Abstract
For pathogens to survive in the human oral cavity, they must identify a suitable niche in the complex multispecies biofilm that exists on oral tissues. The periodontal pathogen Porphyromonas gingivalis adheres to Streptococcus gordonii by interacting with a specific region of the streptococcal SspB polypeptide, designated BAR. However, it does not adhere to Streptococcus mutans, which expresses SpaP, a highly conserved homolog of SspB. Comparison of the predicted secondary structure of BAR with the corresponding region of SpaP suggested that the substitution of Asn for Gly1182 and Val for Pro1185 in SspB may confer a unique local structure that is not conserved in SpaP. A synthetic peptide of 26 amino acids that encompassed residues 1167 to 1193 of SspB promoted avid adherence of P. gingivalis, whereas a peptide derived from the region corresponding to BAR in SpaP was inactive. Substitution of Gly1182 and Pro1185 for Asn1182 and Val1185 in SspB by site-specific mutation generated proteins that were predicted to assume an SpaP-like secondary structure, and the purified proteins did not promote P. gingivalis adherence. Furthermore, Enterococcus faecalis strains expressing the site-specific mutants did not support adherence of P. gingivalis cells. In contrast, P. gingivalis adhered efficiently to E. faecalis strains expressing intact SspB or SspB-SpaP chimeric proteins containing BAR. These results suggest that a region of SspB consisting of 26 amino acids is sufficient to mediate the adherence of P. gingivalis to S. gordonii and that the species specificity of adherence arises from its interaction with a discrete structural determinant of SspB that is not conserved in SpaP.
Porphyromonas gingivalis is regarded as one of the primary pathogens contributing to adult periodontitis, one of the most common infectious diseases of adults (16, 34). In the human oral cavity, this organism resides in a complex mixed-species biofilm that forms on the tooth surface and in the periodontal pocket (5, 14, 28, 31). However, the specific mechanisms utilized by P. gingivalis to establish and maintain itself in the oral biofilm are not fully understood. Early events leading to biofilm development on oral tissues involve the interaction of gram-positive commensal organisms, e.g., streptococci and Actinomyces spp., with the salivary pellicle coating the tissue surface (10, 14). These primary colonizing organisms then provide an attachment substrate for the ordered accumulation of other gram-positive and gram-negative bacterial species, including Fusobacterium nucleatum and periodontal pathogens such as P. gingivalis (15, 30, 32). Thus, colonization of the developing oral biofilm by P. gingivalis likely involves its adherence to various antecedent bacteria such as the oral streptococci and/or F. nucleatum. The interaction of P. gingivalis with primary colonizing organisms such as streptococci may also be important in the invasion of dentinal tubules by P. gingivalis (21).
The adherence of P. gingivalis to Streptococcus gordonii appears to occur through a protein-protein interaction requiring the SspB polypeptide of S. gordonii (17, 18) and the minor fimbrial component of P. gingivalis (3). Indeed, expression of the sspB gene in Enterococcus faecalis resulted in a transformed cell that was capable of promoting P. gingivalis adherence (17). The SspB polypeptide is a multifunctional surface protein of S. gordonii and is a member of the highly conserved antigen I/II (27) family of cell surface proteins that are expressed by virtually all streptococci that inhabit the human oral cavity (22). SspB is 1,500 residues in length and contains seven structural domains that are well conserved in all antigen I/II polypeptides. However, despite the high level of primary sequence identity and the conservation of structural domains among the various streptococcal antigen I/II proteins, P. gingivalis interacts with oral streptococci in a species-specific manner, adhering to S. gordonii cells but not to Streptococcus mutans. The species specificity of adherence arises through the differential binding of P. gingivalis to various antigen I/II proteins. Thus, SspB supports adherence of P. gingivalis cells, whereas the related antigen I/II polypeptide of S. mutans, designated SpaP, does not. Using a series of chimeric SspB-SpaP proteins, Brooks et al. (2) showed that the region encompassing residues 1167 to 1250 of SspB (designated BAR for SspB adherence region) was required for the in vitro adherence of P. gingivalis to S. gordonii cells. However, BAR exhibits 65% primary sequence identity with the corresponding sequences of SpaP, and it is not known how P. gingivalis selectively recognizes and binds only to BAR.
In this report, we compared the predicted secondary structures of BAR and the corresponding sequences of SpaP. This comparison suggested that SspB may possess a structural motif that is not conserved in SpaP. A synthetic peptide representing the putative motif promoted adherence of P. gingivalis, whereas the corresponding SpaP peptide did not. Furthermore, site-specific mutation of specific amino acids in SspB generated polypeptides that were predicted to assume an SpaP-like secondary structure. These proteins did not promote P. gingivalis adherence either in purified form or when expressed by recombinant E. faecalis cells. These results suggest that a region of SspB consisting of 26 amino acids is sufficient to mediate the adherence of P. gingivalis to S. gordonii and that the species specificity of adherence arises from the interaction of P. gingivalis with a discrete structural determinant of SspB that is not conserved in SpaP.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. P. gingivalis 33277 was grown at 37°C in Trypticase soy broth supplemented with 1 g of yeast extract, 5 mg of hemin. and 1 mg of menadione per liter under anaerobic conditions of 85% N2, 10% H2, and 5% CO2. When necessary, P. gingivalis 33277 was metabolically labeled by including [3H]thymidine (10 μCi per ml) in the culture medium. Labeling was carried out for 24 h. S. gordonii DL1, S. mutans KPSK2, and E. faecalis strains were grown aerobically without shaking at 37°C in Trypticase peptone broth supplemented with 5 g of yeast extract and 5 g of glucose per liter. Escherichia coli DH5α was grown aerobically at 37°C in L broth. Where necessary, the culture media above were supplemented with 100 μg of ampicillin (for transformed E. coli strains) or 15 μg of chloramphenicol (for transformed E. faecalis strains) per ml to maintain plasmids. Bacterial cell density was determined using a Klett-Summerson photometer at 600 nm.
TABLE 1.
Bacterial strains
| Strain | Reference | Comments |
|---|---|---|
| P. gingivalis 33277 | ATCC type strain | |
| S. gordonii DL1a | 25 | |
| S. mutans KPSK2b | Oral clinical isolate | |
| E. faecalis | ||
| S161b | Oral clinical isolate | |
| SspB | 6 | Expresses S. gordonii SspB |
| Spac4 | This study | Expresses hybrid SspB-SpaP polypeptide Spac4 |
| Spac5 | This study | Expresses hybrid SspB-SpaP polypeptide Spac5 |
| pAM401 | 34 | Contains shuttle vector pAM401 |
Kindly provided by Howard F. Jenkinson.
Kindly provided by Burton Rosan.
Construction of strains and plasmids.
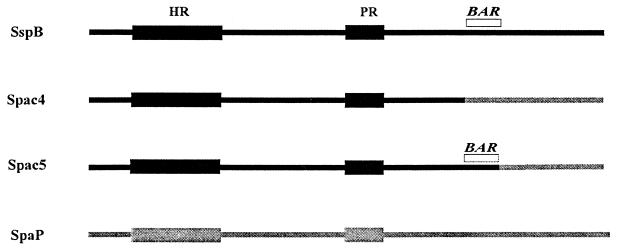
The E. coli/E. faecalis shuttle vector pAM401 was described previously by Wirth et al. (34). Construction of the spac4 and spac5 hybrid genes encoding the chimeric SspB-SpaP polypeptides, shown in Fig. 1, was described previously (2). Briefly, this was accomplished by digesting pEB5, a pUC19 plasmid possessing the entire sspB gene (7) (accession number J05418) with XbaI-BamHI or HpaI-BamHI to remove residues 1167 to 1500 or 1250 to 1500, respectively. A DNA fragment encoding the corresponding C-terminal residues of SpaP was amplified from pPAc7, a pUC19 plasmid containing spaP (9). The sequences of the oligonucleotide primers used for the amplification reactions to generate the appropriate gene fragments for the construction of Spac4 and Spac5 were described previously (2). Briefly, primers 9 and 10 as described by Brooks et al. (2) were used to obtain the portion of spaP required for the spac4 construct; primers 4 and 11 (2) were utilized to generate spac5. The resulting PCR products encoding the C-terminal sequences of SpaP were digested with the appropriate restriction enzymes, purified from agarose gels, and ligated to the digested pEB5 plasmids generated above. Recombinant plasmids containing the chimeric constructs were obtained from E. coli DH5α transformants and sequenced to confirm the successful incorporation of the spaP gene fragments. Transfer of the chimeric genes into pAM401 was accomplished in two steps. The genes were first excised from pUC19 by digestion with EcoRI and BamHI and ligated into pBluescript IISK. The resulting plasmids were subsequently digested with EagI and SalI and ligated into pAM401 digested with the same restriction enzymes. Plasmids pAM401Spac4 and pAM401Spac5 were transformed into E. faecalis S161 by electroporation, and recombinants were grown on Todd-Hewitt agar (Difco) containing 15 μg chloramphenicol per ml. Expression of Spac4 and Spac5 was confirmed by colony blots using polyclonal anti-SspB antibodies (7).
FIG. 1.
Structure of chimeric SspB-SpaP polypeptides Spac4 and Spac5. Spac4 is composed of SspB residues 1 to 1166 fused to residues 1167 to 1500 of SpaP. Spac5 contains residues 1 to 1250 of SspB fused to residues 1251 to 1500 from SpaP. The region of SspB encompassing residues 1167 to 1250 (BAR) mediates the in vitro adherence of P. gingivalis cells to S. gordonii (2). Conserved alanine-rich and proline-rich repetitive domains of SspB and SpaP are labeled HR and PR, respectively.
Construction of the SspB site-specific mutants was carried out on pBluescript-Spac4 and pBluescript-Spac5 (see above) using the QuikChange site-directed mutagenesis kit (Stratagene) essentially as described by the manufacturer. Forward (F) and reverse (R) oligonucleotide primers used for the site-specific mutations Asn/Gly1182 and Val/Pro1185 and the double mutation (dbl_mut) were as follows: N/G1182F, 5′-TTGAAGAAAGCTGGCATTACTGTTAAGG-3′; N/G1182R, 5′-CTTAACAGTAATGCCAGCTTTCTTCAAC-3′; V/P1185F, 5′-GCTAACATTACTCCTAAGGGTGCTTTCC-3′; V/P1185R, 5′-GGAAAGCACCCTTAGGAGTAATGTTAGC-3′; dbl_mutF, 5′-GCTGGCATTACTCCTAAGGGTGCTTTCC-3′; and dbl_mutR, 5′-GGAAAGCACCCTTAGGAGTAATGCCAGC-3′.
Amplification conditions used for mutagenesis were 95°C for 30 s for a single cycle, followed by 18 cycles of 95°C for 30 s, annealing for 1 min at 55°C, and extension at 68°C for 16 min. Subsequent to the amplification reaction, samples were digested at 37°C for 1 h with DpnI and transformed into E. coli XL1-Blue. Successful mutations were confirmed by sequencing plasmids obtained from the transformed colonies. The SspB protein was purified from the appropriate site-specific mutants by gel filtration and anion exchange chromatography as described by Demuth et al. (7).
Adherence of P. gingivalis to synthetic peptide and SspB polypeptides.
Peptides representing residues 1167 to 1193 of BAR and SpaP were synthesized by BioSynthesis (Lewisville, Tex.). Purity of the peptides was assessed by high-pressure liquid chromatography to be ≥95% for each. Adherence of P. gingivalis to synthetic peptides or to purified protein samples was determined by depositing serial twofold dilutions of each peptide sample (0.3 to 2.5 μg/ml) onto a nitrocellulose filter in a vacuum dot blot apparatus. The membrane was subsequently blocked with phosphate-buffered saline containing 0.02% Tween 20 (PBS-Tween 20) for 1 h and reacted with 108 3H-labeled P. gingivalis cells (5 × 104 cpm) for 2 h with shaking at 25°C. After washing four times with PBS-Tween 20, the filters were dried, and the number of bound cells was determined by scintillation spectroscopy.
Adherence of P. gingivalis to E. faecalis expressing SspB polypeptides was determined using a nitrocellulose blot assay described previously (15). Briefly, enterococci were suspended in buffered KCl (5 mM KCl, 2 mM K2PO4, 1 mM CaCl2, pH 6.0), and 108 bacteria were deposited on nitrocellulose paper in a dot blot apparatus. The blot was washed three times in KCl containing 0.1% Tween 20 (KCl-Tween). The adsorbed bacteria were subsequently incubated for 2 h at room temperature with [3H]thymidine-labeled P. gingivalis suspended in KCl-Tween. After washing to remove unbound organisms, the experimental areas of the nitrocellulose were excised, and adherence was quantified by scintillation spectroscopy.
Secondary-structure analysis.
The deduced sequences of BAR and the corresponding region encompassing amino acid residues 1167 to 1250 of SpaP were obtained from the published nucleotide sequences of the sspB and spaP genes from S. gordonii strain M5 (8) and S. mutans strain MT8148 (24), respectively. Secondary-structure analyses were carried out with the algorithm of Chou and Fassman using the ProtPlot program of Ross and Golub (26).
RESULTS
Structural comparison of BAR and SpaP.
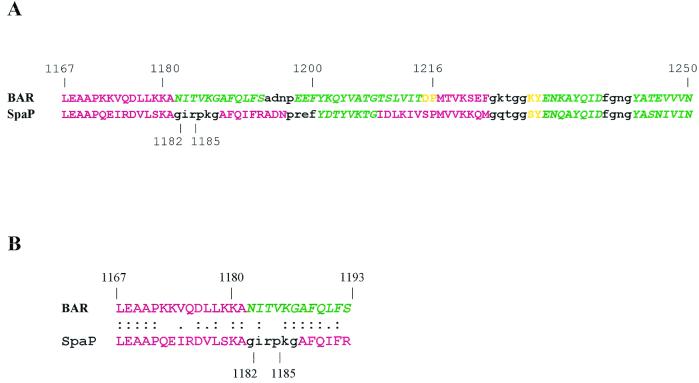
Our previous in vitro adherence studies indicated that P. gingivalis adhered only to S. gordonii and little accumulation occurred on S. mutans KPSK2. In order to further understand the mechanism governing the species specificity of P. gingivalis adherence, we compared the primary sequence and the predicted secondary structure of BAR (residues 1167 to 1250 of SspB) with the corresponding region in the SpaP polypeptide. We hypothesized that amino acid residues of BAR that are not conserved in SpaP may confer a specific structural element that is recognized and bound by P. gingivalis. The primary sequences of BAR and the corresponding portion of SpaP are very similar (8), exhibiting 65% identity (53 of 82 residues). An additional nine conservative amino acid substitutions occur. The predicted secondary structures of BAR and SpaP are compared in Fig. 2A. This comparison shows that SspB and SpaP are predicted to assume very similar secondary structures between residues 1216 and 1250 and that BAR and SpaP also share a putative N-terminal α-helix extending from residues 1167 to 1181. The major structural difference occurs between residues 1180 and 1200. Here, three α-helix-breaking residues, Gly1182, Pro1185, and Gly1187, exist in SpaP, and a β-turn is predicted (shown in black in Fig. 2A), followed by a second α-helix. Of these three residues, only Gly1187 is conserved in BAR. As a result, no β-turn is predicted to occur. Instead, BAR is predicted to form a β-sheet. This suggests that local structural differences may exist in BAR and SpaP despite the high overall conservation of primary sequence. To address this possibility, our subsequent studies concentrated on the portion of BAR between residues 1167 and 1193, shown aligned with the corresponding region of SpaP in Fig. 2B.
FIG. 2.
(A) Secondary-structure predictions distinguish BAR from the corresponding sequences of SpaP. Regions of the two sequences predicted to assume an α-helical configuration are shown in red; β-sheet is represented by green italics; random coil is indicated in yellow; and β-turns are shown in lowercase black. (B) Sequence similarity of the synthetic BAR and SpaP peptides. Identical residues are indicated with colons, and conservative substitutions are shown with single dots. The predicted secondary structures of the peptides are color coded as described for panel A.
P. gingivalis adherence to synthetic BAR and SpaP peptides.
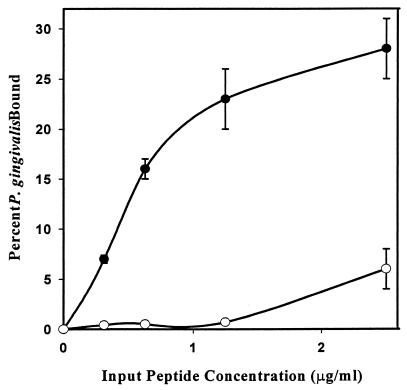
To determine if the region of BAR identified in the structural comparison above plays a functional role in the interaction of P. gingivalis with SspB, the adherence of P. gingivalis to immobilized synthetic peptides corresponding to residues 1167 to 1193 of BAR and SpaP (shown in Fig. 2B) was analyzed. As shown in Fig. 3, the synthetic BAR peptide promoted adherence of P. gingivalis cells in a dose-dependent fashion. Approximately 25% of the P. gingivalis cells adhered to the BAR peptide at an input peptide concentration of 1.25 μg/ml, whereas adherence to the SpaP peptide under these conditions was virtually undetectable. At input peptide concentrations higher than 1.25 μg/ml, the slope of the binding curve was equivalent for the BAR and SpaP peptides, suggesting that a nonspecific association with P. gingivalis was occurring at these higher peptide concentrations. In contrast, P. gingivalis clearly distinguished between BAR and SpaP and preferentially bound to the BAR peptide at the lower input peptide concentrations. This result is consistent with our hypothesis that P. gingivalis interacts with a specific structural motif in BAR that is not conserved in SpaP.
FIG. 3.
A synthetic peptide comprising residues 1167 to 1193 of BAR promotes adherence of P. gingivalis. Adherence of P. gingivalis to the synthetic BAR (●—●) and SpaP (○—○) peptides was determined by incubating immobilized peptide (0.3 to 2.5 μg/ml) with 3H-labeled P. gingivalis cells (108 total cells) for 2 h with shaking at 25°C. Bound cells were determined by scintillation spectroscopy. Error bars represent standard deviation, n = 3.
Site-specific mutagenesis of BAR.
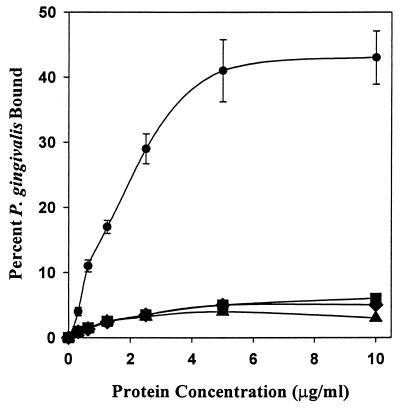
We next sought to determine if the nonconserved residues Asn1182 and Val1185 are essential for P. gingivalis adherence to the full-length SspB polypeptide. To accomplish this, site-specific mutations were introduced into the sspB gene to generate full-length SspB proteins in which Asn1182 and Val1185 were replaced with the corresponding residues of SpaP, Gly1182 and Pro1185. In addition, a construct possessing both substitutions, Asn/Gly1182 and Val/Pro1185, was synthesized. Secondary-structure predictions for each of the resulting mutant polypeptides indicated that the Val/Pro1185 substitution and the double mutation induced a β-turn in BAR encompassing residues 1182 to 1187, similar to the predicted structure for SpaP. The Asn/Gly1182 mutation did not induce a predicted β-turn, but extended the initial α-helix of BAR to residue 1193. Thus, each of the site-specific mutations influenced the predicted secondary structure of BAR, and for two of the constructs, the new secondary structure resembled that of SpaP. To assess the functional properties of the SspB site-specific mutants, the adherence of P. gingivalis to each of the polypeptides was compared with the full-length SspB and SpaP polypeptides. As shown in Fig. 4, P. gingivalis bound poorly to each of the mutant proteins; only the intact SspB protein promoted significant levels of adherence. To discount the possibility that the loss of function resulted from a gross structural change caused by the mutations, we determined if the SspB site-specific mutants were capable of interacting with saliva using a dot blot binding assay described previously (8). For each polypeptide, saliva-binding activity was unaffected by the site-specific mutations (not shown), suggesting that the mutations specifically influence adherence to P. gingivalis and do not cause a generalized loss of SspB function.
FIG. 4.
Site-specific mutations in BAR block P. gingivalis adherence to streptococci. Adherence of P. gingivalis to native SspB protein (●—●) and the site-specific mutants N/G1182 (▪—▪), V/P1185 (⧫—⧫), and N/G1182: V/P1185 (▴—▴) was carried out as described previously for the synthetic peptides. Substitution of Pro for Val1185, alone or in combination with a mutation of Gly for Asn1182, resulted in a sequence with a predicted secondary structure identical to that of SpaP. The substitution of Gly for Asn1182 by itself is not predicted to induce a β-turn, but generates a putative α-helix extending to Ser1193. Error bars represent standard deviation, and all experiments were carried out in triplicate.
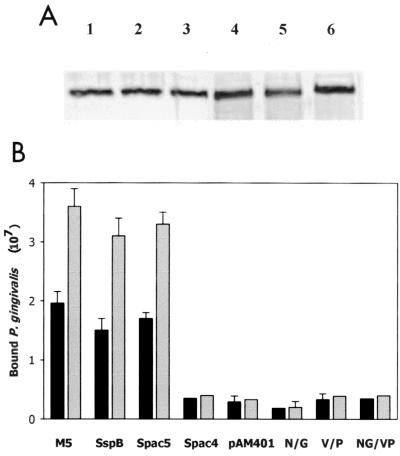
To determine if the mutations influenced the adherence of P. gingivalis to intact cells, the genes containing the site-specific mutations were transferred into the shuttle vector pAM401 and expressed in E. faecalis. To ensure that the overall level of expression of the polypeptides in each of the recombinant E. faecalis strains was similar, aliquots of the recombinant cultures were analyzed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and reacted with anti-SspB antibodies. As shown in Fig. 5A, all of the strains expressed similar levels of immunoreactive protein. The transformed cells were then tested for adherence to P. gingivalis. As shown in Fig. 5B, intact S. gordonii M5 as well as E. faecalis strains expressing SspB and Spac5 adhered to P. gingivalis in a dose-dependent manner. Consistent with our in vitro data for purified proteins, the strains expressing the SspB site-specific mutations or Spac4 promoted adherence at approximately the level of the negative control strain containing pAM401 without a streptococcal insert. Thus, our results suggest that the species specificity of P. gingivalis adherence to oral streptococci may arise from its recognition of a specific structural motif encompassing residues 1167 to 1193 that is present in SspB but is not conserved in SpaP.
FIG. 5.
(A) Western blot of SspB polypeptides expressed by E. faecalis SspB, E. faecalis Spac5, E. faecalis Spac4, E. faecalis N/G1182, E. faecalis V/P1185, and E. faecalis NG/VP (lanes 1 to 6, respectively). Protein was obtained by extraction of 109 cells for 5 min in boiling SDS. Samples were electrophoresed on 10% PAGE gels, blotted onto nitrocellulose, and reacted with polyclonal anti-SspB antibodies. (B) Adherence of P. gingivalis to S. gordonii M5 or recombinant E. faecalis strains expressing the SspB, Spac5, or Spac4 protein or the site-specific mutants of SspB. Adherence was determined as described in Materials and Methods using 4 × 107 (black bars) or 8 × 107 (gray bars) input P. gingivalis cells. Error bars represent standard deviation, n = 3.
DISCUSSION
Long-term survival of bacteria in the oral cavity requires that the organisms adhere to a tissue surface or identify a suitable niche in the complex multispecies biofilm that exists on human oral tissues. However, the molecular mechanisms utilized by many oral organisms to recognize and distinguish appropriate environmental niches in the oral biofilm are not well understood. Brooks et al. (2) showed that P. gingivalis adhered to streptococci in a species-specific manner by interacting with SspB, a member of the antigen I/II family of streptococcal surface proteins (11). A region of approximately 80 residues of SspB, designated BAR, was shown to be essential for P. gingivalis adherence (2). However, the molecular basis for species specificity of adherence was not explained by Brooks et al. Indeed, BAR exhibits approximately 65% sequence identity with S. mutans SpaP, which does not promote adherence. Our work now shows that a synthetic peptide representing a subregion of BAR (residues 1167 to 1193 of SspB) mediates adherence of P. gingivalis in vitro, suggesting that only a portion of BAR is sufficient to confer on P. gingivalis the ability to adhere to S. gordonii. Furthermore, comparing the sequence of the synthetic peptide with the corresponding sequence of SpaP highlighted two specific amino acid residues that may confer a unique structural motif in SspB. Site-specific mutation of these two SspB residues to the corresponding SpaP amino acids generated SspB polypeptides which were predicted to be structurally similar to SpaP and which did not mediate adherence of P. gingivalis. Thus, the species specificity of P. gingivalis adherence appears to arise from the recognition of a discrete structural motif of SspB that is not conserved in the highly related SpaP protein. This suggests that nonconserved residues that reside within a region of antigen I/II that exhibits significant sequence similarity appear to be functionally important.
Within the human oral cavity, the specificity of P. gingivalis interactions with streptococci may be important for identifying a suitable environmental niche in the growing oral biofilm. For example, the specific adherence to S. gordonii may represent a mechanism by which P. gingivalis avoids colonizing sites that are rich in acid-tolerant bacteria such as S. mutans. These organisms may generate and thrive in an acidic local environment in the biofilm, and such conditions are not physiologically favorable for P. gingivalis (33). In contrast, the metabolism of arginine by S. gordonii plays a role in maintaining a neutral or slightly basic environment (4, 23). In addition, other organisms which coadhere well with S. gordonii, e.g., Fusobacterium nucleatum (14), have also been suggested to maintain a neutral local environment (33). Consistent with this, P. gingivalis is known to coaggregate with F. nucleatum (1, 13, 29), and mixed-species biofilms containing both streptococci and F. nucleatum support higher populations of P. gingivalis (1). This suggests that P. gingivalis may adhere to a community of multiple compatible organisms and its adherence to S. gordonii represents just one of a series of distinct interbacterial interactions contributing to the successful colonization of the developing oral biofilm by P. gingivalis. The preferential adherence to S. gordonii rather than S. mutans may also explain in part the clinical observation that adult periodontitis does not usually occur simultaneously with coronal dental caries caused by S. mutans (12). The species specificity of adherence may also be relevant in the pathogenesis of root canal infections. Love et al. have shown that antigen I/II polypeptides bind to collagen I and are required for streptococcal invasion of dentin (19, 20). Interestingly, P. gingivalis penetrates dentin only in conjunction with S. gordonii and not in the presence of wild-type S. mutans, an SspB-deficient mutant of S. gordonii, or the SspB-deficient strain complemented with a plasmid capable of expressing SpaP (19, 20).
The primary sequences and the overall structural organization of members of the streptococcal antigen I/II family of polypeptides are well conserved, yet our results suggest that individual antigen I/II proteins may be functionally distinct. Indeed, functional specificity among the streptococcal antigen I/II proteins has also been demonstrated in their interactions with a mucin-like salivary glycoprotein (SAG). For example, both S. gordonii and S. mutans interact with SAG in a lectin-like reaction mediated by antigen I/II. The binding of SAG by intact S. gordonii cells and purified SspB is blocked by neuraminidase treatment of SAG, suggesting that a sialic acid-containing carbohydrate constituent is recognized. In contrast, the interaction of S. mutans cells or purified SpaP is unaffected by neuraminidase treatment and appears to require carbohydrates containing fucose (9). Interestingly, the functional domains of SspB and SpaP that are involved in their interaction with SAG reside within a highly conserved portion of the polypeptides. This suggests that other functional properties of antigen I/II proteins may also be primarily dependent on local structural motifs that are specific to individual polypeptides and which arise from nonconserved amino acids residing within a conserved structural framework.
In summary, we have shown that a discrete structural motif of SspB mediates the species-specific adherence of P. gingivalis to S. gordonii. The unique local structure in SspB is not conserved in SpaP and may arise from specific amino acid residues that are not conserved in the SpaP sequence. The specificity of P. gingivalis adherence with oral streptococci may represent a mechanism which contributes to colonization of the oral biofilm and may also be relevant in the pathogenesis of P. gingivalis infections.
ACKNOWLEDGMENTS
We thank Yoonsuk Park, Whasun Chung, and Ozlem Yilmaz for technical assistance and E. E. Golub and D. Malamud for critical review of the manuscript.
This collaborative work was supported by Public Health Service grants DE12750, DE12505, and DE13061 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Bradshaw D J, Marsh P D, Watson G K, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66:4729–4732. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks W, Demuth D R, Gil S, Lamont R J. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect Immun. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung W O, Demuth D R, Lamont R J. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect Immun. 2000;68:6758–6762. doi: 10.1128/iai.68.12.6758-6762.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curran T M, Ma Y, Rutherford G C, Marquis R E. Turning on and turning off the arginine deiminase system in oral streptococci. Can J Microbiol Immunol. 1998;44:1078–1085. doi: 10.1139/cjm-44-11-1078. [DOI] [PubMed] [Google Scholar]
- 5.Cutler C W, Kalmar J R, Genco C A. Pathogenic strategies of the oral anaerobe Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51. doi: 10.1016/s0966-842x(00)88874-5. [DOI] [PubMed] [Google Scholar]
- 6.Demuth D R, Berthold P, Leboy P S, Golub E E, Davis C A, Malamud D. Saliva-mediated aggregation of Enterococcus faecalis transformed with a Streptococcus sanguis gene encoding the Ssp-5 surface antigen. Infect Immun. 1989;57:1470–1475. doi: 10.1128/iai.57.5.1470-1475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demuth D R, Davis C A, Corner A M, Lamont R J, Leboy P S, Malamud D. Cloning and expression of a Streptococcus sanguis surface antigen that interacts with a human salivary agglutinin. Infect Immun. 1988;56:2484–2490. doi: 10.1128/iai.56.9.2484-2490.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demuth D R, Golub E E, Malamud D. Streptococcal-host interactions: structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J Biol Chem. 1990;265:7120–7126. [PubMed] [Google Scholar]
- 9.Demuth D R, Lammey M S, Huck M, Lally E T, Malamud D. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb Pathog. 1990;9:199–211. doi: 10.1016/0882-4010(90)90022-i. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons R J. Bacterial adhesion to oral tissues: a model for infectious diseases. J Dent Res. 1989;68:750–760. doi: 10.1177/00220345890680050101. [DOI] [PubMed] [Google Scholar]
- 11.Jenkinson H F, Demuth D R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 12.Kinane D F, Jenkins W M, Adonogianaki E, Murray G D. Cross-sectional assessment of caries and periodontitis risk within the same subject. Community Dent Oral Epidemiol. 1991;19:78–81. doi: 10.1111/j.1600-0528.1991.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 13.Kinder S, Holt S C. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J Bacteriol. 1993;175:840–847. doi: 10.1128/jb.175.3.840-850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolenbrander P E, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamont R J, Hersey S G, Rosan B. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol Immunol. 1992;7:193–197. doi: 10.1111/j.1399-302x.1992.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 16.Lamont R J, Jenkinson H F. Life below the gumline: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamont R J, Bevan C A, Gil S, Persson R E, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 18.Lamont R J, Gil S, Demuth D R, Malamud D, Rosan B. Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiol. 1994;140:867–872. doi: 10.1099/00221287-140-4-867. [DOI] [PubMed] [Google Scholar]
- 19.Love R M, Chandler N P, Jenkinson H F. Penetration of smeared or nonsmeared dentine by Streptococcus gordonii. Int Endod J. 1996;29:2–12. doi: 10.1111/j.1365-2591.1996.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 20.Love R M, McMillan M D, Jenkinson H F. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun. 1997;65:5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love R M, McMillan M D, Park Y, Jenkinson H F. Coinvasion of dentinal tubules by Porphyromonas gingivalis and Streptococcus gordonii depends upon the binding specificity of streptococcal antigen I/II adhesin. Infect Immun. 2000;68:1359–1365. doi: 10.1128/iai.68.3.1359-1365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J K-C, Kelly C G, Munro G H, Whiley R A, Lehner T. Conservation of the gene encoding streptococcal antigen I/II in oral streptococci. Infect Immun. 1991;59:2686–2690. doi: 10.1128/iai.59.8.2686-2694.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Rutherford G C, Curran T M, Reidmiller J S, Marquis R E. Membrane locus and pH sensitivity of paraben inhibition of alkali production by oral streptococci. Oral Microbiol Immunol. 1999;14:244–249. doi: 10.1034/j.1399-302x.1999.140408.x. [DOI] [PubMed] [Google Scholar]
- 24.Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Cloning of a surface protein antigen gene from serotype c Streptococcus mutans. Mol Microbiol. 1989;3:673–678. doi: 10.1111/j.1365-2958.1989.tb01811.x. [DOI] [PubMed] [Google Scholar]
- 25.Pakula R, Walczak W. On the nature of competence of transformable streptococci. J Gen Microbiol. 1963;31:125–133. doi: 10.1099/00221287-31-1-125. [DOI] [PubMed] [Google Scholar]
- 26.Ross A M, Golub E. A computer graphics program system for protein structure representation. Nucleic Acids Res. 1988;16:1801–1812. doi: 10.1093/nar/16.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell M W, Bergmeier L A, Zander E D, Lehner T. Protein antigens of Streptococcus mutans: purification and properties of a double antigen and its protease-resistant component. Infect Immun. 1980;28:486–493. doi: 10.1128/iai.28.2.486-493.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder H E, Listgarten M. The gingival tissues: the architecture of periodontal protection. Periodontology. 1997;13:91–120. doi: 10.1111/j.1600-0757.1997.tb00097.x. [DOI] [PubMed] [Google Scholar]
- 29.Shaniztki B, Hurwitz D, Smorodinsky N, Ganeshkumar N, Weiss E I. Identification of a Fusobacterium nucleatum PK1594 galactose-binding adhesin which mediates coaggregation with periopathogenic bacteria and hemagglutination. Infect Immun. 1997;65:5231–5237. doi: 10.1128/iai.65.12.5231-5237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slots J, Gibbons R. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in the colonization of the mouth and periodontal pockets. Infect Immun. 1978;19:254–264. doi: 10.1128/iai.19.1.254-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Socransky S S, Haffajee A. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 32.Stinson M W, Safulko K, Levine M J. Adherence of Porphyromonas gingivalis to Streptococcus sanguis in vitro. Infect Immun. 1991;59:102–108. doi: 10.1128/iai.59.1.102-108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi N, Saito K, Schachtele C F, Yamada T. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 1997;12:323–328. doi: 10.1111/j.1399-302x.1997.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 34.Wirth R, An F Y, Clewell D B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]