Summary
As a clinical vaccine, lipid nanoparticle (LNP) mRNA demonstrated potent and broad antibody responses, leading to speculation about its potential for antibody discovery. Here, we developed RAMIHM, a highly efficient strategy for developing fully human monoclonal antibodies that employs rapid mRNA immunization of humanized mice followed by single B cell sequencing (scBCR-seq). We immunized humanized transgenic mice with RAMIHM and generated 15 top-ranked clones from peripheral blood, plasma B, and memory B cell populations, demonstrating a high rate of antigen-specificity (93.3%). Two Omicron-specific neutralizing antibodies with high potent and one broad-spectrum neutralizing antibody were discovered. Furthermore, we extended the application of RAMIHM to cancer immunotherapy targets, including a single transmembrane protein CD22 and a multi-transmembrane GPCR target, GPRC5D, which is difficult for traditional protein immunization methods. RAMIHM-scBCR-seq is a broadly applicable platform for the rapid and efficient development of fully human monoclonal antibodies against an assortment of targets.
eTOC
Ren et al. develop a highly efficient strategy (RAMIHM) to generate fully human mAbs, and applied it to generate several potent and specific SARS-CoV-2 neutralizing monoclonal antibodies against six Omicron sublineages. RAMIHM also demonstrates applicability to cancer and immunology targets such as CD22 and GPRC5D.
Graphical Abstract
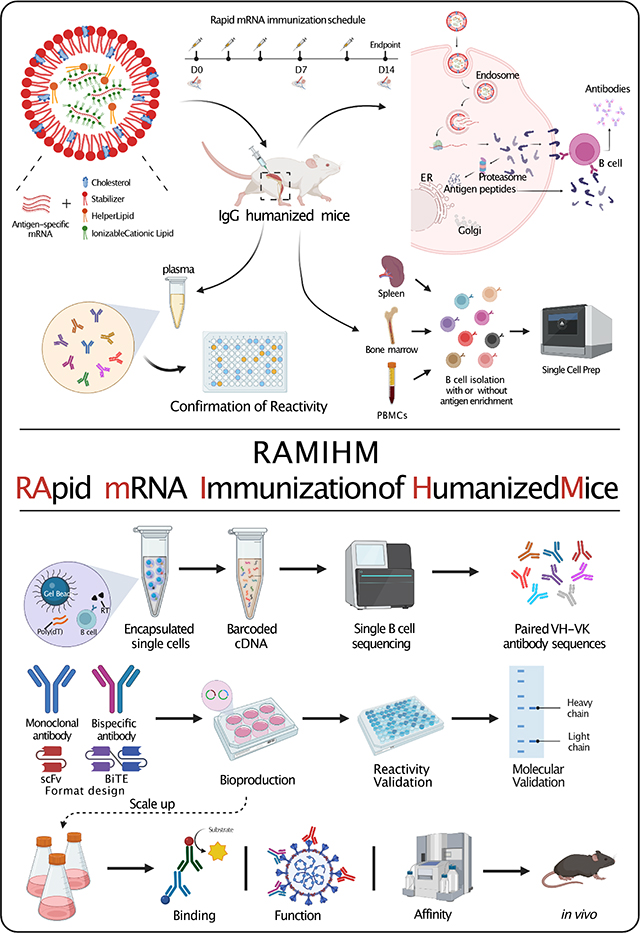
Introduction
Monoclonal antibodies (mAbs) are widely used as predominant therapeutics for diverse diseases, offering exquisite specificity and affinity for specific targets on both cell-surface and secreted proteins. Since the FDA approved the first clinical mAb in the 1970s, over 100 mAbs have been approved by the agency, with the majority targeting soluble proteins and single transmembrane proteins1. Although certain soluble/single-transmembrane proteins have been successfully used in antibody-based therapies, there are a very limited number of such proteins. Consequently, it is imperative to explore complex multi-transmembrane protein families, including ion channels and G protein-coupled receptors (GPCRs), which are among the most commonly exploited therapeutic targets, contributing over a third of the FDA approvals2–5. Significant features of approved GPCR-modulating drugs are nearly all small molecules or peptides, only two GPCR mAbs to date have been approved by the FDA, namely erenumab and mogamulizumab6,7.
Moreover, viral transmembrane proteins have become important anti-viral therapeutic targets since the emergence of COVID-19. The spike protein of COVID-19 pathogen SARS-CoV-2 displays high mutation rates, and each mutation can affect functional properties or alter infectivity. Several “variants of concern” (VOCs) of SARS-CoV-2 have emerged during the past three years and led to continuous waves of global pandemic due to spike mutations that enhance virus transmissibility, antigenicity, and immune evasion. The Omicron variant is one of the most recent SARS-CoV-2 variants, containing over 30 mutations and deletions in its spike protein, including 17 within the receptor binding domain (RBD) as compared to ancestral SARS-CoV-2 variants. A number of studies have proven that the early BA.1 sublineage of Omicron renders most previously authorized monoclonal antibodies ineffective8,9, causing them to be no longer recommended in the COVID-19 treatment guidelines for patients with Omicron infections. Despite widespread vaccinations, certain populations are still vulnerable to the diseases, and hospitalizations and deaths still occur daily due to a high rate of breakthrough infections and frequent reinfections in the high-risk populations. As a consequence, rapidly developing next-generation antibodies that retain potency against Omicron are required when the effectiveness of current vaccines and approved antibody therapies is threatened10.
In this study, we presented a platform for monoclonal antibody discovery and named RApid mRNA Immunization of Humanized Mice (RAMIHM), a highly efficient strategy that utilizes rapid mRNA immunization of humanized mice followed by single B cell sequencing (scBCR-seq) to allow efficient development of fully human monoclonal antibodies. The RAMIHM involves the use of high doses of antigen-specific LNP-mRNA vaccines to frequently immunize immunoglobulin (Ig) humanized mice in 2 weeks, allowing isolation of antigen-specific high-affinity monoclonal antibodies. Customized single cell BCR sequencing (scBCR-seq) is performed on these immunized mice to acquire the paired human variable region sequences from enriched B cell clonotypes, to allow immediate recombinant generation of potent and specific fully human antibodies. To rapidly demonstrate the RAMIHM as a proof-of-principle, we directly performed the RAMIHM with Omicron BA.1 spike-encapsulated LNP-mRNA vaccines 11. Combined with the use of customized single cell BCR sequencing (scBCR-seq) to obtain the human variable region sequences from Omicron BA.1 RBD enriched B cell clonotypes. To explore the rapid development of mAbs for cancer immunotherapy, we further employed RAMIHM on current clinical cancer targets. We generated human CD22 full-length-encapsulated LNP-mRNA and RAMIHM with scBCR-seq to obtain the human variable region sequences from human CD22-extracellular domain (ECD) enriched B cell clonotypes. To extend the applicability of RAMIHM on challenging cancer targets such as GPCRs 12, we applied RAMIHM on GPRC5D to identify the paired human variable region sequences from IgG isotype positive B cell clonotypes.
Results
Development of RApid mRNA Immunization of Humanized Mice (RAMIHM), a highly efficient strategy to identify fully human monoclonal antibodies
To date, a two-dose SARS-CoV-2 mRNA-based vaccination strategy has been demonstrated to effectively induce humoral and cellular immunity to SARS-CoV-2, including the ancestral virus (ancestral, reference, wildtype (WT), Wuhan-1, or WA-1, identical sequences), and its VoCs such as the Delta variant 13,14. However, several recent studies demonstrated that the Omicron BA.1 subvariant has substantial changes in its genome, especially the spike protein (Fig. S1A), and illustrated dramatically decreased neutralizing titers in convalescent or vaccinated recipients, causing waning immunity and massive breakthrough infections 15–18. Importantly, nearly all antibodies initially developed against the ancestral virus have substantially dropped, or completely lost, the neutralization ability against Omicron 8,19–21. Therefore, next-generation neutralizing antibodies are needed quickly. To combat the rapidly evolving VoCs, we thus developed an antibody discovery approach named RAMIHM, with repetitive intramuscular injections using high doses of LNP-mRNA, followed by B cell isolation, antigen enrichment, and single B cell sequencing (Fig. 1A). We applied this directly with Omicron BA.1-spike-encoding LNP-mRNA to induce Omicron-specific immune responses for isolation of Omicron BA.1-targeting mAbs.
Figure 1. Evaluation of Omicron BA.1-RAMIHM humanized mice, and rapid characterization of fully human Omicron BA.1-specific monoclonal antibodies.
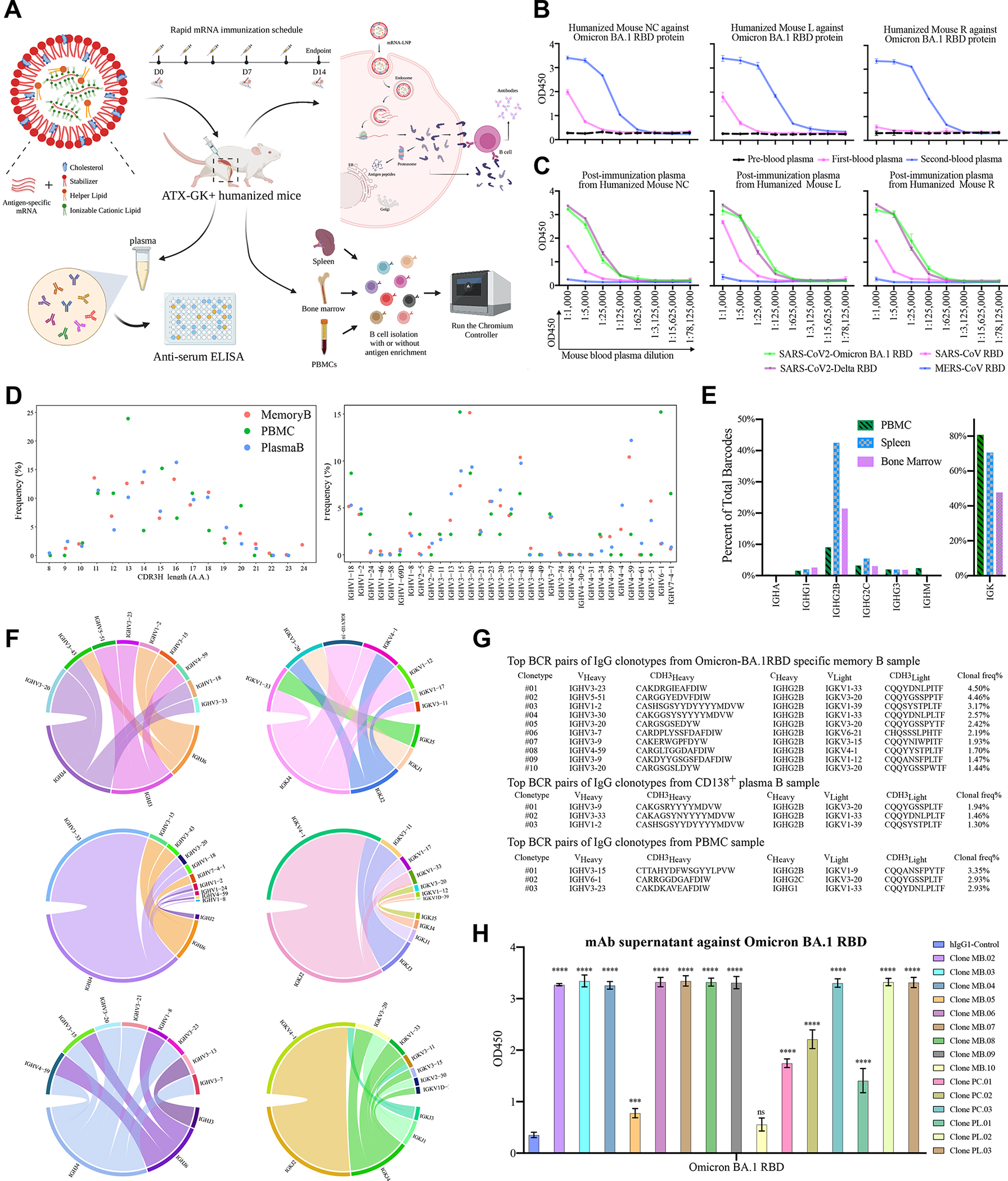
A, Schematic illustration of the humoral immune responses induced by RAMIHM and antibody discovery. Three humanized mice were repeatedly immunized with Omicron LNP-mRNA as an immunogen. 10 μg of Omicron LNP-mRNA were given to each mouse on day0, day2, day4, and day7, followed by 20 μg of Omicron LNP-mRNA injected on day11. Retro-orbital blood was collected on day0, day7, and day14. Plasma was isolated from the blood for downstream experiments. Spleen, bone marrow and PBMCs were collected for downstream antibody library preparation and antibody sequences were characterized by single B cell sequencing.
B, Antibody titer determination in plasma samples. The Omicron-BA.1RAMIHA immunized mice were defined as Mouse NC (no ear clipping), Mouse L (left ear clipping), and Mouse R (right ear clipping) based on ear clipping. All plasma samples were serially 5-fold diluted from 1:1000 and assayed by a direct coating ELISA with Omicron BA.1 RBD protein-coated plate. Error bars represent the mean ± SEM of triplicates with individual data points in plots.
C, Antibody cross-reactivity measurement. All post-immunized plasma samples (2nd blood) were serially 5-fold diluted from 1:1000 and assayed by a direct coating ELISA with a selected pan-CoV-RBD proteins coated plate. Error bars represent the mean ± SEM of triplicates with individual data points in plots.
D, B cell characterization by customized scBCR-seq profiling. Left panel, Distribution of heavy chain complementarity-determining region 3 (HCDR3) length in each B cell group (Memory B, Plasma B, and PBMC) from Omicron BA.1-RAMIHM mice. Right panel, distributions of heavy chain V-segment in each B cell group (Memory B, Plasma B, and PBMC) from Omicron BA.1-RAMIHM mice. Total number of single cells sequenced with BCRs (Memory B library, n = 2,646; Plasma B library, n = 617; PBMC library, n = 239; Total n = 3,502).
E, Ig class distributions of Omicron BA.1-RAMIHM mice’s clonotypes. Distribution and frequency analysis of immunoglobulin isotypes usage in the spleen, bone marrow, and PBMC from Omicron BA.1-RAMIHM mice.
F, Distribution of top10 heavy- and light-chain V/J segment recombination. Chord diagrams (circos plots) showing the distribution of top10 heavy- and light-chain V and J gene-segment recombination obtained in each representative library. Interconnecting lines indicate the relationship between antibodies that share V and J gene segments at both IGH and IGL. Top to bottom: Memory B library, PBMC library, and Plasma B library.
G, Single B cell variable chains for antibody cloning. Variable (V) genes and CDR3 lengths for paired heavy- and light-chains of top-enriched clones to SARS-CoV-2 Omicron BA.1 from single BCR sequencing.
H, ELISA of mAbs supernatant binding specificity against Omicron BA.1 RBD protein. All full-length mAb clones from single BCR sequencing and control were evaluated against Omicron BA.1 RBD protein coated on the ELISA plate and binding activity was recorded at an optical density (OD) of 450 nm. Triplicate data points (n = 3 each).
In this figure:
Data are shown as mean ± SEM plus individual data points in dot plots.
Statistics: One-way ANOVA was used to assess statistical significance. Each mAb clone was compared to the control. Multiple testing correction was made to correct the p values. Two-sided tests were performed. The p-values are indicated in the plots. Statistical significance labels: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. Non-significant comparisons are not shown unless otherwise noted as n.s., not significant.
Source data and additional statistics for experiments are in supplemental excel file(s).
See also Figures S1–S3, S7
Using Omicron BA.1-specific LNP-mRNAs that contain lipid nanoparticle formulated mRNA encoding the HexaPro engineered full length of Omicron BA.1 spike glycoprotein (Methods), we first characterized the biophysical integrity of these LNP-mRNAs (Fig. S1B, S1C), and validated the expression of functional Omicron BA.1 spike protein surface expression via human ACE2 (hACE2) staining of LNP-mRNA transfected HEK293 cells (Fig. S1D). Next, we performed the administration of four 10 μg doses and one 20 μg dose of Omicron BA.1 specific-mRNA LNP in 3 IgG-humanized mice and collected retro-orbital blood samples from each humanized mouse before and after booster immunization. Blood samples were labeled as pre-, 1st-, or 2nd immunization draw depending on the collection sequence (Fig. 1B). Antibody titers were measured using serial plasma dilutions on ELISA plates coated with recombinant Omicron BA.1 RBD protein. Binding activity was visualized using anti-mouse IgG antibodies at 450 nm optical density (OD). Three sequential plasma samples showed increasing vaccine-elicited antibody responses during each blood collection (Fig. 1B, S1D). All post-immunized plasma samples (2nd blood) exhibited significant reactivity to the recombinant SARS-CoV-2 Omicron BA.1 RBD protein (Fig. 1C). In addition, all these samples also showed strong cross-reactivity to recombinant SARS-CoV-2 Delta RBD protein, and intermediately cross-reactivity to recombinant SARS-CoV RBD protein, but no cross-binding to recombinant MERS-CoV RBD protein (Fig. 1C). Together, these results demonstrated Omicron BA.1-specific rapid mRNA immunization (Omicron BA.1-RAMIHM) elicited strong anti-Omicron plasma in all IgG humanized mice within 2 weeks, which also contains broader reactive antibodies against other variant and coronavirus species such as SARS-CoV-2 Delta and SARS-CoV.
Customized single-cell BCR sequencing (scBCRseq) mapped the IgG clonal repertoires of Omicron BA.1-RAMIHM animals
To obtain Omicron BA.1 RBD-specific B cells, we collected them from spleen, lymph nodes, bone marrow, and whole blood from an Omicron BA.1-RAMIHM mouse, and collected three different types of B cells (memory B cells, plasma B cells, and peripheral blood mononuclear cells) by using different isolation procedures (Methods), for B cell repertoire mapping and reactive BCR identification via scBCR-seq. To prepare the memory B cells enriched library, we enriched memory B cells by immunomagnetic negative selection and used 25 μg of recombinant Omicron BA.1 RBD proteins as bait to obtain Omicron BA.1 RBD specific-memory B cells from pre-enriched memory B cells (Memory B library). To generate a plasma B cell enriched library, we applied an anti-mouse CD138+ plasma cell isolation kit to isolate CD138+ plasma B cells from freshly isolated raw bone marrow cells (Plasma B library). To generate a peripheral blood mononuclear cells library, we isolated peripheral blood mononuclear cells (PBMCs) from whole blood by centrifugation following the PBMC isolation method (PBMC/Peripheral B library). Subsequently, we counted and subjected 10,000 fresh cells from above each B cell for single-cell BCR library preparation and sequencing, respectively. After sequencing, we analyzed 3,502 Omicron BA.1 RBD-specific B cells, and eventually obtained 2,558 paired antibody sequences. To further decipher Omicron BA.1 RAMIHM-elicited B cell clonal expansion on a single-cell level, we first characterized the dominant B cell clonotypes based on the number of cells observed and frequencies of identical CDR3 region for both heavy and light chains in pairs. By analyzing the BCR repertoires, we mapped the landscape of BCR populations in Memory B, Plasma B and Peripheral B/PBMC from the Omicron BA.1-RAMIHM immunized mouse (Fig. S2, Dataset S1, Supplemental Codes).
The Omicron BA.1 RBD-specific antibodies had a relative enrichment for IGVH3-7, IGVH3-15, IGVH3-20, IGVH3-23, IGVH3-30, IGVH3-33, IGHV3-43, and IGVH4-59, analyzed from 3 individual BCR libraries (Fig. 1D, Dataset S1). A range of lengths between 8–24 aa was observed for these BCR CDRH3s (Fig. 1D, Dataset S1). Interestingly, a large portion of IgG2B-expressing B cells was identified from three B cell type isolations (Fig. 1E), a signature of potential involvement of Th2 cells in B cells maturation and class switch in these mice undergoing the Omicron BA.1-RAMIHM procedure. With the analysis of heavy chain (IGH) and light chain (IGK) parings, we also mapped out the overall (Fig. S3), enriched and top 10 heavy-and light-chain V/J segment recombination patterns in these B cell populations (Fig. 1F, Dataset S1). In summary, scBCRseq data mapped the clonal repertoires and revealed enriched IgG clonotypes in the peripheral blood, plasma B cell, and memory B cell populations from the Omicron BA.1-RAMIHM humanized mouse.
Identification of Omicron-specific functional mAb clones from top-ranked paired human Ig chains of Omicron BA.1-RAMIHM animals
To test whether the most enriched BCRs in these B cell populations are Omicron BA.1-reactive, we selected a panel of BCRs for recombinant mAb expression, including 3 top clones from peripheral blood, 3 top ones from plasma B cell populations, and 10 top clones from memory B cell populations (Fig. 1G). To functionally analyze the antibody response to SARS-CoV-2 Omicron BA.1 RBD, we cloned paired heavy- and light-variable segments into human IgG1 expression vectors, and used the Expi293F mammalian expression system to produce selected mAbs. Thereafter, we used SARS-CoV-2 Omicron BA.1 RBD-specific ELISA to determine antibody binding by using transfected culture supernatants that contain secreted antibodies (Fig. 2A). As a result, almost all (14/15 reactive) of the top-enriched antibody clones collected from peripheral blood, plasma B cell, and memory B cell populations are recognized by recombinant Omicron BA.1 RBD proteins. Out of those positive mAbs, ten of the selected clones showed potent binding affinity (Fig. 1H, Dataset S2). These findings indicated that RAMIHM is a highly effective approach for generating and isolating antigen-specific mAbs.
Figure 2. Biophysical and functional characterization of lead clones of Omicron BA.1-specific antibodies.
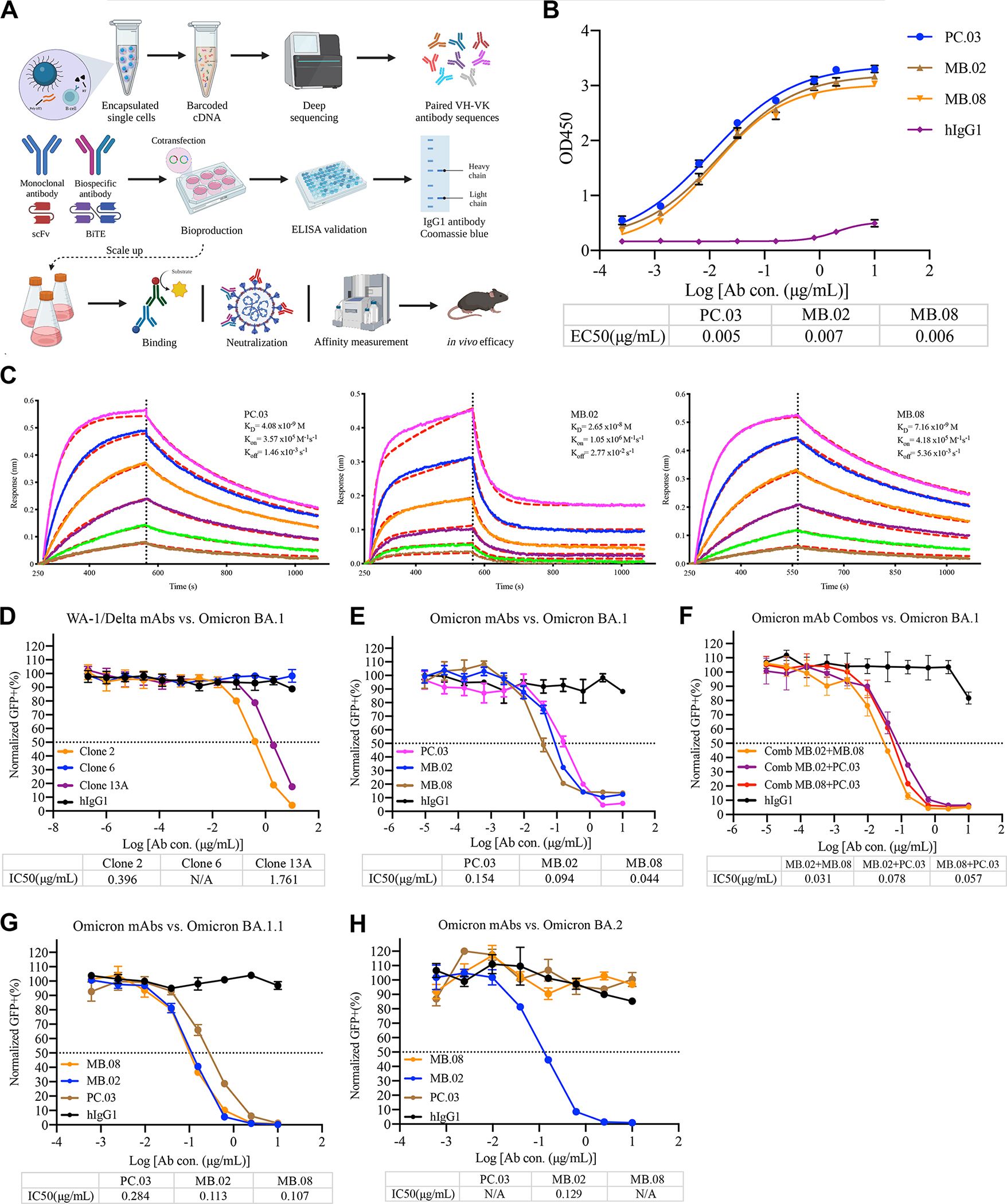
A, Schematic illustration of human IgG1 mAb reconstruction, production, and validation.
B, Graph shows leading Omicron mAbs reactivity. The ELISA EC50 values were calculated by Prism V8.0 software using a four-parameter logistic curve fitting approach. Error bars represent the mean ± SEM of triplicates with individual data points in plots.
C, Binding characteristics of the neutralizing mAbs determined by using BLI. Purified mAbs were immobilized onto an AHC sensor, and all measurements were performed by using a serial 2-fold dilution of soluble SARS-CoV-2 Omicron BA.1 RBD, starting from 50 nM (Magenta) to 1.56 nM (Brown). Global fit curves are shown as red dashed lines. The vertical black dotted dashed lines indicate the transition between the association and disassociation phases.
D, Neutralization assay of SARS-CoV-2 Omicron BA.1 pseudovirus by WA-1/Delta mAbs. The graph shows the normalized relative GFP signals for detection of HEK293T cells expressing hACE2, 24 h after infection with SARS-CoV-2 Omicron BA.1 pseudovirus, in the presence of an increasing concentration of indicated WA-1/Delta mAbs.
E, Neutralization assay of SARS-CoV-2 Omicron BA.1 pseudovirus by leading Omicron mAbs. The graph shows the normalized relative GFP signals for detection of HEK293T cells expressing hACE2, 24 h after infection with SARS-CoV-2 Omicron BA.1 pseudovirus, in the presence of an increasing concentration of indicated Omicron mAbs.
F, Neutralization assay of SARS-CoV-2 Omicron BA.1 pseudovirus by leading Omicron mAb combinations. The graph shows the normalized relative GFP signals for detection of HEK293T cells expressing hACE2, 24 h after infection with SARS-CoV-2 Omicron BA.1 pseudovirus, in the presence of an increasing concentration of indicated Omicron mAb combinations (MB.02+MB.08, MB.08+PC.03, MB.02+PC.03).
G, Neutralization assay of SARS-CoV-2 Omicron BA.1.1 pseudovirus by leading Omicron mAbs. The graph shows the normalized relative GFP signals for detection of HEK293T cells expressing hACE2, 24 h after infection with SARS-CoV-2 Omicron BA.1.1 pseudovirus, in the presence of an increasing concentration of indicated Omicron mAbs.
H, Neutralization assay of SARS-CoV-2 Omicron BA.2 pseudovirus by leading Omicron mAbs. The graph shows the normalized relative GFP signals for detection of HEK293T cells expressing hACE2, 24 h after infection with SARS-CoV-2 Omicron BA.2 pseudovirus, in the presence of an increasing concentration of indicated Omicron mAbs.
The IC50 values were calculated by Prism V8.0 software using a four-parameter logistic curve fitting approach. The dashed line indicated a 50% reduction in viral infectivity.
Error bars represent the mean ± SEM of triplicates with individual data points in plots.
Source data and additional statistics for experiments are in supplemental excel file(s).
See also Figures S1–S4
To further characterize highly potent functional mAbs, we recombinantly expressed these 15 mAb candidate clones in a mammalian system and tested their neutralization ability against the Omicron BA.1 subvariant. By screening the mAbs from culture supernatants by neutralizing assay using a spike-based SARS-CoV-2 Omicron pseudovirus system, we found 3 clones with obvious neutralization activity against Omicron BA.1 pseudovirus (Fig. S4A–B). We chose these top 3 clones (named PC.03, MB.02, and MB.08) for further development and characterization.
Characterization of fully human lead clones with strong binding to Omicron BA.1 RBD
We then purified the three leading antibody clones, PC.03, MB.02, and MB.08, by affinity chromatography using Protein A beads and examined purity by SDS-PAGE (Fig. S1E). Thereafter, purified antibodies were tested for SARS-CoV-2 Omicron BA.1 RBD reactivity by ELISA and monitored real-time association and dissociation to recombinant SARS-CoV-2 Omicron BA.1 RBD proteins using the Octet system. The ELISA titration result of the lead mAb clones vs. recombinant SARS-CoV-2 Omicron BA.1 RBD proteins showed that these three mAb clones have EC50s at the level of ~0.01 μg/mL, suggesting that these mAbs can indeed tightly bind to Omicron BA.1 RBD (EC50< 8 ng/mL for all 3 clones) (Fig. 2B). Octet results with his-tag Omicron BA.1 RBD antigen immobilization displayed ultra-strong binding (KD at 0.8 nM for MB.02, and KD <1 pM for PC.03 and MB.08) (Fig. S1F). The avidity effect due to multivalent binding may have contributed to the results. Therefore, we performed the reverse Octet assay with antibody immobilization to measure the single-mAb binding affinity, and we found the affinity between these clones to Omicron BA.1 is within the low nanomolar range (Fig. 2C). These KD values showed that the binding strengths of the 3 lead mAbs are stronger than that of hACE2 with Omicron BA.1 RBD (31.4±11.62 nM) 22. Noted that most of the approved or EUA mAbs have much weaker binding with Omicron BA.1 RBD 23,24 (summarized in Table. S2). To further determine whether these leading mAbs compete for similar epitopes, we performed epitope binning experiments by Octet using an in-tandem assay. The results have revealed that PC.03, MB.02, and MB.08 likely share overlapping epitopes (Fig. S4C).
Further characterization of fully human lead neutralization mAb clones against other circulating Omicron subvariants
We then performed neutralization assays for the 3 lead mAbs in purified form, along with other mAbs. We previously identified and developed several potent and specific mAbs against the ancestral virus and the Delta variant, namely clone 2, clone 6, and clone 13A25. In a pseudovirus neutralization assay, we found that while clone2 and clone13A can still neutralize the Omicron BA.1 subvariant, the potency is significantly reduced (by 1–2 orders of magnitude in terms of IC50 values, at 0.396 and 1.761 μg/mL for clone2 and clone13A, respectively) (Fig. 2D), a phenomenon similar to other mAbs developed against the ancestral spike 19,20. In contrast, all three leading clones, PC.03, MB.02, and MB.08, potently neutralized the Omicron BA.1 subvariant, with IC50 values at 0.154 μg/mL (PC.03), 0.094 μg/mL (MB.02), and 0.044 μg/mL (MB.08) (Fig. 2E, Fig. S4D). The neutralization potency of the 3 lead Omicron BA.1-specific mAb clones is much stronger than those of our prior mAbs and those under prior regulatory approval or EUAs (Table. S2). These 3 mAbs however showed no neutralization against the Delta variant (Fig. S4E), further suggesting that they are Omicron BA.1-specific.
To test if these clones can be used in combination, we performed neutralization assays by combining two clones. Interestingly, despite epitope overlap, these mAb clones can still slightly improve each other’s neutralization capacity, with the most effective combination being an antibody cocktail of MB.02 + MB.08 (IC50 = 0.031 μg/mL) against pseudotyped SARS-CoV-2 Omicron variant (Fig. 2F). Meanwhile, to test if these Omicron mAbs can be targeted on other Omicron sublineages (BA.1.1, BA.2, BA.2.12.1, BA.3, and BA.5), we conducted a pseudovirus neutralization assay. The findings suggested that all three leading clones can maintain potency to Omicron BA.1.1, with IC50 values at 0.284 μg/mL (PC.03), 0.113 μg/mL (MB.02), and 0.107 μg/mL (MB.08) (Fig. 2G). However, two (PC.03 and MB.08) of the top clones showed no neutralization to Omicron BA.2, and only MB.02 retains potency to Omicron BA.2, with IC50 values at 0.129 μg/mL (PC.03) (Fig. 2H). Moreover, MB.02 also maintains potency against current circulating Omicron subvariants, with IC50 values at 0.433 μg/mL (BA.2.12.1), 0.131 μg/mL (BA.3), and 0.395 μg/mL (BA.5). In summary, these potent neutralizing mAbs showed that they have high affinity vs Omicron BA.1 RBD, and strong potency in pseudovirus neutralization, which are at least 2 orders of magnitude more potent than existing clinically approved or authorized SARS-CoV-2 mAbs. The findings revealed that the leading clone (MB.02) had the capacity to neutralize all tested pseudoviruses of Omicron sublineages.
Application of RAMIHM on CD22
To better understand the breadth of the RAMIHM, we next focused on cancer surface antigens and selected CD22 as a target. CD22, a single-pass transmembrane protein (Fig. 3A), is a well-known therapeutic target for the treatment of B cell malignancies and systemic autoimmune diseases 26–28. To rapidly obtain fully human paired variable sequences of CD22-specific antibodies, we firstly designed CD22-specific LNP-mRNAs. The open reading frame of CD22 was flanked by a 5’untranslated region (UTR), 3’UTR, and polyA tail. Then, we encapsulated the transcribed CD22 mRNA into lipid nanoparticles to generate CD22 LNP-mRNAs and proceeded to characterize the immunogenicity of CD22 LNP-mRNA in vivo. Four 10 μg doses and one 20 μg dose of CD22 specific mRNA-LNPs were administered to 2 IgG-humanized mice with retro-orbital blood samples collected from each humanized mouse before and after booster immunization. Blood samples were labeled as pre-, 1st-, or 2nd immunization draws depending on the collection sequence. Antibody titers were measured using a cell staining assay with 1:200 plasma dilution, and binding activity was determined using flow cytometry. Three sequential plasma samples showed increasing LNP-mRNA-elicited antibody responses during each blood collection (Fig. 3B).
Figure 3. Evaluation of CD22-RAMIHM humanized mice and rapidly identified fully human CD22-specific monoclonal antibodies.
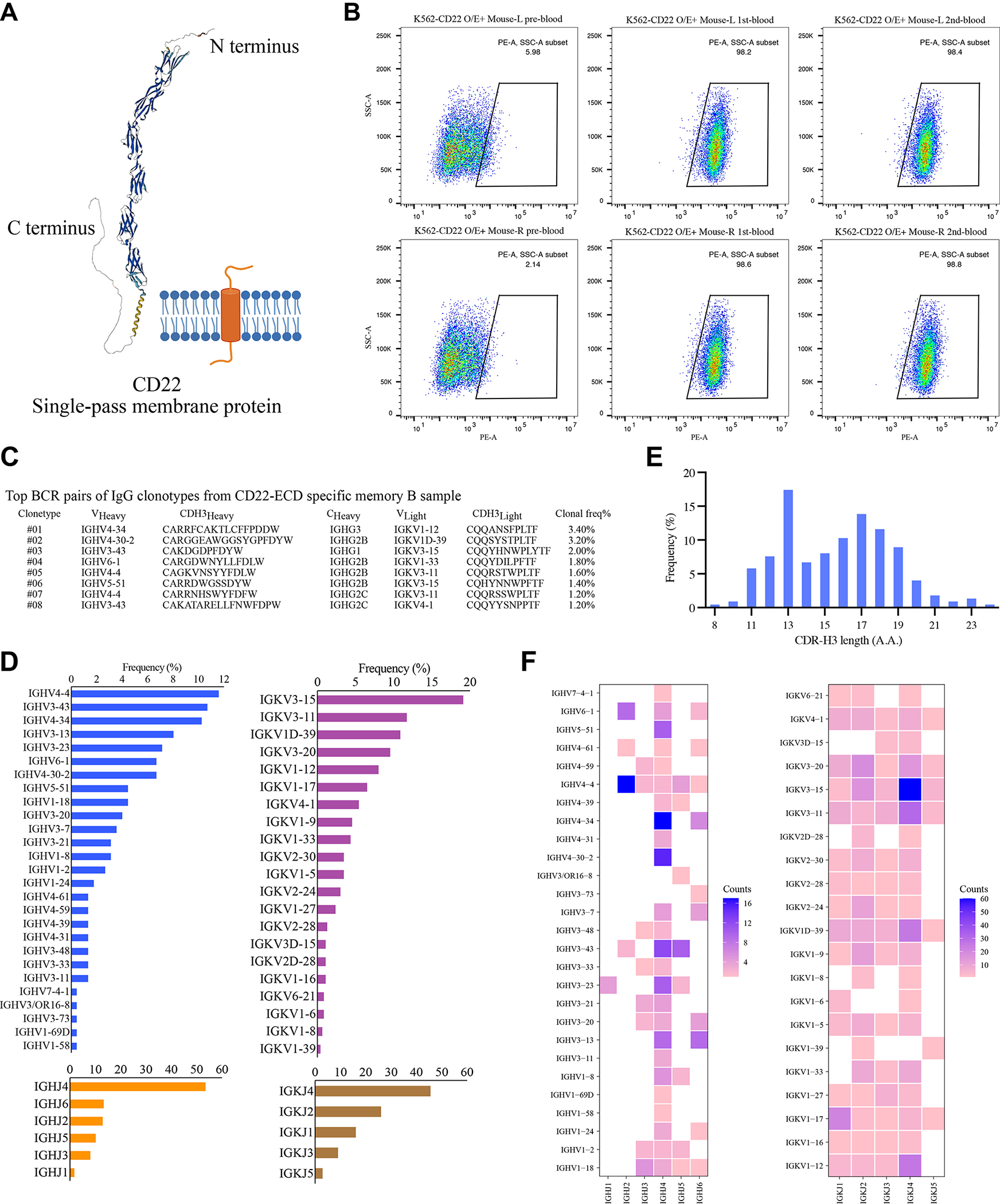
A, The structure of human CD22 protein predicted (UniProt #060926) by AlphaFold Protein Structure Database.
B, CD22 antibody titer measurement in plasma samples. All plasma samples were diluted into 1:200, then incubated with K562-CD22 O/E cells and detected by flow cytometry. Data from two CD22-RAMIHM mice were shown.
C, Single B cell variable chains for antibody cloning. Variable (V) genes and CDR3 lengths for paired heavy- and light-chains of top-enriched clones to CD22 from single BCR sequencing.
D, Distribution of heavy chain complementarity-determining region 3 (HCDR3) length in memory B cells from CD22-RAMIHM mouse.
F, Global frequencies of IGHV, IGHJ, IGKV, and IGKJ genes usage in CD22-RAMIHM mouse.
G, Numbers, and diversifications of IGHV/IGHJ, IGKV/IGKJ gene combination usage in CD22-RAMIHM mouse.
See also Figures S5, S7
To acquire CD22-ECD binding B cells, we collected spleen and lymph nodes from a CD22-RAMIHM mouse, memory B cells were enriched by immunomagnetic negative selection, and 25 μg of recombinant CD22-ECD proteins were used as bait to isolate CD22-ECD binding memory B cells from pre-enriched memory B cells. Subsequently, 10,000 positive memory B cells were counted and subjected to single-cell BCR library preparation and sequencing. The clonotype enrichment was calculated based on the number of cells observed with identical pairs of heavy and light chains by analyzing the BCR repertoires. We mapped the landscape of BCR populations in CD22-RAMIHM mice (Fig. 3C). Meanwhile, the scBCR-seq data allowed us to further decipher potential germline genes associated with the response to CD22 LNP-mRNA immunization. We found that IGHV4-4, IGHV3-43, and IGHV4-34 were the top 3 relative enriched IGHV genes, and IGKV3-15 was the top1 enriched IGKV gene in CD22-RAMIHM mouse (Fig. 3D, Dataset S1). In particular, we noticed that the IGHJ4 germline gene is more frequently used than the other five IGHJ genes (Fig. 3D, Dataset S1), and a range of HCDR3 lengths between 8–24 aa was observed (Fig. 3E, Dataset S1). Furthermore, to provide a more detailed landscape of germline gene expression, we determined the IGHV/IGHJ and IGKV/IGKJ combination usage. The IGHV/IGHJ and IGKV/IGKJ combination usage showed that IGHV4-4/IGHJ2, IGHV4-34/IGHJ4, IGHV4-30-2/IGHJ4, and IGKV3-15/IGKJ4 had greater expression in CD22-RAMIHM mouse (Fig. 3F, Dataset S1). These data demonstrated that RAMIHM can be rapidly applied to cancer immunotherapy targets such as CD22 without the need for recombinant protein antigen expression.
Application of RAMIHM on GPRC5D
GPRCD is an orphan G protein-coupled receptor, which has been identified as an intriguing clinical target for immunotherapy of multiple myeloma in BCMA antigen-escaped models 29. It remains, however, technically challenging to obtain the natural conformation of GPRC5D proteins. Only a short N terminus and three small extracellular loops (ECLs) (Fig. 4A) could be used as immunogens to generate peptide-specific antibodies. Furthermore, GPRC5D-overexpressing cells as immunogens combined with phage display screening have been reported to identify scFvs for CAR-T application 30, which is limited by high-quality and well-validated phage display libraries. To demonstrate the applicability of RAMIHM in GPCR antibody development, we rapidly designed a GPRC5D-specific mRNA, where the open reading frame of GPRC5D flanked by 5’untranslated region (UTR), 3’UTR, and polyA tail. Then, we encapsulated the transcribed GPRC5D mRNA into LNP to generate GPRC5D LNP-mRNA and proceeded to characterize the immunogenicity in vivo. Four 10 μg doses and one 20 μg dose of GPRC5D specific mRNA-LNPs were administered to 2 IgG-humanized mice. Retro-orbital blood samples were collected from each humanized mouse before and after booster immunization. Blood samples were labeled as pre-, 1st-, or 2nd immunization draw depending on the collection sequence. Antibody titers were measured using a cell staining assay with 1:200 plasma dilution, and binding activity was determined using flow cytometry. Three sequential plasma samples showed increasing vaccine-elicited antibody responses during each blood collection (Fig. 4B).
Figure 4. Evaluation of GPRC5D-RAMIHM humanized mice and rapidly identified fully human GPRC5D-specific monoclonal antibodies.
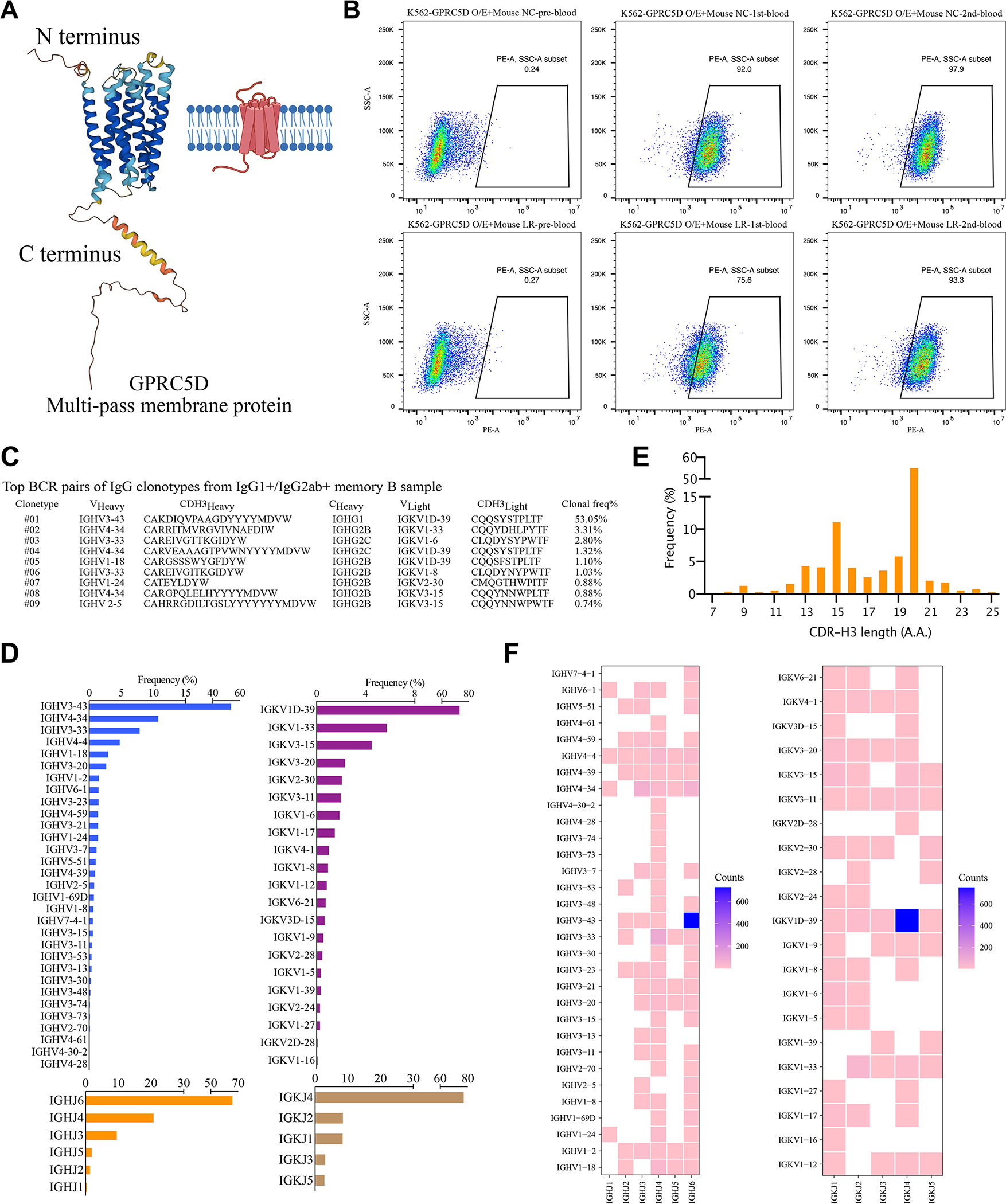
A, The structure of human GPRC5D protein predicted (UniProt #Q9NZD1) by AlphaFold Protein Structure Database.
B, GPRC5D antibody titer measurement in plasma samples. All plasma samples were diluted into 1:200, then incubated with K562-GPRC5D O/E cells and detected by flow cytometry. Data from two CD22-RAMIHM mice were shown.
C, Single B cell variable chains for antibody cloning. Variable (V) genes and CDR3 lengths for paired heavy- and light-chains of top-enriched clones to GPRC5D from single BCR sequencing.
D, Distribution of heavy chain complementarity-determining region 3 (HCDR3) length in memory B cells from GPRC5D-RAMIHM mouse.
F, Global frequencies of IGHV, IGHJ, IGKV, and IGKJ genes usage in GPRC5D-RAMIHM mouse.
G, Numbers, and diversifications of IGHV/IGHJ, IGKV/IGKJ gene combination usage in GPRC5D-RAMIHM mouse.
See also Figures S5–S7
To acquire GPRC5D-specific B cells, we collected spleen and lymph nodes from a GPRC5D-RAMIHM mouse. Memory B cells were enriched by immunomagnetic negative selection and stained with mouse IgG1 and IgG2ab antibodies to isolate IgG isotype positive memory B cells from pre-enriched memory B cells. Subsequently, 10,000 positive memory B cells were counted and subjected to single-cell BCR library preparation and sequencing. The clonotype enrichment was calculated based on the number of cells observed with identical pairs of heavy- and light chains. By analyzing the BCR repertoires, we mapped the landscape of BCR populations in the GPRC5D-RAMIHM mouse (Fig. 4C, Dataset S1). Meanwhile, the scBCR-seq data allowed us to further decipher potential germline genes associated with the response to GPRC5D LNP-mRNAs immunization. Notably, one dominant IgG1 clone was observed. IGHV3-43 and IGKV1D-39 genes had highest frequencies in the GPRC5D-RAMIHM mouse (Fig. 4D, Dataset S1). Furthermore, 24 of 31 IGHV genes were paired with IGHJ6 (Fig. 4F, Dataset S1), suggest the IGHJ6 gene was highly engaged by GPRC5D-RAMIHM immunization. Meanwhile, the IGHJ6 gene is usually longer (has 3–5 more amino acids) than the other five IGHJ genes 31, resulting in a long HCDR3 length in GPRC5D-RAMIHM mouse compared with the above CD22-RAMIHM mouse (Fig. 4E, Dataset S1). These data demonstrated that RAMIHM can be rapidly applied to multi-transmembrane targets such as GPRC5D without the need for recombinant protein antigen expression.
To validate binding activity of those top enriched IgG1/IgG2ab+ clonotypes, we again cloned paired heavy- and light-variable segments into human IgG1 expression vectors, and used the Expi293F mammalian expression system to produce individual clones of mAbs. We used cell staining assay to determine antibody binding by using transfected culture supernatants that contain secreted antibodies. As a result, almost all of the top enriched IgG1/IgG2ab+ antibody clones (8/9, 90%) recognize GPRC5D-overexpressed K562 cells, with varying levels of cells stained positive (Figure S6), potentially due to different binding affinities. Out of these positive mAbs, two clones (clone 2 and clone 8) showed the most potent binding affinity (Figure S6). These data again validated that RAMIHM is a highly effective approach for rapid development of target-specific human mAbs against challenging targets.
Evaluation of background BCR repertoire
To further evaluate technical features of RAMAHM, we characterized the baseline of B cell clonal levels in non-RAMAHM-immunized humanized mouse. We collected spleen, bone marrow, and whole blood from a non-RAMAHM-immunized humanized mouse, followed by previously established processes to isolate desired B cell subsets (memory B, plasma B, PBMC) for comparison. Then, 10,000 fresh cells from each B cell subset were subjected to single-cell BCR library construction and sequencing. However, the library construction reaction can not amplify the IgGH and IgK repertoires from the cDNA of the non-immunized humanized mouse’s PBMC library, indicating that there were rare or undetectable IgG+ B cells in the blood if the mouse is not immunized. Meanwhile, the different density of PCR bands was amplified from different B cell libraries, indirectly reflecting the natural baseline levels of IgG+ B cells in different organs in non-immunized mouse (Fig. S7A, Dataset S1). Furthermore, though scBCRseq data, we analyzed and compared the natural frequencies of IGHV, IGHJ, IGKV, and IGKJ gene usage between the memory B library and plasma B library. The results revealed that there is no strong correlation between the two B cell libraries, and no significant pre-enriched B cell clonotypes were observed in non-immunized humanized mouse (Fig. S7C–D). IGHJ4 and IGKJ4 genes were preferentially engaged in non-immunized humanized mouse, which led to a similar distribution and scope of HCDR3 length between the two samples (Fig. S7B). Subsequently, to further investigate the patterns of antibody isotype distribution between non-RAMAHM mouse and Omicron BA.1 RAMAHM mouse. We compared the two sets of scBCRseq data and found IgG2B was abundantly expressed in response to Omicron BA.1 RAMAHM, where nearly ~40% of IgG2B clonotypes were elevated in Omicron BA.1-enriched memory B library, and ~20% ones were elevated in CD138-enriched plasma B library, indicating the naïve B cells underwent isotype switching during Omicron BA.1 RAMAHM induction (Fig. S7E). Additionally, the IGHV/IGHJ combination usage in Omicron BA.1 RAMAHM mouse showed significantly different patterns from those of non-immunized humanized mouse (Fig. S7F). A total of 63 distinct IGHV/IGHJ combinations were discovered in the Omicron-BA.1-enriched memory B library and 22 distinct ones were observed in the CD138-enriched plasma B library, reflecting increased clonal expansion and diversification of B cells against Omicron BA.1 during RAMAHM induction (Fig. S7F).
Discussion
GPCRs are multispan transmembrane proteins with important biological and pathological roles. Therefore, it is hard to isolate the full length of GPCRs with native conformations using current protein purification methods. In addition, the N-terminus and small extracellular loops (ECLs) of GPCRs are the most commonly materials that used as immunogens for regular anti-GPCRs antibody generation. However, employing those disordered truncated peptides in current antibody discovery platforms (such as mouse immunization and in vitro display techniques) often leads to generating peptide-specific antibodies that are unable to recognize the native form of the target proteins. The mRNA-based vaccines against SARS-CoV-2 can successfully elicit broad and potent antibody responses when administered intramuscularly, indicating that LNP-encapsulated mRNA can be used to achieve protein expression and thereby antigen-specific immunization in vivo. This method could overcome a major obstacle in developing mAbs against challenging targets like multi-transmembrane proteins.
To provide a simple and better platform for antibody discovery, we developed a highly effective animal immunization approach (RAMIHM) combined with high-throughput customized single-cell BCR sequencing. In contrast with the current established antibody discovery platform, RAMIHM enables us to isolate antigen-specific antibodies within 2 weeks, offering the opportunity to rapidly respond to the potential risks of emerging infectious viruses or variants. Meanwhile, RAMIHM does not need human samples and is fully controllable in the laboratory. Most importantly, RAMIHM is faster than conventional protein-based immunization methods and highly maintains the target protein in a natural conformation, thereby enhancing immunogenicity and antigenicity and generating antibodies with in vivo affinity maturation process (Table. S1). Overall, the mAbs developed by RAMIHM are fully human and compatible with downstream drug development and/or translational studies.
To quickly provide countermeasures to upcoming VoCs such as the Omicron variant. We rapidly identified 15 anti-Omicron BA.1 mAbs from Ig humanized mice by RAMIHM within 2 weeks. Among those mAbs, three of them were further validated as potent neutralizing mAbs. MB.08 showed the highest binding capacity (KD = 7 nM) and strongest neutralizing ability against pseudotyped viruses of SARS-CoV-2 Omicron BA.1 (IC50 = 44 ng/mL), and MB.02 exhibited a broad-spectrum neutralizing capacity to adequately cover all major sublineages of the Omicron variant (BA.1, BA.1.1, BA.2, BA.2.12.1, BA.3, BA.5). Meanwhile, an antibody cocktail combining MB.08 with MB.02 exhibited enhanced SARS-CoV-2 Omicron neutralization potency (IC50 = 30 ng/mL) compared to individual clones, although the epitope binning experiment results suggested that MB.08 might bind to sites in Omicron spike RBD that are similar to those found in PC.03 and MB.02. Furthermore, we continued to utilize RAMIHM on cancer immunotherapy targets. We observed that antigen-specific IgG-positive-antibody titers continued to increase during the first week after RAMIHM onset and remained at a peak level until the endpoint. Combined with high-throughput scBCR sequencing, we were able to examine B cell clonotype enrichment and efficiently identify the paired antibodies sequences from RAMIHM-treated humanized mice. Another significant advantage of RAMIHM is that fully human mAb candidates against a specific target can be developed from as few as one single humanized mouse. In brief, summary, RAMIHM can serve as a versatile platform broadly applicable to antibody discovery against emerging pathogens or other therapeutic targets.
Limitations of the study
RAMIHM uses mRNA to encode antigen. This study used mRNAs encoding full-length transmembrane proteins, which can induce immune responses to all possible regions of the protein, including intracellular domains. Thus, the antibodies reacting to those domains might not be desired. Specific domains can be encoded in future studies. RAMIHM requires IgG humanized mice, which currently has limited availability. Finally, scBCR sequencing occasionally detects unpaired BCR clones, with only IgH or IgK/IgL sequence(s).
STAR Methods
Resource availability
Lead contact
Requests for further information or reagents should be directed to the lead contact and corresponding author, Sidi Chen (sidi.chen@yale.edu)
Data and code availability
All data generated or analyzed during this study are included in this article and its supplementary information files. Specifically, source data and statistics for non-high-throughput experiments are provided in a supplementary table excel file. The ATX humanized mice are available via Alloy Therapeutics.
NGS data have been deposited at GEO and are publicly available as of the date of publication. Accession number (GSE203030) is listed in the key resources table.
CCBKEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibody | ||
| Goat anti-human IgG(H+L)/HRP | Fisher Scientific | Cat # 31412 |
| Goat anti-mouse IgG(H+L)/HRP | Fisher Scientific | Cat # A-10677 |
| PE–anti-human FC antibody | Biolegend | Cat#409304 |
| Anti-SARS-CoV-2 mAb (Clone 2) | Sidi Chen Lab | Peng et al. 2022 |
| Anti-SARS-CoV-2 mAb (Clone 6) | Sidi Chen Lab | Peng et al. 2022 |
| Anti-SARS-CoV-2 mAb (Clone 13A) | Sidi Chen Lab | Peng et al. 2022 |
| Anti-Omicron mAb (MB.02) | Sidi Chen Lab | This study |
| Anti-Omicron mAb (MB.08) | Sidi Chen Lab | This study |
| Anti-Omicron mAb (PB.03) | Sidi Chen Lab | This study |
| Bacterial and Virus Strains | ||
| B.1.1.529 variant (Omicron) pseudovirus | Sidi Chen Lab | This study |
| B.1.617 variant (Delta) pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.1.1 pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.2 pseudovirus | Sidi Chen Lab | Fang et al. 2022 |
| Omicron sub-variant BA.2.12.1pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.3 pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.5 pseudovirus | Sidi Chen Lab | This study |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DPBS | Kline | Cat#14190144 |
| RPMI 1640 Medium | Gibco | Cat#11875-093 |
| Fetal Bovine Serum | Sigma Aldrich | Cat#F4135-500ML |
| DMEM | Kline | Cat#11995065 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122 |
| Glutamax | Med School | Cat#35050061 |
| TWEEN-20 | Sigma-Aldrich | Cat# P1379 |
| 50TS microplate washer | Fisher Scientific | Cat#BT50TS16 |
| ACK Lysing Buffer | Lonza | Cat#BP10-548E |
| Gibson Assembly Master Mix - 50 rxn | NEB | Cat#E2611L |
| HiscribeTM T7 ARCA mRNA Kit (with tailing) | NEB | Cat#E2060S |
| Phusion Flash High-Fidelity PCR Master Mix | ThermoFisher | Cat#F548L |
| E-Gel™ Low Range Quantitative DNA Ladder | ThermoFisher | Cat#12373031 |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28706 |
| EndoFree® Plasmid Maxi Kit | Qiagen | Cat#12362 |
| Quant-iT™ RiboGreen™ RNA Assay Kit | ThermoFisher | Cat#R11490 |
| Tetramethylbenzidine substrate | Biolegend | Cat#421101 |
| RNeasy® Plus Mini Kit | Qiagen | Cat#74134 |
| Library Construction Kit, 16 rxns | 10X Genomics | Cat#1000190 |
| Live/Dead aqua fixable stain | Thermofisher | Cat#L34976 |
| BSA | Fisher Scientific | BP1600-100 |
| 100 μm cell strainer | Corning | Cat#352360 |
| 40 μm cell strainer | Corning | Cat#352340 |
| Bovine Serum Albumin | Sigma Aldrich | Cat#A9418-100G |
| EDTA | Kline | Cat#AB00502-01000 |
| Tris-Cl pH 8.5 | TENOVA | Cat#15085 |
| N1-Methylpseudouridine-5’-Triphosphate - (N-1081) | TriLink (NC) | Cat#N-1081-1 |
| Sucrose | Thomas | Cat#C987K85 (EA/1) |
| Lymphoprep | STEMCELL | Cat # 07851 |
| Memory B Cell Isolation Kit, mouse | Miltenyi Biotec | Cat#130-095-838 |
| CD138+ Plasma Cell Isolation Kit, mouse | Miltenyi Biotec | Cat#130-095-530 |
| rProtein A Sepharose Fast Flow | GE healthcare | Cat#17-1279-01 |
| Octet Anti-Penta-HIS (HIS1K) Biosensors | SARTORIUS | Cat#18-5120 |
| Octet Anti-Human Fc Capture (AHC) Biosensors | SARTORIUS | Cat#18-5060 |
| SARS-COV RBD protein (His-tag) | Fisher Scientific | Cat#50-196-4017 |
| SARS-COV2 Delta RBD protein (His-tag) | SINO | Cat#40592-V08H90 |
| SARS-COV2 Omicron RBD protein (His-tag) | SINO | Cat#40592-V08H121 |
| MERS-COV RBD protein (His-tag) | Fisher Scientific | Cat#50-201-9463 |
| ACE2 protein, Human, Recombinant (His Tag) | SINO | Cat # 10108-H08H |
| ACE2 protein, Human, Recombinant (mFc Tag) | SINO | Cat # 10108-H05H |
| Chromium Next GEM Single Cell 5’ Kit v2, 16 rxns PN-1000263 | 10X Genomics | Cat#PN-1000263 |
| Chromium Next GEM Chip K Single Cell Kit, 16 rxns PN-1000287 | 10X Genomics | Cat#PN-1000287 |
| CD22-ECD protein (His-tag) | SINO | Cat#11958-HNAH |
| Experimental Models: Cell Lines | ||
| HEK293T | ThermoFisher | Catalog Number: R70007 |
| K562-CD22-O/E-GFP | Sidi Chen Lab | This study |
| K562-GPRC5D-O/E-GFP | Sidi Chen Lab | This study |
| Experimental Models: Organisms/Strains | ||
| ATX-GK+ Human Transgenic Mouse | Alloy | N/A |
| Oligo name and sequence | ||
| Various oligos (See Table S3 - supplemental oligos file) | Sidi Chen Lab | This study |
| Recombinant DNA | ||
| pcDNA3.1 plasmids | Addgene | Cat# V790-20 |
| pHIVNLGagPol | Schmidt et al., 2020 | Gift from Dr Bieniasz’ lab |
| pCCNanoLuc2AEGFP | Schmidt et al., 2020 | Gift from Dr Bieniasz’ lab |
| pSARS-CoV-2 SΔ19 | Schmidt et al., 2022 | Gift from Dr Bieniasz’ lab |
| pFZ47 (B.1.1.529 variant (6P)) | Sidi Chen Lab | Fang et al. 2022 |
| pVP29b (B.1.617 variant (6P)) | Sidi Chen Lab | Peng et al. 2022 |
| pCCNanoLuc2AEGFP plasmid | Schmidt et al. | Gift from Dr Bieniasz’ lab |
| Polyethylenimine | POLYSCIENCES INC | Cat#24765-1 |
| pFZ57 (Omicron BA.2 subvariant (6P)) | Sidi Chen Lab | Fang et al. 2022 |
| pPR158 (Omicron BA.1.1 subvariant (6P)) | Sidi Chen Lab | This study |
| pPR185 (Omicron BA.2.12.1 subvariant (6P)) | Sidi Chen Lab | This study |
| pPR186 (Omicron BA.3 subvariant (6P)) | Sidi Chen Lab | This study |
| pPR187 (Omicron BA.5 subvariant (6P)) | Sidi Chen Lab | This study |
| pZF46 (B.1.1.529 Omicron spike hexapro mRNA) | Sidi Chen Lab | Fang et al. 2022 |
| pPR128 (CD22-mRNA) | Sidi Chen Lab | This study |
| pPR109 (GPRC5D-mRNA) | Sidi Chen Lab | This study |
| Software and Algorithms | ||
| FlowJo software 9.9.6 | FlowJo, LLC | https://www.flowjo.com |
| GraphPad Prism 8.0 | GraphPad Software Inc | https://www.graphpad.com/scientific-software/prism/ |
| Pymol | Schrödinger | http://www.pymol.org/ |
| Cell Ranger v3.1.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Loupe V(D)J Browser | 10X Genomics | https://support.10xgenomics.com/single-cell-vdj/software/visualization/latest/installation |
| R | R project | https://www.r-project.org |
| plyr R package | Wickham. (2011). Journal of Statistical Software | http://www.jstatsoft.org/v40/i01/ |
| dplyr R package | Wickham et al., (2021). dplyr: A Grammar of Data Manipulation. R package version 1.0.7 | https://CRAN.R-project.org/package=dplyr |
| ggplot2 R package | Wickham. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York | https://ggplot2.tidyverse.org |
| stringr R package | Hadley Wickham (2019). stringr: Simple, Consistent Wrappers for Common String Operations. R package version 1.4.0 | https://CRAN.R-project.org/package=stringr |
| Circlize R package | Gu et al., Bioinformatics, 2014 | https://cran.r-project.org/package=circlize |
| Pheatmap R package | Kolde, 2019 | https://cran.r-project.org/package=pheatmap |
| Deposited Data | ||
| Single cell BCR-seq | This study | GEO: GSE203030 |
| Flow data | This study | Mendeley Data, DOI: 10.17632/4dcgs8mvzx.3 |
| Other | ||
Flow data have been deposited at Mendeley Data and are publicly available as of the date of publication. DOI (doi: 10.17632/4dcgs8mvzx.1) is listed in the key resources table.
Code: This paper does not report original code. This study uses adapted version of existing computational pipelines. Computer codes related to this study is provided as a zip file in supplemental information.
Additional Supplemental Items are available from Mendeley Data at DOI:10.17632/4dcgs8mvzx.3 https://data.mendeley.com/datasets/4dcgs8mvzx/3
The Mendeley Data link includes all Excel tables, Data items, and ZIP files.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon reasonable request.
Materials availability
Materials availability Statement
Materials described in this study will be made available, either via public repository, or upon reasonable request with a Material Transfer Agreement.
Key resources table (KRT)
KRT is provided with details of resource availability in a supplemental file.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon reasonable request.
Experimental model and subject details
This study uses ATX-GK mice.
Health/immune status
Mice (both sex) are healthy before immunization. LNP-mRNA immunization was performed for RAMIHM.
Whether subjects were involved in previous procedures
No.
Whether the subject is drug or test naïve.
Yes.
Housing and husbandry conditions of experimental animals
Mice were housed in standard vivarium condition at YARC, with free access to water and food, ambient room temperature (approximately 22 degree Celsius) and humidity-controlled. Mice were maintained on a 14h:10h light/dark cycle (07:00 to 21:00 light on). Mouse health checks were performed regularly. The general health of for all the mice in this study is in good condition (BAR: bright, alert and responsive) before the related experiments started. Mice, both female and male, aged 8–20 weeks were used for experiments.
Method Details
Rapid mRNA immunization of humanized mice
The full-length Omicron spike sequence used in mRNA immunization was based on two North American patients identified on Nov23rd, 2021. The LNP-mRNA was generated as previously described11. Humanized mice with human IgG and IgK transgene knock-ins (ATX-GK, Alloy Therapeutics) were used for rapid mRNA immunization, according to an accelerated (two-week) vaccination schedule. Pre-immune plasma was collected from the mice before the initiation of immunization. The mice were primed with intramuscular injection of 10 μg Omicron LNP-mRNA and boosted on days 2, 4, and 7 with the same dose as prime. On day 11, three days before sacrifice, mice received a final boost with 20 μg Omicron LNP-mRNA. All mice were retro-orbital bled on days 7, and 14, and anti-plasma titers were evaluated using an immunoassay as described below.
ELISA analysis for plasma and mAb supernatant binding to Omicron RBD protein
Plasma was extracted from the surface layer by using SepMate-15 tubes with Lymphoprep gradient medium (StemCell Technologies) after centrifugation at 1200g for 20 minutes. Afterward, antibody titers in plasma against Omicron RBD were evaluated using a direct coating ELISA. 384-well microtiter plate (Corning) was coated with 3 μg/ml of Omicron RBD recombinant protein (Sino Biological 40592-V08H121) in PBS at 4°C overnight. The plate was washed with standard wash buffer PBS-T (PBS containing 0.05% Tween 20) and blocked with blocking buffer (PBS containing 0.5% BSA) for 1 hour at room temperature (RT). Either serially diluted plasma samples or mAbs supernatant were added to the plate and incubated for 1 hour at RT. Wells were then washed and incubated with secondary goat anti-mouse IgG labeled with HRP (Fisher, Cat# A-10677) at 1:2500 dilution in a blocking buffer for 1h at RT. Thereafter, wells were developed using TMB substrate (Biolegend, 421101) according to the manufacturer’s protocol. The reactions were terminated with 1M H3PO4 after 20 minutes of incubation at RT and optical density (OD) was measured by a spectrophotometer at 450 nm (PerkinElmer EnVision 2105).
Humanized mice B cell isolation and purification
Three sets of single B cells were collected: PBMC sample, Omicron RBD-specific memory B cell sample, and CD138+ plasma B cell sample. PBMC cells were isolated from fresh whole blood by using SepMate-15 tubes with Lymphoprep gradient medium (StemCell Technologies) after centrifugation at 1200g for 20 minutes. Poured top layer solution that contained PBMCs from SepMate tubes to a new falcon tube and washed once with PBS+2%FBS, resuspended with PBS, and stored on ice until use.
According to the manufacturer’s protocol, Omicron RBD-specific memory B cells were isolated from pre-enriched memory B cells by magnetic positive selection (Miltenyi Biotec, 130-095-838). Briefly, the spleen and lymph nodes were gently homogenized and red blood cells were lysed in ACK lysis buffer (Lonza). The remaining cells were washed with PBS with 2%FBS and filtered through with a 50ml falcon tube. Thereafter, memory B cells were labeled with a memory B cell biotin-antibody cocktail combined with anti-biotin microbeads and isolated using a magnetic rack. Enriched memory B cells were eluted and mixed with 25ug of Omicron RBD recombinant protein with his tag and incubated for 30mins on ice. After incubation, the complex was washed and then incubated with anti-his-APC antibodies and anti-APC microbeads. The final antigen-enrichment B cells were eluted in PBS and stored on ice until use.
Plasma B cells were collected by fragmenting and rinsing bone marrows with PBS containing 2% FBS. Non-plasma cells were labeled with a biotin-conjugated antibody cocktail combined with anti-biotin microbeads and separated using a magnetic rack according to the manufacturer’s protocol (Miltenyi Biotec, 130-092-530). Purified plasma B cells were eluted and sequentially incubated with CD138 microbeads for an additional 15 minutes at 4°C. The final CD138+ plasma B cells were eluted in PBS and stored on ice until use.
Single-cell VDJ sequencing and data analysis
For each above collection, 10,000 cells were loaded on a Chromium Next GEM Chip K Single Cell Kit. Single-cell lysis and cDNA first-strand synthesis was performed using Chromium Next GEM Single Cell 5’ Kit v2 according to the manufacturer’s protocol. The barcoded single-strand cDNA was isolated via a Dynabeads MyOne SILANE bead cleanup mixture. The cDNA was amplified by 14 PCR cycles and purified via SPRI bead cleanup (X0.6) according to the manufacturer’s protocol. For BCR repertoire libraries, 2 μL of amplified cDNA underwent two rounds of Target Enrichment using nested custom primer pairs specific for BCR constant regions. The target enrichments for the heavy chain and light chain were performed in separate reactions. After each PCR reaction, the PCR products were subjected to double-sided size selection with SPRI bead cleanup (X0.6 followed by X0.8) The primers were designed by Alloy biotechnologies and synthesized by KECK.
25 ng of each target enrichment PCR product was combined, and used for library preparation, consisting of fragmentation, end repair, A-tailing, adaptor ligation (Library Construction Kit), and sample index PCR (Dual Index Kit TT Set A) according to the manufacturer’s instructions. The final library was profiled and quantified using the D1000 ScreenTape assay (Agilent) for the TapeStation system. Libraries were sequenced by paired-end sequencing (26 × 91 bp) on an Illumina Miseq. All libraries were targeted for sequencing depth of 5,000 raw read pairs per cell.
For bioinformatics analysis, BCL data were converted to demultiplexed FASTQ files by the Illumina Miseq controller. These files were processed by using Cell Ranger v6.0.1 with default settings to align the reads to customized germline V and J gene references. The custom references were created by combining mouse constant genes along with human V(D)J genes. The consensus amino acid sequences of top-enriched clonotypes from each collection were selected by using the Loupe V(D)J Browser and cDNA sequences were synthesized for further molecular cloning and recombinant antibody expression.
In vitro generation of recombinant mAbs
The cDNAs of paired heavy- and light-chains from top-enriched IgG clonotypes were codon-optimized and respectively subcloned into human IgG1 expression vectors, based on Gibson assembly, to generate recombinant mAbs. mAbs were produced by transient transfection into Expi293F™ cells with equal amounts of paired heavy- and light-chain expression vectors using the ExpiFectamine 293 transfection kit according to the manufacturer’s protocol (Thermo Fisher). Five days post antibody expression, the secreted mAbs from cultured cells were collected and purified by affinity chromatography on rProtein A Sepharose Fast Flow beads according to the manufacturer’s instructions (Cytiva). Eluted mAbs were further purified by size exclusion chromatography (SEC) using a Superdex 200 Increase 5/150 GL column (Cytiva), and the column was pre-equilibrated in 1x DPBS (Thermo). The purified mAbs were examined by running SDS-PAGE and kept in −80°C until needed.
Omicron pseudovirus generation and neutralization assay
Omicron pseudovirus was generated by using a modified method from a previously described study. Briefly, a full-length Omicron spike gene was constructed into GFP encoding (pCCNanoLuc2AEGFP) human immunodeficiency vector backbone, then Omicron spike protein expression vectors were combined with HIV-1 structural corresponding plasmids and co-transfected into HEK-293T cells with PEI (1mg/ml, PEI MAX, Polyscience). Two-day post-transfection, viral supernatants were harvested, collected, filtered, and aliquoted to use in assays.
Neutralization assays were performed by incubating pseudovirus with serial dilutions of mAbs. 10,000 cells/well of HEK-293T-hACE2 cells were seeded in a 96-well plate, 24 hours before assay. mAbs supernatant/purified mAbs were serially diluted in DMEM media with 10% FBS and incubated with an equal volume of purified Omicron pseudovirus at 37°C for 1 hour. Thereafter, the virus-antibody mixture was added in triplicate onto HEK-293T-hACE2 cells and grown at 37°C for an additional 24 hours. Then, infected cells were counted and determined by evaluating GFP expression after 24 hours of exposure to a virus-antibody mixture using Attune NxT Acoustic Focusing Cytometer (Thermo Fisher). Half-maximal inhibitory concentration (IC50) for mAbs was calculated with a four-parameter logistic regression using GraphPad Prism (GraphPad Software Inc.).
Antibody binding kinetics, epitope mapping by bio-layer interferometry (BLI)
Antibody binding kinetics for anti-Omicron RBD mAbs were evaluated by BLI on an Octet RED96e instrument (FortéBio) at RT. 25 ng/ul of purified mAbs were captured on an AHC biosensor (Sartorius, 18–5060). The baseline was recorded for 60 s in a running buffer (PBS, 0.02% Tween-20, and 0.05% BSA, pH 7.4). Followed by sensors that were subjected to an association phase for 300 s in wells containing Omicron RBD with his tag protein diluted in the buffer. In the dissociation phase, the sensors were immersed in the running buffer for 500 s. The dissociation constants KD, and kinetic constants Kon and Koff were calculated by FortéBio data analysis software.
For epitope mapping, two different antibodies were sequentially injected and monitored for binding activity to determine whether the two mAbs recognized separate or closely-situated epitopes by an in-tandem approach on OCTET RED. Briefly, SARS-CoV-2 RBD-His recombinant protein (Sino Biological 40592-V08H121) was diluted with PBS to 20 μg/mL and was captured by anti-Penta-His (HIS1K) sensors (Sartorius, 18–5120). The primary antibody was diluted to 150 nM with running buffer in wells. Sensors were firstly subjected to an association phase for 500 s, and the response value was recorded. The sensors were then exposed to the secondary antibody mixture, and the response value was recorded again. Competition tolerance was calculated by the percentage increase in response after the secondary antibody was added. The column indicates the primary antibody, and the row indicates secondary antibodies. Competition tolerance of less than 25% indicates a high possibility of the closely-situated epitope.
Antibody clone cellular binding testing
To validate binding ability of those top enriched IgG1/IgG2ab+ clonotypes, paired heavy- and light-variable segments were cloned into human IgG1 expression vectors, transfected into the Expi293F cell mammalian expression system to produce individual clones of mAbs. Target overexpression cells (e.g. Omicron S in HEK293 cells, GPRC5D in K562 cells) were generated either by lentiviral cDNA expression, or by transfection or electroporation of respective mRNAs. Transfected culture supernatants that contain secreted antibodies were used as primary Ab in cell staining assay to determine antibody binding on cell surface of target overexpression cells. For flow staining, 200 μl of each transfected culture supernatant that contains secreted antibodies was co-cultured with target over-expressed cells for 1 hour at 4°C. Thereafter, the cells were washed twice with MACS buffer, then incubated with anti-hIgG-PE antibodies for additional 30 mins on ice. The binding activity was determined using flow cytometry.
Chemical compounds
No original chemical compounds such as small molecule or peptide were generated. Standard chemical compounds had been sourced from commercial vendors as described in method details. For biomolecules such as antibody constructs, identity is based on sequence and the constructs are sequence verified.
Oligo sequences
Oligo sequences related to this study are provided in Table S3.
Quantification and statistical analysis
Standard statistics
Standard statistical methods were applied to non-high-throughput experimental data. Prism (GraphPad Software) and RStudio were used for these analyses. The statistical significance was labeled as follows: n.s., not significant; * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001. The source data, statistical analyses, statistical details of experiments can be found in figure legends and/or supplementary Excel tables. Additional information can be found in the supplemental excel tables.
Schematic illustrations
Schematic illustrations were created with Affinity Designer or BioRender.
Replication, randomization, blinding and reagent validations
Sample size: Sample size determination was performed according to similar work in the field.
Replicate experiments have been performed for key data shown in this study.
Replication: Biological or technical replicate samples were randomized where appropriate. In animal experiments, mice were randomized by cage, sex and littermates.
Binding: Experiments were not blinded. It is unnecessary for animal immunization for antibody production to be blinded.
Antibodies and dilutions: Commercial antibodies used for various experiments are described in methods, with typical dilutions noted. For custom Antibodies generated in this study, dilutions were often serial titrations (i.e. a number of dilutions as specified in each figure). Commercial antibodies were validated by the vendors, and re-validated in house as appropriate. Custom antibodies were validated by specific antibody-antigen interaction assays, such as ELISA. isotype controls were used for antibody validations. Eukaryotic cell lines: Cell lines are from various sources as described in methods. Cell lines were authenticated by original vendors, and re-validated in lab as appropriate. All cell lines tested negative for mycoplasma. No commonly misidentified lines involved.
Animals and other organisms: Laboratory animals: M. musculus, ATX strain (Alloy Tx).
Additional Resources
Information on additional resources relevant to this study is provided at KRT.
Supplementary Material
Table S1. Comparison of current therapeutic monoclonal antibody discovery technologies, related to all figures
Table S3. Oligos used in this study, related to all figures
Dataset S2. Source data and statistics, related to all figures.
Source data and statistics provided in an excel file
Dataset S3. Supplemental Codes
Supplemental Codes. Zip file of scripts for analysis of single cell BCR-seq, related to figures 1, 3, 4
Highlights.
RAMIHM is an efficient strategy to rapidly generate fully human monoclonal antibodies.
RAMIHM generates human SARS-CoV-2 antibodies against multiple Omicron sublineages.
RAMIHM is applicable to cancer and immunology targets such as CD22 and GPRC5D.
Significance.
Development of fully human, potent and specific monoclonal antibodies (mAbs) has profound research and clinical implications. Existing antibody development methods have their own limitations. Here, we developed RAMIHM, a highly efficient strategy that allows us to generate fully human mAbs within several weeks using LNP-mRNA immunization followed by scBCR-seq. This approach allows us to, first bypass the protein antigen purification step for difficult multi-transmembrane proteins; and secondly, immunize humanized animals with natural conformation of antigen expressed from the hosts’ cells in vivo. Our study demonstrated that RAMIHM can be a versatile platform for antibody discovery against emerging pathogens and other therapeutic targets.
Acknowledgments
We thank various members from our labs for discussions and support. We thank staff from various Yale core facilities (Keck, YCGA, HPC, YARC, CBDS, and others) for technical support. We thank Drs. Tsemperouli, Karatekin, Lin, and others for providing equipment and related support. We thank various support from the Department of Genetics; Institutes of Systems Biology and Cancer Biology; Dean’s Office of Yale School of Medicine and the Office of Vice Provost for Research.
Footnotes
Declaration of Interests
Yale filed a patent application on the technology. SC is a (co)founder of Cellinfinity Bio, EvolveImmune Tx, Chen Consulting (SCC) and Chen Tech (SCTH), all unrelated to the study. Other authors declare no competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Mullard A (2021). FDA approves 100th monoclonal antibody product. Nat Rev Drug Discov 20, 491–495. 10.1038/d41573-021-00079-7. [DOI] [PubMed] [Google Scholar]
- 2.Yang D, Zhou Q, Labroska V, Qin S, Darbalaei S, Wu Y, Yuliantie E, Xie L, Tao H, Cheng J, et al. (2021). G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Target Ther 6, 7. 10.1038/s41392-020-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Dong S, and Xu F (2018). Structural and Druggability Landscape of Frizzled G Protein-Coupled Receptors. Trends Biochem Sci 43, 1033–1046. 10.1016/j.tibs.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Ruat M, Hoch L, Faure H, and Rognan D (2014). Targeting of Smoothened for therapeutic gain. Trends Pharmacol Sci 35, 237–246. 10.1016/j.tips.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. (2010). Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071. 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullard A (2018). FDA approves second GPCR-targeted antibody. Nat Rev Drug Discov 17, 613. 10.1038/nrd.2018.153. [DOI] [PubMed] [Google Scholar]
- 7.Dolgin E (2018). First GPCR-directed antibody passes approval milestone. Nat Rev Drug Discov 17, 457–459. 10.1038/nrd.2018.103. [DOI] [PubMed] [Google Scholar]
- 8.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, et al. (2021). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M, Kruger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer AS, Winkler MS, et al. (2022). The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 185, 447–456 e411. 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cele S, Jackson L, Khan K, Khoury DS, Moyo-Gwete T, Tegally H, Scheepers C, Amoako D, Karim F, Bernstein M, et al. (2021). SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 11.Fang Z, Peng L, Filler R, Suzuki K, McNamara A, Lin Q, Renauer PA, Yang L, Menasche B, Sanchez A, et al. (2022). Omicron-specific mRNA vaccination alone and as a heterologous booster against SARS-CoV-2. Nat Commun 13, 3250. 10.1038/s41467-022-30878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchings CJ, Koglin M, and Marshall FH (2010). Therapeutic antibodies directed at G protein-coupled receptors. MAbs 2, 594–606. 10.4161/mabs.2.6.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, Stowe J, Tessier E, Groves N, Dabrera G, et al. (2021). Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med 385, 585–594. 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naranbhai V, Garcia-Beltran WF, Chang CC, Mairena CB, Thierauf JC, Kirkpatrick G, Onozato ML, Cheng J, St Denis KJ, Lam EC, et al. (2021). Comparative immunogenicity and effectiveness of mRNA-1273, BNT162b2 and Ad26.COV2.S COVID-19 vaccines. J Infect Dis. 10.1093/infdis/jiab593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejnirattisai W, Shaw RH, Supasa P, Liu C, Stuart AS, Pollard AJ, Liu X, Lambe T, Crook D, Stuart DI, et al. (2022). Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet 399, 234–236. 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Peng P, Cao X, Wu K, Chen J, Wang K, Tang N, and Huang AL (2022). Increased immune escape of the new SARS-CoV-2 variant of concern Omicron. Cell Mol Immunol 19, 293–295. 10.1038/s41423-021-00836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreno JM, Alshammary H, Tcheou J, Singh G, Raskin A, Kawabata H, Sominsky L, Clark J, Adelsberg DC, Bielak D, et al. (2021). Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 18.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, San JE, Cromer D, Scheepers C, Amoako DG, et al. (2021). Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dejnirattisai W, Huo J, Zhou D, Zahradnik J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, et al. (2022). SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Iketani S, Guo Y, Chan JF, Wang M, Liu L, Luo Y, Chu H, Huang Y, Nair MS, et al. (2021). Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2. Nature. 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 21.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, et al. (2021). Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 22.Han P, Li L, Liu S, Wang Q, Zhang D, Xu Z, Han P, Li X, Peng Q, Su C, et al. (2022). Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, Pinto D, VanBlargan LA, De Marco A, di Iulio J, et al. (2022). Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602, 664–670. 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCallum M, Czudnochowski N, Rosen LE, Zepeda SK, Bowen JE, Walls AC, Hauser K, Joshi A, Stewart C, Dillen JR, et al. (2022). Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science, eabn8652. 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng L, Hu Y, Mankowski MC, Ren P, Chen RE, Wei J, Zhao M, Li T, Tripler T, Ye L, et al. (2022). Monospecific and bispecific monoclonal SARS-CoV-2 neutralizing antibodies that maintain potency against B.1.617. Nat Commun 13, 1638. 10.1038/s41467-022-29288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Y, Cai H, Li C, Deng B, Song W, Ling Z, Hu G, Yang Y, Niu P, Meng G, et al. (2021). A novel full-human CD22-CAR T cell therapy with potent activity against CD22(low) B-ALL. Blood Cancer J 11, 71. 10.1038/s41408-021-00465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorner T, Shock A, and Smith KG (2012). CD22 and autoimmune disease. Int Rev Immunol 31, 363–378. 10.3109/08830185.2012.709890. [DOI] [PubMed] [Google Scholar]
- 28.Xiao X, Ho M, Zhu Z, Pastan I, and Dimitrov DS (2009). Identification and characterization of fully human anti-CD22 monoclonal antibodies. MAbs 1, 297–303. 10.4161/mabs.1.3.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mailankody S, Devlin SM, Landa J, Nath K, Diamonte C, Carstens EJ, Russo D, Auclair R, Fitzgerald L, Cadzin B, et al. (2022). GPRC5D-Targeted CAR T Cells for Myeloma. N Engl J Med 387, 1196–1206. 10.1056/NEJMoa2209900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith EL, Harrington K, Staehr M, Masakayan R, Jones J, Long TJ, Ng KY, Ghoddusi M, Purdon TJ, Wang X, et al. (2019). GPRC5D is a target for the immunotherapy of multiple myeloma with rationally designed CAR T cells. Sci Transl Med 11. 10.1126/scitranslmed.aau7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Yan Q, Luo K, He P, Hou R, Zhao X, Wang Q, Yi H, Liang H, Deng Y, et al. (2022). Analysis of B Cell Receptor Repertoires Reveals Key Signatures of the Systemic B Cell Response after SARS-CoV-2 Infection. J Virol 96, e0160021. 10.1128/JVI.01600-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Comparison of current therapeutic monoclonal antibody discovery technologies, related to all figures
Table S3. Oligos used in this study, related to all figures
Dataset S2. Source data and statistics, related to all figures.
Source data and statistics provided in an excel file
Dataset S3. Supplemental Codes
Supplemental Codes. Zip file of scripts for analysis of single cell BCR-seq, related to figures 1, 3, 4
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplementary information files. Specifically, source data and statistics for non-high-throughput experiments are provided in a supplementary table excel file. The ATX humanized mice are available via Alloy Therapeutics.
NGS data have been deposited at GEO and are publicly available as of the date of publication. Accession number (GSE203030) is listed in the key resources table.
CCBKEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibody | ||
| Goat anti-human IgG(H+L)/HRP | Fisher Scientific | Cat # 31412 |
| Goat anti-mouse IgG(H+L)/HRP | Fisher Scientific | Cat # A-10677 |
| PE–anti-human FC antibody | Biolegend | Cat#409304 |
| Anti-SARS-CoV-2 mAb (Clone 2) | Sidi Chen Lab | Peng et al. 2022 |
| Anti-SARS-CoV-2 mAb (Clone 6) | Sidi Chen Lab | Peng et al. 2022 |
| Anti-SARS-CoV-2 mAb (Clone 13A) | Sidi Chen Lab | Peng et al. 2022 |
| Anti-Omicron mAb (MB.02) | Sidi Chen Lab | This study |
| Anti-Omicron mAb (MB.08) | Sidi Chen Lab | This study |
| Anti-Omicron mAb (PB.03) | Sidi Chen Lab | This study |
| Bacterial and Virus Strains | ||
| B.1.1.529 variant (Omicron) pseudovirus | Sidi Chen Lab | This study |
| B.1.617 variant (Delta) pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.1.1 pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.2 pseudovirus | Sidi Chen Lab | Fang et al. 2022 |
| Omicron sub-variant BA.2.12.1pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.3 pseudovirus | Sidi Chen Lab | This study |
| Omicron sub-variant BA.5 pseudovirus | Sidi Chen Lab | This study |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DPBS | Kline | Cat#14190144 |
| RPMI 1640 Medium | Gibco | Cat#11875-093 |
| Fetal Bovine Serum | Sigma Aldrich | Cat#F4135-500ML |
| DMEM | Kline | Cat#11995065 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122 |
| Glutamax | Med School | Cat#35050061 |
| TWEEN-20 | Sigma-Aldrich | Cat# P1379 |
| 50TS microplate washer | Fisher Scientific | Cat#BT50TS16 |
| ACK Lysing Buffer | Lonza | Cat#BP10-548E |
| Gibson Assembly Master Mix - 50 rxn | NEB | Cat#E2611L |
| HiscribeTM T7 ARCA mRNA Kit (with tailing) | NEB | Cat#E2060S |
| Phusion Flash High-Fidelity PCR Master Mix | ThermoFisher | Cat#F548L |
| E-Gel™ Low Range Quantitative DNA Ladder | ThermoFisher | Cat#12373031 |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28706 |
| EndoFree® Plasmid Maxi Kit | Qiagen | Cat#12362 |
| Quant-iT™ RiboGreen™ RNA Assay Kit | ThermoFisher | Cat#R11490 |
| Tetramethylbenzidine substrate | Biolegend | Cat#421101 |
| RNeasy® Plus Mini Kit | Qiagen | Cat#74134 |
| Library Construction Kit, 16 rxns | 10X Genomics | Cat#1000190 |
| Live/Dead aqua fixable stain | Thermofisher | Cat#L34976 |
| BSA | Fisher Scientific | BP1600-100 |
| 100 μm cell strainer | Corning | Cat#352360 |
| 40 μm cell strainer | Corning | Cat#352340 |
| Bovine Serum Albumin | Sigma Aldrich | Cat#A9418-100G |
| EDTA | Kline | Cat#AB00502-01000 |
| Tris-Cl pH 8.5 | TENOVA | Cat#15085 |
| N1-Methylpseudouridine-5’-Triphosphate - (N-1081) | TriLink (NC) | Cat#N-1081-1 |
| Sucrose | Thomas | Cat#C987K85 (EA/1) |
| Lymphoprep | STEMCELL | Cat # 07851 |
| Memory B Cell Isolation Kit, mouse | Miltenyi Biotec | Cat#130-095-838 |
| CD138+ Plasma Cell Isolation Kit, mouse | Miltenyi Biotec | Cat#130-095-530 |
| rProtein A Sepharose Fast Flow | GE healthcare | Cat#17-1279-01 |
| Octet Anti-Penta-HIS (HIS1K) Biosensors | SARTORIUS | Cat#18-5120 |
| Octet Anti-Human Fc Capture (AHC) Biosensors | SARTORIUS | Cat#18-5060 |
| SARS-COV RBD protein (His-tag) | Fisher Scientific | Cat#50-196-4017 |
| SARS-COV2 Delta RBD protein (His-tag) | SINO | Cat#40592-V08H90 |
| SARS-COV2 Omicron RBD protein (His-tag) | SINO | Cat#40592-V08H121 |
| MERS-COV RBD protein (His-tag) | Fisher Scientific | Cat#50-201-9463 |
| ACE2 protein, Human, Recombinant (His Tag) | SINO | Cat # 10108-H08H |
| ACE2 protein, Human, Recombinant (mFc Tag) | SINO | Cat # 10108-H05H |
| Chromium Next GEM Single Cell 5’ Kit v2, 16 rxns PN-1000263 | 10X Genomics | Cat#PN-1000263 |
| Chromium Next GEM Chip K Single Cell Kit, 16 rxns PN-1000287 | 10X Genomics | Cat#PN-1000287 |
| CD22-ECD protein (His-tag) | SINO | Cat#11958-HNAH |
| Experimental Models: Cell Lines | ||
| HEK293T | ThermoFisher | Catalog Number: R70007 |
| K562-CD22-O/E-GFP | Sidi Chen Lab | This study |
| K562-GPRC5D-O/E-GFP | Sidi Chen Lab | This study |
| Experimental Models: Organisms/Strains | ||
| ATX-GK+ Human Transgenic Mouse | Alloy | N/A |
| Oligo name and sequence | ||
| Various oligos (See Table S3 - supplemental oligos file) | Sidi Chen Lab | This study |
| Recombinant DNA | ||
| pcDNA3.1 plasmids | Addgene | Cat# V790-20 |
| pHIVNLGagPol | Schmidt et al., 2020 | Gift from Dr Bieniasz’ lab |
| pCCNanoLuc2AEGFP | Schmidt et al., 2020 | Gift from Dr Bieniasz’ lab |
| pSARS-CoV-2 SΔ19 | Schmidt et al., 2022 | Gift from Dr Bieniasz’ lab |
| pFZ47 (B.1.1.529 variant (6P)) | Sidi Chen Lab | Fang et al. 2022 |
| pVP29b (B.1.617 variant (6P)) | Sidi Chen Lab | Peng et al. 2022 |
| pCCNanoLuc2AEGFP plasmid | Schmidt et al. | Gift from Dr Bieniasz’ lab |
| Polyethylenimine | POLYSCIENCES INC | Cat#24765-1 |
| pFZ57 (Omicron BA.2 subvariant (6P)) | Sidi Chen Lab | Fang et al. 2022 |
| pPR158 (Omicron BA.1.1 subvariant (6P)) | Sidi Chen Lab | This study |
| pPR185 (Omicron BA.2.12.1 subvariant (6P)) | Sidi Chen Lab | This study |
| pPR186 (Omicron BA.3 subvariant (6P)) | Sidi Chen Lab | This study |
| pPR187 (Omicron BA.5 subvariant (6P)) | Sidi Chen Lab | This study |
| pZF46 (B.1.1.529 Omicron spike hexapro mRNA) | Sidi Chen Lab | Fang et al. 2022 |
| pPR128 (CD22-mRNA) | Sidi Chen Lab | This study |
| pPR109 (GPRC5D-mRNA) | Sidi Chen Lab | This study |
| Software and Algorithms | ||
| FlowJo software 9.9.6 | FlowJo, LLC | https://www.flowjo.com |
| GraphPad Prism 8.0 | GraphPad Software Inc | https://www.graphpad.com/scientific-software/prism/ |
| Pymol | Schrödinger | http://www.pymol.org/ |
| Cell Ranger v3.1.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Loupe V(D)J Browser | 10X Genomics | https://support.10xgenomics.com/single-cell-vdj/software/visualization/latest/installation |
| R | R project | https://www.r-project.org |
| plyr R package | Wickham. (2011). Journal of Statistical Software | http://www.jstatsoft.org/v40/i01/ |
| dplyr R package | Wickham et al., (2021). dplyr: A Grammar of Data Manipulation. R package version 1.0.7 | https://CRAN.R-project.org/package=dplyr |
| ggplot2 R package | Wickham. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York | https://ggplot2.tidyverse.org |
| stringr R package | Hadley Wickham (2019). stringr: Simple, Consistent Wrappers for Common String Operations. R package version 1.4.0 | https://CRAN.R-project.org/package=stringr |
| Circlize R package | Gu et al., Bioinformatics, 2014 | https://cran.r-project.org/package=circlize |
| Pheatmap R package | Kolde, 2019 | https://cran.r-project.org/package=pheatmap |
| Deposited Data | ||
| Single cell BCR-seq | This study | GEO: GSE203030 |
| Flow data | This study | Mendeley Data, DOI: 10.17632/4dcgs8mvzx.3 |
| Other | ||
Flow data have been deposited at Mendeley Data and are publicly available as of the date of publication. DOI (doi: 10.17632/4dcgs8mvzx.1) is listed in the key resources table.
Code: This paper does not report original code. This study uses adapted version of existing computational pipelines. Computer codes related to this study is provided as a zip file in supplemental information.
Additional Supplemental Items are available from Mendeley Data at DOI:10.17632/4dcgs8mvzx.3 https://data.mendeley.com/datasets/4dcgs8mvzx/3
The Mendeley Data link includes all Excel tables, Data items, and ZIP files.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon reasonable request.


