Abstract
This article summarises the activities of the Bacterial Viruses Subcommittee of the International Committee on Taxonomy of Viruses for the period of March 2021−March 2022. We provide an overview of the new taxa proposed in 2021, approved by the Executive Committee, and ratified by vote in 2022. Significant changes to the taxonomy of bacterial viruses were introduced: the paraphyletic morphological families Podoviridae, Siphoviridae, and Myoviridae as well as the order Caudovirales were abolished, and a binomial system of nomenclature for species was established. In addition, one order, 22 families, 30 subfamilies, 321 genera, and 862 species were newly created, promoted, or moved.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-022-05694-2.
Introduction
Here, we present taxonomic changes submitted to the Bacterial Viruses Subcommittee (BVS) of the International Committee on Taxonomy of Viruses (ICTV) in 2021, assessed and approved at the ICTV Executive Committee meeting (EC53, July 2021), and ratified in 2022 by membership vote [1]. The new taxonomy release (#37) can be found on the ICTV website (https://ictv.global/) and is available for download as two Excel files, the Master Species List (MSL37) and the Virus Metadata Resource (VMR37), which links each species with an exemplar virus genome and accession number.
Abolishment of the families Myoviridae, Podoviridae, and Siphoviridae and the order Caudovirales
The most drastic change in phage classification officialised with this release is the abolishment of the morphology-based families Myoviridae, Podoviridae, and Siphoviridae and the removal of the order Caudovirales, which is replaced by the class Caudoviricetes to group all tailed bacterial and archaeal viruses with icosahedral capsids and a double-stranded DNA genome. The submission of the taxonomy proposal (2021.001B) followed the publication of a manuscript describing a roadmap for phage taxonomy by the Vice Chair, former Chair, and current Chair of the BVS [2]. This change was necessary, given multiple independent assessments that these morphology-based families are polyphyletic and do not accurately reflect shared evolutionary histories [3–8].
The process of accommodating all members of the three morphology-based families into new, genomically coherent families has begun. Creating new families requires at least two genera, but ideally more, to enable definition of demarcation criteria based on core gene sets and phylogenetic analysis. While this leaves some taxa as “unclassified” at the levels of family and order within the class Caudoviricetes (summarised in Supplementary Table S1), it allows for the creation of new genome-based taxa that better reflect the diversity and genomic relationships of these abundant and diverse viruses in the future. At the same time, we recognise the importance of morphological (non-taxonomic) identifiers such as "podovirus", "myovirus", or "siphovirus". These terms can be used freely to reflect these distinctive features and retain their historical reference; however, after the 2022 ratification vote, they do not have any formal taxonomic meaning/significance.
Change to freeform binomial species name format
In 2020, the ICTV voted to mandate the binomial format for the naming of virus species, where the genus name and a species epithet together form a unique species name, consistent with other taxonomies in biology [9, 10]. This change was applied to 2,532 species of bacterial viruses with input from each study group chair and the members of the BVS through an online consultation. The BVS elected to implement a fully freeform format for binomial naming in which the study groups and taxonomy proposal authors were responsible for the final format of the species names. A brief description of how this binomial format was applied to bacterial viruses follows.
In most cases, the species epithet was directly inherited from the exemplar virus name. For example, the species Escherichia virus T4 was renamed to Tequatrovirus T4, reflecting the name of its exemplar phage T4 (also called Enterobacteria phage T4, phage T4, or Escherichia phage T4). Certain study groups and proposal authors elected to define new species epithets, such as the Leviviricetes and Crassvirales Study Groups, who proposed latinised binomials for all species. For example, the model phage MS2 was assigned to the species Emesvirus zinderi [11], the prototypical crAssphage was assigned to the species Carjivirus communis, and the first isolated representative of the order Crassvirales, Bacteroides phage crAss001, was assigned to the species Kehishuvirus primarius [12].
The BVS implemented a number of additional orthographic rules for the creation of new species epithets that are derived directly from the phage name. Where virus names consisted only of numerals, the first letter of the genus names, followed by a “v” (virus) was prepended to the species epithet. For example, Pseudomonas phage 14 − 1 is the exemplar virus of species Pbunavirus pv141. The species epithet can be composed of a mixture of alphanumeric characters in lower and upper case. For future species epithets, we would request that only lowercase letters are used. Where existing phage names had a mixture of upper and lower case letters, these were retained to facilitate recognition of similar virus names such as P2 and p2 by the bacterial virus research community.
Where the species epithet consisted of letters only, the consensus of the BVS was to only permit lowercase letters. For example, Bacillus phage Deep Blue is the exemplar virus of the species Caeruleovirus deepblue.
Overview of taxonomy changes
Ninety-five taxonomy proposals were submitted to the BVS Chair and approved by the Executive Committee in 2021. The number of proposals and taxa created precludes detailed individual descriptions, but they are summarised in Supplementary Table S1, and all individual proposals are available from the ICTV website (https://ictv.global/files/proposals/approved?fid=4467#block-teamplus-page-title). Herein, we provide an overview of changes (Table 1) and a brief summary of proposals resulting in the creation of higher-ranking taxa, including new orders and families.
Table 1.
Summary of taxonomic changes for bacterial viruses for Master Species List 37, ratified in 2022. No changes were made at the class, phylum, kingdom, or realm level.
| Species | Genus | Subfamily | Family | Order | |
|---|---|---|---|---|---|
| Abolished | 5 | 2 | 0 | 3 | 1 |
| New | 804 | 257 | 26 | 21 | 1 |
| Moved or promoted | 58 | 64 | 4 | 1 | 0 |
| Renamed | 2736 | 1 | 0 | 0 | 0 |
| Current total | 3601 | 1199 | 98 | 47 | 4 |
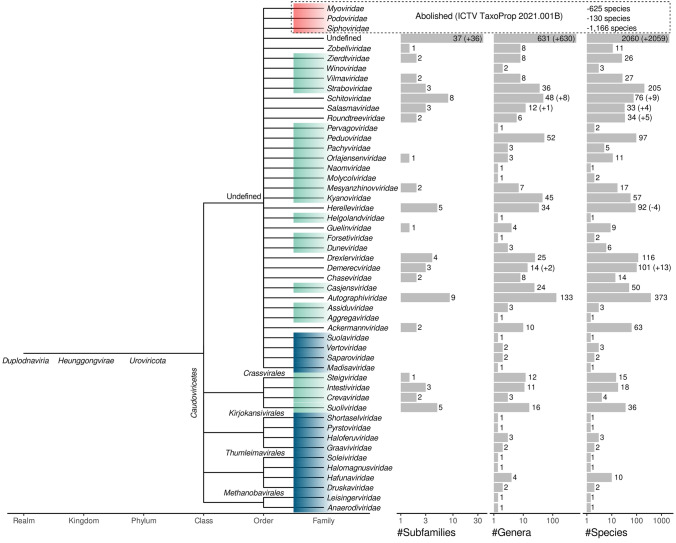
The class Caudoviricetes now contains 14 families assigned to four orders. Three of these orders encompass viruses infecting archaea [1, 13]. A further 33 families have been established but not yet been assigned to an order, in addition to 37 subfamilies and 631 genera that have yet to be classified at the rank of family or order (Fig. 1).
Fig. 1.
Cladogram depicting the taxonomic structure of the order Caudoviricetes. New bacterial and archaeal virus families created in 2021 are highlighted in green or blue, respectively, and those abolished are in red. Numbers of subfamilies, genera, and species are depicted as histograms adjacent to each family. Numbers in parentheses indicate changes recorded from ratified taxonomic proposals. Subfamilies and genera moved from the abolished families Myoviridae, Podoviridae, and Siphoviridae are detailed in Supplementary Table S1.
New taxa in the class Caudoviricetes
Twenty-two new families were delineated in the class Caudoviricetes, 21 of which were newly established (four within the new order Crassvirales) and one of which was promoted from the level of subfamily (Table 1). Additional taxa (Fig. 1) were established in the class Caudoviricetes by the Archaeal Viruses Subcommittee [1, 13] but are not discussed in this report.
Order Crassvirales
The newly established order Crassvirales, named after the computer program crAss [14], which was used in the discovery of its founding member, comprises dsDNA tailed bacteriophages with genome sizes ranging from 83 to 106 kbp (Taxonomy Proposal 2021.021B.A.Crassvirales). Very few representatives of the order have been isolated in culture. The majority of species have been described based on analysis of metagenomically assembled genomes (MAGs) extracted from gut-associated, soil, marine, and industrial environments [15–18]. Members of the order Crassvirales are especially prevalent in mammalian gut microbiomes and were isolated from, or predicted to infect, bacteria of the phylum Bacteroidota (more specifically, order Bacteroidales in the mammalian gut) [19–21].
The first representative to be discovered and reported in the literature was crAssphage, a ubiquitous and highly prevalent member of the human gut virome [19, 22]. Metagenomic sequencing reads mapped to the prototypical crAssphage from up to 73% of the human faecal metagenomic datasets, accounting for up to 90% of the reads in virus-like-particle-enriched metagenomes or 22% of the reads of the total metagenomes in extreme cases. With other members of the order included, these viruses are detectable in 98% of faecal viromes from Western cohorts and in 77% of human faecal viromes worldwide. In 8% of cases, crAss-like phages accounted for > 50% of the virome, making this order the most abundant group of viruses of the human virome [16].
The current genome-based taxonomy of the order Crassvirales includes four families (Intestiviridae, Crevaviridae, Suoliviridae, and Steigviridae), 11 subfamilies, 42 genera, and 73 species. Cultured representatives only exist for the families Intestiviridae (species Jahgtovirus secundus represented by Bacteroides phage crAss002) and Steigviridae (species Kehishuvirus primarius represented by Bacteroides phage crAss001, Wulfhauvirus bangladeshii by Bacteroides phages DAC15 and DAC17, and Akihdevirus balticus by Cellulophaga phage phi14:2). Carjivirus communis, the species comprising the prototypical crAssphage, currently remains without cultured isolates.
Family Peduoviridae
The family Peduoviridae is the formalisation of P2-like phages consisting of tailed, non-enveloped viruses that contain a set of six orthologous core genes. This group of viruses was previously a subfamily (Peduovirinae) in the abolished family Myoviridae but warranted elevation to family status when the morphology-based classification of viruses was abolished and genomic analysis revealed that the diversity among members of the Peduovirinae (created in 2011; Taxonomy Proposal 2009.012a-qB) is similar to that of other recently defined families in the class Caudoviricetes. The family includes arguably the best-studied example of a class of temperate phage commonly found in genomes of Gram-negative bacteria, P2 (isolated in 1951 by Giuseppe Bertani [23]), after which the family is named.
Family Mesyanzhinovviridae
The family Mesyanzhinovviridae is named in honour of the late Vadim V. Mesyanzhinov (Shemyakin-Ovchinnikov Institute Moscow). The family encompasses seven genera comprising temperate and lytic siphoviruses infecting strains of the eubacterial genera Bordetella, Pseudomonas, and Xanthomonas. It consists of two subfamilies – Bradleyvirinae, named for David Edward Bradley, known for his research into the ultrastructure of bacteriophages (genera: Abidjanvirus, Bosavirus, Xooduovirus, and Epaquintavirus) and Rabinowitzvirinae, named for Genia Rabinowitz, one of the first scientists to study bacteriophages of Bacillus pyocyaneus sp. (genera: Vojvodinavirus and Yuavirus) – and the orphan genus Keylargovirus.
Families Straboviridae and Kyanoviridae
The family Straboviridae is named after the Greek philosopher Strabo. The new family encompasses the previous “T4-like” phages classified within the abolished family Myoviridae. The family Straboviridae now consists of 35 genera, with 11 newly created genera (genera: Kagamiyamavirus, Angelvirus, Bragavirus, Mosugukvirus, Mylasvirus, Roskildevirus, Jiangsuvirus, Chrysonvirus, Cinqassovirus, Gualtarvirus, and Carettavirus). There are 21 genera within the subfamilies Tevenvirinae (genera: Dhakavirus, Gaprivervirus, Gelderlandvirus, Jiaodavirus, Kagamiyamavirus, Kanagawavirus, Karamvirus, Moonvirus, Mosigvirus, Mosugukvirus, Roskildevirus, Tegunavirus, Tequatrovirus, and Winklervirus), Emmerichvirinae (genera: Ishigurovirus and Ceceduovirus), and Twarogvirinae (genera: Lasallevirus, Hadassahvirus, Acajnonavirus, and Zedzedvirus), with a further 14 floating genera (Angelvirus, Biquartavirus, Bragavirus, Carettavirus, Chrysonvirus, Cinqassovirus, Gualtarvirus, Jiangsuvirus, Krischvirus, Mylasvirus, Pseudotevenvirus, Schizotequatrovirus, Slopekvirus, and Tulanevirus). The genera Schizotequatrovirus, Slopekvirus, Pseudotevenvirus, and Krischvirus have been removed from the subfamily Tevenvirinae, as the members of these genera are not monophyletic with other members of the subfamily.
The family Kyanoviridae is named after the ancient Greek word Kyanos for “cyan/dark blue”. The family includes “T4-like” phages of the myovirus morphotype, with all representatives to date infecting cyanobacteria. While related to members of the Straboviridae, they are distant relatives. As such, and to be consistent with other viral families, they have been separated into a family, as they form a monophyletic group. The relationship between Straboviridae, Kyanoviridae, and other families at higher taxonomic levels has yet to be resolved. Currently there are no subfamilies, and the family consists of 26 genera previously of the abolished Myoviridae (Cymopoleiavirus, Charybdisvirus, Anaposvirus, Nodensvirus, Vellamovirus, Kanaloavirus, Bellamyvirus, Salacisavirus, Mazuvirus, Atlauavirus, Libanvirus, Ahtivirus, Namakavirus, Pontusvirus, Ronodorvirus, Leucotheavirus, Thetisvirus, Thaumasvirus, Tefnutvirus, Neptunevirus, Palaemonvirus, Nerrivikvirus, Brizovirus, Nereusvirus, Acionnavirus, and Biquartavirus) and 20 new genera (Sedonavirus, Gibbetvirus, Chalconvirus, Potamoivirus, Lipsvirus, Lowelvirus, Makelovirus, Haifavirus, Nilusvirus, Sokavirus, Macariavirus, Greenvirus, Ormenosvirus, Shandvirus, Emcearvirus, Galenevirus, Glaucusvirus, Alisovirus, Neritesvirus, and Bristolvirus).
FamilyNaomviridae
The family Naomviridae consists of the single genus Noahvirus. The etymology of the family name comes from the Hebrew meaning of Naomi, “pleasant one/good one”. Current members of the genus Noahvirus have the defining characteristic of replacing deoxythymidine with deoxyuridine within their genomic DNA [24].
Family Casjensviridae
The family Casjensviridae, named in honour of Sherwood R. Casjens (University of Utah), comprises the four existing genera (Ahduovirus, Chivirus, Nazgulvirus and Sanovirus) and 20 new genera (Broinstvirus, Cenphatecvirus, Dunedinvirus, Enchivirus, Fengtaivirus, Gediminasvirus, Gwanakrovirus, Jacunavirus, Kokobelvirus, Lavrentievavirus, Maxdohrnvirus, Newforgelanevirus, Phobosvirus, Redjacvirus, Salvovirus, Seodaemunguvirus, Sharonstreetvirus, Yonseivirus, and Zhonglingvirus) of flagellotrophic siphoviruses, including Salmonella phage Chi (assigned to the species Chivirus chi) [25].
Family Vilmaviridae
The family Vilmaviridae was named after clusters V, L, and M, defined in the Actinobacteriophage Database [26]. The family is composed of two subfamilies, Lclasvirinae (genera: Bromdenvirus, Bronvirus, Faithunavirus, and Lumosvirus) and Mclasvirinae (genera: Bongovirus and Reyvirus) and two unassigned genera, Kumaovirus and Wildcatvirus. The L and M cluster viruses are all temperate siphoviruses, while those belonging to cluster V are strictly lytic Mycobacterium siphophages.
Family Orlajensenviridae
The family Orlajensenviridae formalises the classification of the cluster EE Microbacterium phages defined in the Actinobacteriophage Database [26]. The family is composed of a single subfamily, Pelczarvirinae, named after Michael Joseph Pelczar, which includes three genera: Paopuvirus, Bonaevitaevirus, and Efekovirus, which share 20 orthologous core genes encoding structural, morphogenesis, and DNA-binding proteins. The family is named after Sigurd Ola-Jensen, who was responsible for the initial isolation and taxonomy of the bacterial genus Microbacterium.
Bacterial viruses infecting members of the bacterial class Flavobacteriia are classified within nine families
Nine new families were established, based on comparative and phylogenetic analysis of isolated bacteriophages infecting members of the bacterial class Flavobacteriia and supported by complete coding sequences from metagenomes. Each of the families forms a monophyletic group in VirClust [27] and VICTOR [28], and their members share at least eight core genes [28]. Members of these families infect bacterial hosts of the genera Polaribacter, Tenacibaculum, Cellulophaga, Winogradskyella, and Olleya.
Two families are named after the islands Helgoland and Dune of the Helgoland Archipelago in the North Sea, where many of these bacterial viruses were isolated: Helgolandviridae, which contains a single genus (Leefvirus), and Duneviridae, which contains three genera (Ingelinevirus Labanvirus and Unahavirus). The family Forsetiviridae is named after the god Forseti, associated with the island Hegoland in Nordic mythology, and consists of a single genus, Freyavirus. Derived from the Latin word pervagus, translated as “widely roaming”, the family Pervagoviridae is composed of a single genus, Callevirus. Pachyviridae, named after the Greek word pakhús for “thick”, consists of three genera (Baltivirus, Bacelvirus, and Gundelvirus). Molycolviridae is named after the two species in the genus Mollyvirus: Mollyvirus molly and Mollyvirus colly. Three genera (Nekkelsvirus, Cebadecemvirus, and Cellubavirus) are classified within the family Assiduviridae, named after the Latin word assiduous, meaning “constant” or “regular”. The family Aggregaviridae, so named because the host Olleya sp. are commonly found on aggregates, consists of a single genus, Harrekavirus. Lastly, the family Winoviridae is named after the host genus Winogradskyella and contains two genera, Peternellavirus and Pippivirus.
FamilyZierdtviridae
The family Zierdtviridae, named after Charles Henry Zierdt, contains two subfamilies: Emilbogenvirinae (genera: Sukkupivirus, Kablunavirus, Foxborovirus, Pleakleyvirus, Skysandvirus, and Gruunavirus) and Toshachvirinae (genera: Chunghsingvirus and Ceetrepovirus), comprising lytic Gordonia phages from cluster CR and Corynebacterium phages of cluster EN defined in the Actinobacteriophage Database [26]. The subfamilies were named after Sheila Toshach and Emil Bogen. Members of the family share a set of 18 core genes.
Conclusions and future directions
The past year has seen substantial changes in the taxonomy of bacterial viruses, as the BVS and ICTV move towards a more coherent and unified system of virus classification based on genome-level relationships. For bacterial viruses, a roadmap and guidance for genomic classification have been published with recommendations for analysis of the genomes of these diverse and ubiquitous viruses [2, 29, 30]. The abolishment of the order Caudovirales and the families Myoviridae, Siphoviridae, and Podoviridae has resulted in a significant number of floating subfamilies and genera. To resolve this, concerted efforts by the BVS and the wider community are required to establish new taxa at the ranks of family and order that accurately reflect the evolutionary relationships of these viruses. With more research, greater coverage of viral diversity, and the development of new and more-sensitive approaches for sequence analysis, we are confident that robust consensus approaches for the delineation of higher taxa will emerge.
The creation of the order Crassvirales and nine families of viruses infecting members of the bacterial class Flavobacteriia serves to illustrate the significance of virome datasets for establishing demarcation criteria for taxonomic classification. The inclusion of viruses identified from metagenomic data is essential to capture the true diversity of bacterial viruses and will undoubtedly lead to the expansion of existing taxa and the creation of new ones in the future. However, for inclusion within the taxonomic framework, it is necessary to incorporate appropriate checks to demonstrate that such sequences represent the complete coding region of the genome [31].
The classification of bacterial viruses unquestionably relies upon the combined efforts of the research community, and the past year has seen increasing numbers of proposals submitted from outside the BVS and its study groups. We strongly encourage such initiatives. Interested researchers can always contact the BVS and study groups for information, consultation, and engagement with the taxonomic framework. While metagenomic virus discovery efforts continue to uncover and catalogue the diversity of the virosphere, taxonomic classification will inevitably lag behind the submission of new genome sequences to databases. We hope that through the engagement of the wider community and the continued development of new tools and approaches for sequence analysis, this gap will decrease substantially in the near future.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Acknowledgements
We would like to acknowledge the assistance provided by colleagues at NCBI, in particular Igor Tolstoy.
Author contribution
All authors contributed to the creation or assessment of taxonomy proposals. DT and EMA wrote the manuscript with input from AMK, ANS, ADM, BED, CL, HMO, LZ, MMP, PA-Z, RAE, and RKA. All authors evaluated and approved the final version.
Funding
B.E.D. was supported by the European Research Council (ERC) Consolidator Grant 865694: DiversiPHI, the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2051 – Project-ID 390713860, the Alexander von Humboldt Foundation in the context of an Alexander von Humboldt Professorship funded by the German Federal Ministry of Education and Research, and the European Union’s Horizon 2020 Research and Innovation Program, under the Marie Skłodowska-Curie Actions Innovative Training Networks grant agreement no. 955974 (VIROINF). Work by J.R.B. was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health. R.A.E was supported by an award from the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institute of Health RC2DK116713 and an award from the Australian Research Council, DP220102915. R.L. is supported by the research grant PHAGEFORCE from the KU Leuven. C.L. is supported by the Research Foundation - Flanders (FWO Grant 12D8623N) and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2051 – Project-ID 390713860. V.M. is supported by the Russian state-funded project For ICBFM SB RAS, Grant 121031300043-8. H.M.O. was supported by the University of Helsinki and Academy of Finland by funding for FINStruct and Instruct Centre Finland, Instruct-ERIC. M.M.P. acknowledges funding from the Sigrid Jusélius Foundation and the Academy of Finland (grant 331627). A.N.S is supported by a Wellcome Trust Research Career Development Fellowship [220646/Z/20/Z] and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No 101001684). E.M.A. gratefully acknowledges funding by the Biotechnology and Biological Sciences Research Council (BBSRC); this research was funded by the BBSRC Institute Strategic Programme Gut Microbes and Health BB/R012490/1 and its constituent projects BBS/E/F/000PR10353 and BBS/E/F/000PR10356.
Declarations
Conflict of interest
All authors are current members of the Bacterial Viruses Subcommittee and/or Archaeal Viruses Subcommittee of the International Committee on Taxonomy of Viruses (ICTV).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dann Turner, Email: dann2.turner@uwe.ac.uk.
Andrey N. Shkoporov, Email: andrey.shkoporov@ucc.ie
Cédric Lood, Email: cedric.lood@kuleuven.be.
Andrew D. Millard, Email: adm39@leicester.ac.uk
Bas E. Dutilh, Email: bedutilh@gmail.com
Poliane Alfenas-Zerbini, Email: palfenas@ufv.br.
Leonardo J. van Zyl, Email: vanzyllj@gmail.com
Ramy K. Aziz, Email: raziz1@gmail.com
Hanna M. Oksanen, Email: Hanna.oksanen@helsinki.fi
Minna M. Poranen, Email: Minna.poranen@helsinki.fi
Andrew M. Kropinski, Email: phage.canada@gmail.com
Jakub Barylski, Email: jakub.barylski@gmail.com.
J Rodney Brister, Email: jamesbr@ncbi.nlm.nih.gov.
Nina Chanisvili, Email: Nina.chanishvili@gmail.com.
Rob A. Edwards, Email: robert.edwards@flinders.edu.au
François Enault, Email: Francois.enault@univ-bpclermont.fr.
Annika Gillis, Email: annika.gillis@gmail.com.
Petar Knezevic, Email: petar.knezevic@dbe.uns.ac.rs.
Mart Krupovic, Email: mart.krupovic@gmail.com.
Ipek Kurtböke, Email: IKurtbok@usc.edu.au.
Alla Kushkina, Email: a.kushkina@gmail.com.
Rob Lavigne, Email: rob.lavigne@kuleuven.be.
Susan Lehman, Email: susan.lehman@fda.hhs.gov.
Malgorzata Lobocka, Email: lobocka@ibb.waw.pl.
Cristina Moraru, Email: liliana.cristina.moraru@uol.de.
Andrea Moreno Switt, Email: andrea.moreno@uc.cl.
Vera Morozova, Email: Vera_morozova@ngs.ru.
Jesca Nakavuma, Email: jesca.nakavuma@gmail.com.
Alejandro Reyes Muñoz, Email: a.reyes@uniandes.edu.co.
Jānis Rūmnieks, Email: j.rumnieks@biomed.lu.lv.
BL Sarkar, Email: bl_sarkar@hotmail.com.
Matthew B. Sullivan, Email: mbsulli@gmail.com
Jumpei Uchiyama, Email: uchiyama@okayama-u.ac.jp.
Johannes Wittmann, Email: jow12@dsmz.de.
Tong Yigang, Email: tong.yigang@gmail.com.
Evelien M. Adriaenssens, Email: evelien.adriaenssens@quadram.ac.uk
References
- 1.Walker PJ, Siddell SG, Lefkowitz EJ, et al. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022) Arch Virol. 2022;167:2429–2440. doi: 10.1007/s00705-022-05516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner D, Kropinski AM, Adriaenssens EM. A Roadmap for Genome-Based Phage Taxonomy. Viruses. 2021;13:506. doi: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiewsakun P, Adriaenssens EM, Lavigne R, et al. Evaluation of the genomic diversity of viruses infecting bacteria, archaea and eukaryotes using a common bioinformatic platform: steps towards a unified taxonomy. J Gen Virol. 2018;99:1331–1343. doi: 10.1099/jgv.0.001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barylski J, Enault F, Dutilh BE, et al. Analysis of Spounaviruses as a Case Study for the Overdue Reclassification of Tailed Phages. Syst Biol. 2020;69:110–123. doi: 10.1093/sysbio/syz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iranzo J, Krupovic M, Koonin EV. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio. 2016;7:e00978–e00916. doi: 10.1128/mBio.00978-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence JG, Hatfull GF, Hendrix RW. Imbroglios of viral taxonomy: Genetic exchange and failings of phenetic approaches. J Bacteriol. 2002;184:4891–4905. doi: 10.1128/JB.184.17.4891-4905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol. 2008;25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- 8.Low SJ, Džunková M, Chaumeil P-A, et al. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat Microbiol. 2019;4:1306–1315. doi: 10.1038/s41564-019-0448-z. [DOI] [PubMed] [Google Scholar]
- 9.Siddell SG, Walker PJ, Lefkowitz EJ, et al. Binomial nomenclature for virus species: a consultation. Arch Virol. 2020;165:519–525. doi: 10.1007/s00705-019-04477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zerbini FM, Siddell SG, Mushegian AR et al (2022) Differentiating between viruses and virus species by writing their names correctly. Arch Virol 1–4. 10.1007/s00705-021-05323-4 [DOI] [PMC free article] [PubMed]
- 11.Callanan J, Stockdale SR, Adriaenssens EM et al (2021) Leviviricetes: expanding and restructuring the taxonomy of bacteria-infecting single-stranded RNA viruses. Microb Genomics 7. 10.1099/mgen.0.000686 [DOI] [PMC free article] [PubMed]
- 12.Postler TS, Rubino L, Adriaenssens EM et al Guidance for creating individual and batch latinized binomial virus species names.Journal of General Virology103:001800. 10.1099/jgv.0.001800 [DOI] [PMC free article] [PubMed]
- 13.Liu Y, Demina TA, Roux S, et al. Diversity, taxonomy, and evolution of archaeal viruses of the class Caudoviricetes. PLoS Biol. 2021;19:e3001442. doi: 10.1371/journal.pbio.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutilh BE, Schmieder R, Nulton J, et al. Reference-independent comparative metagenomics using cross-assembly: CrAss. Bioinformatics. 2012;28:3225–3231. doi: 10.1093/bioinformatics/bts613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown BP, Chopera D, Havyarimana E, et al. crAssphage genomes identified in fecal samples of an adult and infants with evidence of positive genomic selective pressure within tail protein genes. Virus Res. 2021;292:198219. doi: 10.1016/j.virusres.2020.198219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerin E, Shkoporov A, Stockdale SR, et al. Biology and Taxonomy of crAss-like Bacteriophages, the Most Abundant Virus in the Human Gut. Cell Host Microbe. 2018;24:653–664e6. doi: 10.1016/j.chom.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Siranosian BA, Tamburini FB, Sherlock G, Bhatt AS (2020) Acquisition, transmission and strain diversity of human gut-colonizing crAss-like phages. Nat Commun 11. 10.1038/s41467-019-14103-3 [DOI] [PMC free article] [PubMed]
- 18.Yutin N, Makarova KS, Gussow AB, et al. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat Microbiol. 2018;3:38–46. doi: 10.1038/s41564-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutilh BE, Cassman N, McNair K, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hryckowian AJ, Merrill BD, Porter NT, et al. Bacteroides thetaiotaomicron-Infecting Bacteriophage Isolates Inform Sequence-Based Host Range Predictions. Cell Host Microbe. 2020;28:371–379e5. doi: 10.1016/j.chom.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shkoporov AN, Khokhlova EV, Fitzgerald CB, et al. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat Commun. 2018;9:4781. doi: 10.1038/s41467-018-07225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards RA, Vega AA, Norman HM, et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat Microbiol. 2019;4:1727–1736. doi: 10.1038/s41564-019-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertani G. STUDIES ON LYSOGENESIS I: The Mode of Phage Liberation by Lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rihtman B, Puxty RJ, Hapeshi A, et al. A new family of globally distributed lytic roseophages with unusual deoxythymidine to deoxyuridine substitution. Curr Biol. 2021;31:3199–3206e4. doi: 10.1016/j.cub.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrix RW, Ko C-C, Jacobs-Sera D, et al. Genome Sequence of Salmonella Phage χ. Genome Announcements. 2015;3:e01229–e01214. doi: 10.1128/genomeA.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell DA, Hatfull GF. PhagesDB: the actinobacteriophage database. Bioinformatics. 2017;33:784–786. doi: 10.1093/bioinformatics/btw711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moraru C (2021) VirClust – a tool for hierarchical clustering, core gene detection and annotation of (prokaryotic) viruses. 2021.06.14.448304 [DOI] [PMC free article] [PubMed]
- 28.Meier-Kolthoff JP, Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen A, Millard A. Phage genome annotation: where to begin and end. PHAGE. 2021;2:183–193. doi: 10.1089/phage.2021.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner D, Adriaenssens EM, Tolstoy I, Kropinski AM. Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation. Phage. 2021;2:170–182. doi: 10.1089/phage.2021.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutilh BE, Varsani A, Tong Y, et al. Perspective on taxonomic classification of uncultivated viruses. Curr Opin Virol. 2021;51:207–215. doi: 10.1016/j.coviro.2021.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.