Abstract
Probiotics exert anti-obesity effects in high-fat diet (HFD) obese mice, but there are few studies on anti-obesity using heat-killed probiotics. Here, we investigated the effect of heat-killed Lactiplantibacillus plantarum K8 (K8HK) on the anti-differentiation of 3T3-L1 preadipocytes and on anti-obesity in HFD mice. K8HK decreased triglyceride (TG) accumulation in 3T3-L1 cells. Specifically, 1 × 109 CFU/mL K8HK showed the greatest anti-obesity effect, while the same concentration of live L. plantarum K8 (K8 Live) showed cytotoxicity. K8HK increased suppressor of cytokine signaling (SOCS)-1, which might affect the JAK2-STAT3 signaling pathway activated during differentiation. As a result, the levels of transcription factors of adipogenesis such as Peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer binding protein α (C/EBPα) decreased in K8HK-treated cells. We also observed a decrease in the lipogenic enzymes and fatty acid binding protein 4 (FABP4). In the mouse study, oral ingestion of K8 Live and K8HK showed weight reduction and decrease in blood TG content at 12 weeks of feeding. In addition, TG synthesis was suppressed in liver and adipose tissues, and genes related to fat metabolism were suppressed. This study suggests that K8HK could be a good material to prevent obesity by inhibiting adipogenesis genes related to fat metabolism.
Keywords: Anti-obesity, Lactiplantibacillus plantarum K8, Heat-killed, Negative regulators, 3T3-L1, High-fat diet
1. Introduction
Obesity refers to a condition in which metabolic disorders are induced due to an increase in excessive body fat and causes complex diseases such as cardiovascular diseases, diabetes, high blood pressure, and certain cancers [1,2,3,4]. Obesity is generally caused by eating too much and moving too little. Consuming high amounts of energy, particularly fat and sugars, but the energy is not consumed through proper physical activity, the body stores the surplus energy in the form of fat [5]. In addition to environmental factors, the pathophysiology of obesity involves the interaction of several factors, such as changes in central nervous system (CNS)-endocrine signaling, socioeconomic, genetic, and intrinsic factors. Alterations in the gut microbiota also play an important role in the pathophysiology of obesity [6]. Microorganisms in the human gut affect the absorption, breakdown and storage of nutrients and have potential effects on host physiology [7]. Imbalance of the gut microbiome, caused by dietary or environmental changes, promote overgrowth of pathogenic organisms that may play an important role in the pathology of obesity [8]. In contrast, a healthy balance of gut microbiota may play a role in alleviating or preventing obesity [9,10].
Probiotics have physiological functions to control obesity. For example, probiotics modulate mechanisms for body weight, gut microbiome, and inflammation [11]. Wang et al. examined five lactobacillus strains including L. casei NCU011054 group, L. fermrntum NCU0413, L. acidophilus NCU433, L. rhamnosus NCU2217, and L. paracasei NCU622 with a dosage of 108 colony forming units (CFU)/mL. All lactobacillus strains alleviated obesity in HFD mice through inhibition of fat accumulation and lipid metabolism and regulation of the contents of leptin and adiponectin [12]. Lactobacillus strains have been known to help with intestinal health. In particular, L. plantarum regulates the balance of gut microbiota [13]. L. plantarum K8 that was isolated from Kimchi, a traditional Korean fermented food, is a representative probiotic strain and has been shown to be effective in alleviating various diseases. L. plantarum K8 was isolated from the Korean traditional fermented food Kimchi and we previously demonstrated the effects of L. plantarum K8, L. plantarum K8 lysates, and lipoteichoic acid (LTA) isolated from L. plantarum K8 cell wall on immune regulation and skin health [14,15,16,17]. Both lysate and LTA contribute to the maintenance of homeostasis of the immune system by reducing the production of inflammatory cytokines [18,19]. Although there are many anti-obesity research papers using lactobacillus, there are insufficient studies on heat-treated lactobacillus. In the current study, we examined the effect of heat-killed L. plantarum K8 (K8HK) on the differentiation of 3T3-L1 preadipocytes and fat accumulation by measuring the levels of adipose-related genes and intracellular triglyceride (TG). Furthermore, the anti-obesity effect of live L. plantarum K8 (K8 Live) and K8HK was examined in mice induced to obesity by high-fat diet (HFD).
2. Materials and methods
2.1. Preparation of K8 Live and K8HK
L. plantarum K8 (KCTC 10887BP) isolated from Kimchi, a traditional Korean fermented food, was cultured in MRS broth (BD Biosciences, CA, San Jose, USA) at 37 °C overnight. Cells were centrifuged at 8000 rpm for 8 min, and the pellet was washed with Dulbecco’s Phosphate-Buffered Saline (D-PBS). K8 Live was prepared by re-suspending in D-PBS at a concentration of 1011 colony forming units (CFU)/mL for subsequent experiments. The number of CFU per OD per mL is estimated by multiplying colony count by dilution multiple. K8HK was prepared based on the method described previously [20]. After dissolving 1011 CFU/mL K8 Live in DPBS, it was boiled in a water bath at 80 °C for 20 min and then cooled at room temperature (RT). A portion of the prepared K8HK was spread on the MRS plate to confirm that all bacteria were killed.
2.2. In vitro studies
2.2.1. 3T3-L1 cell culture and differentiation
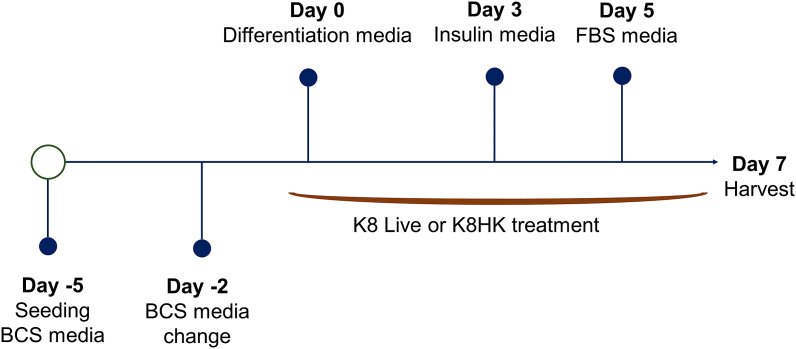
3T3-L1 cells, a mouse pre-adipocyte cell line, were maintained in Dulbecco′s Modified Eagle′s Medium (DMEM) with 10% heat-inactivated bovine calf serum (BCS) and 100 μg/mL penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator. Cells were cultivated onto 6-well plates and maintained in BCS media for 5 days. At day 0, 3T3-L1 cells were added to the first differentiation medium (DMEM, 10% FBS, 1 μM Dexamethasone, 517 μM 3-Isobuthyl-1-methylxanthine, 5 μg/mL Insulin). After 72 h, the cells were grown in DMEM containing 10% fetal bovine serum (FBS) supplemented with insulin (5 μg/mL). After additional incubation for 48 h, the cells were grown in DMEM containing 10% FBS. Cells were harvested for analysis at day 7. K8 Live and K8HK were treated by dilution in an appropriate medium on days 0, 2, 4, and 6, which is the period of media replacement (Fig. 1).
Fig. 1.
3T3-L1 cell differentiation and K8 Live or K8HK treatment process.
2.2.2. Cell viability assay
K8 Live and K8HK were prepared in DPBS solution at a concentration of 109 CFU/mL 3T3-L1 cells were examined with increasing concentrations of K8 Live and K8HK for 7 days during differentiation. Cell viability was measured by EZ-Cytox cell viability assay kit (Daeil Lab Service, Cheongwon, Korea) according to the manufacturer’s instructions. Absorbance was detected at a wavelength of 590 nm using a microplate reader EL800 (Biophotometer, Eppendorf, Hamburg, Germany).
2.2.3. Oil red O staining
After differentiation of 3T3-L1 cells treated with K8 Live or K8HK, cells were washed with PBS and fixed with 3.7% formaldehyde solution for 20 min at RT. The formaldehyde was removed by washing with DW, and the cells were dried at RT. Cells were stained with Oil red O for 20 min at RT and washed 5 times with DW. Stained images for each dish were captured using a Ti-E Inverted Fluorescence Microscope (Nikon, NY, USA). After DW was removed from each well and dried completely, 500 μL isopropanol was added. After 10 min, absorbance was detected at a wavelength of 520 nm using a microplate reader EL800 (Biophotometer, Eppendorf) to examine the lipid contents extracted from Oil red O-stained cells.
2.2.4. Quantification of TG content
A TG-S kit (Asanpharm, Seoul, Korea) was used for quantification of TG content according to the manufacturer’s instructions. 3T3-L1 cells were treated with K8HK for 7 days during differentiation. After cells were washed with PBS, 200 μL hypotonic buffer (20 mM Tris (pH 7.5), 5 mM MgCl2, 5 mM CaCl2, 1 mM DTT, 1 mM EDTA, 0.1% Triton X-100) was added. Supernatant was mixed with reagent from the TG-S kit, and absorbance was detected at a wavelength of 550 nm using a microplate reader EL800 (Biophotometer, Eppendorf).
2.3. mRNA and protein analysis
2.3.1. Real-time polymerase chain reaction (PCR)
For real-time PCR, total RNA was extracted from 3T3-L1 cells or white adipose tissue (WAT) and liver extracts using RNAiso Plus (TaKaRa Bio Inc., Shiga, Japan), following the manufacturer’s instructions. cDNA was synthesized using a PrimeScript™ RT Master Mix (TaKaRa) according to the manufacturer’s instructions. Real-time PCR was performed using CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) and SYBR Premix Ex Taq™ II (TaKaRa). The primer sets used in this study were listed in Table 1. The target gene expression was normalized to that of mGAPDH.
Table 1.
Primer sequences used in this study.
| Genes | Sequences 5′ to 3′ | References | |
|---|---|---|---|
| C/EBPα | Forward | TTACAACAGGCCAGGTTTCC | In this study |
| Reverse | GGCTGGCGACATACAGTACA | In this study | |
| PPAR-γ | Forward | TTTTCAAGGGTGCCAGTTTC | In this study |
| Reverse | AATCCTTGGCCCTCTGAGAT | In this study | |
| FABP4 | Forward | ATGATCATCAGCGTAAATGG | In this study |
| Reverse | TTGCTGGCACTACAGAATGC | In this study | |
| FAS | Forward | TTGCTGGCACTACAGAATGC | In this study |
| Reverse | AACAGCCTCAGAGCGACAAT | In this study | |
| SCD-1 | Forward | CATCGCCTGCTCTACCCTTT | In this study |
| Reverse | GAACTGCGCTTGGAAACCTG | In this study | |
| CPT-1α | Forward | CCAGGCTACAGTGGGACATT | In this study |
| Reverse | GAAGAGCCGAGTCATGGAAG | In this study | |
| PGC1α | Forward | GTCAACAGCAAAAGCCACAA | In this study |
| Reverse | GTGTGAGGAGGGTCATCGTT | In this study | |
| ACC | Forward | GCCTCTTCCTGACAAACGAG | In this study |
| Reverse | TAAGGACTGTGCCTGGAACC | In this study | |
| GAPDH | Forward | TGCTGACAATCTTGAGTGAG | In this study |
| Reverse | GTCGTGGAGTCTACTGGTGT | In this study |
2.3.2. Western blot analysis
3T3-L1 cells were lysed with 2x Laemmli sample buffer and boiled at 100 °C for 5 min. After centrifuging (12,000 rpm, 10 min) at 4 °C, the supernatants were collected. Samples were separated with 10% (w/v) or 12% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a Glycine/Tris/SDS buffer (25 mM Tris, 250 mM Glycine, 0.1% SDS). Proteins were transferred onto polyvinylidene fluoride (PVDF) membranes for 2 h at 100 V. The membranes were blocked with blocking buffer (20 mM Tris-HCl, 150 mM NaCl, 0.05% Tween-20, 5% skim milk) for 2 h at RT and washed 3 times with TBS-T (20 mM Tris-HCl, 150 mM NaCl, 0.05% Tween-20). The membrane was treated with primary antibodies of CCAAT/enhancer binding protein α (C/EBPα) (sc-365318), suppressor of cytokine signaling (SOCS)-1 (sc-518018), β-actin (sc-47778), p-STAT3 (sc-8059) (Santa Cruz Biotechnology, Dallas, TX, USA), Peroxisome proliferator-activated receptor γ (PPAR-γ) (ab45036), fatty acid binding protein 4 (FABP4) (ab92501) (AbCam, Cambridge, England), and phospho-janus kinase (JAK) 2 (Tyr1007/1008, Cell Signaling Technology, Danvers, MA, USA) diluted with TBS-T for 2 h at RT and then washed 3 times with TBS-T. The membrane was treated with secondary antibodies of HRP-conjugated anti-rabbit (sc-2357) and anti-mouse (sc-47778, Santa Cruz Biotechnology) diluted in TBS-T for 2 h at RT and then washed 5 times with TBS-T. The membrane was treated with enhanced chemiluminescence (ECL) reagent and exposed to X-ray film to detect target protein band. The expression of -actin was used as the internal loading control.
2.4. Animal handling and ethical approval
The research protocol has been registered at https://preclinicaltrials.eu (International Register of Preclinical Trials Protocols) with No. PCTE0000338. The mice were cared for and used in accordance with guidelines of the animal ethics committee of Kyung Hee University. Specific approval for the mouse experiments was obtained for protocol (KHUASP(GC)-18-033, 2018) by the Department of Laboratory Animal, Institutional Animal Care and Use Committee at Global campus of Kyung Hee University (Yongin, Korea). Male C57BL/6 (n = 24), 6 weeks old, were obtained from Narabiotech (Seoul, Korea). The number of mice required for the experiment was calculated through G power (http://www.biomath.info/power/index.html). Mice were cared in individual cages at 24 ± 2 °C and 50 ± 10% moisture condition, and were stabilized by supplying nutritionally balanced rodent food (Central Lab. Animal Inc. Korea) and sterilized water. All reasonable efforts were made to ameliorate suffering, including use of anesthesia for painful procedures. During the experimental period, six mice were fed a normal diet (Narabiotech) that provided 66% of energy as carbohydrates, 20.65% as proteins, and 5.77% as fats. Six mice were fed for 10 weeks with a high-fat diet (Narabiotech) that provided 60% of energy as fats, 20% as proteins, and 20% as carbohydrates. Six mice per group were fed K8 Live and K8HK (1 × 1010 CFU/mouse) starting from 2 weeks before initiating the high-fat diet. K8 Live and K8HK dissolved in DPBS were administered orally to mice using an oral gavage needle (2.5 cm/24 G) and cared for over a total of 12 weeks. Changes in body weight of mice were measured at 1-week intervals.
2.5. In vivo studies of K8 Live and K8HK on anti-obesity effect
2.5.1. Quantification of TG content in mouse serum
Whole blood was collected under anesthesia from mice through orbital sinus. After collection of the whole blood, it was allowed to clot by leaving it undisturbed at RT for 30 min. Serum was collected after centrifugation (12,000 rpm 30 min). The TG-S kit (Asanpharm) was used for quantification of mouse serum TG content according to the manufacturer’s instruction.
2.5.2. Isolation of WAT and liver
Mice were sacrificed by exposure to 6% CO2 for 10 min. WAT and liver collection was performed as described previously [21,22]. After dissect out the entire white adipose tissue (WAT) and liver from mice, 100–500 mg of tissue was covered with >10 vol of 10% formalin to tissue and left at 4 °C for a minimum of 72 h. The extracted WAT and liver were photographed and weighed individually. For mRNA quantitative analysis, 2–3 small pieces of tissue was cut from different WAT and liver lobes and dissolved in RNAiso Plus (TaKaRa).
2.5.3. mRNA and protein expression in white adipose tissue (WAT) and livers
After extracting WAT and liver, a portion of tissue was cut and dissolved in RNAiso Plus (TaKaRa) for mRNA quantitative analysis. Tissues were homogenized and stored in deep freezer for one day. Real-time PCR was performed to measure mRNA levels. For protein quantitative analysis, WATs and livers were homogenized in PRO-PREPTM (iNtRON Biotech, Korea). After adding the protease inhibitor, homogenized tissue samples were centrifuged for 10 min at 14,000 rpm. Supernatant was collected and the protein was quantified through Bradford assay. Supernatant was dissolved in 2x Laemmli sample buffer, and SDS-PAGE and Western blot analysis were performed.
2.5.4. Hematoxylin and eosin (H&E) staining of WAT
H&E staining of mouse WAT was performed as described previously [23]. After WAT was isolated, tissues were fixed in 10% buffered formalin and embedded in paraffin wax. The paraffin-embedded tissues were sectioned to a thickness of 5 μm. WAT tissues were then stained with hematoxylin nd eosin. Representative images of WAT were shown.
2.5.5. Oil red O-staining of liver tissue
Liver sections were perfused with PBS for 10 min and fixed in 4% paraformaldehyde for 5 min. The isolated livers were embedded in Tissue-Tek OCT compound and frozen at −70 °C. All samples were sectioned using a cryostat at −20 °C, and six consecutive 5 μm thick sections were cut from the tissues. Sections were stained with Oil red O and counterstained with hematoxylin. To visualize the targets in the lesion, fluorescence-labeled antibodies were used and observed at an excitation of 488 nm.
2.6. Statistical analysis
All the experiments were performed at least three times (triplicates). The data shown are representative results as mean ± SD. Statistical analyses were conducted with an unpaired two-tailed t-test, one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) post hoc test, or two-way ANOVA. The differences were considered significant at p < 0.05. GraphPad Prism 5 software (Ver. 5.01) was used for the analysis (GraphPad Software, Inc., San Diego, CA, USA).
3. Results
3.1. Cytotoxicity effects of K8 Live and K8HK in 3T3-L1 adipocytes
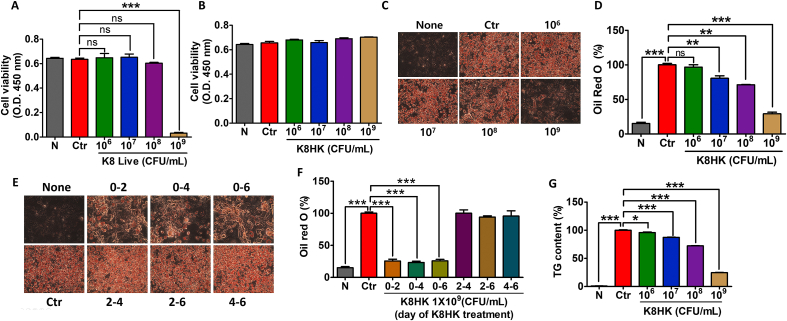
To examine the cytotoxicity of L. plantarum K8 on 3T3-L1 adipocytes, K8 Live and K8HK were applied during 3T3-L1 differentiation at concentrations of 1 × 106, 1 × 107, 1 × 108, and 1 × 109 CFU/mL. Cell viability decreased when treated with 1 × 109 CFU/mL K8 Live (Fig. 2A). However, no cytotoxicity was observed after treatment with 1 × 109 CFU/mL K8HK (Fig. 2B). The difference in cytotoxicity between K8 Live and K8HK is thought to be caused by postbiotics possessed by the K8 Live or by genes in 3T3-L1 cells that are affected by these substances. Transcript changes in live and dead cells of L. plantarum K8 and changes in the transcriptome of 3T3-L1 cells treated with K8 Live and K8HK were investigated (SupplementaryData). There was a difference in apoptosis-related gene expression of 3T3-L1cells by K8HK compared to K8 Live, suggesting that some live lactic acid bacteria can induce cell death due to unknown action or factors. To exclude cytotoxicity, subsequent experiments were performed with 1 × 109 CFU/mL K8HK.
Fig. 2.
K8HK inhibited intracellular lipid accumulation in 3T3-L1 adipocytes. 3T3-L1 adipocytes were treated with (A) live L. plantarum K8 (K8 Live) during differentiation and (B) heat-killed L. plantarum K8 (K8HK), and cell viability was examined by WST-1 assay. (C) Cells were stained with Oil red O after K8HK treatment during differentiation (day 0 to day 7). (D) Graph showing lipid accumulation in 3T3-L1 cells measured with Oil red O. (E) Cells were treated with appropriate media containing 1 × 109 K8HK on days 0–2, 0–4, 0–6, 2–4, 2–6, or 4–6. Control cells) were not treated with K8HK. Cells were stained with Oil red O. (F) The quantity of lipids stained by Oil red O was measured at 492 nm. (G) Triglyceride (TG) content was measured after treatment with 1 × 109 K8HK during differentiation. The data displayed are the mean ± SD. Statistical analysis was conducted with unpaired two-tailed t-test (A) or one-way ANOVA (D, F, G). *p < 0.05, **p < 0.01, ***p < 0.001. N, untreated cells; Ctr, differentiated positive control cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. K8HK inhibited intracellular lipid accumulation in 3T3-L1 adipocytes
To examine the intracellular TG content, cells were stained with Oil red O, and the amount of lipid was visualized (Fig. 2C). The amount of TG accumulation in untreated cells (None) was about 15% compared to that of the positive control (Ctr), which induced by differentiation media. When K8HK was applied, the amount of TG accumulation decreased in a dose-dependent manner. At 1 × 109 CFU/mL concentration, TG accumulation was reduced by 30% compared to the positive control (Fig. 2D). The red droplets in the cells decreased as the concentration of K8HK increased, suggesting that K8HK inhibited intracellular accumulation of lipid in a dose-dependent manner.
The relationship between TG accumulation and timing of K8HK treatment during differentiation was investigated. Compared to the control group in which only differentiation was induced without K8HK treatment, intracellular TG accumulation was significantly reduced when cells were treated with a differentiation medium containing K8HK for 2, 4, and 6 days, respectively, after the start of differentiation induction (day 0). On the other hand, TG accumulation did not decrease when K8HK was treated between 2 and 4 days, 2–6 days, and 4–6 days after the start of differentiation induction (Fig. 2E). Quantifying red droplets verified reduced TG accumulation only when K8HK was applied at the start of differentiation (Fig. 2F). These data suggest that K8HK is involved in the early phase of the differentiation period. As in Oil Red O staining, the TG content increased by the control was decreased in a K8HK-dose-dependent manner (Fig. 2G). Specifically, when 1 × 109 CFU/mL K8HK was applied, the TG content decreased to 25% of that of the control. These data suggest that K8HK helps inhibit TG accumulation in cells.
3.3. Effects of K8HK on variation of lipid metabolism-related genes in 3T3-L1 adipocytes
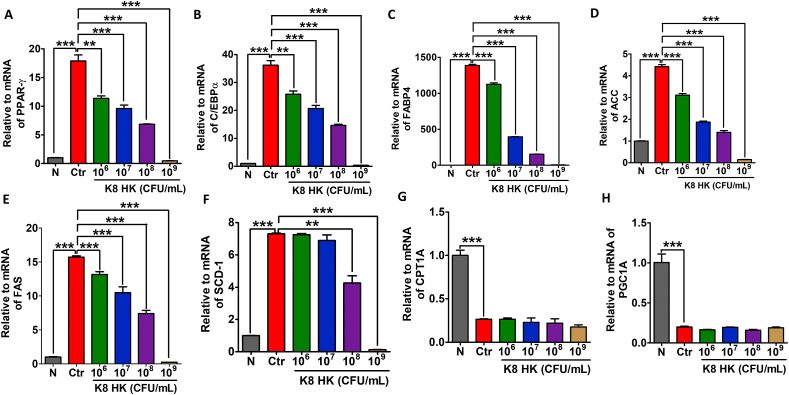
To examine the variation of adipose metabolism-related genes, real-time PCR was performed after differentiation of 3T3-L1 adipocytes treated with K8HK. Fig. 3A and B shows decreasing mRNA levels of transcription factors such as PPAR-γ and C/EBPα in a K8HK-dose-dependent manner, respectively. In particular, when cells were treated with 1 × 109 CFU/mL K8HK, the mRNA levels were nearly absent. PPAR-γ and C/EBPα induce FABP4 and lipogenic enzyme as transcription factors that play an important role in the conversion of glucose to TG. The mRNA level of FABP4, which plays a role in fatty acid uptake, decreased in a K8HK-dose-dependent manner, probably due to decreased expression of PPAR-γ and C/EBPα by K8HK (Fig. 3C). Lipogenic enzymes such as acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), and Stearoyl-CoA desaturase-1 (SCD-1) decreased in a K8HK-dose-dependent manner, suggesting that TG synthesis was reduced in those cells (Fig. 3D, E, and 3F). Next, fat oxidation-related genes such as Carnitine palmitoyltransferase 1 (CPT-1) and Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) were examined. CPT1 is responsible for translocating fatty acids from the cytosol to the mitochondrial matrix where fatty acid oxidation occurs [24]. PGC-1α coactivator plays a crucial role in maintaining lipid balance by participating in numerous metabolic processes such as Krebs cycle, β-oxidation, oxidative phosphorylation, and electron transport chain [25]. When cells were treated with K8HK, there was no change in the mRNA level of CPT-1 or PGC1α, indicating that K8HK does not affect fat oxidation (Fig. 3G and H). These results suggest that K8HK does not inhibit obesity by inhibition of fat oxidation but rather suppresses obesity by inhibiting adipogenesis.
Fig. 3.
Effects of K8HK on the variation of lipid metabolism-related genes in 3T3-L1 adipocytes. 3T3-L1 cells were treated with the indicated dose of K8HK during differentiation, and mRNA levels of (A) PPAR-γ, (B) C/EBPα, (C) FABP4, (D) ACC, (E) FAS, (F) SCD-1, (G) CPT1A, and (H) PGC1A were examined by real-time PCR. mRNA was normalized to mouse glyceraldehyde-3-phosphate (mGAPDH). The data displayed are the mean ± SD. Statistical analysis was conducted with one-way ANOVA. **p < 0.01, ***p < 0.001.
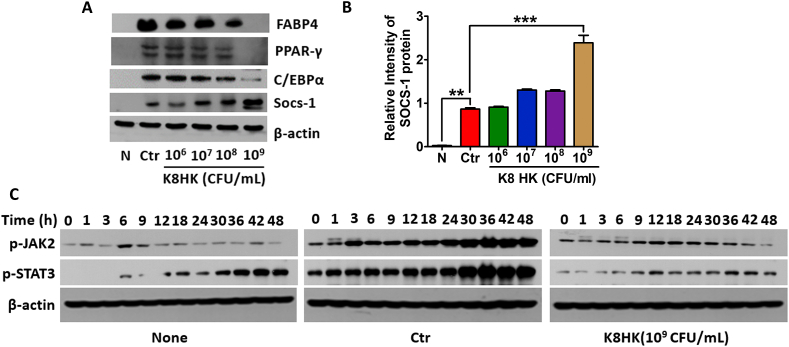
3.4. Negative regulators are involved in inhibition of lipid metabolism-related genes in K8HK-treated 3T3-L1 adipocytes
The protein level of a fat metabolism-related factor was confirmed by Western blot. As shown in Fig. 4A, FABP4 and its transcription factors such as C/EBPα and PPAR-γ were decreased in a K8HK-dose-dependent manner. When cells were treated with 1 × 109 CFU/mL K8HK, proteins were decreased to the level of untreated cells. On the other hand, transcriptome analysis revealed that the negative regulators including IRAK-M, TNFAIP3, SOCS-1, and SOCS-3 were increased in K8HK- and K8 Live-treated 3T3-L1 cells (Table 2). SOCS-1 is a negative regulator of JAK-STAT signaling pathway [26]. Unlike down-regulation of lipid metabolism-related genes, SOCS-1 was increased in a K8HK-dose-dependent manner (Fig. 4A and B). Western blot analysis showed increased phosphorylation of JAK2 and STAT3 up to 48 h in the differentiated control cells but not in the K8HK-treated cells (Fig. 4C). Considering the known mechanism of SOCS-1 and the present findings, the data suggest that K8HK-mediated inhibition of lipid metabolism-related gene expression was mediated by SOCS-1-mediated suppression of the JAK-STAT signaling pathway.
Fig. 4.
SOCS-1 was involved in inhibition of lipid metabolism-related genes in K8HK-treated 3T3-L1 adipocytes. (A) 3T3-L1 cells were treated with the indicated dose of K8HK during differentiation, and lipid metabolism-related proteins and SOCS-1 expression were examined by Western blot. (B) Relative intensity of SOCS-1 was calculated using Image J software. (C) Cells were treated with 1 × 109 CFU/mL K8HK for the indicated times, and phospho-JAK2 and phospho-STAT3 protein levels were examined by Western blot. β-actin was used as a loading control. Statistical analysis was conducted with one-way ANOVA. **p < 0.01, ***p < 0.001. Find the uncropped WB image version in Supplementary Material 2.
Table 2.
Regulators identified by transcriptome analysis.
| Gene symbol | K8 livea | K8HKa | Entrez ID | Description |
|---|---|---|---|---|
| Cyld | 1.438 | 0.986 | 74256 | cylindromatosis (turban tumor syndrome) |
| Gsk3b | 1.489 | 1.226 | 56637 | glycogen synthase kinase 3 beta |
| Irak3 | 13.885 | 7.118 | 73914 | interleukin-1 receptor-associated kinase 3 |
| Otud5 | 1.116 | 1.163 | 54644 | OTU domain containing 5 |
| Pcbp2 | 1.066 | 1.049 | 18521 | poly (rC) binding protein 2 |
| Rnf125 | 7.449 | 3.216 | 67664 | ring finger protein 125 |
| Sigirr | 1.000 | 1.000 | 24058 | single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| Socs1 | 2.258 | 2.612 | 12703 | suppressor of cytokine signaling 1 |
| Socs2 | 1.056 | 0.558 | 216233 | suppressor of cytokine signaling 2 |
| Socs3 | 4.830 | 3.590 | 12702 | suppressor of cytokine signaling 3 |
| Tnfaip3 | 16.282 | 15.317 | 21929 | tumor necrosis factor, alpha-induced protein 3 |
| Tnip2 | 1.619 | 1.963 | 231130 | TNFAIP3 interacting protein 2 |
| Tnip3 | 2.016 | 1.011 | 414084 | TNFAIP3 interacting protein 3 |
| Tollip | 1.104 | 0.943 | 54473 | toll interacting protein |
| Trim30a | 0.862 | 0.936 | 20128 | tripartite motif-containing 30 A |
Fold variation in K8 live- and K8HK-treated 3T3-L1 as compared to untreated cells.
3.5. K8 Live and K8HK have effects on body weight loss
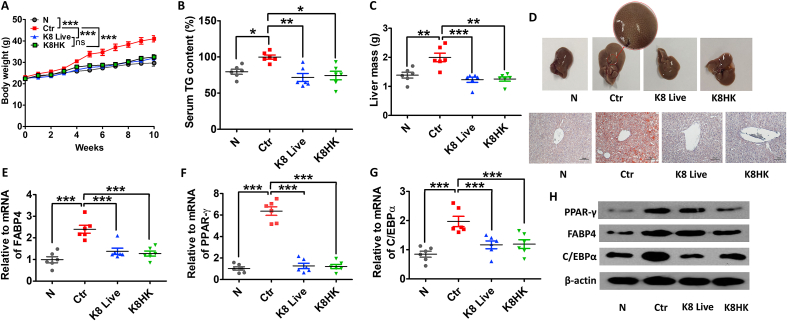
Unlike in vitro experiments, both K8 Live and K8HK have shown weight loss effects without mouse mortality. Mice in the high-fat diet (HFD) group gained weight by 10 weeks. The average weight at 10 weeks was about 40 g. On the other hand, the K8 Live- and K8HK-fed groups showed an average body weight of 30 g at 10 weeks, similar to that of normal diet mice (Fig. 5A), suggesting that oral intake of K8 Live or K8HK can help with weight loss. The TG content was increased in the HFD group compared to the normal diet group, but it was significantly decreased in K8 Live- and K8HK-fed HFD groups (Fig. 5B). In order to confirm the effects of K8 Live and K8HK on the liver of HFD mice, the weight and histology of the liver were examined. Fatty liver was formed in livers isolated from HFD mice, which showed increased liver size and fat accumulation. The average weight of HFD mice liver was about 2 g compared to 1.2 g in K8 Live- and K8HK-fed mice liver, a 40% weight loss (Fig. 5C). Livers of HFD mice were enlarged compared to those of normal diet-fed mice, and the livers of K8 Live- and K8HK-fed HFD mice were smaller than those of HFD control mice (Fig. 5D, upper panel). When the livers were stained with Oil-red O, initiation of fatty liver formation was observed in HFD mice. However, fat staining was not seen in livers isolated from K8 Live- and K8HK-fed HFD mice, and formation of fatty liver was suppressed in both groups (Fig. 5D, lower panel). These data suggest that K8 Live and K8HK can prevent fatty liver by inhibiting the accumulation of fat in the liver. The mRNA expression of a lipid metabolism-related gene in liver was examined. mRNA expression of FABP4 and its transcription factors of PPARγ and C/EBPα in the control was significantly increased compared to N group. However, expression was significantly decreased in mice fed K8 Live and K8HK (Fig. 5E–G). The protein levels of these factors in the liver exhibited similar patterns (Fig. 5H).
Fig. 5.
Effects of K8 Live and K8HK on body weight loss. C57BL/6 (5 weeks old, 6 mice per group) were fed a 60% high fat diet (HFD) for 10 weeks following 2 weeks of normal diet. Experimental mice were orally administered K8 Live (1 × 1010 CFU/mouse) +HFD (indicated as K8 Live) and K8HK (1 × 1010 CFU/mouse) +HFD (indicated as K8HK) for 12 weeks. The control group (Ctr) and the no treatment group (N) were orally administered HFD and normal diet, respectively, for 12 weeks. (A) The change of body weight was examined. (B) Serum TG content was examined using the TG-S kit. (C) Liver mass was examined after extraction. (D) Photographs showing extracted livers. mRNA levels of FABP4 (E), PPAR-γ (F), and C/EBPα (G) were examined by real-time PCR after extraction of total RNA from the liver tissues. (H) Protein levels of lipid metabolism-related genes were examined by Western blot after dissolving the liver tissues. GAPDH and β-actin were used as controls. The data displayed are the mean ± SD. Statistical analysis was conducted with two-way ANOVA (A) or one-way ANOVA (B, C, E, F, and G). *p < 0.05, **p < 0.01, ***p < 0.001. Find the uncropped WB image version in Supplementary Material 2.
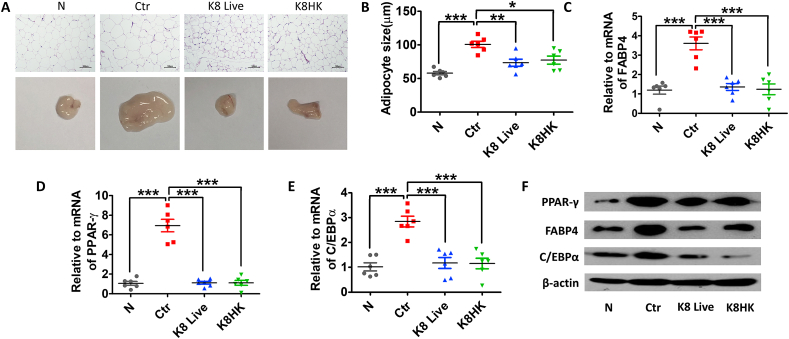
3.6. K8 Live and K8HK inhibit adipocyte growth
Next, we examined the change of gonadal adipose tissue. The size of the adipose tissue isolated from the HFD-fed control mice was morphologically much larger than that of the normal diet-fed group. However, adipocytes were smaller in the K8 Live- and K8HK-fed groups than in the HFD group. (Fig. 6A). The size of adipocytes was about 100 μm in the HFD control group, but about 70 μm in the HFD group supplied with K8 Live and K8HK (Fig. 6B), suggesting that intake of K8 Live or K8HK can help suppress fat production in adipose tissue. Similar to in the liver, mRNA expression of FABP4, PPARγ and C/EBPα in adipocytes isolated from K8 Live- and K8HK-fed HFD mice was decreased compared to HFD control group (Fig. 6C–E). Protein levels of these genes in adipocytes showed a pattern similar to that of mRNA (Fig. 6F).
Fig. 6.
K8 Live and K8HK inhibit adipocyte growth. (A) Appearance and histological analysis of gonadal adipose tissue after feeding K8 Live or K8HK with or without HFD for 12 weeks. Gonadal adipose tissues were stained with hematoxylin and eosin (H & E). (B) The adipocyte size was measured by Image J. mRNA levels of FABP4 (C), PPAR-γ (D), and C/EBPα (E) were examined by real-time PCR after extraction of total RNA from the gonadal adipose tissues. (F) Protein levels of lipid metabolism-related genes were examined by Western blot after dissolving the gonadal adipose tissues. GAPDH and β-actin were used as controls. The data displayed are the mean ± SD. Statistical analysis was conducted with one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001. Find the uncropped WB image version in Supplementary Material 2.
4. Discussion
Lactobacillus strains such as L. plantarum, L. sakei, and L. gasseri have been studied for their anti-obesity effects. For example, L. plantarum LMT1-48 down-regulated lipogenic genes such as PPAR-γ, HSL, SCD-1, and FAT/CD36 in the liver, which resulted in reduction of liver weight and liver TG content [27]. L. sakei also has anti-obesity effects. One randomized, double-blind, placebo-controlled, clinical trial involved 114 adults with a body mass index (BMI) ≥25 kg/m2 revealed that administration of 5 × 109 CFU of L. sakei decreased body fat mass by 0.2 kg, while the placebo group increased by 0.6 kg [28]. L. gasseri BNR17 increased fatty acid oxidation-related gene expression while it lowered fatty acid synthesis-related gene expression. The expression of glucose transporter-4 (GLUT4) was elevated in groups fed BNR17, which can affect anti-obesity as well as insulin reduction [29]. A probiotic mixture of lactobacillus strains improved lipid metabolism and gut microbiota structure in HFD mice [30]. However, there seem to be few reports on cytotoxicity caused by probiotics.
Recent researches on probiotics are expanding from live bacteria to dead cells (parabiotics) and metabolites (postbiotics) produced form probiotics. Parabiotics are microbial cells in which probiotics are inactivated. Parabiotics have many advantages over probiotics, such as clear chemical structures, safe dose parameters, and longer shelf life, and thus have the potential to replace probiotics [31]. This study was conducted to find out whether heat-treated probiotics showed similar efficacy to live probiotics and could replace live bacteria. In in vitro experiments, the cytotoxicity of dermal keratinocytes (HaCaT cells) and adipocytes (3T3-L1) was induced within 24 h after application of a high concentration of probiotics (over 1 × 108 CFU/mL) (data not shown). On the other hand, heat-killed probiotics at the same concentration showed no cytotoxicity. A more precise study should be carried out, but cytotoxicity by K8 Live seems to be related to changes in cell death-related genes expressed in 3T3-L1 cells by stimulation by L. plantarum K8. The expression pattern of these cytotoxicity-related genes differs by administration of K8 Live and K8HK. Unlike live cells, heat-killed probiotics show anti-obesity efficacy similar to that of live cells but without inducing cytotoxicity. Moreover, since probiotics are ‘living bacteria,’ they are sensitive to temperature and can be killed during distribution. In this experiment, if K8 Live and K8HK were used at a concentration lower than 1 × 109 CFU/mL, the difference in activity between the two could be clearly identified. Determining activity at low concentrations may also help lower the cost of production of future products.
Previous study has shown that heat-killed L. plantarum L-137 strain (HK L-137) alleviated adipose tissue inflammation by inhibition of several inflammation-related genes such as F4/80, CD11c, and IL-1β. HK L-137 also decreased the level of plasma glucose, cholesterol, alanine aminotransferase (ALT), and aspartate transaminase (AST) in diet-induced obesity mice [32]. On the other hand, in the experiment using K8HK, changes in genes related to adipogenesis were observed to elicit anti-obesity mechanism. Inactivation of JAK2/STAT3 by negative regulators including SOCS-1 can restrict transcription factors such as PPAR-γ and C/EBPα, and consequently control adipogenesis caused by FABP4 and lipogenic enzymes. In another studies, Kim et al. showed that L. plantarum K8 lysate (LAB-P) has anti-obesity effect. LAB-P inhibited the accumulation of intracellular lipids, which resulted in the alleviation of obesity [33,34]. This phenomenon coincided with inhibition of JAK2-STAT3/5 and AMPKα by LAB-P in adipocytes. These studies suggest that different formulations of probiotics can achieve the desired results without toxicity and can be used to make products of various formulations.
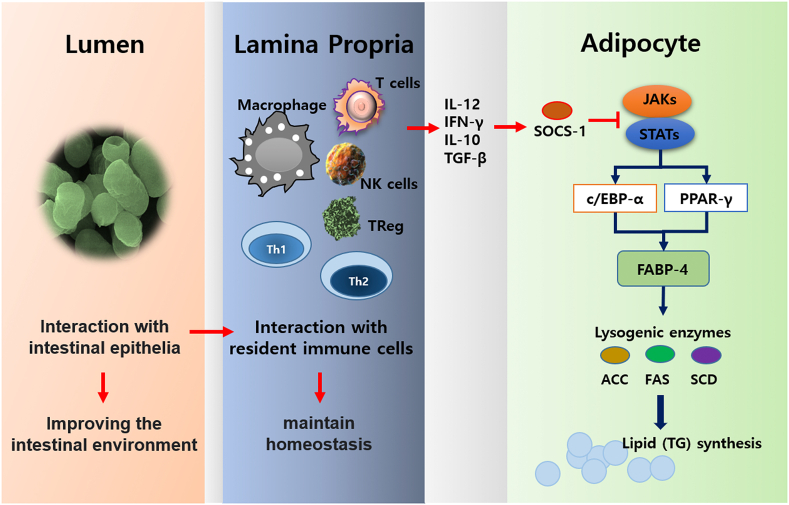
Unlike live bacteria, Heat-killed bacteria induces activity in the intestine without settling. This makes it possible to maintain immune homeostasis through direct contact with immune cells present in the lamina propria. In this interaction, cytokines secreted from immune cells stimulate adipocytes to increase the expression of SOCS-1, which inhibits the JAK-STAT pathway (Fig. 7). Through transcriptome analysis, we found that K8HK induced negative regulators such as SOCS-1, SOCS-3, IRAK-M, and TNFAIP3, which negatively regulate JAK-STAT signaling [35]. Although this study only confirmed the possible involvement of SOCS-1, negative regulators such as IRAK-3, TNFAIP3, and SOCS-3 are also thought to be involved in the regulation of lipid metabolism. The downregulation of JAK2-STAT3 signaling caused by negative regulators decreased the activation of transcription factors PPARγ and C/EBPα. Eventually, lipogenic enzyme and FABP4 decreased and resulted in reduction of TG synthesis in 3T3-L1 cells treated with K8HK. There are few studies related to the correlation between negative regulators and obesity alleviation. In our experiments, it was observed that some of the negative regulators were changed during obesity induction. Studies of negative regulators may improve our understanding of obesity formation mechanisms.
Fig. 7.
Schematic diagram. In the intestine, K8 Live or K8HK improves the intestinal environment through cooperation with enterocytes. This result enhances the ability of immune cells in the lamina propria to maintain homeostasis and induces the expression of appropriate negative regulators for stimulation by external factors. For example, SOCS-1 can inhibit lysogenic enzyme-mediated fat synthesis by blocking JAK-STAT signaling in adipocytes.
5. Conclusions
Our data shows that heat-treated probiotic K8HK increases the expression of negative regulators including SOCS-1 by acting on 3T3-L1 cells. SOCS-1 and possibly other negative regulators might inhibit the activation of the JAK2-STAT3 pathway and down-regulate the genes PPARγ, C/EBPα, and FABP4 involved in obesity formation, inhibiting intracellular TG synthesis and preventing fatty liver. Some probiotics have been shown to have anti-obesity properties. However, live probiotics have limitations in distribution and storage. In order to reduce these disadvantages, recently, studies on parabiotics are increasing. Since K8HK is a parabiotics inactivated through heat treatment, it has low cytotoxicity, can be used in development of various formulations, and has higher stability in the distribution process than do live probiotics. Thus, K8HK can be developed as a food supplement that can replace live probiotics.
Author contribution statement
Kyoung Ok Jang: Performed the experiments; Wrote the paper.
Jung Seo Choi, Kyeong Hun Choi, Seongjae Kim: Performed the experiments.
Hangeun Kim: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Dae Kyun Chung: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This research was financially supported by the Ministry of Trade, Industry, and Energy (MOTIE), Korea, under the “Infrastructure Support Program for Industry Innovation” (P0014714) supervised by the Korea Institute for Advancement of Technology (KIAT).
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no conflict of interest.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e12926.
Contributor Information
Hangeun Kim, Email: hkim93@khu.ac.kr.
Dae Kyun Chung, Email: dkchung@khu.ac.kr.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Park J., Morley T.S., Kim M., Clegg D.J., Scherer P.E. Obesity and cancer--mechanisms underlying tumour progression and recurrence. Nat. Rev. Endocrinol. 2014;10(8):455–465. doi: 10.1038/nrendo.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jokinen E. Obesity and cardiovascular disease. Minerva Pediatr. 2015;67(1):25–32. [PubMed] [Google Scholar]
- 3.Seravalle G., Grassi G. Obesity and hypertension. Pharmacol. Res. 2017;122:1–7. doi: 10.1016/j.phrs.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Malone J.I., Hansen B.C. Does obesity cause type 2 diabetes mellitus (T2DM)? Or is it the opposite? Pediatr. Diabetes. 2019;20(1):5–9. doi: 10.1111/pedi.12787. [DOI] [PubMed] [Google Scholar]
- 5.Patel J.J., Rosenthal M.D., Miller K.R., Codner P., Kiraly L., Martindale R.G. The critical Care obesity paradox and implications for nutrition support. Curr. Gastroenterol. Rep. 2016;18(9):45. doi: 10.1007/s11894-016-0519-8. [DOI] [PubMed] [Google Scholar]
- 6.Pigeyre M., Yazdi F.T., Kaur Y., Meyre D. Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin. Sci. (Lond.) 2016;130(12):943–986. doi: 10.1042/CS20160136. [DOI] [PubMed] [Google Scholar]
- 7.Gentile C.L., Weir T.L. The gut microbiota at the intersection of diet and human health. Science. 2018;362(6416):776–780. doi: 10.1126/science.aau5812. [DOI] [PubMed] [Google Scholar]
- 8.Turroni F., Ventura M., Buttó L.F., Duranti S., O'Toole P.W., Motherway M.O., van Sinderen D. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. 2014;71(2):183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John G.K., Mullin G.E. The gut microbiome and obesity. Curr. Oncol. Rep. 2016;18(7):45. doi: 10.1007/s11912-016-0528-7. [DOI] [PubMed] [Google Scholar]
- 10.Fischer N., Relman D.A. Clostridium difficile, aging, and the gut: can microbiome rejuvenation keep us young and healthy? J. Infect. Dis. 2018;217(2):174–176. doi: 10.1093/infdis/jix417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobyliak N., Virchenko O., Falalyeyeva T. Pathophysiological role of host microbiota in the development of obesity. Nutr. J. 2016;15:43. doi: 10.1186/s12937-016-0166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Zhang B., Hu J., Nie S., Xiong T., Xie M. Intervention of five strains of Lactobacillus on obesity in mice induced by high-fat diet. J. Funct.Foods. 2020;72:104078. doi: 10.1016/j.jff.2020.104078. [DOI] [Google Scholar]
- 13.Yue Y., He Z., Zhou Y., Ross R.P., Stanton C., Zhao J., Zhang H., Yang B., Chen W. Lactobacillus plantarum relieves diarrhea caused by enterotoxin-producing Escherichia coli through inflammation modulation and gut microbiota regulation. Food Funct. 2020;11(12):10362–10374. doi: 10.1039/d0fo02670k. [DOI] [PubMed] [Google Scholar]
- 14.Kim J.Y., Kim H., Jung B.J., Kim N.R., Park J.E., Chung D.K. Lipoteichoic acid isolated from Lactobacillus plantarum suppresses LPS-mediated atherosclerotic plaque inflammation. Mol. Cell. 2013;35(2):115–124. doi: 10.1007/s10059-013-2190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong Y.F., Kim H., Kim H.R., Gim M.G., Chung D.K. Different immune regulatory potential of Lactobacillus plantarum and Lactobacillus sakei isolated from Kimchi. J. Microbiol. Biotechnol. 2014;24(12):1629–1635. doi: 10.4014/jmb.1406.06062. [DOI] [PubMed] [Google Scholar]
- 16.Kim H., Kim H.R., Kim N.R., Jeong B.J., Lee J.S., Jang S., Chung D.K. Oral administration of Lactobacillus plantarum lysates attenuates the development of atopic dermatitis lesions in mouse models. J. Microbiol. 2015;53(1):47–52. doi: 10.1007/s12275-015-4483-z. [DOI] [PubMed] [Google Scholar]
- 17.Ahn J.E., Kim H., Chung D.K. Lipoteichoic acid isolated from Lactobacillus plantarum maintains inflammatory homeostasis through regulation of Th1- and Th2-induced cytokines. J. Microbiol. Biotechnol. 2019;29(1):151–159. doi: 10.4014/jmb.1809.09001. [DOI] [PubMed] [Google Scholar]
- 18.Kim H.G., Kim N.R., Gim M.G., Lee J.M., Lee S.Y., Ko M.Y., Kim J.Y., Han S.H., Chung D.K. Lipoteichoic acid isolated from Lactobacillus plantarum inhibits lipopolysaccharide-induced TNF-alpha production in THP-1 cells and endotoxin shock in mice. J. Immunol. 2008;180(4):2553–2561. doi: 10.4049/jimmunol.180.4.2553. [DOI] [PubMed] [Google Scholar]
- 19.Kim G., Choi K.H., Kim H., Chung D.K. Alleviation of LPS-induced inflammation and septic shock by Lactiplantibacillus plantarum K8 lysates. Int. J. Mol. Sci. 2021;22(11):5921. doi: 10.3390/ijms22115921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li N., Russell W.M., Douglas-escobar M., Hauser N., Lopez M., Neu J. Live and heat-killed Lactobacillus rhamnosus GG: effects on proinflammatory and anti-inflammatory cytokines/chemokines in gastrostomy-fed infant rats. Pediatr. Res. 2009;66(2):203–207. doi: 10.1203/PDR.0b013e3181aabd4f. [DOI] [PubMed] [Google Scholar]
- 21.Sun W., Jiang Y., He F. Extraction and proteome analysis of liver tissue interstitial fluid. Methods Mol. Biol. 2011;728:247–257. doi: 10.1007/978-1-61779-068-3_16. [DOI] [PubMed] [Google Scholar]
- 22.Tan P., Pepin É., Lavoie J.L. Mouse adipose tissue collection and processing for RNA analysis. JoVE. 2018;131:57026. doi: 10.3791/57026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parlee S.D., Lentz S.I., Mori H., MacDougald O.A. Quantifying size and number of adipocytes in adipose tissue. Methods Enzymol. 2014;537:93–122. doi: 10.1016/B978-0-12-411619-1.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M., Wang K., Liao X., Hu H., Chen L., Meng L., Gao W., Li Q. Carnitine palmitoyltransferase system: a new target for anti-inflammatory and anticancer therapy? Front. Pharmacol. 2021;12:760581. doi: 10.3389/fphar.2021.760581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Supruniuk E., Mikłosz A., Chabowski A. The implication of PGC-1α on fatty acid transport across plasma and mitochondrial membranes in the insulin sensitive tissues. Front. Physiol. 2017;8:923. doi: 10.3389/fphys.2017.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liau N., Laktyushin A., Lucet I.S., Murphy J.M., Yao S., Whitlock E., Callaghan K., Nicola N.A., Kershaw N.J., Babon J.J. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 2018;9(1):1558. doi: 10.1038/s41467-018-04013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi W.J., Dong H.J., Jeong H.U., Ryu D.W., Song S.M., Kim Y.R., Jung H.H., Kim T.H., Kim Y.H. Lactobacillus plantarum LMT1-48 exerts anti-obesity effect in high-fat diet-induced obese mice by regulating expression of lipogenic genes. Sci. Rep. 2020;10(1):869. doi: 10.1038/s41598-020-57615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim S., Moon H.H., Shin C.M., Jeong D., Kim B. Effect of Lactobacillus sakei, a probiotic derived from Kimchi, on body fat in Koreans with obesity: a randomized controlled study. Endocrinol. Metab. (Seoul) 2020;35(2):425–434. doi: 10.3803/EnM.2020.35.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang J.H., Yun S.I., Park M.H., Park J.H., Jeong S.Y., Park H.O. Anti-obesity effect of Lactobacillus gasseri BNR17 in high-sucrose diet-induced obese mice. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0054617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Liu F., Lu J., Shi J., Guan J., Yan F., Li B., Huo G. Probiotic mixture of Lactobacillus plantarum strains improves lipid metabolism and gut microbiota structure in high fat diet-fed mice. Front. Microbiol. 2020;11:512. doi: 10.3389/fmicb.2020.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nataraj B.H., Ali S.A., Behare P.V., Yadav H. Postbiotics-parabiotics: the new horizons in microbial biotherapy and functional foods. Microb. cell fact. 2020;19(1):168. doi: 10.1186/s12934-020-01426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshitake R., Hirose Y., Murosaki S., Matsuzaki G. Heat-killed Lactobacillus plantarum L-137 attenuates obesity and associated metabolic abnormalities in C57BL/6 J mice on a high-fat diet. Biosci. Microbiota. Food Heal. 2021;40(2):84–91. doi: 10.12938/bmfh.2020-040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H., Lim J.J., Shin H.Y., Suh H.J., Choi H.S. Lactobacillus plantarum K8-based paraprobiotics suppress lipid accumulation during adipogenesis by the regulation of JAK/STAT and AMPK signaling pathways. J. Funct.Foods. 2021;87:104824. doi: 10.1016/j.jff.2021.104824. [DOI] [Google Scholar]
- 34.Lim J.J., Jung A.H., Suh H.J., Choi H.S., Kim H. Lactiplantibacillus plantarum K8-based paraprobiotics prevents obesity and obesity-induced inflammatory responses in high fat diet-fed mice. Food Res. Int. 2022;155:111066. doi: 10.1016/j.foodres.2022.111066. [DOI] [PubMed] [Google Scholar]
- 35.Skljarevski S., Barner A., Bruno-Murtha L.A. Preventing avoidable central line-associated bloodstream infections: implications for probiotic administration and surveillance. Am. J. Infect. Control. 2016;44(11):1427–1428. doi: 10.1016/j.ajic.2016.07.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.