Abstract
NMR spectroscopy and matrix assisted laser desorption ionization mass spectrometry imaging (MALDI MSI) are both commonly used to detect large numbers of metabolites and lipids in metabolomic and lipidomic studies. We have demonstrated a new workflow, highlighting the benefits of both techniques to obtain metabolomic and lipidomic data, which has realized for the first time the combination of these two complementary and powerful technologies. NMR spectroscopy is frequently used to obtain quantitative metabolite information from cells and tissues. Lipid detection is also possible with NMR spectroscopy, with changes being visible across entire classes of molecules. Meanwhile, MALDI MSI provides relative measures of metabolite and lipid concentrations, mapping spatial information of many specific metabolite and lipid molecules across cells or tissues. We have used these two complementary techniques in combination to obtain metabolomic and lipidomic measurements from triple-negative human breast cancer cells and tumor xenograft models. We have emphasized critical experimental procedures that ensured the success of achieving NMR spectroscopy and MALDI MSI in a combined workflow from the same sample. Our data show that several phospholipid metabolite species were differentially distributed in viable and necrotic regions of breast tumor xenografts. This study emphasizes the power of combined NMR spectroscopy–MALDI imaging to advance metabolomic and lipidomic studies.
Keywords: cancer, imaging, MALDI, mass spectrometry, NMR, spectroscopy
1 |. INTRODUCTION
NMR spectroscopy and mass spectrometry (MS) are two commonly used analytical techniques for the study of metabolomics and lipidomics. Each of these techniques has advantages and disadvantages. Ideally, the goal would be to obtain quantitative and spatial information on as many unique metabolites and lipids as possible. Currently, technology does not exist to achieve this goal in a single experiment. However, with appropriate sample preparation protocols and experimental pipelines, we were able to obtain much of this information by using the powerful combination of NMR spectroscopy and mass spectrometry imaging (MSI).
1.1 |. NMR spectroscopy for metabolomics and lipidomics
Advancements in NMR instrumentation in the early 1980s enabled the emergence of high-resolution (HR) NMR techniques, which eventually resulted in 1H NMR spectroscopy being widely used in metabolomics and lipidomics.1,2 Since then, ex vivo NMR metabolomics has been applied in cancer,3,4 agriculture,5,6 plant biology,7,8 and systems biology,9,10 among others. In NMR metabolomic and lipidomic studies, tissues or cells are typically subjected to extraction protocols to isolate metabolites and lipids prior to NMR spectroscopy measurements.2,11–13
1H NMR spectra of biofluids, i.e., urine and blood plasma, or of extracted cells or tissues can contain NMR signals from hundreds of metabolites at high enough concentrations to be above the detection limit.14–16 Each metabolite typically carries several protons, giving rise to several proton signals per metabolite at different chemical shifts (Figure 1). In addition, through spin–spin coupling, proton signals are frequently split into multiplets. Both of these factors contribute to significant overlap of 1H NMR signals from different metabolites and lipids in 1H NMR spectra of extracted biofluids, cells, and tissues. In instances of significant overlap of metabolite and lipid signals, two-dimensional (2D) NMR spectroscopy pulse sequence experiments may be required to assign all metabolites,17,18 including J-resolved,15,19–21 DQF-COSY,22,23 TOCSY,18,19 and HSQC,18,23,24 among others. The addition of the second spectral dimension allows for definitive identification and improved spectral resolution in metabolomic and lipidomic studies; however, 2D NMR spectroscopy experiments are significantly more time consuming and complex than one-dimensional (1D) experiments. In lipidomic studies, it is possible to use single-pulse 1D 1H NMR spectroscopy experiments because the extracted lipids are dissolved in organic solvents. Water-soluble metabolites, however, require more complex pulse sequences in 1D 1H NMR spectroscopy, including water suppression techniques due to the presence of residual water.19,25,26 For 1D 1H NMR spectroscopy, various studies on biofluids have also explored the use of pulse sequence design with spin echo CPMG pulse trains to perform relaxation edited NMR spectroscopy for improved detection of metabolites.27–30
FIGURE 1.
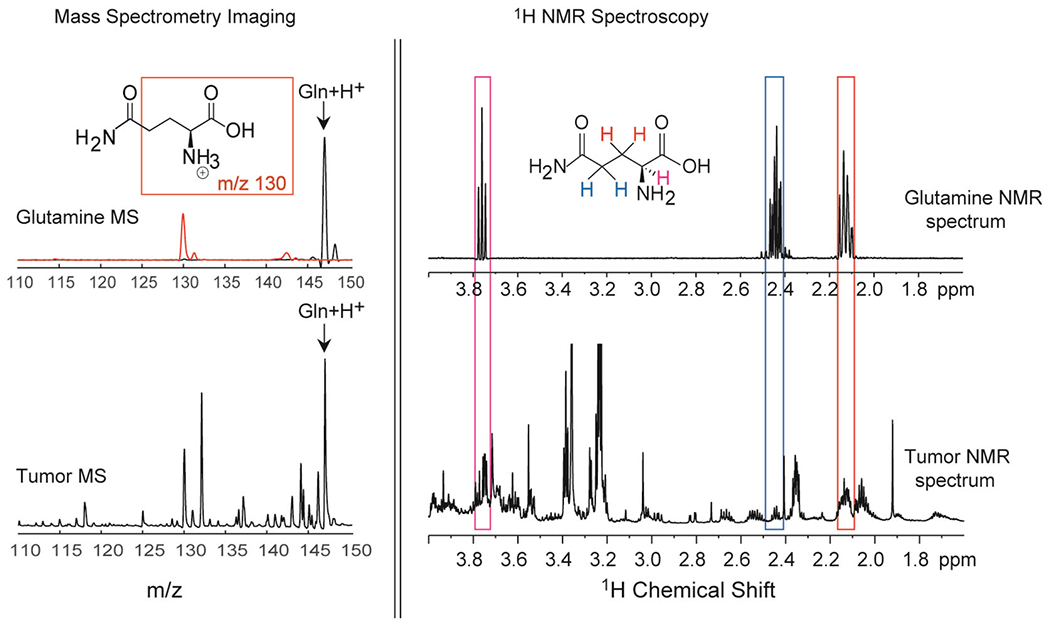
Representative spectra from MALDI imaging and 1H NMR spectroscopy of pure Gln, as well as Gln detected in MDA-MB-231 breast tumor xenografts. MALDI imaging (left) produced spectra based on the m/z of molecular ions, in this case [Gln + H+], which was confirmed using tandem MS (top left, red spectral line). Tandem MS detected the main fragment of Gln at m/z 130. 1H NMR spectroscopy (right) produced a read out of specific protons based on their chemical environments, which are color-coded in red, blue and pink to match boxes of the same colors that outline the corresponding proton NMR signals.
The introduction of high-resolution magic angle spinning (HR-MAS) NMR spectroscopy has opened the possibility of using solid tissues for ex vivo NMR metabolomic studies, allowing for non-destructive NMR measurements. In HR-MAS NMR metabolomics, 10–15 mg of solid tissue is loaded into the NMR spectrometer, measured under magic angle spinning and cooling to 4 °C, which can then be used for further analysis such as histopathology,31 and, perhaps in future studies, MSI. HR-MAS NMR spectroscopy has been explored in a number of studies on various cancer tissues32–38 and gallstones,39 among others. Recent studies have tested HR-MAS coil spinning NMR spectroscopy for the use of smaller tissue samples,40 and combined slice localization with HR-MAS NMR spectroscopy for improved spatial resolution of metabolites.41 While innovations in HR-MAS strive to achieve spatially localized NMR spectroscopic metabolomics, typical sample sizes of 10–15 mg of tissue will inevitably contain heterogeneous regions of various cell types, microenvironments and histologies in the same sample. We therefore set out to test a novel workflow that combines 1H NMR spectroscopy for metabolomics and lipidomics with matrix assisted laser desorption ionization (MALDI) time-of-flight (TOF) MSI.
1.2 |. MALDI-TOF MSI for spatially resolved metabolomics and lipidomics
MS is an analytical technique that measures the mass-to-charge ratio (m/z) of molecules (referred to as analytes),42 thereby using a wide variety of ionization techniques and frequently coupled with liquid or gas chromatography.42,43 MSI detects spatially resolved MS from tissue surfaces. While this study focuses on MALDI imaging, there are review articles summarizing other ionization techniques used for MSI, including SIMS44–46 and DESI,47,48 as well as mass analyzers such as FTICR and ion traps.49 First reported in 1997,50 MALDI-TOF MSI utilizes soft ionization51 to analyze specific molecular classes, including metabolites, lipids, glycans, tryptic or native peptides, and intact proteins, directly from tissue sections. In recent years, fast commercial MALDI-TOF imaging instruments have become available, making MALDI-TOF MSI applications feasible in various biomedical fields.52,53 MALDI-TOF MSI utilizes matrix-coating to assist in the ionization of analytes from the tissue surface.54,55 A UV laser pulse ablates the sample and leads to the desorption of matrix and analytes from the sample surface. These energized molecules then undergo ionization, mainly through proton exchange, and are pulled across an electric field before entering a high-vacuum, field-free flight tube that separates them based on their flight time, which is dependent on their m/z. Based on the time required for the analytes to reach the detector, the m/z of the molecule, including the mass change resulting from ionization, is calculated, and displayed as a spectrum. Since MALDI-TOF produces mostly singly charged species, the m/z is often simplified to directly refer to the molecular mass. For imaging experiments, discrete MALDI-TOF MS spectra are acquired across the tissue by means of a moving stage, where each mass spectrum is encoded with its corresponding x- and y-coordinates, which can then be displayed as relative heat maps, i.e., one image per m/z value.54 MALDI-TOF MSI is currently used across a multitude of fields including plant biology,56–58 clinical and translational research,54,59,60 glycomics,61,62 cancer metabolism,63–65 and microbiology.66,67
Polar lipids are well detected by MALDI-TOF MSI, and lipid imaging and structural lipid identification is possible directly from tissue.68 The lipid region of the mass spectrum between m/z 400 and 1200 is densely populated by both negative (cardiolipins, gangliosides, phosphatidylinositols, and sulfatides, among others) and positive (phosphatidylcholines (PtdChos or PCs), phosphatidylethanolamines (PtdEs or PEs), sphingolipids, and ceramides, among others) ion mode.69 However, not all lipid classes ionize equally well, and multiple different lipid species can overlap within the same peak, also depending on the mass resolution of the MALDI-TOF MSI instrument used.68,69 Lipids generally fragment into their head group and fatty acids for relatively straightforward identification by tandem MS.70 Unambiguous structural identification of the double bond positions in fatty acids is also possible but requires specialized approaches.71
Metabolite imaging can be challenging with MALDI-TOF MSI due to the wide range of chemical structures, stabilities, and ionization efficiencies, also owing to their low molecular weight.72,73 Some metabolites are stable and ionize relatively well.74,75 High-energy metabolites, including adenosine triphosphate (ATP), can break down during sample preparation or during the experiment.76 Neurotransmitters including acetylcholine and serotonin are difficult to ionize; however, novel nanoparticle-based ionization approaches77 and derivatization agents78 have been developed to overcome this barrier. An important factor to consider in MALDI-TOF MSI is matrix overlap. Most matrices used for MALDI imaging are small molecules that produce a significant amount of signal in the mass range of m/z < 500, which can overlap with or suppress ions from small molecule metabolites.79
While hundreds of analytes are typically observed within one MALDI-TOF imaging experiment, there are several important considerations for matrix selection, spectral analysis, and data interpretation. First, depending on the observed m/z range, matrix peaks will be part of the spectrum. Hence, one of the problems inherent to MALDI-TOF MSI is spectral noise due to limited separation of matrix from analytes prior to arrival at the mass detector. Ionization is matrix dependent, and not all molecules will ionize in the same matrix (for a matrix overview see Table 1). For targeted imaging of drug molecules or other analytes of interest, it is important to ensure that there will be no matrix overlap with the analytes of interest prior to imaging. The second consideration is that isobaric peaks are possible. In these instances, more complex techniques, such as derivatization or ion mobility, are necessary to separate overlapping peaks. In general, in positive ion mode, H+ is the most common adduct observed (Figure 1); however, it is possible to observe other adducts, including Na+ and K+. Specialized methods are available to exchange H+ for Na+ to shift the m/z from MW +1 to MW +23, which can be achieved by using a sodium wash.75 Tandem MS and/or high mass resolution measurements are necessary to identify specific molecules. Specialized MALDI MSI experiments are required to obtain quantitative MALDI imaging data.60
TABLE 1.
Commonly used MALDI imaging matrices for metabolite and lipid imaging
| Matrix | Full name | Structure | Neutral mass | Ionization mode | Uses1 |
|---|---|---|---|---|---|
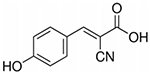
| CHCA | α-Cyano-4-hydroxycinnamic acid |
|
189.17 | Positive | Small metabolites, peptides, bacterial profiling, drugs |
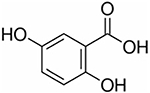
| DHB | 2,5-Dihydroxybenzoic acid |
|
154.12 | Negative | Neutral lipids, phospholipids, small metabolites, small proteins |
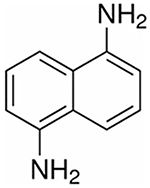
| DAN | 1,5-Diaminonaphthalene |
|
158.20 | Positive, negative | Phospholipids, drugs |
| 9-AA | 9-Aminoacridine |
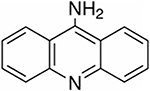
|
194.23 | Negative | Phospholipids |
| nH | Norharmane |
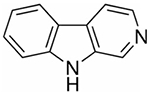
|
168.19 | Negative | Small metabolites, oligosaccharides, glycolipids |
1.3 |. NMR spectroscopy–MALDI imaging workflow integration
For multimodal workflows, it is important to know prior to tissue freezing that both NMR spectroscopy and MALDI imaging will be performed (Figure 2). Traditional MS, as well as most NMR metabolomic and lipidomic studies, are performed with snap-frozen freeze-clamped tissues, which are not suitable for MALDI imaging as the spatial information within the tissue is lost during freeze-clamping. The primary benefit of using MALDI imaging is to measure spatially and morphologically relevant metabolomic and lipidomic information in tissue; therefore, maintaining the original location of molecules within the tissue is of major importance. Instead, tissues for MALDI imaging need to be frozen in liquid nitrogen vapor, if possible without embedding, followed by cryosectioning. For this study, tumor tissue was cut in half, with one half frozen in liquid nitrogen vapor for MALDI imaging and histology and the other half snap-frozen in a clamp for grinding and dual-phase extraction for NMR spectroscopy. Tissues of which two or more organs exist in the same animal—e.g., the kidneys—allow for taking one organ for MSI and the other organ for NMR spectroscopy. Additional, more detailed “how to” information for performing cancer cell and tumor xenograft studies that combine NMR spectroscopy and MALDI imaging is provided in the Supporting Information.
FIGURE 2.
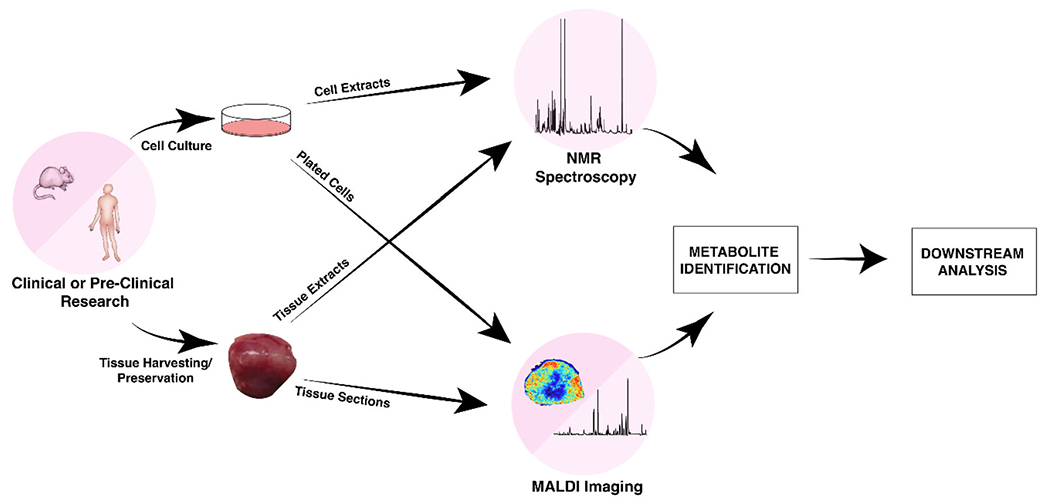
Representative workflows for multimodal pipeline using NMR spectroscopy and MALDI-TOF MSI from the same cell or tissue sample
2 |. MATERIALS AND METHODS
2.1 |. Materials, chemicals, reagents
All materials and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted. All reagents were used without further purification.
2.2 |. Cell culture of human breast cancer cells
The human breast cancer cell line MDA-MB-231 (ATCC catalog number CRM-HTB-26, RRID:CVCL_0062) was obtained from the American Type Culture Collection (ATCC, Manassas, MD, USA). Cells were not used beyond 10 passages after thawing to avoid genetic changes and adaptations. Cell lines tested negative for mycoplasma using a MycoDtect kit (Greiner Bio-One, Monroe, NC, USA) and were authenticated by STR profiling, as is routinely done every six months. Cells were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS). Cells were maintained at 37 °C with 5% CO2 in a humid environment in an incubator.
2.3 |. Orthotopic human breast tumor xenografts in mice
Animal experiments were performed in compliance with the Johns Hopkins University School of Medicine Animal Care and Use Committee (ACUC) guidelines. Approximately 2 × 106 MDA-MB-231 cells were subcutaneously injected into the fourth right mammary fat pad of female athymic nu/nu mice (Taconic Biosciences, Rensselaer, NY, USA) purchased at 4–6 weeks of age. Tumor volumes were measured with standard calipers and calculated using the formula (length × width × depth)/2. Mice were sacrificed at 8 weeks following orthotopic inoculation.
2.4 |. Tissue harvesting and preservation
Animals were sacrificed with CO2 and cervical dislocation. Tumors were harvested and cut into two equal-sized halves. Half of the tumor was flash-frozen in liquid nitrogen with freeze clamping for NMR spectroscopy and the other half was frozen in liquid nitrogen vapor for MALDI imaging. All tissues were stored frozen at −80 °C until sample preparation and measurement.
2.5 |. Dual-phase extraction and 1H NMR spectroscopy
Cells were trypsinized with 0.25% trypsin–ethylenediaminetetraacetic acid (EDTA) solution, and dead cells were stained using trypan blue before cell counts were obtained from viable cells. The dual-phase extraction method (methanol:chloroform:water = 1:1:1) was performed as previously described.12,13,80 The aqueous fractions were freeze-dried and dissolved in D2O containing 0.24 × 10−6 mol 3-(trimethylsilyl)propionic-2,2,3,3,-d4 acid (TSP) as a reference to enable chemical shift calibration and metabolite quantification. Lipid fractions were dried under a gentle stream of nitrogen and dissolved in deuterated chloroform (CDCl3) and methanol-d4 (CD3OD) in a 2:1 ratio.
Fully relaxed high-resolution (HR) 1H NMR spectroscopy was performed using a Bruker Avance-III 750 MHz spectrometer with a 5 mm TXI probe (Bruker BioSpin, Billerica, MA, USA). For aqueous metabolites, water-suppressed spectra were acquired using a 1D NOESY pulse sequence (spectral width 15 495.86 Hz, 64k data points, relaxation delay 10 s, 64 scans, eight dummy scans, receiver gain 40.3, and mixing time 80 ms). 1D 1H NMR spectra of lipid phases were obtained using a one-pulse sequence. Experimental parameters include a spectral width of 20.65 ppm, 64k time domain data points, relaxation delay of 10 s and 64 scans.
Metabolites were quantified using TopSpin (Bruker BioSpin). Aqueous metabolites were quantified as [fmol/cell] or [nmol/mg] tumor tissue according to the formula , where ITSP refers to the intensity of the TSP peak, Imet corresponds to the intensity of the metabolite peak being quantified, H refers to the number of protons at the chemical shift being quantified, [TSP] refers to the TSP concentration, and C refers to the number of cells present in the lysate being analyzed. Lipids were quantified in absolute units (A.U.) per cell or per mg tumor tissue according to the formula for cells and for tumor tissue, where ITMS refers to the signal intensity of the TMS peak. All experiments were performed in triplicate.
2.6 |. MALDI imaging
All indium tin oxide (ITO) slides (Delta Technologies, Loveland, CO, USA) were checked for conductivity prior to use and washed with sonication in hexane (10 min) and ethanol (10 min). Slides were dried and for cell imaging; slides were coated with poly-L-lysine.81 Slides were cleaned with ethanol prior to placing them with the coated side up in a 10 cm tissue culture dish. Trypsinized cells in medium (100 μL) were pipetted onto the slide. Cells were allowed to adhere to the slide for 15 min at 37 °C prior to the addition of 15 mL of medium. After 24 h, the cells on the slide were removed from the medium and bright field images were taken with a ×10 objective using a Nikon Eclipse TS100 microscope with Nikon DS-Fi3 digital camera. The slide was then placed on ice until matrix spraying. Prior to matrix spraying, the cell-covered slide was washed with HPLC-grade water (1 min). Pelleted cells were trypsinized and spun down to remove trypsin and medium. These cells were frozen in a cryomold at −80°C to form a block. More specific experimental details are provided in the supporting information.
For tumor imaging, two primary orthotopic MDA-MB-231 tumor xenografts were mounted with Shandon M-1 embedding medium (Fisher Scientific, Waltham, MA, USA), cryosectioned on a Leica 1860 UV cryostat (Wetzlar, Germany) at 10 μm thickness, and thaw-mounted onto ITO slides. Slides were placed in a vacuum desiccator (Chemglass, Vineland, NJ, USA) for 10 min prior to spraying to equilibrate to room temperature. For all imaging experiments, 1,5-diaminonaphthalene (1,5-DAN, 10 mg/mL) in 50% acetonitrile with 0.2% trifluoroacetic acid (TFA) was sprayed using an HTX M5 sprayer (HTX Technologies, Chapel Hill, NC, USA) with the following parameters: 30 °C nozzle temperature, eight passes, 0.1 mL/min flow rate, 1,200 mm/min velocity, and 2.5 mm track spacing. The final matrix density was 2.13 × 10−6 mg/mm2. MALDI imaging was performed on a Bruker RapifleX MALDI TOF/TOF instrument (Bruker Daltonics, Billerica, MA, USA) in reflectron positive mode with raster width of 100 μm and a 50 μm laser spot size with 200 laser shots per pixel. For cell imaging, a raster width of 5 μm and 5 μm single laser spot size with 200 laser shots per pixel was used. Three separate slide-grown cell samples were generated and imaged from subsequent passages of MDA-MB-231 cells. Two separate cell pellets were sectioned onto slides and two consecutive sections were imaged. Two of the three tumors used for 1H NMR spectroscopy experiments were MALDI imaged with two consecutive sections imaged for each of the two tumors as technical repeats. Representative and skyline spectra for each imaging run can be found in the Supporting Information (Figures S1–S6). A third section per tumor was imaged on the Bruker scimaX MALDI imaging instrument for high mass resolution data. On tissue MS/MS was performed after MALDI imaging on an adjacent tumor section with the same instrument in TOF/TOF geometry in profiling mode using a single-beam laser geometry with 54 μm resulting field with 4000 laser shots using argon collision induced dissociation (CID). All measurements were completed with a ±1 Da isolation window. Annotated tandem MS spectra can be found in the supporting information (Figures S7–S20). An adjacent section of tumor tissue was subjected to hematoxylin and eosin (H&E) staining.
High mass resolution data was collected on a Bruker 7 T scimaX MALDI imaging instrument using the small laser size setting with 15% power in positive ion mode and continuous accumulation of stored ions. Data was acquired using the following three mass ranges: m/z 140 to 220, m/z 158 to 358, and m/z 690 to 820.
Identification was performed using the Human Metabolome Database (hmdb.ca) for metabolites and LipidMaps (lipidmaps.org) for lipids (Table S1). High mass resolution data from the experiments on the Bruker scimaX instrument were peak picked and input into the corresponding database. Matches were confirmed with the MS/MS data acquired on the Bruker RapifleX instrument (Table S1).
All MALDI imaging data sets were background corrected and smoothed using flexImaging (v5.0, Bruker Daltonics). Images were generated in flexImaging. Data was imported into SCiLS Lab software (v2019B, Bruker Daltonics) using the flexImaging parameters. MALDI images of tumor sections were segmented, and all spectra were aligned in SCiLS Lab software. Segmentation analysis was used to determine global spectral changes by pixel in MALDI imaging data sets. This is an unsupervised method, which uses a k-means bisecting algorithm.82,83 Peak alignment was used as a standard post-processing method to ensure that any minor drifts in m/z during the imaging run were aligned to be identified as a single peak rather than multiple peaks.84
3 |. RESULTS
3.1 |. 1H NMR spectroscopy of MDA-MB-231 breast cancer cells and tumor xenografts
Quantification of 1H NMR spectra of aqueous extracts (Figure 3) revealed that the highest metabolite concentrations in MDA-MB-231 breast cancer cells were glucose (Gluc), lactate (Lac), myo-inositol (MyoI), and phosphocholine (pCho). Several amino acids were also observed at lower concentrations. We detected high levels of saturated fatty acids (saturated −[CH2]n−), phosphatidylcholines (PtdCho), and phosphatidylinositols (PtdI) in the corresponding lipid extract spectra of MDA-MB-231 cells (Figure 4). Metabolites (Figure 3) and lipids (Figure 4) were also quantified by 1H NMR spectroscopy of extracted MDA-MB-231 breast tumor xenografts grown in female athymic nude mice. Overall, these quantitative metabolite and lipid data showed that triple-negative human MDA-MB-231 breast cancer cells displayed similar metabolic behaviors when grown in cell culture as compared with orthotopic growth in mammary fat pads of female athymic nude mice (Figures 3, 4). Nevertheless, some minor differences existed, including decreased glutamate (Glu), glutamine (Gln), and pCho, and increased α-ketoglutarate (α-KG) and alanine (Ala), in tumors versus cells: blue bars indicate decreases and red bars indicate increases relative to Gluc in the same sample. However, for direct comparisons of metabolite and lipid concentrations between cell and tumor extracts obtained from the same cell line, it is important to consider that tumor tissue extracts measured using 1H NMR spectroscopy encompass heterogeneous mixtures of necrotic and viable cancer cells and host cells. This emphasizes the necessity of introducing spatially resolved analytical techniques such as MALDI MSI. Moreover, metabolic requirements differ among conditions of cell culture and solid tumor growth, as the latter result in various microenvironmental conditions, including hypoxia, acidosis, and nutrient deprivation.85–88
FIGURE 3.
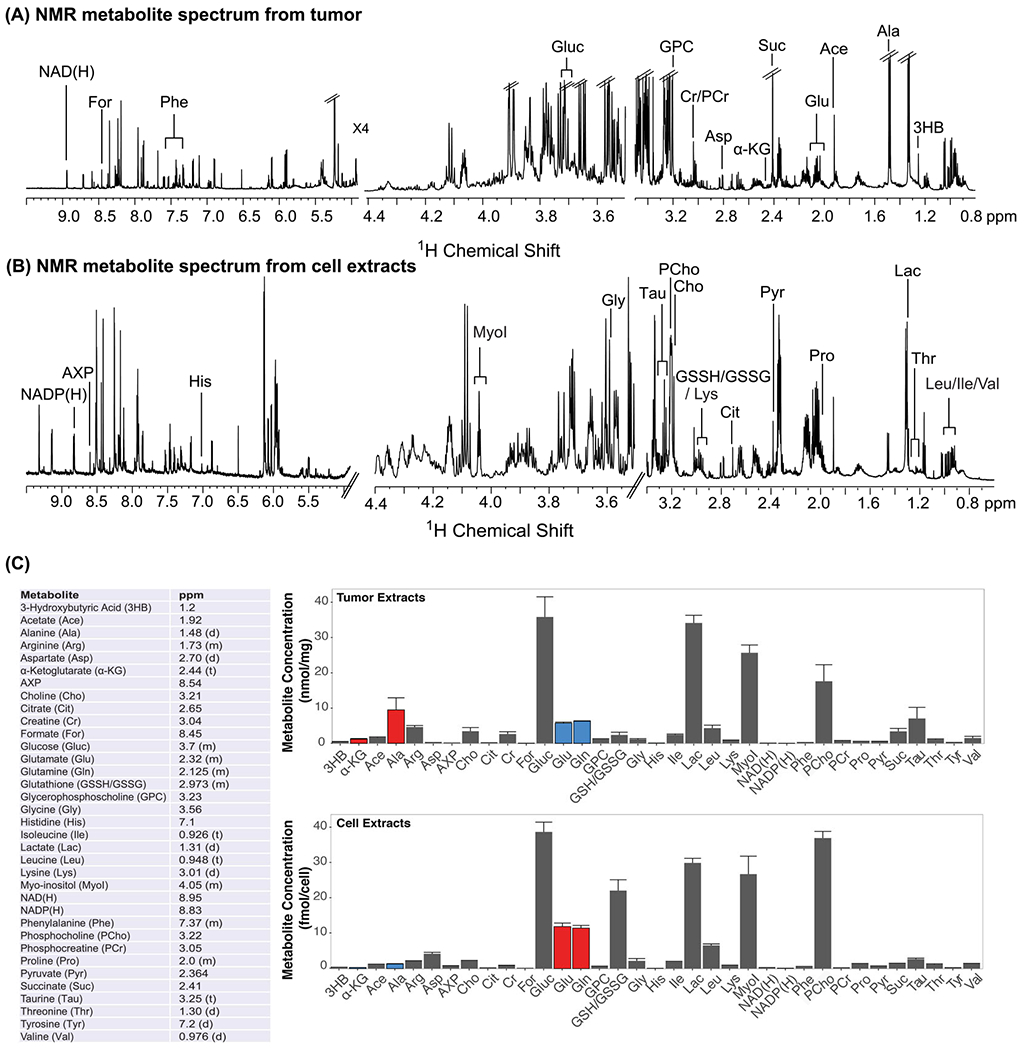
A, Representative metabolite 1H NMR spectra from MDA-MB-231 tumor xenografts. B, Representative metabolite1H NMR spectra from MDA-MB-231 cell extracts. C, Metabolites detected with 1H NMR spectroscopy in MDA-MB-231 cell and tumor xenografts and the chemical shift used for quantification are shown in the table. Average metabolite quantifications are shown from three biological repeats of tumor (top) and cell (bottom) extracts. Error bars represent standard error. While overall metabolic behavior was similar in MDA-MB-231 cells grown in culture as compared with orthotopic tumor growth, some differences existed, i.e., decreased Glu, Gln, pCho, and increased α-KG and Ala in tumors versus cells (blue bars indicate decreases, red bars indicate increases in tumors versus cells, relative to Gluc in the same sample. Significant changes were determined by a two-tailed t-test, with p < 0.5 considered significant.
FIGURE 4.
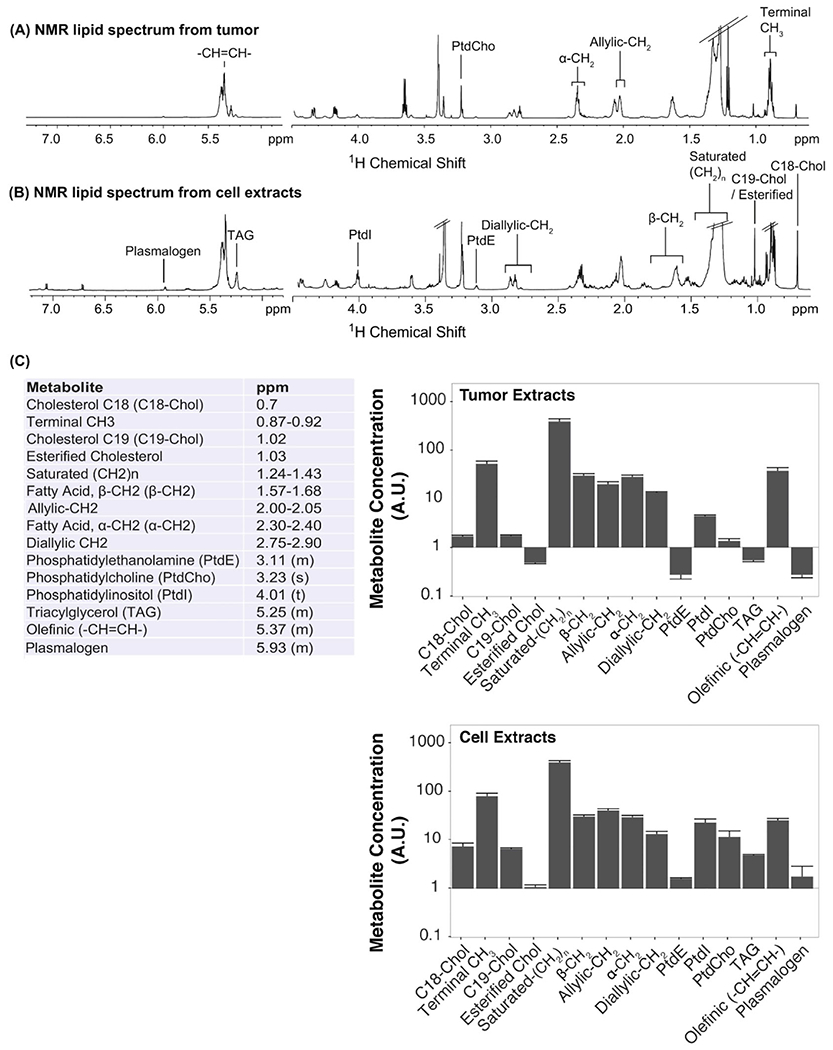
A, Representative lipid 1H NMR spectra from MDA-MB-231 tumor xenografts. B, Representative lipid 1H NMR spectra from MDA-MB-231 cell extracts. C, Lipids detected with 1H NMR spectroscopy in both MDA-MB-231 cell and tumor xenograft experiments and the chemical shift used for quantification. Average lipid quantifications are shown from three biological repeats of tumor (top) and cell (bottom) extracts. Error bars represent standard error.
3.2 |. MALDI imaging of MDA-MB-231 breast cancer cells and tumor xenografts
Breast cancer cells, directly grown on the slide or pelleted, and breast tumor sections were MALDI imaged using the same 1,5-DAN matrix preparation to detect the same molecules in both the cells and tumors. We were particularly interested in choline metabolism, which informed our choice of using the 1,5-DAN matrix as it allows for MALDI imaging of choline metabolites and choline-containing phospholipids in a single experiment. For cell imaging, pelleted cells had higher signal in MALDI imaging for all metabolites than slide-grown cells because of the higher number of cells in the pelleted cells, as evident in the corresponding H&E-stained microscopic images (Figure 5). Pelleting of cells allowed for more washing of the cell pellet, to remove any residual medium from the cell pellet prior to MALDI imaging, which reduced the background signal from medium, than was possible for slide-grown cells due to some loss of cells. As a consequence of less washing, the MALDI images of slide-grown cells contained more artifacts in the metabolite region than did those of pelleted cells (Figure 5). While MDA-MB-231 cells will stay mostly adhered to the slide with washing, some cells may not. Washing the slide-grown cells may also impact structure and cause delocalization of some analytes. We identified pCho and glycerophosphocholine (GPC) in MALDI imaging data of MDA-MB-231 cells and tumor sections (Figures S8 and S9, Table S1). pCho (m/z 184.1) and GPC (m/z 258.1) displayed excellent signal intensity and unique spatial distributions within the viable tumor region when compared with the corresponding H&E-stained microscopic images (Figure 5). Two polyamines, spermidine (m/z 146.2) and spermine (m/z 203.3), were identified in all acquired MALDI imaging data (Figure 5, Figures S7 and S10, Table S1). Spermidine was detected at low intensity in the slide-grown cells and had good signal intensity in both pelleted cells and viable tumor regions. Spermine was observed in both types of cell preparation and in the tumor, in which it localized to the rim region of the tumor. All identified metabolites described above were localized to cells as evident when comparing MALDI images with the corresponding H&E-stained microscopic images (Figures 4 and 5).
FIGURE 5.
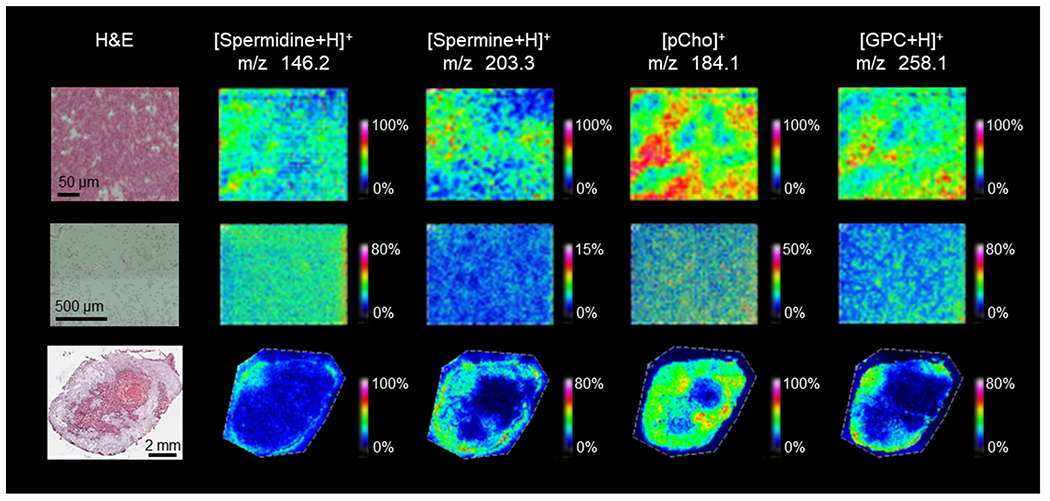
Metabolites identified in MDA-MB-231 cell pellets (top row), slide-grown cells (middle row) and tumor xenograft sections (bottom row) in MALDI imaging experiments with corresponding H&E images (left). Metabolites were identified using a combination of high mass resolution data (Supporting Table S1) and tandem MS data (Supporting Figures S7–S10). Images shown represent the entire imaged area with a mass filter at the reported m/z with a selection window of ±0.05 Da.
We also analyzed the lipids from the same MALDI imaging experiments on the two breast cancer cell preparations and tumor tissue sections. The spectral region of m/z 600 to 900 contained mostly phospholipids. Figure 6 shows identified phospholipids that were detected in slide-grown cells and tumor sections (pelleted cells not shown). We identified 17 unique phospholipids constituting 10 different m/z by using a combination of high mass resolution MALDI imaging and tandem MS experiments (Figures S11–S20, Table S1). Seven m/z consisted of two isobaric lipids each (one PC plus one PE in each case) at exactly the same mass, and the presence of both was confirmed using tandem MS and analysis of their fragmentation patterns. In future studies, it may be possible to separately MALDI image these isobaric m/z using tandem MSI to visualize fragments unique to each lipid. As shown in Figure 6, all identified lipids were present in different spatial distributions within the viable tumor region, except for SM 34:1;O2 (m/z 725.6), which was located exclusively in the necrotic region. To visualize spectrally different regions, the MALDI imaging data from a representative MDA-MB-231 breast tumor section was imported into SCiLS Lab for segmentation analysis (Figure 7). When compared with the corresponding H&E-stained microscopy data (Figure 7A), the segmentation map (Figure 7B) clearly shows that the segmented tumor region shown in yellow colocalizes with the necrotic region, while the segmented green region colocalizes with the viable region. Two representative regions of interest (ROIs) were generated from the segmentation map within the necrotic (red circle, yellow region) and the viable (blue circle, green region) tumor to display global changes associated with necrosis as compared with viable tumor. The necrotic tumor tissue demonstrated overall higher signal intensities in the 600 to 750 m/z region and significantly decreased signal intensities in the 750 to 900 m/z region (Figure 7C), as well as reflecting the changes in identified metabolite and phospholipid species (see Figures 5 and 6).
FIGURE 6.
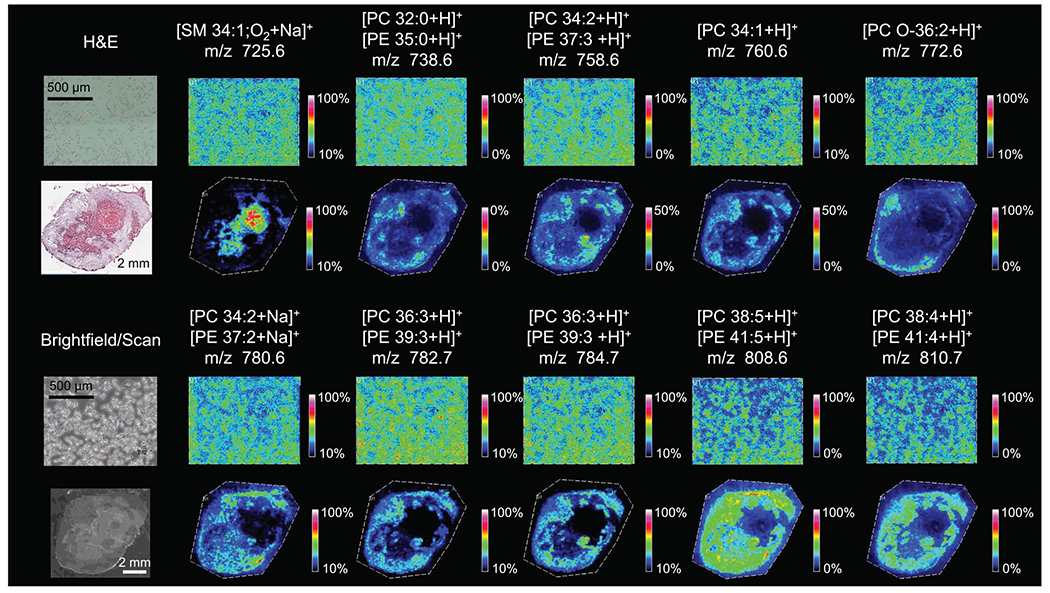
Phospholipids identified in slide-grown MDA-MB-231 cells (top and third rows) and MDA-MB-231 tumor xenograft sections (second and fourth rows) in MALDI imaging experiments with corresponding H&E stain and bright field images. Phospholipids were identified using a combination of high mass resolution data (Supporting Table S1) and tandem MS data (Supporting Figures S11–S20). For images where two phospholipids are reported, high mass resolution data were unable to distinguish between the two lipids and there is evidence for fragments from both species in the tandem MS experiments. Images shown represent the entire imaged area with a mass filter at the reported m/z with a selection window of ±0.05 Da.
FIGURE 7.
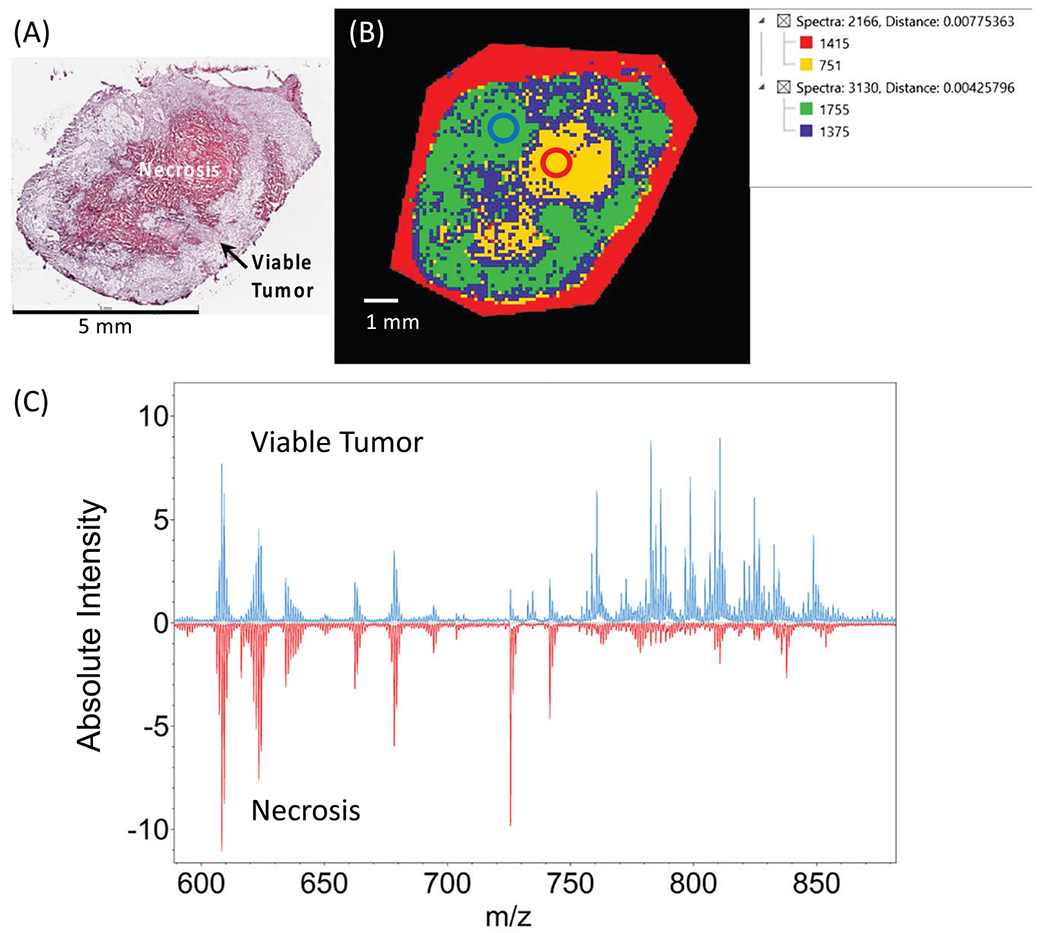
A, B, H&E stain from orthotopic MDA-MB-231 breast tumor xenograft section (A) with SCiLS Lab segmentation map from the same tumor section (B). C, MALDI spectra from the labeled regions of interest circled in B. Average spectra from viable tumor region (blue circle in B) are displayed in blue. Average spectra from necrotic tumor region (red circle in B) are displayed in red.
3.3 |. Complementary analysis of 1H NMR spectroscopy and MALDI imaging data
Most metabolites observed in the MALDI imaging data of MDA-MB-231 breast cancer cells and tumor sections were detected in the corresponding 1H NMR spectra as well, but frequently with overlap with other signals or as combined molecular groups. While generally not noted or reported in breast cancer 1H NMR spectroscopy data, polyamines have been detected at 1.78 ppm in other types of cancer.89 The polyamine multiplet at 1.78 ppm encompasses all polyamines including spermine and spermidine within the sample, and was not quantified in the 1H NMR spectra of breast cancer cells and tumor xenografts in our study due to significant overlap with other metabolite signals (Figure 3). We were however able to identify spermine and spermidine in the corresponding MALDI imaging data (Figure 5). In terms of spatial information, both spermine and spermidine were detected in viable tumor regions and were absent from necrotic regions (Figure 5).
Choline metabolites were also detected in both types of spectral data set, i.e., 1H NMR spectra and MALDI imaging of MDA-MB-231 cells and tumors (Figures 3 and 5). The choline metabolite peaks Cho (3.21 ppm), pCho (3.22 ppm), and GPC (3.23 ppm) were identified and quantified as singlets, arising from nine chemically and magnetically equivalent methyl group protons each in 1H NMR spectroscopy data (Figure 3). These choline metabolites were partially observed in the corresponding MALDI imaging data as well. pCho was the most abundant choline metabolite in 1H NMR spectra of MDA-MB-231 cell and tumor xenograft samples and was also observed by MALDI imaging. However, pCho detected by MALDI imaging at m/z 184.1 probably originated from fragmented PtdCho headgroups as well as the water-soluble intracellular pCho metabolite pool, making biological interpretation difficult (Figure 5). We observed GPC at 3.23 ppm in 1H NMR spectra as well as in MALDI imaging spectra at m/z 258.1, exclusively localizing to the viable regions of the tumor, demonstrating the ideal scenario in which we can quantify GPC concentration from 1H NMR spectra (Figure 3) and observe spatial information from the MALDI imaging data (Figure 5).
Lipid analysis of MDA-MB-231 breast cancer cells and tumor xenografts by combined 1H NMR spectroscopy and MALDI imaging revealed complementary global and specific molecular information. 1H NMR spectroscopy data detected global changes in specific lipid classes (Figure 4), while MALDI imaging provided spatial distributions of specific lipids in MDA-MB-231 cells and tumor sections (Figure 6). We observed spatial distributions of several unique PC and PE species in MALDI imaging from MDA-MB-231 breast cancer cells and tumor sections (Figure 6), whereas in the 1H NMR spectroscopy data all PE species were detected as one combined multiplet signal at 3.11 ppm and all PC species as one combined singlet signal at 3.23 ppm. The combined multimodal pipeline using 1H NMR spectroscopy and MALDI imaging from the same sample provided enhanced molecular information from the cells and tissues under investigation.
4 |. DISCUSSION
Our study demonstrates the power of combining 1H NMR spectroscopy and MALDI imaging from the same cell or tissue sample, which was shown for MDA-MB-231 breast cancer cells and tumor xenografts. We obtained an unprecedented amount of molecular information from a single tissue sample, revealing important metabolomic and lipidomic features of breast cancer cells and tumor tissue. This combined pipeline enabled us to measure quantitative and spatial information of specific metabolites and individual lipids from the same cell or tissue sample. The presented study is a starting point for additional methodological directions that can be pursued in similar multimodal pipelines of combined NMR spectroscopy and MALDI imaging. Future directions in multimodal NMR spectroscopy–MALDI imaging pipelines could explore different cell and tissue types, MALDI matrices (Table 1), and NMR nuclei.
The presented data emphasizes that the strength of 1H NMR spectroscopy is the quantification of a plethora of metabolites from different pathways including glycolysis, tricarboxylic acid (TCA) cycle, amino acids, redox metabolism, choline metabolism, and creatine, among many others, in one measurement. The comparison between 1H NMR spectra of MDA-MB-231 breast cancer cells versus MDA-MB-231 tumor xenografts revealed metabolic differences probably arising from hypoxia, acidosis, necrosis, and nutrient deprivation in the tumor, which were not occurring in the cultured cells.85–88 GPC was optimally detected by NMR spectroscopy–MALDI imaging, allowing for quantitative measurements of its concentration, as well as detecting its localization to specific areas of the viable tumor region. Elevated GPC in certain parts of the viable tumor region could result from slow acidosis, which has previously been shown to increase tumor GPC levels.90 GPC is an important metabolite generated from PtdCho breakdown in membrane choline phospholipid metabolism.91 Altered choline metabolism is a well established hallmark of cancer and an active area of research in many types of cancer.91 Several patient studies have reported that in vivo MRS detection of choline metabolites in tumors may be able to predict response to treatment early on during the course of treatment.92–96 Molecular studies have revealed that several enzymes in choline metabolism, including choline kinase,97–100 various phospholipases,11,101–104 and glycerophosphodiesterases,12,105 are differentially expressed, activated, and drive oncogenic signaling pathways in several cancers,91,106 providing potential targets for treatment.91
In addition to GPC, various phospholipids in the PtdCho and PtdE groups were primarily detected in viable tumor regions and depleted in necrotic tumor regions, while also displaying somewhat different regional distributions within the viable region. This could be due to down-regulation or degradation of these phospholipids in hypoxic and necrotic tumor regions, which is in good agreement with our previous studies.63,107 Ethanolamine metabolism, which gives rise to a plethora of different membrane PtdE species, was shown to be activated in several cancers alongside the activated choline metabolism.108,109 The addition of MALDI imaging, whose strength is phospholipid detection, to studying the lipid compartment of choline and ethanolamine metabolism in cancer will provide novel insights into these important metabolic pathways in cancer.
In our combined 1H NMR spectroscopy–MALDI imaging data sets, we were able to identify two polyamines, spermine and spermidine, in MDA-MB-231 cells and tumor xenografts, which is in good agreement with previous studies.110 Polyamines in breast cancer have been linked to both estrogen-dependent and estrogen-independent cancer growth, and hold promise as biomarkers and treatment targets.111 Our MALDI imaging data revealed that spermine and spermidine were detected in the viable tumor region, particularly at the rim of the tumor. These findings merit further exploration of the role of polyamines in breast cancer cell growth.
While our study was performed on preclinical samples, the same pipeline could be useful for patient samples, including biopsies and surgical tissues. Such patient samples are precious resources for research studies, and in many cases only small amounts of tissue and small sample sizes can be obtained. The use of HR-MAS instead of tissue extractions for 1H NMR spectroscopy would allow for subsequent histopathology on the same tissue, maximizing tissue use.35 While tissue samples can first be measured by HR-MAS spectroscopy and then sectioned for both histopathology and MALDI imaging,112 the separation of metabolomic and lipidomic data in NMR spectroscopy is only realized by dual-phase extraction approaches, as HR-MAS spectroscopy typically detects overlapping metabolite and lipid spectra.16 It is also important to note that spatial information from HR-MAS spectroscopy is currently limited to slice localization approaches.41 Spatially resolved metabolomic and lipidomic studies would be particularly useful in studying field effects of cancers on adjacent normal tissue.113
Combined NMR spectroscopy–MALDI imaging workflows could also advance preclinical and clinical studies of liver disease. Liver tissue has been well studied by both MALDI imaging and NMR spectroscopy.114–121 Liver disease, such as cirrhosis or fatty liver disease, as well as liver-based toxicity, profoundly impacts metabolic and lipidomic pathways.117,118,122,123 Utilizing a combination of ex vivo NMR spectroscopy and MALDI imaging would allow for in-depth region-specific molecular analysis of liver samples. The presented dual-modality pipeline could also benefit infectious disease studies on host-response and drug resistance. Moreover, ex vivo NMR spectroscopy has been a primary method to analyze biofluids, including blood and bronchoalveolar lavage samples.124,125 As MALDI imaging quickly approaches single-cell analysis, the power of examining cell-specific molecules and pathways is undeniable126 and could be applied to various populations of cells from biofluids.
The presented NMR spectroscopy–MALDI imaging pipeline would also be useful for targeted analysis of specific molecules. In this study, an unsupervised, discovery-based approach was pursued to reveal molecules of interest, whereas targeted studies investigate specified molecules, such as drugs and drug metabolites. One of the primary applications of MALDI imaging is in detecting drug distributions in tissue from treated animals or human subjects.128,129 NMR metabolomics has been used to study the effects of drugs on metabolism in various applications.4,80 The ability to quantitatively measure drug concentrations and their spatial distributions and correlate these to metabolomic and lipidomic data would enhance our mechanistic understanding of drug treatments and response to treatment.
5 |. CONCLUSIONS AND OUTLOOK
We have shown that NMR spectroscopy and MALDI imaging are complementary techniques which enhance each other to quantify and localize metabolites and lipids in cells and tissues. We have developed a workflow to harvest and prepare cells and tissues for a combined pipeline using these two techniques. 1H NMR spectroscopy is the preferred technique for quantitative measurements of metabolites. MALDI imaging allows for spatially resolved metabolomic and lipidomic analysis, while quantification is limited owing to differences in ionization efficiency.
Both NMR spectroscopy and MALDI imaging are rapidly evolving fields in which instrumentation technology is driving analytical performance. For NMR spectroscopy, higher field strengths improve spectral resolution, and advanced probe design and pulse sequence development further enhance detection sensitivity129 Imaging mass spectrometers currently undergo advancements in MALDI source design, implementation of MALDI-2 technology for improved spatial resolution, and addition of ion mobility for increased spectral separation.130,131 Establishing core facilities for NMR spectroscopy and MALDI imaging enables greater access of the scientific community to these expensive technologies that require high levels of expertise.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Johns Hopkins Applied Imaging Mass Spectrometry (AIMS) Core Facility at the Johns Hopkins University School of Medicine and the Bruker Daltonics Applications laboratory in Billerica, MA. We would also like to acknowledge the National Institutes of Health grants R01 CA213492, R01 CA213428, R01 CA264901, and S10 OD030500 and the Sol Goldman Cancer Research Center Pilot Fund for funding.
Funding information
National Institutes of Health, Grant/Award Numbers: R01 CA213492, R01 CA213428, R01 CA264901, S10 OD030500; Sol Goldman Cancer Research Center Pilot Fund
Abbreviations:
- 1D
one dimensional
- 2D
two dimensional
- Ala
alanine
- Cho
choline
- Gln
glutamine
- GPC
glycerophosphocholine
- Glu
glutamate
- H&E
hematoxylin and eosin
- HR
high resolution
- HR-MAS
high-resolution magic angle spinning
- ITO
indium tin oxide
- MALDI
matrix assisted laser desorption ionization
- MS
mass spectrometry
- MSI
mass spectrometry imaging
- m/z
mass-to-charge ratio
- pCho
phosphocholine
- PtdE or PE
phosphoethanolamine
- PtdCho or PC
phosphatidylcholine
- PtdI
phosphatidylinositol
- TOF
time of flight
- TSP
3-(trimethylsilyl)propionic-2,2,3,3,-d4 acid
- α-KG
α-ketoglutarate
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Nicholson JK, Wilson ID. High resolution proton magnetic resonance spectroscopy of biological fluids. Prog Nucl Magn Reson Spectrosc. 1989;21(4/5):449–501. doi: 10.1016/0079-6565(89)80008-1 [DOI] [Google Scholar]
- 2.Tyagi RK, Azrad A, Degani H, Salomon Y. Simultaneous extraction of cellular lipids and water-soluble metabolites: evaluation by NMR spectroscopy. Magn Reson Med. 1996;35(2):194–200. doi: 10.1002/mrm.1910350210 [DOI] [PubMed] [Google Scholar]
- 3.Seierstad T, Roe K, Sitter B, et al. Principal component analysis for the comparison of metabolic profiles from human rectal cancer biopsies and colorectal xenografts using high-resolution magic angle spinning 1H magnetic resonance spectroscopy. Mol Cancer. 2008;7(1):33–46. doi: 10.1186/1476-4598-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan KW, Jiang L, Cheng M, et al. CEST-MRI detects metabolite levels altered by breast cancer cell aggressiveness and chemotherapy response. NMR Biomed. 2016;29(6):806–816. doi: 10.1002/nbm.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johns CW, Lee AB, Springer TI, et al. Using NMR-based metabolomics to monitor the biochemical composition of agricultural soils: a pilot study. Eur J Soil Biol. 2017;83:98–105. doi: 10.1016/j.ejsobi.2017.10.008 [DOI] [Google Scholar]
- 6.Tian H, Lam SM, Shui G. Metabolomics, a powerful tool for agricultural research. Int J Mol Sci. 2016;17(11):1871 doi: 10.3390/ijms17111871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauthe SK, Sharma RJ, Aqil F, Gupta RC, Singh IP. Quantitative NMR: an applicable method for quantitative analysis of medicinal plant extracts and herbal products. Phytochem Anal. 2012;23(6):689–696. doi: 10.1002/pca.2375 [DOI] [PubMed] [Google Scholar]
- 8.Ott K-H, Araníbar N, Singh B, Stockton GW. Metabonomics classifies pathways affected by bioactive compounds. Artificial neural network classification of NMR spectra of plant extracts. Phytochemistry. 2003;62(6):971–985. doi: 10.1016/S0031-9422(02)00717-3 [DOI] [PubMed] [Google Scholar]
- 9.Rosato A, Tenori L, Cascante M, De Atauri Carulla PR, Martins dos Santos VAP, Saccenti E. From correlation to causation: analysis of metabolomics data using systems biology approaches. Metabolomics. 2018;14(4):37 doi: 10.1007/s11306-018-1335-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weckwerth W. Metabolomics: an integral technique in systems biology. Bioanalysis. 2010;2(4):829–836. doi: 10.4155/bio.09.192 [DOI] [PubMed] [Google Scholar]
- 11.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64(12):4270–4276. doi: 10.1158/0008-5472.can-03-3829 [DOI] [PubMed] [Google Scholar]
- 12.Cao MD, Cheng M, Rizwan A, et al. Targeting choline phospholipid metabolism: GDPD5 and GDPD6 silencing decrease breast cancer cell proliferation, migration, and invasion. NMR Biomed. 2016;29(8):1098–1107. doi: 10.1002/nbm.3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao MD, Dopkens M, Krishnamachary B, et al. Glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5) expression correlates with malignant choline phospholipid metabolite profiles in human breast cancer. NMR Biomed. 2012;25(9):1033–1042. doi: 10.1002/nbm.2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foxall PJ, Parkinson JA, Sadler IH, Lindon JC, Nicholson JK. Analysis of biological fluids using 600 MHz proton NMR spectroscopy: application of homonuclear two-dimensional J-resolved spectroscopy to urine and blood plasma for spectral simplification and assignment. J Pharm Biomed Anal. 1993;11(1):21–31. doi: 10.1016/0731-7085(93)80145-Q [DOI] [PubMed] [Google Scholar]
- 15.Lindon JC, Nicholson JK, Everett JR. NMR spectroscopy of biofluids. Annu Rep NMR Spectrosc. 1999;38:1–88. [Google Scholar]
- 16.Mori N, Wildes F, Takagi T, Glunde K, Bhujwalla ZM. The tumor microenvironment modulates choline and lipid metabolism. Front Oncol. 2016;6:262. doi: 10.3389/fonc.2016.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wüthrich K. NMR of proteins and nucleic acids. Europhys News. 1986;17(1):11–13. doi: 10.1051/epn/19861701011 [DOI] [Google Scholar]
- 18.Gunther H. NMR Spectroscopy: Basic Principles, Concepts, and Applications in Chemistry. Wiley; 1995. [Google Scholar]
- 19.Lindon JC, Nicholson JK, Holmes E. The Handbook of Metabonomics and Metabolomics. Elsevier; 2007. [Google Scholar]
- 20.Ludwig C, Viant MR. Two-dimensional J-resolved NMR spectroscopy: review of a key methodology in the metabolomics toolbox. Phytochem Anal. 2010;21(1):22–32. doi: 10.1002/pca.1186 [DOI] [PubMed] [Google Scholar]
- 21.Parsons HM. Optimised Spectral Processing and Lineshape Analysis in 2-Dimensional J-Resolved NMR Spectroscopy Based Metabolomics. PhD. University of Birmingham; 2010. [Google Scholar]
- 22.Ernst RR, Bodenhausen G, Wokaun A. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Clarendon; 1990. [Google Scholar]
- 23.Nicholson JK, Holmes E, Lindon JC. The Handbook of Metabonomics and Metabolomics. Elsevier; 2007. [Google Scholar]
- 24.Kalinowski HO, Berger S, Braun S, Griffiths L. Carbon-13 NMR spectroscopy: Wiley, Chichester, 1988 (ISBN 0-471-91306-5). 776 pp. Price £75.00. Anal Chim Acta. 1989;221:367–368. doi: 10.1016/S0003-2670(00)81981-9 [DOI] [Google Scholar]
- 25.Smallcombe SH, Patt SL, Keifer PA. WET solvent suppression and its applications to LC NMR and high-resolution NMR spectroscopy. J Magn Reson A. 1995;117(2):295–303. doi: 10.1006/jmra.1995.0759 [DOI] [Google Scholar]
- 26.Liu M, Mao X-A, Ye C, Huang H, Nicholson JK, Lindon JC. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J Magn Reson. 1998;132(1):125–129. doi: 10.1006/jmre.1998.1405 [DOI] [Google Scholar]
- 27.Lucas LH, Larive CK, Wilkinson PS, Huhn S. Progress toward automated metabolic profiling of human serum: comparison of CPMG and gradient-filtered NMR analytical methods. J Pharm Biomed Anal. 2005;39(1–2):156–163. doi: 10.1016/j.jpba.2004.09.060 [DOI] [PubMed] [Google Scholar]
- 28.Mazzei P, Piccolo A, Nugnes L, Mascolo M, De Rosa G, Staibano S. Metabolic profile of intact tissue from uterine leiomyomas using high-resolution magic-angle-spinning 1H NMR spectroscopy. NMR Biomed. 2010;23(10):1137–1145. doi: 10.1002/nbm.1540 [DOI] [PubMed] [Google Scholar]
- 29.Tang H, Wang Y, Nicholson JK, Lindon JC. Use of relaxation-edited one-dimensional and two dimensional nuclear magnetic resonance spectroscopy to improve detection of small metabolites in blood plasma. Anal Biochem. 2004;325(2):260–272. doi: 10.1016/j.ab.2003.10.033 [DOI] [PubMed] [Google Scholar]
- 30.Singh M, Soni VK, Mishra R, Kurur ND. Relaxation editing using long-lived states and coherences for analysis of mixtures. Anal Chem. 2016;88(6):3004–3008. doi: 10.1021/acs.analchem.6b00050 [DOI] [PubMed] [Google Scholar]
- 31.Cheng LL, Chang IW, Louis DN, Gonzalez RG. Correlation of high-resolution magic angle spinning proton magnetic resonance spectroscopy with histopathology of intact human brain tumor specimens. Cancer Res. 1998;58(9):1825–1832. [PubMed] [Google Scholar]
- 32.Ben Sellem D, Elbayed K, Neuville A, et al. Metabolomic characterization of ovarian epithelial carcinomas by HRMAS-NMR spectroscopy. J Oncol. 2011;2011:174019 doi: 10.1155/2011/174019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng LL, Anthony DC, Comite AR, Black PM, Tzika AA, Gonzalez RG. Quantification of microheterogeneity in glioblastoma multiforme with ex vivo high-resolution magic-angle spinning (HRMAS) proton magnetic resonance spectroscopy. Neuro-Oncology. 2000;2(2):87–95. doi: 10.1093/neuonc/2.2.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng LL, Chang IW, Smith BL, Gonzalez RG. Evaluating human breast ductal carcinomas with high-resolution magic-angle spinning proton magnetic resonance spectroscopy. J Magn Reson. 1998;135(1):194–202. doi: 10.1006/jmre.1998.1578 [DOI] [PubMed] [Google Scholar]
- 35.DeFeo EM, Cheng LL. Characterizing human cancer metabolomics with ex vivo 1H HRMAS MRS. Technol Cancer Res Treat. 2010;9(4):381–391. doi: 10.1177/153303461000900407 [DOI] [PubMed] [Google Scholar]
- 36.CuM R, AnS B, Gil AM, et al. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. J Proteome Res. 2009;9(1):319–332. doi: 10.1021/pr9006574 [DOI] [PubMed] [Google Scholar]
- 37.Wright AJ, Fellows GA, Griffiths JR, Wilson M, Bell BA, Howe FA. Ex-vivo HRMAS of adult brain tumours: metabolite quantification and assignment of tumour biomarkers. Mol Cancer. 2010;9(1):66 doi: 10.1186/1476-4598-9-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Li C, Nie X, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6(7):2605–2614. doi: 10.1021/pr070063h [DOI] [PubMed] [Google Scholar]
- 39.Sharma RK, Sonkar K, Sinha N, et al. Gallstones: a worldwide multifaceted disease and its correlations with gallbladder carcinoma. PLoS ONE. 2016; 11(11):e0166351 doi: 10.1371/journal.pone.0166351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong A, Jiménez B, Li X, et al. Evaluation of high resolution magic-angle coil spinning NMR spectroscopy for metabolic profiling of nanoliter tissue biopsies. Anal Chem. 2012;84(8):3843–3848. doi: 10.1021/ac300153k [DOI] [PubMed] [Google Scholar]
- 41.Vonhof EV, Piotto M, Holmes E, Lindon JC, Nicholson JK, Li JV. Improved spatial resolution of metabolites in tissue biopsies using high-resolution magic-angle-spinning slice localization NMR spectroscopy. Anal Chem. 2020;92(17):11516–11519. doi: 10.1021/acs.analchem.0c02377 [DOI] [PubMed] [Google Scholar]
- 42.Silverstein RM, Webster FX, Kiemle DJ. Spectrometric Identification of Organic Compounds. Wiley; 2005. [Google Scholar]
- 43.McLafferty FW. A century of progress in molecular mass spectrometry. Annu Rev Anal Chem. 2011;4(1):1–22. doi: 10.1146/annurev-anchem-061010-114018 [DOI] [PubMed] [Google Scholar]
- 44.Massonnet P, Heeren RMA. A concise tutorial review of TOF-SIMS based molecular and cellular imaging. J Anal At Spectrom. 2019;34(11):2217–2228. doi: 10.1039/C9JA00164F [DOI] [Google Scholar]
- 45.Agüi-Gonzalez P, Jähne S, Phan NTN. SIMS imaging in neurobiology and cell biology. J Anal At Spectrom. 2019;34(7):1355–1368. doi: 10.1039/C9JA00118B [DOI] [Google Scholar]
- 46.Senoner M, Unger WES. SIMS imaging of the nanoworld: applications in science and technology. J Anal At Spectrom. 2012;27(7):1050–1068. doi: 10.1039/C2JA30015J [DOI] [Google Scholar]
- 47.Swales JG, Hamm G, Clench MR, Goodwin RJA. Mass spectrometry imaging and its application in pharmaceutical research and development: a concise review. Int J Mass Spectrom. 2019;437:99–112. doi: 10.1016/j.ijms.2018.02.007 [DOI] [Google Scholar]
- 48.Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013;32(3):218–243. doi: 10.1002/mas.21360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLaney K, Phetsanthad A, Li L. Advances in high-resolution MALDI mass spectrometry for neurobiology. Mass Spectrom Rev. 2022;41(2):194–214. doi: 10.1002/mas.21661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i [DOI] [PubMed] [Google Scholar]
- 51.Spengler B. Mass spectrometry imaging of biomolecular information. Anal Chem. 2015;87(1):64–82. doi: 10.1021/ac504543v [DOI] [PubMed] [Google Scholar]
- 52.Spraggins JM, Caprioli RM. High-speed MALDI-TOF imaging mass spectrometry: rapid ion image acquisition and considerations for next generation instrumentation. J Am Soc Mass Spectrom. 2011;22(6):1022–1031. doi: 10.1007/s13361-011-0121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Basu SS, Regan MS, Randall EC, et al. Rapid MALDI mass spectrometry imaging for surgical pathology. NPJ Precis Oncol. 2019;3(1):17. doi: 10.1038/s41698-019-0089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaysse P-M, Heeren RMA, Porta T, Balluff B. Mass spectrometry imaging for clinical research—latest developments, applications, and current limitations. Analyst. 2017;142(15):2690–2712. doi: 10.1039/C7AN00565B [DOI] [PubMed] [Google Scholar]
- 55.Hosseini S, Martinez-Chapa SO. Fundamentals of MALDI-ToF-MS Analysis: Applications in Bio-Diagnosis, Tissue Engineering and Drug Delivery. Springer; 2017. doi: 10.1007/978-981-10-2356-9. [DOI] [Google Scholar]
- 56.Li Y, Shrestha B, Vertes A. Atmospheric pressure infrared MALDI imaging mass spectrometry for plant metabolomics. Anal Chem. 2008;80(2):407–420. doi: 10.1021/ac701703f [DOI] [PubMed] [Google Scholar]
- 57.Kaspar S, Peukert M, Svatos A, Matros A, Mock H-P. MALDI-imaging mass spectrometry–an emerging technique in plant biology. Proteomics. 2011;11(9):1840–1850. doi: 10.1002/pmic.201000756 [DOI] [PubMed] [Google Scholar]
- 58.Boughton BA, Thinagaran D, Sarabia D, Bacic A, Roessner U. Mass spectrometry imaging for plant biology: a review. Phytochem Rev. 2016;15(3):445–488. doi: 10.1007/s11101-015-9440-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ly A, Longuespée R, Casadonte R, et al. Site-to-site reproducibility and spatial resolution in MALDI–MSI of peptides from formalin-fixed paraffin-embedded samples. Proteom Clin Appl. 2019;13(1):1800029. 10.1002/prca.201800029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barry JA, Ait-Belkacem R, Hardesty WM, et al. Multicenter validation study of quantitative imaging mass spectrometry. Anal Chem. 2019;91(9):6266–6274. doi: 10.1021/acs.analchem.9b01016 [DOI] [PubMed] [Google Scholar]
- 61.Blaschke CRK, Black AP, Mehta AS, Angel PM, Drake RR. Rapid N-glycan profiling of serum and plasma by a novel slide-based imaging mass spectrometry workflow. J Am Soc Mass Spectrom. 2020;31(12):2511–2520. doi: 10.1021/jasms.0c00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heijs B, Potthoff A, Soltwisch J, Dreisewerd K. MALDI-2 for the enhanced analysis of N-linked glycans by mass spectrometry imaging. Anal Chem. 2020;92(20):13904–13911. doi: 10.1021/acs.analchem.0c02732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chughtai K, Jiang L, Greenwood TR, Glunde K, Heeren RMA. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor models. J Lipid Res. 2013;54(2):333–344. doi: 10.1194/jlr.M027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersen MK, Høiem TS, Claes BSR, et al. Spatial differentiation of metabolism in prostate cancer tissue by MALDI-TOF MSI. Cancer Metab. 2021;9(1):9 doi: 10.1186/s40170-021-00242-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersen MK, Krossa S, Høiem TS, et al. Simultaneous detection of zinc and its pathway metabolites using MALDI MS imaging of prostate tissue. Anal Chem. 2020;92(4):3171–3179. doi: 10.1021/acs.analchem.9b04903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott AJ, Flinders B, Cappell J, et al. Norharmane matrix enhances detection of endotoxin by MALDI-MS for simultaneous profiling of pathogen, host and vector systems. Pathog Dis. 2016;74(8):ftw097 doi: 10.1093/femspd/ftw097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, Dorrestein PC. Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging. Mol Biosyst. 2008;4(6):562–570. doi: 10.1039/B720018H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goto-Inoue N, Hayasaka T, Zaima N, Setou M. Imaging mass spectrometry for lipidomics. BBA Mol Cell Biol Lipids. 2011;1811(11):961–969. doi: 10.1016/j.bbalip.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 69.Sparvero LJ, Amoscato AA, Dixon CE, et al. Mapping of phospholipids by MALDI imaging (MALDI-MSI): realities and expectations. Chem Phys Lipids. 2012;165(5):545–562. doi: 10.1016/j.chemphyslip.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annu Rev Biochem. 2011;80(1):301–325. doi: 10.1146/annurev-biochem-060409-092612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Porta Siegel T, Ekroos K, Ellis SR. Reshaping lipid biochemistry by pushing barriers in structural lipidomics. Angew Chem Int Ed. 2019;58(20):6492–6501. doi: 10.1002/anie.201812698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calvano CD, Monopoli A, Cataldi TRI, Palmisano F. MALDI matrices for low molecular weight compounds: an endless story? Anal Bioanal Chem. 2018;410(17):4015–4038. doi: 10.1007/s00216-018-1014-x [DOI] [PubMed] [Google Scholar]
- 73.Rzagalinski I, Volmer DA. Quantification of low molecular weight compounds by MALDI imaging mass spectrometry—a tutorial review. BBA Proteins Proteom. 2017;1865(7):726–739. doi: 10.1016/j.bbapap.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 74.Fujimura Y, Miura D. MALDI mass spectrometry imaging for visualizing in situ metabolism of endogenous metabolites and dietary phytochemicals. Metabolites. 2014;4(2):319–346. doi: 10.3390/metabo4020319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaurand P, Thomas A, Patterson NH, Laveaux Charbonneau J. Orthogonal organic and aqueous-based washes of tissue sections to enhance protein sensitivity by MALDI imaging mass spectrometry. J Mass Spectrom. 2013;48(1):42–48. doi: 10.1002/jms.3114 [DOI] [PubMed] [Google Scholar]
- 76.Blatherwick EQ, Svensson CI, Frenguelli BG, Scrivens JH. Localisation of adenine nucleotides in heat-stabilised mouse brains using ion mobility enabled MALDI imaging. Int J Mass Spectrom. 2013;345-347:19–27. doi: 10.1016/j.ijms.2013.02.004 [DOI] [Google Scholar]
- 77.McLaughlin N, Bielinski TM, Tressler CM, Barton E, Glunde K, Stumpo KA. Pneumatically sprayed gold nanoparticles for mass spectrometry imaging of neurotransmitters. J Am Soc Mass Spectrom. 2020;31(12):2452–2461. doi: 10.1021/jasms.0c00156 [DOI] [PubMed] [Google Scholar]
- 78.Shariatgorji M, Nilsson A, Fridjonsdottir E, et al. Comprehensive mapping of neurotransmitter networks by MALDI-MS imaging. Nat Methods. 2019;16(10):1021–1028. doi: 10.1038/s41592-019-0551-3 [DOI] [PubMed] [Google Scholar]
- 79.Gusev AI, Wilkinson WR, Proctor A, Hercules DM. Improvement of signal reproducibility and matrix/comatrix effects in MALDI analysis. Anal Chem. 1995;67(6):1034–1041. doi: 10.1021/ac00102a003 [DOI] [Google Scholar]
- 80.Cheng M, Rizwan A, Jiang L, Bhujwalla ZM, Glunde K. Molecular effects of doxorubicin on choline metabolism in breast cancer. Neoplasia. 2017;19(8):617–627. doi: 10.1016/j.neo.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ly A, Buck A, Balluff B, et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat Protoc. 2016;11(8):1428–1443. doi: 10.1038/nprot.2016.081 [DOI] [PubMed] [Google Scholar]
- 82.Alexandrov T, Becker M, Guntinas-Lichius O, Ernst G, von Eggeling F. MALDI-imaging segmentation is a powerful tool for spatial functional proteomic analysis of human larynx carcinoma. J Cancer Res Clin Oncol. 2013;139(1):85–95. doi: 10.1007/s00432-012-1303-2 [DOI] [PubMed] [Google Scholar]
- 83.McCombie G, Staab D, Stoeckli M, Knochenmuss R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal Chem. 2005;77(19):6118–6124. doi: 10.1021/ac051081q [DOI] [PubMed] [Google Scholar]
- 84.Boskamp T, Lachmund D, Casadonte R, et al. Using the chemical noise background in MALDI mass spectrometry imaging for mass alignment and calibration. Anal Chem. 2020;92(1):1301–1308. doi: 10.1021/acs.analchem.9b04473 [DOI] [PubMed] [Google Scholar]
- 85.Dias AS, Almeida CR, Helguero LA, Duarte IF. Metabolic crosstalk in the breast cancer microenvironment. Eur J Cancer. 2019;121:154–171. doi: 10.1016/j.ejca.2019.09.002 [DOI] [PubMed] [Google Scholar]
- 86.Jin M-Z, Jin W-L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther. 2020;5(1):166 doi: 10.1038/s41392-020-00280-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muir A, Danai LV, Vander Heiden MG. Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis Model Mech. 2018;11(8):dmm035758 doi: 10.1242/dmm.035758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinez-Outschoorn U, Sotgia F, Lisanti MP. Tumor microenvironment and metabolic synergy in breast cancers: critical importance of mitochondrial fuels and function. Semin Oncol. 2014;41(2):195–216. doi: 10.1053/j.seminoncol.2014.03.002 [DOI] [PubMed] [Google Scholar]
- 89.Serkova NJ, Gamito EJ, Jones RH, et al. The metabolites citrate, myo-inositol, and spermine are potential age-independent markers of prostate cancer in human expressed prostatic secretions. Prostate. 2008;68(6):620–628. doi: 10.1002/pros.20727 [DOI] [PubMed] [Google Scholar]
- 90.Galons J-P, Job C, Gillies RJ. Increase of GPC levels in cultured mammalian cells during acidosis. A 31P MR spectroscopy study using a continuous bioreactor system. Magn Reson Med. 1995;33(3):422–426. doi: 10.1002/mrm.1910330317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11(12):835–848. doi: 10.1038/nrc3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wijnen JP, Jiang L, Greenwood TR, van der Kemp WJM, Klomp DWJ, Glunde K. 1H/31P polarization transfer at 9.4 tesla for improved specificity of detecting phosphomonoesters and phosphodiesters in breast tumor models. PLoS ONE. 2014;9(7):e102256 doi: 10.1371/journal.pone.0102256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klomp DW, van de Bank BL, Raaijmakers A, et al. 31P MRSI and 1H MRS at 7 T: initial results in human breast cancer. NMR Biomed. 2011;24(10):1337–1342. doi: 10.1002/nbm.1696 [DOI] [PubMed] [Google Scholar]
- 94.Meisamy S, Bolan PJ, Baker EH, et al. Adding in vivo quantitative 1H MR spectroscopy to improve diagnostic accuracy of breast MR imaging: preliminary results of observer performance study at 4.0 T. Radiology. 2005;236(2):465–475. doi: 10.1148/radiol.2362040836 [DOI] [PubMed] [Google Scholar]
- 95.Haddadin IS, McIntosh A, Meisamy S, et al. Metabolite quantification and high-field MRS in breast cancer. NMR Biomed. 2009;22(1):65–76. doi: 10.1002/nbm.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meisamy S, Bolan PJ, Baker EH, et al. Neoadjuvant chemotherapy of locally advanced breast cancer: predicting response with in vivo 1H MR spectroscopy—a pilot study at 4 T. Radiology. 2004;233(2):424–431. doi: 10.1148/radiol.2332031285 [DOI] [PubMed] [Google Scholar]
- 97.de Molina AR, Sarmentero-Estrada J, Belda-Iniesta C, et al. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8(10):889–897. doi: 10.1016/S1470-2045(07)70279-6 [DOI] [PubMed] [Google Scholar]
- 98.Miyake T, Parsons SJ. Functional interactions between choline kinase α, epidermal growth factor receptor and c-Src in breast cancer cell proliferation. Oncogene. 2012;31(11):1431–1441. doi: 10.1038/onc.2011.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ramírez de Molina A, Gutiérrez R, Ramos MA, et al. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002;21(27):4317–4322. doi: 10.1038/sj.onc.1205556 [DOI] [PubMed] [Google Scholar]
- 100.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65(23):11034–11043. doi: 10.1158/0008-5472.CAN-05-1807 [DOI] [PubMed] [Google Scholar]
- 101.Tunset HM, Feuerherm AJ, Selvik L-KM, Johansen B, Moestue SA. Cytosolic phospholipase A2 alpha regulates TLR signaling and migration in metastatic 4T1 cells. Int J Mol Sci. 2019;20(19):4800 doi: 10.3390/ijms20194800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gadiya M, Mori N, Cao MD, et al. Phospholipase D1 and choline kinase-α are interactive targets in breast cancer. Cancer Biol Ther. 2014;15(5):593–601. doi: 10.4161/cbt.28165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Y, Rodrik V, Foster DA. Alternative phospholipase D/mTOR survival signal in human breast cancer cells. Oncogene. 2005;24(4):672–679. doi: 10.1038/sj.onc.1208099 [DOI] [PubMed] [Google Scholar]
- 104.Iorio E, Ricci A, Bagnoli M, et al. Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer Res. 2010;70(5):2126–2135. doi: 10.1158/0008-5472.CAN-09-3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stewart JD, Marchan R, Lesjak MS, et al. Choline-releasing glycerophosphodiesterase EDI3 drives tumor cell migration and metastasis. Proc Natl Acad Sci U S A. 2012;109(21):8155–8160. doi: 10.1073/pnas.1117654109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beloueche-Babari M, Jackson LE, Al-Saffar NMS, Workman P, Leach MO, Ronen SM. Magnetic resonance spectroscopy monitoring of mitogen-activated protein kinase signaling inhibition. Cancer Res. 2005;65(8):3356–3363. doi: 10.1158/10.1158/0008-5472.CAN-03-2981 [DOI] [PubMed] [Google Scholar]
- 107.Amstalden van Hove ER, Blackwell TR, Klinkert I, Eijkel GB, Heeren RMA, Glunde K. Multimodal mass spectrometric imaging of small molecules reveals distinct spatio-molecular signatures in differentially metastatic breast tumor models. Cancer Res. 2010;70(22):9012–9021. doi: 10.1158/0008-5472.CAN-10-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Negendank W, Li CW, Padavic-Shaller K, Murphy-Boesch J, Brown TR. Phospholipid metabolites in 1H-decoupled 31P MRS in vivo in human cancer: implications for experimental models and clinical studies. Anticancer Res. 1996;16(3b):1539–1544. [PubMed] [Google Scholar]
- 109.Shah T, Krishnamachary B, Wildes F, Wijnen JP, Glunde K, Bhujwalla ZM. Molecular causes of elevated phosphoethanolamine in breast and pancreatic cancer cells. NMR Biomed. 2018;31(8):e3936 doi: 10.1002/nbm.3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kremmer T, Pälyi I, Daubner D, et al. Comparative studies on the polyamine metabolism and DFMO treatment of MCF-7 and MDA-MB-231 breast cancer cell lines and xenografts. Anticancer Res. 1991;11(5):1807–1813. [PubMed] [Google Scholar]
- 111.Davidson NE, Hahm HA, McCloskey DE, Woster PM, Casero RA. Clinical aspects of cell death in breast cancer: the polyamine pathway as a new target for treatment. Endocr Relat Cancer. 1999;6(1):69–73. doi: 10.1677/erc.0.0060069 [DOI] [PubMed] [Google Scholar]
- 112.Spur EM, Decelle EA, Cheng LL. Metabolomic imaging of prostate cancer with magnetic resonance spectroscopy and mass spectrometry. Eur J Nucl Med Mol Imaging. 2013;40(Suppl 1):S60–S71. doi: 10.1007/s00259-013-2379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dinges SS, Vandergrift LA, Wu S, et al. Metabolomic prostate cancer fields in HRMAS MRS-profiled histologically benign tissue vary with cancer status and distance from cancer. NMR Biomed. 2019;32(10):e4038 doi: 10.1002/nbm.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bharti S, Jaiswal V, Ghoshal U, et al. Metabolomic profiling of amoebic and pyogenic liver abscesses: an in vitro NMR study. Metabolomics. 2011;8(4):540–555. doi: 10.1007/s11306-011-0339-7 [DOI] [Google Scholar]
- 115.Bollard ME, Contel NR, Ebbels TMD, et al. NMR-based metabolic profiling identifies biomarkers of liver regeneration following partial hepatectomy in the rat. J Proteome Res. 2009;9(1):59–69. doi: 10.1021/pr900200v [DOI] [PubMed] [Google Scholar]
- 116.Duarte IF, Stanley EG, Holmes E, et al. Metabolic assessment of human liver transplants from biopsy samples at the donor and recipient stages using high-resolution magic angle spinning 1H NMR spectroscopy. Anal Chem. 2005;77(17):5570–5578. doi: 10.1021/ac050455c [DOI] [PubMed] [Google Scholar]
- 117.Embade N, Mariño Z, Diercks T, et al. Metabolic characterization of advanced liver fibrosis in HCV patients as studied by serum 1H-NMR spectroscopy. PLoS ONE. 2016;11(5):e0155094 doi: 10.1371/journal.pone.0155094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Genangeli M, Heijens AMM, Rustichelli A, et al. MALDI-mass spectrometry imaging to investigate lipid and bile acid modifications caused by lentil extract used as a potential hypocholesterolemic treatment. J Am Soc Mass Spectrom. 2019;30(10):2041–2050. doi: 10.1007/s13361-019-02265-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kampa JM, Sahin M, Slopianka M, et al. Mass spectrometry imaging reveals lipid upregulation and bile acid changes indicating amitriptyline induced steatosis in a rat model. Toxicol Lett. 2020;325:43–50. doi: 10.1016/j.toxlet.2020.02.007 [DOI] [PubMed] [Google Scholar]
- 120.ščupáková K, Soons Z, Ertaylan G, et al. Spatial systems lipidomics reveals nonalcoholic fatty liver disease heterogeneity. Anal Chem. 2018;90(8):5130–5138. doi: 10.1021/acs.analchem.7b05215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tripathi P, Bala L, Saxena R, Yachha SK, Roy R, Khetrapal CL. 1H NMR spectroscopic study of blood serum for the assessment of liver function in liver transplant patients. J Gastrointest Liver Dis. 2009;18(3):329–336. [PubMed] [Google Scholar]
- 122.Martinez-Granados B, Morales JM, Rodrigo JM, et al. Metabolic profile of chronic liver disease by NMR spectroscopy of human biopsies. Int J Mol Med. 2011;27(1):111–117. doi: 10.3892/ijmm.2010.563 [DOI] [PubMed] [Google Scholar]
- 123.Waters NJ, Holmes E, Waterfield CJ, Farrant RD, Nicholson JK. NMR and pattern recognition studies on liver extracts and intact livers from rats treated with α-naphthylisothiocyanate. Biochem Pharmacol. 2002;64(1):67–77. doi: 10.1016/s0006-2952(02)01016-x [DOI] [PubMed] [Google Scholar]
- 124.Garg M, Misra MK, Chawla S, Prasad KN, Roy R, Gupta RK. Broad identification of bacterial type from pus by 1H MR spectroscopy. Eur J Clin Invest. 2003;33(6):518–524. doi: 10.1046/j.1365-2362.2003.01148.x [DOI] [PubMed] [Google Scholar]
- 125.Halouska S, Fenton RJ, Barletta RG, Powers R. Predicting the in vivo mechanism of action for drug leads using NMR metabolomics. ACS Chem Biol. 2012;7(1):166–171. doi: 10.1021/cb200348m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.ščupáková K, Balluff B, Tressler C, et al. Cellular resolution in clinical MALDI mass spectrometry imaging: the latest advancements and current challenges. Clin Chem Lab Med. 2020;58(6):914–929. doi: 10.1515/cclm-2019-0858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011;3(21):2427–2441. doi: 10.4155/bio.11.232 [DOI] [PubMed] [Google Scholar]
- 128.Rubakhin SS, Jurchen JC, Monroe EB, Sweedler JV. Imaging mass spectrometry: fundamentals and applications to drug discovery. Drug Discov Today. 2005;10(12):823–837. doi: 10.1016/S1359-6446(05)03458-6 [DOI] [PubMed] [Google Scholar]
- 129.Moser E, Laistler E, Schmitt F, Kontaxis G. Ultra-high field NMR and MRI—the role of magnet technology to increase sensitivity and specificity. Front Phys. 2017;5:33: doi: 10.3389/fphy.2017.00033 [DOI] [Google Scholar]
- 130.Spraggins JM, Djambazova KV, Rivera ES, et al. High-performance molecular imaging with MALDI trapped ion-mobility time-of-flight (timsTOF) mass spectrometry. Anal Chem. 2019;91(22):14552–14560. doi: 10.1021/acs.analchem.9b03612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Niehaus M, Soltwisch J, Belov ME, Dreisewerd K. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat Methods. 2019;16(9):925–931. doi: 10.1038/s41592-019-0536-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


