Abstract
Edwardsiella tarda is responsible for hemorrhagic septicemia (edwardsiellosis) in fish and also causes diseases in higher vertebrates such as birds, reptiles, and mammals, including humans. Interactions of E. tarda with blue gourami phagocytes were studied by light microscopy as well as by adherence, intracellular replication, and superoxide anion assays. Both nonopsonized virulent (PPD130/91 and AL9379) and avirulent (PPD125/87 and PPD76/87) bacteria could adhere to and survive and replicate within phagocytes, while only opsonized virulent strains replicated within the phagocytes. Furthermore, only avirulent E. tarda elicited a higher rate of production of reactive oxygen intermediates (ROIs) by phagocytes, indicating that they were unable to avoid and/or resist reactive oxygen radical-based killing by the fish phagocytes. TnphoA transposon mutagenesis was used to construct a library of 200 alkaline phosphatase (PhoA+) fusion mutants from a total of 182,000 transconjugants derived from E. tarda PPD130/91. Five of these mutants induced more ROI production in phagocytes than the wild-type strain. Two mutants had lower replication ability inside phagocytes and moderately higher 50% lethal dose values than the wild-type strain. Sequence analysis revealed that three of these mutants had insertions at sequences having homology to PhoS, dipeptidase, and a surface polymer ligase of lipid A core proteins of other pathogens. These three independent mutations might have changed the cell surface characteristics of the bacteria, which in turn induced phagocytes to produce increased ROIs. Sequences from two other mutants had no homology to known genes, indicating that they may be novel genes for antiphagocytic killing. The present study showed that there are differences in the interactions of virulent and avirulent E. tarda organisms with fish phagocytes and PhoA+ fusion mutants that could be used successfully to identify virulence genes. The information elucidated here would help in the development of suitable strategies to combat the disease caused by E. tarda.
Edwardsiella tarda, the causative agent of edwardsiellosis in fish, is responsible for extensive losses in both freshwater and marine aquaculture. E. tarda infection of many commercially important cultured and wild fish has been reported, namely, channel catfish, eels, mullet, Chinook salmon, flounder, carp, tilapia, and striped bass (46). It has a wide host range, thus causing infection in higher vertebrates such as birds, reptiles (51), and mammals (49), including humans (37). In fish, it causes septicemia with extensive skin lesions, affecting internal organs such as the liver, kidney and spleen and muscle. These bacteria systemically avoid host defense mechanisms, thereby rapidly proliferating within the host and causing death.
Pathogenesis of E. tarda is multifactorial, and many potential virulent factors have been suggested, namely dermatotoxins (47), antiphagocyte killing (1), hemolysins (19), serum resistance, and the ability to invade epithelial cells (21, 28). Although both virulent and avirulent strains were able to invade cultured cells in vitro, only the virulent strain could enter fish in large numbers via mucus, gills, and the gastrointestinal tract (27) and multiply inside various internal organs, causing death. Janda and coworkers (21) also found that pathogenicity of E. tarda did not correlate with plasmid content, chemotactic motility, serum resistance, and expression of selected enzyme activities. However, very little is known about the roles of these factors in disease occurrence.
Serum- and phagocyte-mediated killing represents the two major defense mechanisms of nonspecific immunity in fish (7, 11). Resistance to these mechanisms by pathogens leads to infection. Most virulent fish pathogens such as Aeromonas hydrophila (25, 26) and Renibacterium salmoninarum (4) are resistant to both serum- and phagocyte-mediated killing. Bacterial pathogens use different strategies for subversion of phagocyte defenses. For example, Salmonella spp. are known to withstand low-pH conditions within phagolysosomes and to resist oxidative stress, nutrient limitations, and antimicrobial peptides such as defensins (15). Mycobacterium and Leishmania produce oxygen radical scavengers to resist oxidative burst and also prevent phagolysosomal fusion (22), while Listeria monocytogenes survives by escaping from the phagosome into the cytoplasm (44). The reactive oxygen intermediate (ROI) pathway is one of the oxygen-dependent pathways used by phagocytes to fight against microbial infection (33). Virulent bacteria may avoid or resist ROIs to overcome this type of killing.
The present study was carried out to investigate the interaction of E. tarda with fish phagocytes, their ability to adhere and survive and replicate within phagocytes and to resist and/or avoid ROIs produced by phagocytes. We have also used TnphoA transposon tagging to study the genes responsible for resisting phagocyte-mediated killing. Proper understanding of these mechanisms can help in the development of suitable strategies to overcome disease caused by E. tarda.
MATERIALS AND METHODS
Bacterial strains and media.
Two virulent (PPD130/91 and AL9379) and two avirulent (PPD125/87 and PPD76/87) E. tarda strains were chosen for this study; their characteristics were reported previously (28). The 50% lethal dose (LD50) values of E. tarda PPD130/91, AL9379, PPD76/87, and PPD125/87 were 105.2, 105.9, >107.4, and >107.4, respectively. All these strains except PPD76/87 were serum resistant. Cultures were routinely grown at 25°C in tryptic soy agar (TSA) (Difco) or tryptic soy broth (TSB) (Difco). Stock cultures were maintained at −80°C as a suspension in supplemented TSB containing 25% (vol/vol) glycerol. When required, the antibiotics ampicillin (AMP), neomycin (NEO), and colistin (COL) (all from Sigma) were added at the final concentrations of 50, 50, and 12.5 μg/ml, respectively.
Phagocyte isolation.
Healthy blue gourami (Trichogaster trichopterus Pallas) were obtained from a commercial fish farm and maintained in well-aerated, dechlorinated water at 25 ± 2°C. Phagocytes were isolated from the head kidney of naïve gourami and purified following the procedure of Secombs (42). Purified phagocytic cells (1 × 106 to 2 × 106 cells/well) were allowed to adhere to 24-, 48-, or 98-well tissue culture plates (Falcon; Becton Dickinson Labware) in fetal calf serum- supplemented L-15 medium (Leibovitz; Sigma). After 2 h of incubation at 25°C in a 5% (vol/vol) CO2 atmosphere, cells were washed twice using Hanks balanced salt solution (HBSS) (Sigma) to remove unattached cells. The remaining monolayer consisted of phagocytes (4 × 105 to 5 × 105 cells/well) and was infected with E. tarda at a multiplicity of infection of 1:1 in all the experiments.
Adherence-plus-internalization assay using counts of viable bacteria.
The adherence-plus-internalization assays using counts of viable bacteria were performed as described before (13, 50). This assay quantifies the total number of live bacteria bound to the outside of and internalized by the phagocytes, as well as the bacteria nonspecifically bound to the culture dish wells. The adherence rates were calculated from the mean number of bacteria in two wells in triplicate experiments. For opsonization of bacteria prior to the adherence assays, washed E. tarda cells were resuspended in 50% fresh gourami serum in HBSS, incubated for 30 min at 25°C, then washed once in HBSS, and added to the phagocytic cells as described above.
Microscopic count of adherent-plus-internalized bacteria using Giemsa stain.
Glass coverslips were placed into each well of the 24-well tissue culture plate, and the wells were seeded with blue gourami phagocytes and incubated for 2 h at 25°C in a 5% (vol/vol) CO2 atmosphere as described above. Thirty minutes after infection, the phagocytes were washed three times with HBSS and later stained with Giemsa stain (Merck) for 30 min. After being washed three times with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4 at pH 7.2), the stained samples were then examined under an Axiovert 25 CFL inverted microscope (Carl-Zeiss) at 100× magnification. Photographs were taken with Kodak color ISO200 film. The adherence rate was expressed as the number of adhering bacteria per 100 phagocytes after 200 phagocytes per coverslip were counted. The adherence rate was calculated from the mean number of phagocytes from two coverslips in triplicate experiments. This assay quantifies the total number of bacteria (both live and dead) bound to the outside of and internalized by the phagocytes.
Intracellular replication assay.
The intracellular replication assay was performed as described by Leung and Finlay (24) with the following modifications. Thirty minutes after infection, phagocytes were washed once with HBSS and then incubated for 1.5 h in fetal calf serum-supplemented fresh L-15 medium with 100 μg of gentamicin/ml. This treatment killed extracellular bacteria but did not affect the viability of intracellular organisms. Infected phagocytes were then washed three times in HBSS and incubated with L-15 medium. The intracellular population of bacteria was assayed at 2, 3.5, 5, and 6.5 h. The supernatant was removed, and 0.1 ml of 1% (vol/vol) Triton X-100 solution was added to the infected phagocyte monolayer. This was followed by a 1-min incubation, which released intracellular bacteria, and the bacteria were assayed as described for the adherence assay. The relative bacterial population was calculated by dividing the intracellular population of E. tarda at 3.5, 5.0, and 6.5 h by the population at 2.0 h. Values were calculated from the mean of the bacterial population of two wells in triplicate experiments.
Superoxide anion assay.
Thirty minutes after infection of phagocytes with bacteria in a 96-well tissue culture plate, 100 μl of nitroblue tetrazolium (1 mg/ml) (Sigma) was added to each well, and the monolayers were further incubated at 25°C for 30 min in a 5% (vol/vol) CO2 atmosphere. The reaction was arrested with 100% methanol, followed by two washes with 70% methanol. After the plate was dried for 1 min, 120 μl of 2 M potassium hydroxide and 140 μl of dimethyl sulfoxide were added. Optical density was measured with a microplate reader (Bio-Tek Instruments) at 630 nm, and the values were represented as the mean ± the standard error of the mean (SEM) of the counts of quadruple wells. The production of ROIs was from one set of representative results taken from one of the three independent experiments.
Transposon mutagenesis.
E. tarda PPD130/91 (recipient; Colr) and Escherichia coli SM10λpir(pJM703.1::TnphoA) (donor; Ampr Neor) cultures were statically grown in TSB at 25 and 37°C, respectively. Conjugative transfer of the suicide plasmid pJM703.1::TnphoA was performed by plate mating. The bacterial cell ratio of E. coli to E. tarda was adjusted to 4:1, with a total of about 107 CFU of both donor and recipient during mating. After 24 h of mating on TSA plates at 25°C, the cells were harvested and resuspended in 3 ml of PBS. Appropriate dilutions were plated on TSA supplemented with NEO, COL, and 5-bromo-3-chloro-indolyl phosphate (Sigma) (40 μg ml−1) (TSANCX) to select for transconjugants of E. tarda. Blue PhoA+ fusion clones were purified by streaking on TSANCX plates.
Characterization of transposon mutants.
TnphoA mutants were characterized by standard procedures for their ability to grow in TSA, TSB, and phosphate-limiting medium (PLM); production of superoxide anions, hemolysin, and catalase; replication within phagocytes; ability to withstand low-pH conditions; survival in serum; and adherence and internalization into fish epithelial cells. Briefly, growth of mutants in TSB and TSA was recorded after 24 h of incubation. Growth under phosphate-limiting conditions was examined by culturing the bacterial cells in a modified defined minimal medium (8) with the phosphate salts replaced by 3 μM Na2HPO4 and the pH adjusted to 7.0 with 30 mM HEPES. Hemolysin production was determined by inoculating bacteria onto TSA with 5% (vol/vol) heparinized gourami whole blood. A clear zone around a bacterial colony indicates hemolysin production. Catalase production was assayed by the hydrogen peroxide inhibition zone test as described previously (53). Zones of inhibition were visualized after bacteria were placed on sterile Whatman 3MM disks (0.6 cm in diameter) containing 10 μl of 2, 20, and 200 mM H2O2 and incubated overnight at 25°C. The pH sensitivity of mutants was characterized by estimating their ability to grow in low-pH conditions (16). Overnight bacterial cultures were inoculated into TSB, grown for 3 h to obtain 108 CFU/ml, and inoculated into TSB at pH 5.8 ± 0.1. Afterward, bacterial growth was monitored over a period of 24 h. Survival of the mutants in serum was calculated by dividing the number of viable bacteria after serum treatment by the initial number of bacteria before serum treatment. Washed bacteria (108 CFU/ml) were incubated in 50% (vol/vol) of fresh gourami serum for 1 h. A value of >1 was scored serum resistant; a value of <1 was considered serum sensitive (50). Determination of the adherence and internalization of mutants to carp epithelial cells (epithelial papillosum of carp Cyprinus carpio [EPC]) was performed following the protocol of Wang and coworkers (50).
DNA manipulations and Southern hybridization.
Bacterial genomic DNA was extracted according to the Genome DNA kit manual (BIO 101). Plasmid DNA was extracted using QIAprep mini columns (Qiagen). Restriction endonuclease digestion was accomplished by standard methods (41). Southern blotting was performed to characterize the transposon mutants with the BluGene Non-Radioactive Nucleic Acid Detection system (Gibco-BRL). Transfer of the DNA to nylon membranes (GeneScreen; NEM Research Products) and hybridization conditions were in accordance with standard methods (41). Genomic DNA from E. tarda PPD130/91 and its mutants were digested with EcoRV, hybridized with a HindIII-digested 14-dATP-biotinylated (Bio-Nick Labeling System; Gibco-BRL) pJM703.1 plasmid probe, and then visualized with streptaviridin-alkaline phosphate conjugate (BluGene Nonradioactive Nucleic Acid Detection System; Gibco-BRL).
Cloning chromosomal segments flanking TnphoA insertions, genome walking, and DNA sequencing.
BamHI fragments of mutant genomic DNA flanking the transposon were cloned into pBluescript SK(+) (Ampr) vector and transformed into E. coli Top10F′-competent cells (Clontech). Transformants, bearing TnphoA and flanking E. tarda chromosomal sequences, were selected by their ability to grow on Luria-Bertani (LB) agar containing AMP and NEO. Later these clones were sequenced. To obtain full-length sequences of all the mutants, a genome walker library of wild-type E. tarda PPD130/91digested with EcoRV and StuI was constructed according to the procedure described in the Universal Genome Walker kit (Clontech). PCR amplification was performed with primers specific to known upstream nucleotide sequences of mutants 153, 348, 387, and 392 were synthesized by GENSET (Singapore Biotech), and the adapter primer 1 (AP1) was used as the second primer. PCR was carried out with Advantage Tth Polymerase Mix (Clontech), and the cycling parameters were as follows: 7 cycles of 10 s at 94°C, 30 s at 59°C, and 3 min at 67°C; and 37 cycles of 10 s at 94°C, 30 s at 56°C, and 3 min at 67°C. The amplified fragments were gel eluted with the Qiaquick gel extraction kit (Qiagen), cloned into the pGEMT Easy vector system (Promega), transformed into E. coli Top 10F′-competent cells, and sequenced using AP1- and mutant-specific primers.
DNA sequencing was carried out with an Applied Biosystems PRISM 377 automated DNA sequencer by the dye termination method. The ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit was used (Applied BioSystems). Sequence assembly and further editing were done with DNASIS DNA analysis software (Hitachi Software). BLASTN, BLASTX, and FASTA sequence homology analyses were performed by using the National Center for Biotechnology Information BLAST network service.
LD50 studies.
Naive blue gourami of approximately 14 g each were obtained from commercial fish farms and acclimatized for more than 1 month. Three groups of 10 fish each were injected intramuscularly with 0.1 ml of PBS-washed bacterial cells adjusted to the required concentrations. Fish were monitored for mortality for 7 days, and LD50 values were calculated by the method of Reed and Muench (39).
Statistical analysis.
All data were expressed as means ± SEM. The data were analyzed by one-way analysis of variance and a Duncan multiple-range test (SAS software; SAS Institute). Values of P < 0.05 were considered significant.
RESULTS
Adherence and replication kinetics of E. tarda inside fish phagocytes.
Phagocyte-mediated killing is one of the major nonspecific defense barriers in fish. The abilities of E. tarda to adhere to and survive and multiply inside the fish phagocytes were investigated. The adherence-plus-internalization ability of the E. tarda strains was estimated by direct microscopic counts and counts of viable bacteria (Table 1). All the virulent (PPD130/91 and AL9379) and avirulent (PPD125/87 and PPD76/87) strains adhered to and were ingested by the phagocytes. Under nonopsonized conditions, both direct microscopic counts and counts of viable bacteria showed the highest percentage of adherence to phagocytes by PPD125/87, followed by AL9379, PPD76/87, and PPD130/91. Light microscopic observation of Giemsa-stained phagocytes revealed that PPD125/87 had the highest number of bacteria adhering to the phagocytes after 30 min of infection, while PPD130/91 had the lowest (Fig. 1A and B).
TABLE 1.
E. tarda strains and their percentage of adherence to phagocytes
| E. tarda strain | Inoculum (105) | Adherence at
25°Ca
|
|||
|---|---|---|---|---|---|
| Viable count
(%)b
|
Direct microscopic count
(%)c
|
||||
| Nonopsonized | Opsonized | Nonopsonized | Opsonized | ||
| PPD130/91 | 7.3 ± 1.3 | 8.8 ± 0.5 Aα | 11.7 ± 0.8 Bα | 10.9 ± 1.4 Aγ | 14.4 ± 1.6 Aγ |
| AL9379 | 8.2 ± 1.3 | 22.9 ± 0.6 Cα | 38.2 ± 5.2 Dβ | 47.8 ± 2.0 Cγ | 68.1 ± 3.1 BCδ |
| PPD76/87 | 6.4 ± 1.0 | 12.5 ± 0.6 Bα | 0.02 ± 0.01 Aβ | 37.7 ± 2.6 Bγ | 78.9 ± 6.1 Cδ |
| PPD125/87 | 9.8 ± 1.5 | 33.0 ± 2.3 Dα | 21.0 ± 1.6 Cβ | 57.1 ± 1.9 Dγ | 65.1 ± 0.7 Bδ |
Within each column, values followed by different letters (A, B, C, or D) are significantly different (P < 0.05). Similarly, within each row, values followed by different Greek letters (α and β for viable counts and γ and δ for direct counts) are significantly different (P < 0.05).
The viable count is the percentage of inoculum bacteria still adherent 30 min postinfection and after being washed three times. Values were recorded as the mean ± SEM for three trials in duplicate wells.
The direct microscopic count is the number of adherent bacteria (both live and dead) per 100 phagocytes 30 min postinfection. Values were recorded as the mean ± SEM for three trials in duplicate coverslips.
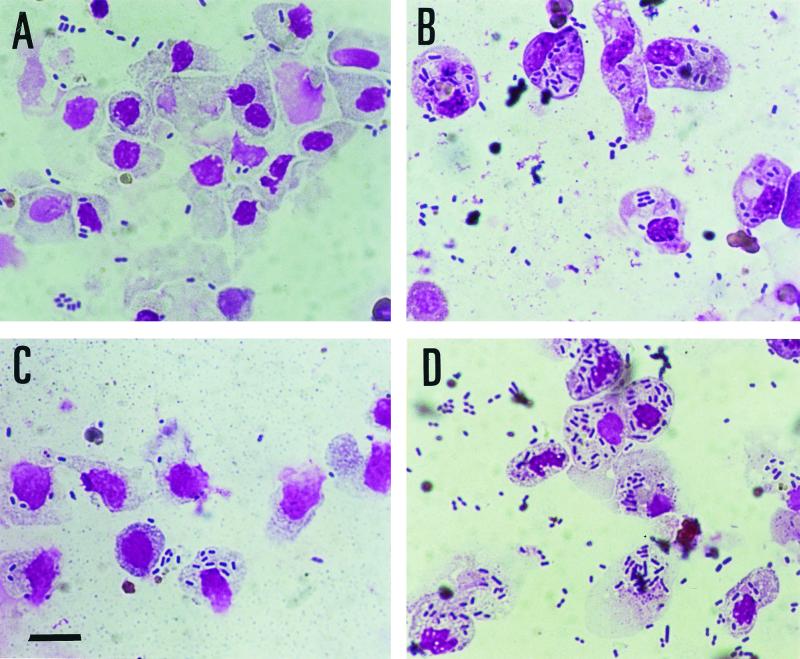
FIG. 1.
Giemsa-stained bright-field micrographs of blue gourami phagocytes infected with nonopsonized E. tarda PPD130/91 (A) and PPD125/87 (B) and opsonized PPD130/91 (C) and PPD125/87 (D) after 30 min of infection. Bar, 10 μm.
When E. tarda strains were opsonized with naive gourami serum, adherence to phagocytes increased significantly (P < 0.05) for all the strains except PPD130/91, as observed under light microscopy (Fig. 1C and D; Table 1). However, the percentage adherence by estimated count of viable bacteria decreased for the opsonized avirulent strains (PPD125/87 and PPD76/87), whereas it increased in the case of the opsonized virulent strain AL9379, but not for PPD130/91. For the adherence assay using counts of viable bacteria, controls were included for bacteria binding directly to culture plates. The rate of nonspecific binding ranged from 5.7 to 10% of the counts of viable bacteria and was insignificant compared to the rates of adherence-plus-internalization to phagocytes.
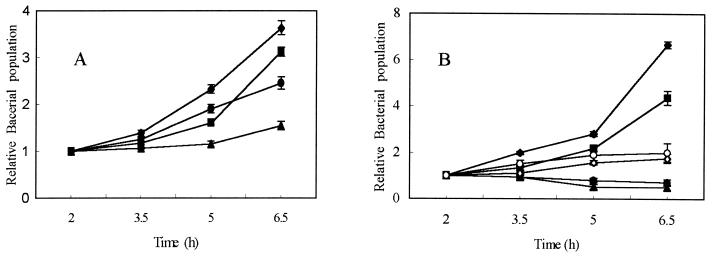
Both virulent and avirulent strains of E. tarda were able to survive and replicate within fish phagocytes, but at varying rates (Fig. 2). Nonopsonized E. tarda PPD130/91, AL9379, and PPD125/87 replicated faster than PPD76/87 until 5 h of incubation; after 6.5 h of incubation, nonopsonized virulent strains (PPD130/91 and AL9379) multiplied 1.5 to 2.5 times faster (Fig. 2A) than the avirulent ones (PPD125/87 and PPD76/87). However, when opsonized, only virulent strains could multiply within phagocytes (Fig. 2B). Opsonized virulent strains could multiply significantly faster (P < 0.05) than the nonopsonized virulent strains (Fig. 2). As infection time increased, the bacterial count of the opsonized avirulent strains was reduced, indicating that the bacteria were killed within the phagocytes.
FIG. 2.
Kinetics of intracellular multiplication of E. tarda in blue gourami phagocytes. Nonopsonized (A) and opsonized (B) E. tarda strains (PPD130/91 (♦), PPD125/87 (●), AL9379 (▪), PPD76/87 (▴), P1 (⋄), and 348 (○) were used. P1 and 348 are TnphoA mutants of E. tarda PPD130/91. The ratio of intracellular bacteria at various times was normalized to the number of bacteria at 2 h. Values represent the mean ± SEM of the bacteria counted in two wells in triplicate experiments.
Effect of E. tarda on ROI production.
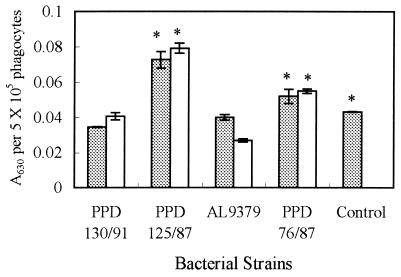
ROI production is one of the several strategies used by phagocytes to kill microorganisms. Therefore, it was of interest to examine whether E. tarda could overcome or resist respiratory burst activity. Figure 3 shows that gourami phagocytes produced significantly (P < 0.05) fewer ROIs when they were infected with virulent strains (PPD130/91 and AL9379) than with avirulent ones (PPD125/87 and PPD76/87). Opsonization did not change the ROI values for E. tarda PPD125/87, PPD76/87, and PPD130/91. However, opsonized E. tarda AL9379 produced fewer ROIs than nonopsonized strains.
FIG. 3.
ROIs produced by blue gourami phagocytes after being infected with various nonopsonized (░⃞) or opsonized (□) E. tarda strains. Control, the basal level of ROI production by phagocytes that are not infected with the bacteria. Results were expressed as the mean ± SEM from quadruplicate experiments. Data represented by bars with asterisks differ significantly from results obtained with E. tarda PPD130/91 (P < 0.05).
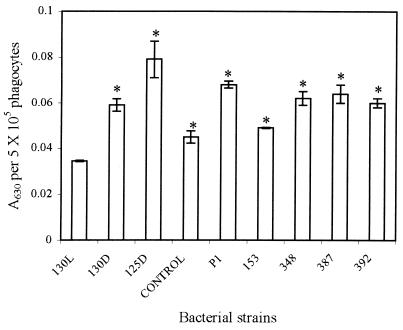
Since virulent E. tarda PPD130/91 could not induce a high rate of ROI production, we were interested in knowing whether this organism was actively suppressing oxidative responses or simply failing to stimulate phagocyte respiratory burst activity. To examine this, we infected the fish phagocytes with live and heat-killed (100°C for 5 min) PPD130/91 and PPD125/87 and monitored the ROI production. Our results indicate that the heat-killed virulent PPD130/91 (Fig. 4) could induce significantly higher numbers of ROIs than live virulent PPD130/91 (Fig. 3), but there was no difference for killed and live avirulent strain PPD125/87 (Fig. 3 and 4).
FIG. 4.
ROIs produced by blue gourami phagocytes infected with various strains of E. tarda. Results were expressed as the mean ± SEM from quadruplicate experiments. Data represented by bars with asterisks differ significantly from results obtained with live E. tarda PPD130/91 (P < 0.05). L, live bacteria; D, heat-killed bacteria; Control, without any bacteria. P1, 153, 348, 387, and 392 are TnphoA mutants of E. tarda PPD130/91.
Isolation and characterization of TnphoA mutants.
We have obtained a library of 200 alkaline phosphatase (PhoA+) fusion mutants from a total of 182,000 transconjugants derived from E. tarda PPD130/91. These NEO- and COL-resistant transconjugants were obtained at a frequency of 7.0 × 10−3 from several matings, with a spontaneous mutation frequency of <4.0 ×10−8 and <5.5 ×10−7 for E. tarda PPD130/91 and E. coli SM10λpir(pJM 703.1::TnphoA), respectively. TnphoA mutagenesis was carried out to target the secreted and surface proteins of E. tarda, which may influence the ROI production by the phagocytes upon infection. Two hundred PhoA+ fusion mutants were screened, and 5 (mutants P1, 153, 348, 387, and 392) were found to significantly increase ROI production compared to the wild type (E. tarda PPD130/91) (Fig. 4). All these mutants were ampicillin sensitive, suggesting that there was no suicide plasmid retention and integration.
These five isolates were further characterized by several different assays, such as growth on TSA and in TSB and PLM, intracellular replication within phagocytes, adherence to and internalization into EPC, production of catalase and hemolysin, and serum survival assay (Table 2). Growth of all the mutants except P1 was comparable to that of the wild-type strain in both TSB and PLM. Mutant P1 formed smaller colonies on TSA than the other mutants and the wild-type strain, indicating growth deficiency. Two of the mutants (P1 and 348) were found to have a decreased ability to replicate within phagocytes (Fig. 2B) compared to the wild-type strain and were also sensitive to low-pH conditions. All five mutants could produce catalase and hemolysin and had the ability to survive in naïve gourami serum, similar to the wild type. The ability of these mutants to adhere to and internalize into EPC cells was assayed. Although all the mutants could adhere to the EPC cells at rates similar to that of the wild type, their ability to internalize the cells was slightly reduced (Table 2).
TABLE 2.
Characterization of TnphoA mutants of E. tarda PPD130/91
| Parameter |
E. tarda
straina
|
|||||
|---|---|---|---|---|---|---|
| PPD130/91 | P1 | 153 | 348 | 387 | 392 | |
| Growthb in: | ||||||
| TSA | ++ | + | ++ | ++ | ++ | ++ |
| TSB | ++ | + | ++ | ++ | ++ | ++ |
| PLM | ++ | + | ++ | ++ | ++ | + |
| ROI productionc | + | ++ | ++ | ++ | ++ | ++ |
| Intracellular replication in phagocytesd | ++ | + | ++ | + | ++ | ++ |
| pH sensitivitye | R | S | R | S | R | R |
| Serum resistance | 1.9 ± 0.3 A | 1.3 ± 0.4 A | 1.2 ± 0.7 A | 1.6 ± 0.4 A | 1.6 ± 0.4 A | 1.7 ± 0.4 A |
| LD50 | 105.2 | 106.0 | 105.7 | 105.4 | 106.2 | 105.5 |
| Adherence to EPC cells (%) | 5.2 ± 0.2 A | 4.37 ± 0.4 A | 5.9 ± 0.6 A | 4.6 ± 0.7 A | 3.8 ± 0.2 A | 4.2 ± 0.4 A |
| Internalization into EPC cells (%) | 7.3 ± 0.6 A | 1.9 ± 0.6 B | 3.6 ± 0.8 B | 4.3 ± 1.2 B | 2.6 ± 0.7 B | 2.7 ± 0.5 B |
Within each row, values followed by different letters (A or B) are significantly different (P < 0.05).
+, little growth; ++, significant growth.
++, elevated production of ROI compared to that of the wild type (+).
+, decreased rate of intracellular replication compared to the that of wild type (++).
R and S, resistance and sensitivity to low pH (5.8 ± 0.1), respectively.
The virulence of the mutants was determined by LD50 values for blue gourami injected intramuscularly with different concentrations of bacteria. It was found that all the mutants had relatively higher LD50 levels than the wild type (Table 2). Of the five different mutants, P1 and 387 were less virulent than the wild-type strain, with LD50 values of 106.0 and 106.2, respectively. Southern blotting was also carried out to confirm the presence of TnphoA insertions in the genomic DNA of all the mutants. All five mutants had a single transposon insertion in their genomic DNA (data not shown). The hybridized fragments of all the mutants had a size of more than 8.0 kb. No band was found for the wild-type (PPD130/91) genomic DNA.
Sequence analysis.
Genes interrupted by the TnphoA transposon insertion in the mutants were sequenced (Table 3). Analysis of the gene sequences obtained showed that all the mutants had insertions at different loci. Predicted open reading frames of those sequences were used to search the National Center for Biotechnology Information database for sequences showing homology. Sequence data from mutant P1 showed an 86% similarity to the PhoS of S. enterica serovar Typhimurium (expectation value [E] = 2e−37) over a stretch of 88 amino acids. It also had high similarity (85%) to the periplasmic phosphate binding protein, PstS, of E. coli (9e−3). Transposon insertion in mutant 153 occurred in sequences having 30% identity to putative lipid A core-surface polymer ligase WaaL of Klebsiella pneumoniae. The sequence-flanking transposon in genomic DNA of mutant 392 had 48% homology to that of the dipeptidase of S. enterica serovar Dublin (E = 7e−93) over a stretch of 499 amino acids. All the above mutants had insertions at the amino acid positions 89, 195, and 211, respectively. Two other mutants, 348 and 387, did not have any significant homology to known genes in the GenBank.
TABLE 3.
Sequence analysis of the potential virulence genes interrupted by TnphoA in various transposon mutants of E. tarda PPD130/91
| Mutant (accession no.) | Sequence obtained (bp) | Predicted protein amino acidsa | Homolog protein (homolog accession no.) | Eb | % Identityb |
|---|---|---|---|---|---|
| P1 (AF326581) | 365 | 88+ | Phosphate binding protein (phoS) of S. enterica serovar Typhimurium (AF020805) | 2e − 37 | 86 |
| Periplasmic phosphate binding protein (PstS) of E. coli (L10328) | 9e − 3 | 85 | |||
| 153 (AF326578) | 1,019 | 377 | Lipid A core-surface polymer ligase (WaaL) of K. pneumoniae (AF146532) | 3e − 19 | 30 |
| 348 (AF326580) | 800 | 95 | No homolog | NA | NA |
| 387 (AF326582) | 500 | 90+ | No homolog | NA | NA |
| 392 (AF326579) | 1,551 | 499 | Dipeptidase homolog of S. enterica serovar Dublin (AF060858) | 7e − 93 | 48 |
+, partial sequence of the predicted protein.
NA, not applicable.
DISCUSSION
Interactions of E. tarda with phagocytes.
Adherence, invasion and intracellular replication in host cells such as phagocytes are important for pathogenesis by intracellular pathogens (14). In the present study, we looked into the interaction between E. tarda and fish phagocytes. Adherence count by microscopy, which estimates the total number of viable and dead bacteria adhering to phagocytes, showed large numbers of nonopsonized avirulent E. tarda (PPD125/87 and PPD76/87) organisms binding to and ingested by the phagocytes compared to virulent strain PPD130/91 (Table 1; Fig. 1A and B). Virulent strain AL9379 had significantly higher binding and/or ingestion by phagocytes than PPD130/91, indicating that these two virulent strains interact with phagocytes in different ways. Ling and coworkers (28) also reported a significantly decreased ability of PPD130/91 to adhere to EPC cells than of other three isolates.
Adherence and phagocytosis are influenced by opsonization and cell surface structures of the pathogen (2). Serum has complement proteins, which coat the bacteria, thereby making them easily recognizable by the receptors present on the phagocytes leading to phagocytosis (55). Opsonization will enhance uptake as well as killing by the phagocytes and will simulate the actual bacterium-phagocyte interactions, for better understanding of bacterial pathogenesis. Many researchers have proved the role of the C3-dependent complement mechanism present in fish serum for phagocytosis of pathogens by fish macrophages (31, 35, 40). A similar complement system may be present in gourami serum, enabling phagocytes to recognize E. tarda. Direct microscopic observations showed a significantly higher number of opsonized E. tarda organisms adhering to and also within the phagocytes than the nonopsonized strains, except for PPD130/91 (Fig. 1C and D; Table 1). Interestingly, the adherence assessment carried out by a count of viable bacteria, which provides only a count of live bacteria adhering to/or ingested by phagocytes, showed that only the opsonized virulent strain AL9379 had increased adherence compared to nonopsonized bacteria, while both opsonized avirulent strains PPD125/87 and PPD76/87 had lower percentages of adherence (Table 1). This indicates that the phagocytes may be killing the opsonized avirulent strains. Moreover, PPD76/87 is also serum sensitive (28), contributing to a considerably lower levels of viable bacteria.
The above variation in the number of viable bacteria, after a 30-min infection for adherence to and/or ingestion by phagocytes, led us to study the ability of these bacterial isolates to survive and multiply within fish phagocytes. An intracellular replication assay of nonopsonized virulent and avirulent E. tarda organisms revealed that they could enter, survive, and replicate in phagocytes (Fig. 2A). Similar results were also obtained by Leung and coworkers (26) when phagocytes isolated from tilapia were infected with virulent and avirulent A. hydrophila. The ability of these E. tarda strains to multiply within phagocytes clearly suggests that E. tarda is an intracellular pathogen. Ling and coworkers (28) also demonstrated the ability of these strains to effectively multiply within EPC cells. However, opsonized avirulent E. tarda (PPD125/87 and PPD76/87) lacked the ability to multiply within the phagocytes, while the opsonized virulent strains (PPD130/91 and AL9379) could successfully proliferate (Fig. 2B). Gordon and coworkers (17) have also reported significantly increased binding, intracellular trafficking, and killing of Streptococcus pneumoniae by human alveolar macrophages after opsonization of the bacteria with serum.
Bacterial pathogens may abuse phagocytes to invade and spread within the host system. Therefore, an enhanced uptake by opsonization can actually become harmful to the host if the bacterium is virulent and has special mechanisms to subvert the host macrophage defenses (22, 44). In the present study, only opsonized virulent E. tarda strains (PPD130/91 and AL9379) were able to replicate intracellularly, indicating that they were able to overcome the defense barrier and cause infection. Another fish pathogen, R. salmoninarum, also had the ability to proliferate intracellularly within rainbow trout macrophages upon opsonization (4). Our earlier studies of infection kinetics of E. tarda in vivo with a fish model also showed that only virulent PPD130/91 had a sequential increase in its numbers inside the hematopoietic organs such as the kidney, liver, and spleen, whereas the avirulent PPD125/87 population decreased in the postinfection period within these organs (27). The present in vitro experiments carried out with phagocytes also showed a similar trend when the bacteria were opsonized.
ROI production by phagocytes.
The microbicidal mechanisms within the phagocyte can be broadly classified as oxygen dependent and independent (2, 12, 55). Very little is known about the different mechanisms used by fish phagocytes. However, the oxygen-dependent mechanism has been demonstrated in many different fish, namely tilapia, catfish, blue gourami, rainbow trout, and salmon (3, 4, 26, 56). In the present study, virulent E. tarda strains induced lower rates of ROI production than avirulent strains (Fig. 3), indicating that they have the ability to circumvent the formidable array of antibacterial defenses by failing to trigger the respiratory burst and thereby surviving and replicating inside the phagocyte. Some bacteria such as S. enterica serovar Typhi (32), Brucella abortus (23), Haemophilus somnus (9), Mycobacterium leprae (20), Erysipelothrix rhusiopathiae (43), and R. salmoninarum (3) also failed to trigger the oxidative burst, resulting in successful intracellular survival. In contrast, virulent strains of A. hydrophila increased the chemiluminiscence production (measure of respiratory burst activity) compared to that of avirulent strains (26), showing that they might be using a different strategy to subvert the defense mechanisms of fish.
Several studies have suggested that intracellular pathogens have evolved effective mechanisms for resisting or avoiding phagocytic microbicidal activity, either by inhibiting or neutralizing the production of oxygen metabolites or by preventing interaction with these substances (34, 36, 52). Experiments carried out here revealed that only live E. tarda PPD130/91 could reduce ROI production, compared to heat-killed PPD130/91, while live and heat-killed PPD125/87 did not induce any significant changes in ROI production (Fig. 4). These results indicate that live E. tarda PPD130/91 may simply have failed to induce ROI production by being unrecognizable. The phagocytes, on the other hand, recognize heat-killed PPD130/91, since heat killing may have damaged the cellular characteristics of the bacteria. A higher induction of ROIs by PPD125/87 infection may occur because of the lack of special cell surface characteristics that help to avoid respiratory burst activities. The other possibility may be that virulent E. tarda produces enzymes such as superoxide dismutase (SOD) which quench the reactive oxygen metabolites. Yamada and Wakabayashi (54) have shown that both virulent and avirulent E. tarda strains can produce the SOD enzyme; phylogenetic analysis based on sequences of SOD genes reveals that virulent and avirulent E. tarda strains belong to two separate classes. Other fish pathogens such as Aeromonas salmonicida, R. salmoninarum, and Photobacterium damselae subsp. piscicida are known to resist oxidative killing by producing SOD (5, 6), thereby withstanding the harsh phagocytic environment. Further investigation using isogenic mutants of SOD is required to clarify the role of SOD in pathogenesis.
Transposon mutagenesis and characterization of mutants.
To further understand the genetic mechanism involved, 200 PhoA+ mutants of PPD130/91(Neor Colr) were generated by transposon mutagenesis. TnphoA mutants of E. tarda PPD130/91 were screened for their ability to induce higher ROI production by phagocytes than the wild-type PPD130/91. Five of the mutants (P1, 153, 348, 387, and 392) had the ability to induce a higher level of ROIs than the wild-type strain (Fig. 4). Mutants P1 and 348 were pleiotrophic in nature, having a lower replication ability inside phagocytes and also a sensitivity to lower pH (Table 2). The other three mutants were comparable to the wild-type strain with respect to all the characteristics except for ROI production and internalization in EPC cells.
Mutant P1 had transposon insertion in the gene homologous to the phoS (pstS) gene of S. enterica serovar Typhimurium, which is shown to be a macrophage-inducible gene (48). Mutant P1 showed a significant increase in ROI production by phagocytes compared to the wild type. Daigle and coworkers (10) found that insertion of TnphoA in the pstC gene of E. coli could cause changes in the production of surface polysaccharides of the bacteria. Since the pstC gene is present in the downstream of pstS gene as part of the pstSCAB-phoU operon, insertion of transposon in the pstS gene causes changes in the production of surface polysaccharides and enables the phagocytes to recognize mutant P1 and trigger a higher production of ROI. In the present study, we also noticed that mutant P1 was growth deficient on TSA, in TSB, and also in PLM (Table 2), indicating that Pi uptake and assimilation is an important process for its growth. Furthermore, a lower Pi inside the phagocytes would have reduced mutant P1's ability to multiply efficiently within them. Lucas and coworkers (30) also suggested that low-Pi conditions could directly or indirectly repress the functions of some invasion genes in S. enterica serovar Typhimurium. Mutant P1 also showed significantly lower internalization into EPC cells than the wild type. Since the pstSCAB-phoU operon in other bacteria such as E. coli is a member of the Pho regulon, which is involved in phosphate uptake and assimilation, it may control various functions rendering mutant P1 pleiotrophic. This may also control the expression of some important factors that are involved in the degradation of ROI, thus causing an increase in ROI production when phagocytes were infected with this mutant.
Mutant 153 had an insertion in the gene having homology to the putative lipid A core-surface polymer ligase WaaL of K. pneumoniae. This gene is known to play a major role in the biosynthesis of lipopolysaccharide (LPS); any mutation in this gene may disrupt LPS synthesis, thereby changing the bacterial cell surface structure. A sequence analysis of mutant 392 showed high similarity to dipeptidase homolog of the S. enterica serovar Dublin. The dipeptidase gene localizes in the periplasmic space and plays a role in peptidoglycan metabolism in Salmonella (18). Outer membrane proteins, peptidoglycan, and LPSs are on the bacterial cell surface and often aid in enhancement or prevention of host immune system activation. Several cell surface factors such as the outer membrane protein (porin) of Neisseria gonorrhoeae (29), capsule-deficient mutants of E. rhusiopathiae (43), aggregation substance of Enterococcus faecalis (45), melanin pigment of Burkholderia cepacia (57), and LPS and lipid A of B. abortus (38) have failed to either induce or suppress oxidative bursts of phagocytes.
In conclusion, insertions of transposons in the phoS (mutant P1), lipid A (mutant 153) and didpeptidase (mutant 392) genes would have altered the cell surface properties of E. tarda or the biosynthesis of a second substance that degraded the ROIs. These changes could have made these mutants readily recognizable to the phagocytes, thus increasing ROI production. Sequence analyses of mutants 348 and 387 did not yield any homology to known genes in GenBank. These may be novel genes controlling some of the important functions of E. tarda in overcoming phagocyte-mediated killing. The present study clearly demonstrated the ability of virulent E. tarda to adhere to and survive and replicate within the blue gourami phagocytes. Virulent E. tarda failed to induce oxidative bursts, indicating that this may be one of the mechanisms used by them to overcome phagocytic killing. We have also showed that TnphoA transposon tagging can be effectively used to “fish out” genes responsible for virulence in E. tarda. The information elucidated here will help in the development of suitable strategies to combat the disease caused by E. tarda.
ACKNOWLEDGMENTS
We are grateful to National University of Singapore for providing the research grant for this work.
We are thankful to John Grizzle from Auburn University for providing us with E. tarda AL9379 and T. Ngiam and H. Loh from Agri-food and the Veterinary Authority of Singapore for providing us with the rest of the E. tarda isolates. We also acknowledge J. A. Matthew, who carried out the mutagenesis of E. tarda PPD130/91.
REFERENCES
- 1.Ainsworth A J, Chen D X. Differences in the phagocytosis of four bacteria by channel catfish neutrophils. Dev Comp Immunol. 1990;14:201–209. doi: 10.1016/0145-305x(90)90091-r. [DOI] [PubMed] [Google Scholar]
- 2.Auger M J, Ross J A. The biology of macrophage. In: Lewis C E, McGee O D, editors. The macrophage: the natural immune system. Oxford, England: Oxford University Press; 1991. pp. 215–263. [Google Scholar]
- 3.Bandin I, Ellis A E, Barja J L, Secombs C J. Interaction between rainbow trout macrophages and Renibacterium salmoninarum in vitro. Fish Shellfish Immunol. 1993;3:25–33. [Google Scholar]
- 4.Bandin I, Rivas C, Santos Y, Secombs C J, Barja J C, Ellis A E. Effect of serum factors on the survival of Renibacterium salmoninarumwithin rainbow trout macrophages. Dis Aquat Org. 1995;23:221–227. [Google Scholar]
- 5.Barnes A C, Balebona M C, Horne M T, Ellis A E. Super oxide dismutase and catalase in Photobacterium damselae subsp. piscicidaand their roles in resistance to reactive oxygen species. Microbiology. 1999;145:483–494. doi: 10.1099/13500872-145-2-483. [DOI] [PubMed] [Google Scholar]
- 6.Barnes A C, Horne M T, Ellis A E. Effect of iron on expression of superoxide dismutase by Aeromonas salmonicidaand associated resistance to superoxide anion. FEMS Microbiol Lett. 1996;142:19–26. [Google Scholar]
- 7.Blazer V S. Piscine macrophage function and nutritional influences: a review. J Aquat Anim Health. 1991;3:77–86. [Google Scholar]
- 8.Collins L, Thune R L. Development of a defined minimal medium for the growth of Edwardsiella ictaluri. Appl Environ Microbiol. 1996;62:848–852. doi: 10.1128/aem.62.3.848-852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czuprynski, C. J., and H. L. Hamilton. 1985. Bovine neutrophils ingest but not kill Haemophilus somnus in vitro. 50:431–436. [DOI] [PMC free article] [PubMed]
- 10.Daigle F, Fairbrother J M, Harel J. Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia colistrain causing septicemia in pigs. Infect Immun. 1995;63:4924–4927. doi: 10.1128/iai.63.12.4924-4927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalmo R A, Ingebrigtsen K, Bogwald J. Non-specific defence mechanisms in fish, with particular reference to the reticuloendothelial system. J Fish Dis. 1997;20:241–273. [Google Scholar]
- 12.Ellis A E. Immunity to bacteria in fish. Fish Shellfish Immunol. 1999;9:291–308. [Google Scholar]
- 13.Elsinghorst E A. Measurement of invasion by gentamicin resistance. Methods Enzymol. 1994;236:405–420. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- 14.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster J W, Spector M P. How Salmonellasurvive against odds. Annu Rev Microbiol. 1995;49:145–174. doi: 10.1146/annurev.mi.49.100195.001045. [DOI] [PubMed] [Google Scholar]
- 16.Foster J W, Hall H K. Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol. 1990;172:771–778. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon S B, Irving G R B, Lawson R A, Lee M E, Read R C. Intracellular trafficking and killing of Streptococcus pneumoniaeby human alveolar macrophages are influenced by opsonins. Infect Immun. 2000;68:2286–2293. doi: 10.1128/iai.68.4.2286-2293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilbert F, del Portillo F G, Groisman E A. A periplasmic d-alanyl-d-alanine dipeptidase in the gram-negative bacterium Salmonella enterica. J Bacteriol. 1999;181:2158–2165. doi: 10.1128/jb.181.7.2158-2165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirono I, Tange N, Aoki T. Iron regulated hemolysin gene from Edwardsiella tarda. Mol Microbiol. 1997;24:851–856. doi: 10.1046/j.1365-2958.1997.3971760.x. [DOI] [PubMed] [Google Scholar]
- 20.Holzer T J, Nelson K E, Schauf V, Crispen R G, Andersen R. Mycobacterium lepraefails to stimulate phagocytic cell super oxide anion generation. Infect Immun. 1986;51:514–520. doi: 10.1128/iai.51.2.514-520.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janda J M, Abbott S L, Kroske-Bystrom S, Cheung W K W, Powers C, Kokka R P, Tamura K. Pathogenic properties of Edwardsiellaspecies. J Clin Microbiol. 1991;29:1997–2001. doi: 10.1128/jcm.29.9.1997-2001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufmann S H E, Reddehase M J. Infection of phagocytic cells. Curr Opin Immun. 1989;2:43–49. doi: 10.1016/0952-7915(89)90096-4. [DOI] [PubMed] [Google Scholar]
- 23.Kruetzer D L, Dreyfus L A, Robertson D C. Interaction of polymorphonuclear leukocytes with smooth and rough strains of Brucella abortis. Infect Immun. 1979;23:737–742. doi: 10.1128/iai.23.3.737-742.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung K Y, Finlay B B. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung K Y, Yeap I V, Lam T J, Sin Y M. Serum resistance is a good indicator of virulence in Aeromonas hydrophilastrains isolated from diseased fish in South-East Asia. J Fish Dis. 1995;18:511–518. [Google Scholar]
- 26.Leung K Y, Low K W, Lam T J, Sin Y M. Interaction of fish pathogen Aeromonas hydrophila with tilapia, Oreochromis aureus(Steindachner), phagocytes. J Fish Dis. 1995;18:435–447. [Google Scholar]
- 27.Ling S H M, Wang X H, Lim T M, Leung K Y. Green fluorescent protein-tagged Edwardsiella tardareveals portal of entry in fish. FEMS Microbiol Lett. 2001;194:239–243. doi: 10.1111/j.1574-6968.2001.tb09476.x. [DOI] [PubMed] [Google Scholar]
- 28.Ling S H M, Wang X H, Xie L, Lim T M, Leung K Y. Use of green flourescent protein (GFP) to track the invasion pathways of Edwardsiella tarda in in vivo and in vitrofish models. Microbiology. 2000;146:7–19. doi: 10.1099/00221287-146-1-7. [DOI] [PubMed] [Google Scholar]
- 29.Lorenzen D R, Gunther D, Pandit J, Rudel T, Brandt E, Meyer T F. Neisseria gonorrhoeaeporin modifies the oxidative burst of professional phagocytes. Infect Immun. 2000;68:6215–6222. doi: 10.1128/iai.68.11.6215-6222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucas R L, Lostroh C P, Dirusso C C, Spector M P, Wanner B L, Lee C A. Multiple factors independently regulate hilA and invasion gene expression in Salmonella entericaserovar Typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama H, Yano T, Yamakawa T, Nakao M. Opsonic effect of the third complement component (C3) of carp (Cyprinus carpio) on phagocytosis by neutrophils. Fish Shellfish Immun. 1992;2:69–78. [Google Scholar]
- 32.Miller R M, Garbus J, Hornick R B. Lack of enhanced oxygen consumption by polymorphonuclear leukocytes on phagocytosis of virulent Salmonella typhi. Science. 1972;175:1010–1011. doi: 10.1126/science.175.4025.1010. [DOI] [PubMed] [Google Scholar]
- 33.Miller R A, Britigan B E. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathan C F, Nakagawara A. Role of reactive oxygen intermediates in macrophage killing of intracellular pathogens: a review. In: Mizuna D, Cohn Z A, Takeya K, Ishida N, editors. Self defense mechanisms: role of macrophages. Tokyo, Japan: Elsevier Biomedical Press; 1981. pp. 279–291. [Google Scholar]
- 35.Nonaka M, Iwaki M, Nakai C, Nozaki M, Kaidoh T, Nonaka M, Natsume-Sakai S, Takahashi M. Purification of a major serum protein of rainbow trout (Salmo gairdneri) homologus to the third component of mammalian complement. J Biol Chem. 1984;259:6327–6333. [PubMed] [Google Scholar]
- 36.Parson R D, Harcus J L, Symes P H, Romito R, Donowitz G R. Failure of the phagocytic oxidative response to protect monocyte derived macrophages from infection by Leishmania donovani. J Immunol. 1982;129:1282–1286. [PubMed] [Google Scholar]
- 37.Plumb J A. Edwardsiella septicaemia. In: Inglis V, Roberts R J, Bromage N R, editors. Bacterial diseases of fish. Oxford, England: Blackwell Scientific; 1993. pp. 61–79. [Google Scholar]
- 38.Rasool O, Freer E, Moreno E, Jarstarand C. Effect of Brucella abortuslipopolysaccharide on oxidative metabolism and lysozyme release by human neutrophils. Infect Immun. 1992;60:1699–1702. doi: 10.1128/iai.60.4.1699-1702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed L J, Muench H. A simple method of estimating fifty percent end points. Am J Hygiene. 1938;27:493–497. [Google Scholar]
- 40.Rose A S, Levin R P. Complement-mediated opsonization and phagocytosis of Renibacterium salmoninarum. Fish Shellfish Immun. 1992;2:223–240. [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Secombs C J. Isolation of salmonid macrophages and analysis of their killing activity. In: Stolen J S, Fletcher T C, Anderson D P, Roberson B S, Van Muiswinkel W B, editors. Techniques in fish immunology. Fair Haven, N.J: SOS Publications; 1990. pp. 137–154. [Google Scholar]
- 43.Shimoji Y, Yokomizo Y, Mori Y. Intracellular survival and replication of Erysipelothrix rhusiopathiaewithin murine macrophages: failure of induction of the oxidative burst of macrophages. Infect Immun. 1996;64:1789–1793. doi: 10.1128/iai.64.5.1789-1793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Speert D P. Macrophages in bacterial infection. In: Lewis C E, McGee O D, editors. The macrophage: the natural immune system. Oxford, England: Oxford University Press; 1991. pp. 215–263. [Google Scholar]
- 45.Submuth D S, Silberhorn A M, Wirth R, Susa M, Marre R, Rozszinski E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecaliswithin human macrophages and suppresses respiratory burst. Infect Immun. 2000;68:4900–4906. doi: 10.1128/iai.68.9.4900-4906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thune R L, Stanley L A, Cooper R K. Pathogenesis of gram-negative bacterial infections in warm water fish. Annu Rev Fish Dis. 1993;3:37–68. [Google Scholar]
- 47.Ullah M A, Arai T. Pathological activities of the naturally occurring strains of Edwardsiella tarda. Fish Pathol. 1983;18:65–70. [Google Scholar]
- 48.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 49.Van Damme L R, Vandepitte J. Isolation of Edwardsiella tarda and Plesiomonas shigelloidesfrom mammals and birds in Zaire. Rev Elev Med Vet Pays Trop. 1984;37:145–151. [PubMed] [Google Scholar]
- 50.Wang X H, Oon H L, Ho G W P, Wong W S F, Lim T M, Leung K Y. Internalization and cytotoxicity are important virulence mechanisms in Vibriofish epithelial cell interactions. Microbiology. 1998;144:2987–3002. doi: 10.1099/00221287-144-11-2987. [DOI] [PubMed] [Google Scholar]
- 51.White F H, Simpson C F, Williams L E. Isolation of aquatic animal species and surface waters in Florida. J Wildl Dis. 1973;9:204–207. doi: 10.7589/0090-3558-9.3.204. [DOI] [PubMed] [Google Scholar]
- 52.Wilson C B V, Tsai V, Remington J. Failure to trigger the oxidative metabolic burst by normal macrophages. Possible mechanism for survival of intracellular pathogens. J Exp Med. 1980;151:328–346. doi: 10.1084/jem.151.2.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X Q, Pan S Q. An Agrobacteriumcatalase is a virulence factor involved in tumorigenesis. Mol Microbiol. 2000;35:407–414. doi: 10.1046/j.1365-2958.2000.01709.x. [DOI] [PubMed] [Google Scholar]
- 54.Yamada Y, Wakabayashi H. Identification of fish pathogenic strains belonging to genus Edwardsiella by sequence analysis of sodB. Fish Pathol. 1999;3:145–150. [Google Scholar]
- 55.Yano T. The nonspecific immune system: humoral defense. In: Iwama G K, Nakanishi T, editors. The fish immune system: organism, pathogen, and environment. New York, N.Y: Academic Press; 1990. pp. 105–157. [Google Scholar]
- 56.Yin Z, Lam T J, Sin Y M. Cytokine-mediated antimicrobial immune response of catfish, Clarias gariepinus, as a defence against Aeromonas hydrophila. Fish Shellfish Immunol. 1997;7:93–104. [Google Scholar]
- 57.Zughaier S M, Ryley H C, Jackson S K. A melanin pigment purified from an epidemic strain of Burkholderia cepaciaattenuates monocyte respiratory burst activity by scavenging superoxide anion. Infect Immun. 1999;67:908–913. doi: 10.1128/iai.67.2.908-913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]