Abstract
In vitro models of liver (patho)physiology, new technologies, and experimental approaches are progressing rapidly. Based on cell lines, induced pluripotent stem cells or primary cells derived from mouse or human liver as well as whole tissue (slices), such in vitro single- and multicellular models, including complex microfluidic organ-on-a-chip systems, provide tools to functionally understand mechanisms of liver health and disease. The International Society of Hepatic Sinusoidal Research (ISHSR) commissioned this working group to review the currently available in vitro liver models and describe the advantages and disadvantages of each in the context of evaluating their use for the study of liver functionality, disease modeling, therapeutic discovery, and clinical applicability.
Keywords: Hepatic Sinusoid, Mechanobiology, Omics, Bioengineering, Cirrhosis, NAFLD, NASH
Abbreviations used in this paper: CLD, chronic liver disease; EC, endothelial cell; ECM, extracellular matrix; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HSC, hepatic stellate cell; LoC, liver-on-a-chip; LSEC, liver sinusoidal endothelial cell; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PCLS, precision cut liver slices; PDGF, platelet-derived growth factor; PNPLA3, patatin-like phospholipase domain containing 3; sc-RNA Seq, single-cell RNA sequencing; snRNA-seq, single-nuclei RNA sequencing; 2D, 2-dimensional; 3D, 3-dimensional
Summary.
This review provides a glimpse on the most advanced methods for studying liver sinusoidal biology in vitro to uncover liver disease pathophysiology focusing on liver-on-a-chip and microfluidic devices, liver scaffolds and matrices, mechanobiological cues, spheroids/organoids, liver slices and omics.
Liver disease represents one of the leading causes of death worldwide, and the incidence of some pathologies, such as nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), and liver cancer, continues to increase.1 Despite years of research, liver diseases still have limited treatment options in the clinic. This paucity of treatments is explained partly by the limitations of traditional in vitro tools and animal models that do not accurately mimic the clinical pathophysiology of diseases and have a low accuracy for drug discovery purposes. Indeed, several studies have shown that traditional cell culture methodologies do not reflect the complexity of a human liver in vivo and thus cannot predict drug sensitivity. In contrast, animal models differ in biology compared with human pathologies, which explains why promising therapies tested in animal models often fail when tested in human beings and, unfortunately, the field of hepatology has numerous recent examples of failures in clinical phases.2 With the advent of precision medicine, which offers much hope for individual patient outcomes, there is increased demand for robust and patient-specific tools to better improve our understanding and treatment of complex and multifactorial diseases such as liver diseases. Advances in vascular biology, microfluidics, and bioengineering have led to the development of sophisticated in vitro models that could fill this gap (Figure 1). In addition, omics techniques provide further insight to preclinical research in hepatology. In this review, we discuss the benefits and limitations of advanced in vitro research techniques that presently are being applied to the study of liver diseases and further critique how these tools may provide insight into the prediction of patient responses to a therapy.
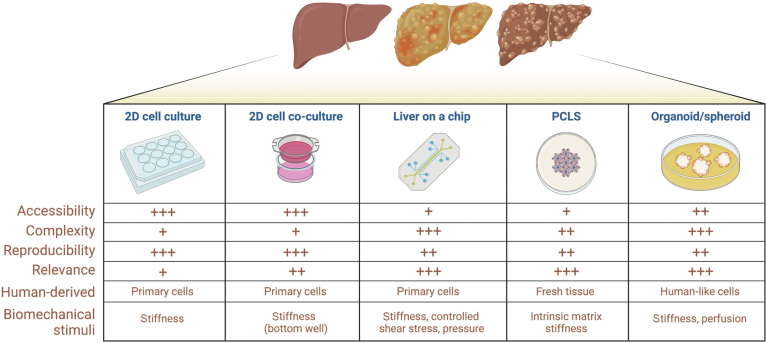
Figure 1.
Schematic view of mostly used in vitro models in hepatology. +, low; ++, medium; +++, high.
Liver-on-a-Chip and Microfluidic Devices
During the 21st century, the development of biology-inspired devices aimed at mimicking the sinusoidal niche integrating microfluidics led to the rapidly evolving liver-on-a-chip (LoC) technology.3 The design of these in vitro liver-resembling tools, which have been reviewed extensively by Ortega-Ribera et al4 and is out of the scope of the present review, is inspired in sinusoidal cell biology, architecture, and hemodynamics, but materialized under each research teams’ eyes in terms of appearance, size, fabrication procedures, costs, and microfluidics integration, leading to significant variation in the finalized product.
The latest advances in the field include chronic liver disease–specific devices, LoC models designed to study key pathophysiological processes in the development of liver disease, and to understand the interconnection with other organs-on-chip to better depict liver functions and systemic implications (Figure 2). Multi-organ chips, for instance, liver-, adipose-, and gut-on-a-chip connected, may be particularly suitable to understand organ-crosstalk in chronic liver disease (CLD), such as NAFLD/NASH or cholangiopathies.5
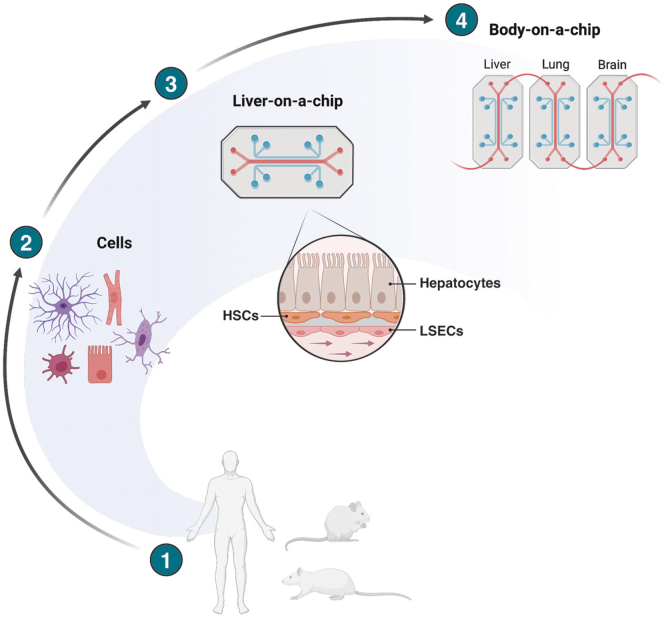
Figure 2.
Fromsingle cells to liver-on-a-chip and body on-a-chip.
In recent years, disease-focused LoC devices mimicking some of the landmark etiological characteristics of CLD have been developed. Fat accumulation in hepatocytes, occurring in NAFLD/NASH, has been represented in LoC under the combination of glucose and free fatty acids (usually a 2:1 ratio of oleic and palmitic acid).6 Antifibrotic compounds such as obeticholic acid, elafibranor,7 pioglitazone, or metformin8,9 showed promising results in reducing lipid droplets in these in vitro settings. Indeed, the anti-NASH agent lanifibranor efficiently reduced hepatocytic lipid accumulation,10 and improved human hepatocyte and hepatic stellate cell (HSC) phenotype11 in a LoC model but not in 2-dimensional (2D) cell cultures, supporting the specific value of multicellular LoC devices over traditional monocell cultures. Alcohol-associated liver disease has been addressed in several publications, focusing on its impact in sinusoidal cell biology during development12 or recovery from alcohol (abstinence) either with perfused spheroid13 or layered cultures.14 Importantly, Ortega-Prieto et al15 developed a model for hepatitis B virus (HBV) long-term infection in primary human hepatocytes that recapitulates virus–host interactions and its associated immune effectors. LoC devices using primary cells isolated through standardized protocols from preclinical models of CLD and from patients' liver tissue also have been developed.16,17 These specialized LoC settings may widen the current knowledge on disease dynamics and provide potential applicability as in vitro preclinical models for drug screening.
Even though LoC complexity has increased outstandingly since the initial models/prototypes, the intricacy of the whole liver still is under-represented. In this regard, several scientists brought the attention and focused their studies on specific processes or structures. For example, the essential features of the bile duct containing primary mouse cholangiocytes,18 the unique vasculature organization of the liver,19 the sinusoidal zonation within LoC devices,20 neutrophil recruitment and interaction with liver sinusoidal endothelial cells (LSECs) after lipopolysaccharide stimulation,21 or drug metabolism and toxicity22 now are embedded in available LoC systems.
Moreover, CLD has been reported extensively as a systemic syndrome with major extrahepatic implications.23 Therefore, the combination of LoC devices now has evolved to the extent that disease-specific models are being combined with others such as intestine, brain, kidney,24, 25, 26 or even metastasis niche-on-chip models27 to re-create body-on-chip structures to further study the gut–liver–brain axis, systemic drug clearance, or exosome communication between the liver and the tumor microenvironment. However, alongside these advances in multi-organ and etiology-centered approaches, a lack of consensus in cellular sources and mechanobiological cues within the various LoC models remain as unsolved challenges in the field.
Liver Scaffolds, Matrices, and Other Substrates
A key component for the engineering of in vitro liver models is the development of the appropriate scaffold/matrix that recapitulates the hepatic microenvironment well enough to result in realistic functional cells. Several factors affect the efficiency of a scaffold as a support for liver cell growth and function including porosity, pore size, biomechanical properties, and the scaffold design. To simulate the microenvironment of natural extracellular matrix (ECM), substrate design and biomechanical properties are of great significance; hence, bioinspired and biomimetic approaches have been explored to model the healthy or damaged liver (Figure 3). Double layers of collagen have been used for years as a well-established 2D in vitro model for sandwich cultures of hepatocytes.28 Recent studies have identified, and characterized, the hepatic matrisome comprising ECM signatures beyond collagen that potentially can provide matrix for in vitro systems to study liver diseases.29,30
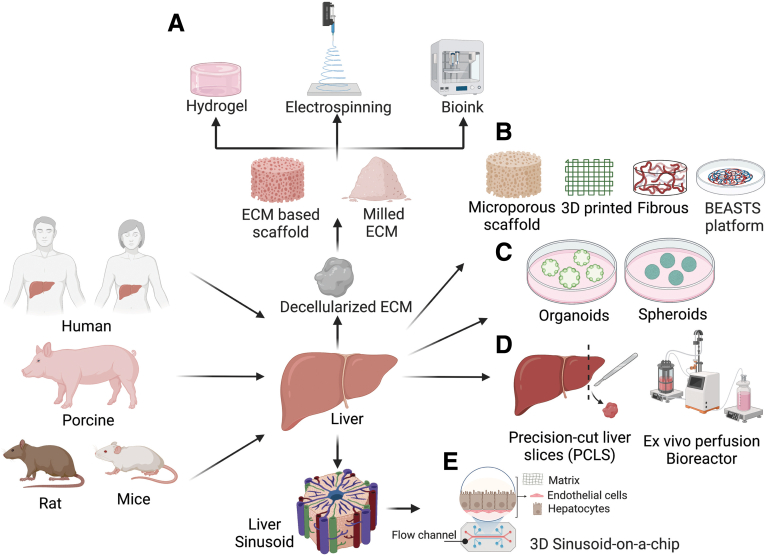
Figure 3.
Schematic overview of in vitro and ex vivo liver models using (A) natural scaffolds including hydrogels, fiber-like structures generated by electrospinning, or bioinks, and (B) synthetic scaffolds such as microporous, 3D fibrous, or bioengineered platforms to generate (C) organoids/spheroids, (D) PCLS and bioreactors, and (E) 3D liver sinusoid on a chip. Natural scaffolds obtained from hepatic tissues from various sources such as human, porcine, and rat undergo decellularization using detergents. Synthetic scaffolds including polymer and hydrogel based in combination with novel methods such as 3D printing provides the capability for tuning the properties of the material to re-create the liver microenvironment at stages of disease progression. Current 2D and 3D hepatic models include organoids and spheroids, tissue-based approaches such as PCLS and ex vivo bioreactors, and liver-on-a-chip micropatterned co-culture models. BEASTS, Bio-Engineered Adhesive Siloxane substrate with Tunable Stiffness; ECM, extracellular matrix.
Scaffolds have been fabricated using natural polymers such as gelatin, elastin, silk fibroin, chitosan, chitin, fibrin, and fibrinogen, or synthetic polymers such as polylactic acid, poly(glycolic acid), polyhydroxyalkanoate, and poly (lactic-co-glycolic acid).31 Modified versions of biomaterials such as collagen-incorporated poly (lactic-co-glycolic acid) have resulted in enhancement of hepatocyte survival and functions likely owing to an increase in the bioactivity of the newly developed scaffolds.32,33 Similarly, natural polymers such as silk fibroin have been modified with arginyl-glycyl-aspartic acid (RGD, an integrin-based cell adhesion motif), which has been reported to support the growth of functional hepatocyte clusters.34 The modification with RGD also may support attachment of LSECs, known as endothelization of materials.35 Efficient spheroid cultures of hepatocytes have been reported on highly porous hydrogel scaffolds composed of alginate and galactosylated chitosan.36 In addition, synthetic polymer thin films–based scaffolds allow organized hepatocyte culture and patterned co-culture of hepatocytes with nonparenchymal cells.37, 38, 39 Recent interest in mechanical signaling has led to the development of scaffolds that can re-create liver stiffness in physiological and pathologic conditions. In this context, heparin hydrogel has been developed to modulate stiffness and has shown that hepatocytes cultured on a softer heparin hydrogel (10 kPa) retained 5 times higher levels of albumin production compared with those on a stiffer heparin gel (110 kPa) after 5 days.40 Primary hepatocytes grown on modified polyacrylamide gels with cell-adhesive ligands are shown to reduce albumin production and impair hepatocyte function with increasing stiffness.41,42 Desai et al43 used polyacrylamide gels to tune the substrate stiffness and showed that fibrotic levels of stiffness significantly inhibit hepatocyte-specific functions in part through the inhibition of the hepatocyte nuclear factor 4α transcriptional network mediated via the Rho/Rho-associated protein kinase pathway. An innovative platform called bio-engineered adhesive siloxane substrate with tunable stiffness based on a polydimethyl siloxane substrate in combination with polyelectrolyte multilayer film-coating technology was developed to engineer mechanically tunable substrates mimicking physiologic and pathologic liver stiffness.44, 45, 46, 47, 48 More recently, 3-dimensional (3D) bioprinting has emerged for precise spatial positioning of both cells and biomaterials or bioinks such as alginate together in 3D complex geometries and providing mechanical support.49,50 Nguyen et al51 have bioprinted hepatocytes and nonparenchymal cells in 3D architecture and developed models of drug-induced liver injury. Recent studies have printed liver cells along with a liver decellularized ECM bioink, creating an environment for maximal cellular function.52, 53, 54 With 3D bioprinting, vascular and biliary fluidic channels also have been created successfully in the LoC device format.53 3D bioprinting of spheroids and organoids represent the next level of technological advancement for creating the highly complex liver architecture.55
Liver Spheroids and Organoids
As described earlier, over the past century, 2D cell cultures have been used as common in vitro models to study cellular responses to stimulation and allowing the construction of low-cost, simple, and well-accepted models of liver disease. However, they do not precisely reflect the true physiological state of cells in vivo owing to the absence of structural, mechanical, and biochemical cues, as well as the interaction between cells and extracellular matrices.56 To overcome this limitation, novel 3D cell culture platforms including liver spheroid and organoid cultures are being created to better mimic the in vivo conditions.57, 58, 59, 60, 61 3D spheroids are produced via self-assembly, in which monodispersed cells form 3D microtissues called multicellular spheroids, and mimic natural processes that occur during embryogenesis, morphogenesis, and organogenesis.62 3D organoids derive from either pluripotent stem cells, neonatal tissue stem cells, or adult stem cells/adult progenitors, in which cells spontaneously self-organize into properly differentiated functional cell types and progenitors, resembling their in vivo counterparts and recapitulating at least some functions of the organ.63
In the field of studying liver diseases, recent innovation of hepatic 3D spheroids also offer a promising application via combination of 3D printing–based techniques and HepG2 liver spheroid culture models to develop in situ quantitative evaluation and high-throughput monitoring of drug-induced hepatotoxicity.64 HepG2 cell-laden hydrogel constructs were 3D printed in the shape of a cross on the mini-9-well plate, which showed the HepG2 liver spheroids embedded in the gelatin-alginate hydrogel. On the 6th day of culture, HepG2 liver spheroids exposed to varying concentrations of troglitazone and nefazodone were used to predict hepatotoxicity. This model provided a promising tool for screening and characterization of hepatotoxicity in a 3D spheroid-embedded hydrogel system that more closely resembles conditions in vivo.
In 2013, Takebe et al59 described the in vitro generation of 3D liver bud organoids from human induced pluripotent stem cell–derived hepatic endoderm cells co-cultured with endothelial and mesenchymal lineages. Interestingly, when these liver buds were ectopically transplanted at various sites including the cranium, subrenal capsule, and distal and proximal-mesentery in immunodeficient mice they were able to rescue the drug-induced lethal liver failure model.61,65 These studies have provided a promising new approach to study regenerative medicine and to translate these techniques for treating patients with end-stage liver failure.65 Furthermore, single-cell RNA sequencing (scRNA-seq) data from human liver bud organoids showed several aspects of heterotypic interlineage communication and organ development.66 Interestingly, Shinozawa et al67 reported a simple, robust, and high-throughput human liver organoid system to measure bile transport activity by live fluorescent imaging with large-scale screening and multiplexed readouts. By using this system, the study analyzed the pathology of drug-induced liver injury and provided the possibility of assessing varying drug susceptibilities based on individual polymorphism at organoid resolution. These approaches are undergoing rapid developments, allowing establishment of human organoids from adult/fetal human liver or pluripotent stem cells and modeling different liver diseases.68, 69, 70, 71 Different types of liver organoid models from mice, human beings, dogs, and cats now are available for several monogenic liver diseases such as Alagille syndrome, cystic fibrosis, primary sclerosing cholangitis, Wilsons disease, HBV infection, steatosis, or liver cancer, among others.72, 73, 74, 75, 76, 77, 78 The generation of organoids from adult patient liver tissues also retains the genetic background of the individual patient, thus creating patient-specific disease models and enabling in-depth investigations of pathogenesis mechanisms underlying genetic diseases and cancer. In conclusion, induced liver buds and liver organoids provide a platform for cell-based therapy, liver disease models, and drug screening that satisfy the demands of both basic and translational biomedical research.
Tissue-Based Approaches
Precision-cut liver slices (PCLS) are a native liver–like ex vivo model with intact intercellular and cell-matrix interactions.79 PCLS systems use ex vivo liver explants with a well-defined thickness, and, in comparison with the primary hepatocytes that are short-lived and lose much of their function in culture, PCLS cultures have been maintained for 15 days under optimal conditions. Hepatocytes in slices retain their membrane and intracellular polarization, in contrast to isolated hepatocytes, which lose their anatomic polarity after isolation. PCLS cultures have been established both from murine and human livers (Figure 3).
Human tissue for PCLS are obtained from explanted, resected, or nontransplantable tissues from liver tumor patients undergoing transplantation or liver resection. Liver slices also can be obtained from patients with severe fibrosis and cirrhosis undergoing transplantation. These usually are obtained using Krumdieck (now Alabama) tissue slicer to make liver slices with a diametric from 5–8 mm,79,80 and a thickness of 250–350 μm.81 These slices then are cultured with William’s E medium in regular tissue plates in either static, dynamic, or bioreactor-based culture systems. In static conditions, PCLS cultures have a shorter lifespan (24–48 hours) resulting from hypoxia and increased cell death. To minimize hypoxic death, strategies such as the use of synthetic oxygen carriers, rocking or shaking cultures, or perfusion bioreactors have been used to provide better perfusion of oxygen and media components.82,83 One study reported PCLS ex vivo cultures with sustained viability for over a 2-week period on a rocking platform.84 Through microarray profiling of purified individual cells, this study illustrated that all liver cells undergo changes in their gene expression profiles until day 4 of PCLS cultures, however, these changes seem to be stabilized from day 4 until day 15.
Recently, a study cultured PCLS on a bioreactor platform at a flow rate of approximately 18 μL/s, which imparted functional longevity to the system for approximately 6 days without any hepatocellular stress or fibrogenesis.83 Using this system, the study also successfully modeled ex vivo liver fibrogenesis using transforming growth factor β1 and platelet-derived growth factor (PDGF) stimulation. In another similar culture platform, primary hepatocytes or liver stem cells were cultured on ECM discs developed from a decellularized porcine85 or rat liver.86
Human PCLS cultures have proven indispensable for modeling of liver diseases and to the study of transport, metabolism, and biotransformation of drugs, toxins, and xenobiotics in both normal and diseased conditions.87, 88, 89 They also have been used to study ischemia/reperfusion damage in rodents and to evaluate the efficacy, specificity, and toxicity of virus-mediated gene therapy agents.80,90
With improved technological advancements and culture longevity, PCLS cultures of patient-specific tissues offers enormous potential for the characterization of patient-specific liver cellular heterogeneity and for the screening of novel antifibrotic and antitumorigenic drugs.
Mimicking the Sinusoidal Mechanobiology In Vitro
LSECs, the second most abundant cell type in the liver, are key players in maintaining hepatic homeostasis.91 Importantly, LSECs differ from classic vasculature endothelium because they lack an organized basement membrane and have cytoplasm that is penetrated by open fenestrae, making the hepatic microvascular endothelium discontinuous.92 LSEC behavior is largely regulated by shear stress and mechanical stretch induced by blood perfusion and liver microenvironment stiffness changes derived from deposition of ECM.93, 94, 95, 96 The effect of these varying mechanical cues on LSECs is particularly interesting, however, this has not been explored extensively until recently. The use of in vitro culture models of LSECs with microfluidic setups showed the effects of shear stress–derived effects. In a pioneering work from the Sessa and Groszmann laboratories, the investigators showed that LSECs respond to increasing shear stress in the microenvironment by increasing nitric oxide (NO) synthesis.97 Subsequent work defined the upstream signaling pathways, including the induction of the transcription factor Krüppel-like factor 2, in both healthy and cirrhotic LSECs.98 A recent study by Hilscher et al99 further elucidated the role of the mechanosensitive pathways in LSECs that drive recruitment of circulating blood cells contributing to portal hypertension and fibrogenesis. Using a flexcell device, cyclic biaxial stretch on murine LSECs was modeled, and showed transcriptional up-regulation of several chemotactic cytokines, neutrophil-extracellular trap activation from the recruited neutrophils, and microthrombi formation contributing to fibrosis. More recently, a LoC device with microfluidics was used to mimic physiological and pathologic pressures on primary LSECs culture.100 Transcriptomic analysis showed the detrimental effect of increased pressure on the LSEC phenotype and allowed identification of LSEC-derived pressure-related genes as noninvasive biomarkers for portal hypertension. Altogether these data show that mechanical cues can cause angiocrine and phenotypic changes in LSECs, leading to rapid alteration of HSC phenotype and fibrogenesis.101
In fibrotic livers, microvasculature remodeling contributes to increased ECM deposition and the consequent increase in intrahepatic vascular resistance.92 Olsen et al102 showed that increased stiffness induced activation of HSCs. Juin et al103 showed that increased ECM matrix rigidity increased the number of podosomes (actin-rich structures involved in motility and proteolysis) formed in LSECs, suggesting that the cells responded to mechanical stress, however, the effect on LSEC function was not explored. Impairment of hepatocyte and stellate cell function in response to high stiffness has been described previously in the literature.41,43,102,104 In the context of liver-specific endothelial cells, a recent publication showed that LSECs also dedifferentiate in high-stiffness conditions, losing their capacity to produce vasoactive mediators such as NO and becoming capillarized as shown by the loss of their characteristic fenestrae. Interestingly, the investigators pointed out the tension between the cytoskeleton and the nuclear shape as a fundamental process transducing the sensing of stiffness into phenotypical responses in all sinusoidal cells.105 Importantly, this study also showed that cirrhotic liver cells improve their phenotype when cultured in a healthy (nonstiff) environment, suggesting potential new avenues of therapy development. In an unpublished work, Kidambi et al confirmed that LSECs are responsive to stiffness resulting in rapid capillarization (loss in fenestrae), loss of hyaluronic acid endocytosis, and higher cell adhesion molecules.46
These advanced in in vitro experiments point to an interesting and underexplored area of the role of mechanical stimuli on sinusoidal biology during physiological and pathologic conditions. The key to unlocking the potential therapeutic avenues for sinusoidal dysfunction from these in vitro findings will be to integrate the data with in vivo functions.
“Omics” for the Study of Liver Cells
Advances in omics methods have led to discoveries in liver biology and pathology at the cellular, tissue, and system levels. These methods also have facilitated holistic insight into CLD in the clinical setting, and are generating noninvasive diagnostic modalities for the distinct stages of liver diseases. This multi-omics approach consists of tracing the flow of information from transcriptomics, proteomics, metabolomics, scRNA-seq, single-nucleus analysis, and interactomics. The key findings of these techniques are summarized herein.
Transcriptomics refers to the quantitative assessment of all coding and noncoding RNA transcripts and reflects cellular transcriptional activity. Transcriptomic profiling has resulted in various predictive modalities involving gene expression parameters, targeted measurements, and micro RNA (miRNA) panels with increased functionality in different chronic liver diseases.106,107 Several studies have identified miR-122 as a potential diagnostic biomarker for CLDs. Most of them have shown that miR-122 alone or in combination with other miRNAs (eg, miR-1290, miR-27, miR-192, miR-34, miR-99a) can accurately predict the presence of NAFLD or NASH, but they all perform inadequately when trying to differentiate NAFLD from NASH.108, 109, 110 A recent study performed a comprehensive transcriptomic analysis of primary LSECs during the progression of cirrhosis in which specific molecular signatures, novel biomarkers, and therapeutic targets associated with LSECs dedifferentiation were delineated.111
Proteomics refers to the investigation of proteomes—all proteins expressed by a cell. Several studies have investigated the hepatic proteome alone or in combination with the blood proteome, both in animal models or in human beings with CLDs, aiming to answer fundamental pathophysiological questions.112,113 Krahmer et al112 assessed the levels and cellular distribution of 6000 liver proteins and 16,000 phosphopeptides in the liver of mice developing hepatic steatosis owing to a high-fat diet. This work produced important fundamental information about the reorganization of organelles, lipid accumulation, and cellular dysfunction that occurs with nutrient overload. Xue et al113 identified almost 220 proteins that are significantly different in patients with NAFLD compared with obese metabolically healthy individuals. The proteins that were identified to be increased in CLD were those involved in peroxisome proliferator-activated receptor signaling and ECM receptor interactions whereas the ones that were reduced were localized mainly in mitochondria and involved with oxidative phosphorylation. Expanding on complications of the disease, the proteome of specific cells also has been examined. Vu et al114 compared the proteome of hepatocyte monoculture and hepatocytes in organotypic rat liver models, and showed that in a 3D liver model the predominant proteomic phenotype supports fatty acid metabolism and when hepatocytes are cultured in monoculture the proteome shifts to favor glucose metabolism. In addition, they observed an increase in structural and migratory proteins (signaling hepatocyte dedifferentiation) in hepatocyte monoculture, highlighting the need for cell–cell and cell–ECM interactions for maintenance of functional hepatocytes. Lao et al115 performed a proteomic analysis between normal and dedifferentiated LSECs. Dedifferentiation and loss of fenestrae in LSECs precedes the onset of fibrosis and is considered a crucial event in the pathology of liver diseases.92,116,117 A comparison of the normal and dedifferentiated LSECs showed that in dedifferentiated LSECs the most enriched functional categories of proteins were those related to nucleotide, organic acid metabolism, oxidative stress, small molecular and lipid metabolism, cell death regulation, and endocytosis, while those down-regulated by dedifferentiation were transcription regulation, actin cytoskeleton reorganization, cell migration, immune system process, ribosome biogenesis, apoptotic process, angiogenesis, glycerophospholipid metabolism, and cellular lipid metabolism.
Metabolomics refers to the investigation of small molecules and metabolic products, such as amino acids, fatty acids, and carbohydrates. An increasing number of studies have begun to study liver-specific metabolomics in the context of disease using both primary cells and cell culture models. Kim et al118 analyzed and compared metabolites in fetal and adult hepatocytes from human donors. They identified 211 metabolites in the hepatocytes. Specifically, the metabolites in the glycolysis/glyconeogenesis pathway, tricarboxylic acid cycle, and urea cycle were lower in fetal hepatocytes than in adult hepatocytes. Li et al119 used nuclear magnetic resonance–based metabolomics to investigate the metabolic alterations in hepatocytes caused by HBV infection. They showed that HBV infection contributed to hepatocellular carcinoma (HCC) by up-regulation of the glutamine-fructose-6-phosphate amidotransferase 1-activated hexosamine biosynthesis and choline kinase α-activated phosphatidylcholine biosynthesis. Yue et al120 showed using the nuclear magnetic resonance (NMR)-based metabolomic approach that HBV protein (HBx) disrupted the metabolism of glucose, lipids, and amino acids, especially nucleic acids. Min Hae et al121 performed metabolic profiling on Huh7 cells with patatin-like phospholipase domain containing 3 (PNPLA3) small interfering RNA silencing and overexpression using gas chromatography–mass spectrometry and liquid chromatography–mass spectrometry metabolic profiling to investigate their role in HCC. Silencing of the PNPLA3 gene resulted in a decrease in amino acid metabolism, suggestive of a catabolic response with extensive protein breakdown. Among the lipids, there was an increase in the levels of myoinositol, cysteine sulfinic acid, polyunsaturated fatty acids, lysolipids, and sphingolipids Overexpression of PNPLA3 mirrored metabolic changes in the opposite direction with an increase in the levels of cholesterol and lactic acid with a shift to anaerobic metabolism. Some of the metabolic signatures associated with the presence of PNPLA3 risk allele such as high cholesterol levels, very low density lipoproteins levels, and so forth, also have been associated with cardiovascular disease in patients with NAFLD.122 These and other studies123 explain how the use of omics approaches could help to unravel novel phenotype and pathogenesis mechanisms associated with the presence of genetic polymorphisms in complex human liver diseases.
Single-cell, single-nuclei transcriptomics using next-generation transcript sequencing (scRNA-seq/snRNA-seq) is now emerging as a powerful tool to profile cell-to-cell variability on a genomic scale with broad implications for both basic and clinical research.124 In a mouse model of liver fibrosis induced by CCl4, Krenkel et al125 used freshly isolated HSCs for scRNA-seq and found that activation of HSCs and their transdifferentiation toward collagen-secreting myofibroblasts split into heterogeneous populations, characterized by α-smooth muscle actin, collagens, or immunologic markers, while resting HSCs formed a homogenous population characterized by high PDGF-receptor expression. A similar scRNA-seq study using CCl4 to induce advanced liver cirrhosis identified 6 clusters of liver endothelial cell (EC) populations including 3 clusters of LSECs that associated with zone-specific transcriptomic changes, 2 clusters of vascular ECs, and 1 cluster of lymphatic ECs.126 Hepatotoxicity induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin also showed the diversity of liver cells through the identification of 11 subtypes following pericentral, midzonal, and periportal hepatocyte subpopulations by snRNA-seq, which was a more feasible technique than scRNA-seq in terms of application to frozen samples.127 Recently, scRNA-seq was used to characterize mouse embryos on embryonic days 7.5 to 10.5, and Lotto et al128 provided a comprehensive atlas of liver cell lineage detailing the divergence of vascular and sinusoidal endothelia, hepatoblast specification, and the emergence of a distinct migratory hepatomesenchymal cell type. The most developed scRNA-seq data set likely is that established by key immune cell populations in the liver, particularly from mouse models of NAFLD/NASH.129 A series of elegant studies using scRNA-seq has provided unprecedentedly granular insights into hepatic immune cell heterogeneity, showing striking alterations, particularly in myeloid cells and macrophages in liver diseases,130, 131, 132, 133, 134 and into related extrahepatic compartments such as bone marrow135 or adipose tissue.136
Regarding the cellular landscape of the human liver, scRNA-seq also has shown the physiological heterogeneity of human liver cells,137,138 the fibrotic niche of human liver cirrhosis including the identification of pathogenic subpopulations of TREM2+CD9+ macrophages, atypical chemokine receptor 1+, and plasmalemma vesicle–associated protein+ ECs and PDGF-receptor α+ collagen-producing myofibroblasts,139 and the immune microenvironment in the context of HCC.140 Although scRNA-seq/snRNA-seq remains an expensive and time-consuming technique that requires skilled bioinformatics support, it is a valuable tool to characterize liver function and gene expression dynamics during liver disease, as well as to identify prognostic markers or signatures, and to facilitate discovery of new therapeutic targets.141 A key challenge for all mentioned omics techniques is accurate data integration. For instance, the most granular insight into single-cell transcriptomes by scRNA-seq/snRNA-seq techniques comes at the expense of isolating the cells (or nuclei) out of their cellular context141; therefore, spatially resolved modalities (eg, multiplex immunostaining, spatial transcriptomics, imaging mass cytometry) are needed to complement these findings.142 This has been shown convincingly for immune cell populations in which not only the immune cell phenotype (or single-cell transcriptome), but also their location within the hepatic microenvironment, determines their most likely function during liver diseases.143
Conclusions and Future Directions
As described earlier, in recent years there has been a great advance in the availability and utility of in vitro systems for the study of pathophysiology of the liver. Today, we have a wide range of possibilities to better understand the behavior of cells and tissues in the laboratory, which combine harmoniously with those observations obtained in animal models. Although progress in the field of translational hepatology is evident, we must continue working to create more complete, reliable, and cost-effective systems of human liver diseases. We herein summarize some of the avenues of work that we should develop through collaborative multidisciplinary work, combining the academic and private sectors.
Liver-on-a-chip systems, which already reflect the multicellularity of the liver, should be improved by incorporating biomechanical stimuli typical of the disease under study, such as a specific matrix or sinusoidal pressure, and potentially the relevant immune cells. In addition, the incorporation of biochemical or biological parameter sensors would be of great help for real-time cellular analysis in response to new drugs.
The great potential of 3D liver systems, which currently use mostly matrices of natural origin, has the great advantage of simulating the ECM of the human liver but, at the same time, complicates its standardization and global use. The development and validation of matrices with defined composition, perhaps including the most abundant components in adequate ratios, could assist with expanding their use. Similarly, experimental variables that mimic the biomechanics of the sinusoid (shear stress, pressure, stiffness, and so forth) also should be standardized, thus facilitating the comparison of results from different research groups.
The use of PCLS allows an understanding of the hepatic response to new compounds, but only for a limited period of time. It would be very beneficial to improve the viable incubation time, perhaps by combining several in vitro systems including slices, and the use of tissue from liver disease patients/models.
The field of omics applied to hepatology, and to the rest of biomedical disciplines, is immense and it is difficult to ensure the currency of literature and use of the most advanced techniques. Analysis at the single-cell level, which already is being prototyped using fixed tissue, will transform what we know today as spatial omics. However, tissue cartography requires significant financial investment and excellent experimental design. Therefore, public–private consortiums that include basic scientists and physicians would be of great interest for the sake of advancing knowledge.
Overall, the techniques described in this review and those that are on the horizon can greatly assist to understand liver diseases, develop new therapies, and foster personalized medicine in hepatology. Of course, we need to combine them in a virtuous way, including tissue/cells of human origin whenever possible, and improving the way we mimic human diseases in vitro. If future work is developed further by multidisciplinary teams, success is assured.
Acknowledgments
The authors acknowledge the International Society for Hepatic Sinusoidal Research (www.ishsr.net) for commissioning this review article and for continuously fostering research in the field of basic and translational hepatology. Figures 1-3 of this work were created with biorender.com.
Footnotes
Conflicts of interest These authors disclose the following: Frank Tacke’s laboratory received research grants from Gilead, Allergan, Bristol-Myers Squibb, and Inventiva; and Jordi Gracia-Sancho’s laboratory received research funding from Gilead, Inventiva, Conatus, Surrozen, GAT Tx, and BrudyLab. The remaining authors disclose no conflicts.
Funding Supported by ASEAN grant CRD/2019/000120 from the Department of Science and Technology, India (S.K.), by National Institutes of Health grants 1R01AA027189-01A1 and P20 GM104320 to the Nebraska Center for the Prevention of Obesity Diseases, grant P20 GM113126 to the Nebraska Center for Integrated Biomolecular Communication, and a Nebraska Research Initiative-systems grant (S.K.). Supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science JSPS-22K08083 and 22H03061, and a grant for the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (AMED-JP22fk0210107) (L.T.T.T.). Funded by US Public Health Service grant R01 DK111677 from the National Institute of Diabetes and Digestive and Kidney Diseases, grant R01AA025907 from the National Institute on Alcohol Abuse and Alcoholism, and US Veterans Administration grant I01BX005093 from the Biomedical Laboratory Research and Development (N.N.). Supported by the McKnight Philanthropic Bequest and National Health and Medical Research Council (V.C.C.); by National Natural Science Foundation of China grant 82030021 (W.-F.X.); by German Research Foundation grants DFG SFB/TRR 296 and CRC1382 (project-ID 403224013), and the German Ministry of Education and Research (BMBF DEEP-HCC consortium) (F.T.); and by Instituto de Salud Carlos III grant FIS PI20/00220 (co-funded by the European Union), the CIBEREHD, Swiss National Science Funds 320030_189252, the Novartis Foundation for Medical-Biological Research, and the Foundation Suisse Contre le Cancer du Foie (J.G.-S.).
References
- 1.Karlsen T.H., Sheron N., Zelber-Sagi S., et al. The EASL–Lancet Liver Commission: protecting the next generation of Europeans against liver disease complications and premature mortality. Lancet. 2022;399:61–116. doi: 10.1016/S0140-6736(21)01701-3. [DOI] [PubMed] [Google Scholar]
- 2.Nevzorova Y.A., Boyer-Diaz Z., Cubero F.J., Gracia-Sancho J. Animal models for liver disease – a practical approach for translational research. J Hepatol. 2020;73:423–440. doi: 10.1016/j.jhep.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Polidoro M.A., Ferrari E., Marzorati S., et al. Experimental liver models: from cell culture techniques to microfluidic organs-on-chip. Liver Int. 2021;41:1744–1761. doi: 10.1111/liv.14942. [DOI] [PubMed] [Google Scholar]
- 4.Ortega-Ribera M., Yeste J., Villa R., Gracia-Sancho J. In: Nanoengineered biomaterials for regenerative medicine. Mozafari M., Rajadas J., Kaplan D., editors. Elsevier; 2018. Nanoengineered biomaterials for the treatment of liver diseases; pp. 417–441. [Google Scholar]
- 5.Shroff T., Aina K., Maass C., et al. Studying metabolism with multi-organ chips: new tools for disease modelling, pharmacokinetics and pharmacodynamics. Open Biol. 2022;12 doi: 10.1098/rsob.210333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassan S., Sebastian S., Maharjan S., et al. Liver-on-a-chip models of fatty liver disease. Hepatology. 2020;71:733–740. doi: 10.1002/hep.31106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du K., Li S., Li C., et al. Modeling nonalcoholic fatty liver disease on a liver lobule chip with dual blood supply. Acta Biomater. 2021;134:228–239. doi: 10.1016/j.actbio.2021.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Teng Y., Zhao Z., Tasnim F., et al. A scalable and sensitive steatosis chip with long-term perfusion of in situ differentiated HepaRG organoids. Biomaterials. 2021;275 doi: 10.1016/j.biomaterials.2021.120904. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Wang H., Deng P., et al. Modeling human nonalcoholic fatty liver disease (NAFLD) with an organoids-on-a-chip system. ACS Biomater Sci Eng. 2020;6:5734–5743. doi: 10.1021/acsbiomaterials.0c00682. [DOI] [PubMed] [Google Scholar]
- 10.Lefere S., Puengel T., Hundertmark J., et al. Differential effects of selective- and pan-PPAR agonists on experimental steatohepatitis and hepatic macrophages. J Hepatol. 2020;73:757–770. doi: 10.1016/j.jhep.2020.04.025. [DOI] [PubMed] [Google Scholar]
- 11.Boyer-Diaz Z., Aristu-Zabalza P., Andrés-Rozas M., et al. Pan-PPAR agonist lanifibranor improves portal hypertension and hepatic fibrosis in experimental advanced chronic liver disease. J Hepatol. 2021;74:1188–1199. doi: 10.1016/j.jhep.2020.11.045. [DOI] [PubMed] [Google Scholar]
- 12.Deng J., Chen Z., Zhang X., et al. A liver-chip-based alcoholic liver disease model featuring multi-non-parenchymal cells. Biomed Microdevices. 2019;21:57. doi: 10.1007/s10544-019-0414-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee J., Choi B., No D.Y., et al. A 3D alcoholic liver disease model on a chip. Integr Biol (Camb) 2016;8:302–308. doi: 10.1039/c5ib00298b. [DOI] [PubMed] [Google Scholar]
- 14.Nawroth J.C., Petropolis D.B., Manatakis D.V., et al. Modeling alcohol-associated liver disease in a human Liver-Chip. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega-Prieto A.M., Skelton J.K., Wai S.N., et al. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat Commun. 2018;9:682. doi: 10.1038/s41467-018-02969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortega-Ribera M., Fernández-Iglesias A., Illa X., et al. Resemblance of the human liver sinusoid in a fluidic device with biomedical and pharmaceutical applications. Biotechnol Bioeng. 2018;115:2585–2594. doi: 10.1002/bit.26776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Igleasias A., Ortega Ribera M., Guixé-Muntet S., Gracia-Sancho J. 4 In 1: antibody-free protocol for isolating the main hepatic cells from healthy and cirrhotic single rat livers. J Cell Mol Med. 2019;23:877–886. doi: 10.1111/jcmm.13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Y., Khandekar G., Llewellyn J., et al. A bile duct-on-a-chip with organ-level functions. Hepatology. 2020;71:1350–1363. doi: 10.1002/hep.30918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ya S., Ding W.G., Li S., et al. On-chip construction of liver lobules with self-assembled perfusable hepatic sinusoid networks. ACS Appl Mater Interfaces. 2021;13:32640–32652. doi: 10.1021/acsami.1c00794. [DOI] [PubMed] [Google Scholar]
- 20.Kang Y.B.A., Eo J., Mert S., et al. Metabolic patterning on a chip: towards in vitro liver zonation of primary rat and human hepatocytes. Sci Rep. 2018;8:8951. doi: 10.1038/s41598-018-27179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Y., Li N., Yang H., et al. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab Chip. 2017;17:782–794. doi: 10.1039/c6lc01374k. [DOI] [PubMed] [Google Scholar]
- 22.Jang K.J., Otieno M.A., Ronxhi J., et al. Reproducing human and cross-species drug toxicities using a liver-chip. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aax5516. eaax5516. [DOI] [PubMed] [Google Scholar]
- 23.Tsochatzis E.A., Bosch J., Burroughs A.K. Liver cirrhosis. Lancet. 2014;383:1749–1761. doi: 10.1016/S0140-6736(14)60121-5. [DOI] [PubMed] [Google Scholar]
- 24.Theobald J., Ghanem A., Wallisch P., et al. Liver-kidney-on-chip to study toxicity of drug metabolites. ACS Biomater Sci Eng. 2018;4:78–89. doi: 10.1021/acsbiomaterials.7b00417. [DOI] [PubMed] [Google Scholar]
- 25.De Gregorio V., Telesco M., Corrado B., et al. Intestine-liver axis on-chip reveals the intestinal protective role on hepatic damage by emulating ethanol first-pass metabolism. Front Bioeng Biotechnol. 2020;8:163. doi: 10.3389/fbioe.2020.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wogram E., Svoboda D., Communal C., et al. Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci Adv. 2021;7 doi: 10.1126/sciadv.abd1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J., Lee C., Kim I., et al. Three-dimensional human liver-chip emulating premetastatic niche formation by breast cancer-derived extracellular vesicles. ACS Nano. 2020;14:14971–14988. doi: 10.1021/acsnano.0c04778. [DOI] [PubMed] [Google Scholar]
- 28.Dunn J.C.Y., Tompkins R.G., Yarmush M.L. Hepatocytes in collagen sandwich: evidence for transcriptional and translational regulation. J Cell Biol. 1992;116:1043–1053. doi: 10.1083/jcb.116.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arteel G.E., Naba A. The liver matrisome – looking beyond collagens. JHEP Rep. 2020;2 doi: 10.1016/j.jhepr.2020.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W., Desert R., Ge X., et al. The matrisome genes from hepatitis B–related hepatocellular carcinoma unveiled. Hepatol Commun. 2021;5:1571–1585. doi: 10.1002/hep4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur S., Tripathi D.M., Venugopal J.R., Ramakrishna S. Advances in biomaterials for hepatic tissue engineering. Curr Opin Biomed Eng. 2020;13:190–196. [Google Scholar]
- 32.Brown J.H., Das P., DiVito M.D., et al. Nanofibrous PLGA electrospun scaffolds modified with type I collagen influence hepatocyte function and support viability in vitro. Acta Biomater. 2018;73:217–227. doi: 10.1016/j.actbio.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das P., DiVito M.D., Wertheim J.A., Tan L.P. Collagen-I and fibronectin modified three-dimensional electrospun PLGA scaffolds for long-term in vitro maintenance of functional hepatocytes. Mater Sci Eng C. 2020;111 doi: 10.1016/j.msec.2020.110723. [DOI] [PubMed] [Google Scholar]
- 34.Janani G., Nandi Samit K., Mandal Biman B. Functional hepatocyte clusters on bioactive blend silk matrices towards generating bioartificial liver constructs. Acta Biomater. 2018;67:167–182. doi: 10.1016/j.actbio.2017.11.053. [DOI] [PubMed] [Google Scholar]
- 35.Bartneck M., Topuz F., Tag C.G., et al. Molecular response of liver sinusoidal endothelial cells on hydrogels. Mater Sci Eng C. 2015;51:64–72. doi: 10.1016/j.msec.2015.02.045. [DOI] [PubMed] [Google Scholar]
- 36.Lou R., Xie H., Zheng H., et al. Alginate-based microcapsules with galactosylated chitosan internal for primary hepatocyte applications. Int J Biol Macromol. 2016;93:1133–1140. doi: 10.1016/j.ijbiomac.2016.09.078. [DOI] [PubMed] [Google Scholar]
- 37.Kidambi S., Yarmush R.S., Novik E.R., et al. Oxygen-mediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc Natl Acad Sci U S A. 2009;106:15714–15719. doi: 10.1073/pnas.0906820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidambi S., Sheng L., Yarmush M.L., et al. Patterned co-culture of primary hepatocytes and fibroblasts using polyelectrolyte multilayer templates. Macromol Biosci. 2007;7:344–353. doi: 10.1002/mabi.200600205. [DOI] [PubMed] [Google Scholar]
- 39.Kidambi S., Lee I., Chan C. Controlling primary hepatocyte adhesion and spreading on protein-free polyelectrolyte multilayer films. J Am Chem Soc. 2004;126:16286–16287. doi: 10.1021/ja046188u. [DOI] [PubMed] [Google Scholar]
- 40.You J., Park S.A., Shin D.S., et al. Characterizing the effects of heparin gel stiffness on function of primary hepatocytes. Tissue Eng Part A. 2013;19:2655–2663. doi: 10.1089/ten.tea.2012.0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen A.A., Khetani S.R., Lee S., et al. Modulation of hepatocyte phenotype in vitro via chemomechanical tuning of polyelectrolyte multilayers. Biomaterials. 2009;30:1113–1120. doi: 10.1016/j.biomaterials.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Semler E.J., Lancin P.A., Dasgupta A., Moghe P.V. Engineering hepatocellular morphogenesis and function via ligand-presenting hydrogels with graded mechanical compliance. Biotechnol Bioeng. 2005;89:296–307. doi: 10.1002/bit.20328. [DOI] [PubMed] [Google Scholar]
- 43.Desai S.S., Tung J.C., Zhou V.X., et al. Physiological ranges of matrix rigidity modulate primary mouse hepatocyte function in part through hepatocyte nuclear factor 4 alpha. Hepatology. 2016;64:261–275. doi: 10.1002/hep.28450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan V., Moeun Y., Kidambi S. Exploring interactions between primary hepatocytes and non-parenchymal cells on physiological and pathological liver stiffness. Biology (Basel) 2021;10:408. doi: 10.3390/biology10050408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kidambi S. In: Mueller, S. (eds) Liver Elastography. Springer; 2020. Stiffness and hepatocytes function in vitro; pp. 645–660. [Google Scholar]
- 46.Natarajan V., Moeller M., Casey C.A., Harris Edward N. Matrix stiffness regulates liver sinusoidal endothelial cell function mimicking responses in fatty liver disease. bioRxiv. 2020;27 [Google Scholar]
- 47.Ganesan M., Dagur R.S., Makarov E., et al. Matrix stiffness regulate apoptotic cell death in HIV-HCV co-infected hepatocytes: importance for liver fibrosis progression. Biochem Biophys Res Commun. 2018;500:717–722. doi: 10.1016/j.bbrc.2018.04.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natarajan V., Berglund E.J., Chen D.X., Kidambi S. Substrate stiffness regulates primary hepatocyte functions. RSC Adv. 2015;5:80956–80966. doi: 10.1039/C5RA15208A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y., Kang K., Jeong J., et al. Three-dimensional (3D) printing of mouse primary hepatocytes to generate 3D hepatic structure. Ann Surg Treat Res. 2017;92:67–72. doi: 10.4174/astr.2017.92.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H., Sun L., Pang Y.H., et al. Three-dimensional bioprinted hepatorganoids prolong survival of mice with liver failure. Gut. 2021;70:567–574. doi: 10.1136/gutjnl-2019-319960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen D.G., Funk J., Robbins J.B., et al. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma A., Rawal P., Tripathi D.M., et al. Upgrading hepatic differentiation and functions on 3d printed silk-decellularized liver hybrid scaffolds. ACS Biomater Sci Eng. 2021;7:3861–3873. doi: 10.1021/acsbiomaterials.1c00671. [DOI] [PubMed] [Google Scholar]
- 53.Lee H., Chae S., Kim J.Y., et al. Cell-printed 3D liver-on-a-chip possessing a liver microenvironment and biliary system. Biofabrication. 2019;11 doi: 10.1088/1758-5090/aaf9fa. [DOI] [PubMed] [Google Scholar]
- 54.Lee H., Han W., Kim H., et al. Development of liver decellularized extracellular matrix bioink for three-dimensional cell printing-based liver tissue engineering. Biomacromolecules. 2017;18:1229–1237. doi: 10.1021/acs.biomac.6b01908. [DOI] [PubMed] [Google Scholar]
- 55.Rawal P., Tripathi D.M., Ramakrishna S., Kaur S. Prospects for 3D bioprinting of organoids. Bio-Des Manuf. 2021;4:627–640. [Google Scholar]
- 56.Duval K., Grover H., Han L.H., et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 57.Ramaiahgari S.C., Den Braver M.W., Herpers B., et al. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch Toxicol. 2014;88:1083–1095. doi: 10.1007/s00204-014-1215-9. [DOI] [PubMed] [Google Scholar]
- 58.Sato K., Zhang W., Safarikia S., et al. Organoids and spheroids as models for studying cholestatic liver injury and cholangiocarcinoma. Hepatology. 2021;74:491–502. doi: 10.1002/hep.31653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takebe T., Zhang R.R., Koike H., et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat Protoc. 2014;9:396–409. doi: 10.1038/nprot.2014.020. [DOI] [PubMed] [Google Scholar]
- 60.Sekine K., Takebe T., Taniguchi H. Liver regeneration using cultured liver bud. Methods Mol Biol. 2017;1597:207–216. doi: 10.1007/978-1-4939-6949-4_15. [DOI] [PubMed] [Google Scholar]
- 61.Takebe T., Sekine K., Kimura M., et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Achilli T.M., Meyer J., Morgan J.R. Advances in the formation, use and understanding of multi-cellular spheroids. Expert Opin Biol Ther. 2012;12:1347–1360. doi: 10.1517/14712598.2012.707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huch M., Koo B.K. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 64.Hong S., Song J.M. A 3D cell printing-fabricated HepG2 liver spheroid model for high-content in situ quantification of drug-induced liver toxicity. Biomater Sci. 2021;9:5939–5950. doi: 10.1039/d1bm00749a. [DOI] [PubMed] [Google Scholar]
- 65.Takebe T., Sekine K., Enomura M., et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 66.Camp J.G., Sekine K., Gerber T., et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 67.Shinozawa T., Kimura M., Cai Y., et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell-derived organoids. Gastroenterology. 2021;160:831–846.e10. doi: 10.1053/j.gastro.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ouchi R., Togo S., Kimura M., et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30:374–384.e6. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hendriks D., Artegiani B., Hu H., et al. Establishment of human fetal hepatocyte organoids and CRISPR–Cas9-based gene knockin and knockout in organoid cultures from human liver. Nat Protoc. 2021;16:182–217. doi: 10.1038/s41596-020-00411-2. [DOI] [PubMed] [Google Scholar]
- 70.Sampaziotis F., Muraro D., Tysoe O.C., et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science. 2021;371:839–846. doi: 10.1126/science.aaz6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roos F.J.M., van Tienderen G.S., Wu H., et al. Human branching cholangiocyte organoids recapitulate functional bile duct formation. Cell Stem Cell. 2022;29:776–794.e13. doi: 10.1016/j.stem.2022.04.011. [DOI] [PubMed] [Google Scholar]
- 72.Guan Y., Xu D., Garfin P.M., et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight. 2017;2 doi: 10.1172/jci.insight.94954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ogawa M., Ogawa S., Bear C.E., et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 74.Soroka C.J., Assis D.N., Alrabadi L.S., et al. Bile-derived organoids from patients with primary sclerosing cholangitis recapitulate their inflammatory immune profile. Hepatology. 2019;70:871–882. doi: 10.1002/hep.30470. [DOI] [PubMed] [Google Scholar]
- 75.Nantasanti S., Spee B., Kruitwagen H.S., et al. Disease modeling and gene therapy of copper storage disease in canine hepatic organoids. Stem Cell Rep. 2015;5:895–907. doi: 10.1016/j.stemcr.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nie Y.Z., Zheng Y.W., Miyakawa K., et al. Recapitulation of hepatitis B virus–host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine. 2018;35:114–123. doi: 10.1016/j.ebiom.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kruitwagen H.S., Oosterhoff L.A., Vernooij I.G.W.H., et al. Long-term adult feline liver organoid cultures for disease modeling of hepatic steatosis. Stem Cell Rep. 2017;8:822–830. doi: 10.1016/j.stemcr.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Broutier L., Mastrogiovanni G., Verstegen M.M.A., et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Graaf I.A.M., Olinga P., De Jager M.H., et al. Preparation and incubation of precision-cut liver and intestinal slices for application in drug metabolism and toxicity studies. Nat Protoc. 2010;5:1540–1551. doi: 10.1038/nprot.2010.111. [DOI] [PubMed] [Google Scholar]
- 80.Dewyse L., Reynaert H., van Grunsven L.A. Best practices and progress in precision-cut liver slice cultures. Int J Mol Sci. 2021;22:7137. doi: 10.3390/ijms22137137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kartasheva-Ebertz D., Gaston J., Lair-Mehiri L., et al. Adult human liver slice cultures: modelling of liver fibrosis and evaluation of new anti-fibrotic drugs. World J Hepatol. 2021;13:187–217. doi: 10.4254/wjh.v13.i2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koch A., Saran S., Tran D., et al. Murine precision-cut liver slices (PCLS): a new tool for studying tumor microenvironments and cell signaling ex vivo. Cell Commun Signal. 2014;12:73. doi: 10.1186/s12964-014-0073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paish H.L., Reed L.H., Brown H., et al. A bioreactor technology for modeling fibrosis in human and rodent precision-cut liver slices. Hepatology. 2019;70:1377–1391. doi: 10.1002/hep.30651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu X., Roberto J.B., Knupp A., et al. Precision-cut human liver slice cultures as an immunological platform. J Immunol Methods. 2018;455:71–79. doi: 10.1016/j.jim.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lang R., Stern M.M., Smith L., et al. Three-dimensional culture of hepatocytes on porcine liver tissue-derived extracellular matrix. Biomaterials. 2011;32:7042–7052. doi: 10.1016/j.biomaterials.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Schneeberger K., Sánchez-Romero N., Ye S., et al. Large-scale production of LGR5-positive bipotential human liver stem cells. Hepatology. 2020;72:257–270. doi: 10.1002/hep.31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Othman A., Ehnert S., Dropmann A., et al. Precision-cut liver slices as an alternative method for long-term hepatotoxicity studies. Arch Toxicol. 2020;94:2889–2891. doi: 10.1007/s00204-020-02861-9. [DOI] [PubMed] [Google Scholar]
- 88.Bigaeva E., Gore E., Simon E., et al. Transcriptomic characterization of culture-associated changes in murine and human precision-cut tissue slices. Arch Toxicol. 2019;93:3549–3583. doi: 10.1007/s00204-019-02611-6. [DOI] [PubMed] [Google Scholar]
- 89.Brugger M., Laschinger M., Lampl S., et al. High precision-cut liver slice model to study cell-autonomous antiviral defense of hepatocytes within their microenvironment. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dubray B.J., Conzen K.D., Upadhya G.A., et al. Novel in vitro model for studying hepatic ischemia-reperfusion injury using liver cubes. Surgery. 2012;152:247–253. doi: 10.1016/j.surg.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gracia-Sancho J., Marrone G., Fernández-Iglesias A. Hepatic microcirculation and mechanisms of portal hypertension. Nat Rev Gastroenterol Hepatol. 2019;16:221–234. doi: 10.1038/s41575-018-0097-3. [DOI] [PubMed] [Google Scholar]
- 92.Gracia-Sancho J., Caparrós E., Fernández-Iglesias A., Francés R. Role of liver sinusoidal endothelial cells in liver diseases. Nat Rev Gastroenterol Hepatol. 2021;18:411–431. doi: 10.1038/s41575-020-00411-3. [DOI] [PubMed] [Google Scholar]
- 93.Ayajiki K., Kindermann M., Hecker M., et al. Intracellular pH and tyrosine phosphorylation but not calcium determine shear stress-induced nitric oxide production in native endothelial cells. Circ Res. 1996;78:750–758. doi: 10.1161/01.res.78.5.750. [DOI] [PubMed] [Google Scholar]
- 94.Kang N. Mechanotransduction in liver diseases. Semin Liver Dis. 2020;40:84–90. doi: 10.1055/s-0039-3399502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piecha F., Peccerella T., Bruckner T., et al. Arterial pressure suffices to increase liver stiffness. Am J Physiol Gastrointest Liver Physiol. 2016;311:G945–G953. doi: 10.1152/ajpgi.00399.2015. [DOI] [PubMed] [Google Scholar]
- 96.Mueller S. Does pressure cause liver cirrhosis? The sinusoidal pressure hypothesis. World J Gastroenterol. 2016;22:10482–10501. doi: 10.3748/wjg.v22.i48.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shah V., Haddad F.G., Garcia-Cardena G., et al. Liver sinusoidal endothelial cells are responsible for nitric oxide modulation of resistance in the hepatic sinusoids. J Clin Invest. 1997;100:2923–2930. doi: 10.1172/JCI119842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gracia-Sancho J., Russo L., García-Calderó H., et al. Endothelial expression of transcription factor Kruppel-like factor 2 and its vasoprotective target genes in the normal and cirrhotic rat liver. Gut. 2011;60:517–524. doi: 10.1136/gut.2010.220913. [DOI] [PubMed] [Google Scholar]
- 99.Hilscher M.B., Sehrawat T., Arab J.P., et al. Mechanical stretch increases expression of CXCL1 in liver sinusoidal endothelial cells to recruit neutrophils, generate sinusoidal microthombi, and promote portal hypertension. Gastroenterology. 2019;157:193–209.e9. doi: 10.1053/j.gastro.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ortega-Ribera M., Gibert-Ramos A., Abad-Jordà L., et al. Increased sinusoidal pressure influences LSEC mechanosensing pathways uncovering specific plasma biomarkers of portal hypertension. Hepatology. 2021;74(S1):157–1288. doi: 10.1016/j.jhepr.2023.100722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu L., You Z., Yu H., et al. Mechanotransduction-modulated fibrotic microniches reveal the contribution of angiogenesis in liver fibrosis. Nat Mater. 2017;16:1252–1261. doi: 10.1038/nmat5024. [DOI] [PubMed] [Google Scholar]
- 102.Olsen L., Bloomer S.A., Chan E.P., et al. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am J Physiol Gastrointest Liver Physiol. 2011;301:G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Juin A., Planus E., Guillemot F., et al. Extracellular matrix rigidity controls podosome induction in microvascular endothelial cells. Biol Cell. 2013;105:46–57. doi: 10.1111/boc.201200037. [DOI] [PubMed] [Google Scholar]
- 104.Dou C., Liu Z., Tu K., et al. P300 acetyltransferase mediates stiffness-induced activation of hepatic stellate cells into tumor-promoting myofibroblasts. Gastroenterology. 2018;154:2209–2221.e14. doi: 10.1053/j.gastro.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guixé-Muntet S., Ortega-Ribera M., Wang C., et al. Nuclear deformation mediates liver cell mechanosensing in cirrhosis. J HEP Rep. 2020;2 doi: 10.1016/j.jhepr.2020.100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baselli Guido A., Dongiovanni P., Rametta R., et al. Liver transcriptomics highlights interleukin-32 as novel NAFLD-related cytokine and candidate biomarker. Gut. 2020;69:1855–1866. doi: 10.1136/gutjnl-2019-319226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fujiwara N., Kubota N., Crouchet E., et al. Molecular signatures of long-term hepatocellular carcinoma risk in nonalcoholic fatty liver disease. Sci Transl Med. 2022;14 doi: 10.1126/scitranslmed.abo4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Chang, Ampuero J., Gil-Gómez A., et al. miRNAs in patients with non-alcoholic fatty liver disease: a systematic review and meta-analysis. J Hepatol. 2018;69:1335–1348. doi: 10.1016/j.jhep.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 109.Pirola C.J., Fernández Gianotti T., Castaño G.O., et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Estep M., Armistead D., Hossain N., et al. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32:487–497. doi: 10.1111/j.1365-2036.2010.04366.x. [DOI] [PubMed] [Google Scholar]
- 111.Manicardi N., Fernández-Iglesias A., Abad-Jordà L., et al. Transcriptomic profiling of the liver sinusoidal endothelium during cirrhosis reveals stage-specific secretory signature. Cancers (Basel) 2021;13:2688. doi: 10.3390/cancers13112688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krahmer N., Najafi B., Schueder F., et al. Organellar proteomics and phospho-proteomics reveal subcellular reorganization in diet-induced hepatic steatosis. Dev Cell. 2018;47:205–221. doi: 10.1016/j.devcel.2018.09.017. e7. [DOI] [PubMed] [Google Scholar]
- 113.Yuan X., Sun Y., Cheng Q., et al. Proteomic analysis to identify differentially expressed proteins between subjects with metabolic healthy obesity and non-alcoholic fatty liver disease. J Proteomics. 2020;221 doi: 10.1016/j.jprot.2020.103683. [DOI] [PubMed] [Google Scholar]
- 114.Vu L.T., Orbach S.M., Ray W.K., et al. The hepatocyte proteome in organotypic rat liver models and the influence of the local microenvironment. Proteome Sci. 2017;15:12. doi: 10.1186/s12953-017-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lao Y., Li Y., Hou Y., et al. Proteomic analysis reveals Dab2 mediated receptor endocytosis promotes liver sinusoidal endothelial cell dedifferentiation. Sci Rep. 2017;7 doi: 10.1038/s41598-017-13917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deleve L.D. Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology. 2015;61:1740–1746. doi: 10.1002/hep.27376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gibert-Ramos A., Sanfeliu-Redondo D., Aristu-Zabalza P., et al. The hepatic sinusoid in chronic liver disease: the optimal milieu for cancer. Cancers (Basel) 2021;13:5719. doi: 10.3390/cancers13225719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim S.-R., Kubo T., Kuroda Y., et al. Comparative metabolome analysis of cultured fetal and adult hepatocytes in humans. J Toxicol Sci. 2014;39:717–723. doi: 10.2131/jts.39.717. [DOI] [PubMed] [Google Scholar]
- 119.Li H., Zhu W., Zhang L., et al. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci Rep. 2015;5:8421. doi: 10.1038/srep08421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yue D., Zhang Y., Cheng L., et al. Hepatitis B virus X protein (HBx)-induced abnormalities of nucleic acid metabolism revealed by 1 H-NMR-based metabonomics. Sci Rep. 2016;6 doi: 10.1038/srep24430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Min Hae K., Sookoian S., Pirola C.J., et al. Metabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in Huh-7 hepatoma cells. Am J Physiol Gastrointest Liver Physiol. 2014;307:G66–G76. doi: 10.1152/ajpgi.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martínez-Arranz I., Bruzzone C., Noureddin M., et al. Metabolic subtypes of patients with NAFLD exhibit distinctive cardiovascular risk profiles. Hepatology. 2022;76:1121–1134. doi: 10.1002/hep.32427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alonso C., Fernández-Ramos D., Varela-Rey M., et al. Metabolomic identification of subtypes of nonalcoholic steatohepatitis. Gastroenterology. 2017;152:1449–1461. doi: 10.1053/j.gastro.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saliba Antoine E., Westermann A.J., Gorski S.A., Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krenkel O., Hundertmark J., Ritz T., et al. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells. 2019;8:503. doi: 10.3390/cells8050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Su T., Yang Y., Lai S., et al. Single-cell transcriptomics reveals zone-specific alterations of liver sinusoidal endothelial cells in cirrhosis. Clin Mol Gastroenterol Hepatol. 2021;11:1139–1161. doi: 10.1016/j.jcmgh.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nault R., Fader K.A., Bhattacharya S., Zacharewski T.R. Single-nuclei RNA sequencing assessment of the hepatic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Clin Mol Gastroenterol Hepatol. 2021;11:147–159. doi: 10.1016/j.jcmgh.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lotto J., Drissler S., Cullum R., et al. Single-cell transcriptomics reveals early emergence of liver parenchymal and non-parenchymal cell lineages. Cell. 2020;183:702–716.e14. doi: 10.1016/j.cell.2020.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ramachandran P., Matchett K.P., Dobie R., et al. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat Rev Gastroenterol Hepatol. 2020;17:457–472. doi: 10.1038/s41575-020-0304-x. [DOI] [PubMed] [Google Scholar]
- 130.Blériot C., Barreby E., Dunsmore G., et al. A subset of Kupffer cells regulates metabolism through the expression of CD36. Immunity. 2021;54:2101–2116. doi: 10.1016/j.immuni.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 131.Remmerie A., Martens L., Thoné T., et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity. 2020;53:641–657. doi: 10.1016/j.immuni.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Seidman J.S., Troutman T.D., Sakai M., et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity. 2020;52:1057–1074. doi: 10.1016/j.immuni.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tran S., Baba I., Poupel L., et al. Impaired Kupffer cell self-renewal alters the liver response to lipid overload during non-alcoholic steatohepatitis. Immunity. 2020;53:627–640.e5. doi: 10.1016/j.immuni.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 134.Daemen S.A., Gainullina A., Kalugotla G., et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krenkel O., Hundertmark J., Abdallah A.T., et al. Myeloid cells in liver and bone marrow acquire a functionally distinct inflammatory phenotype during obesity-related steatohepatitis. Gut. 2020;69:551–563. doi: 10.1136/gutjnl-2019-318382. [DOI] [PubMed] [Google Scholar]
- 136.Jaitin D.A., Adlung L., Thaiss C.A., et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178:686–698. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Payen V.L., Arnaud L., Niki Alevra S., et al. Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.MacParland S.A., Liu J.C., Ma X.Z., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ramachandran P., Dobie R., Wilson-Kanamori J.R., et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Q., He Y., Luo N., et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845.e20. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 141.Saviano A., Henderson N.C., Baumert T.F. Single-cell genomics and spatial transcriptomics: discovery of novel cell states and cellular interactions in liver physiology and disease biology. J Hepatol. 2020;73:1219–1230. doi: 10.1016/j.jhep.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guillot A., Tacke F. Location, location, location — spatial insight into hepatic macrophage populations. Nat Rev Gastroenterol Hepatol. 2022;19:281–282. doi: 10.1038/s41575-022-00600-2. [DOI] [PubMed] [Google Scholar]
- 143.Guilliams M., Bonnardel J., Haest B., et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396.e38. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]