORF) in a position downstream of the CNLAC1 leader sequence by using divergent PCR and Extend Mix (Stratagene) and the primers GCCGCCCCTAGGATATCGAAGGTATACTCTCT and GCCGCCCCTAGGGCTTTCGCCAGCCCTGAT, followed by endonuclease digestion with AvrII, religation, transformation, and recovery in E. coli. The GFP ORF of pFRED25 (a generous gift of A. Stauber) (32) was amplified by means of PCR using the primers GCCGCCCCTAGGTAGCAAAGGAGAAGAACTCTT and GCCGCCCCTAGGGTTGTACAGTTCATCCATGC, followed by digestion with AvrII and ligation into a compatible site within the CNLAC1 ORF to produce pGFP-CNLAC. Sequence of the GFP-tagged laccase construct pGFP-CNLAC was verified by automated sequencing (CRC-DNA Sequencing Facility, Chicago, Ill.). pGFP-CNLAC was electroporated into H99 cells and selected on hygromycin-containing media as previously described (9). Two transformants were selected and shown by Southern blotting to have the construct present as an episome. The transformants were grown on yeast-peptone-dextrose (YPD) agar for 2 days and derepressed for laccase activity by 2 days of growth on asparagine agar, without glucose (pH 7.0), as previously described (20) and examined for epifluorescence after being embedded in soft agarose as previously described (2). Microscopy was performed with a Zeiss 510 laser confocal microscope and transformants were observed after excitation with 450 to 490 nm of light with a 520-nm cutoff filter. Cells were also observed at other wavelengths to monitor background autofluorescence.
Biochemical localization studies of cryptococcal laccase.
Cells were derepressed for laccase production in the same way as for electron microscopy, and cell walls were prepared with a protocol similar to to that of Kollar et al. (16). Briefly, cells were broken by agitation with 0.45-μm-diometer glass beads for a total of 2 min with a Braun homogenizer (B. Braun Biotech, Allentown, Pa.). Microscopic observation of broken cells revealed only cellular debris with few intact cells. Cell suspensions were assayed for laccase activity, carbohydrate, and protein; cell components were then subjected to centrifugation at 16,000 × g for 30 min. Fractured cell pellets were washed three times with 10 mM sodium phosphate buffer, pH 6.5, followed by sequential extraction with 1 M NaCl, 6 M urea, and 1% sodium dodecyl sulfate (SDS) in 10 mM sodium phosphate, pH 6.5. Salt-urea-SDS-extracted fractured cell pellets were then washed twice in distilled water and once with 10 mM sodium citrate, pH 5.8, and digested with 40 mg of glucanase per ml from Trichoderma harzianum (Sigma) at 37°C for 2 h. Digested cell pellets were then subjected to microcentrifugation (16,000 × g for 30 min) or ultracentrifugation (100,000 × g for 4 h). Alternatively, salt-urea-SDS-extracted fractured cell pellets were incubated with either 30 mM sodium hydroxide, 100 mM dithiothreitol (DTT), or 2% β-mercaptoethanol at 37°C for 30 min. Laccase enzyme was assayed with epinephrine as previously described (1 U of enzyme was defined as 0.001 A475 of activity in 30 min at 37°C and was expressed as the mean of three determinations ± the standard deviation) (37). Carbohydrate was assayed by a phenol-sulfuric acid method (11) and was expressed as glucose equivalents. Protein was determined by a Bio-Rad (Hercules, Calif.) assay. Western blotting was performed using an SDS–10% polyacrylamide gel electrophoresis (PAGE) gel, anti-laccase MAb clone G3P4D3 at 2.6 μg/ml, and horseradish peroxide-labeled anti-mouse antibody at a dilution of 1:2,500 (Sigma) and was visualized with Pico luminescent developer (Pierce, Rockford, Ill.). according to the manufacturer's directions.
RESULTS
Production of anti-laccase MAbs.
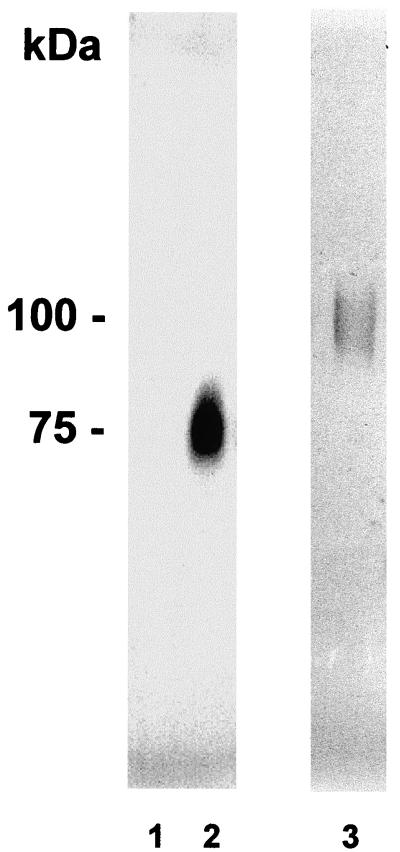
To localize laccase in C. neoformans cells, we generated MAbs from splenocytes of mice immunized with recombinant enzyme expressed in P. pastoris. Hybridoma clones were screened by ELISA. Two hybridomas were recovered producing a MAb (IgG1) to laccase, of which one (G3P4D3) recognized both full-length recombinant P. pastoris-expressed laccase and an E. coli-expressed 196-amino-acid N-terminal fragment. A second clone (G3P2H11) recognized full-length laccase but not the N-terminal fragment (data not shown). Western blotting using the G3P4D3 clone showed no bands when laccase was repressed (Fig. 1, lane 1) and a single band when laccase was derepressed (Fig. 1, lane 2).
FIG. 1.
Western blots of whole-cell suspensions of C. neoformans. Western blot analyses were performed with C. neoformans cells suspensions (20 μg of carbohydrate) from cells either glucose repressed (lane 1) or derepressed in the absence of glucose (lane 2) or 100 ng of recombinant laccase (lane 3) and blots were developed with the G3P4D3 clone MAb.
Localization of cryptococcal laccase by immunoelectron microscopy.
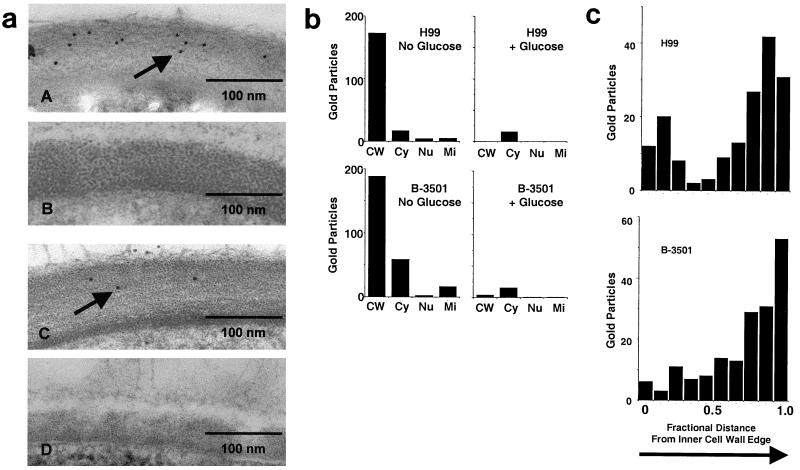
A representative serotype A strain of C. neoformans (H99; ATCC 208821) and a serotype D strain (B-3501; ATCC 34873) were derepressed for laccase production by incubation in the absence of glucose and processed for immunoelectron microscopy. As shown in Fig. 2aA, multiple 5-μm-diameter gold particles were observed in the cell walls of derepressed cells of H99, but not in glucose-repressed H99 cells (Fig. 2aB). Previous reports have shown laccase transcription and enzyme activity to be completely repressed by glucose (37). Gold particles were also found to stain cell walls of derepressed cells of serotype D cells (Fig. 2aC) but not glucose-repressed cells (Fig. 2aD). As shown in Fig. 2b, sampling seven random cryptococcal cells from each of the four incubation conditions showed a predominance of gold particles in the cell walls of derepressed H99 and B-3501 cells (H99, 173 particles; B-3501, 189 particles) versus only a few particles in the cytoplasm (H99, 17; B-3501, 59) and less in other structures such as the nucleus (H99, 4; B-3501, 3) or mitochondria (H99, 3; B-3501, 17). Glucose-repressed cells showed only small amounts of binding throughout the cell with no one organelle predominating. In addition, no gold particle staining was evident with any of the cell preparations after incubation with gold-labeled secondary antibody alone (data not shown). Adjusting particle counts for a cross-sectional area of cell organelles in each of the four groups of seven cells yielded a mean particle density of 22.9 ± 7.5 particles/pm2 in the cell walls of H99 cells and 2.2 ± 1.3 particles/pm2 in the interior of H99, and 17.4 ± 8.8 particles/pm2 in B-3501 cell walls and 1.9 ± 1.3 particles/pm2 in the interior (P < 0.005 between cell wall and interior particle densities in each case). These data indicate that laccase is localized to the cell wall under these growth conditions. In addition, the fractional distance from the inner edge of the cell wall to the outer edge was measured for each cell wall gold particle from each of the seven derepressed cells of H99 and B-3501, and a histogram was constructed using 0.1 fractional intervals. As shown in Fig 2C, laccase appeared to be predominantly located at the outer cell wall, indicated by the greater quantities of gold particles in the outer regions. This outer cell wall distribution of laccase is the first data showing polarity of the cell wall of C. neoformans.
FIG. 2.
Immuno electron microscopy of cell walls of C. neoformans. (a) H99 (A and B) or B-3501 (C and D) cells that expressed laccase (A and C) or were glucose repressed (B and D) were subjected to immunoelectron microscopy with laccase MAbs. Arrows point to 5-μm gold particles. (b) Gold particles from seven random cells of each group were counted and localized to the cell wall (CW), cytoplasm (Cy), nucleus (Nu), or mitochondria (Mi). The fractional distance from the inner edge of the cell wall to the outer edge was measured for each cell wall gold particle from seven cells of H99 and B-3501, and a histogram was constructed using 0.1 fractional intervals (the inner wall edge was designated as 0 distance).
Localization of cryptococcal laccase by epifluorescence of GFP-tagged laccase.
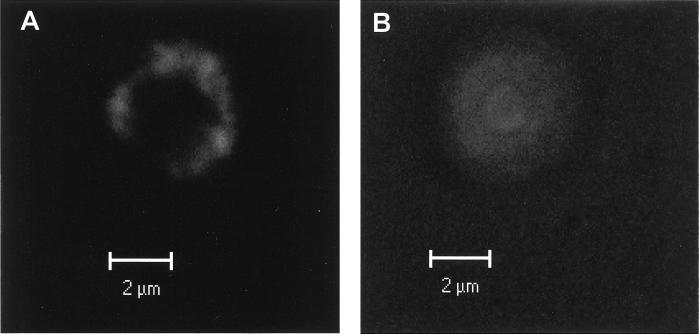
To demonstrate the localization of laccase in live cells of C. neoformans, a GFP (32) was used to tag recombinant laccase expressed in C. neoformans strain H99. GFP was inserted into the ORF of laccase in the N-terminal region downstream from the leader sequence cleavage site, such that it would not interfere with cellular trafficking. Two laccase-GFP transformants and wild-type H99 colonies were selected, grown on YPD agar for 2 days, derepressed for laccase activity by a 2-day incubation on asparagine agar without glucose (pH 7.0) as previously described (20), and examined for epifluorescence. Microscopy was performed with a Zeiss 510 laser confocal microscope, and cells were observed after excitation with 450 to 490 nm of light and with a 520-nm cutoff filter. As shown in Fig. 3A, transformation of H99 with the laccase-GFP fusion protein yielded a recombinant laccase showing a strong signal that localized to the cell periphery. In contrast, untransformed wild-type H99 showed no specific cellular localization and only weak autofluorescence at these wavelengths that required a high-gain setting to visualize. In addition, this low-level autofluorescence was evident at numerous wavelengths such as excitation at 568 nm and emission 610 nm, whereas the fluorescence shown in Fig. 3A was evident only at specific GFP-related wavelengths. While the limited resolution of light microscopy does not allow a differentiation between a cell wall and a membrane localization, these data provide independent confirmation of a peripheral localization of laccase of C. neoformans.
FIG. 3.
Localization of GFP-tagged laccase by epifluorescence. GFP-laccase transformants of H99 (A) and wild-type H99 (B) were derepressed in the absence of glucose and examined for epifluorescence of GFP with a Zeiss 510 laser confocal microscope.
Cellular fractionation and detection of laccase enzyme activity.
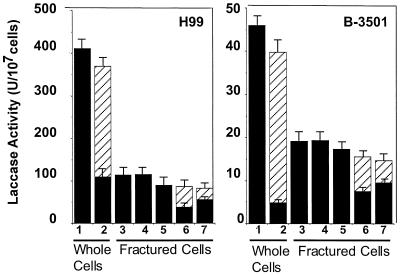
To corroborate the above data indicating a cell wall location of laccase, cryptococcal cells expressing laccase were fractured to determine if enzyme activity was localized to the cell wall fraction. Laccase was derepressed in representative serotype A (H99) and serotype D (B-3501) cells of Cryptococcus by incubation of freshly grown cells on asparagine agar plates in the absence of glucose. Whole-cell assays of laccase from washed cells showed significant levels of enzyme activity (H99, 429 ± 33 U/107 cells; B-3501, 46 ± 4 U/107 cells) (Fig. 4, lane 1). Digestion of intact cells with a glucanase preparation from the fungus T. harzianum widely used to digest cryptococcal cell walls (34) results in the solubilization of significant amounts of laccase activity (H99, 260 ± 20 U/107 cells; B-3501, 35 ± 4 U/107 cells) (Fig. 4, lanes 1 and 2). The glucanase preparation was not found to contain significant laccase activity at the concentrations used. Trypan blue staining of digested cells showed <5% uptake by intact cells, suggesting that the enzyme was removed from the cell wall without disruption of the cell membrane. Incomplete removal of laccase was most likely due to incomplete digestion of the cell wall typical of stationary-phase cells, indicated by retained osmotic stability of the cells in 1% SDS (data not shown). To study the nature of the laccase-cell wall attachment, laccase-expressing cryptococcal cells were fractured with a Braun homogenizer. A degradative carbohydrate assay was used to estimate the number of disintegrated cells contained in a given preparation by reference to the carbohydrate content of intact cells counted by hemocytometer (for H99, 1 μg of carbohydrate = 1.24 × 105 cells; for B-3501, 1 μg of carbohydrate = 1.1 × 105 cells). Disintegration of cryptococcal cells resulted in a significant loss of laccase activity (H99, 114 ± 6 U/107 cells; B-3501, 19 ± 1 U/107 cells) versus that in whole cells, presumably due to the vigorous shaking required to break the stationary-phase cryptococcal cell walls. After centrifugation, essentially all laccase activity was found in the pellet (H99, 115 ± 4 U/107 cells; B-3501, 19 ± 1 U/107 cells) (Fig. 4, lane 4), and no enzyme activity could be detected in the supernatant. After extraction of the disintegrated cell pellet sequentially with 1 M NaCl, 6 M urea, and 1% SDS, virtually all of the laccase activity in the disintegrated cells was found in the cell pellet (H99, 89 ± 4 U/107 cells; B-3501, 17.3 ± 0.6 U/107 cells) (Fig. 4, lane 5), suggesting a very strong association with the carbohydrate cell wall. Again, digestion of the disrupted cell wall pellet with glucanase resulted in solubilization of significant amounts of laccase activity evident in the supernatant after microfugation (H99, 43 ± 1 U/107 cells; B-3501, 7.9 ± 0.2 U/107 cells) (Fig. 4, lane 6) or ultracentrifugation (H99, 23 ± 1 U/107 cells; B-3501, 5.2 ± 0.2 U/107 cells) (Fig. 4, lane 7), suggesting that the laccase-cell wall attachment is dependent on intact cell wall carbohydrate.
FIG. 4.
Enzymatic activity of laccase in C. neoformans. H99 and B-3501 cells were laccase derepressed in the absence of glucose, and the enzyme activity of the whole cells was measured before (lane 1) and after (lane 2) digestion with glucanase (lane 2). After disintegration of cells, total extract was measured for laccase activity (lane 3) as well as laccase activity in the pellet after centrifugation (lane 4). Fractured cells were extracted sequentially with 1 M NaCl, 6 M urea, and 1% SDS, and the enzyme in the pellet was assayed (lane 5). Fractured cell pellets were then subjected to digestion by glucanase (lanes 6 and 7) and laccase was assayed after microfugation (lane 6) and ultracentrifugation ▨ (lane 7). Enzyme activity was measured in the pellet (■) and/or the supernatant (▨) as indicated.
Western blots of laccase from cellular fractions of C. neoformans.
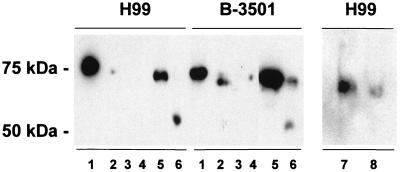
To investigate the nature of the laccase-cell wall association in more detail, Western blot analysis with a MAb to laccase was performed to assess the location of laccase after exposure of the cell wall to conditions that result in loss of laccase activity. As shown in Fig. 5, lane 1, boiling of disintegrated cells in 1% SDS (without reducing agents) resulted in solubilization of laccase from the cell wall, as evidenced by the ability of the protein to migrate into the SDS-PAGE gel as a 75-kDa glycosylated protein similar to that of the purified enzyme (37). While the supernatants of fractured cells did not contain laccase activity, small amounts of laccase were detectable in these fractions by Western blot analysis (Fig. 5, lane 2) and may represent a cytosolic laccase precursor. Without being boiled, laccase in the fractured cell pellet that had been washed in buffer (Fig. 5, lane 3) or from fractured cell pellets extracted with 1 M NaCl, 6 M urea, and 1% SDS did not migrate into the SDS-PAGE gel after exposure to sample buffer containing 1% SDS (lane 4), similar to experiments described above which showed that enzyme activity was not extractable with these room temperature extraction procedures. However, subjecting the salt-urea-SDS-extracted cell wall preparation to boiling in 1% SDS before SDS-PAGE solubilized the laccase, suggested an easily hydrolyzable cell wall attachment (Fig. 5, lane 6). Since numerous covalently attached cell wall proteins in yeast are extractable with sodium hydroxide (22), these conditions were used to test if laccase could be extracted under these conditions. As shown in Fig. 5, lane 7, laccase was solubilized with dilute sodium hydroxide, which also resulted in limited proteolysis of the protein. Further investigation of the laccase cell wall attachment in H99 cells that showed that laccase could also be solubilized with either room temperature DTT or β-mercaptoethanol (Fig. 5, lanes 7 and 8), suggesting a role for a disulfide or thioester bond in the laccase-cell wall anchor.
FIG. 5.
SDS-PAGE Western blot of laccase of cellular fractions of C. neoformans. Laccase-containing cryptococcal cell suspension from either H99 or B-3501 cells were either boiled in 1% SDS (lane 1) or centrifuged, and the supernatant (lane 2) and the pellet (lane 3) were separately subjected to electrophoresis without boiling. Alternatively, centrifuged fractured cell pellets were extracted sequentially with 1 M NaCl, 6 M urea, and 1% SDS and either subjected to electrophoresis without boiling (lane 4) or with boiling in 1% SDS (lane 5). Salt-urea-SDS-extracted fractured cell pellets were additionally treated with 30 mM NaOH at 37°C for 30 min (lane 6) or 100 mM DTT (lane 7) or 2% β-mercaptoethanol (lane 8) and then electrophoresed without boiling. Each lane contained approximately 20 μg of carbohydrate; in addition, lanes 1 and 2 contained 10 μg of protein.
DISCUSSION
In the present study, a MAb to C. neoformans laccase was prepared and used to localize the enzyme to the cell wall of the yeast in representative serotype A (H99) and D (B-3501) strains by immunoelectron microscopy. The cell wall is the same location where melanin has been localized in cells grown in vitro (36). An interesting finding was that laccase also appeared to localize predominately to the outer region of the cell wall, which suggests polarity in the cryptococcal cell wall. Cells grown in glucose did not show significant immunoreactivity and were used as negative controls, since laccase is transcriptionally repressed in the presence of glucose (37). In addition, a GFP-tagged laccase construct was also found to express fluorescent protein that localized to the periphery in live cells. In addition, recovery of laccase activity in cell wall preparations after extraction with 1 M NaCl, 6 M urea, and 1% SDS suggests that laccase is not predominately a cytosolic or membrane-bound protein in its mature form. Solubilization of laccase activity from intact cells with glucanase enzymes and from SDS-extracted fractured cell pellets further supports a cell wall-associated laccase.
Determining the location of laccase expression is important for understanding the mechanism by which this enzyme contributes to virulence. Yeast cell walls typically allow small molecules to diffuse freely through the lattice of the cell wall (25), and the C. neoformans capsule appears permeable even to large molecules such as immunoglobulins (17, 30). Thus, a cell wall localization indicates that laccase is in a position outside the plasma membrane where it can interact more directly with extracellular substances and host immune cell products without the need for ancillary membrane or cytosolic transporters. For example, oxidation of catecholamines on the exterior portion of the cell wall lessens the exposure to oxidized dopamine products, which have been shown to be potently cytotoxic in other systems (33). An extracellular location also removes the need for dopamine transporters into and out of the cell. In addition, production of catecholamine oxidation products in the extracellular space (rather than in a specialized structure such as a melanosome in mammalian cells) may explain why formation of cell wall melanin in vitro requires very high concentrations of dopamine (100 mg/liter; [36, 38]), which may not be optimal in the brain where lower concentrations of dopamine are found (1 to 7 mg/liter [13]). Indeed, the chemical identity of the catecholamine oxidation condensation products in vivo has been a matter of great controversy because of their unusual properties (21, 23, 24). Besides the oxidative effects on catecholamines, laccase also has been proposed to exert a fungal protective effect from host macrophages by virtue of its newly described iron oxidase activity (20). Reduced FeII is required by macrophages to produce toxic oxygen metabolites, and the oxidation of iron by laccase thus competes with the host cell oxidation machinery. A cell wall laccase thus allows greater access to transient iron species, thereby maximizing its effect against host effector cells. It is important to note that cells in the present study were grown at approximately neutral pH (6.5 to 7.0). Previous reports by laboratories including our own have noted a minor and variable amount of soluble laccase in addition to an insoluble component, but these reports used cells derepressed at a very acidic pH of approximately 4.5 (14, 37). This acidic pH is similar to that found in the phagolysosome of macrophages after ingestion of either live or heat-killed C. neoformans (19). Thus, the ability to alter laccase cellular properties by pH effects could be utilized by immunocompetent macrophages to alter laccase-associated properties which could, in turn, reduce virulence.
A cell wall location for laccase may also facilitate the survival of the organism in the environment. Recently, C. neoformans var. neoformans has been found to inhabit the hollows of living trees (18). Laccase is produced by many of the white-rot basidiomycete fungi such as Coriolus versicolor that inhabit the same environment and is thought to serve in the degradation of lignin in wood pulp (15). Since lignin polymers are large molecules that would have great difficulty traversing a cell membrane, a peripheral location of laccase may facilitate degradation of these polymers. Since these molecules can be quite large, location in the outer portion of the cell wall in C. neoformans may be critical to efficient degradation of these polymers. An additional benefit to placing laccase in the outer cell wall may be to lessen possible toxic effects of the iron oxidase activity of laccase. In yeast, reduced iron is the substrate of the iron transporter Fet3p, which is provided by an ancillary plasma membrane iron reductase (1). Indeed, iron reductase activity has also been demonstrated in C. neoformans (26). Placing laccase in the outer cell wall rather than in the plasma membrane may lessen competition of laccase with the plasma membrane iron reductase and therefore interfere less with iron uptake in the yeast.
The yeast cell wall is a highly complex structure whose major components are β-1,3-and β-1,6-glucans with a number of mannoproteins attached to the glucan network (for a review, see reference 12). Cell wall proteins may be loosely associated, such as the WI-1 protein of Blastomyces dermatitidis, which is extracted by washing with distilled H2O (3). A second group of cell wall proteins are resistant to extraction with boiling SDS and can be extracted only with glucanase preparations. All proteins described thus far of the second group contain glycosylphosphatidylinositol (GPI)-anchoring signals at their C termini and are covalently linked to the glucan wall through a remnant of a GPI anchor containing five α-linked mannosyl residues (16). A third group of proteins appears more heterogeneous; they can be released from the cell wall by hot SDS, β-mercaptoethanol, or by dilute sodium hydroxide and are exemplified by the Pir proteins of Saccharomyces, but the structure of attachment has not been described (22). The laccase of C. neoformans appears to fall into the third category of cell wall proteins. This enzyme was resistant to solubilization by 1 M NaCl, 6 M urea, or 1% SDS at room temperature, which disrupts ionic, hydrophobic, and membrane-associated interactions. It is doubtful the laccase-cell wall linkage is dependent on a GPI anchor, since there are no hydrophobic regions within the C terminus of the laccase ORF to which a GPI anchor would be attached (37). However, boiling cells in SDS results in the removal of enzyme and suggests the involvement of an easily hydrolyzable covalent bond. The size of the SDS-solubilized enzyme (75 kDa) corresponds to that of a highly glycosylated soluble form of the purified enzyme isolated as a minor fraction from cells grown at low pH and suggests that the cell wall-bound enzyme is similarly glycosylated. A role for a disulfide or perhaps a thioester linkage is also suggested by the ability of room temperature DTT or β-mercaptoethanol to solubilize cell wall-associated laccase. Further studies are currently underway to chemically characterize the laccase-cell wall anchor.
ACKNOWLEDGMENTS
J.G.-R. is supported by NIH award T32-AI07501. A.C. is supported by NIH awards AI33774, AI3342, and HL-59842. P.R.W. is supported by NIH grants AI45995 and AI38258 and a grant from the American Lung Association.
We are grateful to M. L. Chen, who performed the confocal microscopy examinations, and Matthew Scharff and Susan Bulh, for their technical support in the generation of the hybridomas. We are also grateful to E. Cabib, W. Walden, and J. Kaplan for helpful discussions.
REFERENCES
- 1.Askwith C C, deSilva D, Kaplan J. Molecular biology of iron acquisition in Saccharomyces cerevisiae. Mol Microbiol. 1996;20:27–34. doi: 10.1111/j.1365-2958.1996.tb02485.x. [DOI] [PubMed] [Google Scholar]
- 2.Banta L M, Robinson J S, Klionsky D J, Emr S D. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandhorst T, Klein B. Cell wall biogenesis of Blastomyces dermatitilidis: evidence for a novel mechanism of cell surface localization of a virulence-associated adhesin via extracellular release and reassociation with cell wall chitin. J Biol Chem. 2000;275:7925–7934. doi: 10.1074/jbc.275.11.7925. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y C, Kwon-Chung K J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chasakes S, Tyndall R L. Pigment production by Cryptococcus neoformans from para- and ortho-di-phenols: effect of the nitrogen source. J Clin Microbiol. 1975;1:509–514. doi: 10.1128/jcm.1.6.509-514.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi V, Flynn T, Niehaus W G, Wong B. Stress tolerance and pathogenic potential of a mannitol mutant of Cryptococcus neoformans. Microbiology. 1996;142:937–943. doi: 10.1099/00221287-142-4-937. [DOI] [PubMed] [Google Scholar]
- 7.Chen S C, Muller M, Zhou J Z, Wright L C, Sorrell T C. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J Infect Dis. 1997;175:414–420. doi: 10.1093/infdis/175.2.414. [DOI] [PubMed] [Google Scholar]
- 8.Cox G M, Mukherjee J, Cole G T, Casadevall A, Perfect J R. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox G M, Toffaletti D L, Perfect J R. Dominant selection system for use in Cryptococcus neoformans. J Med Vet Mycol. 1996;34:385–391. [PubMed] [Google Scholar]
- 10.de StGroth S F, Scheidegger D. Production of monoclonal antibodies: strategy and tactics. J Immunol Methods. 1980;35:1–21. doi: 10.1016/0022-1759(80)90146-5. [DOI] [PubMed] [Google Scholar]
- 11.Dubois M, Giles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–358. [Google Scholar]
- 12.Fleet G H. Composition and structure of yeast cell walls. Curr Top Med Mycol. 1985;1:24–56. doi: 10.1007/978-1-4613-9547-8_2. [DOI] [PubMed] [Google Scholar]
- 13.Hadesman R, Wiesner R H, Go V L W, Tyce G M. Concentrations of 3,4-dihydroxyphenylalanine and catecholamines and metabolites in brain in an anhepatic model of hepatic encephalopathy. J Neurochem. 1995;65:1166–1175. doi: 10.1046/j.1471-4159.1995.65031166.x. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda R, Shinoda T, Morita T, Jacobson E S. Characterization of a phenol oxidase from Cryptococcus neoformans var. neoformans. Microbiol Immunol. 1993;37:759–764. doi: 10.1111/j.1348-0421.1993.tb01702.x. [DOI] [PubMed] [Google Scholar]
- 15.Kawai S, Umezawa T, Higuchi T. Degradation mechanisms of phenolic beta-1 lignin substructure model compounds by laccase of Coriolus versicolor. Arch Biochem Biophys. 1988;262:99–110. doi: 10.1016/0003-9861(88)90172-5. [DOI] [PubMed] [Google Scholar]
- 16.Kollar R, Reinhold B B, Petrakova E, Yeh H J C, Ashwell G, Drgonova J, Kapteyn J C, Klis F M, Cabib E. Architecture of the yeast cell wall. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 17.Kozel T R, Pfrommer G S, Guerlain A S, Highison B A, Highison G J. Role of the capsule in phagocytosis of Cryptococcus neoformans. Rev Infect Dis. 1988;10(Suppl. 2):S436–S439. doi: 10.1093/cid/10.supplement_2.s436. [DOI] [PubMed] [Google Scholar]
- 18.Lazera M S, Salmito Cavalcanti M A, Londero A T, Trilles L, Nishikawa M M, Wanke B. Possible primary ecological niche of Cryptococcus neoformans. Med Mycol. 2000;38:379–383. doi: 10.1080/mmy.38.5.379.383. [DOI] [PubMed] [Google Scholar]
- 19.Levitz S M, Nong S H, Seetoo K F, Harrison T S, Speizer R A, Simons E R. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect Immun. 1999;67:885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Tewari R P, Williamson P R. Laccase protects Cryptococcus neoformans from antifungal activity of alveolar macrophages. Infect Immun. 1999;67:6034–6039. doi: 10.1128/iai.67.11.6034-6039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Wakamatsu K, Ito S, Williamson P R. Catecholamine oxidative products, but not melanin, are produced by Cryptococcus neoformans during neuropathogenesis in mice. Infect Immun. 1999;67:108–112. doi: 10.1128/iai.67.1.108-112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mrsa V, Tanner W. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of the Pir protein family) in stability of the Saccharomyces cerevisiae cell wall. Yeast. 1999;15:813–820. doi: 10.1002/(SICI)1097-0061(199907)15:10A<813::AID-YEA421>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Nosanchuk J D, Rosas A L, Lee S C, Casadevall A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet. 2000;355:2049–2050. doi: 10.1016/S0140-6736(00)02356-4. [DOI] [PubMed] [Google Scholar]
- 24.Nosanchuk J D, Valadon P, Feldmesser M, Casadevall A. Melanization of Cryptococcus neoformans in murine infection. Mol Cell Biol. 1999;19:745–750. doi: 10.1128/mcb.19.1.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novick P, Osmond B C, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyhus K J, Wilborn A T, Jacobson E S. Ferric iron reduction by Cryptococcus neoformans. Infect Immun. 1997;65:434–438. doi: 10.1128/iai.65.2.434-438.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odom A, Muir S, Lim E, Toffaletti D L, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinner R W, Hajjeh R A, Powderly W G. Prospects for preventing cryptococcosis in persons infected with human immunodeficiency virus. Clin Infect Dis. 1995;21(Suppl. 1):S103–S107. doi: 10.1093/clinids/21.supplement_1.s103. [DOI] [PubMed] [Google Scholar]
- 29.Polacheck I, Hearing V J, Kwon-Chung K J. Biochemical studies of phenoloxidase and utilization of catecholamines in Cryptococcus neoformans. J Bacteriol. 1982;150:1212–1220. doi: 10.1128/jb.150.3.1212-1220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosas A L, Nosanchuk J D, Feldmesser M, Cox G M, McDade H C, Casadevall A. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect Immun. 2000;68:2845–2853. doi: 10.1128/iai.68.5.2845-2853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staib F. Cryptococcus neoformans and Guizotia abyssinica (syn. G. oleifera D.C.) (farbreadktion für C. neoformans) Z Hyg. 1962;148:466–475. [Google Scholar]
- 32.Stauber R H, Horie K, Carney P, Hudson E A, Tarasova N I, Gaitanaris G A, Pavlakis G N. Development and applications of enhanced green fluorescent protein mutants. BioTechniques. 1998;24:462–471. doi: 10.2144/98243rr01. [DOI] [PubMed] [Google Scholar]
- 33.Tse D C S, McCreery L R, Adams R N. Potential oxidative pathways of brain catecholamines. J Med Chem. 1976;19:37–41. doi: 10.1021/jm00223a008. [DOI] [PubMed] [Google Scholar]
- 34.Varma A, Kwon-Chung K J. Rapid method to extract DNA from Cryptococcus neoformans. J Clin Microbiol. 1991;29:810–812. doi: 10.1128/jcm.29.4.810-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Aisen P, Casadevall A. Melanin, melanin “ghosts,” and melanin composition in Cryptococcus neoformans. Infect Immun. 1996;64:2420–2424. doi: 10.1128/iai.64.7.2420-2424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson P R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson P R, Wakamatsu K, Ito S. Melanin biosynthesis in Cryptococcus neoformans. J Bacteriol. 1998;180:1570–1572. doi: 10.1128/jb.180.6.1570-1572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]