Abstract
The intestinal tract is a vital organ responsible for digestion and absorption in the human body and plays an essential role in pathogen invasion. Compared with other traditional models, gut-on-a-chip has many unique advantages, and thereby, it can be considered as a novel model for studying intestinal functions and diseases. Based on the chip design, we can replicate the in vivo microenvironment of the intestine and study the effects of individual variables on the experiment. In recent years, it has been used to study several diseases. To better mimic the intestinal microenvironment, the structure and function of gut-on-a-chip are constantly optimised and improved. Owing to the complexity of the disease mechanism, gut-on-a-chip can be used in conjunction with other organ chips. In this review, we summarise the human intestinal structure and function as well as the development and improvement of gut-on-a-chip. Finally, we present and discuss gut-on-a-chip applications in inflammatory bowel disease (IBD), viral infections and phenylketonuria. Further improvement of the simulation and high throughput of gut-on-a-chip and realisation of personalised treatments are the problems that should be solved for gut-on-a-chip as a disease model.
Keywords: Gut-on-a-chip, disease model, inflammatory bowel disease, SARS-CoV-2, phenylketonuria
Introduction
The digestive, absorption and barrier functions of the small intestine are closely related to its unique structural features, such as villi and microvilli structures, mucus layers and periodic peristalsis.1 –3 The large surface area of the villi enhances absorption in the small intestine.4 The mucus layer is the first significant barrier between the small intestine and external world.5 Periodic peristalsis can aid digestion and absorption and promote the transfer of waste. Once the intestinal cells become abnormal, intestinal stem cells can repair the intestine via rapid proliferation and differentiation.6 Additionally, intestinal microorganisms play an important role in intestine functions.7 –12 These processes are essential for maintaining intestinal homoeostasis.13 –16
Animal models, such as those of mice and pigs, are famous for studying intestinal diseases.17 Due to differences in species, some animals cannot be used to study human diseases. Moreover, the use of animals in research is controversial. Compared to animal models, culturing cells in vitro to study conditions exhibits the advantages of convenience, low cost and no ethical issues.18 However, traditional culturing cells in vitro models usually lack in vivo characteristics such as fluid flow, periodic peristalsis, crosstalk between host and microorganism and crosstalk between tissues.19,20 Therefore, it is critical to reconstruct this complexity by using an in vitro model.
With the development of micromanufacturing and 3D printing technologies, gut-on-a-chip provides a new method for studying intestinal diseases in vitro.3,21,22 Based on intestinal functions, gut-on-a-chip introduces modules with different parts, such as an injection pump for fluid flow and a pressure system for mechanical deformation.1,23 –25 Many modules, including the trans-epithelial electrical resistance (TEER) module, pH module and metabolite analysis module, have been introduced to detect cell growth on a chip in real-time.26 Therefore, the concept of ‘multi-organs-on-a-chip’ can aid in providing new insights into diseases and has also been proposed to examine certain conditions involving multiple organs.27 –31
In this review, the vital structures and functions of the human intestine in gut-on-a-chip simulations is summarised. Moreover, the development of a gut-on-a-chip is presented by summarising studies on gut-on-a-chip in the literature. We also discuss some of the current applications of gut-on-a-chip and the future direction of development. We believe that the simulation ability and high throughput of gut-on-a-chip are key to gut-on-a-chip as a disease model.
Human intestinal structure and function
Human intestinal structure
The human intestine includes the small and large segments. The small intestine is an integral part of the digestive system that can break down and absorb most nutrients. It is a massive organ with an average length of 3–5 m and can be divided into the duodenum, jejunum and ileum. The large intestine includes the caecum, appendix, colon, rectum and anal canal. Unlike the small intestine, it has a shorter length but much larger lumen.32,33
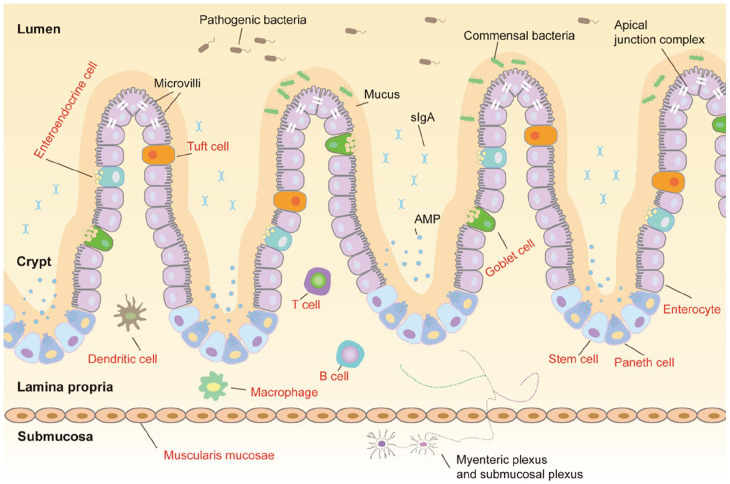
The intestinal mucosa, the innermost layer of the intestine, includes a layer of polarised columnar epithelial cells and subepithelial region that contains the lamina propria, enteric nervous system (ENS), connective tissue and muscular layers.34 The epithelium incorporates enterocytes, goblet cells, enteroendocrine cells, tuft cells, Paneth cells and intestinal stem cells (Figure 1).14,35 Enterocytes are primarily responsible for nutrient absorption. Enteroendocrine cells secrete various gastrointestinal hormones. It has been determined that tuft cells, as receptors, play an essential role in anti-parasite infection.36 Goblet cells can synthesise and release mucin, while Paneth cells can synthesise antimicrobial peptides (AMP). Given the ability of intestinal stem cells, the renewal rate of intestinal epithelial cells is rapid, and the cells last for only 3–5 days.37,38 The apical junction complex consists of tight junctions, adherent junctions and desmosomes (Figure 1). These structures confer mechanical strength on the intestinal epithelial barrier and regulate paracellular permeability.39 The mucus layer above the epithelial barrier separates the luminal contents from the intestinal epithelial cells. Mucus consists of water and glycosylated proteins, termed as mucins, which are O-linked glycan-attached glycoproteins anchored to the intestinal epithelial layer.40 The lamina propria contains many immune cells (Figure 1), including macrophages and dendritic cells.34
Figure 1.
Schematic diagram of the intestinal mucosa. The intestinal structure can be divided into mucus, villi, crypt, lamina propria, muscularis mucosae and submucosa from the outside to the inside. Symbiotic bacteria in mucus play an essential role in intestinal barrier function. The epithelium includes many types of cells, including enterocytes, goblet cells, enteroendocrine cells, tuft cells, Paneth cells and intestinal stem cells. Similar to many vital parts, the lamina propria also contains many immune cells. Tight junctions, adherent junctions and desmosomes are the main components of the apical junction complex. The ENS resides in the submucosa, consisting of two major plexuses: the myenteric plexus and submucosal plexus.
The ENS is the most significant part of the peripheral nervous system (PNS), which differs in size, morphology, composition and complexity from the rest of the PNS.41 Moreover, the ENS originates from neural crest cells that colonise the gut during intrauterine life. The ENS is an intertwined network of neurones and glial cells (Figure 1) consisting of two significant plexuses: the myenteric plexus and submucosal plexus.34 Many neurones are present in the ENS, approximately 200–600 million in humans. Approximately 20 types of intestinal neurones can be defined with slightly different numbers in different regions.42
Microbiota of the intestine
The intestine is a complex ecosystem that contains a wide variety of microorganisms under anaerobic conditions. The human intestine contains approximately 1014 microbial cells, including bacteria (the vast majority), viruses (5.8%), archaea (0.8%) and eukaryotes (0.5%).43 The composition of the intestinal microbiota is influenced by host genetics, diet and environmental factors.44 Hence, the diversity of the gut microbiota is highly dynamic and differs for each human individual and changes during lifetime.
The intestinal microbiota interacts directly with the host by producing a diverse reservoir of metabolites obtained from exogenous or endogenous substances.40 Many intestinal diseases are associated with a decreased diversity of the intestinal microbiota. However, aberrant intestinal microbiota are not only associated with intestinal diseases, such as Crohn’s disease and ulcerative colitis, but also with non-intestinal diseases, such as obesity, type 2 diabetes mellitus, rheumatoid arthritis and neurological and psychiatric disorders.45 –47 However, it remains unclear whether the imbalance of intestinal microbiota causes or is a consequence of the disease.48
The commensal intestinal microbial species can fight evading pathogens by producing antimicrobials, such as bacteriocins and certain metabolites, competing for luminal nutrients and attachment sites and producing signalling molecules that can modulate the gene expression of other bacteria. In return, the human host provides a substrate for microbiota.43 Commensal microbes benefit from the nutrient-rich intestinal environment. The microbiota produces hundreds of proteins and metabolites, including phenolic metabolites and short-chain fatty acids (SCFAs). These metabolites modulate crucial host functions, including nutrient processing, maintenance of energy homoeostasis and immune system development.49 –52
Digestive and absorption function of the intestine
The stomach receives and stores food for several hours and secretes acids and enzymes to facilitate digestion. During this time, the smooth muscles of the stomach contract and relax to mix and break down food into smaller particles that are then processed further in the duodenum.53,54
The duodenum is the initial portion of the small intestine where absorption begins. Pancreatic enzymes (a complex mixture of proteases, amylases and lipases) interact with other digestive enzymes produced by the inner wall of the small intestine to break down food components. Before reaching the jejunum, bicarbonate is secreted into the duodenum to neutralise stomach acid, maintaining a pH of approximately 6–7 in the small intestine, which is suitable for digestion of proteins, carbohydrates and fats.55 The primary function of the jejunum is to absorb sugar, amino acids and fatty acids. The ileum absorbs any remaining nutrients that are not absorbed by the duodenum or jejunum, particularly vitamin B12, as well as bile acids that continue to be recycled.32,33 Moreover, the colon, the last part of the digestive system, reabsorbs the remaining water from the indigestible contents and prepares the luminal contents for elimination.56 Fermentation is a vital function of the large bowel or colon and is considered as the process by which anaerobic bacteria break down carbohydrates into short-chain fatty acids, gases (hydrogen, methane and carbon dioxide) and other metabolites.57 In general, the intestinal microbiota plays an essential role in the digestive and absorption functions of the intestine.47
Barrier function of the intestine
The intestine constructs three types of barriers: physical, chemical and immunological barriers. The physical and chemical barriers spatially segregate gut microbiota in the intestinal lumen and immune cells in the lamina propria. These two barriers can prevent conflicts between the intestinal microbiota and host immune cells, resulting in intestinal inflammation.52 Furthermore, immunological barriers can protect the intestine via the powerful immune functions of immune cells.
Mucus prevents microbiota and large molecules from contacting the epithelial cells, but simultaneously allows the passage of small molecules.56 Intestinal mucus is an organised glycoprotein network with a host-specific glycan structure. However, the maturation and function of the mucus layer are strongly influenced by intestine microbiota.58 –60 The mucus layer in the colon is composed of inner and outer layers, and the intestinal microbiota is confined to the outer layer. Conversely, the mucous layer diffuses in the small intestine and does not form a double layer.43,61 Additionally, the apical junction complex can physically hamper microbial invasion via the paracellular pathway.62
Microbe-associated signals can induce the expression of defensins in enterocytes and antimicrobial factors in Paneth cells, leading to the production of antimicrobial peptides. Furthermore, these signals can also stimulate the maturation of B and T cells. In this case, B cells produce more IgA, and serum amyloid A-dependent T helper 17 cells improve their differentiation ability, which leads to innate immune defence mechanisms for fighting infections.63,64
Development and improvement of gut-on-a-chip
Researchers developed many models, such as the intestinal ring, intestinal segment, everted intestinal sac, Boyden Chamber and Transwell, to examined the complex structure and function of the intestinal tract. In a previous study, the advantages and disadvantages of traditional intestinal and gut-on-a-chip models have been extensively discussed.14 Compared with traditional intestinal models, gut-on-a-chip, based on a microfluidic chip, exhibits certain advantages, such as providing similar fluid velocity and peristalsis, over traditional models. These advantages have led to the rapid development of gut-on-a-chip in recent years, which has become a powerful tool for examining the intestinal tract. However, several limitations of the gut-on-a-chip model, such as complex fabrication processes, strict operating procedures and small chip capacity, must be improved. This in turn can lead to an increase in simulation of in vivo conditions.
Development of gut-on-a-chip
In 2012, Kim et al. used microfluidic system engineering to develop a mechanically active ‘human lung-on-a-chip’ model that can exhibit cyclic breathing motions. Given the advantages of this model, they explored whether a similar in vitro model of the human intestine, which can replicate the key features in the intestine, can be developed. Thus, they first proposed the concept ‘gut-on-a-chip’ and used this model to examine the co-culture of Caco-2 cells with living intestinal microbes.65
The ‘gut-on-a-chip’ contains upper and lower microchannels that are separated by a porous Polydimethylsiloxane (PDMS) membrane. On the left and right sides of the channel, vacuum chambers are used to exert cyclic mechanical strain to mimic peristaltic motion. The process of making an intestinal chip has been described previously,66 and Figure 2(a) depicts a schematic diagram of the general process. They determined that the application of physiological fluid flow and shear stress can promote accelerated intestinal epithelial cell differentiation, formation of 3D villi-like structures and increased intestinal barrier function. The addition of cyclic mechanical strain further enhances these responses. Moreover, they cultured Lactobacillus rhamnosus GG (LGG) on the apical surface of Caco-2 cells monolayer for up to 100 h and determined that ‘gut-on-a-chip’ supports the growth of microbial flora without compromising human cell viability. The authors also showed reprogramming of human intestinal epithelial cell lines to undergo spontaneous villus morphogenesis and small intestinal differentiation.67 Therefore, the proposed model is highly successful because it can effectively recapitulate many complex functions of the human intestine.
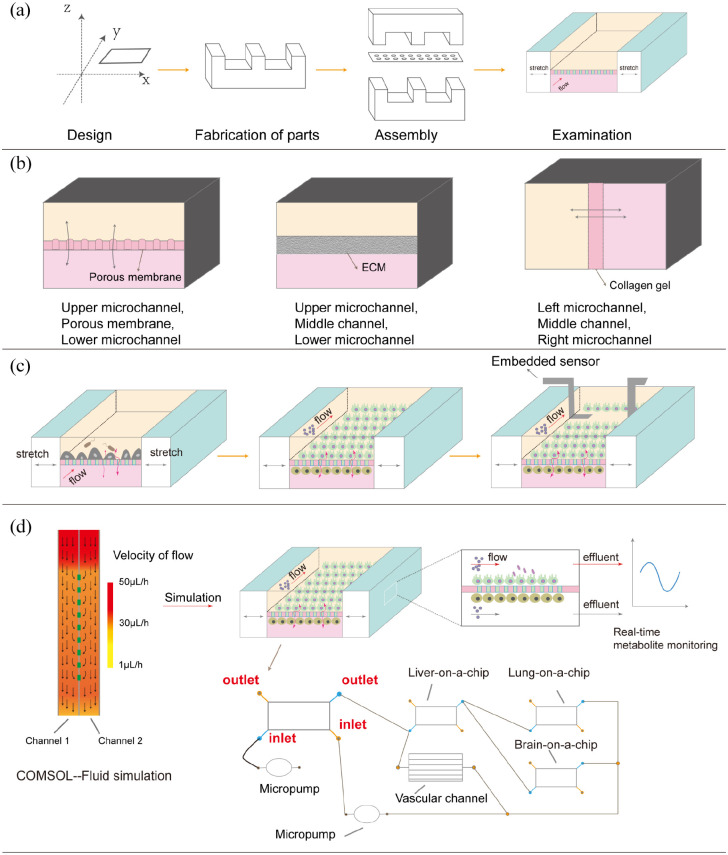
Figure 2.
Schematic diagram showing the current development of gut-on-a-chip. (a) The process of making an organ-on-a-chip generally includes: Design, fabrication of parts, assembly and examination. (b) Gut-on-a-chip can be architecturally classified into three types, the first type one contains porous membrane, and the last two use other materials (extracellular matrix and collagen gel) as opposed to porous membrane. (c) The development of a single gut-on-a-chip. Initially, gut-on-a-chip was only able to simply culture monolayer cells. Gradually, gut-on-a-chip was able to co-culture a variety of cells and bacteria. Currently, there are a lot of sensors embedded in gut-on-a-chip, which can be analysed in real time. (d) Schematic diagram of the application of gut-on-a-chip at present. The gut-on-a-chip model is designed according to the anatomical knowledge and then applied to evaluate whether the flow velocity and shear force are in line with the physiological conditions in the human body. To make the experimental process more in line with human physiological conditions, gut-on-a-chip has been used in conjunction with other organ chips (including lung-on-a-chip, liver-on-a-chip and brain-on-a-chip). Simultaneously, each organ chip can use embedded sensors to analyse various metabolites in real time.
In a follow-up study, Kim et al. analysed as to how probiotic and pathogenic bacteria, lipopolysaccharides, immune cells, inflammatory cytokines, vascular endothelial cells and mechanical forces individually and in combination contribute to intestinal inflammation, villus injury and compromise epithelial barrier function. The results showed that the microfluidic gut-on-a-chip device can be used to create human intestinal disease models to further examine intestinal pathophysiology.66,68,69 Based on the aforementioned studies, it can be observed that gut-on-a-chip is a complex system composed of many types of structural and functional units in a modular way.70 In other words, the complexity of the system depends on the number and functionality of modules that should be explored. The aforementioned articles provide an essential idea to use gut-on-a-chip to study intestinal and non-intestinal diseases in the future.
Improvement of gut-on-a-chip
Since the concept of gut-on-a-chip was proposed, many scholars have performed significant studies based on gut-on-a-chip and introduced many practical modules (Table 1). Gut-on-a-chip can be classified into three types according to their structure (Figure 2(b)). The first type of gut-on-a-chip consists of an upper microchannel, a lower microchannel and a porous membrane in the middle. The second type of gut-on-a-chip consists of an upper microchannel, a lower microchannel and a middle channel (usually filled with an extracellular matrix). The third type of gut-on-a-chip consists of a left microchannel, right microchannel and middle channel filled with collagen gel. To further improve the ability of gut-on-a-chip to simulate the in vivo microenvironment, cells with different functions, concentration gradient of oxygen and scaffolds were added to the gut-on-a-chip. HT-29 cells, displaying the properties of goblet cells, were co-cultured with Caco-2 cells with low expression of mucus proteins.71 –75 In previous studies, human primitive intestinal epithelial cells obtained from biopsies can overcome the limitations of using Caco-2 cells.76 –78 Human pulmonary microvascular endothelial cells, human umbilical vein endothelial cells (HUVECs), intestinal subepithelial myofibroblasts and peripheral blood mononuclear cells (PBMCs) were also added to the system to provide cellular crosstalk.71,79 –82 Shim et al.83 incorporated a collagen scaffold mimicking human intestinal villi into a microfluidic device, and thereby, providing cells with a 3D tissue structure and fluidic shear.
Table 1.
Characteristics of gut-on-a-chip in different kinds of literature.
| Structure | Flow rate | Cell type | Microorganism type | Mechanical strain | Characteristics |
|---|---|---|---|---|---|
| Upper microchannel, Porous membrane, Lower microchannel | 30 μL/h, (40 μL/h) | Caco-2 | Lactobacillus rhamnosus GG | 10% strain, 0.15 Hz | Mimicking the intestinal peristalsis |
| Co-culture with bacteria65 | |||||
| 30 μL/h | Caco-2 | N/A | 10% strain, 0.15 Hz | Mimicking the intestinal peristalsis67 | |
| Using direct current to measure the TEER values in the chip84 | |||||
| 50 μL/h | Caco-2 | Bifidobacterium adolescentis (DSM 20083), Eubacterium hallii (DSM 17630) | 10% strain, 0.15 Hz | Oxygen Gradient | |
| Co-culture with the obligate anaerobic gut microbiome | |||||
| Computational simulation in COMSOL85 | |||||
| 25 μL/h, 30 μL/h, 40 μL/h, 50 μL/h, 400 μL/h, 6000 μL/h | Caco-2 | N/A | N/A | Containing four cell culture chambers and NC porous membrane86 | |
| Glass-based chip87 | |||||
| The embedded electrodes for measuring the TEER88 | |||||
| Using clinical IBD patient cells89 | |||||
| Introduction of a collagen scaffold and using gravity flow device83 | |||||
| 50 μL/h | Caco-2 | Faecal microbiome | 5% strain, 0.15 Hz | Convoluted design of microchannels | |
| Using organoid-derived epithelial cells | |||||
| Computational simulation in COMSOL90 | |||||
| 60 μL/h | Caco-2, HT-29 MTX | Synthetic biotic strain (SYN5183) | 10% strain, 0.15 Hz | Analysis of effluent from compartments73 | |
| 30 μL/h, (60 μL/h) | Caco-2 | Coxsackievirus B1 | 10% strain, 0.15 Hz | Studying viral infections91 | |
| 50 μL/h, (200 μL/h) | Caco-2, HT-29, PBMCs, HUVECs | SARS-COV-2 strain 107 | N/A | Introduction a variety of cells | |
| Building a human intestinal SARS-CoV-2 infection model80 | |||||
| Upper microchannel, Middle channel, Lower microchannel | Unknown | Caco-2 | N/A | N/A | Allowing membrane-free co-culture |
| Real-time imaging | |||||
| High throughput | |||||
| Measuring the TEER values | |||||
| Using interval rocker92 | |||||
| Left microchannel, Middle channel, Right microchannel | 21 μL/h | Caco-2 HUVECs | Lactiplantibacillus plantarum HY7715 probiotic, Bifidobacterium animalis spp. lactis HY8002 probiotic | N/A | Osmosis-driven fluidic flow |
| Allowing membrane-free co-culture | |||||
| The embedded electrodes for measuring the TEER79 |
COMSOL: A fluid simulation software; HUVECs: human umbilical vein endothelial cells; IBD: inflammatory bowel disease; PBMCs: peripheral blood mononuclear cells; TEER: trans-epithelial electrical resistance.
The flow rates, cell types, microorganism types and mechanical strains shown in the table can be adjusted independently according to the objective of the study.
Owing to the structure of the intestine, anaerobic gut bacteria play a vital role in human health and disease. It is technically challenging to examine the interactions of oxygen-sensitive bacteria with oxygen-requiring intestinal epithelial cells in vitro.48 Walsh et al.93 developed a microfabricated device to generate stable and repeatable defined oxygen gradients from 0% to 4% partial pressure O2 to emulate the steep oxygen gradient at the colon wall. Shin et al. developed an anoxic–oxic interface-on-a-chip by leveraging a modified human gut-on-a-chip. Furthermore, the results demonstrated a controlled oxygen gradient in the lumen-capillary transepithelial interface by flowing anoxic and oxic culture media in various physiological environments.85
Many studies focused on improving the throughput of gut-on-a-chip. Guo et al.86 developed a biomimetic human gut-on-a-chip with four culture chambers to model drug metabolism in the intestine. Beaurivage et al.92 developed a robust high-throughput 3D gut-on-a-chip model containing 40 individual microfluidic chips to investigate the features of inflammatory bowel disease (IBD).
To improve and faster monitor cell growth in gut-on-a-chip, many scholars adopted trans-epithelial electrical resistance, a widely used parameter to characterise the quality of the barrier function of epithelial and endothelial cell monolayers. The internal structure of a single gut-on-a-chip begins to change from simple to complex (Figure 2(c)). Odijk et al.84 presented a mathematical model to enhance the fidelity of TEER measurements in microfluidic organs-on-chips, and their study illustrated the differences measured in TEER between microfluidic chips and Transwell systems. Van der Helm et al.88 embedded electrodes in gut-on-a-chip and proposed novel methods for combining impedance spectroscopy with electrical stimulation to measure the cell layer barrier function and detect changes in villus differentiation. Additionally, microchannel modelling can be performed with COMSOL Multiphysics (COMSOL Inc., USA) software to conduct fluidic flow modelling for designing a model that is more consistent with human in vivo condition.79,90,93 However, PDMS, the preferred material for developing gut-on-a-chip, is unsuitable for certain applications because of its gas permeability and capacity to absorb small hydrophobic molecules.72,87 Therefore, many materials are also being used to replace PDMS, including Polyethylene terephthalate, Polycarbonate and Polyester.78,94 Owing to the growing demand for gut-on-a-chip, an increasing number of sensors have been introduced, including pH sensors, SCFA biosensors and cytokine biosensors.95 The combination of gut-on-a-chip and other organ chips, such as liver-on-a-chip, brain-on-a-chip and lung-on-a-chip, is another way of examining enteric diseases further.26,76,96 –98 Different organ chips can be connected in same or different layers via pipes. By appropriately adding a micropump to the pipeline, the flow rate of the fluid can satisfy the requirements of each organ chip. Vascular channels that simulate blood flow can be added between organs, which stabilises metabolism between organs in the device.76 Hence, a novel multi-organ-on-a-chip model, combined with gut-on-a-chip, liver-on-a-chip, brain-on-a-chip and lung-on-a-chip models (Figure 2(d)), is proposed in this study. However, experimental data must be compared with anatomical knowledge to ensure the accuracy of multi-organ chips.
When designing microphysiological systems (MPSs), there are usually three basic elements to consider: anatomy, physiology and cell sources.99,100 Through a unique understanding of these three basic elements, we can design an in vitro model that is highly similar to the human body. First, for anatomy, we should understand the basic structure of the intestinal tract (such as the type and number of intestinal cells and morphology of intestinal villi). Second, for physiology, we should understand the biological and abiotic factors that exist in the intestinal tract. These factors include pathogenic and symbiotic bacteria, extracellular matrix (ECM) that supports cell growth, intestinal pressure, shear forces due to intestinal fluid flow and external physical stimulation of the intestinal tract. Communication and interactions between the intestinal tract and other organs are also important abiotic factors. Under low shear stress and cyclic strain, the columnar epithelium polarises rapidly and spontaneously grows into folds that recapitulate the structure of the intestinal villi. Simultaneously, cells can form a high integrity barrier, which cannot be realised via traditional cell culture.65,101 However, the shear stress and cyclic strain can have different values owing to different diseases; therefore, these parameters should be constantly verified with anatomical knowledge in the experiment. Finally, for cell sources, we can select cells from different sources for accurate replication of diseases according to different diseases for providing a better scheme for personalised treatment. Therefore, through a comprehensive consideration of these three basic elements, gut-on-a-chip will be more accurate than the traditional model, and the experimental results obtained using the gut-on-a-chip can be easily repeated.102 The gut-on-a-chip can realise real-time control of the experimental variables using microfluidic devices, and the visual observation and analysis of the experimental process can be realised using a variety of embedded sensors. Based on the many advantages mentioned above, research and application of gut-on-a-chip can be rapidly developed in the future.
Disease models using human gut-on-a-chip
Gut-on-a-chip for studying IBD
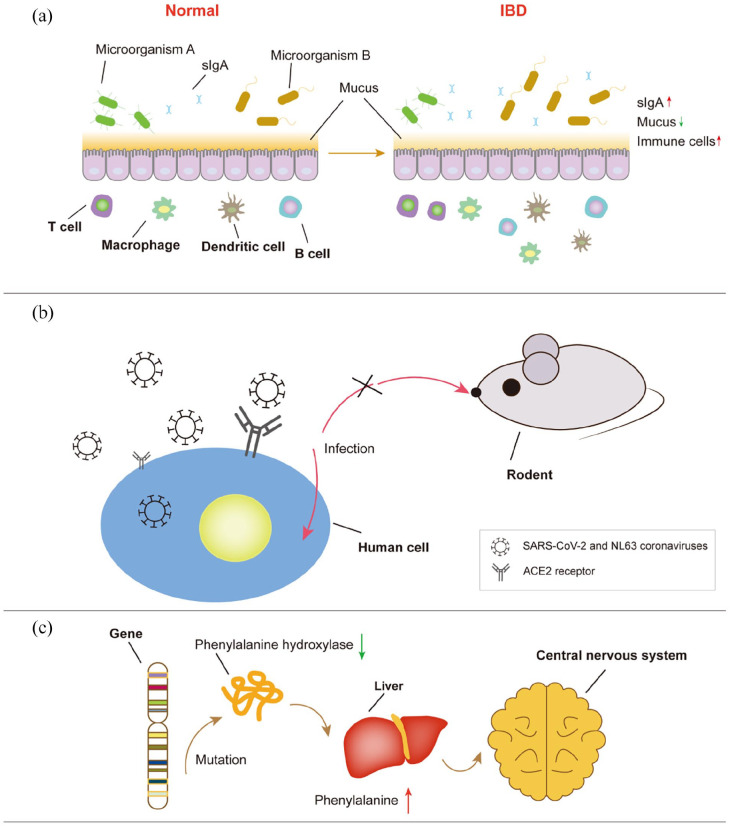
Inflammatory bowel disease, including ulcerative colitis and Crohn’s disease, is a devastating chronic inflammation of the human intestine (Figure 3(a)). However, its exact aetiology remains unknown. Reduced production of mucus and antimicrobial peptides has been observed in some IBD patients.52 Generally, it is considered to be due to interactions between the environment, heredity, infection, immunity and mental factors. However, it is difficult to confirm the interactions and contributing factors that play a crucial role in the development of IBD. Therefore, it is essential to develop an in vitro IBD model that can recapitulate the contributing factors to the maximum possible extent and reconstruct the structure and microenvironment of the intestine. Moreover, gut-on-a-chip can satisfy these requirements via microfluidic control and modules with different functions.103 Furthermore, it is easy to obtain the effluent of gut-on-a-chip for analysing metabolic activity, which can aid in understanding the mechanism of IBD.
Figure 3.
Schematic diagram of the mechanism of three diseases related to the gut. (a) Changes in intestinal structure and composition in IBD patients. The type and quantity of intestinal microorganisms in patients change. Compared with earlier, mucus secretion decreases; on the contrary, the number of immune cells increases. (b) A schematic diagram of invasion of two types of epidemic coronaviruses of the human body. These coronaviruses recognise angiotensin-converting enzyme 2 (ACE2) receptor to infect human cells. Given that rodents do not possess this receptor, they are not suitable for applications involving the infection of these viruses. (c) A schematic diagram of the primary mechanism of Phenylketonuria (PKU). Gene mutation reduces the activity of phenylalanine hydroxylase, resulting in a large accumulation of phenylalanine in the liver. Eventually, these physiological processes will have toxic effects on the nervous system.
Beaurivage et al. demonstrated the application of a robust high-throughput gut-on-a-chip model to investigate the hallmarks of IBD. They applied an optimised immune-relevant cytokine trigger that mimicked the effect of Escherichia coli-activated dendritic cells (DCs) on intestinal epithelial cells (IECs) to mimic inflammatory characteristics in this model. Furthermore, they determined that TPCA-1, an anti-inflammatory compound, can prevent inflammation in gut-on-a-chip, which demonstrated the validity of this model for drug discovery purposes.92
However, when stimulated, Caco-2 cells do not express some of the major inflammatory cytokines involved in IBD. Furthermore, patients with IBD often lack the necessary regulatory mechanisms and exhibit abnormal activation of certain types of immune cells, leading to a persistent inflammatory state. Beaurivage et al. integrated IECs derived from human intestinal organoids with monocyte-derived macrophages on a gut-on-a-chip. They used lipopolysaccharide and interferon-gamma to induce IBD hallmarks, leading to the activation and increased cytokine production in human intestinal organoids and macrophages. Under microfluidic conditions, they determined that the transcriptome of gut-on-a-chip resembled that of a normal adult human colon in vivo. In this study, TPCA-1 played a similar role in preventing inflammation.104
Recently, Yoon et al. used gut-on-a-chip to culture IBD patient cells with and without peptide-hydrogel treatment to validate the synergistic actions of peptides and hydrogels used to treat IBD. The data showed that peptide-hydrogel treatment for 96 h induced significant structural recovery of IBD patient cells in gut-on-a-chip, supported by improved villi formation and ZO-1 expression.89
It is also a new method to examine IBD based on the interaction between the microbiota and IBD. Significant variations in intestinal microbiota have been associated with IBD. The intestines of IBD patients show relatively lower bacterial diversity, particularly the loss of anaerobic bacteria.105 Some studies have suggested that an altered intestinal microbiome can be considered as the core of IBD. However, it remains unclear whether dysbiosis precedes disease development or is a by-product of the disease.81,106 Some clinical trials have shown that faecal microbiota transplantation (FMTs) can contribute to a positive outcome in IBD. Based on these data, we can take advantage of gut-on-a-chip and FMTs to find a promising treatment for IBD.
Gut-on-a-chip for studying infection of virus
Conventional methods for studying infections include the use of transformed cell lines, primary tissue-derived human cells, stem cells and animal models. Although animal models are one of the most popular models in studies involving infections, some animal models are unsuitable for examining viruses associated with humans (Figure 3(b)). Rodents are evolutionarily distant from humans. Additionally, there are ethical issues associated with experiments involving rodents.91 Furthermore, conventional cell cultures exhibit other problems such as differences in gene profiles, epigenetics and functions with natural tissues. However, the limited source of primary cells and lack of a microenvironment similar to that of the human body are also problems that cannot be ignored in traditional cell culture.96
Villenave et al. used coxsackievirus B1 as a prototype enterovirus strain to analyse human enterovirus infection and replication using a human gut-on-a-chip. They determined high coxsackievirus B1 replication efficiencies in gut-on-a-chip, which almost completely destroyed the villi within 24 h after infection.91
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and human coronavirus NL63 (HCoV-NL63) recognise angiotensin-converting enzyme 2 receptors in cells for attacking the human body. According to statistics, a large number of patients infected with these coronaviruses exhibit gastrointestinal symptoms. Some viral marker proteins are found in the human gastrointestinal (GI) tract. Therefore, the epithelial lining of the GI tract has been suggested as a potential transmission route and target of SARS-CoV-2 infection.107 Furthermore, it was discovered that the gut microbiota influences the occurrence and development of lung diseases and are, in turn, perturbed by the respiratory virus infection.108
Bein et al. used the gut-on-a-chip to examine host cellular and inflammatory responses to infection with NL63 coronavirus. They determined that cells cultured in gut-on-a-chip significantly increased angiotensin-converting enzyme 2 (ACE2) protein levels when compared to those cultured statically. Furthermore, under the infection of NL63, gut-on-a-chip showed certain characteristics of inflammation, such as loss of barrier function, increased cytokine production and recruitment of circulating peripheral blood mononuclear cells. Moreover, gut-on-a-chip infected with NL63 was used to test the antiviral effects of nafamostat and remdesivir, indicating that gut-on-a-chip can also aid as a human preclinical model for studying coronavirus.109
Guo et al. used SARS-COV-2 strain 107 and a gut-on-a-chip to model the mechanism of the virus in infecting the intestine. Caco-2 and HT-29 cells were co-cultured in the upper channel, and HUVECs were cultured with PBMCs in the lower channel. Destruction of intestinal villi, decreased integrity of the intestinal barrier, abnormal mucus secretion and activation of the immune response were observed on the chip infected with SARS-CoV-2. Guo et al. determined that intestinal mucin secretion changes from a concentrated distribution to a dispersed distribution after viral infection. Although transcriptomic analysis demonstrated significant alterations in the intestinal epithelium and endothelium in RNA and protein metabolism pathways, cell cycle regulation and oxidative phosphorylation, similar to the clinical manifestations of COVID-19, intestinal epithelial cells were more susceptible to SARS-CoV-2 infection than endothelial cells.80
Although using a gut-on-a-chip to study SARS-CoV-2 infection can be a practical approach, organs in the human body do not exist in isolation, and infectious diseases often have systemic pathological symptoms.96 Zhang et al.110 developed a human alveolar infection model of SARS-CoV-2 using an organ chip. Therefore, combining gut-on-a-chip with lung-on-a-chip to develop a ‘multi-organs-on-a-chip’ can contribute to a better understanding of SARS-CoV-2.
Gut-on-a-chip for studying phenylketonuria
Phenylketonuria (PKU) is a genetic disease characterised by a metabolic disorder of phenylalanine, which has toxic effects on the central nervous system (Figure 3(c)). PKU is generally treated with reasonable doses of phenylalanine for normal growth and other nutrients to prevent nutritional deficiency.
Microbes can respond to environmental signals within the human body to metabolise many compounds, including potentially toxic compounds. Some studies suggested that bacterially delivered phenylalanine (Phe) ammonia lyase is a potential therapy for PKU. Moreover, Escherichia coli Nissle does not colonise humans and is not present in the faeces of humans a week after ingestion. Isabella et al. constructed SYNB1618, a Phe-degrading derivative of Escherichia coli Nissle, to create a biotherapeutic agent that is expected to be suitable for treating PKU. Two pathways for Phe degradation were engineered in Escherichia coli Nissle. The results showed that SYNB1618 might have an excellent therapeutic effect on this disease as it can consume Phe in the human gastrointestinal tract, which defines a strategy for the translation of live bacterial therapeutics to treat metabolic disorders.111 Similarly, Nelson et al.73 used SYN5183, a synthetic live biotherapeutic, to study the treatment of PKU. They determined that SYN5183 resulted in dose-dependent increases in the biomarker trans-cinnamic acid and a corresponding 26.9% decrease in systemic Phe.
Discussion
In general, gut-on-a-chip technology is rapidly developing. Scholars examined and improved the primary conditions of gut-on-a-chip, such as oxygen concentration gradients, cell types, microorganisms and production materials, in a relatively short time. In recent years, research on gut-on-a-chip has focused on improving the flux and efficiency of gut-on-a-chip, monitoring the chip in real time and realising the combination of different organ chips. However, it is still difficult to fully reconstruct intestinal structure and function in vitro. To better design gut-on-a-chip, the use of fluid simulation software for simulating the channel of the chip is also a popular method. Nevertheless, there is no need to add every element to gut-on-a-chip, which should depend on the subject. Furthermore gut-on-a-chip should exhibit a high degree of simulation, but should not be overly complex. A basic module of gut-on-a-chip, which has the characteristics of a particular flow rate and regular peristalsis, can be developed. Subsequently, the module can be modified and adjusted based on the research such as bacteria and cytokines. More importantly, the development of modularisation is beneficial for the development of chips and reduces their cost.
Given the microfluidic system of gut-on-a-chip, the related symptoms of IBD can be easily induced by adding certain triggers (such as lipopolysaccharide and interferon-gamma). Similarly, microfluidic systems can also be used to change many variables that affect IBD to gain an in-depth understanding of the role of each variable and relationship between each variable. Furthermore, given that the materials for preparing gut-on-a-chip are highly transparent, the cell morphology can be observed and recorded directly using a microscope. However, gut-on-a-chip has a disadvantage in that it cannot directly study the impact of psychological factors on the condition of patients with IBD, which also has a particular impact on IBD. Gut-on-a-chip can efficiently study the two predominant problems of IBD (genetic susceptibility and immune abnormality). Therefore, it is expected that gut-on-a-chip can be used to realise personalised treatment of patients with IBD.
Given the particularity of viral infection, gut-on-a-chip provides a new method for studying specific viruses. Gut-on-a-chip can be used to cultivate human-related cells to study the infection of some viruses, enhance the credibility of the experiment and avoid ethical problems. More importantly, gut-on-a-chip can be easily combined with other organ chips, and thereby, the infection of certain viruses in different organs or parts can be more easily understood. In the face of the recent COVID-19 epidemic, it may be a breakthrough to study SARS-CoV-2 using the gut-on-a-chip.
Engineered bacteria represent a new type of therapy that uses synthetic biological tools. Gut-on-a-chip can realise co-culture between cells and microorganisms for a relatively long time. Based on microfluidic control, it is relatively easy to collect metabolites produced by the gut-on-a-chip system, which can be used to study drug metabolism. Therefore, gut-on-a-chip provides a powerful platform for studying the use of biotherapeutic agents to treat PKU. With respect to the treatment of PKU, improving the high throughput of gut-on-a-chip and using the chip for personalised therapy can be the future direction.
Conclusion
To address the complexity of intestinal diseases, the structure and function of gut-on-a-chip are constantly optimised via computer simulations and cell experiments. To date, several practical modules and functions have been introduced. Therefore, the gut-on-a-chip has served as a powerful platform for studying the treatment of IBD, viral infection and phenylketonuria. Owing to the flexibility of the chip, factors that affect a disease or the interaction between them can be examined. Different modules can be adopted for diseases with different mechanisms to build gut-on-a-chip. Furthermore, models can be developed for some diseases with individual differences by adjusting parameters in some modules, such as taking primary cells from patients and adjusting the flow rate. However, intestinal diseases also affect not only the intestine but also other organs. Hence, ‘multi-organs-on-a-chip’ presents another method for examining intestinal diseases in the future. Chips with high simulation, high throughput, multiorgan nature, real-time detection and other characteristics can potentially become powerful disease models.
Footnotes
Author contributions: Changxiu Xian performed the literature review and wrote the original draft. Jiaxin Zhang sorted out the table and figures of this article. Suqing Zhao provided support for review and editing. Xiang-Guang Li was responsible for the study concept, performed critical revision and editing of the manuscript for important intellectual content and obtained funding. All authors approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (32002213) and the Basic and Applied Basic Research Foundation of Guangdong Province (2022A1515011180 and 2019A1515110205).
ORCID iD: Xiang-Guang Li  https://orcid.org/0000-0002-1374-1444
https://orcid.org/0000-0002-1374-1444
References
- 1. Pimenta J, Ribeiro R, Almeida R, et al. Organ-on-chip approaches for intestinal 3D in vitro modeling. Cell Mol Gastroenterol Hepatol 2022; 13: 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Neill JD, Pinezich MR, Guenthart BA, et al. Gut bioengineering strategies for regenerative medicine. Am J Physiol Gastrointest Liver Physiol 2021; 320: G1–G11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hewes SA, Wilson RL, Estes MK, et al. In vitro models of the small intestine: engineering challenges and engineering solutions. Tissue Eng Part B Rev 2020; 26: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fois CAM, Le TYL, Schindeler A, et al. Models of the gut for analyzing the impact of food and drugs. Adv Healthc Mater 2019; 8: e1900968. [DOI] [PubMed] [Google Scholar]
- 5. Hagen SJ. Mucosal defense: gastroduodenal injury and repair mechanisms. Curr Opin Gastroenterol 2021; 37: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siwczak F, Loffet E, Kaminska M, et al. Intestinal stem cell-on-chip to study human host-microbiota interaction. Front Immunol 2021; 12: 798552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moossavi S, Arrieta MC, Sanati-Nezhad A, et al. Gut-on-chip for ecological and causal human gut microbiome research. Trends Microbiol 2022; 30: 710–721. [DOI] [PubMed] [Google Scholar]
- 8. Fusco F, Perottoni S, Giordano C, et al. The microbiota-gut-brain axis and epilepsy from a multidisciplinary perspective: clinical evidence and technological solutions for improvement of in vitro preclinical models. Bioeng Transl Med 2022; 7: e10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anjum M, Laitila A, Ouwehand AC, et al. Current perspectives on gastrointestinal models to assess probiotic-pathogen interactions. Front Microbiol 2022; 13: 831455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sardelli L, Perottoni S, Tunesi M, et al. Technological tools and strategies for culturing human gut microbiota in engineered in vitro models. Biotechnol Bioeng 2021; 118: 2886–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Puschhof J, Pleguezuelos-Manzano C, Clevers H. Organoids and organs-on-chips: insights into human gut-microbe interactions. Cell Host Microbe 2021; 29: 867–878. [DOI] [PubMed] [Google Scholar]
- 12. Kim MH, van Noort D, Sung JH, et al. Organ-on-a-chip for studying gut-brain interaction mediated by extracellular vesicles in the gut microenvironment. Int J Mol Sci 2021; 22: 13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pizarroso NA, Fuciños P, Gonçalves C, et al. A review on the role of food-derived bioactive molecules and the microbiota-gut-brain axis in satiety regulation. Nutrients 2021; 13: 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Chen M, Zhao S, et al. Intestinal models for personalized medicine: from conventional models to microfluidic primary intestine-on-a-chip. Stem Cell Rev Rep 2022; 18: 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harriman R, Lewis JS. Bioderived materials that disarm the gut mucosal immune system: potential lessons from commensal microbiota. Acta Biomater 2021; 133: 187–207. [DOI] [PubMed] [Google Scholar]
- 16. Garcia-Gutierrez E, Cotter PD. Relevance of organ(s)-on-a-chip systems to the investigation of food-gut microbiota-host interactions. Crit Rev Microbiol 2022; 48: 463–488. [DOI] [PubMed] [Google Scholar]
- 17. Mittal R, Woo FW, Castro CS, et al. Organ-on-chip models: implications in drug discovery and clinical applications. J Cell Physiol 2019; 234: 8352–8380. [DOI] [PubMed] [Google Scholar]
- 18. Preksha G, Yesheswini R, Srikanth CV. Cell culture techniques in gastrointestinal research: methods, possibilities and challenges. Indian J Pathol Microbiol 2021; 64: S52–S57. [DOI] [PubMed] [Google Scholar]
- 19. Wang H, Brown PC, Chow ECY, et al. 3D cell culture models: drug pharmacokinetics, safety assessment, and regulatory consideration. Clin Transl Sci 2021; 14: 1659–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moysidou CM, Owens RM. Advances in modelling the human microbiome-gut-brain axis in vitro. Biochem Soc Trans 2021; 49: 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thakare K, Jerpseth L, Pei Z, et al. Bioprinting of organ-on-chip systems: a literature review from a manufacturing perspective. J Manuf Mater Process 2021; 5: 91. [Google Scholar]
- 22. O’Farrell C, Stamatopoulos K, Simmons M, et al. In vitro models to evaluate ingestible devices: present status and current trends. Adv Drug Deliv Rev 2021; 178: 113924. [DOI] [PubMed] [Google Scholar]
- 23. Shin W, Kim HJ. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat Protoc 2022; 17: 910–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maurissen TL, Pavlou G, Bichsel C, et al. Microphysiological neurovascular barriers to model the inner retinal microvasculature. J Pers Med 2022; 12: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao Q, Cole T, Zhang Y, et al. Mechanical strain-enabled reconstitution of dynamic environment in organ-on-a-chip platforms: a review. Micromachines 2021; 12: 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ashammakhi N, Nasiri R, Barros NR, et al. Gut-on-a-chip: current progress and future opportunities. Biomaterials 2020; 255: 120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nitsche KS, Müller I, Malcomber S, et al. Implementing organ-on-chip in a next-generation risk assessment of chemicals: a review. Arch Toxicol 2022; 96: 711–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trapecar M. Multiorgan microphysiological systems as tools to interrogate interorgan crosstalk and complex diseases. FEBS Lett 2022; 596: 681–695. [DOI] [PubMed] [Google Scholar]
- 29. Tang HY, Jiang AJ, Wang XY, et al. Uncovering the pathophysiology of irritable bowel syndrome by exploring the gut-brain axis: a narrative review. Ann Transl Med 2021; 9: 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Picollet-D’hahan N, Zuchowska A, Lemeunier I, et al. Multiorgan-on-a-chip: a systemic approach to model and decipher inter-organ communication. Trends Biotechnol 2021; 39: 788–810. [DOI] [PubMed] [Google Scholar]
- 31. Sung JH, Wang YI, Narasimhan Sriram N, et al. Recent advances in body-on-a-chip systems. Anal Chem 2019; 91: 330–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kahai P, Mandiga P, Wehrle CJ, et al. Anatomy, abdomen and pelvis, large intestine. Treasure Island, FL: StatPearls Publishing, 2022. [PubMed] [Google Scholar]
- 33. Collins JT, Nguyen A, Badireddy M. Anatomy, abdomen and pelvis, small intestine. Treasure Island, FL: StatPearls Publishing, 2022. [PubMed] [Google Scholar]
- 34. Salvo Romero E, Alonso Cotoner C, Pardo Camacho C, et al. The intestinal barrier function and its involvement in digestive disease. Rev Esp Enferm Dig 2015; 107: 686–696. [DOI] [PubMed] [Google Scholar]
- 35. Li XG, Sui WG, Yan HC, et al. The in ovo administration of l-trans pyrrolidine-2,4-dicarboxylic acid regulates small intestinal growth in chicks. Animal 2014; 8: 1677–1683. [DOI] [PubMed] [Google Scholar]
- 36. Xiong Z, Zhu X, Geng J, et al. Intestinal tuft-2 cells exert antimicrobial immunity via sensing bacterial metabolite n-undecanoylglycine. Immunity 2022; 55: 686–700.e687. [DOI] [PubMed] [Google Scholar]
- 37. Li XG, Zhu M, Chen MX, et al. Acute exposure to deoxynivalenol inhibits porcine enteroid activity via suppression of the Wnt/β-catenin pathway. Toxicol Lett 2019; 305: 19–31. [DOI] [PubMed] [Google Scholar]
- 38. Li XG, Wang Z, Chen RQ, et al. LGR5 and BMI1 increase pig intestinal epithelial cell proliferation by stimulating WNT/β-Catenin signaling. Int J Mol Sci 2018; 19: 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. D’Antongiovanni V, Pellegrini C, Fornai M, et al. Intestinal epithelial barrier and neuromuscular compartment in health and disease. World J Gastroenterol 2020; 26: 1564–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jung SM, Kim S. In vitro models of the small intestine for studying intestinal diseases. Front Microbiol 2021; 12: 767038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Niesler B, Kuerten S, Demir IE, et al. Disorders of the enteric nervous system - a holistic view. Nat Rev Gastroenterol Hepatol 2021; 18: 393–410. [DOI] [PubMed] [Google Scholar]
- 42. Furness JB, Callaghan BP, Rivera LR, et al. The enteric nervous system and gastrointestinal innervation: integrated local and central control. Adv Exp Med Biol 2014; 817: 39–71. [DOI] [PubMed] [Google Scholar]
- 43. Schuijt TJ, van der Poll T, de Vos WM, et al. The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol 2013; 21: 221–229. [DOI] [PubMed] [Google Scholar]
- 44. Kastl AJ, Jr, Terry NA, Wu GD, et al. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol Gastroenterol Hepatol 2020; 9: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Generoso JS, Giridharan VV, Lee J, et al. The role of the microbiota-gut-brain axis in neuropsychiatric disorders. Braz J Psychiatry 2021; 43: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng 2019; 3: 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martinez-Guryn K, Hubert N, Frazier K, et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 2018; 23: 458–469.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. von Martels JZH, Sadaghian Sadabad M, Bourgonje AR, et al. The role of gut microbiota in health and disease: in vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 2017; 44: 3–12. [DOI] [PubMed] [Google Scholar]
- 49. Shabbir U, Rubab M, Daliri EB, et al. Curcumin, quercetin, catechins and metabolic diseases: the role of gut microbiota. Nutrients 2021; 13: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martin-Gallausiaux C, Marinelli L, Blottière HM, et al. SCFA: mechanisms and functional importance in the gut. Proc Nutr Soc 2021; 80: 37–49. [DOI] [PubMed] [Google Scholar]
- 51. Diether NE, Willing BP. Microbial fermentation of dietary protein: an important factor in diet–microbe–host interaction. Microorganisms 2019; 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Okumura R, Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med 2017; 49: e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lopez PP, Gogna S, Khorasani-Zadeh A. Anatomy, abdomen and pelvis, duodenum. Treasure Island, FL: StatPearls Publishing, 2022. [PubMed] [Google Scholar]
- 54. Chaudhry SR, Liman MNP, Peterson DC. Anatomy, abdomen and pelvis, stomach. Treasure Island, FL: StatPearls Publishing, 2022. [PubMed] [Google Scholar]
- 55. Li C, Yu W, Wu P, et al. Current in vitro digestion systems for understanding food digestion in human upper gastrointestinal tract. Trends Food Sci Technol 2020; 96: 114–126. [Google Scholar]
- 56. Farré R, Fiorani M, Abdu Rahiman S, et al. Intestinal permeability, inflammation and the role of nutrients. Nutrients 2020; 12: 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang M, Wichienchot S, He X, et al. In vitro colonic fermentation of dietary fibers: fermentation rate, short-chain fatty acid production and changes in microbiota. Trends Food Sci Technol 2019; 88: 1–9. [Google Scholar]
- 58. Yao D, Dai W, Dong M, et al. MUC2 and related bacterial factors: therapeutic targets for ulcerative colitis. EBioMedicine 2021; 74: 103751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut 2020; 69: 2232–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep 2019; 7: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Johansson ME, Phillipson M, Petersson J, et al. The inner of the two muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA 2008; 105: 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Suzuki T. Regulation of the intestinal barrier by nutrients: the role of tight junctions. Anim Sci J 2020; 91: e13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bron PA, Kleerebezem M, Brummer RJ, et al. Can probiotics modulate human disease by impacting intestinal barrier function? Br J Nutr 2017; 117: 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol 2014; 14: 667–685. [DOI] [PubMed] [Google Scholar]
- 65. Kim HJ, Huh D, Hamilton G, et al. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012; 12: 2165–2174. [DOI] [PubMed] [Google Scholar]
- 66. Kim HJ, Lee J, Choi JH, et al. Co-culture of living microbiome with microengineered human intestinal villi in a gut-on-a-chip microfluidic device. J Vis Exp 2016; 114: e54344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim HJ, Ingber DE. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol 2013; 5: 1130–1140. [DOI] [PubMed] [Google Scholar]
- 68. Shin W, Kim HJ. Pathomimetic modeling of human intestinal diseases and underlying host-gut microbiome interactions in a gut-on-a-chip. In: Doh J, Fletcher D, Piel M. (eds) Microfluidics in cell biology, Pt A: microfluidics for multicellular systems. Cambridge, MA: Academic Press, 2018, pp.135–148. [DOI] [PubMed] [Google Scholar]
- 69. Kim HJ, Li H, Collins JJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA 2016; 113: E7–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kang TH, Kim HJ. Farewell to animal testing: innovations on human intestinal microphysiological systems. Micromachines 2016; 7: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Macedo MH, Barros AS, Martínez E, et al. All layers matter: innovative three-dimensional epithelium-stroma-endothelium intestinal model for reliable permeability outcomes. J Control Release 2022; 341: 414–430. [DOI] [PubMed] [Google Scholar]
- 72. Rahman S, Ghiboub M, Donkers JM, et al. The progress of intestinal epithelial models from cell lines to gut-on-chip. Int J Mol Sci 2021; 22: 13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nelson MT, Charbonneau MR, Coia HG, et al. Characterization of an engineered live bacterial therapeutic for the treatment of phenylketonuria in a human gut-on-a-chip. Nat Commun 2021; 12: 2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. de Haan P, Santbergen MJC, van der Zande M, et al. A versatile, compartmentalised gut-on-a-chip system for pharmacological and toxicological analyses. Sci Rep 2021; 11: 4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Santbergen MJC, van der Zande M, Gerssen A, et al. Dynamic in vitro intestinal barrier model coupled to chip-based liquid chromatography mass spectrometry for oral bioavailability studies. Anal Bioanal Chem 2020; 412: 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang L, Wu J, Chen J, et al. Advances in reconstructing intestinal functionalities in vitro: from two/three dimensional-cell culture platforms to human intestine-on-a-chip. Talanta 2021; 226: 122097. [DOI] [PubMed] [Google Scholar]
- 77. Kasendra M, Tovaglieri A, Sontheimer-Phelps A, et al. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci Rep 2018; 8: 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bein A, Shin W, Jalili-Firoozinezhad S, et al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol 2018; 5: 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jeon MS, Choi YY, Mo SJ, et al. Contributions of the microbiome to intestinal inflammation in a gut-on-a-chip. Nano Converg 2022; 9: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guo Y, Luo R, Wang Y, et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull 2021; 66: 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shin W, Hackley LA, Kim HJ. “Good fences make good neighbors”: how does the human gut microchip unravel mechanism of intestinal inflammation? Gut Microbes 2020; 11: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Seiler KM, Bajinting A, Alvarado DM, et al. Patient-derived small intestinal myofibroblasts direct perfused, physiologically responsive capillary development in a microfluidic gut-on-a-chip model. Sci Rep 2020; 10: 3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shim KY, Lee D, Han J, et al. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed Microdevices 2017; 19: 37. [DOI] [PubMed] [Google Scholar]
- 84. Odijk M, van der Meer AD, Levner D, et al. Measuring direct current trans-epithelial electrical resistance in organ-on-a-chip microsystems. Lab Chip 2015; 15: 745–752. [DOI] [PubMed] [Google Scholar]
- 85. Shin W, Wu A, Massidda MW, et al. A robust longitudinal co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Front Bioeng Biotechnol 2019; 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Guo Y, Li Z, Su W, et al. A biomimetic human gut-on-a-chip for modeling drug metabolism in intestine. Artif Organs 2018; 42: 1196–1205. [DOI] [PubMed] [Google Scholar]
- 87. Kulthong K, Duivenvoorde L, Mizera BZ, et al. Implementation of a dynamic intestinal gut-on-a-chip barrier model for transport studies of lipophilic dioxin congeners. RSC Adv 2018; 8: 32440–32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Van der Helm MW, Henry OYF, Bein A, et al. Non-invasive sensing of transepithelial barrier function and tissue differentiation in organs-on-chips using impedance spectroscopy. Lab Chip 2019; 19: 452–463. [DOI] [PubMed] [Google Scholar]
- 89. Yoon HJ, Lee S, Kim TY, et al. Sprayable nanomicelle hydrogels and inflammatory bowel disease patient cell chips for development of intestinal lesion-specific therapy. Bioact Mater 2022; 18: 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shin YC, Shin W, Koh D, et al. Three-dimensional regeneration of patient-derived intestinal organoid epithelium in a physiodynamic mucosal interface-on-a-chip. Micromachines 2020; 11: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Villenave R, Wales SQ, Hamkins-Indik T, et al. Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS One 2017; 12: e0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Beaurivage C, Naumovska E, Chang YX, et al. Development of a gut-on-a-chip model for high throughput disease modeling and drug discovery. Int J Mol Sci 2019; 20: 5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Walsh DI, Dydek EV, Lock JY, et al. Emulation of colonic oxygen gradients in a microdevice. SLAS Technol 2018; 23: 164–171. [DOI] [PubMed] [Google Scholar]
- 94. Xiang Y, Wen H, Yu Y, et al. Gut-on-chip: recreating human intestine in vitro. J Tissue Eng 2020; 11: 2041731420965318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Marrero D, Pujol-Vila F, Vera D, et al. Gut-on-a-chip: mimicking and monitoring the human intestine. Biosens Bioelectron 2021; 181: 113156. [DOI] [PubMed] [Google Scholar]
- 96. Wang Y, Wang P, Qin J. Microfluidic organs-on-a-chip for modeling human infectious diseases. Account Chem Res 2021; 54: 3550–3562. [DOI] [PubMed] [Google Scholar]
- 97. Lucchetti M, Kaminska M, Oluwasegun AK, et al. Emulating the gut-liver axis: dissecting the microbiome’s effect on drug metabolism using multiorgan-on-chip models. Curr Opin Endocr Metab Res 2021; 18: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lee SH, Choi N, Sung JH. Pharmacokinetic and pharmacodynamic insights from microfluidic intestine-on-a-chip models. Expert Opin Drug Metab Toxicol 2019; 15: 1005–1019. [DOI] [PubMed] [Google Scholar]
- 99. Wang K, Man K, Liu J, et al. Microphysiological systems: design, fabrication, and applications. ACS Biomater Sci Eng 2020; 6: 3231–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Blutt SE, Broughman JR, Zou W, et al. Gastrointestinal microphysiological systems. Exp Biol Med 2017; 242: 1633–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang YI, Carmona C, Hickman JJ, et al. Multiorgan microphysiological systems for drug development: strategies, advances, and challenges. Adv Healthc Mater 2018; 7: 1701000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hinman SS, Kim R, Wang Y, et al. Microphysiological system design: simplicity is elegance. Curr Opin Biomed Eng 2020; 13: 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lee J, Choi JH, Kim HJ. Human gut-on-a-chip technology: will this revolutionize our understanding of IBD and future treatments? Expert Rev Gastroenterol Hepatol 2016; 10: 883–885. [DOI] [PubMed] [Google Scholar]
- 104. Beaurivage C, Kanapeckaite A, Loomans C, et al. Development of a human primary gut-on-a-chip to model inflammatory processes. Sci Rep 2020; 10: 21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Khan R, Roy N, Ali H, et al. Fecal microbiota transplants for inflammatory bowel disease treatment: synthetic- and engineered communities-based microbiota transplants are the future. Gastroenterol Res Pract 2022; 2022: 9999925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Devkota S. Big data and tiny proteins: shining a light on the dark corners of the gut microbiome. Nat Rev Gastroenterol Hepatol 2020; 17: 68–69. [DOI] [PubMed] [Google Scholar]
- 107. Donia A, Hassan SU, Zhang X, et al. COVID-19 crisis creates opportunity towards global monitoring & surveillance. Pathogens 2021; 10: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Signore MA, De Pascali C, Giampetruzzi L, et al. Gut-on-chip microphysiological systems: latest advances in the integration of sensing strategies and adoption of mature detection mechanisms. Sens Biosensing Res 2021; 33: 100443. [Google Scholar]
- 109. Bein A, Kim S, Goyal G, et al. Enteric coronavirus infection and treatment modeled with an immunocompetent human intestine-on-a-chip. Front Pharmacol 2021; 12: 718484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang M, Wang P, Luo R, et al. Biomimetic human disease model of SARS-CoV-2-induced lung injury and immune responses on organ chip system. Adv Sci 2021; 8: 2002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Isabella VM, Ha BN, Castillo MJ, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol 2018; 36: 857–864. [DOI] [PubMed] [Google Scholar]