Abstract
Chronic lung diseases, such as chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF), are characterized by regional extracellular matrix (ECM) remodeling which contributes to disease progression. Previous proteomic studies on whole decellularized lungs have provided detailed characterization on the impact of COPD and IPF on total lung ECM composition. However, such studies are unable to determine the differences in ECM composition between individual anatomical regions of the lung. Here, we employ a post-decellularization dissection method to compare the ECM composition of whole decellularized lungs (wECM) and specific anatomical lung regions, including alveolar-enriched ECM (aECM), airway ECM (airECM), and vasculature ECM (vECM), between non-diseased (ND), COPD, and IPF human lungs. We demonstrate, using mass spectrometry, that individual regions possess a unique ECM signature characterized primarily by differences in collagen composition and basement-membrane associated proteins, including ECM glycoproteins. We further demonstrate that both COPD and IPF lead to alterations in lung ECM composition in a region-specific manner, including enrichment of type-III collagen and fibulin in IPF aECM. Taken together, this study provides methodology for future studies, including isolation of region-specific lung biomaterials, as well as a dataset that may be applied for the identification of novel ECM targets for therapeutics.
Graphical Abstract
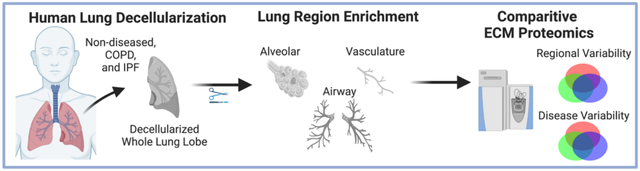
Introduction
The lung is a complex organ with distinct anatomical regions that each play a critical role in respiration. Examples include cartilaginous branching airways that facilitate organ-wide airflow, and thin basement membranes in the distal lung that allow for efficient gas exchange between alveolar and vasculature compartments [1,2]. As the structural and functional needs within these specialized lung regions drastically differ, so too must the protein composition of the extracellular matrix (ECM). In addition to providing structural support to tissues, the ECM is now widely recognized as an important regulator of organ homeostasis, regeneration, and health [3]. Bioactive components, such as proteoglycans, matrix-associated growth factors, and other secreted factors sequestered within the ECM provide region-specific cues that drive localized cell functions, including proliferation, differentiation, and maturation [4–6]. However, dysregulation of the delicate interplay between the ECM and resident cells often leads to detrimental health effects. Within the lung, chronic lung diseases, such as chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF), are hallmarked by spatial abnormalities in ECM structure and composition, which contribute to aberrant tissue regeneration and disease progression [3,7–9]. As such, it is critical to understand the structural and bioactive ECM composition of specific regions within the lung in both normal and disease conditions.
Currently, the majority of studies characterizing the ECM within specific anatomical regions of the lung have relied on targeted methodologies such as histochemical staining or immunohistochemistry (IHC) [as reviewed in 1,2,7]. These studies have provided valuable information including the primary localization of specific principal collagens as well as select bioactive components, including laminins and TGF-β [1,10–15]. Additionally, IHC has been used to assess differences in the lung ECM composition in both COPD and IPF, including the decrease or deposition of type-I collagen (COL1) in COPD or IPF, respectively [7–10,12–16]. However, as these studies utilize selective and targeted methodologies, they are unable to provide a complete and unbiased proteomic characterization of the lung ECM.
With advances in proteomic methodologies, global characterization of the ECM composition of tissues is now possible. With regards to the lung, decellularized lungs from animal or cadaveric human lungs can be assessed by mass spectrometry to provide semi-quantitative characterization of the lung ECM [17–21]. Current studies, including our own, have previously demonstrated that decellularized lungs retain much of the native (i.e. non-decellularized) ECM matrix composition, including larger structural ECM proteins, such as Col1 and type-III collagen (COL3) as well as smaller bioactive ECM proteins, such as basement membrane associated laminins, proteoglycans, and glycoproteins [17,19,20,22]. As such, decellularized lungs from patients with no history of lung disease (ND) and patients with end-stage lung diseases (i.e. COPD and IPF) have been directly compared using semi-quantitative mass spectrometry. Interestingly, while no ECM proteins were significantly altered in abundance between COPD and ND lungs [17], IPF lungs were significantly different from ND lungs [23]. In particular, Booth et al. demonstrated that decellularized IPF lungs displayed an increase in COL3 and microfibirillar-associated proteins as well as a decrease in basement membrane associated laminins [23].
Despite providing valuable insight into global ECM composition in normal and diseased lungs, proteomic studies of decellularized lungs have previously utilized whole lung tissue, and therefore, cannot discriminate the detailed lung ECM composition between different anatomical regions, notably between airways, alveoli, and vasculature. To advance current proteomic studies on decellularized lungs, we here employ a post-decellularization dissection technique prior to mass spectrometric analyses. As such, this work provides novel characterization of ECM composition from whole lung (wECM), airway (airECM), alveoli (aECM), and vasculature ECM (vECM) from ND, COPD, and IPF patients.
Materials and Methods
2.1. Human lung decellularization and dissection.
Human lungs were obtained from University of Vermont (UVM) autopsy services, under appropriate institutional guidelines and HIPAA protections, from twelve patients with either no history of lung disease (ND) (n=6), COPD/emphysema (n=3), or IPF (n=3). Information on patient demographics including age, sex, and pulmonary and smoking history are depicted in Supplemental Table 1. Whole lung lobe decellularization was performed as previously described by our group [17–19,22]. In brief, single lobes from each patient were rinsed with phosphate buffered saline (PBS, Corning) and deionized (DI) water six times to remove excess blood from lungs lobes. Decellularization was performed via perfusion of airways (i.e.main bronchi) and vasculature (pulmonary arteries) using a peristaltic roller pump (Stockert Shiley, SOMA Technologies) at a 2 L/min rate. Lobe perfusion included sequential 2L rinses with 0.1% Triton-X 100 (Sigma), 2% sodium deoxycholate (SDC, Sigma), 1M sodium chloride (NaCl, Sigma), DNase (Sigma), peracetic acid (Sigma), and a DI wash.
Whole decellularized lung lobes were manually dissected to remove airway and vasculature trees (Figure 1). Surgical scissors and forceps were used to expose the most proximal regions of the airways, which were subsequently used as a guide to carefully dissect down the length of the airways towards the most distal tips of airway trees. As expected, decellularized vascular tissue remained in close proximity to the airway regions during the dissection process, yet differed in color and elasticity, making it simple to identify and remove from the airway. Airway samples between ~5–1mm were taken for further processing (~airway generations 2–16) [24]. Alveolar-enriched tissue was identified as the distal tips of the small branching airways < 1mm (~airway generations 17–23). Isolated samples were grouped as whole lung (i.e. non-dissected fractions obtained from the ventral portion of the lung and containing all regions), airways, vasculature, or alveolar-enriched regions. Samples from the individual lungs were frozen at −80°C, lyophilized, and liquid nitrogen milled (Freezer Mill, Spex) to a fine powder. The ECM powders were designated whole lung ECM (wECM), airway ECM (airECM), vasculature ECM (vECM), or alveolar-enriched ECM (aECM) and stored at −20°C until future use.
Figure 1. Schematic of decellularized lung processing for mass spectrometry.
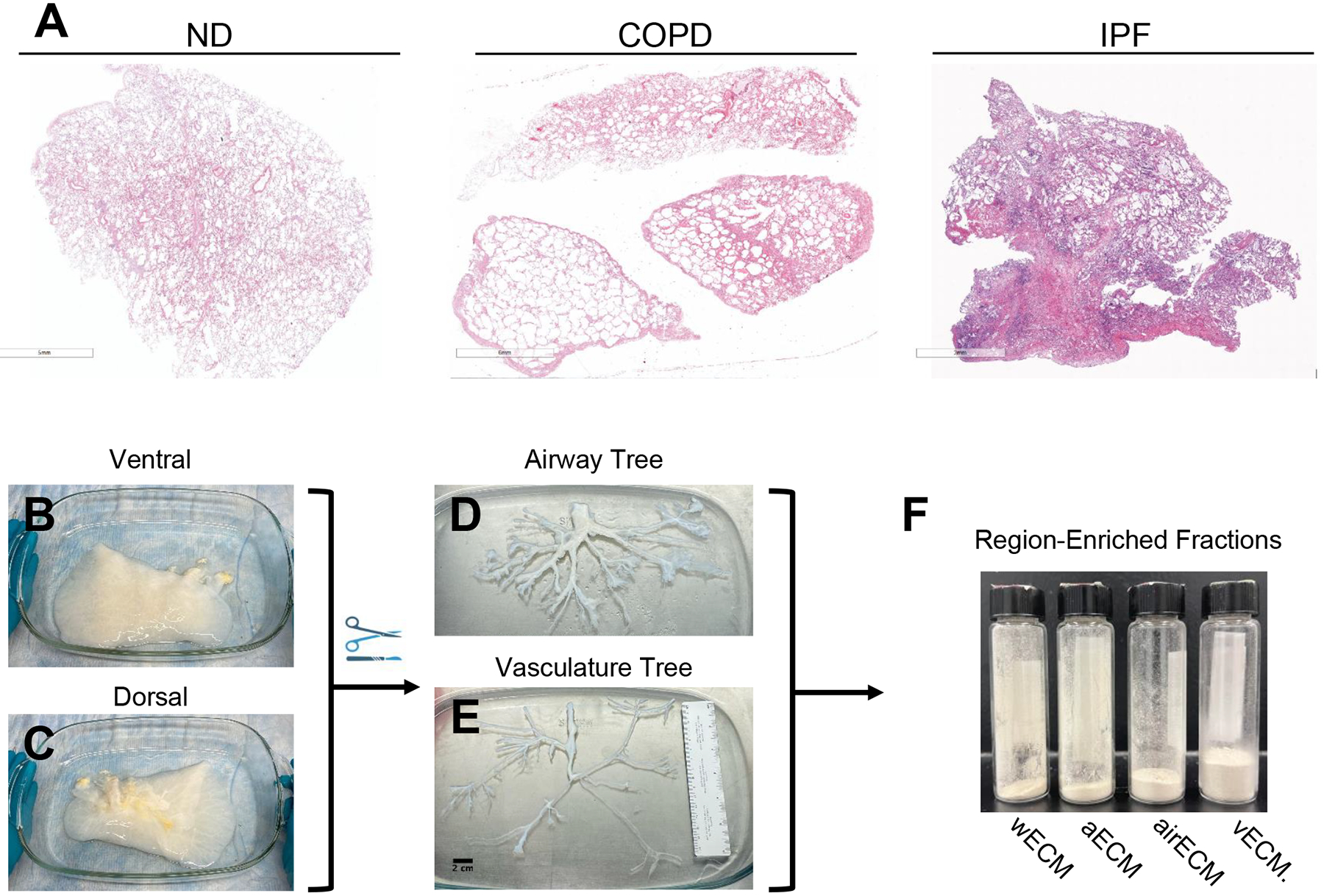
(A) H&E staining of native (non-decellularized) human patient lungs showing lung morphology, including emphysematous regions in COPD patient lung and fibrotic regions in IPF patient lungs. Representative image of decellularized normal lung on ventral (B) and dorsal (C) side. Representative image of isolated airway (D) and vascular (E) trees. (F) Liquid nitrogen milled ECM powders of whole decellularized lung ECM (wECM), as well as ECM from alveolar-enriched (aECM), airway-enriched (airECM), and vasculature-enriched (vECM) regions.
2.2. Validation of human lung decellularization.
Decellularization efficiency was assessed by measurement of residual double-stranded DNA (dsDNA) and by hematoxylin and eosin (H&E) staining, as previously described [18]. In brief, native and decellularized lung fragments (~1mm3) were excised from the distal portion of the lung, fixed in 4% paraformaldehyde, and mounted as 5μm sections for H&E staining. Quantitative dsDNA of lyophilized native and decellularized lung samples was performed using DNeasy Blood & Tissue Kit (Qiagen) and QuantiT PicoGreen dsDNA Assay Kit (Thermo) according to the manufacturer’s protocols. Qualitative assessment of any potential residual dsDNA was performed by gel electrophoresis on an 0.8% agarose gel labelled with SYBR Safe DNA Gel stain (Thermo).
2.3. Mass spectrometry analysis of decellularized ECM powder.
Decellularized ECM powder was resuspended in Mild Laemmli Sample Buffer (MLSB; 2% SDS, 62.5mM Tris-HCl) at a ratio of 1mg/100uL and heated for 30min at 45°C. Samples were centrifuged at 10,000 RCF and supernatant was collected and quantified using a Pierce BCA Protein Assay Kit (Thermo) according to the manufacturer’s protocols. To remove detergent impurities, samples (20–50mg per lane) were run through an SDS-PAGE gel at 80V for 10–15min. Protein bands were excised from the SDS-PAGE gel, fragmented into small cubes of approximately 1mm3, and subjected to destaining and tryptic in-gel digestion as previously described [18].
Tryptic digests were subsequently dried using a SpeedVac and resuspended in 6μL of 0.1% formic acid (FA) in 2.5 % acetonitrile (ACN) (Fisher Chemical). Tryptic peptide samples (2μL) were loaded onto a fused silica microcapillary liquid chromatography (LC) column (15cm long × 100μm inside diameter) packed in-house with C18 reversed-phase resin (1.8μm 120A, UChrom C18). Peptides were separated by applying a gradient of 0–50% Solvent B (0.1% FA in 80% ACN) at a flow rate of 300nL/min for 65min. Peptides were introduced via a nano-electrospray ionization (nano ESI) fitted onto a Thermo Q-Exactive Plus (QE+) mass spectrometer (Thermo Electron, San Jose, CA) that was operated in a Higher-energy C-trap dissociation mode to obtain both MS and tandem MS (MS/MS) spectra.
Mass spectrometry data were acquired in a data-dependent top-10 acquisition mode, using a full MS scan from 350–1600 m/z at 70,000 resolution (automatic gain control [AGC] target, 1e6; maximum ion time [max IT], 100ms; profile mode), and the lock mass function was activated (371.12m/z). Resolution for dd-MS2 spectra was set to 17,500 (AGC target: 1e5) with a maximum ion injection time of 50ms. The normalized collision energy was 27eV. Obtained MS spectra were searched against the Human protein database (UniProt-proteome_UP000005640_human) using Proteome Discoverer 2.5 (Thermo Electron, San Jose, CA). The search parameters permitted a 10ppm peptide MS tolerance and a 0.02Da MS/MS tolerance. Carboxymethylation of cysteines was set as a fixed modification and oxidation of methionine was set as a dynamic modification. For comparative analysis, in parallel, hydroxylation of Proline (+15.995 Da) and oxidation of methionine were set as dynamic modifications. Up to two missed tryptic peptide cleavages were considered, and three maximum dynamic modifications were allowed per peptide. The Fixed Value PSM Validator node was included in the workflow to limit the false positive (FP) rates to less than 1% in the dataset.
Result files from PD were consolidated by Scaffold (version Scaffold 5.1.1, Proteome Software Inc., Portland, OR). Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm with a Scaffold delta-mass correction [25]. Protein identifications were accepted if established at greater than 95.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm [26]. Proteins that contained similar peptides, and could not be differentiated based on MS/MS analysis alone, were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
Proteins were assigned manually assigned to protein identifications groups (ECM, cytoplasm, cytoskeletal, nuclear, membrane-associated, or secreted) in accordance with previously defined lists published by Naba et al. [27]. ECM proteins were further classified as “matrisome” sub-groups (collagens, ECM glycoproteins, ECM regulators, ECM-affiliated proteins, proteoglycans, or secreted factors) [27]. ECM proteins were additionally defined as basement membrane associated proteins in accordance with Jayadev et al. [28].
2.4. Statistics.
Semi-quantitative analysis of individual ECM proteins was performed by normalizing the relative abundance of spectral hits for each individual ECM protein (%) to total spectral hits of ECM proteins within each respective sample. Similarly, ECM protein type (i.e. collagens, ECM glycoproteins, etc.) was normalized as the relative abundance of spectral hits for each respective ECM protein type to total spectral hits of ECM proteins within each respective sample. Due to the non-normal distribution of the results obtained from mass spectrometry, and as the assumptions of one-way ANOVA test were not met, conditions were compared using a non-parametric Wilcoxon/Kruskal-Wallis rank sums test and were considered significant at p<0.05. Following Kruskal-Wallis, multiple pairwise comparisons were made using Dunn’s method for joint ranking. All statistical analyses were performed using JMP Pro 15.0.0. Principle component analysis (PCA) of each respective conditions normalized individual ECM protein quantifications were created using R 4.1.3. Heatmaps depicting the most abundant normalized ECM proteins across respective samples and pie charts depicting relative abundance of ECM protein types were generated using GraphPad Prism 9.4. Non-quantitative analysis was additionally utilized to assess qualitative differences between samples. Venn diagrams produced using BioVenn were generated by pooling all identified ECM proteins from respective conditions [29].
Results
3.1. Isolating specific anatomical regions of decellularized lungs.
Consistent with our previous studies, decellularized ND lungs retained much of the histological appearance of native (i.e. non-decellularized) lungs, while decellularized COPD lungs presented with similar spatially heterogeneous emphysematous changes as seen in native COPD lungs (Figure 1a, Supplemental Figure 1a) [17]. Comparably, decellularized IPF lungs maintained distinct fibrotic and non-fibrotic regions following decellularization (Figure 1a, Supplemental Figure 1a). Minimal residual DNA (<50ng/mg) was observed both quantitatively and qualitatively for all lungs following decellularization (Supplemental Figures 1a,c). Additionally, mass spectrometry analysis of native (i.e. non-decellularized) and decellularized lungs demonstrated overall preservation of matrisome proteins, apart from elastin, but significant loss of other non-matrisomal proteins with decellularization (Supplemental Figure2 2).To isolate anatomically specific regions, each decellularized whole lung lobe was subsequently dissected from the proximal regions of the large airways to the distal-most tips capable of extraction (Figure 1b–e). As lung vasculature parallels the airways in both the proximal and distal regions, we were able to simultaneously dissect out both airway and vasculature trees, providing materials for subsequent assessments of both airway and vasculature ECM (airECM and vECM, respectively) from the same lobe (Figure 1d–e). The distal-most tips of the airway trees, measuring under 1mm in diameter (correlating with generation 17–23 of the airway and alveoli), were further isolated to provide alveolar-enriched ECM (aECM) consisting of a mix of alveolar and pulmonary microcapillary ECM. Isolated regions, as well as non-dissected whole lung ECM (wECM), were subsequently lyophilized and liquid nitrogen milled into an ECM powder for subsequent proteomic analyses (Figure 1f).
3.2. ECM composition of non-diseased decellularized human lungs differs in a region-specific manner.
ND samples were initially assessed to determine respective ECM compositions within wECM as compared to isolated aECM, airECM, and vECM regions (Figure 2a). Mass spectrometry analyses demonstrated a range of proteins in the decellularized lungs (the full list with defined abbreviations is depicted in Supplemental Table 2). Principle component analyses (PCA) demonstrated that samples derived from individual ND lungs demonstrated remarkable regional-specific similarity in proteomic composition (Figure 2b). Similar to previous proteomic studies from our group and others characterizing ECM composition from whole ND human lungs, the most abundant proteins detected across all regions were basement membrane-associated proteins including type-VI collagen 6 (COL6), heparan sulfate proteoglycan-2/perlecan (HSPG2), fibronectin (FN1), and a variety of laminins (Figure 2c) [17,30,31]. Further analyses of the relative proportion and composition of basement membrane-associated proteins extracted from each region, using a defined basement-membrane protein database [28], demonstrated that wECM, aECM, and vECM were each enriched for total basement membrane-associated proteins in comparison to airECM (Figure 2d, Supplemental Figure 5a). Further, wECM and aECM contained an increased proportion of basement-membrane-associated collagens and laminins as compared to airECM and vECM, while airECM contained increased proteoglycans, and vECM contained increased FN1 (Supplemental Fig 5b, Figure 2e). Assessing the relative abundance of individual proteins, airECM was enriched with cartilage-associated collagens (COL2, COL9, COL10), ECM glycoproteins (aggrecan: ACAN), and proteoglycans (chondroadherin: CHAD) while vECM showed increased enrichment for blood vessel-associated ECM proteins and secreted factors, including Fn1, versican (VCAN), and various complement proteins (C1QA, C1QB) (Figure 2e–f). Notably, previous studies have shown that adding an additional database search with proline hydroxylation (Hyp), a common post-translational modification (PTM) within ECM fibrillar collagens, results in a significant increase in spectra identifications of fibrillar collagens (COL1, COL2, COL3) [32]. As such, parallel analyses of wECM, aECM, vECM, and airECM from ND lungs was performed with the addition of Hyp modification for comparison (Supplemental Figure 3). As expected, this evidenced an increase in the relative abundance (%) of fibrillar collagens (COL1, COL3, and COL2 (in airECM)) detected within the ECM composition of each sample as compared to analysis without Hyp modification data set(Supplemental Figure 3a). While detection of most other ECM proteins were unaffected his increase in spectral identifications of fibrillar collagens subsequently resulted in a relative decrease in abundance of several other highly identified ECM proteins (COL6, HSPG2, and ACAN in airECM), (Supplemental Figure 3a). tThe Hyp search also resulted in higher interpatient variability in the fibrillar collagens, notably within COL1A1 abundance, not previously observed (Supplemental Figure 3b; Figure 2c). Further, PCA analysis of complete ECM composition of ND regions using Hyp search resulted in loss of region-specific clusters, suggesting that the addition of these modifications obscured differences of less abundant ECM proteins between different lung regions (Supplemental Figure 3c; Figure 2b). As such, analysis of mass spectral data in the absence of Hyp modification search were utilized for the remainder of this study.
Figure 2. Regional ND Human Lung ECM Composition.
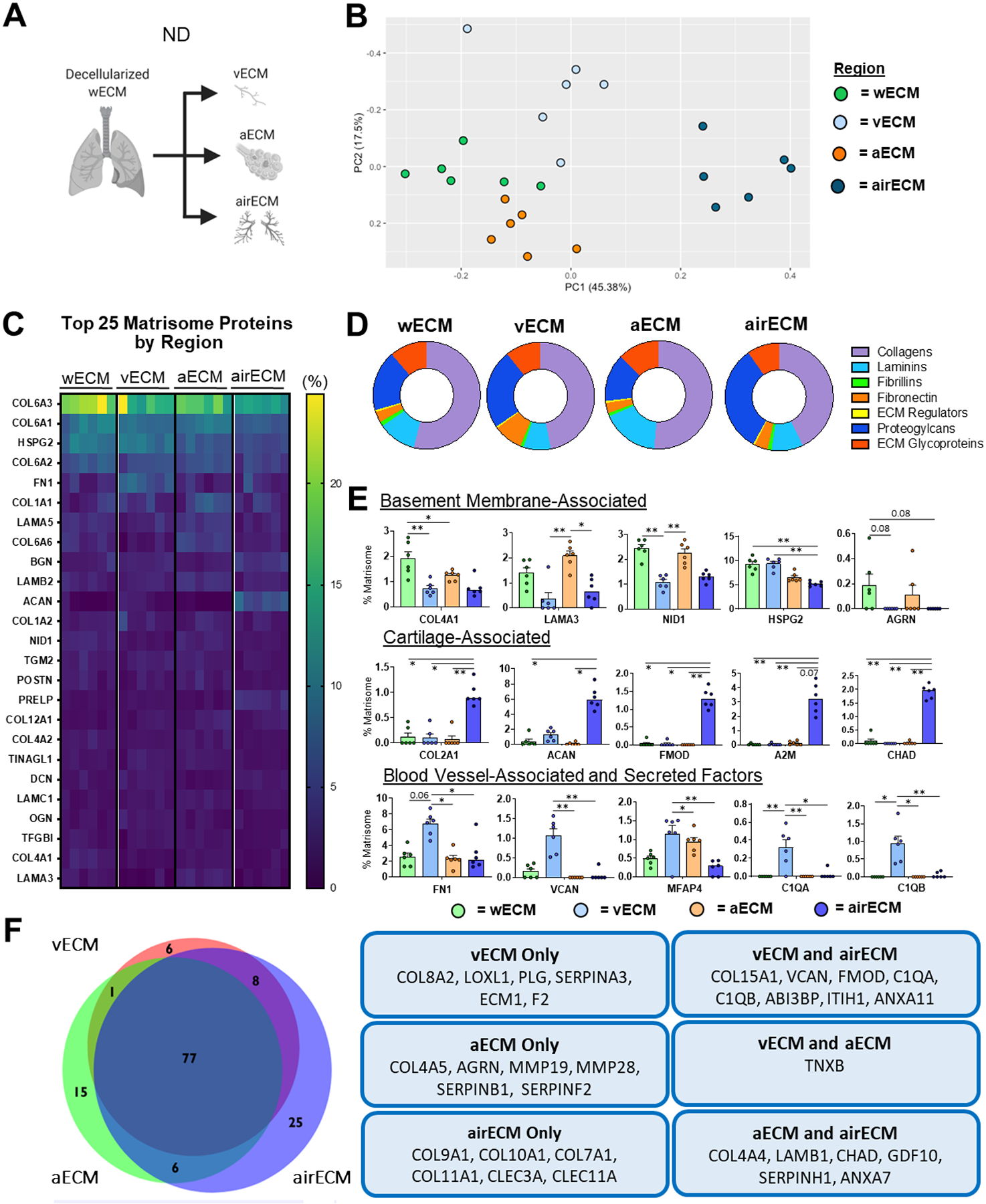
wECM, airECM, aECM, and vECM samples were obtained from six individual ND human lungs for proteomic analysis (n=6). (A) Schematic depicting samples utilized for proteomic analysis. (B) PCA plot demonstrating similarity of ECM protein composition amongst region-specific samples. Each data point represents the relative abundance (%) of all individual ECM proteins (normalized to abundance of total ECM composition within each respective sample). (C) Heatmap of top 25 normalized ECM proteins across all decellularized lung regions.(D) Ratio of mean basement membrane composition from decellularized lung specific regions. (E) Comparison of normalized individual ECM proteins across lung regions. Bars indicate mean +/− SEM, n=6 biological replicates; *p<0.05, **p<0.01 using Dunn’s method. Statistical trends are identified if 0.05≤p<0.1. (F) Qualitative Venn diagram depicting differences in region-specific ECM composition of pooled samples.
3.3. Region-specific ECM composition in diseased lungs.
Unlike ND lungs, PCA analysis of regional ECM protein composition between separate COPD patient lungs did not cluster strongly, suggesting heterogeneity between individual COPD patient lung regions (Figure 3a–b). In particular, wECM and aECM clusters were indistinguishable from one another, suggesting significant similarities in ECM composition between the regions (Figure 3b). Strikingly, the most highly detected ECM proteins amongst all COPD regions were largely consistent with proteins isolated from ND lungs, with the exception of cathepsin G (CTSG), a serine protease involved in elastin degradation which was increased in COPD lungs (Figure 3c) [33]. Further analysis demonstrated that wECM and aECM were both enriched for total amount of basement membrane-associated proteins (Supplemental Figure 5c), showing increased amounts of basement membrane-associated collagens as compared to airECM and vECM (Supplemental Figure 5d, Figure 3d). Similar to ND lungs, airECM demonstrated an enrichment of proteoglycans, including ACAN and fibromodulin (FMOD) (Supplemental Figure 5d, Figure 3e), as well as enrichment in cartilage-associated collagens (COL2, COL9, COL10) (Figure 3e–f). Further, vECM regions were the only regions to express VCAN, a blood vessel-associated proteoglycan, as well as ABI3BP, an ECM glycoprotein associated with COPD severity Figure 3f [34].
Figure 3. Regional COPD Human Lung ECM Composition.
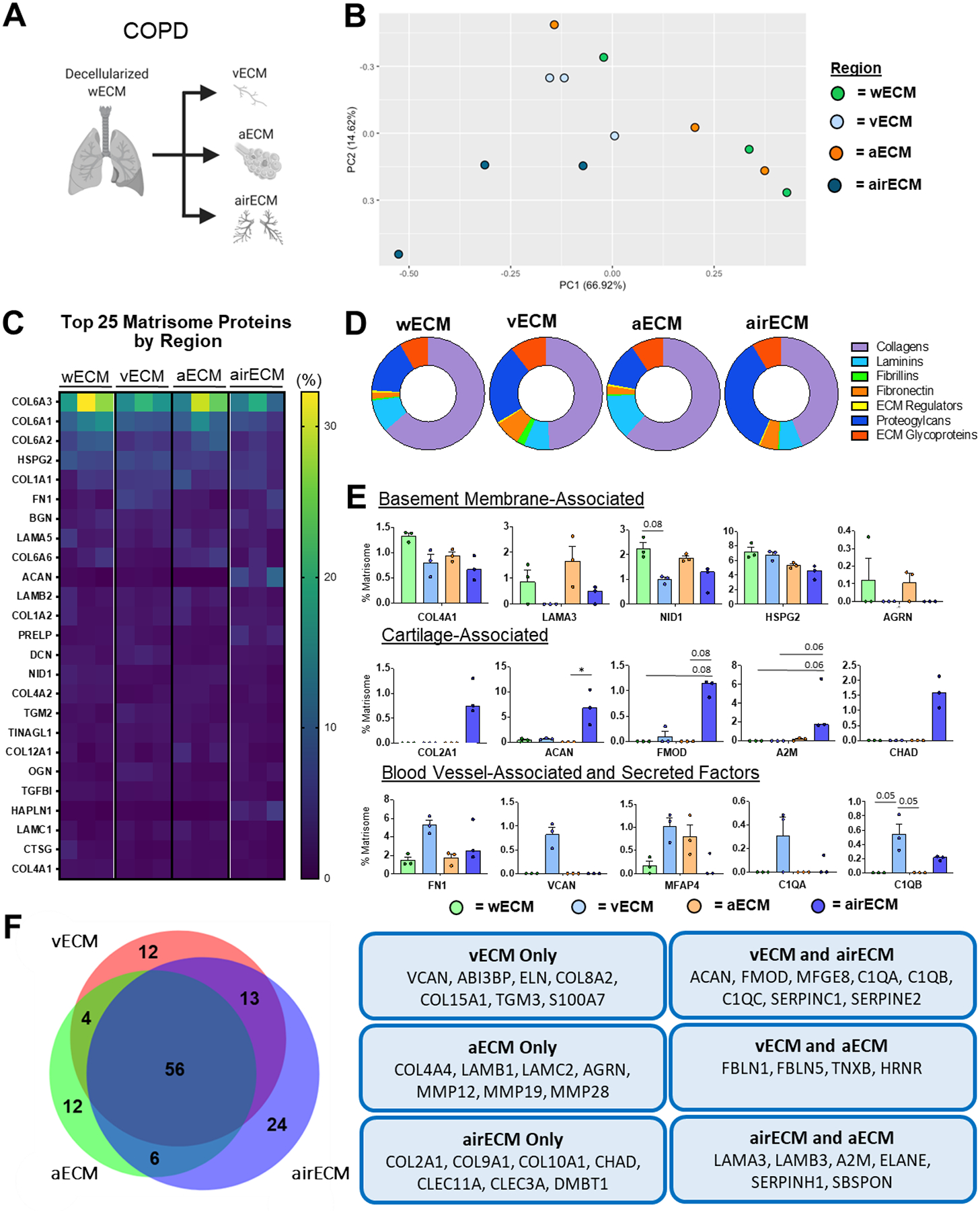
wECM, airECM, aECM, and vECM samples were obtained from three individual COPD human lungs for proteomic analysis (n=3). (A) Schematic depicting samples utilized for proteomic analysis. (B) PCA plot demonstrating similarity of ECM protein composition amongst region-specific samples. Each data point represents the relative abundance (%) of all individual ECM proteins (normalized to abundance of total ECM composition within each respective sample).(C) Heatmap of top 25 normalized ECM proteins across all decellularized lung regions. (D) Ratio of mean basement membrane composition from decellularized lung specific regions. (E) Comparison of normalized individual ECM proteins across lung regions. Bars indicate mean +/− SEM, n=3 biological replicates; *p<0.05, **p<0.01 using Dunn’s method. Statistical trends are identified if 0.05≤p<0.1. (F) Qualitative Venn diagram depicting differences in region-specific ECM composition of pooled samples.
Similar to COPD lung regions, PCA of regional ECM protein composition of IPF patient lungs did not form strong clusters within individual regions (Figure 4a–b). Additionally, wECM, aECM, and vECM all clustered together indistinguishably, suggesting similarities in total ECM composition in these different compartments (Figure 4b). Further, the most abundant ECM proteins from IPF lungs across regions were similar to those of ND and COPD lungs, with the exception of alpha-2-macroglobulin (A2M) and milk fat globule-EGF factor 8 (MFGE8), both of which have been implicated in the emergence and severity of pulmonary fibrosis (Figure 4c) [35,36]. Notably, no differences were observed in total basement membrane-associated proteins between wECM, aECM, and vECM, despite aECM containing significantly more basement-associated proteins than airECM (Supplemental Figure 5e). However, aECM did possess the highest amount of basement-membrane-associated collagens and laminins (Supplemental Figure 5f, Figure 4d). Additionally, consistent with ND and COPD lungs, IPF airECM was enriched with basement-membrane associated proteoglycans, most notably ACAN and FMOD (Supplemental Figure 5f, Figure 4d–e). However, due to the small sample size (n=3) and the expected heterogeneity of IPF samples, only few individual proteins were found to be significant altered in abundance between wECM, aECM, and vECM at the (Figure 4d–e). Notably, aECM was the only region to express MFAP2, a TGF-β binding ECM glycoprotein that has previously been shown to be upregulated in decellularized IPF ECM (Figure 4f) [23].
Figure 4. Regional IPF Human Lung ECM Composition.
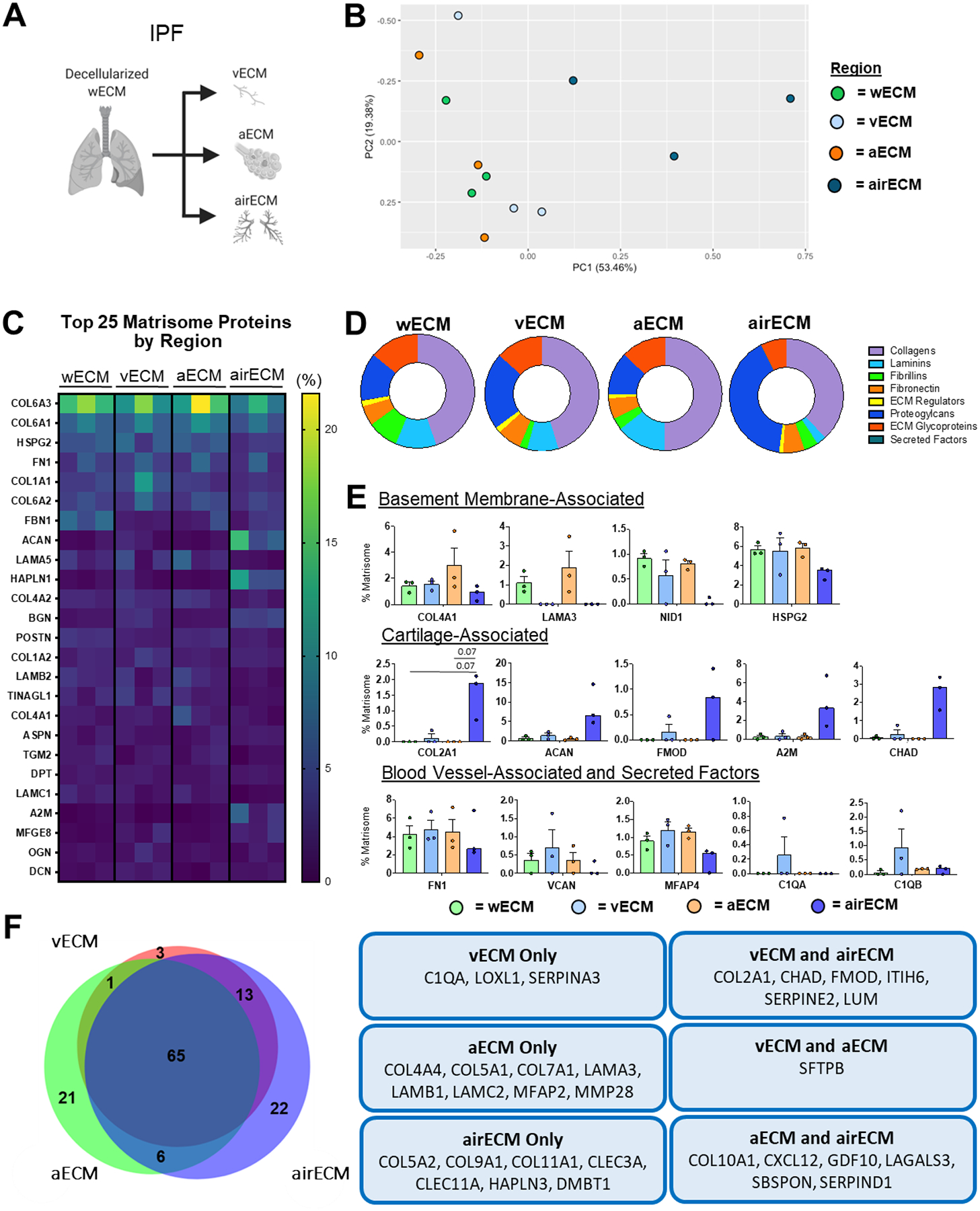
wECM, airECM, aECM, and vECM samples were obtained from three individual IPF human lungs for proteomic analysis (n=3). (A) Schematic depicting samples utilized for proteomic analysis. (B) PCA plot demonstrating similarity of ECM protein composition amongst region-specific samples. Each data point represents the relative abundance (%) of all individual ECM proteins (normalized to abundance of total ECM composition within each respective sample). (C) Heatmap of top 25 normalized ECM proteins across all decellularized lung regions. (D) Ratio of mean basement membrane composition from decellularized lung specific regions. (E) Comparison of normalized individual ECM proteins across lung regions. Bars indicate mean +/− SEM, n=3 biological replicates; *p<0.05, **p<0.01 using Dunn’s method. Statistical trends are identified if 0.05≤p<0.1. (F) Qualitative Venn diagram depicting differences in region-specific ECM composition of pooled samples.
3.4. Region-specific differences between ND and diseased lungs.
PCA demonstrated that wECM composition from ND and IPF lungs formed distinctly separate clusters while wECM from one COPD patient lung, COPD1, clustered more similarly to the ND population (Figure 5a,b; Supplementary Figure 5b). Of particular note, COPD1 wECM was obtained from a patient who quit smoking 36 years prior to death, while COPD2 and COPD3 quit 2 years prior to death or did not quit smoking at all, respectively (Supplementary Table 1). Analysis of the relative abundance of specific total ECM protein types demonstrated an enrichment of overall collagen content in wECM in COPD lungs and of ECM glycoproteins in IPF lungs, respectively (Figure 5c). While no individual collagen peptides were significantly increased in COPD wECM, subunits of Col6 (COL6A1, COL6A2, COL6A3) in COPD wECM showed a trend of enrichment, as demonstrated via IHC in human COPD patient lungs (Figure 5d) [37]. Further, despite a decrease in overall collagen content in IPF wECM compared to ND wECM, COL3A1 was increased, consistent with previous studies of IPF lung ECM composition, as well as a single-cell RNA sequencing study of fibroblasts during severe IPF [23,38]. Within the enrichment of total ECM glycoproteins in IPF wECM, several individual proteins, including nephronectin (NPNT) and tenascin XB (TNXB), were increased, as well as a trending increase in fibrillin-1 (FBN1), as previously demonstrated through mass spectrometry of decellularized IPF lungs (NPNT, FBN1) and IHC of fibrotic human lungs (tenascin), respectively (Figure 5d) [23,39]. Notably, an increase in the matrix metallopeptidase 19 (MMP19), a key regulator of pulmonary fibrosis, was observed in IPF wECM Figure 5d [40]. Interestingly, no ECM proteins were expressed exclusively in COPD wECM, while IPF wECM expressed 16 unique ECM proteins, including secreted factors CXCL12 and CXCL13, which have been implicated in pulmonary fibrosis (Figure 5e) [41,42].
Figure 5. ND vs. Diseased wECM Lung ECM Composition.
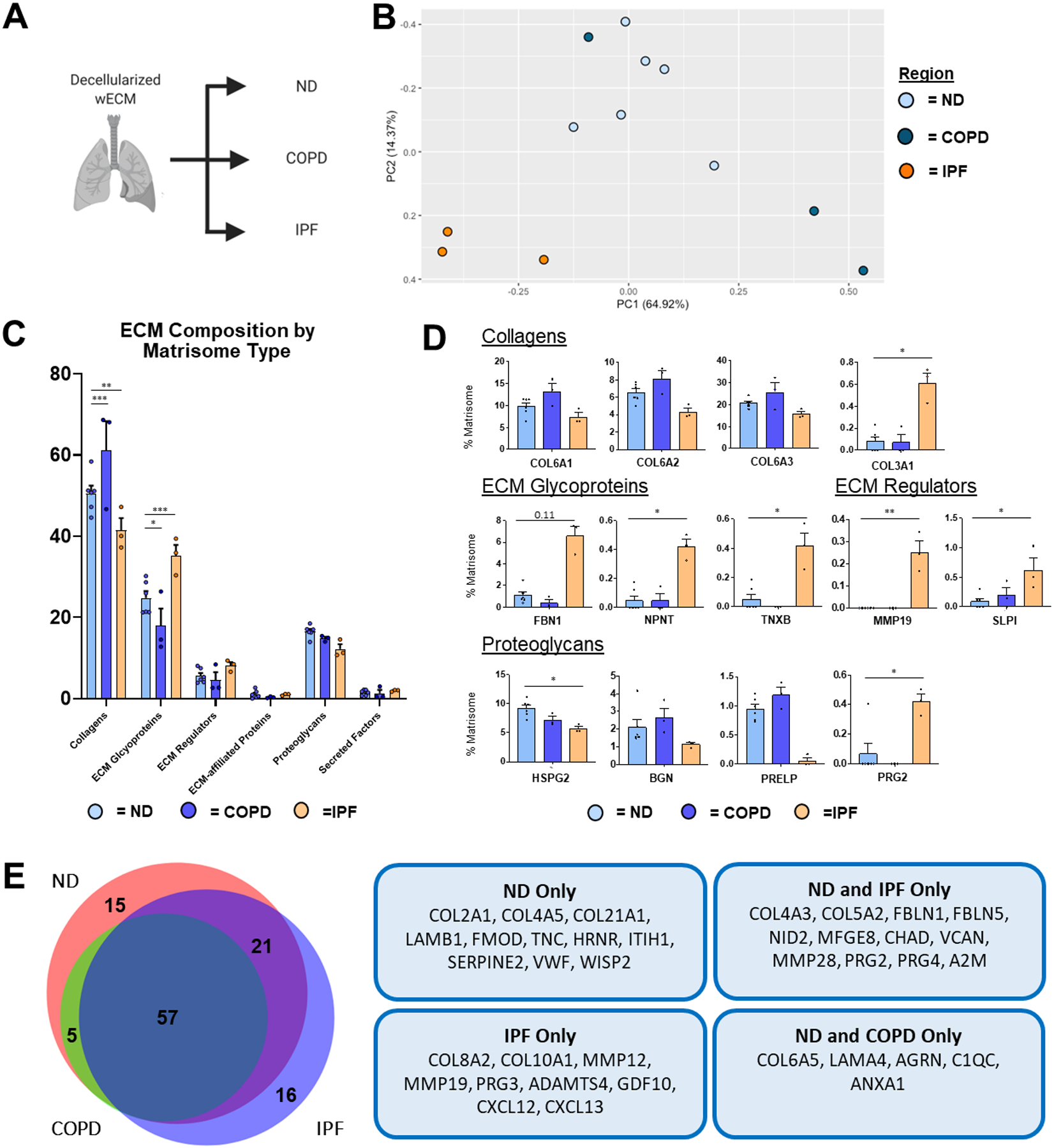
Proteomic analysis of wECM samples obtained from ND (n=6), COPD (n=3), and IPF (n=3) decellularized patient lung lobes. (A) Schematic depicting samples utilized for proteomic analysis. (B) PCA plot demonstrating similarity of wECM protein composition across lung conditions. Each data point represents the relative abundance (%) of all individual wECM proteins (normalized to abundance of total wECM composition) within each respective sample.(C) Relative abundance (%) of wECM protein type (normalized to total wECM composition within each respective sample) within each respective sample). (D) Comparison of normalized individual wECM proteins across lung conditions. Bars in (C,D) indicate mean +/− SEM; *p<0.05, **p<0.01 using Dunn’s method. Statistical trends are identified if 0.05≤p<0.1. (E) Qualitative Venn diagram depicting differences in wECM proteins of pooled samples by condition.
Similar to wECM, analysis of aECM composition by PCA demonstrated distinct clustering of ECM composition of IPF aECM, while a single COPD sample, COPD1 aECM, clustered more similarly to the ND population (Figure 6a–b; Supplementary Figure 5b). Also comparable with wECM results, COPD aECM showed an increase in collagen content compared to ND aECM, while IPF aECM showed a decrease in collagen content (Figure 6c). Additionally, no individual collagen peptides were significantly increased in COPD aECM, despite a trending increase in COL6A1, COL6A2, and COL6A3 (Figure 6d). IPF aECM showed an increase in COL10A1, a cartilage-associated collagen that has been shown to correlate with increased IPF severity, that was not present in either non-diseased aECM or COPD aECM (Figure 6d–e) [43]. Further, COPD aECM showed a decrease in total ECM glycoproteins (Figure 6c), while IPF aECM contained significantly more fibulin-2 (FBLN2), as has previously been described by mass spectrometry of decellularized IPF lungs (Figure 6d) [23]. Interestingly, IPF aECM also possessed increased amounts of several proteoglycans, including hyaluronan and proteoglycan link protein-1 (HAPLN1), proteoglycan-2 (PRG2), and VCAN, similar to observations in studies analyzing the fibrotic foci of IPF lungs (Figure 6d) [23,44]. Similar to analysis of wECM, IPF aECM was the only aECM tissue to contain COL10A1, CXCL12, and CXCL13 (Figure 6e). In addition, aECM from COPD lungs were the only aECM tissues to contain von Willebrand factor (Vwf), a biomarker of COPD (Figure 6e) [45].
Figure 6. ND vs. Diseased aECM Lung ECM Composition.
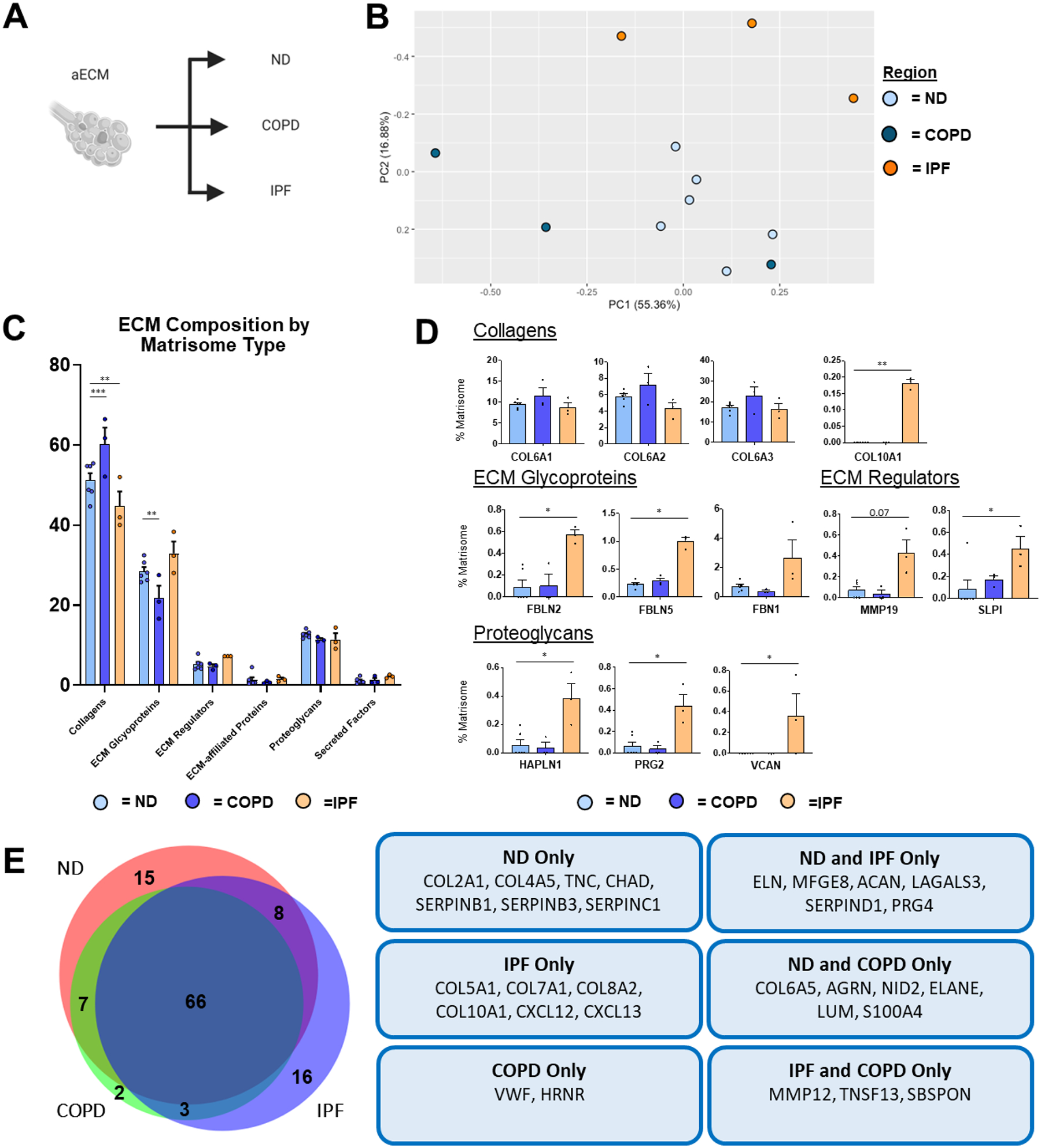
Proteomic analysis of aECM samples obtained from ND (n=6), COPD (n=3), and IPF (n=3) decellularized patient lung lobes. (A) Schematic depicting samples utilized for proteomic analysis. (B) PCA plot demonstrating similarity of aECM protein composition across lung conditions. Each data point represents the relative abundance (%) of all individual aECM proteins (normalized to abundance of total aECM composition) within each respective sample.(C) Relative abundance (%) of aECM protein type (normalized to total aECM composition within each respective sample) within each respective sample). (D) Comparison of normalized individual aECM proteins across lung conditions. Bars in (C,D) indicate mean +/− SEM; *p<0.05, **p<0.01 using Dunn’s method. Statistical trends are identified if 0.05≤p<0.1. (E) Qualitative Venn diagram depicting differences in aECM proteins of pooled samples by condition.
PCA of airECM composition from ND, COPD, and IPF patients demonstrated large interpatient variability, particularly within COPD and IPF airECM, suggesting heterogeneous ECM composition within and between the different samples (Figure 7a–b). Interestingly, no significant changes were seen with regards to the abundance of any major ECM protein types (Figure 7c). However, several differences were observed between ND, COPD, and IPF airECM at the individual protein level (Figure 7d). In particular, basement membrane associated collagen (COL6A6), ECM glycoproteins (LAMC1, nidogen-1: NID1), and proteoglycan (HSPG2) were all significantly less abundant in IPF airECM, in concordance with the dysregulation of the basement membrane in IPF lungs (Figure 7d) [46]. Further, proteases MMP12 and MMP19 were only present in IPF airECM (Figure 7e). Additionally, vitronectin (VTN), a multi-functional glycoprotein located at the bronchial surface was significantly less abundant in COPD airECM [47], while a secreted marker of severe COPD, S100B, was significantly more abundant (Figure 7d) [48].
Figure 7. ND vs. Diseased airECM Lung ECM Composition.
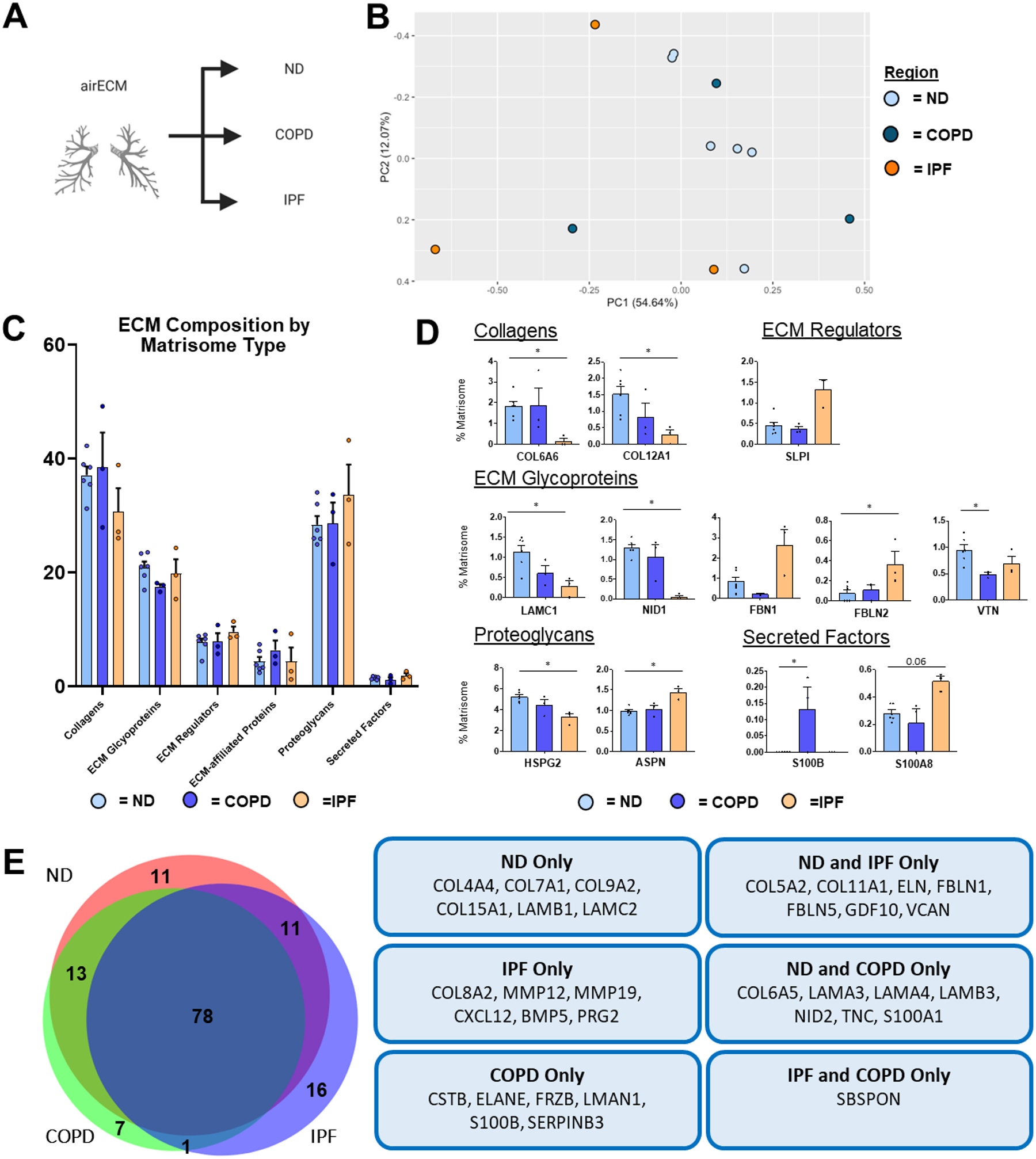
Proteomic analysis of airECM samples obtained from ND (n=6), COPD (n=3), and IPF (n=3) decellularized patient lung lobes. (A) Schematic depicting samples utilized for proteomic analysis. (B) PCA plot demonstrating similarity of airECM protein composition across lung conditions. Each data point represents the relative abundance (%) of all individual airECM proteins (normalized to abundance of total airECM composition) within each respective sample.(C) Relative abundance (%) of airECM protein type (normalized to total airECM composition within each respective sample) within each respective sample). (D) Comparison of normalized individual airECM proteins across lung conditions. Bars in (C,D) indicate mean +/− SEM; *p<0.05, **p<0.01 using Dunn’s method. Statistical trends are identified if 0.05≤p<0.1. (E) Qualitative Venn diagram depicting differences in airECM proteins of pooled samples by condition.
PCA of vECM composition from ND, COPD, and IPF revealed heterogeneous clustering between individual samples, particularly in COPD and IPF (Figure 8a–b). However, samples did cluster together in accordance with disease condition (Figure 8b). Interestingly, similar to airECM, vECM did not show differences in abundance between ECM protein categories (Figure 8c). However, at the individual protein level, collagens COL3A1 and COL8A1, ECM glycoprotein dermatopontin (DPT), and ECM regulators MMP12 and MMP19 were all enriched in IPF vECM, in accordance with previous single-cell RNA sequencing studies of fibroblasts in IPF patients (Figure 8d–e) [38,40]. Interestingly, COPD vECM was significantly enriched in decorin (DCN), a proteoglycan involved in fibrillogenesis, that has previously been shown to be downregulated in COPD airways (Figure 8d) [49].
Figure 8. ND vs. Diseased vECM Lung ECM Composition.
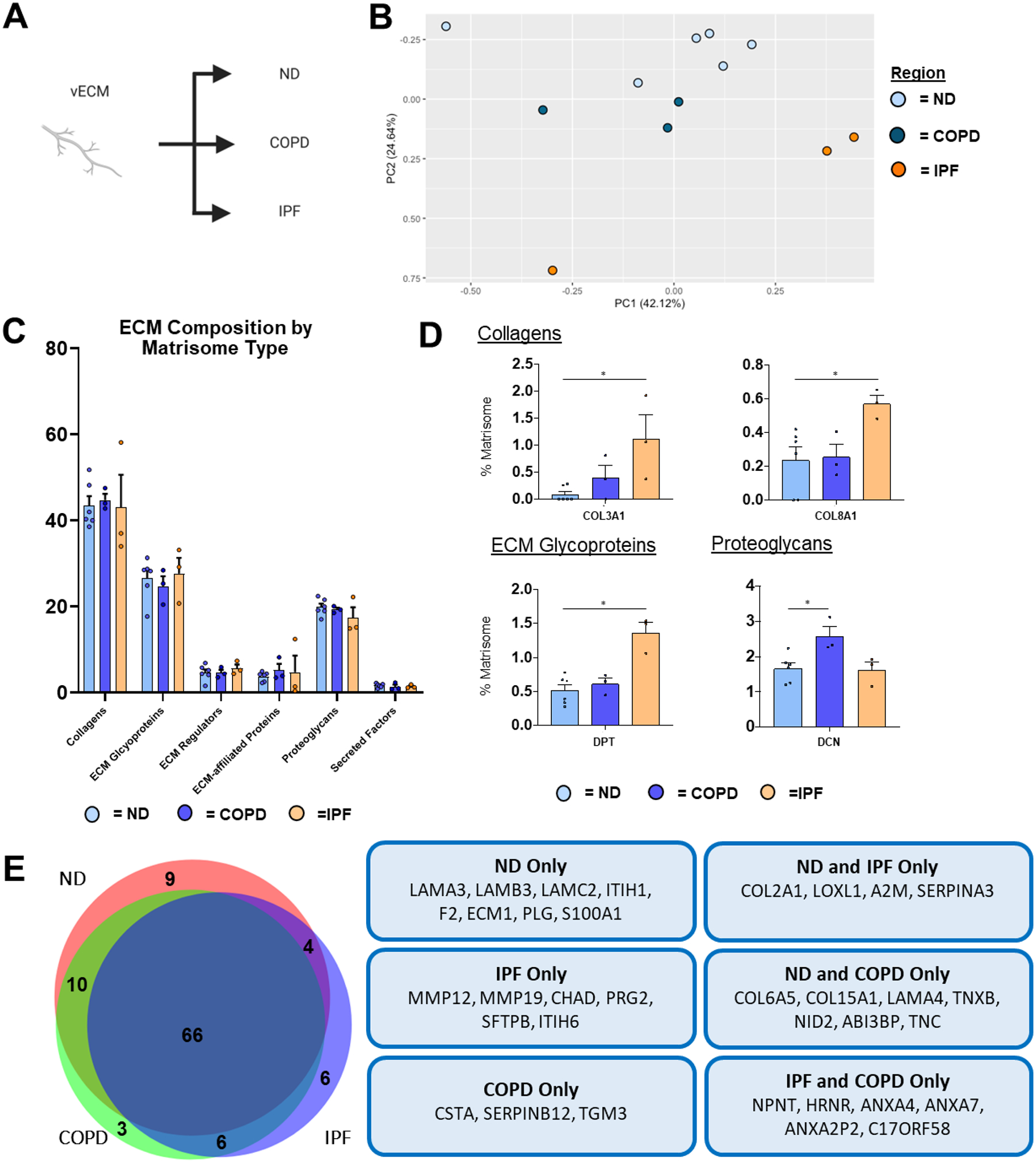
Proteomic analysis of vECM samples obtained from ND (n=6), COPD (n=3), and IPF (n=3) decellularized patient lung lobes. (A) Schematic depicting samples utilized for proteomic analysis. (B) PCA plot demonstrating similarity of vECM protein composition across lung conditions. Each data point represents the relative abundance (%) of all individual vECM proteins (normalized to abundance of total vECM composition) within each respective sample.(C) Relative abundance (%) of vECM protein type (normalized to total vECM composition within each respective sample) within each respective sample). (D) Comparison of normalized individual vECM proteins across lung conditions. Bars in (C,D) indicate mean +/− SEM; *p<0.05, **p<0.01 using Dunn’s method. Statistical trends are identified if 0.05≤p<0.1. (E) Venn diagram depicting differences in vECM proteins of pooled samples by condition.
Discussion
Defining features of several chronic lung diseases, such as COPD and IPF, are irreversible compositional and spatial abnormalities within the lung ECM that propagate disease pathology. Prior studies characterizing the lung ECM in normal lungs, and following chronic lung disease, have largely relied on: (1) targeted IHC that can only characterize select ECM proteins or (2) mass spectrometry approaches that do not discern between different anatomical regions with the lung. Within this study, we have developed a post-decellularization dissection method to strictly characterize the global ECM composition of individual anatomical lung regions (aECM, airECM, and vECM) from both non-diseased and diseased (i.e. COPD and IPF) decellularized human lungs. Further, the region and disease-specific ECM characterized within this study may additionally serve as novel biomaterials for future tissue engineering and ex vivo modeling applications for determining key cell-ECM interactions that contribute to pathological cellular phenotypes.
We demonstrate that isolated anatomical lung regions (airECM, aECM, vECM) present unique proteomic signatures as compared to wECM derived from ND patients (Figure 2). Between regions, airECM exhibited the most distinct ECM prolife from wECM, aECM, and vECM, due to the inclusion of cartilaginous tissue from the upper airways, as indicated by the enrichment of COL2A1, the primary collagen associated with cartilaginous tissues, as well as an array of cartilage-associated proteoglycans, including ACAN, FMOD, and CHAD (Figure 2c–f) [11,50,51]. Further, aECM displayed the most similar ECM composition profile to wECM (Figure 2c–f), which is likely explained by the majority of the lung surface area being occupied by alveolar space and because wECM samples were taken from distal regions of the decellularized lung lobe [52]. Specifically, both aECM and wECM were enriched in key basement membrane proteins, including COL4, HSPG2, NID1, AGRN, and a range of laminins, as compared to airECM and vECM. While vECM was also composed of an increased proportion of basement-membrane associated proteins in comparison to airECM (Supplemental Figure 5a), vECM was also characterized by an increase proportion of blood vessel-associated proteins, including an increased amount of FN1 and the proteoglycan VCAN, confirming successful isolation of vasculature regions [53,54]. Taken together, post-decellularization dissection methods of ND lungs provided reproducible and biologically relevant global proteomic ECM signatures for both wECM and distinct anatomical regions. Notably, however, region specific ECM signatures did not persist in either COPD and IPF conditions, with the exception of airECM, which was still defined by an increased proportion of cartilage-associated collagens and proteoglycans (Figure 3c–f, Figure 4c–f). The lack of diversity between wECM, aECM, and vECM may in part be due to the spatial heterogeneity typical of both COPD and IPF pathology, where only select areas within specific regions of patient lungs display emphysema or fibrosis, respectively. Additionally, analysis of diseased lungs for both COPD and IPF conditions in this study were limited by the amount of available donor human samples.
As observed in previous proteomic studies of decellularized human lungs, we found that the primary subunits of COL6 (COL6A1, COL6A2, COL6A3) were consistently amongst the most abundant ECM proteins identified across all samples independent of lung condition (Figure 2–4) [17,23,30,31,55]. The abundance of COL6 subunits identified by mass spectrometry may be in part due to binding of COL6 to COL1 as well as a variety of elastin-interacting proteins [56–58]. However, despite the acknowledged prevalence of COL6 in the lung, its role in lung development, function, and pathology remains poorly understood [37,59–61]. COL6 is known to be present within both the basement membrane and interstitial matrix of the lung and has been demonstrated to connect these matrices as well as interact directly with resident cells [55,62–64]. Further, COL6 deficiency has been shown to lead to aberrant lung physiology and cellular homeostasis while also demonstrated to be upregulated in COPD [37,65,66]. Therefore, the findings of this study corroborate previous findings indicating the importance of further evaluating the role of COL6 in lung physiology and disease progression
This study further sought to compare the differences in ND ECM composition to both disease conditions, COPD and IPF, within wECM and individual anatomical regions (aECM, airECM, vECM). We found, similar to previous proteomic studies of whole decellularized COPD and IPF lungs, that there were far greater differences between ND and IPF lungs than between ND and COPD lungs at the individual ECM protein level independent of anatomical region [17,23]. Additionally, wECM and aECM regions had the largest differences in ECM composition between ND and COPD or IPF lungs. We observed, in both wECM and aECM, an increase in total collagen between ND and COPD lungs (Figure 5c, Figure 6c). Interestingly, an increase in total collagen in COPD lungs has previously been shown in a proteomic study of native human lungs, where total collagen accounted for 60% of the total ECM composition in COPD and IPF lungs as compared to 47% of the ND control group [67]. However, we also observed a decrease in total collagen in IPF compared to ND lungs in both wECM and aECM (Figure 5c, Figure 6c). As collagen deposition, particularly within the alveolar space, is a hallmark trait of IPF, we expected to see an increase in total collagen within IPF wECM and aECM samples. The lack of enrichment of collagen in IPF samples may, in part, be due to incomplete solubilization of dense fibrotic regions of the IPF lungs or due to collagen crosslinking preventing detection via mass spectrometry.
However, despite the decrease in overall collagen in IPF lungs, we observed an increase in COL3A1 in wECM and vECM (Figure 5d, Figure 8d). Previously, COL3A1 has been identified as upregulated in IPF as demonstrated using proteomics of decellularized whole IPF lungs, single-cell RNA sequencing showing an upregulation of COL3A1 by myofibroblasts during IPF disease progression, and even recently as a potential serum biomarker for progressive disease phenotypes in IPF patients [23,38,68]. As such, this study corroborates the potential importance of COL3A1 in IPF and, as such, further studies should be performed on the impact of COL3A1 in disease progression.
The proportion of ECM glycoproteins within total ECM composition was also altered between ND and disease groups, decreasing between ND and COPD lungs in wECM and aECM, and increasing between ND and IPF in wECM. As ECM glycoproteins provide a vast array of biochemical cues, through attaching glycosaminoglycans and directly binding to cells, the alterations in ECM glycoproteins within disease conditions likely has a significant impact on cell behavior and disease progression [19]. Within the ECM glycoprotein category, we demonstrate an increase in fibulins (FBLN2 and FBLN3) in IPF aECM, similar to previous studies demonstrating increased fibulin gene expression in fibroblasts during progression of IPF.
While this study demonstrates novel characterization of region-specific lung ECM composition in ND and diseases lungs, we acknowledge that the data presented is dependent on several factors including the methods of decellularization, dissection, and mass spectrometry analysis. In particular, we followed our group’s published Triton/SDC decellularization protocol [18]. Similar to other methods, Triton/SDC decellularization has previously been shown to lead to a decrease in lung elastin, which may in part account for the minimal elastin identifications within our dataset [17,69]. In addition, we employed an SDS-solubilization method for ECM proteins prior to mass spectrometry analysis, as previously reported by our group and others [17,18,70,71]. Other methods, such as cyanogen bromide (CNBr) digestion of insoluble proteins following our initial SDS digestion, may have lead to an increase in the solubilization of some (COL1A1, COL1A2, FBN1), but not all (elastin), larger insoluble ECM proteins as previously demonstrated [72]. However, given the coverage of ECM proteins identified using our current protocols we are confident with the methods utilized within this study.
In addition, this study employed dissection of decellularized lungs for the enrichment of region-specific ECM at a macroscopic level in order to obtain samples for both proteomic compositional characterization as well as the collection of biomaterials for future tissue engineering the ex vivo culture applications. As such, follow-up studies that possess greater spatial resolution at the microscopic level, notably laser capture microdissection coupled mass spectrometry (LCM-MS), may provide additional information on region and disease specific ECM composition [44,73]. Further, as both COPD and IPF are spatially heterogenous lungs diseases, we acknowledge the limitation of patient samples (n=3 for both COPD and IPF) within this study. While we did find that analysis of mass spectra using a Hyp modification search parameter led to increased interpatient variability amongst fibrillar collagens (particularly COL1, COL2, and COL3), we demonstrate that our standard analysis (without Hyp modification search parameters) possessed minimal interpatient variability and thus find our methods were suitable for this study. In summary, the methodology presented within this study provide a novel and simple method for isolating specific decellularized lung regions for advanced proteomics of the human lung ECM in non-diseased and diseased lung conditions. We demonstrate that individual lung regions contain specific ECM signatures at both the structural and biochemical ECM protein level. Further, we demonstrate proteomic ECM differences between independent anatomical regions of ND, COPD, and IPF lungs which could provide novel biotherapeutic targets as well as provide a proteomic index for future studies. In addition, ECM powders acquired during the processing of individual samples utilized in this study are amenable to future biomaterials studies, including the formation of region specific ECM-based 3D hydrogels for cell culture [74,75].
Supplementary Material
Acknowledgement
The authors would like to thank Ying Wai Lam for the Vermont Genetics Network for their help and dedication with mass spectrometry. Additionally, the authors would like to thank Biorender.com, which was used for the creation of the graphical abstract and schematics within figures throughout the manuscript. This work was supported by: NHBLI R01 HL127144-01 (DJ Weiss, PI). The Vermont Biomedical Research Network Proteomics Facility (RRID: SCR_018667) is supported through NIH grant P20GM103449 from the INBRE Program of the National Institute of General Medical Sciences
Declaration of interests
Daniel J. Weiss reports financial support was provided by University of Vermont. Daniel J. Weiss reports a relationship with National Heart Lung and Blood Institute that includes: funding grants. None to declare
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CRediT Author Statement
Hoffman et al. Regional and Disease Specific Human Lung Extracellular Matrix Composition jbmt61723
Evan T. Hoffman: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing-original, writing-revision, visualization
Franziska E. Uhl: conceptualization, methodology, validation, investigation, data curation, writing-original, writing-revision, visualization
Loredana Asarian: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing-original, writing-revision, visualization
Bin Deng: formal analysis, investigation, data curation, writing-revision
Chloe Becker: investigation
Juan J. Uriarte: investigation, writing-original, writing-revision
Isaac Downs: investigation
Brad Young: investigation
Daniel J. Weiss: conceptualization, methodology, validation, formal analysis, investigation, resources, data curation, writing-original, writing-revision, visualization, supervision, project administration, funding acquisition
Data Availability
The mass spectral data (raw, PD msf, mzML) and LC-MS/MS and data analysis method required to reproduce these findings are available to download from Supplementary Table 2. The processed data required to reproduce these findings are available to download from Supplementary Table 2. This data has further been uploaded to the MassIVE repository with the following identifiers:
MassIVE accession: MSV000090771
ProteomeXchange accession: PXD038289
DOI: 10.25345/C58911W11
References
- [1].Dunsmore SE, Rannels DE, Extracellular matrix biology in the lung, Am. J. Physiol. Cell. Mol. Physiol 270 (1996) L3–L27. 10.1152/ajplung.1996.270.1.L3. [DOI] [PubMed] [Google Scholar]
- [2].Balestrini JL, Niklason LE, Extracellular matrix as a driver for lung regeneration., Ann. Biomed. Eng 43 (2015) 568–76. 10.1007/s10439-014-1167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhou Y, Horowitz JC, Naba A, Ambalavanan N, Atabai K, Balestrini J, Bitterman PB, Corley RA, Sen Ding B, Engler AJ, Hansen KC, Hagood JS, Kheradmand F, Lin QS, Neptune E, Niklason L, Ortiz LA, Parks WC, Tschumperlin DJ, White ES, Chapman HA, Thannickal VJ, Extracellular matrix in lung development, homeostasis and disease, Matrix Biol. 73 (2018) 77–104. 10.1016/j.matbio.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hynes RO, Naba A, Overview of the matrisome-An inventory of extracellular matrix constituents and functions, Cold Spring Harb. Perspect. Biol 4 (2012). 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hynes RO, The Extracellular Matrix: Not Just Pretty Fibrils, Science (80-.) 326 (2009) 1216–1219. 10.1126/SCIENCE.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hussey GS, Dziki JL, Badylak SF, Extracellular matrix-based materials for regenerative medicine, Nat. Rev. Mater 3 (2018) 159–173. 10.1038/s41578-018-0023-x. [DOI] [Google Scholar]
- [7].Burgstaller G, Oehrle B, Gerckens M, White ES, Schiller HB, Eickelberg O, The instructive extracellular matrix of the lung: basic composition and alterations in chronic lung disease., Eur. Respir. J 50 (2017) 1601805. 10.1183/13993003.01805-2016. [DOI] [PubMed] [Google Scholar]
- [8].Annoni R, Lancaąs T, Tanigawa RY, Matsushita MDM, Fernezlian SDM, Bruno A, Da Silva LFF, Roughley PJ, Battaglia S, Dolhnikoff M, Hiemstra PS, Sterk PJ, Rabe KF, Mauad T, Extracellular matrix composition in COPD, Eur. Respir. J 40 (2012) 1362–1373. 10.1183/09031936.00192611. [DOI] [PubMed] [Google Scholar]
- [9].Herrera J, Henke CA, Bitterman PB, Extracellular matrix as a driver of progressive fibrosis, J. Clin. Invest 128 (2018) 45–53. 10.1172/JCI93557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bateman ED, Turner-Warwick M, Adelmann-Grill BC, Immunohistochemical study of collagen types in human foetal lung and fibrotic lung disease, Thorax. 36 (1981) 645–653. 10.1136/thx.36.9.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu L, Stephens B, Bergman M, May A, Chiang T, Role of Collagen in airway mechanics, Bioengineering. 8 (2021) 1–13. 10.3390/bioengineering8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kuhn C, McDonald JA, The roles of the myofibroblast in idiopathic pulmonary fibrosis: Ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis, Am. J. Pathol 138 (1991) 1257–1265. /pmc/articles/PMC1886011/?report=abstract (accessed June 13, 2022). [PMC free article] [PubMed] [Google Scholar]
- [13].Kuhn C, Boldt J, King TE, Crouch E, Vartio T, McDonald JA, An Immunohistochemical Study of Architectural Remodeling and Connective Tissue Synthesis in Pulmonary Fibrosis, Am. Rev. Respir. Dis 140 (1989) 1693–1703. 10.1164/ajrccm/140.6.1693. [DOI] [PubMed] [Google Scholar]
- [14].Bensadoun ES, Burke AK, Hogg JC, Roberts CR, Proteoglycan deposition in pulmonary fibrosis, Am. J. Respir. Crit. Care Med 154 (1996) 1819–1828. 10.1164/ajrccm.154.6.8970376. [DOI] [PubMed] [Google Scholar]
- [15].Broekelmann TJ, Limper AH, Colby TV, McDonald JA, Transforming growth factor β1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis, Proc. Natl. Acad. Sci. U. S. A 88 (1991) 6642–6646. 10.1073/pnas.88.15.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Madri JA, Furthmayr H, Collagen polymorphism in the lung. An immunochemical study of pulmonary fibrosis, Hum. Pathol 11 (1980) 353–366. 10.1016/S0046-8177(80)80031-1. [DOI] [PubMed] [Google Scholar]
- [17].Wagner DE, Bonenfant NR, Parsons CS, Sokocevic D, Brooks EM, Borg ZD, Lathrop MJ, Wallis JD, Daly AB, Lam YW, Deng B, DeSarno MJ, Ashikaga T, Loi R, Weiss DJ, Comparative decellularization and recellularization of normal versus emphysematous human lungs, Biomaterials. 35 (2014) 3281–3297. 10.1016/j.biomaterials.2013.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Uhl FE, Wagner DE, Weiss DJ, Preparation of decellularized lung matrices for cell culture and protein analysis, in: Methods Mol. Biol, 2017: pp. 253–283. 10.1007/978-14939-7113-8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Uhl FE, Zhang F, Pouliot RA, Uriarte JJ, Rolandsson Enes S, Han X, Ouyang Y, Xia K, Westergren-Thorsson G, Malmström A, Hallgren O, Linhardt RJ, Weiss DJ, Functional role of glycosaminoglycans in decellularized lung extracellular matrix, Acta Biomater. 102 (2020) 231–246. 10.1016/j.actbio.2019.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Calle EA, Hill RC, Leiby KL, Le AV, Gard AL, Madri JA, Hansen KC, Niklason LE, Targeted proteomics effectively quantifies differences between native lung and detergent-decellularized lung extracellular matrices, Acta Biomater. 46 (2016) 91. 10.1016/J.ACTBIO.2016.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li Q, Uygun BE, Geerts S, Ozer S, Scalf M, Gilpin SE, Ott HC, Yarmush ML, Smith LM, V Welham N, Frey BL, Proteomic analysis of naturally-sourced biological scaffolds., Biomaterials. 75 (2016) 37–46. 10.1016/j.biomaterials.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wagner DE, Bonenfant NR, Sokocevic D, DeSarno MJ, Borg ZD, Parsons CS, Brooks EM, Platz JJ, Khalpey ZI, Hoganson DM, Deng B, Lam YW, Oldinski RA, Ashikaga T, Weiss DJ, Three-dimensional scaffolds of acellular human and porcine lungs for high throughput studies of lung disease and regeneration, Biomaterials. 35 (2014) 2664–2679. 10.1016/j.biomaterials.2013.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, Moore BB, Martinez FJ, Niklason LE, White ES, Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation, Am. J. Respir. Crit. Care Med 186 (2012) 866–876. 10.1164/rccm.2012040754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ahookhosh K, Pourmehran O, Aminfar H, Mohammadpourfard M, Sarafraz MM, Hamishehkar H, Development of human respiratory airway models: A review, Eur. J. Pharm. Sci 145 (2020) 105233. 10.1016/j.ejps.2020.105233. [DOI] [PubMed] [Google Scholar]
- [25].Keller A, Nesvizhskii AI, Kolker E, Aebersold R, Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search, Anal. Chem 74 (2002) 5383–5392. 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [26].Nesvizhskii AI, Keller A, Kolker E, Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry, Anal. Chem 75 (2003) 4646–4658. 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- [27].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO, The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices, Mol. Cell. Proteomics 11 (2012). 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jayadev R, Mychel R. Morais, Ellingford JM, Srinivasan S, Naylor RW, Lawless C, Li AS, Ingham JF, Hastie E, Chi Q, Fresquet M, Koudis N-M, Thomas HB, O’keefe RT, Williams E, Adamson A, Stuart HM, Banka S, Smedley D, Sherwood DR, Lennon R, Sherwood D, A basement membrane discovery pipeline uncovers network complexity, new regulators, and human disease associations, BioRxiv. (2021) 2021.10.25.465762. 10.1101/2021.10.25.465762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hulsen T, de Vlieg J, Alkema W, BioVenn - A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams, BMC Genomics. 9 (2008) 1–6. 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schiller HB, Fernandez IE, Burgstaller G, Schaab C, Scheltema RA, Schwarzmayr T, Strom TM, Eickelberg O, Mann M, Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair, Mol. Syst. Biol 11 (2015) 819. 10.15252/msb.20156123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Burgstaller G, Oehrle B, Gerckens M, White ES, Schiller HB, Eickelberg O, The instructive extracellular matrix of the lung: basic composition and alterations in chronic lung disease., Eur. Respir. J 50 (2017) 1601805. 10.1183/13993003.01805-2016. [DOI] [PubMed] [Google Scholar]
- [32].Naba A, Pearce OMT, Del Rosario A, Ma D, Ding H, Rajeeve V, Cutillas PR, Balkwill FR, Hynes RO, Characterization of the Extracellular Matrix of Normal and Diseased Tissues Using Proteomics, J. Proteome Res 16 (2017) 3083–3091. 10.1021/acs.jproteome.7b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gudmann NS, Manon-Jensen T, Sand JMB, Diefenbach C, Sun S, Danielsen A, Karsdal MA, Leeming DJ, Lung tissue destruction by proteinase 3 and cathepsin G mediated elastin degradation is elevated in chronic obstructive pulmonary disease, Biochem. Biophys. Res. Commun 503 (2018) 1284–1290. 10.1016/j.bbrc.2018.07.038. [DOI] [PubMed] [Google Scholar]
- [34].Radder JE, Gregory AD, Leme AS, Cho MH, Chu Y, Kelly NJ, Bakke P, Gulsvik A, Litonjua AA, Sparrow D, Beaty TH, Crapo JD, Silverman EK, Zhang Y, Berndt A, Shapiro SD, Variable susceptibility to cigarette smoke-induced emphysema in 34 inbred strains of mice implicates Abi3bp in emphysema susceptibility, Am. J. Respir. Cell Mol. Biol 57 (2017) 367–375. 10.1165/rcmb.2016-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xia H, Bodempudi V, Benyumov A, Hergert P, Tank D, Herrera J, Braziunas J, Larsson O, Parker M, Rossi D, Smith K, Peterson M, Limper A, Jessurun J, Connett J, Ingbar D, Phan S, Bitterman PB, Henke CA, Identification of a cell-of-origin for fibroblasts comprising the fibrotic reticulum in idiopathic pulmonary fibrosis, Am. J. Pathol 184 (2014) 1369–1383. 10.1016/j.ajpath.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Atabai K, Jame S, Azhar N, Kuo A, Lam M, McKleroy W, DeHart G, Rahman S, Xia DD, Melton AC, Wolters P, Emson CL, Turner SM, Werb Z, Sheppard D, Mfge8 diminishes the severity of tissue fibrosis in mice by binding and targeting collagen for uptake by macrophages, J. Clin. Invest 119 (2009) 3713–3722. 10.1172/JCI40053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Abdillahi SM, Bober M, Nordin S, Hallgren O, Baumgarten M, Erjefält J, Westergren-Thorsson G, Bjermer L, Riesbeck K, Egesten A, Mörgelin M, Collagen VI Is Upregulated in COPD and Serves Both as an Adhesive Target and a Bactericidal Barrier for Moraxella catarrhalis, J. Innate Immun 7 (2015) 506–517. 10.1159/000381213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, DeIuliis G, Januszyk M, Duan Q, Arnett HA, Siddiqui A, Washko GR, Homer R, Yan X, Rosas IO, Kaminski N, Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis, Sci. Adv 6 (2020). 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kaarteenaho-Wiik R, Lakari E, Soini Y, Pollanen R, Kinnula VL, Paakko P, Tenascin expression and distribution in pleural inflammatory and fibrotic diseases, J. Histochem. Cytochem 48 (2000) 1257–1268. 10.1177/002215540004800909. [DOI] [PubMed] [Google Scholar]
- [40].Yu G, Kovkarova-Naumovski E, Jara P, Parwani A, Kass D, Ruiz V, Lopez-Otiń C, Rosas IO, Gibson KF, Cabrera S, Ramiŕez R, Yousem SA, Richards TJ, Chensny LJ, Selman M, Kaminski N, Pardo A, Matrix metalloproteinase-19 is a key regulator of lung fibrosis in mice and humans, Am. J. Respir. Crit. Care Med 186 (2012) 752–762. 10.1164/rccm.201202-0302OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li F, Xu X, Geng J, Wan X, Dai H, The autocrine CXCR4/CXCL12 axis contributes to lung fibrosis through modulation of lung fibroblast activity, Exp. Ther. Med 19 (2020) 1844. 10.3892/etm.2020.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vuga LJ, Tedrow JR, Pandit KV, Tan J, Kass DJ, Xue J, Chandra D, Leader JK, Gibson KF, Kaminski N, Sciurba FC, Duncan SR, C-X-C motif chemokine 13 (CXCL13) is a prognostic biomarker of idiopathic pulmonary fibrosis, Am. J. Respir. Crit. Care Med 189 (2014) 966–974. 10.1164/rccm.201309-1592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bauer Y, Tedrow J, De Bernard S, Birker-Robaczewska M, Gibson KF, Guardela BJ, Hess P, Klenk A, Lindell KO, Poirey S, Renault BR, Rey M, Weber E, Nayler O, Kaminski N, A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis, Am. J. Respir. Cell Mol. Biol 52 (2015) 217–231. 10.1165/rcmb.2013-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Herrera JA, Dingle L, Montero MA, Shah R, V Venkateswaran R, Blaikley JF, Lawless C, Schwartz MA, The IPF fibroblastic focus is an active collagen biosynthesis factory embedded in a distinct extracellular matrix, BioRxiv. (2021) 2021.11.06.467549. 10.1101/2021.11.06.467549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Langholm LL, Rønnow SR, Sand JMB, Leeming DJ, Tal-Singer R, Miller BE, Vestbo J, Karsdal MA, Manon-Jensen T, Increased von willebrand factor processing in COPD, reflecting lung epithelium damage, is associated with emphysema, exacerbations and elevated mortality risk, Int. J. COPD 15 (2020) 543–552. 10.2147/COPD.S235673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Camelo A, Dunmore R, Sleeman MA, Clarke DL, The epithelium in idiopathic pulmonary fibrosis: Breaking the barrier, Front. Pharmacol 4 JAN (2014) 173. 10.3389/fphar.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Salazar-Peláez LM, Abraham T, Herrera AM, Correa MA, Ortega JE, Paré PD, Seow CY, Vitronectin expression in the airways of subjects with asthma and chronic obstructive pulmonary disease, PLoS One. 10 (2015). 10.1371/journal.pone.0119717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li J, Fei GH, The unique alterations of hippocampus and cognitive impairment in chronic obstructive pulmonary disease, Respir. Res 14 (2013) 140. 10.1186/1465-992114-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Merrilees MJ, Hankin EJ, Black JL, Beaumont B, Matrix proteoglycans and remodelling of insterstitial lung tissue in lymphangioleiomyomatosis, J. Pathol 203 (2004) 653–660. 10.1002/path.1577. [DOI] [PubMed] [Google Scholar]
- [50].Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J, Proteoglycans and Glycosaminoglycans, (1999). https://www.ncbi.nlm.nih.gov/books/NBK20693/ (accessed June 17, 2022).
- [51].Shum L, Chondroadherin binds to type II collagen, Arthritis Res. Ther 3 (2001) 72650. 10.1186/ar-2001-72650. [DOI] [Google Scholar]
- [52].Knudsen L, Ochs M, The micromechanics of lung alveoli: structure and function of surfactant and tissue components, Histochem. Cell Biol 150 (2018) 661–676. 10.1007/s00418-018-1747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chelladurai P, Seeger W, Pullamsetti SS, Matrix metalloproteinases and their inhibitors in pulmonary hypertension, (n.d.). 10.1183/09031936.00209911. [DOI] [PubMed] [Google Scholar]
- [54].Kenagy RD, Plaas AH, Wight TN, Versican Degradation and Vascular Disease, Trends Cardiovasc. Med 16 (2006) 209–215. 10.1016/j.tcm.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mereness JA, Mariani TJ, The critical role of collagen VI in lung development and chronic lung disease, Matrix Biol. Plus 10 (2021) 100058. 10.1016/j.mbplus.2021.100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bonaldo P, Russo V, Bucciotti F, Doliana R, Colombatti A, Structural and Functional Features of the α3 Chain Indicate a Bridging Role for Chicken Collagen VI in Connective Tissues, Biochemistry. 29 (1990) 1245–1254. 10.1021/bi00457a021. [DOI] [PubMed] [Google Scholar]
- [57].Finnis ML, Gibson MA, Microfibril-associated glycoprotein-1 (MAGP-1) binds to the pepsin-resistant domain of the α3(VI) chain of type VI collagen, J. Biol. Chem 272 (1997) 22817–22823. 10.1074/jbc.272.36.22817. [DOI] [PubMed] [Google Scholar]
- [58].Specks U, Mayer U, Nischt R, Spissinger T, Mann K, Timpl R, Engel J, Chu ML, Structure of recombinant N-terminal globule of type VI collagen alpha 3 chain and its binding to heparin and hyaluronan., EMBO J. 11 (1992) 4281–4290. 10.1002/j.1460-2075.1992.tb05527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bober M, Enochsson C, Collin M, Mörgelin M, Collagen VI is a subepithelial adhesive target for human respiratory tract pathogens, J. Innate Immun 2 (2010) 160–166. 10.1159/000232587. [DOI] [PubMed] [Google Scholar]
- [60].Specks U, Nerlich A, Colby TV, Wiest I, Timpl R, Increased expression of type VI collagen in lung fibrosis, Am. J. Respir. Crit. Care Med 151 (1995) 1956–1964. 10.1164/ajrccm.151.6.7767545. [DOI] [PubMed] [Google Scholar]
- [61].Jessen H, Hoyer N, Prior TS, Frederiksen P, Rønnow SR, Karsdal MA, Leeming DJ, Bendstrup E, Sand JMB, Shaker SB, Longitudinal serological assessment of type VI collagen turnover is related to progression in a real-world cohort of idiopathic pulmonary fibrosis, BMC Pulm. Med 21 (2021) 382. 10.1186/s12890-021-01684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mereness JA, Bhattacharya S, Wang Q, Ren Y, Pryhuber GS, Mariani TJ, Type VI collagen promotes lung epithelial cell spreading and wound-closure, PLoS One. 13 (2018) e0209095. 10.1371/journal.pone.0209095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cescon M, Gattazzo F, Chen P, Bonaldo P, Collagen VI at a glance, J. Cell Sci 128 (2015) 3525–3531. 10.1242/jcs.169748. [DOI] [PubMed] [Google Scholar]
- [64].Groulx JF, Gagné D, Benoit YD, Martel D, Basora N, Beaulieu JF, Collagen VI is a basement membrane component that regulates epithelial cell-fibronectin interactions, Matrix Biol. 30 (2011) 195–206. 10.1016/j.matbio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- [65].Dassah MA, Almeida D, Hahn R, Bonaldo P, Worgall S, Hajjar KA, Annexin A2 mediates secretion of collagen VI, pulmonary elasticity and apoptosis of bronchial epithelial cells, J. Cell Sci 127 (2014) 828–844. 10.1242/jcs.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mereness JA, Bhattacharya S, Ren Y, Wang Q, Anderson CS, Donlon K, Dylag AM, Haak J, Angelin A, Bonaldo P, Mariani TJ, Collagen VI Deficiency Results in Structural Abnormalities in the Mouse Lung, Am. J. Pathol 190 (2020) 426–441. 10.1016/j.ajpath.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Åhrman E, Hallgren O, Malmström L, Hedström U, Malmström A, Bjermer L, Zhou XH, Westergren-Thorsson G, Malmström J, Quantitative proteomic characterization of the lung extracellular matrix in chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis, J. Proteomics 189 (2018) 23–33. 10.1016/j.jprot.2018.02.027. [DOI] [PubMed] [Google Scholar]
- [68].Jessen H, Hoyer N, Prior TS, Frederiksen P, Karsdal MA, Leeming DJ, Bendstrup E, Sand JMB, Shaker SB, Turnover of type I and III collagen predicts progression of idiopathic pulmonary fibrosis, Respir. Res 22 (2021) 205. 10.1186/s12931-021-01801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Petersen TH, Calle EA, Colehour MB, Niklason LE, Matrix composition and mechanics of decellularized lung scaffolds, Cells Tissues Organs. 195 (2012) 222–231. 10.1159/000324896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bonvillain RW, Danchuk S, Sullivan DE, Betancourt AM, Semon JA, Eagle ME, Mayeux JP, Gregory AN, Wang G, Townley IK, Borg ZD, Weiss DJ, Bunnell BA, A nonhuman primate model of lung regeneration: Detergent-mediated decellularization and initial in vitro recellularization with mesenchymal stem cells, Tissue Eng. - Part A 18 (2012) 2437–2452. 10.1089/ten.tea.2011.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ericsson C, Nistér M, Protein Extraction from Solid Tissue, in: Methods Mol. Biol, Humana Press Inc., 2011: pp. 307–312. 10.1007/978-1-59745-423-0_17. [DOI] [PubMed] [Google Scholar]
- [72].Hill RC, Calle EA, Dzieciatkowska M, Niklason LE, Hansen KC, Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering, Mol. Cell. Proteomics 14 (2015) 961–973. 10.1074/mcp.M114.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Herrera JA, Mallikarjun V, Rosini S, Montero MA, Lawless C, Warwood S, O’Cualain R, Knight D, Schwartz MA, Swift J, Laser capture microdissection coupled mass spectrometry (LCM-MS) for spatially resolved analysis of formalin-fixed and stained human lung tissues, Clin. Proteomics 17 (2020) 24. 10.1186/s12014-020-09287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Link PA, Pouliot RA, Mikhaiel NS, Young BM, Heise RL, Tunable Hydrogels from Pulmonary Extracellular Matrix for 3D Cell Culture, J. Vis. Exp (2017) e55094. 10.3791/55094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Pouliot RA, Young BM, Link PA, Park HE, Kahn AR, Shankar K, Schneck MB, Weiss DJ, Heise RL, Porcine Lung-Derived Extracellular Matrix Hydrogel Properties Are Dependent on Pepsin Digestion Time, Tissue Eng. - Part C Methods 26 (2020) 332–346. 10.1089/ten.tec.2020.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectral data (raw, PD msf, mzML) and LC-MS/MS and data analysis method required to reproduce these findings are available to download from Supplementary Table 2. The processed data required to reproduce these findings are available to download from Supplementary Table 2. This data has further been uploaded to the MassIVE repository with the following identifiers:
MassIVE accession: MSV000090771
ProteomeXchange accession: PXD038289
DOI: 10.25345/C58911W11


