Abstract
Genotoxins cause nascent strand degradation (NSD) and fork reversal during DNA replication. NSD and fork reversal are crucial for genome stability and exploited by chemotherapeutic approaches. However, it is unclear how NSD and fork reversal are triggered. Additionally, the fate of the replicative helicase during these processes is unknown. We developed a biochemical approach to study synchronous, localized NSD and fork reversal using Xenopus egg extracts, and validated this approach with experiments in human cells. We show that replication fork uncoupling stimulates NSD of both nascent strands and progressive conversion of uncoupled forks to reversed forks. Importantly, the replicative helicase remains bound during NSD and fork reversal. Unexpectedly, NSD occurs before and after fork reversal, indicating that multiple degradation steps take place. Overall, our data show that uncoupling causes NSD and fork reversal and elucidate key events that precede fork reversal.
Editor summary:
Kavlashvili et al. use a new in vitro approach to show that uncoupled replication forks can cause fork reversal and nascent strand degradation. Both processes occur without loss of the replisome from DNA and involve multiple degradation steps.
INTRODUCTION
During DNA synthesis genotoxins can cause replication forks to stall. This leads to regression of the replication fork junction and reannealing of parental DNA strands to generate a reversed fork (Extended Data Fig. 1A,i-ii, fork reversal)1-4, which facilitates bypass of DNA lesions and regulates fork progression5,6. Stalling also leads to degradation of both nascent DNA strands by nucleases (nascent strand degradation, NSD)7-9. NSD is thought to target the regressed arm of a reversed fork (Extended Data Fig. 1A,ii-iii) to convert the reversed fork back to a replication fork so that DNA replication can restart (Extended Data Fig. 1A,iii-iv)1-4. NSD contributes to chemotherapeutic approaches10-12 and many important proteins are implicated in fork reversal and NSD1-3,7,9,11,13-17. However, despite the importance of NSD and fork reversal many questions about the underlying mechanisms remain. For example, NSD can be inferred as an initial response to stalling based on the roles of nucleases at replication forks7,18,19, yet NSD can only be observed after prolonged treatment with genotoxic agents7,20 or inactivation of negative regulators8,9,21. Thus, NSD has not formally been demonstrated to be an initial response. Additionally, NSD and fork reversal must be triggered efficiently to promote replication restart but not excessively because this causes genome instability8,11,13,18,21 but it is unclear how fork reversal and NSD are triggered in such a way that this balance is achieved
Fork reversal and NSD can be caused by a variety of genotoxins7,14,22-26. Hydroxyurea, which stimulates both fork reversal and NSD7,8,14,21, stalls replication forks indirectly by depleting cellular nucleotides, resulting in DNA polymerase stalling27. The replicative helicase continues to unwind in the absence of DNA polymerase activity, resulting in replication fork uncoupling28,29. Uncoupling correlates well with fork reversal14, suggesting that uncoupling may cause fork reversal and possibly NSD. However, once the replicative helicase uncouples, it effectively stalls (the helicase slows ~5-10 fold 30,31) raising the possibility that helicase stalling causes fork reversal and NSD. Accordingly, helicase stalling lesions such as topoisomerase poisons and crosslinking agents can also cause fork reversal14,22-24 and NSD7,25,32. Moreover, recent data suggests that neither replication fork uncoupling nor stalling of the replicative helicase is sufficient to trigger NSD unless combined with reactive oxygen species33. Thus, it is unclear whether fork stalling leads to NSD and fork reversal by causing replication fork uncoupling, helicase stalling, or some other event.
Key mechanistic questions about fork reversal and NSD remain unaddressed. One such question is what happens to the replicative helicase during these processes. Strand annealing in the vicinity of the replicative helicase was recently reported to promote its transition from single-stranded to double-stranded DNA34, which would be expected to trigger replicative helicase unloading during fork reversal35. Accordingly, unloading of the replicative helicase is required for fork reversal during one of the pathways for DNA interstrand cross-link repair24. However, the replicative helicase can be detected at replication forks in response to hydroxyurea treatment36, which would be expected to cause fork reversal14 and NSD7. It is therefore possible that the replicative helicase is present during fork reversal and NSD. Another question is whether multiple DNA structures are targeted for degradation. Current models propose that degradation begins at reversed forks1-4, consistent with a requirement for fork reversal during extensive NSD13. However, it is unclear whether degradation continues at replication forks once the regressed arm is lost and whether the same is true during the more limited NSD that occurs during replication restart7.
We used Xenopus egg extracts to develop a synchronous and locus-specific approach to study NSD and fork reversal. NSD induced by this approach involves DNA2, as previously reported, and we corroborate our approach with cellular experiments. NSD degrades both nascent strands, as previously described8, and replication forks are progressively converted to reversed forks. Using this approach, we show that replication fork uncoupling, rather than helicase stalling, elicits both fork reversal and nascent strand degradation. Additionally, the replicative helicase remains bound throughout both fork reversal and NSD. Since replisome removal pathways are not activated, this observation strongly suggests that the replisome remains on single-stranded DNA, which would place fork reversal and NSD take place behind the uncoupled helicase. Furthermore, our data reveal that degradation occurs at replication forks before and after reversal, indicating that two degradation steps occur. Overall, our research identify a trigger for NSD and fork reversal and elucidate key events that precede fork reversal.
RESULTS
NSD is an initial response to fork stalling
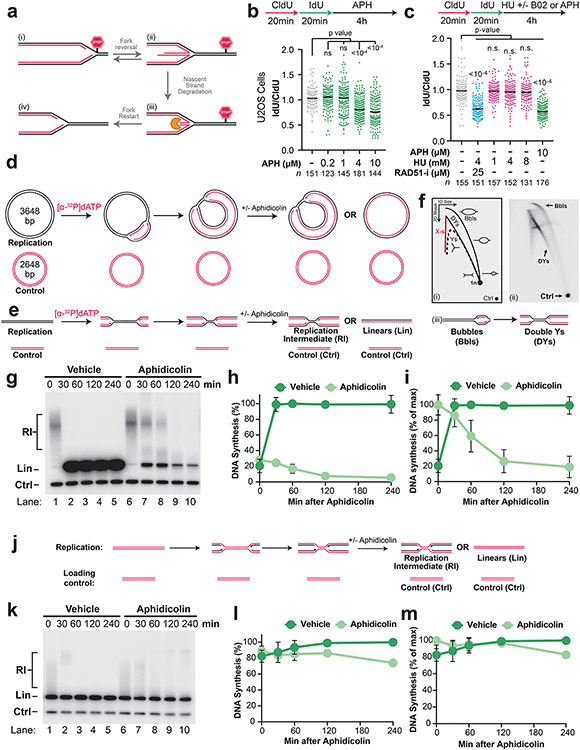
In cells, hydroxyurea (HU) is commonly used to stimulate nascent strand degradation (NSD), which is thought to be a consequence of fork stalling7,8,21,37. However, HU also induces reactive oxygen species, which were reported to be required for NSD33. We therefore tested whether NSD took place following treatment with aphidicolin, which induces fork stalling by inhibition of DNA polymerases14,28,29,38 but does not induce oxidative stress33. Aphidicolin induced NSD in both human cells (Extended Data Fig. 1B-C) and in vitro using Xenopus egg extracts39 (Extended Data Fig. 1D-M). Thus, we conclude that fork stalling by aphidicolin is sufficient to trigger NSD without inactivation of fork protection proteins or induction of oxidative stress, as expected7. Additionally, parental DNA strands were relatively stable (Extended Data Fig. 1J-M) indicating that aphidicolin-induced degradation was not due to formation of DNA breaks40,41. Importantly, NSD was abundant within 60 minutes of adding aphidicolin (Extended Data Fig. 1E-I), which formally demonstrates that NSD is an initial response to fork stalling.
A synchronous biochemical approach to study NSD
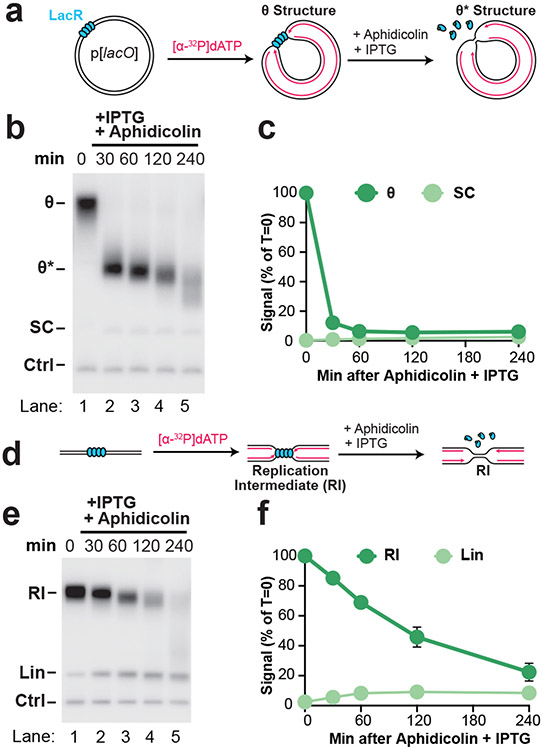
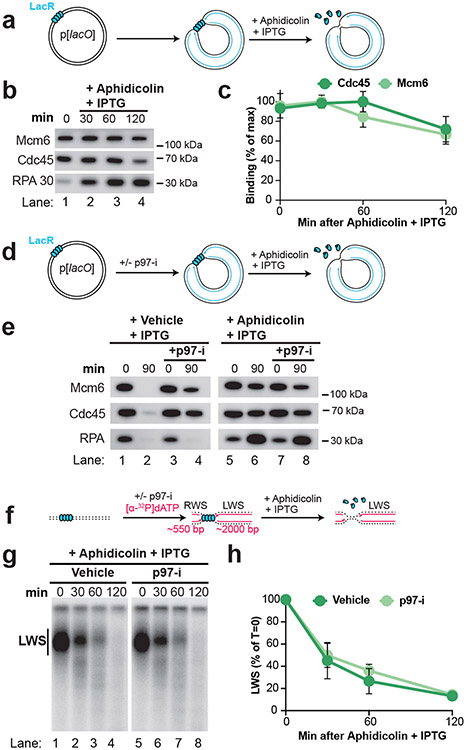
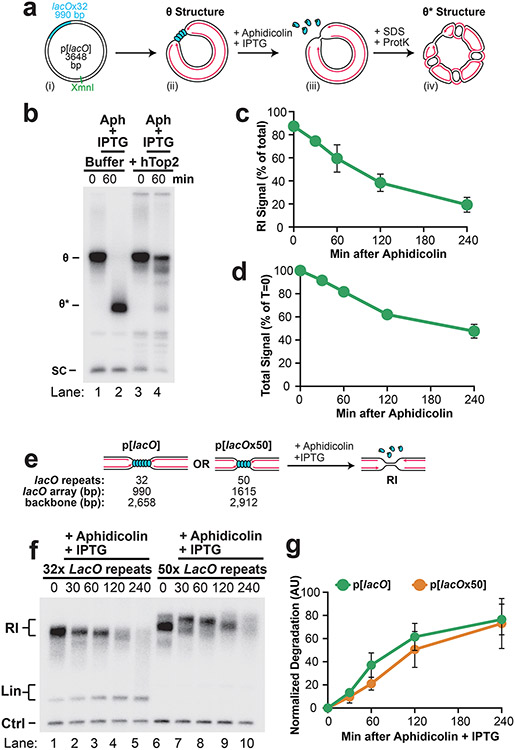
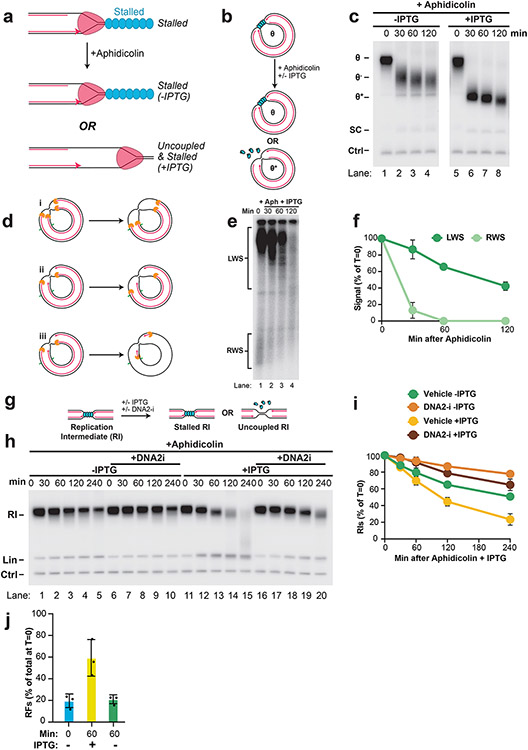
To examine NSD in more detail, we sought to synchronize and localize NSD events. To this end, we localized forks to a lac repressor (LacR) barrier42 before NSD was induced (Fig. 1A). Plasmid DNA harboring a lacO array was pre-incubated with LacR and then replicated in Xenopus egg extracts, which gave rise to θ structures (Fig. 1A; Fig. 1B, lane 1), as expected42. Forks were then stalled with aphidicolin (Fig. 1A) and IPTG was simultaneously added to disrupt the LacR array to allow uncoupling of the replicative helicase, which also occurs following HU treatment in cells28,29. Upon aphidicolin and IPTG treatment, θs were converted to θ* structures (Fig. 1B, lanes 2-5), which arose from reannealing of unwound DNA strands in the detergent-treated samples (Extended Data Fig. 2A,B) and showed that uncoupling occurred. To remove any topological effects, we purified the DNA and performed restriction digests to visualize the replication fork structures (Fig. 1D)42. Replication Intermediates disappeared over time without any appreciable increase in linear products of replication (Fig. 1E, lanes 2-5; Fig. 1F; Extended Data Fig. 2C). Thus, loss of Replication Intermediates was due to degradation of nascent DNA strands. Accordingly, most nascent strand DNA signal was lost (Extended Data Fig. 2D). Importantly, degradation did not appear to be influenced by the proximity of the converging replication forks (Extended Data Fig. 2E-G). Overall, these results show that we have developed a synchronous biochemical approach to induce NSD.
Figure 1: Induction of synchronous and localized nascent strand degradation in vitro.
(A) Plasmid DNA harboring a LacR array was replicated using Xenopus egg extracts and dATP[α-32P] was added to label newly-synthesized DNA strands. Once forks were localized at the LacR barrier, replication was restarted by addition of IPTG, which removed the LacR barrier, and replication forks were stalled by addition of aphidicolin. (B) Samples from (a) were separated on an agarose gel and visualized by autoradiography. As a loading control (Ctrl) the reactions include a fully-replicated plasmid. See also Extended Data Fig. 2A-B. (C) Quantification of θ structures and supercoiled monomers (SC) from (b). Mean ± S.D., n=3 independent experiments. (D) Samples from (a) were digested with XmnI to allow unambiguous identification of replication intermediates (RIs) and linear products of replication (Lin). See Extended Data Fig. 2A,i for a restriction map. (E) Samples from (d) were separated on an agarose gel and visualized by autoradiography. (F) Quantification of RI and Lin structures in (e). Mean ± S.D., n=5 independent experiments. See also Extended Data Fig. 2C-D.
NSD involves the DNA2 exonuclease
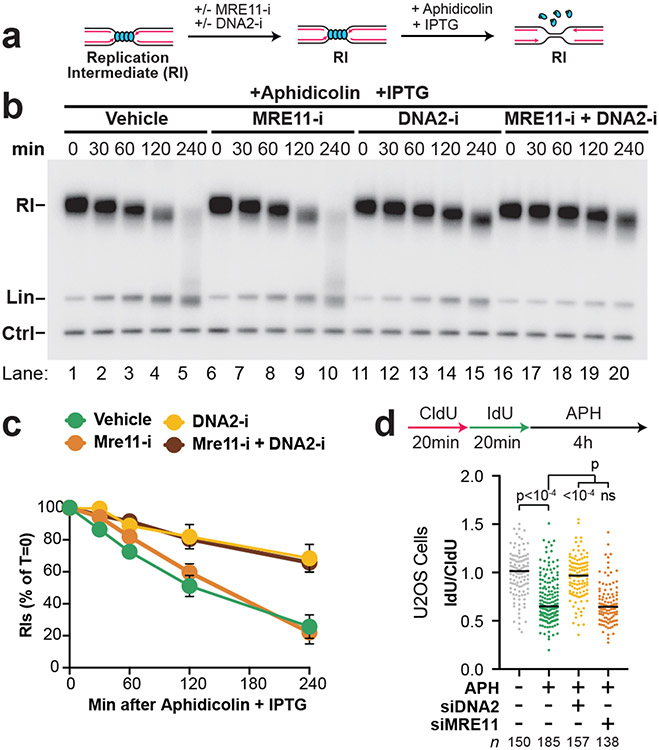
In other settings NSD involves the DNA2 or MRE11 exonucleases, or both (Extended Data Fig. 3A-B)2,7,8,15,37. To address the roles of DNA2 and MRE11 in the NSD we observed, we used the small molecule inhibitors C5 (DNA2-i)43 and Mirin (MRE11-i)44 to inactivate these nucleases in Xenopus egg extracts. We therefore induced NSD (as in Fig. 1) in the presence of either MRE11-i, DNA2-i or both (Fig. 2A). DNA2-i treatment strongly inhibited NSD (Fig. 2B, lanes 1-5,11-15; Fig. 2C) while MRE11-i had no effect either alone (Fig. 2B, lanes 1-10; Fig. 2C) or in combination with DNA2-i (Fig. 2B, lanes 1-5 and 16-20; Fig. 2C). Importantly, both DNA2-i and MRE11-i effectively inhibited their targets (Extended Data Fig. 3C-E) and neither MRE11-i nor DNA2-i prevented uncoupling (Extended Data Fig. 3F-G). These data show that the NSD we observe involves DNA2 but not MRE11, consistent with previous reports7-9,21 (but see also12,37).
Figure 2: Nascent strand degradation involves DNA2 but not MRE11.
(A) Forks were localized to a LacR barrier and NSD was induced by addition of IPTG and aphidicolin in the absence or presence of MRE11-i and DNA2-i. Purified DNA was subjected to restriction digest so that replication fork structures could be visualized as RIs. (B) Samples from (a) were separated on an agarose gel and visualized by autoradiography. See also Extended Data Fig. 3F-G. (C) Quantification of RI signal from (b). Mean ± S.D., n=3 independent experiments. See also Extended Data Fig. 3H-I. (D) U2OS cells were transfected with the indicated siRNA, pulse labeled with CldU and IdU, then treated with aphidicolin. DNA fiber analysis was performed to determine the lengths of IdU and CldU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’. See also Fig. 3K.
Aphidicolin treatment alone was not reported to induce NSD in cells. To investigate the requirement for DNA2 and MRE11 during aphidicolin-induced NSD, U2OS cells were treated with siRNAs targeting DNA2 or MRE11 and DNA fiber analysis was performed to monitor NSD (as in Extended Data Fig. 1B). U2OS cells were pulse-labeled with CldU then IdU followed by treatment with aphidicolin so that loss of the CldU labeled tracks could be monitored as a measurement of NSD. DNA fiber analysis was performed to determine the lengths of CldU and IdU labeled DNA tracks in each condition. Aphidicolin treatment reduced the IdU/CldU ratio, indicating that NSD took place (Fig. 2D). siRNA knockdown of DNA2 prevented NSD while knockdown of MRE11 had no effect (Fig. 2D). The same result was obtained when DNA2-i or MRE11-i was used (Extended Data Fig. 3J) and in HCT116 cells (Extended Data Fig. 3K). Leading and lagging DNA strands cannot be separated by fiber assays, so loss of IdU signal means that both DNA strands were degraded, as previously described8, while restoration of IdU signal means one or both strands were stabilized. Because both DNA strands are degraded but DNA2 only possesses 5’-3’ activity45 another nuclease with 3’-5’ activity is likely to be involved. However, it cannot be FAN146 or SAMHD147 as these nucleases had no effect on NSD (Extended Data Fig. 4). Overall, our data show that aphidicolin induces NSD that involves DNA2 but not MRE11 in both cells and Xenopus egg extracts.
NSD involves fork reversal
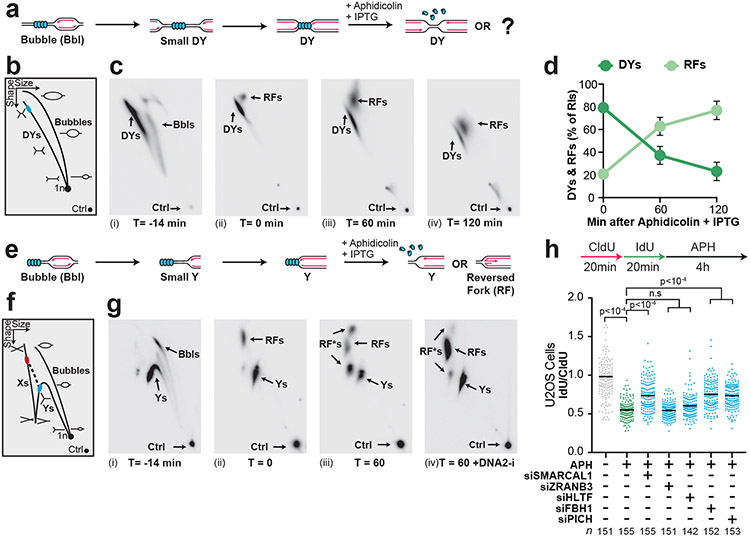
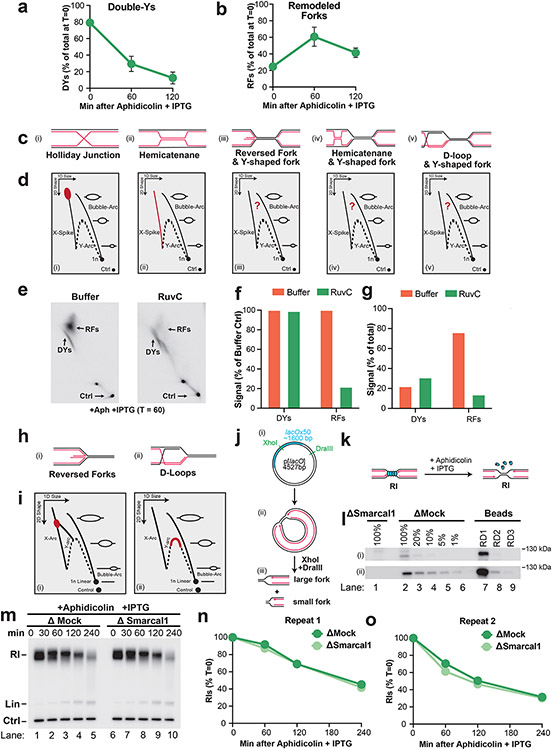
NSD typically involves fork reversal 7,13,21. To test whether this was also the case in our experiments, we performed 2D gel electrophoresis to monitor the DNA structures formed during NSD (Fig. 3A,B). Initially, signal was distributed throughout the bubble and double-Y arcs, due to initiation of replication throughout the plasmid (Fig. 3C,i). Subsequently, a discrete spot formed on the double-Y arc, which arose from forks localized to the LacR barrier (Fig. 3C,ii, DYs). Interestingly, we also observed a small population of replication forks that did not represent canonical replication intermediates and were therefore altered replication fork structures i.e. remodeled forks (Fig. 3C,ii, RFs). During NSD, the abundance of Double-Ys declined (Fig. 3C,ii-iv; Fig. 3D) while remodeled forks increased ~4 fold (Fig. 3C,ii-iv, Fig. 3D), indicating that Double-Ys were converted to remodeled forks. Importantly, the frequency of remodeled forks increased over time indicating progressive conversion of Double-Ys to these species (Fig. 3D). Moreover, the mobility of both Double-Ys and remodeled forks increased over time (Fig. 3C,ii-iv) indicating that both types of structures included degraded molecules. Thus, nascent strand degradation involves progressive remodeling of replication forks and may involve degradation of multiple DNA structures.
Figure 3: Nascent strand degradation involves fork reversal.
(A) Cartoon depicting DNA structures formed during replication of LacR-bound plasmid DNA and induction of NSD. Indicated structures arise from treatment with a restriction enzyme that cuts once. (B) 2-D gel migration pattern of the DNA structures depicted in (a). The blue spot indicates the expected migration of DY structures localized to the LacR array. (C) DNA structures from (a) were analyzed by 2-D gel electrophoresis and visualized by autoradiography. T=0 is when IPTG and aphidicolin were added. DY, bubble (Bbl), and remodeled fork (RF) structures are indicated. (D) Quantification of DY and RF structures from (c) as a percentage of replication intermediates (RIs) at each time point. Mean ± S.D., n=3 independent experiments. See also Extended Data Fig. 5A-B. (E) Cartoon depicting structures from (a) after restriction digest with two different enzymes to release a single fragment. See also Extended Data Fig. 5J. (F) 2-D gel migration pattern of the DNA structures depicted in (e). The blue spot on the Y-arc indicates the expected migration of Y structures localized to the LacR array. The red spot indicates the expected migration of reversed fork structures arising from the Y structures within the black spot. (G) DNA structures from (e) were analyzed by 2-D gel electrophoresis and visualized by autoradiography. T=0 is when IPTG and aphidicolin were added. (H) U2OS cells were transfected with the indicated siRNA. 72 hours later cells were pulse labeled with CldU and IdU, then treated with aphidicolin. DNA fiber analysis was performed to determine lengths of CldU and IdU labeled DNA tracks in each condition. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’.
Treatment with RuvC revealed that the remodeled forks contained either reversed forks or D-loops (Extended Data Fig. 5C-G). Migration of these species is well defined for DNA molecules that contain only a single fork structure (Extended Data Fig. 5H-I)48,49. We therefore performed restriction digests to yield two replication forks of differing sizes (Extended Data Fig. 5J). This allowed us to analyze the larger fork only (Fig. 3E) by 2-D gel electrophoresis (Fig. 3F). Initially, signal was distributed throughout the bubble and Y-arcs, due to initiation of replication throughout the plasmid (Fig. 3G,i), then formed a discrete spot on the Y arc, which arose from forks localized to the LacR barrier (Fig. 3G,ii, Ys). A small population of remodeled forks was observed immediately prior to NSD (Fig. 3G,ii). Importantly, this species migrated mid-way down the X spike (Fig. 3G,ii, RFs) in the expected position for a reversed fork (Extended Data Fig. 5I,i) and not along the Y-arc, as would be expected for a D-loop (Extended Data Fig. 5I,ii). Surprisingly, this species did not increase in abundance during NSD (Fig. 3G,iii, RFs) in contrast to the remodeled forks observed previously (Fig. 3C). Instead, we observed two additional species during NSD that also migrated along the X-spike with a similar mobility to reversed forks (Fig. 3G,iii, RF*s). These species almost completely disappeared following DNA2-i treatment (Fig. 3G,iv), which also led to a corresponding increase in RF signal (Fig. 3G,iv). Thus, these species correspond to reversed forks that are normally degraded by DNA2. Overall, these data show that NSD induced by our approach involves progressive conversion of replication forks to reversed forks, which are then degraded by DNA2.
We next wanted to test whether fork reversal is involved in the aphidicolin-induced NSD we observed in cells (Fig. 2D). We therefore tested the role of fork reversal enzymes. U2OS cells were treated with siRNA targeting SMARCAL1, ZRANB3, HLTF, FBH111, and PICH50, then DNA fiber analysis was performed to monitor NSD. siRNA knockdown of SMARCAL1, FBH1, and PICH reduced NSD (Fig. 3H), indicating that fork reversal is involved in NSD following aphidicolin treatment. Notably, ZRANB3 and HLTF did not affect NSD (Fig. 3H), despite cooperating with NSD in other contexts11. Surprisingly, NSD in Xenopus egg extracts did not involve SMARCAL1 (Extended Data Fig. 5K-O), suggesting other fork reversal enzymes operate instead. We conclude that aphidicolin induces NSD and fork reversal in human cells and Xenopus egg extracts but note that in the latter case only a subset of cellular pathways may operate.
Uncoupling elicits NSD and fork reversal
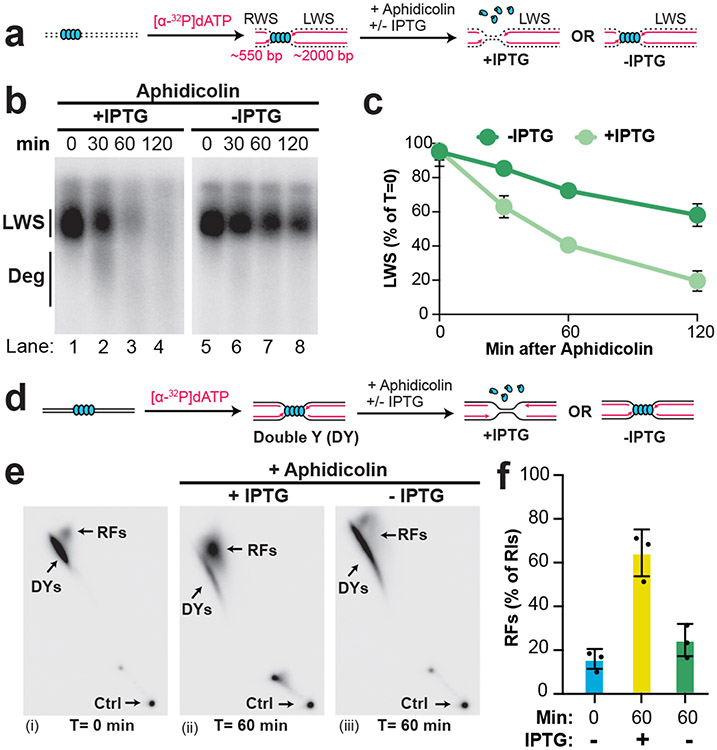
Fork stalling by aphidicolin results in replication fork uncoupling51 and helicase stalling30. We therefore wanted to test which of these events trigger NSD and fork reversal. To directly test this, we localized forks to a LacR barrier, added aphidicolin, and either added or omitted IPTG to facilitate or inhibit helicase uncoupling, respectively (Extended Data Fig. 6A-C). DNA intermediates were then purified, restriction digested, and separated on an agarose gel under denaturing conditions (Fig. 4A). This approach allowed us to monitor the loss of intact nascent strands (Fig. 4A, LWS) as a more direct read-out for the onset of NSD than measurement of Replication Intermediate signal (as in Fig. 1; Fig. 2). Upon addition of IPTG and aphidicolin, intact strands were readily lost (Fig. 4B, LWS; Fig. 4C). Degraded strands could be visualized (Fig. 4B, lane 2, Deg) and both strands were degraded (Extended Data Fig. 6D-F), as expected8. When IPTG was omitted, uncoupling was strongly inhibited (Extended Data Fig. 6B-C) but not blocked, as expected42. Importantly, omission of IPTG caused intact strands to persist (Fig. 4B, lanes 5-8; Fig. 4C). Omission of IPTG also caused Replication Intermediates to persist when total degradation was monitored (as in Fig. 1; Fig. 2), in the absence or presence of DNA2-i (Extended Data Fig. 6G-I). These data show that replication fork uncoupling, independent of helicase stalling, causes NSD.
Figure 4: Uncoupling promotes nascent strand degradation and fork reversal.
(A) Forks were localized to a LacR barrier and aphidicolin was added in the absence or presence of IPTG. Purified DNA was restriction digested to yield leftward strands (LWS) and rightward strands of different sizes. (B) DNA structures from (a) were separated on an alkaline denaturing agarose gel and visualized by autoradiography. Intact and degraded (deg) LWS are indicated. See also Extended Data Fig. 6B-C. (C) Quantification of intact LWS strands from (b). Mean ± S.D., n=3 independent experiments. (D) Samples from (a) were subjected to restriction digest so that replication fork structures could be visualized. (E) Double-Y (DY) and reversed fork (RF) structures from (d) were separated by 2-D gel electrophoresis (as in Fig. 3B-C). (F) Quantification of RFs from (e) as a percentage of replication intermediates (RIs) at each time point. Mean ± S.D., n=3 independent experiments. See also Extended Data Fig. 6J.
We next tested whether fork reversal is also stimulated by replication fork uncoupling. To this end, we performed 2-D gel analysis to monitor the abundance of reversed forks during NSD (as in Fig. 3) in the absence or presence of IPTG (Fig. 4D). Reversed forks accumulated ~4-fold in the presence of IPTG (Fig. 4E,i-ii; Fig. 4F) as previously shown (Fig. 3D). Strikingly, omission of IPTG essentially blocked the accumulation of reversed forks (Fig. 4E,iii; Fig. 4F). Thus, replication fork uncoupling induces fork reversal. Overall, our data definitively show that replication fork uncoupling can stimulate both NSD and fork reversal.
The CMG helicase is retained during NSD and fork reversal
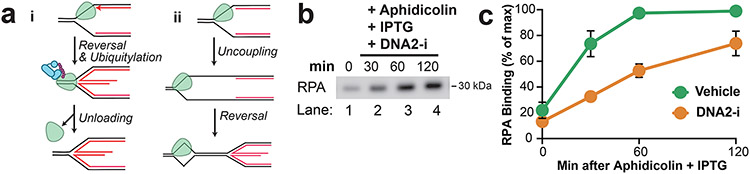
The fate of the CMG helicase during fork reversal and nascent strand degradation is largely elusive. One possibility1 (Extended Data Fig. 7A,i) is that CMG translocates onto double-stranded DNA (dsDNA) during fork reversal34, which would lead to replisome unloading35. Accordingly, CMG unloading has been reported to be required for fork reversal24. Another possibility (Extended Data Fig. 7A,ii) is that the replisome remains associated with the nascent DNA, consistent with long-term retention of the replisome36 under conditions known to cause fork reversal14. To determine the fate of the replisome during NSD and fork reversal, we induced NSD then recovered chromatin at different time points and performed Western blotting to monitor the abundance of replisome components (Fig. 5A). The single-stranded DNA binding protein RPA accumulated and then plateaued (Fig. 5B; Extended Data Fig. 7B), consistent with the presence of high levels of ssDNA from both uncoupling and NSD. Although reannealing of parental DNA strands should lead to a subsequent decrease in RPA signal, this was not observed (Extended Data Fig. 7C). This may be because most of the RPA signal was due to NSD (Extended Data Fig. 7B-C) but could also reflect limited reannealing of parental DNA strands (see discussion). Importantly, levels of the CMG components MCM6 and CDC45 were essentially unaltered for the first 60 minutes (Fig. 5B, lanes 1-3; Fig. 5C) despite most NSD and fork reversal taking place within this time frame (Fig. 3; Fig. 4). Thus, our data shows that the CMG helicase remains bound to DNA during NSD and fork reversal.
Figure 5: The replisome remains associated with DNA during nascent strand degradation.
(A) Forks were localized to a LacR barrier and NSD was induced by addition of IPTG and aphidicolin. At different time points chromatin-bound proteins were recovered. (B) Proteins from (a) were detected by Western Blotting. (C) Quantification of CDC45 and MCM6 from (b). Mean ± S.D., n=3 independent experiments. See also Extended Data Fig. 7B-C. (D) NSD was monitored as in (a) in the absence or presence of p97-i. (E) Proteins from (d) were detected by western blotting. (F) NSD was induced as in (d) and dATP[α-32P] was included to radiolabel nascent DNA strands. Purified DNA was restriction digested to yield leftward strands (LWS) and rightward strands of different sizes. (G) Samples from (f) were separated on an alkaline denaturing agarose gel and visualized by autoradiography. (H) Quantification of LWS from (g). Mean ± S.D., n=3 independent experiments.
Although replisome components were relatively stable at early time points during our experiments, we noticed an ~25% decrease in MCM6 and CDC45 signal by 120 minutes (Fig. 5B, lanes 3-4; Fig. 5C). While this was not enough to account for all NSD events (Fig. 4C), we wondered whether a subset might require replisome unloading. To test this, we monitored the onset of NSD in the presence of a small molecule inhibitor of p97 (p97-i), which inhibits all known replisome unloading pathways 42,52 (Fig. 5D). When replication forks were allowed to complete DNA synthesis in the absence of aphidicolin, p97-i led to retention of MCM6 and CDC45, but not RPA (Fig. 5E, lanes 1-4), consistent with a specific role for p97 in unloading the replicative helicase and not other replication fork components53. When aphidicolin was added, p97-i had little effect on levels of CDC45 or MCM6 (Fig. 5E, lanes 5-8) consistent with replisomes being largely stable at early time points. Accordingly, we monitored the onset of NSD in presence of p97-i (Fig. 5F, as in Fig. 4) and observed no measurable difference in degradation compared to the control (Fig. 5G-H). These data rule out any requirement for replisome unloading in the vast majority of NSD events we observe.
NSD can occur before fork reversal
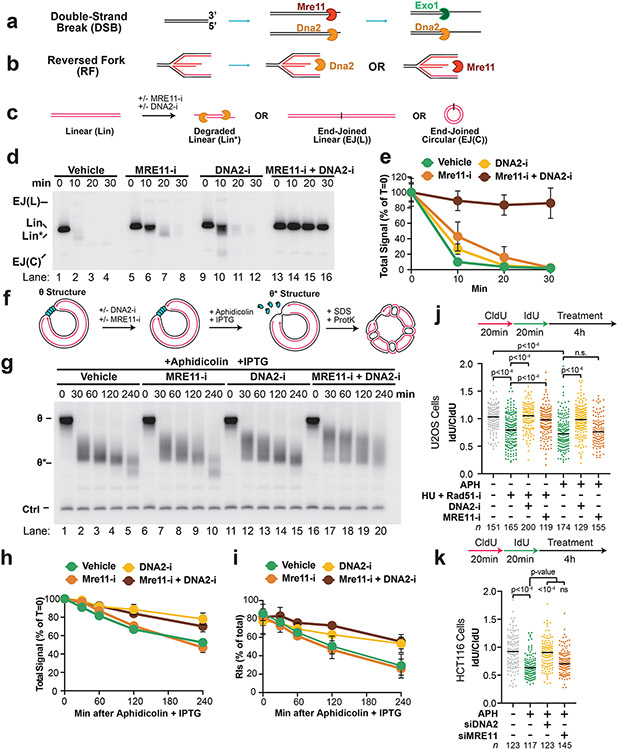
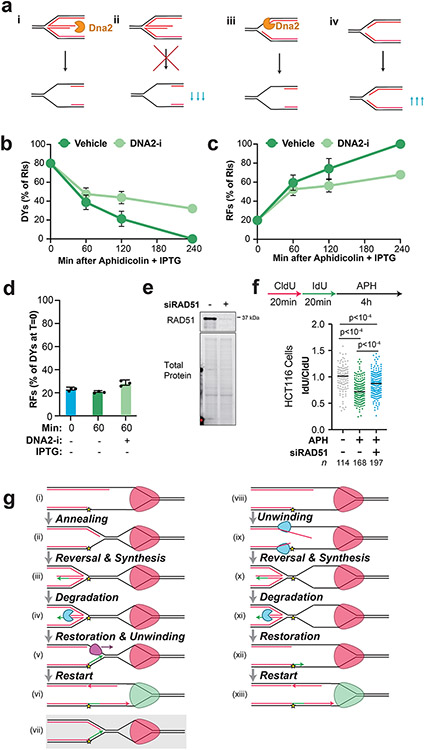
Fork reversal is thought to precede NSD11,13,16,17,21 (Extended Data Fig. 8A,i). However, the mobility of Double-Ys increased over time, indicating that these molecules were degraded (Fig. 3C,ii-iv). Additionally, NSD still took place following knockdown of fork reversal enzymes in human cells (Fig. 3H)7,14. These data suggest that replication forks can be degraded prior to fork reversal (Extended Data Fig. 8A,iii).
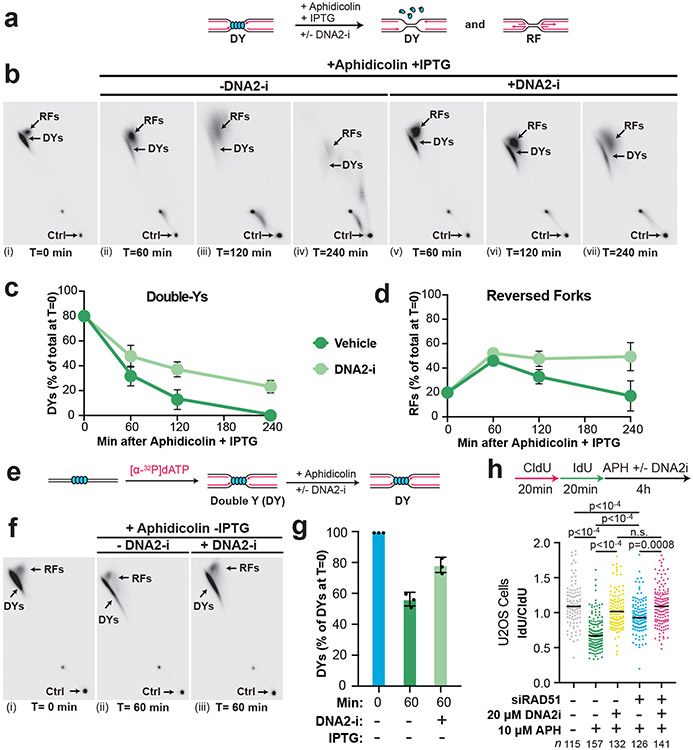
To test whether degradation of Y-shaped forks (double-Ys) arose from degradation of reversed forks (Extended Data Fig. 8A,i) or degradation prior to fork reversal (Extended Data Fig. 8A,iii) we investigated how double-Ys were affected by DNA2-i (as in Fig. 2). If degradation begins at reversed forks and converts them to replication forks (Extended Data Fig. 8A,i) then DNA2-i treatment should lead to a decrease in Y-shaped forks (Extended Data Fig. 8A,ii). Alternatively, if degradation begins at Y-shaped forks (Extended Data Fig. 8A,iii), as suggested by our data (above), then DNA2-i treatment should lead to an accumulation of Y-shaped forks (Extended Data Fig. 8A,iv). To distinguish these possibilities, we induced NSD in the presence or absence of DNA2-i (Fig. 6A) then performed 2D gel analysis (as in Fig. 3A-C) to monitor the signal from double-Ys and reversed forks (Fig. 6B-D). In the vehicle control, double-Y signal decreased over time (Fig. 6B,i-iv, Fig. 6C), as previously observed (Fig. 3D). DNA2-i treatment increased double-Y signal (Fig. 6B,iii,vi, Fig. 6C, Extended Data Fig. 8B), as expected if degradation can initiate at replication forks (Extended Data Fig. 8A,iv). These data cannot be explained by degradation and then restoration of reversed forks because double-Ys were progressively converted to reversed forks (Extended Data Fig. 8B-C). Thus, our data support a model where degradation can occur prior to fork reversal.
Figure 6: DNA2 degrades replication forks and reversed forks.
(A) Forks were localized to a LacR barrier and NSD was induced by addition of IPTG and aphidicolin in the absence or presence of DNA2-i. Purified DNA was subjected to restriction digest so that replication fork structures could be visualized. (B) Double-Y (DY) and reversed fork (RF) structures from (a) were separated by 2-D gel electrophoresis (as in Fig. 3B-C). (C) Quantification of DY structures from (b) as a percentage of total signal at T=0. Mean ± S.D., n=3 independent experiments. See also Extended Data Fig. 8B. (D) Quantification of RF structures from (d) as a percentage of total signal at T=0. Mean ± S.D., n=3 independent experiments. See also Extended Data Fig. 8C. (E) Forks were localized to a LacR barrier and aphidicolin was added in the absence or presence of DNA2-i. IPTG was omitted to retain the intact LacR barrier. (F) Double-Y (DY) and reversed fork (RF) structures from (a) were separated by 2-D gel electrophoresis (as in Fig. 3B-C). (G) Quantification of DY structures from (f) as a percentage of total signal at T=0. Mean ± S.D., n=3 independent experiments. See also Extended Data Fig. 8D. (I) U2OS cells were transfected with control or RAD51 siRNA, pulse labeled with CldU and IdU, then treated with aphidicolin and/or DNA2-i. DNA fiber analysis was performed to determine the lengths of the CldU and IdU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’. See also Extended Data Fig. 8E-F.
We next tested whether Double-Ys could be degraded under conditions where reversed forks did not form. To do so, we monitored NSD under conditions where the LacR array was kept intact (Fig. 6E) so that fork reversal was not induced (as in Fig. 4D-F). Addition of aphidicolin resulted in loss of double-Y signal (Fig. 6Fi-ii, 6G) but did not affect reversed fork abundance (Extended Data Fig. 8D). DNA2-i treatment increased double-Y signal (Fig. 6F,ii-iii, Fig. 6G), but had negligible effect on reversed fork signal (Extended Data Fig. 8D). These data show that replication forks can be degraded without formation of reversed forks.
RAD51 is involved in all reported NSD pathways37 and is thought to be required for fork reversal. Our observations that NSD can occur prior to (Fig. 6A-D) and without (Fig. 6E-G) fork reversal suggested that we might be able to observe RAD51-independent NSD in cells. To test this, U2OS cells were treated with siRNA targeting RAD51 and DNA fiber analysis was performed to monitor NSD. siRNA knockdown of RAD51 reduced, but did not block, NSD (Fig. 6H), consistent with RAD51-independent NSD. The same was also true in HCT116 cells (Extended data Fig. 8F). Furthermore, addition of DNA2-i reduced the amount of NSD following RAD51 knockdown (Fig. 6H). These data (Fig. 6H) are consistent with fork reversal-independent NSD in cells treated with aphidicolin, as observed in Xenopus egg extracts (Fig. 6A-G).
DISCUSSION
We developed an in vitro approach to study nascent strand degradation (NSD) in vertebrates (Fig. 1-3). Using this approach, in combination with cellular experiments, we show that uncoupling causes NSD and fork reversal (Fig. 4), the replicative helicase remains bound through this process (Fig. 5), and degradation occurs both before and after fork reversal (Fig. 6). These data inform a new model for NSD (Fig. 7) and have implications for fork reversal and template switching (Extended Data Fig. 8G). These points are discussed further below.
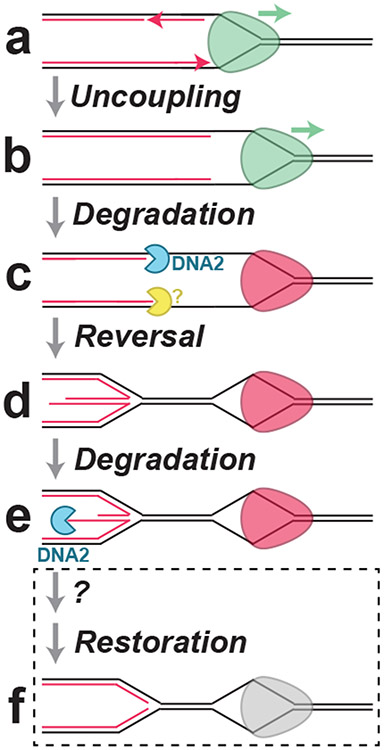
Figure 7: Model for nascent strand degradation in Xenopus egg extracts.
(A) Replicative helicase and DNA polymerase activities are coupled at replication forks. (B) When DNA polymerases stall the replicative helicase continues unwinding, resulting in uncoupling. (C) Uncoupling initially stimulates NSD by DNA2 and one or more other nucleases (yellow). The uncoupled replicative helicase stalls (red). (D) Uncoupling causes replication fork reversal. The replicative helicase remains bound to single-stranded DNA suggesting parental DNA strands are reannealed behind it. (E) The regressed arm undergoes NSD, which requires DNA2. (F) Restoration of the reversed arm is likely to involve an additional step that has not been identified and it is unclear whether the replicative helicase would be active at this point.
How does uncoupling elicit fork reversal and NSD?
Uncoupling stimulates both NSD and fork reversal (Fig.4, Fig. 7A-D). This is consistent with a correlation between uncoupling and replication fork reversal14. However, it is unclear exactly which molecular event involved in uncoupling causes NSD and fork reversal. One possibility is that nascent strands efficiently stimulate fork reversal and NSD but this is typically inhibited by interactions between the replisome and the nascent DNA strands. According to this view, uncoupling would physically separate the replisome from the nascent strands to grant access to fork processing and remodeling enzymes. Alternatively, ssDNA generated by CMG helicase activity during uncoupling may stimulate reannealing of the parental strands and generate the substrate for fork reversal. This would be consistent with other situations where NSD and processing occur at post-replicative gaps9,54-57. It will be important to address whether any of the proteins that modulate post-replicative gap stability independent of fork reversal56,57 are also involved in limiting the initial NSD step we have identified at forks (Fig. 7B-C).
Implications of replicative helicase retention
Replication fork DNA protects the CMG helicase from replisome removal pathways58,59. CMG is retained on DNA (Fig. 5C) during NSD and fork reversal (Fig. 4C,F) indicating that CMG remains at a replication fork structure during these processes. Although CMG can backtrack60, this would involve extensive reannealing of parental DNA, which we did not observe (Fig 1B, Extended Data Fig. 7C). Instead, we favor the idea that fork reversal takes place behind CMG, which would then reside within a single-stranded DNA bubble (Fig. 7C-D). One possibility is that parental DNA strands reanneal in the wake of CMG to generate a second replication fork structure, which then undergoes reversal (Extended Data Fig. 8i-vi). Alternatively, nascent DNA strands might be unwound and directly annealed to each other (Extended Data Fig. 8viii-xiii). We note that FBH1 catalyzes fork reversal15 and exhibits the required ssDNA helicase activity61 needed to support this second model (Extended Data Fig. 8viii-xiii). Notably, the former scenario (Extended Data Fig. 8i-vi) would require a helicase for replication restart (Extended Data Fig. 8v-vii) while the latter would not (Extended Data Fig. 8xi-xii). Our data do not show evidence of extensive re-annealing of parental DNA strands (Fig. 1b, Fig. 5b) but do not rule out the possibility that limited reannealing takes place and is masked by extensive NSD (Extended Data Fig. 7C). Thus, it will be important to determine the molecular events that generate reversed forks. Regardless of how exactly reversed forks are generated, the double-stranded DNA translocase activity of the helicases involved (Fig. 3H)62,63 means that the replisome will ultimately reside within a ssDNA bubble, separated from the reversed fork by a short stretch of duplex DNA (Fig. 7C), although the size of the bubble and duplex vary between the two models (Extended Data Fig. 8Giii,x).
Mechanisms of degradation
DNA2 degrades uncoupled forks prior to fork reversal (Fig. 6, Fig. 7C-D). However, fork reversal did not increase when DNA2 was inhibited (Extended Data Fig. 8c), which is inconsistent with a role for DNA2 in limiting reversed fork formation19. Instead, our results are consistent with a role for DNA2 in degradation of reversed forks (Fig. 6D), as previously reported7. It is possible that DNA2 activity at uncoupled forks supports this function by priming the nascent strands for extensive degradation64 upon fork reversal. It will be important to determine the role of NSD at uncoupled forks and whether it is negatively regulated by any fork protection proteins, as for NSD at reversed forks8,9,21.
We found that both nascent DNA strands are degraded (Extended Data Fig. 1B, Extended Data Fig. 6D-F), as previously reported8. It is likely that 3’-5’ nucleases cooperate with the 5'-3' activity of DNA2 to ensure that both DNA strands are degraded. We did not find roles for FAN146 or SAMHD147 (Extended Data Fig. 4), but WRN7 and EXD265 are alternate possibilities. We also note that reversed forks were completely stable when DNA2 was inhibited (Fig. 6D). This could mean that only 5’-3’45 degradation takes place at the reversed fork or that both strands are degraded by 5’-3’ and 3’-5’ degradation activities that depend on each other. Additional work will be required to decipher how the many nucleases implicated in NSD1 process different replication fork structures.
There are two main cellular pathways for NSD; one dependent on SMARCAL1 and the other dependent on FBH137. Surprisingly, aphidicolin induced NSD that involved both SMARCAL1 and FBH1 (Fig. 3H), suggesting both pathways operate simultaneously. However, HLTF and ZRANB3 normally cooperate with SMARCAL1 during NSD37 but were not required in this case (Fig. 3H), so the situation may be more complex. Importantly, SMARCAL1 was not involved in NSD in Xenopus egg extracts (Extended Data Fig. 5K-O), despite being involved in NSD in a different Xenopus egg extract system16. This suggests that our in vitro approach favors a subset of cellular pathways, as is the case for other types of DNA repair66,67. SMARCAL1 and FBH1 operate in different pathways37 so it is possible that fork reversal using our in vitro approach depends on FBH1. It will be crucial to determine how the proteins involved in NSD are determined in different situations.
Restoration of reversed forks
Replication forks were progressively converted to reversed forks following uncoupling (Fig. 3D). Because reversed forks need to be converted to replication forks to resume DNA synthesis, a restoration event must convert reversed forks to replication forks (Fig. 7E-G). Our experiments block DNA-synthesis so restoration of reversed forks likely involves DNA synthesis. This would be consistent with the reported antagonism between repriming and fork reversal pathways55,68. It will be crucial to determine how reversed forks are converted back to replication forks during replication restart.
METHODS
Statistics and reproducibility
For experiments using Xenopus egg extracts each experiment was repeated twice independently with similar results, unless otherwise indicated.
For DNA fiber assays using human cells at least 100 fibers were counted for each sample. Statistical analyses were performed using Prism. A one-way ANOVA was performed using Dunnett’s multiple comparisons test. P-values are reported in the figures. Non-significant comparisons are indicated as ‘ns’. No statistical methods or criteria were used to estimate sample size or to include/exclude samples. Extended Data Figure 1C Was performed twice independently with similar results. All other DNA fiber assays were performed three times independently with similar results.
Xenopus egg extracts
Xenopus egg extracts were prepared from Xenopus laevis wild-type males and females (Nasco) as previously described69. Animal protocols were approved by Vanderbilt Division of Animal Care (DAC) and Institutional Animal Care and Use committee (IACUC).
Plasmid Construction and Preparation
pJD145, pJD156, and pJD90 were described previously42. To create pJD161 (p[lacOx25]-XhoI-[lacOx25]) DNA oligonucleotides JDO120 (GTACAAGTAAATCAGAGCCAGATTT TTCCTCCTCTCGGAATTGTGAGCGGATAACAATTCCCTCGAGCCAATTGTGAGCGGATAACAATTGGAAGTGCAGAACCAATGCATGCAGGAGATTTGAC) and JDO121 (GT ACGTCAAATCTCCTGCATGCATTGGTTCTGCACTTCCAATTGTTATCCGCTCACAATTGGCTCGAGGGAATTGTTATCCGCTCACAATTCCGAGAGGAGGAAAAATCTGGCTCTGATTTACTT) were annealed to create a DNA fragment that contained a XhoI site flanked by a lacO sequence on each side. The JDO120/JDO121 duplex was then cloned into the BsrGI site of pJD90 (p[lacOx24]). This cloning procedure regenerated the BsrGI site, which was then used as the target for insertion of a BsrGI-BsiWI fragment from pJD90 that contained 24 tandem lacO repeats. The resulting plasmid was then digested with BsrGI and BsiWI to create a fragment containing 50 tandem lacO repeats with a XhoI site in the middle. This fragment was inserted into the BsiWI site of pJD145 to yield pJD161.
DNA replication in Xenopus egg extracts
High Speed Supernatant (HSS) was supplemented with nocodazole (3 ng/μl) and ATP regenerating system (ARS; 20mM phosphocreatine, 2 mM ATP and 5 ng/μl creatine phosphokinase) then incubated at room temperature for 5 minutes. To license plasmid DNA, 1 volume of ‘licensing mix’ was prepared by adding plasmid DNA to HSS at a final concentration of 15 ng/μl, followed by incubation at room temperature for 30 minutes. NucleoPlasmid Extract (NPE), extract was supplemented with ARS, DTT (final concentration: 2 mM), [α-32P]dATP (final concentration: 350 nM) and diluted to 45% in 1X Egg Lysis Buffer (ELB, 250 mM Sucrose, 2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES, pH 7.7). To form a replication barrier, 0.1 volumes of LacR (37 μM) was incubated with 0.05 volumes of plasmid DNA (300 ng/μl) for at least 90 minutes at room temperature42. To initiate replication, 2 volumes of NPE mix were added to 1 volume of Licensing mix. Replication forks were stalled at the LacR-bound lacO array and then released by addition of 0.06 volumes IPTG (10 mM final)42. Reactions were stopped by addition of 30 volumes Extraction Stop Solution (0.5% SDS, 25 mM EDTA, 50 mM Tris-HCl, pH 7.5). Samples were subsequently treated with RNase A (final concentration: 190 ng/μl) and then Proteinase K (909 ng/μl) before either direct analysis by gel electrophoresis or purification of DNA. To purify DNA proteins were first removed by phenol:chloroform extraction, then DNA was precipitated with ethanol (70% (v/v) final) and NaOAc (86 mM final) in the presence of 1 μl glycogen, and finally resuspended in 1-2 volumes 10 mM Tris-HCl (pH 8.0).
For most experiments pJD156 (p[lacOx32]) was used as the template for replication. For the experiments in Fig. 3E-G, pJD161 (p[lacOx25]-XhoI-[lacOx25], also referred to as p[lacOx50]) was used. For Extended Data Fig. 2E-G both pJD156 and pJD161 were used. pJD145 (p[CTRL]) did not contain a lacO array, which allowed it to fully replicate in the presence of LacR. pJD145 was used as a loading control because its small size allowed it to be readily distinguished from pJD156 and pJD161. In most experiments, pJD145 was added to the licensing mix at a final concentration of 1 ng/μl and the plasmid replicated prior to induction of NSD due to the absence of any LacR array. In Extended Data Fig. 1D-M radiolabeled pJD145 was added to NPE at a concentration of 1 ng/μl prior to initiation of DNA replication and the plasmid did not replicate because it had not undergone prior licensing of the DNA.
Aphidicolin from Nigrospora sphaerica (Sigma-Aldrich) was dissolved in DMSO and added to reactions at a final concentration of 330 μM. Mirin (Selleckchem) was dissolved in DMSO and added to reactions at a final concentration of 500 μM. C5 (AOBIOUS) was dissolved in DMSO and added to reactions at a final concentration of 3.5 mM. NMS-873 (Selleckchem) was dissolved in DMSO and added to reactions at a final concentration of 200 μM. For drugs dissolved in DMSO the final reaction concentration of DMSO was 4% (V/V).
Protein Purification
Biotinylated LacR was expressed in Escherichia coli and purified as described previously42.
Western Blotting & Immunodepletions
The following antibodies targeting Xenopus proteins were previously validated by Western blotting of Xenopus egg extracts: RPA51, CDC4551, MCM653, SAMHD147 and FAN170. Antibody (i) targeting Xenopus SMARCAL1 was raised against a peptide of CKRRKIDDYFAL (New England Peptide 3850), affinity purified against its target peptide, and confirmed to immunoprecipitate only a single band that was also recognized by the previously-described Xenopus SMARCAL1 antibody16. Antibody (ii) targeting Xenopus SMARCAL1 was previously affinity purified against its target polypeptide then validated by Western blotting in Xenopus egg extracts16. For Western blotting, the secondary antibody was HRP Goat anti-rabbit (Jackson Immunoresearch 111-035-003) in all cases. The following antibody dilutions were used for Western blotting: CDC45, SMARCAL1 (i) and (ii), SAMHD1, FAN1 (1:10,000); RPA (1:5,000); MCM6 (1:2,000); and HRP Goat anti-rabbit (1:30,000).
Immunodepletions were performed as described71 but with an additional round of depletion. Protein A-coupled magnetic beads (Dynabeads Protein A 10001D (30μg/μl)) were first bound to 0.5μg antibody per 1μg beads. 1.29 volumes of antibody-bound beads were then incubated with 1 volume of NPE or 0.5 volumes HSS for 20minutes at room temperature with end-over-end rotation. This was repeated twice for a total of three rounds of immunodepletion. Depleted extracts were then collected and used for experiments.
Antibody targeting human RAD51 (Abcam AB63801) was obtained from Abcam and validated by Western blotting of HeLa cell lysates. The secondary antibody was IRDye 800CW Goat anti-rabbit (LI-COR 926-32211). RAD51 antibody was used at a dilution of 1:10,000 for Western blotting.
Nascent Strand Degradation assays
To monitor DNA synthesis, samples were separated on a 1% agarose gel at 5 V/cm. Radiolabeled DNA was visualized by phosphorimaging to measure incorporation of radiolabeled nucleotides. Signal was quantified using ImageQuant (GE Healthcare) and ImageJ and normalized to the loading control in each lane.
To monitor NSD, samples were purified and then digested with 0.4 U/μl XmnI in CutSmart Buffer (NEB) for 30 minutes at 37°C and then separated on a 1% agarose gel at 5 V/cm. Radiolabeled DNA was detected by phosphorimaging and Replication Intermediate signal was quantified and normalized to pJD145 signal, which served as a loading control. To calculate normalized degradation in Extended Data Fig. 2G, degradation was calculated as 100 - [Signal (% of T=0)]. Degradation for p[lacOx50] was then multiplied by 2912/2658 to normalize degradation to the size of the plasmid backbone, while degradation for p[lacO] was left unaltered.
To monitor disappearance of intact nascent strands, purified NSD intermediates were digested with 0.4 U/μl AlwnI (NEB) in CutSmart Buffer (NEB) for 1 hour at 37°C. Digest was stopped by adding EDTA to a final concentration of 30 mM. Reaction was then prepared for electrophoresis by adding Alkaline Loading Buffer 6X (EDTA, Ficoll, Bromocresol green, Xylene Cyanol and NaOH (10N)) to a final concentration of 1X. Nascent strands were then separated on a 1.5% denaturing alkaline gel at 1.5 V/cm, which was then neutralized by gentle agitation in 7% TCA solution. Subsequently, radiolabeled DNA was detected by phosphorimaging, as described above.
2D gel electrophoresis
2D gels were performed as described 53 with slight modifications. Briefly, purified DNA was digested with XhoI and DraIII (Fig. 3G) or XmnI (all other gels) and digested DNA was separated on a 0.4% agarose gel at 1 V/cm for 22 hours. A second dimension gel containing 1.2% agarose and 0.3 μg/ml ethidium bromide was cast over the first dimension gel slice and separated at 5 V/cm for 12 hours at 4°C. Radiolabeled DNA was then detected by phosphorimaging as described above.
Nick translation of DNA
To radiolabel parental DNA strands, 60 ng/μl plasmid DNA was resuspended in 1X NEB Buffer 2.1 and treated with 0.5 U/μl Nt.BbvCI at 37°C for 1 hour, followed by heat inactivation at 80°C for 20 minutes. To perform nick translation the reaction was supplemented with 0.5 volumes of dNTPs (12.5 mM each of dCTP, dGTP, dTTP), 3 U/μl E. coli DNA Polymerase 1, and 0.33 mM [α32P]-dATP in 1X NEB Buffer 2.1 and incubated at 16°C for 10 minutes then on ice for 5 minutes. Nick-translated DNA was purified into 10 mM Tris-HCL, pH 8.0 using micro Bio-Spin columns (Bio-Rad) and used for replication and NSD assays.
Plasmid pull downs
Streptavidin M-280 Dynabeads (Invitrogen 11205D) were washed twice with Binding Buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA pH 8, 0.02% Tween-20). Biotinylated LacR was added to the beads (30 pmol per 15 μl beads) and incubated at room temperature for 40 min. After binding, beads were washed twice with Binding Buffer and twice with ELB-BT (10 mM Hepes pH 7.7, 50 mM KCl, 2.5 mM MgCl2, 0.5 M Sucrose, 0.5mg/ml BSA, 0.04% Tween-20). Bead suspensions were stored on ice and at indicated timepoints, 3 μl samples were withdrawn and mixed with beads. Bead and sample suspensions were placed on a rotating wheel and rotated for 30 min at 4°C. Beads were washed twice with ELB-Salts Buffer (10 mM Hepes pH 7.7, 50 mM KCl, 2.5 mM MgCl2, 0.25 mg/ml BSA, 0.03% Tween-20). All residual buffer was removed and beads were resuspended in 30 μl 1X SDS protein sample buffer (3% SDS, 62.5 mM Tris-HCl (pH 6.8), 10% Glycerol, 0.005% Bromophenol blue, 50 mM DTT) then heated at 95°C for 5 minutes. Samples were then resolved on SDS-PAGE gels, transferred onto PVDF membranes and probed with the indicated antibodies.
siRNA knock downs
siRNAs targeting MRE11 (L-009271-00), DNA2 (L-026431-01), RAD51 (L-003530-00), PICH (L-031581-01), HLTF (L-006448-00), FBH1 (L-017404-00), FAN1 (L-020327-00), and SAMHD1 (L-013950-01) were ON-TARGETplus siRNA from Horizon. siRNA targeting ZRANB3 was s38488 from ThermoFisher. SMARCAL1 was targeted using siRNA GCUUUGACCUUCUUAGCAA. All siRNA transfections were performed using DharmaFECT reagents according to the manufacturer’s instructions. Experiments were performed 3 days after transfection.
DNA fiber analysis in human cells
ATCC was the source of U2OS (HTB-96) and HCT116 (CCL-247) cells used in this study. For DNA fiber spreading experiments37, cells were labeled with 20 μM CldU followed by 100 μM IdU for 20 min each, then treated as indicated with the following concentrations of drugs: aphidicolin (10 μM, unless otherwise indicated), Rad51 inhibitor B02 (25 μM), DNA2 inhibitor C5 (20 μM), MRE11 inhibitor Mirin (20 μM), HU (4 mM). Following stretching and fixation on glass slides, DNA was denatured in 2.5 M HCl for 80 min, washed three times with phosphate-buffered saline (PBS), and blocked in 10% goat serum/PBS with 0.1% Triton X-100 for 1 hour. Slides were immunoblotted with 1% (v/v) rat anti-CldU antibody (Abcam ab6326) and 1% (v/v) mouse anti-IdU antibody (BD B44) for 2 hours, then washed 3 times with PBS followed by 0.4% (v/v) Alexa Fluor 594 Goat anti-Rat (Invitrogen A11007) or Alexa Fluor Plus 488 Goat anti-mouse (Invitrogen A32723) secondary antibody. Slides were then washed with PBS 3 times, then mounted with prolong gold without DAPI (Invitrogen P36930). Fibers were imaged using a Nikon Ti-DH immunofluorescence microscope.
Extended Data
Extended Data Fig. 1. Nascent strand degradation is an initial response to fork stalling.
A) A current model for fork reversal and nascent strand degradation. Note that fork reversal enzymes are thought to directly convert reversed forks back to replication forks1-3.
B) U2OS cells were pulse-labeled with CldU then IdU followed by treatment with the indicated concentrations of aphidicolin so that loss of the IdU labeled tracks could be monitored as a measurement of NSD. DNA fiber analysis was performed to determine the lengths of CldU and IdU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’. Although lower concentrations of aphidicolin (0.2-1 μM) did not impact the IdU/CldU ratio higher concentrations (4-10 μM) significantly reduced the IdU/CldU ratio indicating that NSD took place. Thus, we observed a dose-dependent induction of NSD by aphidicolin.
C) U2OS cells were pulse labeled with CldU and IdU, then treated with the indicated concentrations of aphidicolin and hydroxyurea (HU). DNA fiber analysis was performed to determine the lengths of CldU and IdU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’. Co-addition of the RAD51 inhibitor B02 (RAD51-i)4 was also performed as a positive control for degradation in the presence of HU. RAD51-i addition inactivated fork protection and led to extensive degradation in the presence of HU, as previously described 5. In contrast, 1, 4, and 8 mM HU did not result in detectable NSD, consistent with limited degradation in the presence of HU alone6. Notably, the extent of NSD following 10 μM aphidicolin treatment was comparable to the level observed following HU treatment and RAD51-i addition.
D) To stall replication forks in vitro, plasmid DNA was replicated using Xenopus egg extracts and newly-synthesized nascent strands were radiolabeled by inclusion of [α-32P]dATP. In this system, plasmid templates replicate semi-synchronously from a single origin per plasmid7. 6 minutes after initiation, reactions were treated with aphidicolin, to stall DNA synthesis, or vehicle control. As a loading control (Ctrl) the reactions include a smaller plasmid that was radiolabeled prior to the experiment and did not undergo replication.
E) DNA structures from (d) were purified and digested with XmnI, which cuts the plasmid once.
F) DNA taken from (e) at the moment of aphidicolin addition (6 minutes after initiation) was separated by 2-D gel electrophoresis and visualized by autoradiography. The entire Double-Y and Bubble arcs (i) were visible (ii), indicating that DNA molecules of all sizes, and thus at all stages of replication, were present. Note that origin firing generates bubble structures, which are converted to double-Ys once one of the two forks moves beyond the restriction site (iii).
G) Samples from (e) were separated on an agarose gel and visualized by autoradiography. Time after aphidicolin addition is indicated.
H) Quantification of DNA synthesis from (g) normalized to the maximum signal across all time points and conditions. Mean ± S.D., n=3 independent experiments.
I) Quantification of DNA synthesis in (g) normalized to the maximum signal for each condition across all time points. Mean ± S.D., n=3 independent experiments. Following vehicle treatment in (h)-(i), radioactive signal increased and then plateaued, indicating that a single round of replication was completed. In contrast, following aphidicolin treatment in (h)-(i) signal declined, indicating that degradation took place. Importantly, signal was appreciably reduced by 60 minutes, indicating that degradation was an initial response and did not require prolonged fork stalling.
J) To test whether degradation in (h)-(i) corresponded to both strands or nascent strands only, pre-radiolabeled plasmid DNA was replicated using Xenopus egg extracts. 6 minutes after initiation, DNA synthesis was inhibited by the addition of Aphidicolin, as in (e).
K) XmnI digested molecules were separated on an agarose gel and visualized by autoradiography. As a loading control (Ctrl) the reactions include a smaller radiolabeled plasmid that did not undergo replication.
L) Quantification of (k) as in (h). Mean ± S.D., n=3 independent experiments.
M) Quantification of (k) as in (i). Mean ± S.D., n=3 independent experiments. In (l)-(m) parental strand signal was relatively stable and there was little difference between aphidicolin and vehicle treatment. Moreover, there was no appreciable loss of parental strand signal for the first 120 minutes, during which time most of the nascent strands were degraded in (h)-(i). Thus, most signal loss was due to degradation of nascent DNA strands. Overall, (d)-(l) show that degradation of nascent strands is an initial response to fork stalling
Extended Data Fig. 2. Characterization of localized, synchronous nascent strand degradation.
A) Cartoon depicting the source of θ* structures in Fig. 1B. The XmnI site is located 1013 base pairs away from the closest edge of the lacO array.
B) DNA structures from Fig. 1A were treated with human topoisomerase II (hTop2) or buffer control, then separated on an agarose gel and visualized by autoradiography. hTop2 treatment converted θ* signal back to θs, demonstrating that θ* structures are topoisomers of θs. Note that in lane 4 an additional band appears below the θ, suggesting that replication forks undergo remodeling (see Fig. 3 and related discussion in the main text).
C) Quantification of Replication Intermediates (RIs) as a % of total lane signal from Fig. 1E. Mean ± S.D., n=5 independent experiments.
D) Quantification of total lane signal from Fig. 1E. Mean ± S.D., n=5 independent experiments.
E) Plasmid DNA harboring a 32xlacO array (p[lacO]) or a 50xlacO array (p[lacOx50]) was incubated with LacR then replicated in Xenopus egg extracts. Once forks were localized to the LacR barrier, NSD was induced by addition of IPTG and aphidicolin. Purified DNA was subjected to restriction digest so that replication fork structures (RIs) could be visualized.
F) Samples from (e) were separated on an agarose gel and visualized by autoradiography. Note that a mobility shift can readily be detected for p[lacOx50], suggesting that fork remodeling occurs (see Fig. 4 and related main text).
G) Quantification of the amount of DNA degraded in (f). Normalized degradation accounts for the different backbone sizes of the 50x and 32x lacO plasmids. Mean ± S.D., n=3 independent experiments. The 50xlacO array is ~60% larger than the 32xlacO array so if NSD was influenced by the proximity of the replication forks on either side of the lacO array there would be a substantial difference in NSD between p[lacO] and p[lacOx50]. However, the difference in normalized degradation between the two plasmids is negligible. Thus, NSD does not appear to be influenced by the proximity of the two converging forks.
Extended Data Fig. 3. Characterization of MRE11 and DNA2 activity during nascent strand degradation.
A) Cartoon depicting the roles of MRE11 and DNA2 in resection at double-strand breaks.
B) Cartoon depicting the roles of MRE11 and DNA2 in resection at reversed forks.
C) Linear radiolabeled DNA was incubated in Xenopus egg extracts in the presence of MRE11 inhibitor (MRE11-i) or DNA2 inhibitor (DNA2-i) or both. Double-Strand Break (DSB) resection involves both enzymes in Xenopus egg extracts (b) and is blocked by inactivation of both DNA2 and MRE11 8. Thus, if both MRE11-i and DNA2-i effectively inhibit their targets then combination of both drugs should block resection.
D) Samples from (c) were separated on an agarose gel and visualized by autoradiography. In the vehicle control the linear substrate was rapidly degraded (lanes 1-4). We also observed a small number of additional products that arose from end-joining (EJ(L) and EJ(C)). As expected9, DNA2-i and MRE11-i each inhibited resection (lanes 5-8, 9-12), while a combination of both inhibitors almost completely blocked resection (lanes 13-16).
E) Quantification of total signal from (d). Mean ± S.D., n=3 independent experiments.
F) Depiction of the DNA structures generated by Fig. 2A, prior to restriction digest.
G) DNA intermediates from (f) were separated on an agarose gel and visualized by autoradiography. Uncoupled species (θ*) are indicated.
H) Quantification of total lane signal from Fig. 2B. Mean ± S.D., n=3 independent experiments.
I) Quantification of RI signal in Fig. 2B as a percentage of total signal for each lane. Mean ± S.D., n=3 independent experiments.
J) U2OS cells were pulse labeled with CldU and IdU, then treated with either aphidicolin, or hydroxyurea combined with RAD51-i. MRE11-i, DNA2-i, or vehicle control were also added, as indicated. DNA fiber analysis was performed to determine the lengths of IdU and CldU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’. Treatment with Rad51-i compromised fork protection and led to NSD in the presence of HU, as previously reported 5. Additionally, NSD was rescued by either DNA2-i or MRE11-i, as expected 10, confirming that both drugs were able to effectively inhibit their targets. Aphidicolin treatment resulted in NSD (as in Extended Data Fig. 1B) and this was rescued by DNA2-i but not MRE11-i. Thus, NSD caused by aphidicolin treatment results in DNA2-dependent but MRE11-independent degradation.
K) HCT116 cells were transfected with the indicated siRNA. 72 hours later cells were pulse labeled with CldU and IdU, then treated with aphidicolin. DNA fiber analysis was performed to determine the lengths of IdU and CldU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’.
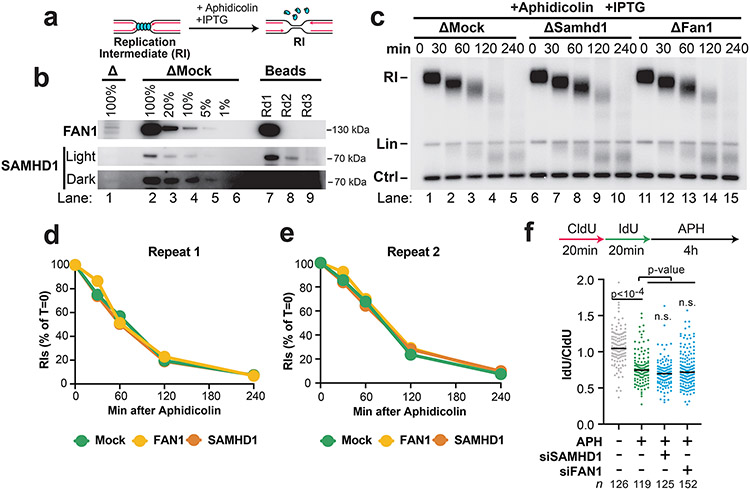
Extended Data Fig. 4. SAMHD1 and FAN1 are not involved in nascent strand degradation.
A) Forks were localized to a LacR barrier and NSD was induced by addition of IPTG and aphidicolin in SAMHD1- and FAN1-immunodepleted Xenopus egg extracts. Purified DNA was subjected to restriction digest so that replication fork structures could be visualized (RIs).
B) Immunodepleted extracts and the corresponding immunoprecipitates (beads) from (a) were analyzed by Western blotting to determine the extent of SAMHD1 and FAN1 immunodepletion. In both cases the immunodepletion went to completion because no detectable protein was recovered by round 3 (compare lanes 7-8 to lane 9). Additionally, at least 95% of protein was removed in each case (compare lanes 1 and 5).
C) Samples from (a) were separated on an agarose gel and visualized by autoradiography.
D) Quantification of RI signal from (c).
E) Quantification of RI signal from an experimental replicate of (c). In both (d) and (e) neither SAMHD1 nor FAN1 depletion altered the rate of degradation, indicating that they do not participate in NSD.
F) U2OS cells were transfected with the indicated siRNA, pulse labeled with CldU and IdU, then treated with aphidicolin. DNA fiber analysis was performed to determine the lengths of CldU and IdU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’. Neither SAMHD1 nor FAN1 affected degradation, consistent with (d)-(e).
Extended Data Fig. 5. Characterization of DNA structures detected by 2-D gel during nascent strand degradation.
A) Quantification of Double-Ys from Fig. 3C expressed as a percentage of total signal at T=0. Mean ± S.D., n=3 independent experiments.
B) Quantification of remodeled forks from Fig. 3C expressed as a percentage of total signal at T=0. Mean ± S.D., n=3 independent experiments.
C) Cartoon indicating the different DNA structures that the remodeled forks in Fig. 3C could correspond to.
D) Expected 2-D gel migration pattern of the structures depicted in (c). The region highlighted in red indicates where each structure is expected to migrate. ‘?’ indicates structures whose expected migration on a 2-D gel is unclear. The remodeled forks (Fig. 3C) did not correspond to canonical Holliday junctions (i) or hemicatenanes (ii). However, these species could have reflected reversed forks (iii), hemicatenanes (iv), or Holliday junctions (v) in the wake of one or both forks.
E) Samples from Fig. 3C,iii were treated with buffer or RuvC, which cleaves reversed forks and Holliday junctions, but not hemicatenanes or other replication fork structures11. RuvC-treated samples were then separated by 2-D gel electrophoresis and visualized by autoradiography.
F) Quantification of DYs and RFs following RuvC treatment in (e) expressed relative to the abundance of each structure in the buffer treated condition. RuvC treatment had essentially no effect on Double-Ys, as expected, but reduced remodeled forks by ~5-fold. Thus, the remodeled forks contain four-way junctions that arise from reversed forks or Holliday junctions i.e. D-loops.
G) Quantification of RFs and DYs from (c) expressed relative to total signal on each gel.
H) Cartoon indicating the structure of a D-loop and a reversed fork.
I) Expected 2-D gel migration pattern of the structures depicted in (h). The region highlighted in red indicates where each structure is expected to migrate.
J) Cartoon indicating the restriction digest strategy used to excise individual replication forks for 2-D gel analysis. The DraIII site is 450 bp away from the nearest edge of the lacO array and 1250 bp away from the XhoI site.
K) Forks were localized to a LacR barrier and NSD was induced by addition of IPTG and aphidicolin in SMARCAL1-immunodepleted Xenopus egg extracts. Purified DNA was subjected to restriction digest so that replication fork structures could be visualized (RIs).
L) Immunodepleted extracts and the corresponding immunoprecipitates (beads) from (k) were analyzed by Western blotting to determine the extent of SMARCAL1 immunodepletion. Immunoblotting was performed using an antibody generated by this study (i) and the previously published antibody (ii)12. No detectable protein was recovered by the final round of depletion (compare lanes 7-8 to lane 9), indicating that the depletion was successful. Additionally, at least 99% of protein was removed (compare lanes 1 and 6 for ii).
M) Samples from (k) were separated on an agarose gel and visualized by autoradiography.
N) Quantification of RI signal from (m).
O) Quantification of RI signal from an experimental replicate of (m). In both (n) and (o) SMARCAL1 immunodepletion had no impact on degradation.
Extended Data Fig. 6. Characterization of stalled and uncoupled replication forks.
A) Cartoon depicting the strategy to test the role of helicase stalling during NSD. If helicase stalling triggers NSD then NSD should be increased in the absence of IPTG. If uncoupling triggers NSD then addition of IPTG should increase NSD.
B) Depiction of the DNA structures generated by Fig. 4A, prior to restriction digest.
C) Samples from (e) were separated on an agarose gel and visualized by autoradiography. θ* species arise from uncoupling. θ− species arise from the low level of uncoupling that occurs even in the presence of LacR, which is not a complete block to helicase progression13.
D) Depiction of different models for degradation of leading and lagging strands during NSD observed in Fig. 4 and the expected effect on degradation of LWS and RWS depicted in Fig. 4A. In (i) both leading and lagging strand are degraded simultaneously and synchronously, which should result in disappearance of RWS before LWS. In (ii) lagging strands only are degraded synchronously, which should result in persistence of ~50% of RWS and LWS until the leading strand are degraded by nuclease activity that initiated at lagging strands of the opposite fork. This would result in biphasic kinetics of degradation. In (iii) lagging strands only are degraded asynchronously, which should result in loss of RWS and LWS at the same rate.
E) Uncropped gel of Fig. 4B lanes 1-4 shown after a longer exposure so that the weaker rightward strands (RWS) could be visualized.
F) Quantification of LWS and RWS from (b). RWS are degraded before LWS, with no evidence of biphasic kinetics, indicating that both strands are degraded simultaneously, as in (a,i). Mean ± S.D., n=3 independent experiments.
G) NSD was induced as in Fig. 4A in the presence or absence of DNA2-i.
H) Samples from (g) were separated on a native agarose gel and visualized by autoradiography.
I) Quantification of RI signal from (h). Mean ± S.D., n=3 independent experiments.
J) Quantification of reversed forks from Fig. 4E expressed as a percentage of total signal at T=0. Mean ± S.D., n=3 independent experiments.
Extended Data Fig. 7. Binding of replication fork proteins during NSD.
A) Cartoon depicting two different models of CMG helicase behavior during fork reversal and NSD. In (i) the CMG helicase translocates onto double-stranded DNA, as suggested1,14, which would result in its removal from DNA15-18. In (ii) the replisome remains on DNA, suggesting that it resides in a ssDNA bubble ahead of the reversed fork.
B) In parallel to Fig. 5A chromatin bound proteins were analyzed in the presence of DNA2-i. RPA was detected by Western Blotting.
C) Quantification of RPA signal from Fig. 5B. and (b). Mean ± S.D., n=3 independent experiments. Between 0 and 60 minutes, when most fork reversal took place (Fig. 3D), most RPA signal was due to DNA2 activity. Thus, most RPA signal during this time was due to DNA2-dependent NSD, which may mask any dissociation of RPA that occurred due to reannealing of parental DNA strands.
Extended Data Fig. 8. Additional degradation steps during NSD.
A) Cartoon of different models to explain a role for DNA2 in degrading Y-shaped forks during NSD and the expected impact of loss of DNA2 activity on Y-shaped forks in each case. (i) depicts a model where DNA2 degrades reversed forks to generate Y-shaped forks. In this model the Y-shaped forks are degraded as consequence of prior DNA2 activity at reversed forks. (ii) depicts the impact of impaired DNA2 activity on (i). Y-shaped forks should decrease in abundance as less degradation of reversed forks should reduce the formation of Y-shaped forks. (iii) depicts a model where DNA2 degrades forks prior to fork reversal. (iv) depicts the impact of impaired DNA2 activity on (iii). Y-shaped forks should increase in abundance due to less degradation of nascent strands, which is the source of the radioactive signal that is measured.
B) Quantification of Double-Ys from Fig. 6B expressed as a percentage of total replication intermediates (RIs). Mean ± S.D., n=3 independent experiments.
C) Quantification of reversed forks from Fig. 6B expressed as a percentage of total replication intermediates (RIs). Mean ± S.D., n=3 independent experiments.
D) U2OS cells treated with control or RAD51 siRNA were lysed and analyzed by western blotting for RAD51. Total protein levels were also determined by stain-free imaging (Bio-Rad).
E) HCT116 cells were transfected with RAD51 siRNA, pulse labeled with CldU and IdU, and then treated with aphidicolin. DNA fiber analysis was performed to determine the lengths of CldU and IdU labeled DNA tracks in each condition.
F) HCT116 cells were transfected with control or RAD51 siRNA, pulse labeled with CldU and IdU, then treated with aphidicolin. DNA fiber analysis was performed to determine the lengths of the CldU and IdU labeled DNA tracks. n indicates the number of DNA fibers measured for each condition. P-values from a one-way ANOVA performed using Dunnett’s multiple comparisons test are reported for each condition. Non-significant comparisons are indicated as ‘ns’. Degradation still occurred even following knockdown of RAD51, consistent with results from U2OS cells in Fig. 6H.
G) Potential models for template switching, based on our new model for NSD (Fig. 7). (i) After encountering a polymerase-blocking lesion (yellow) replication forks uncouple, resulting in a stalled replisome (red). (ii) The replisome is retained on DNA and parental DNA strands anneal to create a second replication fork that is the substrate for fork reversal. This places the replisome within a single-stranded DNA bubble. (iii) The nascent lagging strands serve as template for leading strand synthesis (green). (iv) Exonuclease activity at the extruded DNA end generates a 3’ overhang. (v) restoration of the reversed fork results in a lagging strand gap that allows loading of a 5’-3’ single-stranded DNA helicase that can unwind the re-annealed parental strands. (vi) nascent strands are extended to the replisome, which resumes unwinding (green). (vii) Without exonuclease activity at the extruded DNA end (in (iv)) restoration of the reversed fork would result in a 3-way fully duplex fork structure that could not be readily unwound. (viii) An alternative model of the response replication fork uncoupling by a polymerase-blocking lesion. (ix) Nascent DNA strands are unwound by single-stranded DNA helicases, allowing them to anneal together. (x) The annealed duplex generated in (ix) is the substrate for fork reversal enzymes, which reanneal parental duplex as the nascent DNA strands are extruded. (xi) Exonuclease activity at the extruded end generates a 3’ overhang, as in (iv), but the bubble structure is more extensive and the parental duplex is more limited. (xii) Restoration of the reversed fork removes all parental duplex, provided that only one of the two strands remains intact. (xiii) nascent strands are extended to the replisome, which resumes unwinding.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant R35GM128696 to JMD, ACS grant IRG-15-169-56 to JMD, funding provided by the Breast Cancer Research Foundation to DC, and Vanderbilt-Ingram Cancer Center Support Grant P30CA068485. We thank Vincenzo Costanzo for Xenopus SAMHD1 and SMARCAL1 antibodies as well as for helpful discussion about the work. We thank Johannes Walter for Xenopus FAN1 antibody.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
DATA AVAILABILITY
All data supporting the findings of this study are available within the article and its Supplementary Information files. Source data showing unprocessed and uncropped gel and blot images are provided with this paper.
REFERENCES
- 1.Berti M, Cortez D & Lopes M The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat Rev Mol Cell Biol 21, 633–651, doi: 10.1038/s41580-020-0257-5 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Quinet A, Lemaçon D & Vindigni A Replication Fork Reversal: Players and Guardians. Mol Cell 68, 830–833, doi: 10.1016/j.molcel.2017.11.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rickman K & Smogorzewska A Advances in understanding DNA processing and protection at stalled replication forks. J Cell Biol 218, 1096–1107, doi: 10.1083/jcb.201809012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ait Saada A, Lambert SAE & Carr AM Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair (Amst) 71, 135–147, doi: 10.1016/j.dnarep.2018.08.017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mutreja K et al. ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Rep 24, 2629–2642.e2625, doi: 10.1016/j.celrep.2018.08.019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manosas M, Perumal SK, Croquette V & Benkovic SJ Direct observation of stalled fork restart via fork regression in the T4 replication system. Science 338, 1217–1220, doi: 10.1126/science.1225437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thangavel S et al. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol 208, 545–562, doi: 10.1083/jcb.201406100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlacher K et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542, doi: 10.1016/j.cell.2011.03.041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto Y, Ray Chaudhuri A, Lopes M & Costanzo V Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17, 1305–1311, doi: 10.1038/nsmb.1927 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dias MP, Moser SC, Ganesan S & Jonkers J Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat Rev Clin Oncol, doi: 10.1038/s41571-021-00532-x (2021). [DOI] [PubMed] [Google Scholar]
- 11.Taglialatela A et al. Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol Cell 68, 414–430.e418, doi: 10.1016/j.molcel.2017.09.036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray Chaudhuri A et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387, doi: 10.1038/nature18325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mijic S et al. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun 8, 859, doi: 10.1038/s41467-017-01164-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zellweger R et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol 208, 563–579, doi: 10.1083/jcb.201406099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fugger K et al. FBH1 Catalyzes Regression of Stalled Replication Forks. Cell Rep 10, 1749–1757, doi: 10.1016/j.celrep.2015.02.028 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Kolinjivadi AM et al. Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol Cell 67, 867–881.e867, doi: 10.1016/j.molcel.2017.07.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vujanovic M et al. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol Cell 67, 882–890.e885, doi: 10.1016/j.molcel.2017.08.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaçon D et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat Commun 8, 860, doi: 10.1038/s41467-017-01180-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu J et al. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149, 1221–1232, doi: 10.1016/j.cell.2012.04.030 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Mason JM, Chan YL, Weichselbaum RW & Bishop DK Non-enzymatic roles of human RAD51 at stalled replication forks. Nat Commun 10, 4410, doi: 10.1038/s41467-019-12297-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgs MR et al. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Mol Cell 59, 462–477, doi: 10.1016/j.molcel.2015.06.007 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Ray Chaudhuri A et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 19, 417–423, doi: 10.1038/nsmb.2258 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Berti M et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 20, 347–354, doi: 10.1038/nsmb.2501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amunugama R et al. Replication Fork Reversal during DNA Interstrand Crosslink Repair Requires CMG Unloading. Cell Rep 23, 3419–3428, doi: 10.1016/j.celrep.2018.05.061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iannascoli C, Palermo V, Murfuni I, Franchitto A & Pichierri P The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1-dependent degradation. Nucleic Acids Res 43, 9788–9803, doi: 10.1093/nar/gkv836 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallerga MB, Mansilla SF, Federico MB, Bertolin AP & Gottifredi V Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc Natl Acad Sci U S A 112, E6624–6633, doi: 10.1073/pnas.1508543112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timson J Hydroxyurea. Mutat Res 32, 115–132, doi: 10.1016/0165-1110(75)90002-0 (1975). [DOI] [PubMed] [Google Scholar]
- 28.Couch FB et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 27, 1610–1623, doi: 10.1101/gad.214080.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toledo LI et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103, doi: 10.1016/j.cell.2013.10.043 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Sparks JL et al. The CMG Helicase Bypasses DNA-Protein Cross-Links to Facilitate Their Repair. Cell 176, 167–181.e121, doi: 10.1016/j.cell.2018.10.053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham JE, Marians KJ & Kowalczykowski SC Independent and Stochastic Action of DNA Polymerases in the Replisome. Cell 169, 1201–1213.e1217, doi: 10.1016/j.cell.2017.05.041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickman KA et al. Distinct roles of BRCA2 in replication fork protection in response to hydroxyurea and DNA interstrand cross-links. Genes Dev 34, 832–846, doi: 10.1101/gad.336446.120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somyajit K et al. Homology-directed repair protects the replicating genome from metabolic assaults. Dev Cell 56, 461–477.e467, doi: 10.1016/j.devcel.2021.01.011 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Wasserman MR, Schauer GD, O'Donnell ME & Liu S Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase. Cell 178, 600–611.e616, doi: 10.1016/j.cell.2019.06.032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vrtis KB et al. Single-strand DNA breaks cause replisome disassembly. Mol Cell 81, 1309–1318.e1306, doi: 10.1016/j.molcel.2020.12.039 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dungrawala H et al. RADX Promotes Genome Stability and Modulates Chemosensitivity by Regulating RAD51 at Replication Forks. Mol Cell 67, 374–386.e375, doi: 10.1016/j.molcel.2017.06.023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Krishnamoorthy A, Zhao R & Cortez D Two replication fork remodeling pathways generate nuclease substrates for distinct fork protection factors. Sci Adv 6, doi: 10.1126/sciadv.abc3598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byun TS, Pacek M, Yee MC, Walter JC & Cimprich KA Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19, 1040–1052, doi: 10.1101/gad.1301205 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter J, Sun L & Newport J Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell 1, 519–529, doi: 10.1016/s1097-2765(00)80052-0 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Hanada K et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14, 1096–1104, doi: 10.1038/nsmb1313 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Petermann E, Orta ML, Issaeva N, Schultz N & Helleday T Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37, 492–502, doi: 10.1016/j.molcel.2010.01.021 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dewar JM, Budzowska M & Walter JC The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350, doi: 10.1038/nature14887 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W et al. A Selective Small Molecule DNA2 Inhibitor for Sensitization of Human Cancer Cells to Chemotherapy. EBioMedicine 6, 73–86, doi: 10.1016/j.ebiom.2016.02.043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupré A et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol 4, 119–125, doi: 10.1038/nchembio.63 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Z, Chung WH, Shim EY, Lee SE & Ira G Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994, doi: 10.1016/j.cell.2008.08.037 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaudhury I, Stroik DR & Sobeck A FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol Cell Biol 34, 3939–3954, doi: 10.1128/mcb.00457-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coquel F et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61, doi: 10.1038/s41586-018-0050-1 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Long DT & Kreuzer KN Regression supports two mechanisms of fork processing in phage T4. Proc Natl Acad Sci U S A 105, 6852–6857, doi: 10.1073/pnas.0711999105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunter N & Kleckner N The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70, doi: 10.1016/s0092-8674(01)00430-5 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Tian T et al. The ZATT-TOP2A-PICH Axis Drives Extensive Replication Fork Reversal to Promote Genome Stability. Mol Cell 81, 198–211.e196, doi: 10.1016/j.molcel.2020.11.007 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Walter J & Newport J Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell 5, 617–627, doi: 10.1016/s1097-2765(00)80241-5 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Deng L et al. Mitotic CDK Promotes Replisome Disassembly, Fork Breakage, and Complex DNA Rearrangements. Mol Cell 73, 915–929.e916, doi: 10.1016/j.molcel.2018.12.021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dewar JM, Low E, Mann M, Räschle M & Walter JC CRL2(Lrr1) promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev 31, 275–290, doi: 10.1101/gad.291799.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong RP, García-Rodríguez N, Zilio N, Hanulová M & Ulrich HD Processing of DNA Polymerase-Blocking Lesions during Genome Replication Is Spatially and Temporally Segregated from Replication Forks. Mol Cell 77, 3–16.e14, doi: 10.1016/j.molcel.2019.09.015 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Quinet A et al. PRIMPOL-Mediated Adaptive Response Suppresses Replication Fork Reversal in BRCA-Deficient Cells. Mol Cell 77, 461–474.e469, doi: 10.1016/j.molcel.2019.10.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cong K et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol Cell 81, 3128–3144.e3127, doi: 10.1016/j.molcel.2021.06.011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tirman S et al. Temporally distinct post-replicative repair mechanisms fill PRIMPOL-dependent ssDNA gaps in human cells. Mol Cell 81, 4026–4040.e4028, doi: 10.1016/j.molcel.2021.09.013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Low E, Chistol G, Zaher MS, Kochenova OV & Walter JC The DNA replication fork suppresses CMG unloading from chromatin before termination. Genes Dev 34, 1534–1545, doi: 10.1101/gad.339739.120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deegan TD, Mukherjee PP, Fujisawa R, Polo Rivera C & Labib K CMG helicase disassembly is controlled by replication fork DNA, replisome components and a ubiquitin threshold. Elife 9, doi: 10.7554/eLife.60371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kose HB, Xie S, Cameron G, Strycharska MS & Yardimci H Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase. Nat Commun 11, 3713, doi: 10.1038/s41467-020-17443-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masuda-Ozawa T, Hoang T, Seo YS, Chen LF & Spies M Single-molecule sorting reveals how ubiquitylation affects substrate recognition and activities of FBH1 helicase. Nucleic Acids Res 41, 3576–3587, doi: 10.1093/nar/gkt056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bétous R et al. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26, 151–162, doi: 10.1101/gad.178459.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biebricher A et al. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol Cell 51, 691–701, doi: 10.1016/j.molcel.2013.07.016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paudyal SC, Li S, Yan H, Hunter T & You Z Dna2 initiates resection at clean DNA double-strand breaks. Nucleic Acids Res 45, 11766–11781, doi: 10.1093/nar/gkx830 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nieminuszczy J et al. EXD2 Protects Stressed Replication Forks and Is Required for Cell Viability in the Absence of BRCA1/2. Mol Cell 75, 605–619.e606, doi: 10.1016/j.molcel.2019.05.026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J et al. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol 22, 242–247, doi: 10.1038/nsmb.2956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang J et al. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell 52, 434–446, doi: 10.1016/j.molcel.2013.09.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bai G et al. HLTF Promotes Fork Reversal, Limiting Replication Stress Resistance and Preventing Multiple Mechanisms of Unrestrained DNA Synthesis. Mol Cell 78, 1237–1251.e1237, doi: 10.1016/j.molcel.2020.04.031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods only references
- 69.Lebofsky R, Takahashi T & Walter JC DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol 521, 229–252, doi: 10.1007/978-1-60327-815-7_13 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Klein Douwel D et al. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54, 460–471, doi: 10.1016/j.molcel.2014.03.015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heintzman DR, Campos LV, Byl JAW, Osheroff N & Dewar JM Topoisomerase II Is Crucial for Fork Convergence during Vertebrate Replication Termination. Cell Rep 29, 422–436.e425, doi: 10.1016/j.celrep.2019.08.097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its Supplementary Information files. Source data showing unprocessed and uncropped gel and blot images are provided with this paper.