Abstract
Pathogenesis of Shigella flexneri is dependent on the ability of the bacterium to invade and spread within epithelial cells. In this study, we identified dksA as a gene necessary for intercellular spread in, but not invasion of, cultured cells. The S. flexneri dksA mutant exhibited sensitivity to acid and oxidative stress, in part due to an effect of DksA on production of RpoS. However, an S. flexneri rpoS mutant formed plaques on tissue culture monolayers, thus excluding DksA regulation of RpoS as the mechanism responsible for the inability of the dksA mutant to spread intercellularly. Intracellular analysis of the dksA mutant indicates that it survived and divided within the Henle cell cytoplasm, but the dksA mutant cells were elongated, and some exhibited filamentation in the intracellular environment. Some of the S. flexneri dksA mutant cells showed aberrant localization of virulence protein IcsA, which may inhibit spread between epithelial cells.
Shigella flexneri colonizes the colon and causes bacterial dysentery in humans (13, 27, 37). These bacteria are highly infectious, with as few as 10 cells being sufficient to cause disease in healthy adults (6). This low infectious dose is partially due to a significant resistance to acidic conditions, which permits survival during transit through the acidic conditions encountered in the human stomach (12, 38). In the colon, Shigella cells cross the epithelial layer, attach to the basal surfaces of epithelial cells in a receptor-specific process, and induce their own internalization (29). Following internalization, Shigella cells are temporarily located in a phagocytic vacuole. Lysis of this vacuole occurs within minutes, and Shigella cells initiate replication and cell to cell spread. This process is accompanied by a complex pattern of protein induction and suppression (16). Cell-to-cell spread requires that Shigella protein IcsA (VirG) be targeted to the old pole of the bacterium, where it induces assembly of F-actin by the epithelial cell (2, 25, 26, 39). The process by which S. flexneri localizes IcsA to a polar location is unknown, but the unipolar localization of IcsA when expressed in Escherichia coli indicates that Shigella virulence plasmid proteins are not involved in this process in E. coli (36). Polymerization of actin at one pole is responsible for propelling the bacterium through the host cytoplasm and into a protrusion of a double-membrane barrier between two host cells. Shigella lyses this membrane barrier, and, after being released into the new epithelial cell, the bacteria repeat the process of multiplication and spread. Progressive spread of these bacteria leads to degradation of the epithelium and inflammation, resulting in symptoms of disease. An in vitro model utilizing tissue culture monolayers that mimics the process of S. flexneri invasion, intercellular multiplication, and spread has been developed (14, 22, 31). Bacteria that form plaques on these tissue culture monolayers are capable of performing these intracellular processes. Using this model, we examined TnphoA mutants that were deficient in intracellular multiplication or intercellular spread. One gene, dksA, was identified here to be required for the intercellular spread of S. flexneri.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used here are listed with their sources in Table 1. All strains were stored at −80°C in tryptic soy broth plus 20% glycerol. S. flexneri strains were grown on Congo red agar to screen colonies that bind Congo red (33). E. coli strains were grown in Luria broth (LB) or on Luria agar (L agar). Intracellular salts medium (ISM) was used to mimic the intracellular conditions (16). All strains were routinely cultured at 37°C unless otherwise noted. Antibiotics were used as appropriate at the following concentrations: carbenicillin, 250 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; tetracycline, 12.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| DH5α | E. coli cloning strain | |
| SA100 | S. flexneri wild-type serotype 2a | 34 |
| SA511 | SA100 Cmr | 19 |
| SA710 | SA100 rpoS::kan | This work |
| SA2287 | SA511 dksA::TnphoA | This work |
| SA5287 | SA100 dksA::TnphoA | This work |
| ZK1000 | E. coli W3110 ΔlacU169 tna-2 katF (rpoS)::kan | 3 |
| Plasmids | ||
| pDEB2 | katF (rpoS) in pUC19 | 3 |
| pLR56 | Y. enterocolitica invA in pUC19 | L. Runyen-Janecky |
| pSAM1 | SA100 dksA in pWKS30 | This work |
| pUC-GFP | gfp in pUC19 | S. Seliger |
| pWKS30 | Low-copy-number cloning vector | 45 |
Isolation and identification of the dksA mutation.
TnphoA mutagenesis of wild-type S. flexneri 2a (SA100) Cmr derivative SA511 was performed as described previously (19). Mutants that were invasive but unable to form plaques on HeLa cell monolayers were isolated, and the sites of the insertions were determined by inverse PCR as described below.
The chromosomal DNA from the TnphoA insertion mutants was digested with TaqI restriction endonuclease, which has a site 403 bp downstream from the 5′ end of TnphoA. DNA fragments were circularized by ligation with T4 DNA ligase. The ligation reaction mixtures were purified using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.). PCR was conducted on the ligation reaction mixture using Taq polymerase (Qiagen) in the reaction buffer supplied by the manufacturer and supplemented with 250 μM deoxynucleoside triphosphates and 1 μM concentrations of primers TnphoA-1 (5′CAAAACGGGAAAGGTTCCG) and TnphoA-2 (5′GCTCTGCGTGATTCTCTTAGCG). The reaction conditions were 30 cycles of 94°C for 1 min, 51°C for 1 min, and 72°C for 1 min. The DNA sequences of the inverse PCR products were determined with an ABI Prism 377 DNA sequencer (Perkin-Elmer Co., Applied Biosystems Division).
Cloning of the dksA gene.
The dksA gene of S. flexneri SA100 was amplified by PCR using chromosomal DNA and Pfu polymerase (Stratagene Cloning Systems, La Jolla, Calif.) in the reaction buffer supplied by the manufacturer and supplemented with 250 μM deoxynucleoside triphosphate and 1 μM concentrations of primers dksA-1 (5′AGAACGCAGCCGTATTGAC) and dksA-2 (5′CAGAGAGCCAAAATGAAGC). The reaction conditions were 30 cycles of 95°C for 45 s, 55.9°C for 45 s, and 72°C for 200 s. This PCR fragment was cloned into low-copy-number vector pWKS30 to generate pSAM1.
Construction of bacterial mutants.
The dksA mutation from SA2287 was moved into wild-type SA100 by P1 transduction (28) to generate SA5287. The presence of the TnphoA insertion in dksA was confirmed by PCR. S. flexneri rpoS mutant SA710 was constructed in SA100 by P1 transduction of the rpoS::kan allele from E. coli ZK1000 (3). The presence of the kan insertion in rpoS was confirmed by PCR.
Environmental stress resistance assays.
Resistance to either acid or the oxidizing agent cumene hydroperoxide (CHP) (Sigma Chemical Co.) was determined by a modification of the method of Waterman and Small (46). Bacterial cultures containing the appropriate antibiotics were incubated in LB for 19 h with aeration at 37°C. Bacteria were then exposed to either acid or CHP as follows. In the acid resistance assay, acidic medium was prepared by adjusting LB to pH 2.5 with HCl and filter sterilization. Overnight bacterial cultures were diluted 1:50 into LB, pH 2.5. In the CHP resistance assay, the CHP stock (approximately 80%) was diluted 1:10 in dimethyl sulfoxide. One milliliter of each overnight culture was removed, and 3.8 μl of 8% CHP was added to yield a final concentration of approximately 2 mM CHP. Immediately following exposure to either acid or CHP, cultures were briefly vortexed and an aliquot was removed, diluted in saline, and plated on L agar to determine the initial CFU per milliliter. The remaining culture was incubated without aeration at 37°C. At each time point, an aliquot of the sample was removed, diluted, and plated as described above. L plates were incubated overnight at 37°C, and values of CFU per milliliter were calculated the following day. Percent survival is reported as CFU per milliliter at each time point divided by CFU per milliliter at time zero.
Tissue culture, cell invasion, and plaque assays.
Henle 407 cell monolayers were used in all experiments and were cultured in Earle's minimal essential medium plus 2 mM glutamine plus 10% fetal calf serum (Life Technologies, Grand Island, N.Y.) and incubated in a 5% CO2 atmosphere at 37°C. Invasion assays to examine the ability of S. flexneri to invade and multiply within Henle cells were performed as described by Hale and Formal (14) and as modified by Hong et al. (19). Plaque assays were conducted to assay the intercellular spread of S. flexneri as described by Oaks et al. (31) and as modified by Hong et al. (19).
Phagocytic vacuole escape assay.
Henle cells were grown on glass coverslips according to the invasion assay procedure (19). Approximately 3 h prior to invasion, the medium was removed and replaced with serum-free medium containing 0.015 mg of 5-(and 6)-carboxytetramethylrhodamine succinimidyl ester (Molecular Probes, Eugene, Oreg.)/ml to label Henle cell membrane proteins. Cells were incubated at 37°C for 1 h, washed four times with phosphate-buffered saline (PBS) to remove excess rhodamine probes, and incubated for 1 h in fresh serum-containing medium to conjugate unbound rhodamine probes to serum proteins. The Henle cells were then washed twice with PBS, and fresh medium was added. Bacterial invasion was conducted as described previously (19) using bacteria expressing green fluorescent protein (GFP) encoded by plasmid pUC-GFP. At 1 h postinvasion, the glass coverslips were washed four times with PBS, fixed for 10 min in 4% (wt/vol) paraformaldehyde in PBS, washed twice with water, and sealed inverted on a glass slide. Fluorescent images were visualized by excitation at either 488 or 568 nm on a Leica TCS 4D confocal laser scanning microscope. Images show the excitation from only one wavelength to ensure that the fluorescence signal results from only one signal. Each image was examined by optically sectioning through the eukaryotic cell membrane and cytoplasm to confirm that the bacteria were located intracellularly.
In vitro growth rate.
In vitro growth rate was determined by diluting overnight LB cultures to approximately 106 CFU/ml in 25 ml of LB or ISM and incubating them with aeration at 37°C. At each time point, CFU were determined by diluting and plating.
IcsA localization assay.
IcsA (VirG) localization was determined by modification of the indirect immunofluorescence procedure of van den Bosch et al. (44). For localization of IcsA in vitro, 1 ml of bacteria grown to late logarithmic phase in LB was fixed in 4% paraformaldehyde for 10 min; this was followed by two washes with PBS. The bacteria were resuspended in 100 μl of PBS, and rabbit anti-IcsA antiserum 35 (obtained from Edwin Oaks) was added at a 1:50 dilution. The samples were incubated at room temperature for 1 h, centrifuged, and washed two times with PBS. Samples were resuspended in 100 μl of fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (ICN Pharmaceuticals, Inc., Aurora, Ohio) at a 1:100 dilution. After 1 h of incubation at room temperature, the samples were centrifuged, washed two times with PBS, and resuspended in 20 μl of PBS. One microliter of each sample was air dried on a glass slide and mounted in 90% glycerol–10% PBS containing 1 mg of p-phenylenediamine/ml (pH 8.0).
For localization of IcsA in vivo, invasion assays were done as described above with Henle cells grown on glass coverslips. After 3 h, samples were fixed in 1% paraformaldehyde for 15 min, washed two times with PBS, treated with PBS containing 50 mM NH4Cl for 5 min, and washed two times with PBS. Henle cells were permeabilized with 0.2% Triton X-100 for 10 min and then washed once with PBS for 5 min. Coverslips were incubated cell side down on 50 μl of anti-IcsA antisera diluted 1:50 in PBS containing 1 mg of bovine serum albumin(BSA)/ml for 1 h at room temperature and washed three times for 5 min each with PBS. Coverslips were incubated cell side down on 50 μl of FITC-conjugated goat anti-rabbit antisera diluted 1:100 in PBS–1 mg of BSA/ml for 1 h at room temperature, washed three times for 5 min each with PBS, and mounted in 90% glycerol–10% PBS containing 1 mg of p-phenylenediamine/ml (pH 8.0).
Samples were visualized on a Leica TCS 4D confocal laser scanning microscope equipped with a 488-nm laser, FITC detection filters, and differential interference contrast for phase-contrast imaging.
Nucleotide sequence accession number.
The DNA sequence of the S. flexneri dksA gene has been submitted to the GenBank database under accession no. AF323722.
RESULTS
Isolation of an S. flexneri dksA mutant defective in plaque formation.
A library of S. flexneri TnphoA mutants was previously constructed to identify mutants that were defective in intracellular multiplication or cell-to-cell spread by screening for mutants that were invasive but unable to form plaques on tissue culture monolayers (19). Although TnphoA mutagenesis was used primarily to identify mutations in periplasmic or membrane proteins by screening for PhoA+ colonies, some mutants with little or no activity were isolated for further study. DNA sequencing of inverse PCR products revealed that one mutant that had low alkaline phosphatase activity, SA2287, contained a TnphoA insertion in dksA, a gene originally identified in E. coli as a suppressor of a dnaK mutation. The deduced amino acid sequence of S. flexneri DksA has 98% amino acid identity to the predicted E. coli and Salmonella enterica serovar Typhimurium sequences.
The initial dksA::TnphoA mutation was transduced with phage P1 into a wild-type SA100 background, generating SA5287. This mutant exhibited the same invasive but plaque-deficient phenotype as SA2287 (Table 2). Transformation of SA5287 with plasmid pSAM1, containing the wild-type S. flexneri dksA gene, restored plaque formation in the dksA mutant (Table 2). These results indicated that the dksA mutation was responsible for the plaque-deficient phenotype and that DksA is required for intracellular multiplication or cell-to-cell spread, but not for invasion of cultured cells, by S. flexneri.
TABLE 2.
Comparison of wild-type and mutant S. flexneri in Henle cell monolayers
| Strain | Phenotype | Result of assay for:
|
|
|---|---|---|---|
| Invasiona | Plaque formationb | ||
| SA100 | Wild type | + | + |
| SA2287 | DksA− | + | − |
| SA5287 | DksA− | + | − |
| SA5287/pSAM1 | DksA+ | + | + |
| SA710 | RpoS− | + | + |
+, at least 30% of the Henle cells contained three or more intracellular bacteria in the invasion assay.
+, formation of wild-type-size plaques in Henle cell monolayers; −, pinpoint or no plaques.
Plaque assay of the rpoS mutant.
DksA has been reported to be required for the optimal expression of sigma factor RpoS (47), which regulates the stress and starvation response. Therefore, we postulated that S. flexneri may require DksA for survival of possible environmental stresses experienced in the intracellular environment. To determine whether the defect in plaque formation in the dksA mutant was due to reduced expression of RpoS, we constructed an rpoS mutant by P1 transduction from an E. coli rpoS::kan mutant and tested it for the ability to form plaques. S. flexneri rpoS mutant SA710 formed plaques on tissue culture monolayers (Table 2). Thus, the failure of the dksA mutant to spread on cultured cell monolayers is independent of RpoS.
Acid sensitivity of the dksA and rpoS mutants.
Although the rpoS plaque assay indicated that DksA regulation of RpoS is not required for virulence in Henle cells, this regulation may be important for survival of S. flexneri during passage through the host stomach. S. flexneri is highly resistant to acidic conditions and is capable of surviving at pH 2.5 for several hours (12, 38). This ability has been attributed to expression of RpoS-dependent genes, and an rpoS mutant is extremely acid sensitive (46). Since S. flexneri encounters acidic conditions during transit through the stomach and possibly in other locations within the host, we compared the in vitro acid resistances of dksA and rpoS mutants by examining the survival of stationary-phase cultures in LB at pH 2.5.
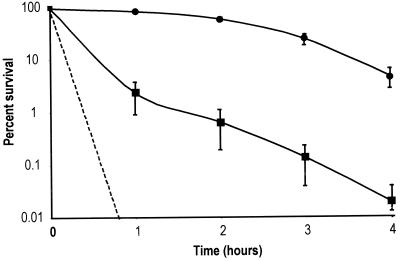
Both the rpoS and dksA mutants were significantly more sensitive to acid than the parental strain. The dksA mutant exhibited a sharp reduction in acid survival during the first hour of incubation in LB at pH 2.5 (Fig. 1) and was approximately 100 times more sensitive than the wild type. The rpoS mutant was even more sensitive to acid than the dksA mutant, exhibiting sensitivity below the detectable level of 0.001% survival. The fact that the rpoS mutant was more acid sensitive than the dksA mutant suggests that rpoS expression was not completely blocked in the dksA mutant. This result was confirmed by Western blot analysis, which showed that RpoS was present, yet below wild-type levels, in the dksA mutant (data not shown).
FIG. 1.
Effect of dksA and rpoS on survival of S. flexneri in acid. Shown are percentages of survival over time in LB-HCl, pH 2.5, for wild-type SA100 (circles) and dksA mutant SA5287 (squares) S. flexneri. The rpoS mutant SA710 (dashed line) was below detectable levels of 0.001% survival at all time points and is plotted as the theoretical maximal level of survival. The averages of three experiments are shown with the standard deviations.
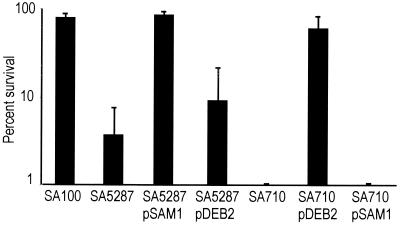
If the decrease in acid resistance in a dksA mutant was due to decreased expression of rpoS, then increased expression of rpoS should restore acid resistance in a dksA mutant. Western blot analysis indicated that RpoS levels were greater in the dksA mutant containing the cloned rpoS gene than in the dksA mutant without the gene or in the wild-type parent strain (data not shown). To test the effect of overexpression of rpoS in the dksA mutant, we examined the acid resistance at 1 h of mutants containing either the dksA or rpoS cloned genes (Fig. 2). Overexpressing rpoS in the dksA mutant was not sufficient to restore acid resistance. The resistance of SA5287/pDEB2 was slightly higher than that of SA5287 (Fig. 2), but the difference was not significant (P = 0.19). Additionally, dksA carrried on a plasmid was insufficient to restore acid resistance in an rpoS mutant (Fig. 2). However, the wild-type dksA gene restored acid resistance in the dksA mutant, and rpoS restored acid resistance in the rpoS mutant.
FIG. 2.
Effect of dksA and rpoS on acid resistance. The indicated strains were exposed to LB, pH 2.5, for 1 h, and the percent survival was determined by plating. The averages of at least three experiments are shown with the standard deviations.
CHP sensitivity of the dksA and rpoS mutants.
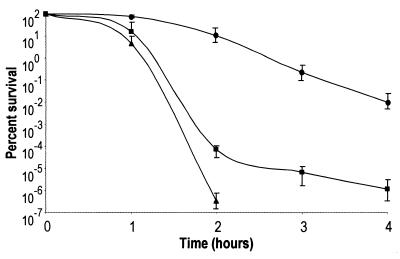
In addition to acid resistance, oxidative stress resistance is another RpoS-dependent environmental response that Shigella may induce in the host environment. To test the sensitivity of the dksA mutant to this stress, we exposed undiluted, stationary-phase cultures to oxidizing agent CHP. All tested strains exhibited sensitivity to oxidative stress, but the dksA and rpoS mutants were significantly more sensitive than the wild type (Fig. 3). As observed with exposure to acid stress, the rpoS mutant exhibited greater sensitivity than the dksA mutant.
FIG. 3.
Effect of dksA and rpoS on survival of S. flexneri in CHP. Shown is percent survival over time in oxidizing agent CHP at an approximately 2 mM concentration. Wild-type SA100 (circles), dksA mutant SA5287 (squares), and rpoS mutant SA710 (triangles) strains of S. flexneri were examined for 4 h. The rpoS mutant was below the detectable levels of 10−7% survival at 3 and 4 h, and therefore these data points do not appear on the graph. The averages of three experiments are shown with the standard deviations.
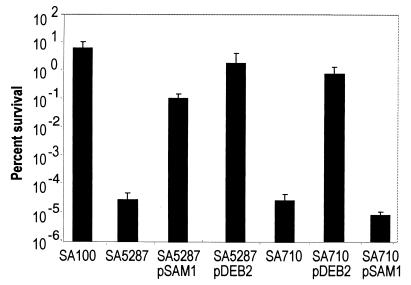
The ability of dksA and rpoS carried on plasmids to restore CHP resistance in the mutant strains was examined at 2 h (Fig. 4). As expected, plasmid-expressed dksA complemented the CHP-sensitive phenotype in the dksA mutant. However, the dksA-expressing plasmid failed to restore CHP resistance in the rpoS mutant. Interestingly, plasmid-expressed rpoS not only complemented sensitivity to oxidative stress in the rpoS mutant but also restored resistance to oxidative stress in the dksA mutant. These results suggest that DksA and RpoS operate in the same pathway during oxidative stress and that DksA is required upstream of RpoS for oxidative resistance.
FIG. 4.
Effect of dksA and rpoS on CHP resistance. The indicated strains were exposed to 2 mM CHP for 2 h, and the percent survival was determined by plating. The averages of at least three experiments are shown with the standard deviations.
Phagocytic vacuole escape of the dksA mutant.
Increased sensitivity of the dksA mutant to acid and oxidative stress appears to be due, at least in part, to the effects of DksA on RpoS expression. However, the fact that rpoS mutant SA710 produced plaques on Henle cell monolayers indicates that DksA operates in an RpoS-independent role in intracellular multiplication or spread of Shigella. To determine the role of DksA in the intracellular environment, we examined the effect of a dksA mutation on the different stages of the intracellular life cycle of S. flexneri. The possible stages include escape from the phagocytic vacuole, intracellular survival and multiplication, and spread to an adjacent epithelial cell.
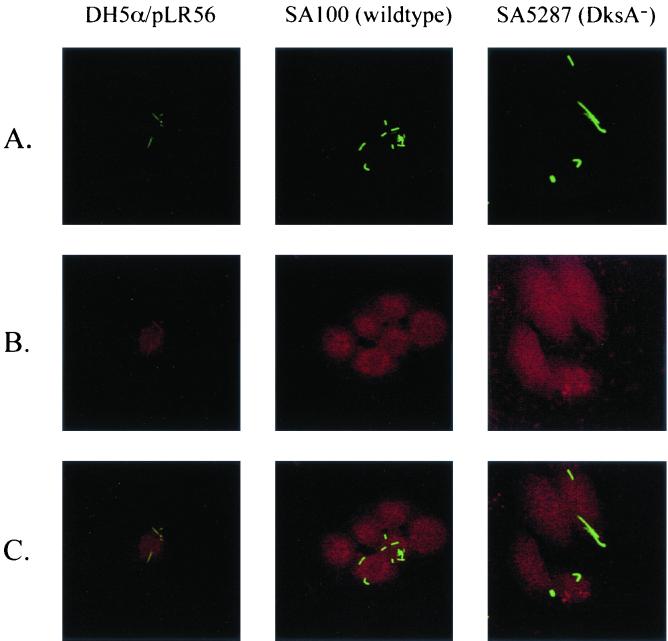
The ability of the dksA mutant to escape the phagocytic vacuole was determined by infecting rhodamine-labeled Henle cells with GFP-expressing bacteria (Fig. 5). The 5-(and 6)-carboxytetramethylrhodamine succinimidyl ester probe used in this study forms peptide bonds with cellular proteins located on the outer surfaces of eukaryotic cell plasma membranes (10, 32). The probe does not cross the plasma membrane and thus does not label intracellular proteins. Phagocytic vacuoles that result from the internalization of the plasma membrane are specifically labeled with the rhodamine probe. Labeling the Henle cells with the rhodamine probe did not affect bacterial invasion (data not shown). Following invasion, we visualized GFP-expressing bacteria and rhodamine-labeled Henle cell membranes by confocal scanning laser microscopy. Intracellular bacteria that were unable to lyse the phagocytic vacuole (E. coli DH5α/pLR56/pUC-GFP) were surrounded by a rhodamine-labeled membrane (Fig. 5). This strain expresses the Yersinia enterocolitica invA gene, which confers on E. coli the ability to attach to Henle cells and be internalized but not to lyse the phagocytic vacuole. In contrast, wild-type S. flexneri cells, which lyse the vacuole, were located in the Henle cell cytoplasm and were not surrounded by a rhodamine-labeled membrane (Fig. 5). Similarly, no colocalization of GFP and rhodamine in Henle cells infected with the dksA mutant was noted (Fig. 5). These results indicate that the dksA mutant is capable of lysing the phagocytic vacuole and gaining access to the host cell cytoplasm.
FIG. 5.
Escape of wild-type and dksA mutant bacteria from the Henle cell vacuole. Henle cells in which the cell membrane had been labeled with 5- and 6-carboxytetramethylrhodamine succinimidyl ester were infected with E. coli DH5α/pLR56 (InvA+), wild-type S. flexneri SA100, or dksA mutant SA5287. Each strain contained plasmid pUC-GFP to provide constitutive production of GFP. (A) GFP-labeled intracellular bacteria. (B) Rhodamine labeling of the Henle cell and vacuole membrane. (C) Overlay of the two images created in Adobe Photoshop.
Growth rate and in vivo multiplication of the dksA mutant.
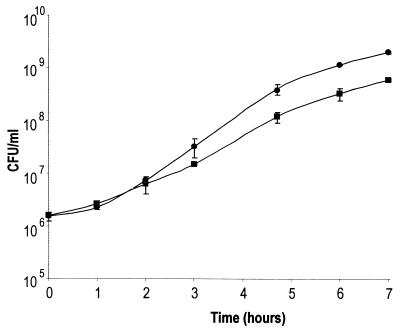
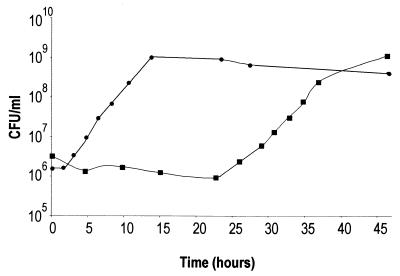
Although the dksA mutant gained access to the Henle cell cytoplasm, the ability of the mutant to grow in the intracellular environment could affect its ability to spread. Initial observations indicated that the dksA mutant exhibited a slightly reduced growth rate in LB at 37°C (Fig. 6) and a very long lag in ISM, which contains ions at concentrations comparable to those found in the eukaryotic cell cytoplasm (Fig. 7). Following the extended lag in ISM, however, the growth rate of the dksA mutant appeared to be approximately the same as that of wild-type S. flexneri (Fig. 7). When the dksA mutant population was subcultured from late-exponential or stationary-phase growth in ISM, these cultures also exhibited a 25-h lag (data not shown). Thus, the growth observed in ISM following the delay was not the result of outgrowth of revertants or mutants that had suppressors of the original dksA mutation.
FIG. 6.
In vitro growth rates of S. flexneri wild-type SA100 (circles) and dksA mutant SA5287 (squares) in LB. The averages of three experiments are shown with the standard deviations.
FIG. 7.
Growth of S. flexneri in ISM. Wild-type (circles) and the dksA mutant (squares) bacteria were grown in ISM, and the numbers of bacteria at each time point were determined by plating. The averages of three experiments are shown.
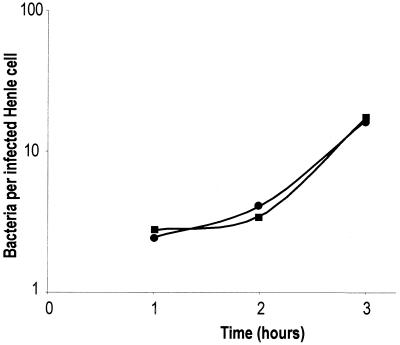
Since the dksA mutant exhibited a long lag in ISM, we examined whether impaired intracellular growth could account for the failure of the mutant to spread intercellularly. Intracellular growth rate was determined by counting the number of intracellular bacteria per infected Henle cell at various times following invasion. In contrast to the delayed onset of growth in ISM in vitro, the dksA mutant multiplied normally within Henle cells, and the average growth rate of the dksA mutant was similar to that of the wild type (Fig. 8).
FIG. 8.
In vivo growth rate of S. flexneri. Wild-type SA100 (circles) and dksA mutant SA5287 (squares) strains of S. flexneri were isolated from Henle cells at the indicated times, and the numbers of intracellular bacteria were determined by plating. The average numbers of intracellular bacteria in three independent experiments are shown.
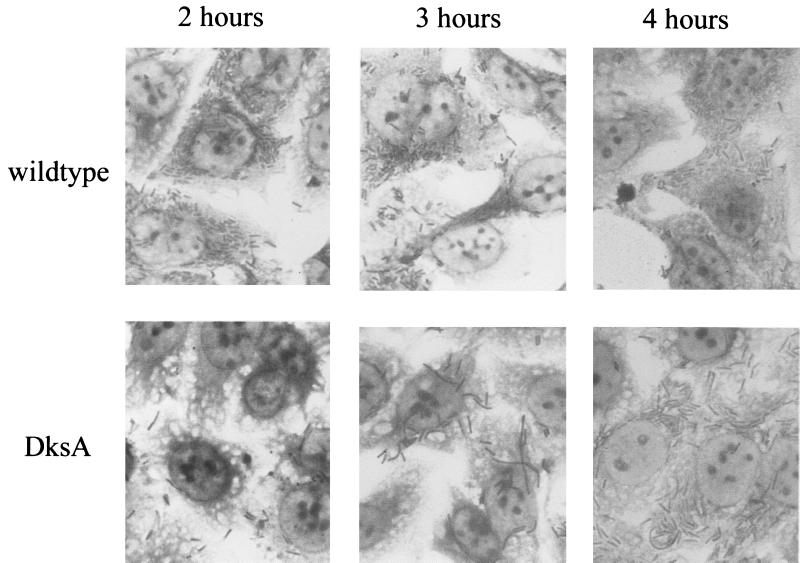
Filamentation of the dksA mutant.
During examination of intracellular growth, we observed elongation and filamentation of the dksA mutant. The dksA mutant bacteria appeared elongated compared to the wild type in the intracellular environment during the first 2 h postinvasion (Fig. 9). At 3 h postinvasion, the elongation was more pronounced and filamentation of some of the dksA mutant cells was observed. Less filamentation was noted at 4 h (Fig. 9), suggesting that this may be a transient phenomenon. Filamentation of the dksA mutant may impede bacterial spread to an adjacent cell, but this phenomenon is unlikely to be solely responsible for the inability of the dksA mutant to form wild-type plaques, since only a portion of the cells showed filamentation and since fewer filaments were observed at 4 h than at 3 h postinvasion.
FIG. 9.
Growth of the S. flexneri dksA mutant in Henle cells. Henle cells infected with wild-type SA100 and dksA mutant SA5287 bacteria were stained with Wright-Giemsa stain and observed at 1,000× magnification at 2, 3, and 4 h postinvasion. Representative images are shown.
IcsA localization in the dksA mutant.
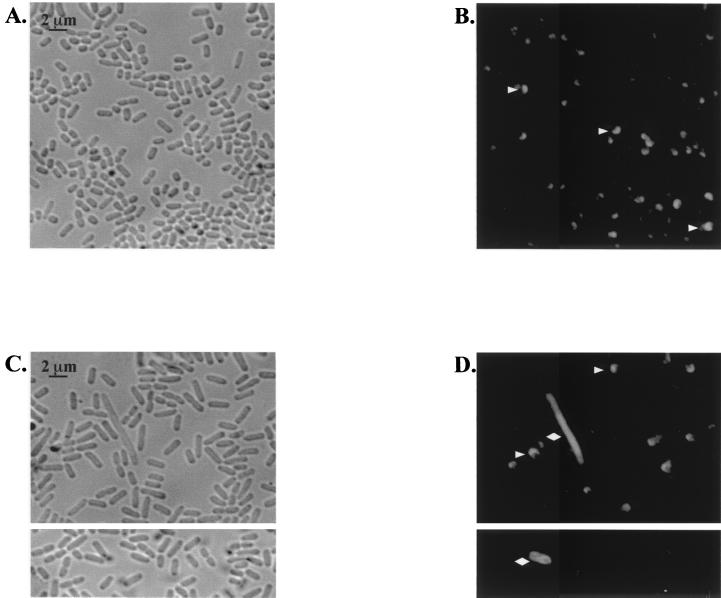
Because the dksA mutant survived and multiplied in Henle cells but did not form wild-type plaques, we examined the localization of IcsA on the bacterial cells. The failure of S. flexneri to express or properly localize IcsA, especially on filamentous bacteria, could impede intercellular spread and the ability to form plaques (11). IcsA was observed at a polar location in wild-type S. flexneri bacteria grown in vitro (Fig. 10B). The dksA mutant grown in broth showed examples of aberrant IcsA surface expression or localization, and fewer mutant cells bound the IcsA antibody than did wild-type bacteria (Fig. 10D and data not shown). IcsA was localized to one pole on some of the mutant bacteria, but others had IcsA located over the entire cell surface or at both poles. The dksA cells grown in vitro, like those growing intracellularly, were elongated compared to the wild type, and some filamentation was observed (Fig. 10C). However, there was no clear correlation between elongation or filamentation and aberrant IcsA localization (Fig. 10C and D, and data not shown).
FIG. 10.
Localization of IcsA on S. flexneri grown in vitro. Phase-contrast and immunofluorescence images of SA100 (A and B) and SA5287 (C and D) followed staining with anti-IcsA are shown. Cells were observed at 1,000× magnification. The SA100 images are from 5-h cultures, and the SA5287 images are from two separate fields at 4 and 5 h. Arrowheads, representative bacteria with polarized IcsA; diamonds, representative bacteria with aberrant localization of IcsA (either partially polarized or nonpolarized).
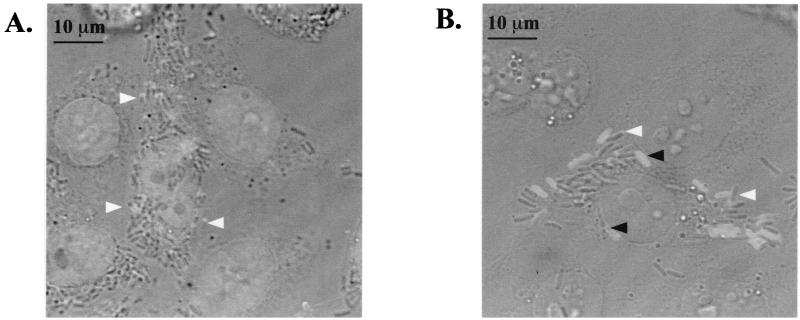
IcsA expression in intracellular bacteria was also examined (Fig. 11). The wild-type bacteria growing within Henle cells showed polar localization of IcsA (Fig. 11A), but the intracellular dksA mutant population was a mixture of cells with polar and nonpolar IcsA localization (Fig. 11B). While DksA may have other important functions in the cell, the requirement for consistent polar localization of IcsA may be sufficient to account for the aberrant plaque formation by the dksA mutant.
FIG. 11.
Localization of IcsA on Shigella growing within Henle cells. Phase-contrast and immunofluorescence images of SA100- (A) and SA5287-infected (B) Henle cells were overlaid using Adobe Photoshop. Cells were observed at 1,000× magnification. White arrowheads, representative bacteria with polarized IcsA; black arrowheads, representative bacteria with aberrant localization of IcsA (either partially polarized or nonpolarized).
DISCUSSION
Here, we identify DksA as a protein necessary for intercellular spread of S. flexneri. DksA appears to influence or function in a variety of pathways in enteric bacteria. The phenotypes include a failure to properly localize IcsA, sensitivity to environmental stress, and cell elongation or filamentation. Two-dimensional gel analysis of the S. flexneri dksA mutant showed that expression of several proteins is altered compared to that for the wild type (data not shown), which is in agreement with a previous report for S. enterica serovar Typhimurium (47). Because the dksA mutant exhibited phenotypic deficiencies in several assays and exhibited altered protein expression relative to that of the wild type, we hypothesize that DksA may function in a central pathway within the cell.
The dksA gene was initially isolated in E. coli as part of a locus that, when overexpressed, could suppress the temperature-sensitive growth of a dnaK deletion strain at 40.5°C (20). The dksA gene (dnaK suppressor) encodes a 17.5-kDa protein that comprises 151 amino acids and that has a pI of 4.87. Construction of an E. coli dksA deletion mutant indicated that dksA is not an essential gene (20). The C terminus of DksA is similar, and possibly related, to that of the F plasmid-encoded TraR protein (5). This region of homology contains a zinc finger domain common to many regulatory proteins, which may give insight into the function of the protein (5). Support for a regulatory role of DksA is provided by two-dimensional gel analysis of proteins expressed by an exponentially growing culture of an S. enterica serovar Typhimurium dksA mutant. There is differential expression of at least 14 proteins compared to expression by the wild type (47).
Genetic analysis indicates that DksA is required for the optimal translation of RpoS (sigma S), and an S. enterica serovar Typhimurium dksA mutant has a reduced amount of RpoS in stationary phase (47). Our Western blot analysis indicates that a similar effect is observed in the S. flexneri dksA mutant (data not shown). RpoS is the sigma factor serving as a master regulator of stationary-phase and stress response gene expression (23), including genes required for survival during low pH (24, 46), oxidative stress (8), UV radiation (42, 43), and hyperosmolarity (17, 18). Thus, the decreased survival in harsh environmental conditions of the dksA mutant may partially be due to decreased levels of RpoS.
The role of RpoS in virulence appears to be dependent on the pathogen as well as the stage of the infection process. RpoS is required for the intracellular survival of Legionella pneumophila in its host amoeboid species (15) as well as for virulence in S. enterica serovar Typhimurium (1, 4, 9, 48). However, the role of RpoS in virulence is rarely due to the regulation of specific virulence genes. For example, fewer E. coli rpoS mutant than wild-type cells are isolated following passage through mouse and bovine hosts (35). This observation is proposed to be due to the significant reduction in acid resistance of the bacterium (35). RpoS has been reported to be important during the initial colonization of S. enterica serovar Typhimurium of the gut-associated lymphoid tissue, but RpoS does not affect the ability of the bacterium to attach to and invade Int-407 cells and J774 macrophage-like cells or the ability to survive in macrophages (30). RpoS has been excluded from any role during the long-term colonization by E. coli of the mouse large intestine (21). Interestingly, the presence of this sigma factor appears to inhibit the virulence of Pseudomonas aeruginosa since an rpoS mutant survives as well as wild-type bacteria in rat lungs yet causes greater damage to lung tissue (40).
DksA is necessary for S. enterica serovar Typhimurium colonization of 3-week-old chicks and for virulence in 1-day-old chicks, which lack a complex intestinal flora (41). There also is a significant increase in the 50% lethal dose values for oral infection of mice by the S. enterica serovar Typhimurium dksA mutant (47). To date, the avirulent phenotype of dksA mutants has only been examined in animal models, and the molecular mechanism responsible has not been investigated. However, it has been proposed that the avirulence of an S. enterica serovar Typhimurium dksA mutant is partially due to DksA regulation of RpoS (47).
Since the dksA mutant has reduced resistance to environmental stresses, it is likely that DksA regulation of RpoS may be required for full virulence of S. flexneri during infection of the host, particularly during passage through the acidic human stomach. However, the ability of the S. flexneri rpoS mutant to form plaques on Henle cell monolayers excluded DksA regulation of RpoS as the molecular mechanism responsible for failure of the dksA mutant to spread in Henle cell monolayers. Additionally, since the rpoS mutant is extremely sensitive to environmental stresses, the results of the rpoS plaque assay suggest that S. flexneri does not experience extreme acid or oxidative stress during invasion, growth in the intracellular environment, or intercellular spread in cultured cells. Thus, the sensitivity of the dksA mutant to environmental stress, even in an RpoS-independent pathway, is not responsible for the inability of the mutant to spread intercellularly. These results are supported by experiments in which we were able to isolate viable dksA mutant bacteria from infected Henle cells (data not shown).
The dksA mutant, like wild-type Shigella, was able to lyse the phagocytic vacuole and is capable of dividing in the intracellular environment. Unlike the wild type, however, the dksA mutant did not show consistent polar location of IcsA. IcsA induces the continuous polymerization of F-actin. This process is responsible for propelling the bacterium into an adjacent epithelial cell. The mechanics of this type of movement necessitate a single focus of IcsA to produce unidirectional forces. Therefore, polymerization of actin around the entire surface or at both poles of the dksA mutant would prevent net movement of the bacterium in one direction through the host cell cytoplasm and thus prevent spread into an adjacent epithelial cell.
The specific mechanism by which DksA affects IcsA localization is unknown, although it may be related to the effect of the dksA mutation on cell division and filamentation. SopA (IcsP), which cleaves IcsA on the entire bacterial surface, may play a role in the polar localization of IcsA (7). However, IcsA, when expressed in E. coli in the absence of SopA, exhibited polar localization (36), indicating that SopA is not necessary for this process in E. coli. IcsA appears to be targeted directly to the old pole of the bacterium in an unknown process in both S. flexneri and E. coli (39). Here, we have identified DksA as a protein necessary for this process. The inability of a dksA mutant to properly localize IcsA likely contributes to the inability of the dksA mutant to form wild-type plaques on tissue culture monolayers.
ACKNOWLEDGMENTS
We gratefully acknowledge Elizabeth Wyckoff for her insight throughout this investigation and for a critical reading of the manuscript. We thank John Mendenhall and Barbara Goettgens for technical assistance with the confocal laser scanning microscope. Finally, we are very grateful to Edwin Oaks for his generous gift of polyclonal rabbit anti-IcsA antiserum 35, which contributed significantly to these results.
This study was supported by grant AI16935 from the National Institutes of Health.
REFERENCES
- 1.Bearson S M D, Benjamin J W H, Swords W E, Foster J W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernardini M L, Mounier J, d'Hauteville H, Coquis-Rondon M, Sansonetti P J. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci USA. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohannon D E, Connell N, Keener J, Tormo A, Espinosa-Urgel M, Zambrano M M, Kolter R. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of ς70. J Bacteriol. 1991;173:4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coynault C, Robbe-Saule V, Norel F. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (ςS) regulon. Mol Microbiol. 1996;22:149–160. doi: 10.1111/j.1365-2958.1996.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 5.Doran T J, Loh S M, Firth N, Skurray R A. Molecular analysis of the F plasmid traVR region: traV encodes a lipoprotein. J Bacteriol. 1994;176:4182–4186. doi: 10.1128/jb.176.13.4182-4186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuPont H L, Levine M M, Hornick R B, Formal S B. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 7.Egile C, d'Hauteville H, Parsot C, Sansonetti P J. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol. 1997;23:1063–1073. doi: 10.1046/j.1365-2958.1997.2871652.x. [DOI] [PubMed] [Google Scholar]
- 8.Eisenstark A, Calcutt M J, Becker-Hapak M, Ivanova A. Role of Escherichia coli rpoS and associated genes in defense against oxidative damage. Free Rad Biol Med. 1999;21:975–993. doi: 10.1016/s0891-5849(96)00154-2. [DOI] [PubMed] [Google Scholar]
- 9.Fang R C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative ς factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller M E, Streger S H, Rothmel R K, Mailloux B J, Hall J A, Onstott T C, Fredrickson J K, Balkwill D L, DeFlaun M F. Development of a vital fluorescent staining method for monitoring bacterial transport in subsurface environments. Appl Environ Microbiol. 2000;66:4486–4496. doi: 10.1128/aem.66.10.4486-4496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg M B, Theriot J A. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc Natl Acad Sci USA. 1995;92:6572–6576. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorden J, Small P L C. Acid resistance in enteric bacteria. Infect Immun. 1993;61:364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale T L. Genetic basis of virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale T L, Formal S B. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect Immun. 1981;32:137–144. doi: 10.1128/iai.32.1.137-144.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hales L M, Shuman H A. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol. 1999;181:4879–4889. doi: 10.1128/jb.181.16.4879-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Headley V L, Payne S M. Differential protein expression by Shigella flexneri in intracellular and extracellular environments. Proc Natl Acad Sci USA. 1990;87:4179–4183. doi: 10.1073/pnas.87.11.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengge-Aronis R. Back to log phase: ςS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 18.Hengge-Aronis R, Lange R, Henneberg N, Fischer D. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J Bacteriol. 1993;175:259–265. doi: 10.1128/jb.175.1.259-265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong M, Payne S M. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol Microbiol. 1997;24:779–791. doi: 10.1046/j.1365-2958.1997.3731744.x. [DOI] [PubMed] [Google Scholar]
- 20.Kang P J, Craig E A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogfelt K A, Hjulgaard M, Sorensen K, Cohen P S, Givskov M. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect Immun. 2000;68:2518–2524. doi: 10.1128/iai.68.5.2518-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaBrec E H, Schneider H, Magnani T J, Formal S B. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J Bacteriol. 1964;88:1503–1518. doi: 10.1128/jb.88.5.1503-1518.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee I S, Lin J, Hall H K, Bearson B, Foster J W. The stationary-phase sigma factor ςS (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol Microbiol. 1995;17:155–167. doi: 10.1111/j.1365-2958.1995.mmi_17010155.x. [DOI] [PubMed] [Google Scholar]
- 25.Lett M C, Sasakawa C, Okada N, Sakai T, Makino S, Yamada M, Komatsu K, Yoshikawa M. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the virG protein and determination of the complete coding sequence. J Bacteriol. 1989;171:353–359. doi: 10.1128/jb.171.1.353-359.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino S, Sasakawa C, Kamata K, Kurata T, Yoshikawa M. A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in S. flexneri 2a. Cell. 1986;46:551–555. doi: 10.1016/0092-8674(86)90880-9. [DOI] [PubMed] [Google Scholar]
- 27.Menard R, Dehio C, Sansonetti P J. Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 1996;4:220–226. doi: 10.1016/0966-842X(96)10039-1. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 29.Nhieu G T V, Sansonetti P J. Mechanism of Shigella entry into epithelial cells. Curr Opin Microbiol. 1999;2:51–55. doi: 10.1016/s1369-5274(99)80009-5. [DOI] [PubMed] [Google Scholar]
- 30.Nickerson C A, Curtiss R., III Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oaks E V, Wingfield M E, Formal S B. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parish C R. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol Cell Biol. 1999;77:499–508. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]
- 33.Payne S M, Finkelstein R A. Detection and differentiation of iron-responsive avirulent mutants on Congo red agar. Infect Immun. 1977;18:94–98. doi: 10.1128/iai.18.1.94-98.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne S M, Niesel D W, Peixotto S S, Lawlor K M. Expression of hydroxamate and phenolate siderophores by Shigella flexneri. J Bacteriol. 1983;155:949–955. doi: 10.1128/jb.155.3.949-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price S B, Cheng C-M, Kaspar C W, Wright J C, DeGraves F J, Penfound T A, Castanie-Cornet M-P, Foster J W. Role of rpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl Environ Microbiol. 2000;66:632–637. doi: 10.1128/aem.66.2.632-637.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandlin R C, Maurelli A T. Establishment of unipolar localization of IcsA in Shigella flexneri 2a is not dependent on virulence plasmid determinants. Infect Immun. 1999;67:350–356. doi: 10.1128/iai.67.1.350-356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sansonetti P J. Molecular and cellular biology of Shigella flexneri invasiveness: from cell assay systems to shigellosis. Curr Top Microbiol Immunol. 1992;180:1–19. doi: 10.1007/978-3-642-77238-2_1. [DOI] [PubMed] [Google Scholar]
- 38.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinhauer J, Agha R, Pham T, Varga A W, Goldberg M B. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol Microbiol. 1999;32:367–377. doi: 10.1046/j.1365-2958.1999.01356.x. [DOI] [PubMed] [Google Scholar]
- 40.Suh S-J, Silo-Suh L, Woods D E, Hassett D J, West S E H, Ohman D E. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner A K, Lovell M A, Hulme S D, Zhang-Barber L, Barrow P A. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect Immun. 1998;66:2099–2106. doi: 10.1128/iai.66.5.2099-2106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuveson R W. The interaction of a gene (nur) controlling near-UV sensitivity and the polA1 gene in strains of E. coli K12. Photochem Photobiol. 1981;33:919–923. doi: 10.1111/j.1751-1097.1981.tb05513.x. [DOI] [PubMed] [Google Scholar]
- 43.Tuveson R W, Jonas R B. Genetic control of near-UV (300–400 nm) sensitivity independent of the recA gene in strains of Escherichia coli K12. Photochem Photobiol. 1979;30:667–676. doi: 10.1111/j.1751-1097.1979.tb07197.x. [DOI] [PubMed] [Google Scholar]
- 44.van den Bosch L, Manning P A, Morona R. Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol Microbiol. 1997;23:765–775. doi: 10.1046/j.1365-2958.1997.2541625.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 46.Waterman S R, Small P L C. Identification of ςS-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 47.Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss III R, Foster J W. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol Microbiol. 1999;34:112–123. doi: 10.1046/j.1365-2958.1999.01581.x. [DOI] [PubMed] [Google Scholar]
- 48.Wilmes-Riesenberg M R, Foster J W, Curtiss R., III An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun. 1997;65:203–210. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]