Abstract
The performance of an automated commercial CRISPR/Cas based technology was evaluated and compared with routine RT-PCR testing to diagnose COVID-19. Suspected and discharged COVID-19 cases were included and tested with CRISPR-based SARS-CoV-2 test and RT-PCR assay using throat swab and sputum specimens. The diagnostic yield was calculated and compared using the McNemar test. A total of 437 patients were included for analysis, including COVID-19 (n = 171), discharged cases (n = 155), and others (n = 111). For the diagnosis of COVID-19, the CRISPR-SARS-CoV-2 test had a sensitivity and specificity of 98.2% (168/171) and 100.0% (266/266), respectively; the RT-PCR test had a sensitivity and specificity of 100.0% (171/171) and 100.0% (266/266), respectively. No significant difference was found in the sensitivity of CRISPR-SARS-CoV-2 and RT-PCR. In conclusion, the CRISPR-SARS-CoV-2 test had a comparable performance with RT-PCR and showed several advantages, such as short assay time, low cost, and no requirement for expensive equipment.
Keywords: CRISPR, RT-PCR, Diagnosis
1. Introduction
Through December 2021, there were 620 million cases of Coronavirus disease (COVID-19) and 6.5 million deaths reported worldwide (https://covid19.who.int/accessed on Oct 16, 2022). Although vaccines are already available, the emergence and rapid transmission of new variants threaten attempts to protect against COVID-19 [1]. In addition, the efficiency of approved drugs (such as molnupiravir) remains to be elucidated [2]. These challenges serve as obstacles to the management of COVID-19. Thus, early diagnosis is helpful for timely interventions and improving health outcomes [[3], [4], [5]].
The host response produces biomarkers that can be used to identify individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [6,7]. For that purpose, assays detecting specific SARS-CoV-2 antigens and antibodies have been developed and are recommended to detect COVID-19 cases with symptom initiation and high viral load [8]. However, several disadvantages of these assays should be noted. For example, the standardization of performing such tests is required to improve diagnostic performance; the performance of antigen testing is highly related to viral load, with a low to modest sensitivity compared to reference nucleic acid amplification testing (NAAT) methods [9]. Serological assays for COVID-19 have similar obstacles in diagnostic performance. In addition, the cross-reactivity caused by vaccination presents another concern [10]. Therefore, further diagnostic development is required to improve testing sensitivity and specificity [11].
As the infectious agent for COVID-19, SARS-CoV-2 is an RNA virus, and thus all available RNA detection tools are applicable to detect the virus. Currently, RT-PCR or NAAT tools remain the first diagnostic methods for SARS-CoV-2 infection [9]. As screening tests, RT-PCR and RT-LAMP have an overall pooled sensitivity of 0.96 (0.93–0.98) and 0.92 (0.85–0.96), respectively [12]. However, false-positive results should be taken into consideration [[12], [13], [14]].
Recently, the clustered regularly interspaced short palindromic repeats (CRISPR) system has been introduced in the diagnosis of infectious diseases, such as tuberculosis [15], metapneumovirus [16], and malaria [17]. Such assays employ the trans cleavage activity of a Cas12/gRNA complex bound to an amplicon recognition sequence to cleave and derepress a quenched fluorescent oligonucleotide reporter that is present at a high concentration. Compared to PCR-based tests, CRISPR-associated proteins (Cas) diagnostic tests can be performed at physiological temperature (even room temperature), offer comparable sensitivity and specificity, can outperform PCR tests, do not require sophisticated equipment or well-established labs, and are less costly [18]. Therefore, the introduction of CRISPR-Cas could significantly impact the field of molecular diagnostics [19]. Indeed, there is a demand for rapid, accurate, and sensitive tests for COVID-19 diagnosis. Previously, several CRISPR methods have been developed for COVID-19 diagnosis. For example, the FnCas9 Editor Linked Uniform Detection Assay (FELUDA) was reported by Azhar et al., with a sensitivity of 100% and specificity of 97% [20]. Similar CRISPR methods has been developed and evaluated by Fasching et al. [21], Mahas et al. [22], and Liu et al. [23]. CRISPR-based assays can also detect single-copy targets [24] and distinguish targets with single base differences [25] to permit sensitive and accurate mapping of SARS-CoV-2 RNA changes in respiratory and non-respiratory samples. For example, a FnCas9-based CRISPR diagnostic was developed for rapid and accurate detection of major SARS-CoV-2 variants [26], and a CRISPR-Cas12a-empowered electrochemical biosensor was evaluated for rapid and ultrasensitive detection of the SARS-CoV-2 Delta variant [27].
In this prospective study, we aimed to assess the usefulness of an automated commercial CRISPR based technology for COVID-19 diagnosis. For a more comprehensive evaluation, routine methods, such as RT-PCR, were also performed and included for a direct comparison among cases with suspected COVID-19.
2. Materials and methods
The study protocol was approved by the Ethical Committee of Shanghai Public Health Clinical Center (No. 2021-E011-02) and Heilongjiang Center for Disease Control and Prevention (No. Hei-CDC 2020-09). All methods were carried out in accordance with relevant guidelines and regulations. Written informed consent was waived due to anonymous data collection, and samples were collected for routine activities.
2.1. Subjects
Cases with suspected COVID-19 between February 2021 and April 2021 were included and evaluated for further analysis. As a study group, COVID-19 cases were diagnosed clinically based on an exposure history of COVID-19 and corresponding symptoms of COVID-19 infection, or confirmed with NAAT, following the guideline issued by the Chinese National Health Commission [28]. The control group included cases with an alternative diagnosis or those discharged after two rounds of consecutively negative COVID-19 RT-PCR results [28].
2.2. Specimen collection
Throat swab and sputum specimens were either collected and sent for SARS-CoV-2 assays immediately, stored at 4 °C for 24 h and −70 °C for longer storage periods, or stored at −80 for analysis on the next day. Briefly, throat swab sampling was performed as follows: a swab (1) was inserted, reaching the posterior pharynx and tonsillar areas; (2) rotated against the tonsillar pillars and posterior oropharynx; and (3) placed into the transport tube and closed. Sputum collection was guided by a trained healthcare professional, and deep cough sputum was collected in a sterile container.
2.3. Principle
The CRISPR-based SARS-CoV-2 (CRISPR-SARS-CoV-2, BioGerm, Shanghai, China) test was performed using an automated system (BG-Nova-X8, BioGerm, Shanghai, China) and designed based on reverse transcription recombinase polymerase amplification (RT-RPA) and CRISPR Cas12a (orthologs from Lachnospiraceae bacterium; Tolo Biotech, Shanghai, China; hereinafter referred to as “LbCas12a”) -mediated detection (limit of detection, 800 copies/ml). The primers are listed in Table 1. The system consisted of an instrument (BG-Nova-X8) and disposable tubes that fully integrated RNA extraction, gene amplification, CRISPR reaction, and detection (Fig. 1A−C). The user simply added the sample to the tube and placed it in the apparatus. A result was provided 30 min later. The CRISPR-SARS-CoV-2 test was performed according to the manufacture's instructions. In parallel, samples were also tested using a RT-PCR kit (limit of detection, 500 copies/ml) purchased from BioGerm (Shanghai, China), which detects the ORF1ab gene and the N gene. The kit has a presion of <5%, and no significant influential evidence or cross reaction was observed for within-run, between-run, within-day, between-day, inter-class, intra-class, within lab, and between-lab precision parameters (more details listed in the instructions). In addition, the assay performance was validated using a SARS-CoV-2 certified reference material (GBW(E)091099, National Reference Material, China NMPA).
Table 1.
Primers for the CRISPR-based SARS-CoV-2 test.
| Upstream primer (5′-3′) | Downstream primer (5′-3′) | CRISPR RNA (5′-3′) | ssDNA | |
|---|---|---|---|---|
| N | N–F: ATGGAAGTCACACCTTCGGGAACGTGGTTG | N-R: GGCTCTGTTGGTGGGAATGTTTTGTATGCG | SARS-CoV2-N-CRISPR RNA: UAAUUUCUACUAAGUGUAGAUAAAGAUCAAGUCAUUUUGCUGAA | TTATT |
| RNP | Rnase P–F: GATCCGCAATAACTCAGCCATCCACATCCG | Rnase P-R: CCCACCAAGAGACAATTACCCCCACCCTCA | Rnase P- CRISPR RNA: AUUUCUACUAAGUGUAGAUAAUUACUUGGGUGUGACCCUGAA |
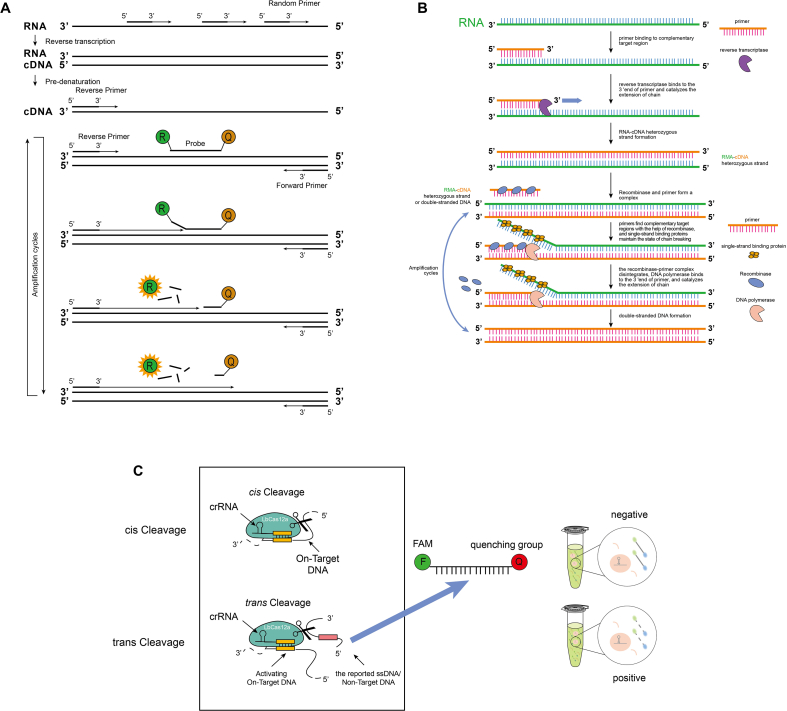
Fig. 1.
Schematics showing the principles of RT-PCR, RPA, and CRISPR/Cas12a detection systems. The commercial SARS-CoV-2 kit (BioGerm, Shanghai, China) is designed based on RPA and CRISPR. (A) RT-PCR: N gene target was amplified and evaluated using real-time Taqman PCR. (B) RPA: At 42 °C, RNA was first reverse transcribed to cDNA using reverse transcriptase. Recombinase proteins form complexes with each primer, which scan cDNA for homologous sequences. DNA polymerase uses designed primers to transcribe the complementary strand of cDNA and forms a double-stranded DNA (dsDNA) template. The recombinase/primer complex binds to dsDNA and promotes strand displacement by the primer at the cognate site, and single stranded binding proteins stabilize the displaced DNA chain. The recombinase disassembles and a strand displacing DNA polymerase binds to the 3′-end of the primers, which elongates the primer. The exponential amplification is then achieved by cyclic repetition of this process. (C) Detector assay: The target sequence is amplified by the RPA reaction, followed by mixing with the CRISPR/LbCas12a reagents for cleavage. LbCas12a utilizes the crRNA as guides to bind cognate DNA sequences. LbCas12a is then activated for on-target DNA cleavage (cis cleavage) and non-target single-stranded DNA cleavage (trans/collateral cleavage). The reported signal-stranded DNA (ssDNA) includes a fluorophore-quencher pair, and the cleavage events will free the fluorophore from its quencher, in effect activating fluorescence that can be measured.
RNA extraction: RNA extraction was carried out using superparamagnetic particles [29] adapted for automation on the platform. Briefly, samples were mixed with magnetic particles and ethanol, and the RNA was then captured by the particles [30]. Subsequently, the particles were immobilized on a magnet. After washing twice with 80% ethanol, the captured nucleic acids were eluted in 10 mM Tris, pH 7.
2.4. CRISPR-SARS-CoV-2 test
The mixture containing the sample, forward, and reverse primers targeting the SARS-CoV-2 N gene and human RNase P, magnesium acetate, and RPA was incubated together. RNA was reverse transcribed into cDNA, and cDNA was then amplified using PCR. The amplification product was then added to the CRISPR reaction mix consisting of CRISPR RNA (crRNA), LbCas12a, and the ssDNA reporter. Finally, the fluorescence signal was recorded and analyzed. According to the manufacturer's instruction, if the RNase P gene and N gene targets were detected, the specimen was considered positive. If the RNase P gene targets were positive and no amplification of the N gene target was found, the specimen was considered negative. Otherwise, an amplication control failure was obsvered and a repeated amplification or specimen collection was required for further analysis.
2.5. Statistical analysis
Sensitivity was calculated as the proportion of CRISPR-SARS-CoV-2-positive cases among the study group. Specificity was calculated as the proportion of CRISPR-SARS-CoV-2-negative cases among the control group. The McNemar test was used to compare the sensitivity of detection of SARS-CoV-2. Data analysis was performed using SPSS version 16.0 software. Statistical significance was indicated by a P-value less than 0.05.
3. Results
3.1. Baseline characteristics
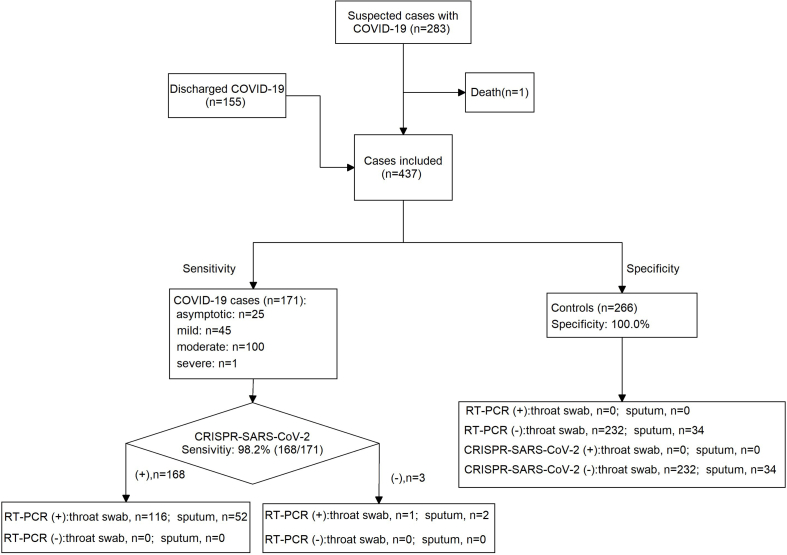
Only one case was excluded due to death during hospitalization and a total of 437 patients were included for final analysis. Of them, 258 cases were male (59.0%). The mean age was 30.0 ± 20.6 years old. Among the 437 patients, 171 patients were diagnosed as COVID-19 positive, 151 patients recovered and met the discharge criteria, and the remaining 115 cases had an alternative disease (Fig. 2), including pneumonia (n = 13), bronchitis (n = 59), upper respiratory tract infection (n = 28), tonsillitis (n = 5), asthmatic bronchitis (n = 5), herpetic angina (n = 2), laryngitis (n = 1), acute gastroenteritis (n = 1), and unknown cause of fever (n = 1). The 171 COVID-19 cases were further classified as asymptotic (n = 25), mild (n = 45), moderate (n = 100), and severe (n = 1). In addition to throat swab, sputum speciments were collected from 54 patients.
Fig. 2.
Flow chart of eligible and included patients for analysis.
3.2. Diagnostic performance
For the CRISPR-SARS-CoV-2 test, the sensitivity and specificity for the diagnosis of COVID-19 were 98.2% (95% CI: 95.0–99.4%; 168/171) and 100.0% (95% CI: 98.6–100.0%; 266/266) (Table 2). In addition, all discharged patients were CRISPR-SARS-CoV-2-negative.
Table 2.
Results of CRISPR-SARS-CoV-2 and RT-PCR tests (throat swab and sputum) among patients with COVID-19.
Including sputum (n = 52).
Including sputum (n = 2).
For the RT-PCR test, the mean CT value was 31.8 ± 4.3 among RT-PCR(+) cases. The sensitivity and specificity for the diagnosis of COVID-19 were 100.0% (95% CI: 97.8–100.0%; 171/171) and 100.0% (95% CI: 98.6–100.0%, 266/266).
Statistical analysis showed that there was no significant difference in the sensitivity of CRISPR-SARS-CoV-2 and RT-PCR for the diagnosis of COVID-19.
3.3. Discordance
Only three cases were reported with a discordant result, all of which were CRISPR-SARS-CoV-2-negative and RT-PCR-positive. This may be explained by the low abundance of N gene levels. Two sequential RT-PCR tests for the same individual had only one positive result for the N gene; another two cases were confirmed with positive results for the ORF1ab gene, without evidence of the N gene.
4. Discussion
Several effector proteins were introduced into the CRISPR system, such as type Ⅱ (Cas9), typeⅤ (Cas14, Cas12a, and Cas12b), and type Ⅵ (Cas13a and Cas13 b). In this study, a commercial CRISPR-SARS-CoV-2 test, which was designed using CRISPR LbCas12a, was evaluated. We found that the kit had equal sensitivity compared to the routine RT-PCR for the diagnosis of COVID-19. To the best of our knowledge, this is the first report investigating a commercial CRISPR based PCR test for SARS-CoV-2 infection. Compared with the routine RT-PCR, this CRISPR method has several advantages, such as low cost, rapid results, and no expensive equipment requirement.
Recent progress in CRISPR research has opened up a new area for NAAT-based pathogen detection [19]. This method for detection of infectious agents has been developed and widely evaluated. Compared with routine PCR methods, CRISPR has several advantages, such as a short turn round time, low cost, easy-to-use, and no requirement of expensive equipment [16,31,32]. For example, CRISPR-Cas12a technology was evaluated in the diagnosis of human papillomavirus, and the data suggested that this tool could achieve results within 35 min with a low detection limit [33]. An integrated assay including RT-RPA and CRISPR Cas12a-mediated detection was evaluated in the diagnosis of HIV-1 infection, showing a sensitivity of 98.95% and specificity of 100%, respectively [34]. Similarly, this combination showed a high sensitivity (93.75%) and specificity (90.63%) for the detection of Yersinia pestis, as reported by You et al. [35]. Jiang et al. evaluated CRISPR-group B Streptococcus (GBS) for the detection of GBS [36]. They demonstrated that the method has a relatively good sensitivity (94.5%), while PCR has a relatively low sensitivity (90.9%) [36]. In addition, this method offered a shorter turnaround time and lower instrument demands than routine PCR-based assays [36].
The global COVID-19 pandemic has escalated the interest in using CRISPR tools for COVID-19 diagnosis. Many CRISPR-based PCR assays have been constructed for the detection of SARS-CoV-2. Several types of CRISPR methods were summarized in a prior review article. The assays described included CRISPR-Cas13a enzyme-, CRISPR-Cas12a enzyme-, and CRISPR-Cas12 b enzyme-mediated DNA detection, all-in-one dual CRISPR-Cas12a assay, and enhanced analysis of nucleic acids with crRNA extensions [37]. However, these tools remain as basic milestones at the developmental statge and very few have been used in clinical settings [38]. Compared to PCR-based methods, CRISPR/Cas-based SARS-CoV-2 assays are very promising due to several characteristics: rapidity, simplicity, and higher sensitivity and specificity [39]. For example, Ooi et al. developed an engineered AsCas12a enzyme based on CRISPR. Their test exhibits a specificity and positive predictive value of 100% and required less time (within 30 min) to obtain restuls [40]. Li et al. published an enhanced strip method based on CRISPR, which was used to visualize the results directly and was 100% concordant with the RT-PCR method [41]. Finally, Hou et al. reported a 100% sensitivity for the diagnostic performance of ‘CRISPR-nCoV’ for SARS-CoV-2 detection [42].
The role of multiple regions of the SARS-CoV-2 genome has been discussed in CRISPR methods. For instance, Ali et al. designed primers targeting N and E genes [43] and found that among the positive samples with qPCR, the N gene assay had a positivity of 86%, and the E gene had a positivity of 38%. Multiple primers targeting several regions of ORF1ab were designed by Lucia et al. and achieved a good detection limit of 10 copies/μL [44]. In our study, although only the N gene was included for analysis, the diagnostic performance of the CRISP-SARS-CoV-2 test had equal sensitivity compared to routine RT-PCR targeting ORF1ab and N genes together. This finding is consistent with a recent meta-analysis that showed that the one target gene method had a high sensitivity of 0.99 (95% CI: 0.93–1.00) and specificity of 1.00 (95% CI: 0.98–1.00) [45].
As previously reported, it is a challenge to detect an extremely low target DNA (lower than 10 nM) using the CRISPR method within less than 1 h [46,47]. Therefore, amplification methods, such as PCR [48,49], LAMP [50], and RPA, have been integrated into CRISPR-based detection systems [19] to enhance its sensitivity. An additional RT step is also required for RNA targets, such as SARS-CoV-2. RT-RPA and RT-LAMP were primarily adopted as the amplification technique for the SARS-CoV-2 nucleic acid, and the corresponding performance showed no significant difference [45]. In our study, RPA was included in the CRISP-SARS-CoV-2 detection system.
Except for the sample preparation time, the operational time for the SARS-CoV-2 RT-PCR assay is approximately 2 h [51]. By comparison, the turnaround time of all Cas12-based studies is reported to be less than 1 h (after excluding sample preparation time). In our study, the CRISPR- SARS-CoV-2 only required approximately 30 min. Hence, this system is rapid compared with routine PCR methods. SARS-CoV-2 assays usually need to be conducted under biological safety level conditions by trained professionals. However, the BG-Nova-X8 platform includes all essential materials for detection, without any need for special laboratory equipment, highly trained personnel, or additional procedures during the test. Therefore, it is cost-effective for both mass production and administration and could decrease human error during needle insertion, improving test reliability.
This study has several limitations that should be noted. First, the dynamic changes of the CRISPR SARS-CoV2 assay could not be monitored, and no additional information is provided for the comparison of critical values between CRISPR (fluorescence intensity) and RT-PCR (ct value) methods. Second, only confirmed cases (RT-PCR positive) were included for analysis, which may have increased the diagnostic yield of the CRISPR SARS-CoV2 assay. Third, although no significant difference was found for the comparison of CRISPR and RT-PCR methods, RT-PCR outperformed the CRISPR method with regards to detection limit. Thus, caution should be paid when interpreting our results.
In conclusion, an RPA coupled with CRISPR-LbCas12a has been commercially introduced for SARS-CoV-2 detection. Our data suggest that the CRISPR-SARS-CoV-2 test has a comparable sensitivity with routine RT-PCR assays, as well as has several merits, such as short assay time, low cost, and no requirement of expensive equipment. In addition, this method provides a valuable reference for the subsequent development of CRISPR technology.
Declarations
Ethics approval
The study protocol was approved by the Ethical Committee of Shanghai Public Health Clinical Center (No. 2021-E011-02) and Heilongjiang Center for Disease Control and Prevention (No. Hei-CDC 2020-09), all methods were carried out in accordance with relevant guidelines and regulations. Written informed consent was waived due to anonymous data collection and samples were collected for routine activities.
Author contribution statement
Jun Xu: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper. Yuanyuan Ma: Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Zhigang Song: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data. Wei Sun: Performed the experiments; Analyzed and interpreted the data. Yi Liu, Chang Shu, Hua Hua, Ming Yang, Qi Liang: Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare no conflict of interest.
Acknowledgements
Not applicable.
Contributor Information
Zhigang Song, Email: songzg@shaphc.org.
Wei Sun, Email: sunwei7979@163.com.
List of abbreviations
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; NAAT: nucleic acid amplification testing; CRISPR: clustered regularly interspaced short palindromic repeats; Cas: CRISPR-associated proteins; RT-RPA: reverse transcription recombinase polymerase amplification; crRNA: CRISPR RNA; GBS: group B Streptococcus.
References
- 1.Rae M. Omicron: a failure to act with a global focus will continue the proliferation of new variants of COVID-19. BMJ. 2021;375:n3095. doi: 10.1136/bmj.n3095. [DOI] [PubMed] [Google Scholar]
- 2.Whitley R. Molnupiravir – a step toward orally bioavailable therapies for COVID-19. N. Engl. J. Med. 2022;386:592–593. doi: 10.1056/NEJMe2117814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi M.Y., Sun S.Q., Zhang W., Zhang X., Xu G.H., Chen X., Su Z.J., Song X.M., Liu L.J., Zhang Y.B., Zhang Y.L., Sun M., Chen Q., Xue Y., Lu H., Yuan W.A., Chen X.R., Lu Y.F. Early therapeutic interventions of traditional Chinese medicine in COVID-19 patients: a retrospective cohort study. J. Integr. Med. 2021;19:226–231. doi: 10.1016/j.joim.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cummings C.L., Miller C.S. COVID-19: how a self-monitoring checklist can empower early intervention and slow disease progression. Environ. Syst. Decis. 2021;41:181–183. doi: 10.1007/s10669-021-09806-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwak P.E., Connors J.R., Benedict P.A., Timen M.R., Wang B., Zhang Y., Youlios S., Sureau K., Persky M.J., Rafeq S., Angel L., Amin M.R. Early outcomes from early tracheostomy for patients with COVID-19. JAMA Otolaryngol. Head Neck Surg. 2021;147:239–244. doi: 10.1001/jamaoto.2020.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali H., Alahmad B., Al-Shammari A.A., Alterki A., Hammad M., Cherian P., Alkhairi I., Sindhu S., Thanaraj T.A., Mohammad A., Alghanim G., Deverajan S., Ahmad R., El-Shazly S., Dashti A.A., Shehab M., Al-Sabah S., Alkandari A., Abubaker J., Abu-Farha M., Al-Mulla F. Previous COVID-19 infection and antibody levels after vaccination. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.778243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y., Wu Y., Ding L., Huang X., Xiong Y. Point-of-care COVID-19 diagnostics powered by lateral flow assay. Trends Anal. Chem. 2021;145 doi: 10.1016/j.trac.2021.116452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brummer L.E., Katzenschlager S., Gaeddert M., Erdmann C., Schmitz S., Bota M., Grilli M., Larmann J., Weigand M.A., Pollock N.R., Mace A., Carmona S., Ongarello S., Sacks J.A., Denkinger C.M. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: a living systematic review and meta-analysis. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson K.E., Altayar O., Caliendo A.M., Arias C.A., Englund J.A., Hayden M.K., Lee M.J., Loeb M., Patel R., El Alayli A., Sultan S., Falck-Ytter Y., Lavergne V., Mansour R., Morgan R.L., Murad M.H., Patel P., Bhimraj A., Mustafa R.A. The infectious diseases society of America guidelines on the diagnosis of COVID-19: antigen testing. Clin. Infect. Dis. 2021:ciab557. doi: 10.1093/cid/ciab557. [DOI] [PubMed] [Google Scholar]
- 10.Ebanks D., Faustini S., Shields A., Parry H., Moss P., Plant T., Richter A., Drayson M. Cross reactivity of serological response to SARS-CoV-2 vaccination with viral variants of concern detected by lateral flow immunoassays. J. Infect. 2021;83:e18–e20. doi: 10.1016/j.jinf.2021.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vengesai A., Midzi H., Kasambala M., Mutandadzi H., Mduluza-Jokonya T.L., Rusakaniko S., Mutapi F., Naicker T., Mduluza T. A systematic and meta-analysis review on the diagnostic accuracy of antibodies in the serological diagnosis of COVID-19. Syst. Rev. 2021;10:155. doi: 10.1186/s13643-021-01689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pu R., Liu S., Ren X., Shi D., Ba Y., Huo Y., Zhang W., Ma L., Liu Y., Yang Y., Cheng N. The screening value of RT-LAMP and RT-PCR in the diagnosis of COVID-19: systematic review and meta-analysis. J. Virol. Methods. 2021;300 doi: 10.1016/j.jviromet.2021.114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler-Laporte G., Lawandi A., Schiller I., Yao M., Dendukuri N., McDonald E.G., Lee T.C. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern. Med. 2021;181:353–360. doi: 10.1001/jamainternmed.2020.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boger B., Fachi M.M., Vilhena R.O., Cobre A.F., Tonin F.S., Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control. 2021;49:21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ai J.W., Zhou X., Xu T., Yang M., Chen Y., He G.Q., Pan N., Cai Y., Li Y., Wang X., Su H., Wang T., Zeng W., Zhang W.H. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microb. Infect. 2019;8:1361–1369. doi: 10.1080/22221751.2019.1664939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian W., Huang J., Wang T., He X., Xu G., Li Y. Visual detection of human metapneumovirus using CRISPR-Cas12a diagnostics. Virus Res. 2021;305 doi: 10.1016/j.virusres.2021.198568. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham C.H., Hennelly C.M., Lin J.T., Ubalee R., Boyce R.M., Mulogo E.M., Hathaway N., Thwai K.L., Phanzu F., Kalonji A., Mwandagalirwa K., Tshefu A., Juliano J.J., Parr J.B. A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping. EBioMedicine. 2021;68 doi: 10.1016/j.ebiom.2021.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsang J., LaManna C.M. Open sharing during COVID-19: CRISPR-based detection tools. CRISPR J. 2020;3:142–145. doi: 10.1089/crispr.2020.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azhar M., Phutela R., Kumar M., Ansari A.H., Rauthan R., Gulati S., Sharma N., Sinha D., Sharma S., Singh S., Acharya S., Sarkar S., Paul D., Kathpalia P., Aich M., Sehgal P., Ranjan G., Bhoyar R.C., Singhal K., Lad H., Patra P.K., Makharia G., Chandak G.R., Pesala B., Chakraborty D., Maiti S. Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens. Bioelectron. 2021;183 doi: 10.1016/j.bios.2021.113207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fasching C.L., Servellita V., McKay B., Nagesh V., Broughton J.P., Sotomayor-Gonzalez A., Wang B., Brazer N., Reyes K., Streithorst J., Deraney R.N., Stanfield E., Hendriks C.G., Fung B., Miller S., Ching J., Chen J.S., Chiu C.Y. COVID-19 variant detection with a high-fidelity CRISPR-Cas12 enzyme. J. Clin. Microbiol. 2022;60 doi: 10.1128/jcm.00261-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahas A., Marsic T., Lopez-Portillo Masson M., Wang Q., Aman R., Zheng C., Ali Z., Alsanea M., Al-Qahtani A., Ghanem B., Alhamlan F., Mahfouz M. Characterization of a thermostable Cas13 enzyme for one-pot detection of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2118260119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Wang H., Zhang L., Lu Y., Wang X., Shen M., Li N., Feng L., Jing J., Cao B., Zou X., Cheng J., Xu Y. Sensitive and rapid diagnosis of respiratory virus coinfection using a microfluidic chip-powered CRISPR/Cas12a system. Small. 2022;18 doi: 10.1002/smll.202200854. [DOI] [PubMed] [Google Scholar]
- 24.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strohkendl I., Saifuddin F.A., Rybarski J.R., Finkelstein I.J., Russell R. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell. 2018;71:816–824.e813. doi: 10.1016/j.molcel.2018.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar M., Gulati S., Ansari A.H., Phutela R., Acharya S., Azhar M., Murthy J., Kathpalia P., Kanakan A., Maurya R., Vasudevan J.S., Ac A.S., Pandey R., Maiti S., Chakraborty D. FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. eLife. 2021;10 doi: 10.7554/eLife.67130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C., Chen Z., Li C., Hao Y., Tang Y., Yuan Y., Chai L., Fan T., Yu J., Ma X., Al-Hartomy O.A., Wageh S., Al-Sehemi A.G., Luo Z., He Y., Li J., Xie Z., Zhang H. CRISPR-Cas12a-empowered electrochemical biosensor for rapid and ultrasensitive detection of SARS-CoV-2 Delta variant. Nano–Micro Lett. 2022;14:159. doi: 10.1007/s40820-022-00888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Commission N.H. Translation: diagnosis and treatment protocol for novel coronavirus pneumonia (trial version 7) Infect. Microbes Dis. 2020;2:48–54. [Google Scholar]
- 29.Fernandez-Montero A., Argemi J., Rodriguez J.A., Arino A.H., Moreno-Galarraga L. Validation of a rapid antigen test as a screening tool for SARS-CoV-2 infection in asymptomatic populations. Sensitivity, specificity and predictive values. EClinicalMedicine. 2021;37 doi: 10.1016/j.eclinm.2021.100954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berensmeier S. Magnetic particles for the separation and purification of nucleic acids. Appl. Microbiol. Biotechnol. 2006;73:495–504. doi: 10.1007/s00253-006-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng W., Peng H., Xu J., Liu Y., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Babiuk S., Zhang H., Le X.C. Integrating reverse transcription recombinase polymerase amplification with CRISPR technology for the one-tube assay of RNA. Anal. Chem. 2021;93:12808–12816. doi: 10.1021/acs.analchem.1c03456. [DOI] [PubMed] [Google Scholar]
- 32.Talwar C.S., Park K.H., Ahn W.C., Kim Y.S., Kwon O.S., Yong D., Kang T., Woo E. Detection of infectious viruses using CRISPR-Cas12-based assay. Biosensors. 2021;11:301. doi: 10.3390/bios11090301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong J., Zhang G., Wang W., Liang L., Li Q., Liu M., Xue L., Tang G. A simple and rapid diagnostic method for 13 types of high-risk human papillomavirus (HR-HPV) detection using CRISPR-Cas12a technology. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-92329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Ao C., Wan Z., Dzakah E.E., Liang Y., Lin H., Wang H., Tang S. A point-of-care rapid HIV-1 test using an isothermal recombinase-aided amplification and CRISPR Cas12a-mediated detection. Virus Res. 2021;303 doi: 10.1016/j.virusres.2021.198505. [DOI] [PubMed] [Google Scholar]
- 35.You Y., Zhang P., Wu G., Tan Y., Zhao Y., Cao S., Song Y., Yang R., Du Z. Highly specific and sensitive detection of Yersinia pestis by portable Cas12a-UPTLFA platform. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.700016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang L., Zeng W., Wu W., Deng Y., He F., Liang W., Huang M., Huang H., Li Y., Wang X., Su H., Pan S., Xu T. Development and clinical evaluation of a CRISPR-based diagnostic for rapid group B Streptococcus screening. Emerg. Infect. Dis. 2021;27:2379–2388. doi: 10.3201/eid2709.200091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Javalkote V.S., Kancharla N., Bhadra B., Shukla M., Soni B., Sapre A., Goodin M., Bandyopadhyay A., Dasgupta S. CRISPR-based assays for rapid detection of SARS-CoV-2. Methods. 2022;203:594–603. doi: 10.1016/j.ymeth.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh B., Datta B., Ashish A., Dutta G. A comprehensive review on current COVID-19 detection methods: from lab care to point of care diagnosis. Sens Int. 2021;2 doi: 10.1016/j.sintl.2021.100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12:582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ooi K.H., Liu M.M., Tay J.W.D., Teo S.Y., Kaewsapsak P., Jin S., Lee C.K., Hou J., Maurer-Stroh S., Lin W., Yan B., Yan G., Gao Y.G., Tan M.H. An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 2021;12:1739. doi: 10.1038/s41467-021-21996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manning B.J., Khan W.A., Peña J.M., Fiore E.S., Boisvert H., Tudino M.C., Barney R.E., Wilson M.K., Singh S., Mowatt J.A., Thompson H.J., Tsongalis G.J., Blake W.J. High-throughput CRISPR-Cas13 SARS-CoV-2 test. Clin. Chem. 2021;68:172–180. doi: 10.1093/clinchem/hvab238. [DOI] [PubMed] [Google Scholar]
- 42.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Gu B., Wang J., Xu T. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M., Hamdan S.M., Pain A., Alofi F.S., Alsomali A., Hashem A.M., Khogeer A., Almontashiri N.A.M., Abedalthagafi M., Hassan N., Mahfouz M.M. iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucia C., Federico P.-B., Alejandra G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv. 2020 2020.2002.2029.971127. [Google Scholar]
- 45.Li X., Zhang H., Zhang J., Song Y., Shi X., Zhao C., Wang J. Diagnostic accuracy of CRISPR technology for detecting SARS-CoV-2: a systematic review and meta-analysis. Expert Rev. Mol. Diagn. 2022;22:655–663. doi: 10.1080/14737159.2022.2107425. [DOI] [PubMed] [Google Scholar]
- 46.Nouri R., Jiang Y., Lian X.L., Guan W. Sequence-specific recognition of HIV-1 DNA with solid-state CRISPR-Cas12a-assisted nanopores (SCAN) ACS Sens. 2020;5:1273–1280. doi: 10.1021/acssensors.0c00497. [DOI] [PubMed] [Google Scholar]
- 47.Teng F., Guo L., Cui T., Wang X.G., Xu K., Gao Q., Zhou Q., Li W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019;20:132. doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S.H., Yu J., Hwang G.H., Kim S., Kim H.S., Ye S., Kim K., Park J., Park D.Y., Cho Y.K., Kim J.S., Bae S. CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR. Oncogene. 2017;36:6823–6829. doi: 10.1038/onc.2017.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S.Y., Cheng Q.X., Wang J.M., Li X.Y., Zhang Z.L., Gao S., Cao R.B., Zhao G.P., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discovery. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 51.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.