Abstract
N6-Methyladenosine (m6A) and its reader, writer, and eraser (RWE) proteins assume crucial roles in regulating the splicing, stability, and translation of mRNA. Aside from m6A, RNA is known to carry many other types of chemical modifications; no systematic investigations, however, have been conducted about the crosstalk between m6A and other modified nucleosides in RNA. Here, we modified our recently established liquid chromatography-parallel-reaction monitoring (LC-PRM) method by incorporating stable isotope-labeled (SIL) peptides as internal or surrogate standards for profiling epitranscriptomic RWE proteins. We were able to detect reproducibly a total of 114 RWE proteins in HEK293T cells with the genes encoding m6A eraser proteins (i.e., ALKBH5, FTO) and the catalytic subunit of the major m6A writer complex (i.e., METTL3) being individually ablated. Notably, eight proteins, including writer proteins for 5-methylcytidine and pseudouridine, were altered by more than 1.5-fold in the opposite directions in HEK293T cells depleted of METTL3 and ALKBH5. Analysis of previously published m6A mapping results revealed the presence of m6A in the corresponding mRNAs for four of these proteins. Together, we integrated SIL peptides into our LC-PRM method for quantifying epitranscriptomic RWE proteins, and our work revealed potential crosstalks between m6A and other epitranscriptomic modifications. Our modified LC-PRM method with the use of SIL peptides should be applicable for high-throughput profiling of epitranscriptomic RWE proteins in other cell types and in tissues.
Graphical Abstract
There has been a surging interest in the field of epitranscriptomics in recent years. N6-Methyladenosine (m6A) in mRNA was first identified in mouse L cells in 1974.1 Recent pioneering work about m6A included transcriptome-wide mapping of m6A,2,3 as well as the discoveries of METTL3 as the catalytic subunit of the major m6A methyltransferase (i.e., writer),4 FTO and ALKBH5 as m6A demethylases (erasers),5,6 and YTHDF family proteins as m6A-binding proteins (readers).7,8 These reader, writer, and eraser (RWE) proteins of m6A assume important roles in modulating the splicing,9 stability,8,10 and translation efficiencies of mRNA.7,11,12
Apart from m6A, the biological functions of N1-methyladenosine (m1A), 5-methylcytidine (m5C), and pseudouridine (Ψ) have also been investigated. In this vein, m1A regulates RNA folding and stability, ribosome biosynthesis, and translation.13–15 m5C modulates the export,16 stability,17 and translation of mRNA.18 Ψ, primarily located in tRNA and rRNA,19 is the most abundant internal modification in cellular RNA, and it affects RNA structure and translation.20,21 Moreover, ribosomes can read through Ψ in the stop codon via unusual base-pairing with tRNA, thereby altering mRNA coding.22
Several recent studies revealed interplays between m6A and other RNA modifications, although the underlying mechanisms remain poorly investigated. For instance, METTL3/METTL14 and NSUN2, which are the core subunits of the m6A writer complex and m5C writer, respectively, act on the 3′ untranslated region (3′-UTR) of p21 mRNA to synergistically enhance its expression.23 YTHDF2, an m6A reader protein, is also capable of binding directly with m5C and m1A in RNA, albeit at lower affinities than that toward m6A.24,25 Moreover, FTO, an eraser of m6A and m6Am,5,26 can also demethylate m1A in tRNA.27 Despite the above-described studies, there has been no systematic investigation about the potential crosstalk between m6A and other RNA modifications.
To fill in the above knowledge gap, we set out to examine how the expression levels of epitranscriptomic RWE proteins are perturbed by genetic depletion of m6A writer and eraser proteins. To this end, we first modified our recently developed LC-PRM method28 by employing a mixture of 48 stable isotope-labeled (SIL) peptides representing 45 RWE proteins in the PRM library as internal standards or surrogate standards (Table S1a). These 45 proteins were chosen based on existing knowledge about RWE proteins and their significances. We also employed this modified method for high-throughput profiling of a total of 152 epitranscriptomic RWE proteins in HEK293T cells and the isogenic cells with the catalytic subunit of the m6A writer complex (i.e., METTL3) and m6A eraser proteins (i.e., ALKBH5 and FTO) being genetically ablated (Figure 1a and Figure S1). We employed HEK293T cells for the experiment because we previously knocked out ALKBH5, FTO, and METTL3 genes in this cell line.29 We chose to focus on these three proteins because ALKBH5 and FTO are the only known erasers of m6A, and METTL3 is the catalytic subunit of the major m6A writer complex. While METTL16 was the other known m6A writer,30 we attempted, but failed, to ablate this gene in HEK293T cells using CRISPR-Cas9, probably because METTL16 is essential for the survival of HEK293T cells.
Figure 1.
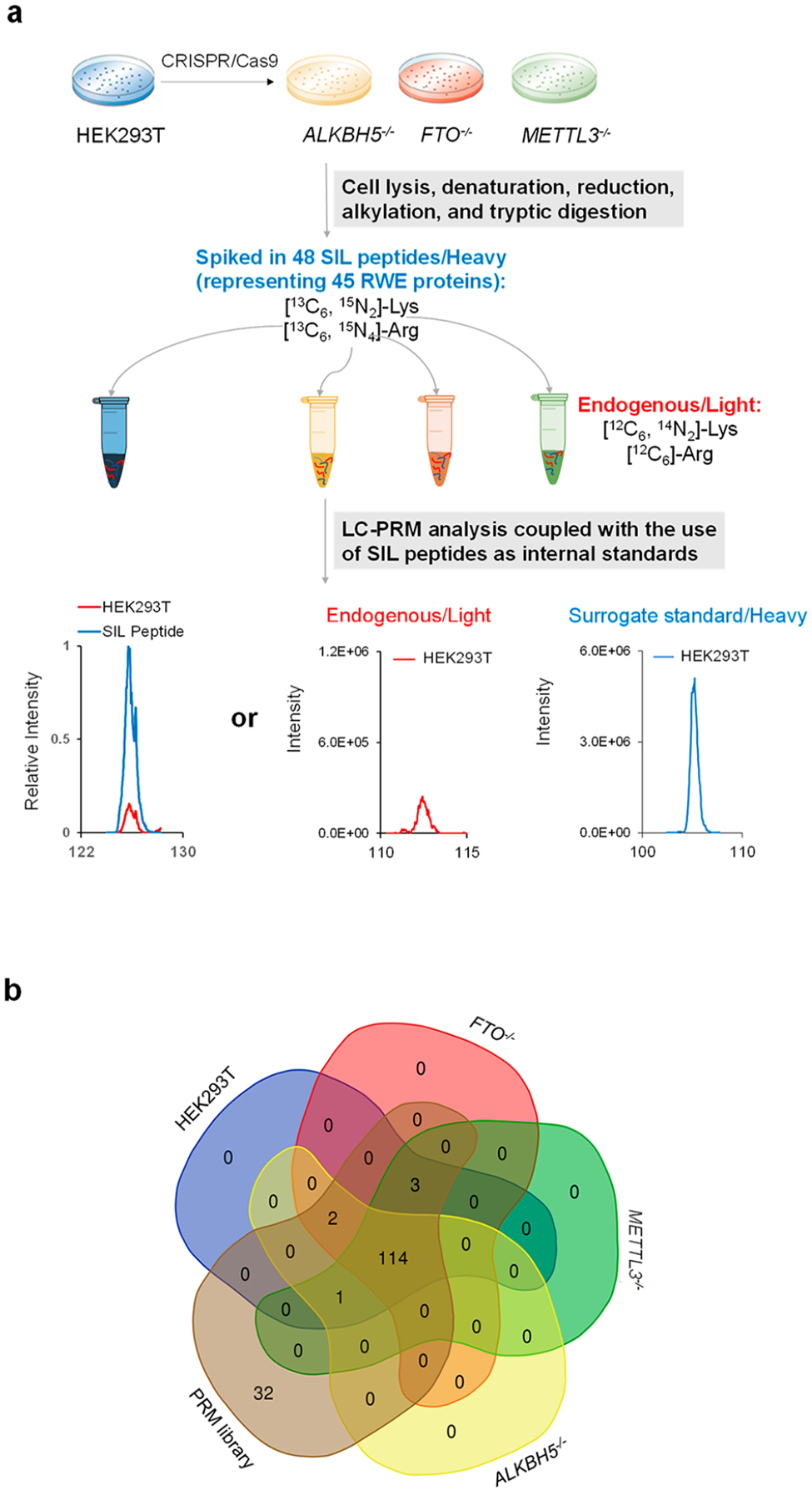
LC-PRM method for uncovering alterations in expression of epitranscriptomic RWE proteins elicited by genetic ablations of m6A writer and eraser proteins. (a) Workflow of LC-PRM analysis coupled with the use of SIL peptides as internal or surrogate standards for profiling epitranscriptomic RWE proteins in HEK293T and the isogenic ALKBH5−/−, FTO−/−, and METTL3−/− cells. (b) A Venn diagram depicting the numbers of quantified RWE proteins in HEK293T, ALKBH5−/−, FTO−/−, and METTL3−/− cells, compared with those deposited in the PRM library. The Venn diagram was designed using the webtool at https://bioinformatics.psb.ugent.be/webtools/Venn/.
We quantified those peptides with the SIL internal standards based on their peak areas relative to those of the corresponding SIL peptides. Those peptides without SIL internal standards were quantified from their peak areas relative to those of the surrogate standards, which were selected based on similar elution times as those of the target peptides.
By using the LC-PRM method together with the use of SIL peptides, we were able to quantify the relative expression levels of 117, 119, and 118 RWE proteins in ALKBH5, FTO, and METTL3 knockout cells with respect to parental HEK293T cells, which represent approximately 78% of the proteins in the PRM library (Figure 1b and Table S1b, c). Figure 2 displays the results from hierarchical clustering analysis of the log2-transformed LC-PRM quantification results for these RWE proteins in ALKBH5−/−, FTO−/−, and METTL3−/− cells relative to parental HEK293T cells. In this vein, a positive peptide identification entails dot product (dotp) value for its fragment ions observed in MS/MS being greater than 0.7 and 4−6 transitions sharing the same retention time. In addition, for those peptides with SIL internal standards, the analytes and the corresponding SIL internal standards must exhibit the same elution time.
Figure 2.
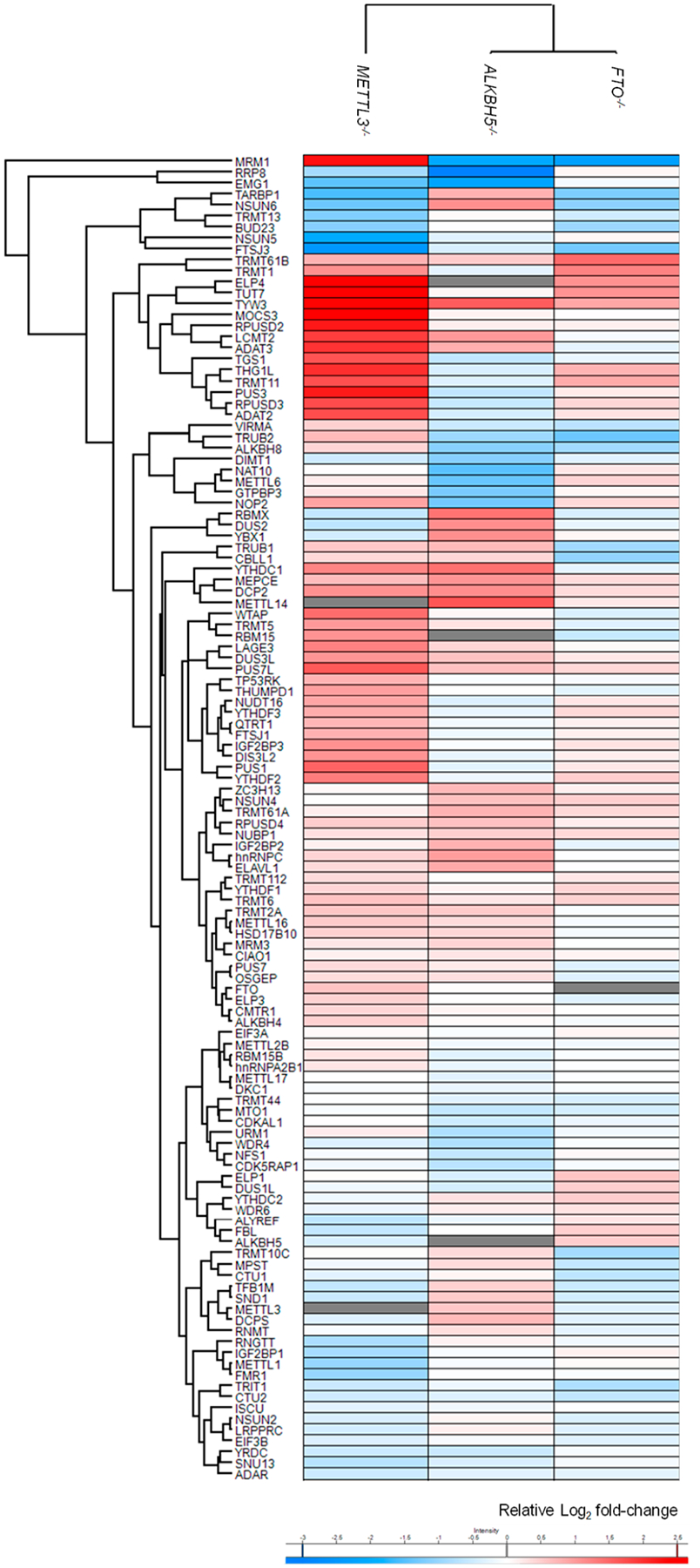
Hierarchical clustering illustrating the log2-transformed expression ratios of RWE proteins in ALKBH5−/− (n = 3), FTO−/− (n = 3), and METTL3−/− (n = 2) cells relative to parental HEK293T cells (n = 3). Hierarchical clustering was generated using Perseus, where red and blue boxes designate up- and down-regulated RWE proteins in the knockout compared to parental HEK293T cells. White boxes illustrate no substantial differences in expression of the corresponding proteins between knockout and HEK293T cells. Gray boxes indicate missing data.
The modified LC-PRM method, coupled with the use of SIL peptides, is efficient, robust, reproducible, and accurate. Compared with SILAC, the utilization of SIL peptides obviates the need of metabolic labeling. In addition, the LC-PRM quantification results of each peptide from different biological replicates of HEK293T and the isogenic ALKBH5, FTO, and METTL3 knockout cells displayed a mean relative standard deviation (RSD) of 12.7% for peptides quantified based on their corresponding SIL internal standards (Table S1d, e) and 16.1% for peptides quantified on the basis of surrogate standards (Table S1f, g). These results demonstrate very good reproducibility of the modified LC-PRM method. It is worth noting that, while the use of SIL surrogate peptides allow for relative quantification of epitranscriptomic RWE proteins, their absolute quantification requires the use of purified SIL peptides as internal standards.
We also examined the quantification accuracy of this approach by conducting Western blot analyses for three proteins, NSUN6, NOP2, and PUS3. We chose these three proteins based on their differential expression in HEK293T and at least one of the knockout backgrounds and their important functions as epitranscriptomic writer proteins. We found that LC-PRM and Western blot analyses yielded consistent quantification results for NSUN6 and NOP2 proteins (Figure 3). On the other hand, our PRM results showed that PUS3 is down-regulated in ALKBH5−/− over parental HEK293T cells, whereas Western blot revealed an up-regulation of the protein in the knockout background. The different quantification results of PUS3 obtained from Western blot and PRM may originate from difference(s) in post-translational modifications of the protein in the two genetic backgrounds, which may affect peptide detection by LC-PRM and/or antigen recognition by the antibody employed in Western blot analysis. In addition, inadequate specificity of the primary antibody used in Western blot analysis may also contribute to the difference.
Figure 3.
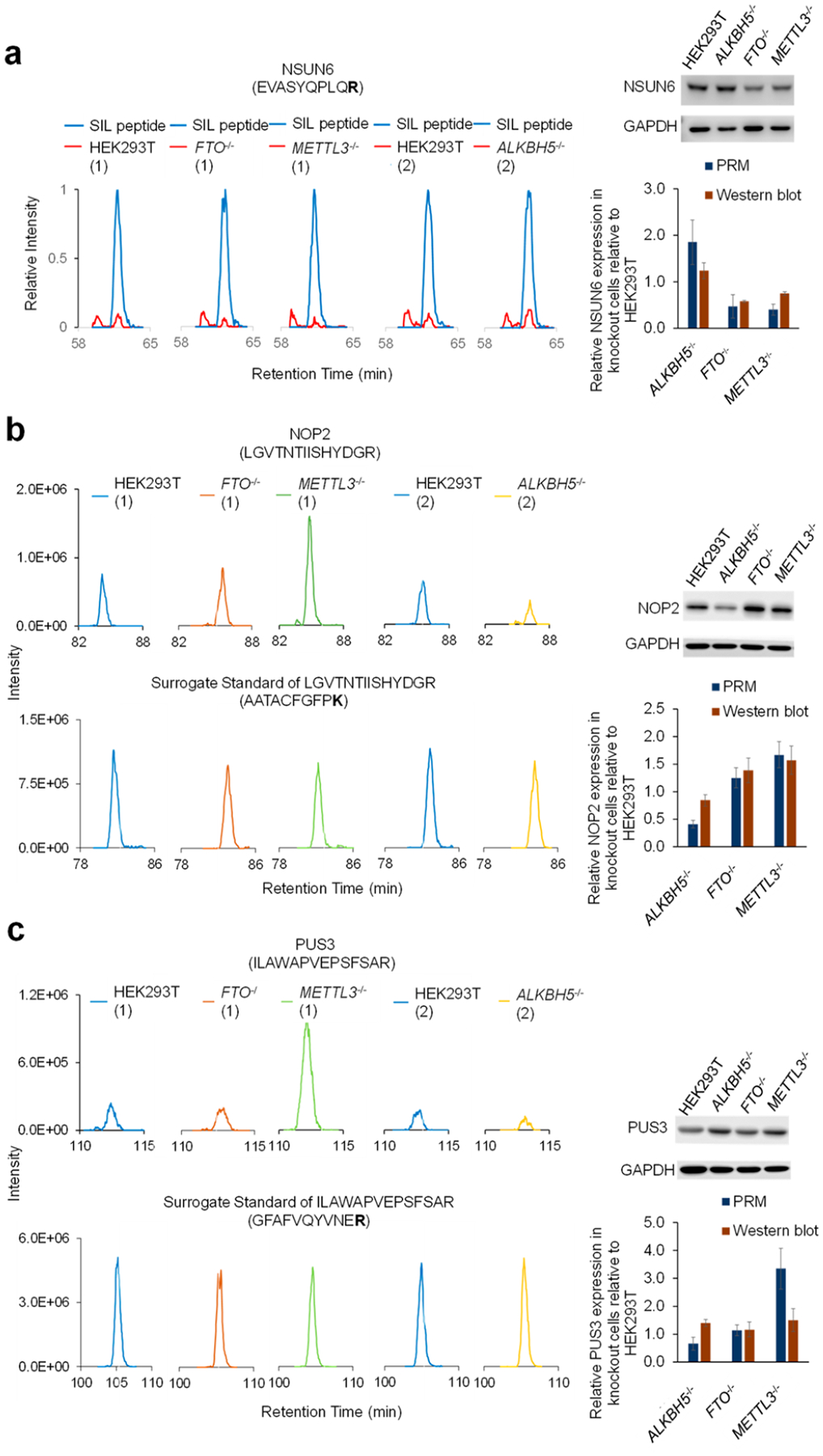
Extracted-ion chromatograms of tryptic peptides EVASYQPLQR from NSUN6 and its corresponding SIL peptide (a), LGVTNTIISHYDGR from NOP2 and its surrogate standard AATACFGFPK (b), ILAWAPVEPSFSAR from PUS3 and its surrogate standard GFAFVQYVNER (c). Shown on the right are the Western blot results (n = 3) of NSUN6 (a), NOP2 (b), and PUS3 (c) proteins in ALKBH5−/−, FTO−/−, and METTL3−/− cells relative to parental HEK293T cells. The proteomic experiments were conducted in two batches, where we assessed the relative expression levels of epitranscriptomic RWE proteins in HEK293T and the isogenic FTO−/− and METTL3−/− cells in the first batch (1) and those in HEK293T and the isogenic ALKBH5−/− cells in the second batch (2).
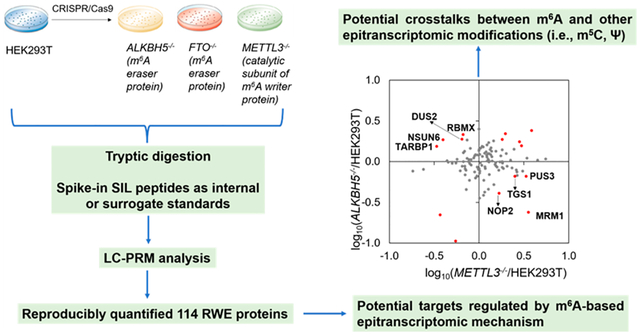
We next sought to identify potential targets regulated by m6A-based epitranscriptomic mechanism(s). RWE proteins with altered expressions of over 1.5-fold in individual knockout backgrounds (i.e., ALKBH5−/−, FTO−/−, or METTL3−/−) relative to parental HEK293T cells are illustrated in Figure 4a. Notably, when compared with parental HEK293T cells, many more proteins exhibit differential expressions in METTL3−/− than in ALKBH5−/− and FTO−/− cells. Among these differentially expressed RWE proteins, four (MRM1, PUS3, NOP2, and TGS1) were down- and up-regulated by at least 1.5-fold in ALKBH5−/− and METTL3−/− cells, respectively, relative to parental HEK293T cells (Figure 4b, left panel). This result suggests that m6A deposited by METTL3 and/or removed by ALKBH5 may promote the decay of mRNA encoding these proteins. Another four RWE proteins (DUS2, TARBP1, NSUN6, and RBMX) were down-regulated by at least 1.5-fold in METTL3−/− cells relative to parental HEK293T (WT) cells, which is associated with their marked up-regulation (by at least 1.5-fold) in ALKBH5−/− over parental HEK293T cells (Figure 4b, left panel). This result indicates that the relevant m6A installed by METTL3 and removed by ALKBH5 in the mRNAs of these genes may increase the stability and/or translation efficiency of these mRNAs. Together, eight RWE proteins, namely, MRM1, PUS3, NOP2, TGS1, DUS2, TARBP1, NSUN6, and RBMX, displayed opposite trends in expression levels in ALKBH5−/− and METTL3−/− cells relative to the isogenic parental HEK293T cells, suggesting that their corresponding mRNAs may be subjected to regulation by an m6A-mediated epitranscriptomic mechanism.
Figure 4.
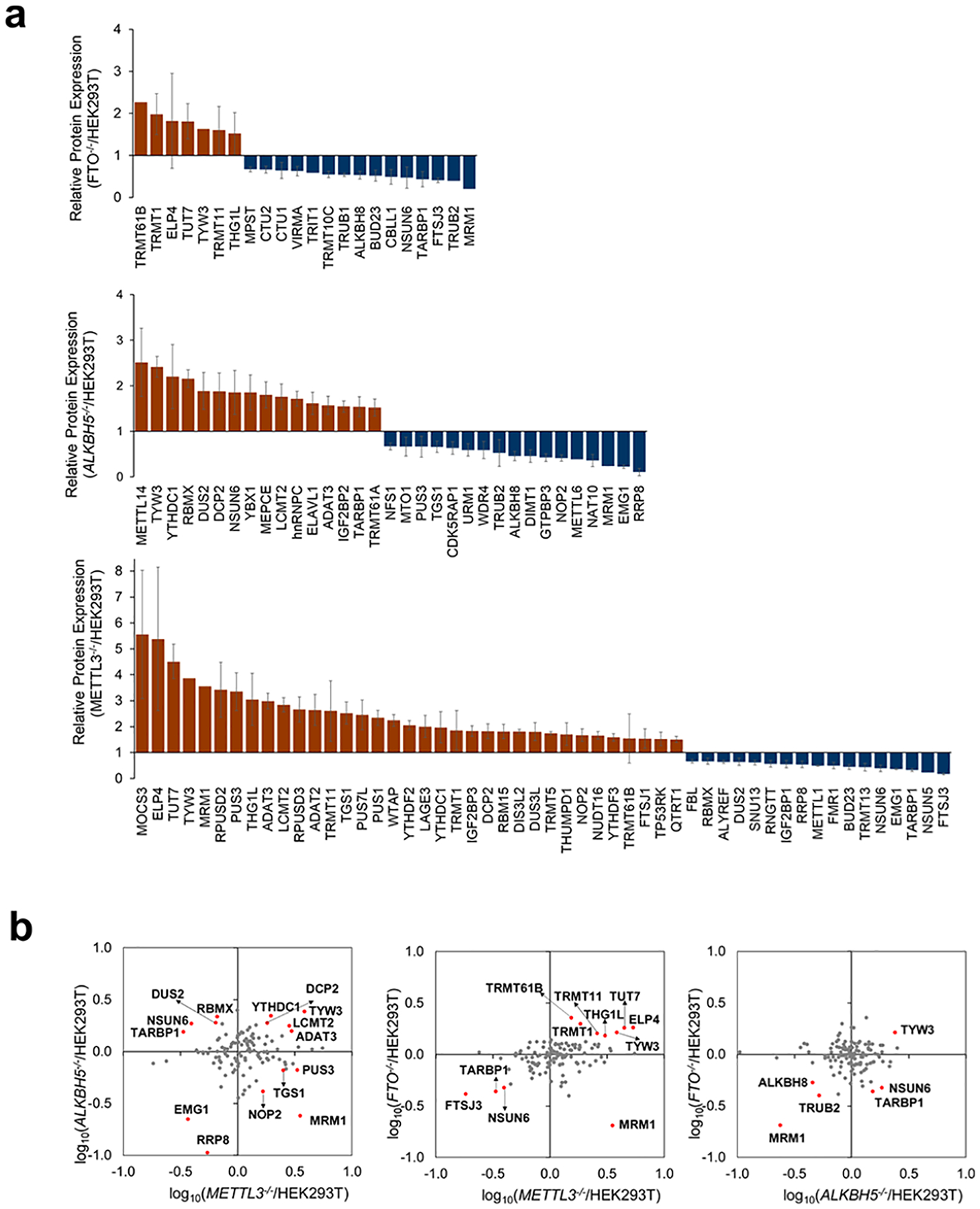
(a) LC-PRM quantification results of RWE proteins in ALKBH5−/− (n = 3), FTO−/− (n = 3), and METTL3−/− (n = 2) cells relative to parental HEK293T cells (n = 3). Only proteins with ratios in knockout/parental cells being greater than 1.5 or less than 0.67 are displayed. The ratio of each peptide representing a specific RWE protein was determined following the procedures described in Materials and Methods in the Supporting Information. (b) Scatter plots depicting the LC-PRM quantification results of RWE proteins in one knockout over HEK293T cells vs another knockout over HEK293T cells. Those RWE proteins with expression fold changes being over 1.5-fold in the knockout cells vs HEK293T cells are labeled in red.
We next asked if these eight RWE proteins could be regulated through an m6A-based epitranscriptomic mechanism. We began by examining the presence of m6A in the mRNAs of these eight genes using a publicly available data set (GSE63753) on single-nucleotide resolution mapping of m6A in HEK293 cells.31 The mapping method is capitalized on UV cross-linking between anti-m6A antibody and m6A-modified mRNA and the resulting C → T mutation at the +1 position of the cross-linked m6A site, or a truncation at the m6A site, induced by reverse transcription.31 As shown in the integrative genomics viewer (IGV) plots, we observed the presence of m6A sites in the mRNA of NOP2, PUS3, TGS1, and RBMX genes in HEK293 cells (Figure S2). In particular, we found m6A sites in the 3′-untranslated region (3′-UTR) and the last exon of NOP2 mRNA and in the last exon, an internal exon, and the 3′-UTR of PUS3, TGS1, and RBMX mRNAs, respectively (Figure S2). The same data set, nevertheless, did not reveal the presence of m6A in the mRNAs of the other four genes in HEK293 cells; the exact reason is unclear, although this could be attributed to the lack of adequate sensitivity of the m6A mapping method.
It is worth noting that several epitranscriptomic RWE proteins exhibited markedly altered (by at least 1.5-fold) expressions in the same direction in ALKBH5−/− and METTL3−/− cells relative to parental HEK293T cells. This could be due to the actions of METTL3 and ALKBH5 on m6A at different sites in mRNA, the involvement of METTL1630 and/or FTO26 in the methylation and demethylation, respectively, of some m6A sites in the mRNAs encoding these proteins, and/or the involvement of different m6A reader proteins in conferring distinct effects on mRNA decay or translation efficiency in ALKBH5−/− and METTL3−/− cells.
Our aforementioned quantitative proteomic data showed that genetic ablations of METTL3 resulted in augmented expression of NOP2, PUS3, and TGS1 proteins, which are accompanied by diminished expression of these proteins in ALKBH5−/− cells. In addition, m6A is present in the mRNA encoding these three proteins, suggesting the role of m6A in promoting the degradation of mRNA encoding these proteins. Hence, we next analyzed previously published photoactivatable ribonucleoside cross-linking and immunoprecipitation (PAR-CLIP)32 data to explore if NOP2, PUS3, and TGS1 are regulated by a m6A reader protein YTHDF2, which is known to stimulate the degradation of mRNA through binding with m6A.8,10 The results showed that transcripts of NOP2 and TGS1, but not PUS3, are YTHDF2 binding targets.
Aside from the aforementioned PUS3, our LC-PRM results showed that other members of the PUS family, namely, PUS1, TRUB1, PUS7, and PUS7L, were increased in METTL3−/− cells by 2.34-, 1.36-, 1.23-, and 2.46-folds, respectively (Figure S3a). Interestingly, by analyzing GSE63753, we identified m6A sites in the next-to-last exon of PUS1, in the last exon of TRUB1, in the 3′UTR of PUS7, and near the stop codon of PUS7L (Figure S3). PUS1 catalyzes the formation of Ψ from uridine at positions 27/28 in the anticodon stem-loop of some tRNAs and at positions 34/36 in intron-containing tRNAs.33,34 PUS3, TRUB1, and PUS7 catalyze Ψ formation in some tRNAs at positions 38/39, 55, and 13, respectively.35–38 Not much is known about the function of PUS7L, where a previous study indicated that it may target positions 13 and/or 35 in tRNAs.39 These results suggest a broad role of METTL3 in modulating ψ biosynthesis in human cells.
We also examined those proteins that are regulated in opposite directions by METTL3 and FTO, another eraser protein of m6A.5 The results also illustrated that MRM1 was down-regulated in FTO−/− cells by more than 1.5-fold and up-regulated in METTL3−/− cells by over 1.5-fold (Figure 4b, middle panel). These findings suggest that reversible methylation at the N6 position of adenosine in the mRNAs of these genes, mediated by METTL3 and FTO, may also modulate the stabilities and translation efficiencies of these mRNAs. Moreover, the expression fold changes of TYW3, ALKBH8, TRUB2, and MRM1 were both up- or down-regulated by over 1.5-fold in FTO−/− and ALKBH5−/− cells relative to parental HEK293T cells (Figure 4b, right panel).
In summary, we modified our recently developed LC-PRM method by incorporating SIL peptides as internal or surrogate standards. By using this modified targeted proteomic method, we were able to commonly quantify 114 RWE proteins, representing 75% of the RWE proteome in the PRM library, in ALKBH5−/−, FTO−/−, METTL3−/− cells, relative to their parental HEK293T cells. NOP2, PUS3, and TGS1 were up-regulated in METTL3−/− cells by over 1.5-fold and down-regulated in ALKBH5−/− cells by at least 0.67-fold, compared with the isogenic parental HEK293T cells. In addition, analysis of previously published m6A mapping results revealed the presence of m6A in mRNA of NOP2, PUS3, TGS1, and RBMX genes. It will be important to examine if the up-regulation of NOP2, PUS3, and TGS1 and down-regulation of RBMX in METTL3−/− cells arise from altered mRNA stabilities and/or the binding of m6A reader proteins, for example, YTHDF1, YTHDF2, and YTHDF3. In this regard, it will also be important to examine how genetic depletion of these m6A reader proteins affects the expression of epitranscriptomic RWE proteins at the proteome-wide scale. Together, we modified our LC-PRM method by employing SIL peptides as internal or surrogate standards, and the method should be applicable for assessing quantitatively the expression levels of epitranscriptomic RWE proteins in tissue samples. The sample preparation workflow for cellular samples reported herein should be readily amenable to tissue samples except that the latter involves an additional step of tissue homogenization. Moreover, our results revealed potential crosstalks between m6A and other RNA modifications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R35 ES031707).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.2c03549.
Experimental procedures, LC-PRM results, and m6A occupancy in transcripts of relevant epitranscriptomic RWE proteins based on previously reported m6A mapping results (PDF)
Table S1: List of SIL peptides used in this study and LC-PRM quantification results (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.2c03549
The authors declare no competing financial interest.
Contributor Information
Tianyu F. Qi, Environmental Toxicology Graduate Program, University of California Riverside, Riverside, California 92521-0403, United States
Xiaochuan Liu, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Feng Tang, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Jiekai Yin, Environmental Toxicology Graduate Program, University of California Riverside, Riverside, California 92521-0403, United States.
Kailin Yu, Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Yinsheng Wang, Environmental Toxicology Graduate Program and Department of Chemistry, University of California Riverside, Riverside, California 92521-0403, United States.
Data Availability Statement
All the LC-MS/MS raw files and Skyline PRM library were deposited to the ProteomeXchange Consortium via the PRIDE40 partner repository with the data set identifier PXD036032.
REFERENCES
- (1).Perry RP; Kelley DE Cell 1974, 1, 37–42. [Google Scholar]
- (2).Dominissini D; Moshitch-Moshkovitz S; Schwartz S; Salmon-Divon M; Ungar L; Osenberg S; Cesarkas K; Jacob-Hirsch J; Amariglio N; Kupiec M; Sorek R; Rechavi G Nature 2012, 485, 201–6. [DOI] [PubMed] [Google Scholar]
- (3).Meyer KD; Saletore Y; Zumbo P; Elemento O; Mason CE; Jaffrey SR Cell 2012, 149, 1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bokar JA; Shambaugh ME; Polayes D; Matera AG; Rottman FM RNA 1997, 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- (5).Jia G; Fu Y; Zhao X; Dai Q; Zheng G; Yang Y; Yi C; Lindahl T; Pan T; Yang Y-G; He C Nat. Chem. Biol 2011, 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zheng G; Dahl JA; Niu Y; Fedorcsak P; Huang CM; Li CJ; Vågbø CB; Shi Y; Wang WL; Song SH; Lu Z; Bosmans RP; Dai Q; Hao YJ; Yang X; Zhao WM; Tong WM; Wang XJ; Bogdan F; Furu K; Fu Y; Jia G; Zhao X; Liu J; Krokan HE; Klungland A; Yang YG; He C Mol. Cell 2013, 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wang X; Zhao BS; Roundtree IA; Lu Z; Han D; Ma H; Weng X; Chen K; Shi H; He C Cell 2015, 161, 1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wang X; Lu Z; Gomez A; Hon GC; Yue Y; Han D; Fu Y; Parisien M; Dai Q; Jia G; Ren B; Pan T; He C Nature 2014, 505, 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Xiao W; Adhikari S; Dahal U; Chen YS; Hao YJ; Sun BF; Sun HY; Li A; Ping XL; Lai WY; Wang X; Ma HL; Huang CM; Yang Y; Huang N; Jiang GB; Wang HL; Zhou Q; Wang XJ; Zhao YL; Yang YG Mol. Cell 2016, 61, 507–519. [DOI] [PubMed] [Google Scholar]
- (10).Zaccara S; Jaffrey SR Cell 2020, 181, 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Meyer KD; Patil DP; Zhou J; Zinoviev A; Skabkin MA; Elemento O; Pestova TV; Qian SB; Jaffrey SR Cell 2015, 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Choe J; Lin S; Zhang W; Liu Q; Wang L; Ramirez-Moya J; Du P; Kim W; Tang S; Sliz P; Santisteban P; George RE; Richards WG; Wong K-K; Locker N; Slack FJ; Gregory RI Nature 2018, 561, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Helm M; Giegé R; Florentz C Biochemistry 1999, 38, 13338–13346. [DOI] [PubMed] [Google Scholar]
- (14).Sharma S; Watzinger P; Kötter P; Entian KD Nucleic Acids Res. 2013, 41, 5428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dominissini D; Nachtergaele S; Moshitch-Moshkovitz S; Peer E; Kol N; Ben-Haim MS; Dai Q; Di Segni A; Salmon-Divon M; Clark WC; Zheng G; Pan T; Solomon O; Eyal E; Hershkovitz V; Han D; Doré LC; Amariglio N; Rechavi G; He C Nature 2016, 530, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yang X; Yang Y; Sun BF; Chen YS; Xu JW; Lai WY; Li A; Wang X; Bhattarai DP; Xiao W; Sun HY; Zhu Q; Ma HL; Adhikari S; Sun M; Hao YJ; Zhang B; Huang CM; Huang N; Jiang GB; Zhao YL; Wang HL; Sun YP; Yang YG Cell Res. 2017, 27, 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Yang Y; Wang L; Han X; Yang WL; Zhang M; Ma HL; Sun BF; Li A; Xia J; Chen J; Heng J; Wu B; Chen YS; Xu JW; Yang X; Yao H; Sun J; Lyu C; Wang HL; Huang Y; Sun YP; Zhao YL; Meng A; Ma J; Liu F; Yang YG Mol. Cell 2019, 75, 1188–1202. [DOI] [PubMed] [Google Scholar]
- (18).Tuorto F; Liebers R; Musch T; Schaefer M; Hofmann S; Kellner S; Frye M; Helm M; Stoecklin G; Lyko F Nat. Struct. Mol. Biol 2012, 19, 900–905. [DOI] [PubMed] [Google Scholar]
- (19).Zhao BS; Roundtree IA; He C Nat. Rev. Mol. Cell Biol 2017, 18, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Han L; Kon Y; Phizicky EM RNA 2015, 21, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jack K; Bellodi C; Landry DM; Niederer RO; Meskauskas A; Musalgaonkar S; Kopmar N; Krasnykh O; Dean AM; Thompson SR; Ruggero D; Dinman JD Mol. Cell 2011, 44, 660–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Fernández IS; Ng CL; Kelley AC; Wu G; Yu Y-T; Ramakrishnan V Nature 2013, 500, 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Li Q; Li X; Tang H; Jiang B; Dou Y; Gorospe M; Wang WJ Cell Biochem. 2017, 118, 2587–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Dai X; Wang T; Gonzalez G; Wang Y Anal. Chem 2018, 90, 6380–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dai X; Gonzalez G; Li L; Li J; You C; Miao W; Hu J; Fu L; Zhao Y; Li R; Li L; Chen X; Xu Y; Gu W; Wang Y Anal. Chem 2020, 92, 1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mauer J; Luo X; Blanjoie A; Jiao X; Grozhik AV; Patil DP; Linder B; Pickering BF; Vasseur JJ; Chen Q; Gross SS; Elemento O; Debart F; Kiledjian M; Jaffrey SR Nature 2017, 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wei J; Liu F; Lu Z; Fei Q; Ai Y; He PC; Shi H; Cui X; Su R; Klungland A; Jia G; Chen J; He C Mol. Cell 2018, 71, 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Qi TF; Miao W; Wang Y Anal. Chem 2022, 94, 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yang Y-Y; Yu K; Li L; Huang M; Wang Y Anal. Chem 2020, 92, 10145–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pendleton KE; Chen B; Liu K; Hunter OV; Xie Y; Tu BP; Conrad NK Cell 2017, 169, 824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Linder B; Grozhik AV; Olarerin-George AO; Meydan C; Mason CE; Jaffrey SR Nat. Methods 2015, 12, 767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hafner M; Landthaler M; Burger L; Khorshid M; Hausser J; Berninger P; Rothballer A; Ascano M Jr.; Jungkamp AC; Munschauer M; Ulrich A; Wardle GS; Dewell S; Zavolan M; Tuschl T Cell 2010, 141, 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wu W; Chen Y; d’Avignon A; Hazen SL Biochemistry 1999, 38, 3538–48. [DOI] [PubMed] [Google Scholar]
- (34).Motorin Y; Keith G; Simon C; Foiret D; Simos G; Hurt E; Grosjean H RNA 1998, 4, 856–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lecointe F; Simos G; Sauer A; Hurt EC; Motorin Y; Grosjean HJ Biol. Chem 1998, 273, 1316–23. [DOI] [PubMed] [Google Scholar]
- (36).Chen J; Patton JR Biochemistry 2000, 39, 12723–30. [DOI] [PubMed] [Google Scholar]
- (37).Zhang W; Pan T Nat. Chem. Biol 2020, 16, 107–108. [DOI] [PubMed] [Google Scholar]
- (38).Kaya Y; Ofengand J RNA 2003, 9, 711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).de Crécy-Lagard V; Boccaletto P; Mangleburg CG; Sharma P; Lowe TM; Leidel SA; Bujnicki JM Nucleic Acids Res. 2019, 47, 2143–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaino JA Nucleic Acids Res. 2019, 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the LC-MS/MS raw files and Skyline PRM library were deposited to the ProteomeXchange Consortium via the PRIDE40 partner repository with the data set identifier PXD036032.


