Abstract
Microbiome is an integral part of the gut and is essential for its proper function. Imbalances of the microbiota can be devastating and have been linked with several gastrointestinal conditions. Current gastrointestinal models do not fully reflect the in vivo situation. Thus, it is important to establish more advanced in vitro models to study host-microbiome/pathogen interactions. Here, we developed for the first time an apical-out human small intestinal organoid model in hypoxia, where the apical surface is directly accessible and exposed to a hypoxic environment. These organoids mimic the intestinal cell composition, structure and functions and provide easy access to the apical surface. Co-cultures with the anaerobic strains Lactobacillus casei and Bifidobacterium longum showed successful colonization and probiotic benefits on the organoids. These novel hypoxia-tolerant apical-out small intestinal organoids will pave the way for unraveling unknown mechanisms related to host-microbiome interactions and serve as a tool to develop microbiome-related probiotics and therapeutics.
Keywords: Intestinal epithelium, epithelial polarity, hypoxia, organoids, gut microbiome
Introduction
The intestinal epithelium is a highly organized, dynamic cell layer with rapid self-renewing capacity.1 Main functions of the intestine include the formation of a physical barrier between the luminal contents and the external environment, nutrient absorption and transport, and regulation of host-microbiome interactions.2 Intestinal epithelial cells are polarized, featuring an apical and a basolateral membrane domain, which are characterized by different biochemical and functional properties. Maintenance of this organization is crucial for the proper function of the intestine. The apical surface is facing the intestinal lumen and is responsible for the uptake of nutrients and the formation of a defensive barrier against pathogens.3 Trillions of bacteria, fungi and other microbes reside in the gut lumen and together create an interconnected community with symbiotic and/or pathogenic relationships. The gut microbiome has a key role in metabolism and training and homeostasis of the immune system regulation and thus a tremendous impact on overall health and disease of its host.4,5 Disruption of the gut microbiome has been associated, amongst others, with inflammatory bowel disease, metabolic diseases, such as diabetes, obesity, and neurodevelopmental disorders.4 –6 Thus, there is a growing demand for advanced in vitro co-culture systems to reveal the mechanistic insights of the complex relationship and interactions between the microbiome and the intestinal epithelium, which often cannot been retrieved from more simple in vitro or in vivo models.
Intestinal organoids are three-dimensional (3D) in vitro models that have shown great potential in modeling intestinal physiology and disease. They recapitulate both the architecture and function of the in vivo tissue closer than traditional 2D culture systems. Specifically, they are structured into crypt and villus domains that surround a central lumen, they have self-renewal capacity, multicellular composition, and they can perform numerous specialized intestinal functions.1,7 Based on their proven capacity to closely resemble the intestinal epithelium, organoids have been used for the study of intestinal microbiota-host interactions.8 –10 Specifically, organoids have been co-cultured with commensal bacteria, such as Lactobacillus11 –13 and non-pathogenic Escherichia coli (E. coli) strains,14 and pathogenic bacteria, such as Cryptosporidium,15 Salmonella enterica serovar Typhimurium,16,17 E. coli strains,17 –22 Clostridioides difficile,23 –25 and Bacteroides thetaiotaomicron.26 In these systems, to bring the microorganisms in contact with the apical surface of the epithelial cells of the organoids, microinjenctions of the bacteria into the central lumen were necessary. Although with this method the microorganisms were placed in a hypoxic environment similar to the in vivo situation, microinjections are technically challenging, time-consuming and require skilled personnel.
As an alternative method to gain access to the apical surface that faces the lumen, researchers developed ways to reverse epithelial polarity. By culturing organoids in suspension, human adult,27,28 human pluripotent stem cell,29 and chicken,30 and porcine31 adult stem cell-derived organoids reversed their polarity in a way that the apical surface is facing outwards to the culture medium. These organoids have been used to study infections by Salmonella Typhimurium, Listeria monocytogenes, influenza A virus strain PR8, Eimeria tenella, and transmissible gastroenteritis virus.28,30,31 It has been shown that organoid models with reversed polarity facilitate such co-cultures since the microorganisms can simply be added to the culture medium. Although these apical-out models are highly valuable, with numerous advantages over microinjections, they do not resemble the low oxygen concentration environment that the apical surface of the intestine in vivo is exposed to. More specifically, in vivo there is an oxygen gradient from the intestinal lumen to the epithelium, ranging from 2% O2 in the lumen to 8% in the crypt area.32 Indeed, the apical surface of the apical-out organoids so far is exposed to approximately 18% O2, since it is in contact with the cell culture medium and the organoids are grown in a standard normoxia (21% O2) incubator.33 The hypoxic intestinal lumen in vivo supports the growth and survival of anaerobic microorganisms, which constitute the predominant bacterial species in the gut.32,34 Thus, co-culture of the current reversed polarity organoids with anaerobic bacteria is not optimal, since the high oxygen levels are harmful to these microorganisms and in some cases even lethal.35,36
In this study, we aimed to establish an apical-out small intestinal organoid model in hypoxic conditions, in order to overcome the aforementioned issues and create an in vitro model that facilitates both the easy access to the apical surface and the survival of anaerobic bacteria. To this end, we adapted our previously described protocol to generate apical-out small intestinal organoids from pluripotent stem cells29 to low oxygen conditions. Specifically, we cultured the organoids in a suspension system that was sustained in a hypoxic environment (5% O2). After assessing the differentiation capacity and functionality of these apical-out hypoxic organoids, we established co-culture systems with the probiotic strains Lactobacillus casei (L. casei) and Bifidobacterium longum (B. longum). These anaerobic bacteria reside in the intestine and have attracted a lot of interest from the food industry over the years because of their health-promoting probiotic benefits (e.g. epithelial barrier integrity and host immune response).37 Since multiple bacterial strains co-reside in the gut, we also performed a triple co-culture of organoids, L. casei, and B. longum. All co-culture experiments showed tighter barrier formation and increased mucin production in the organoids, compared to organoids without bacteria. This innovative, hypoxia tolerant apical-out small intestinal organoid model will be a valuable tool in future to decipher the complex gut-microbiome interactions, which have a great impact on health.
Materials and methods
Maintenance of pluripotent stem cells
The human embryonic stem cell (ESC) line WA09 (H9) was purchased from WiCell. The cells were maintained in feeder-free conditions using mTESR®1 (StemCell Technologies). Every 4–5 days (depending on colony density), the ES cells were passaged onto Matrigel (Corning®)-coated tissue culture dishes.
Fabrication and preparation of microwell arrays
Arrays of U-bottom microwells were fabricated using 50 μm thin polymer films by microthermoforming as previously described.38,39 Each microwell had a diameter of 500 μm and depth of approximately 300 μm, and each array contained 289 microwells. Microwell arrays were sterilized in a graded series of 2-propanol (VWR) (100%–70%–50%–25%–10%) and then washed twice with Dulbecco’s phosphate buffered saline (PBS; Sigma-Aldrich). The microwell arrays were mounted at the bottoms of 24-well plates using elastomeric O-rings (ERIKS).
Differentiation of pluripotent stem cells toward small intestinal organoids
Directed differentiation of ESCs toward intestinal organoids was performed as previously described.29 Briefly, to create embryoid bodies (EBs), ESC colonies were dissociated into single cells with TrypLE™ Express Enzyme (Thermofisher), resuspended in mTesR1 supplemented with Y-27632 (10 μΜ; Tocris) and then seeded in the microwell arrays at a density of 1000 cells/microwell. A 3-day incubation with Activin A (100 ng/mL; Cell guidance systems) in RPMI 1640 (Thermofisher) medium supplemented with increasing concentrations (0%, 0%, 2%, and 2%) of Hyclone defined fetal bovine serum (dFBS; Fisher scientific) promoted the definitive endoderm (DE) differentiation. During the next 4 days, DE spheroids were treated with FGF4 (500 ng/mL; R&D Systems) and CHIR99021 (3 μM; Stemgent) to induce the hindgut formation. The medium was refreshed daily from the sidewalls of the well plates, to avoid disruption of the spheroids.
Differentiation of hindgut spheroids toward small intestinal organoids was performed in two ways in order to promote apical-in or apical-out epithelial polarity. For apical-in organoids, hindgut spheroids were collected and embedded in Matrigel. A 50 μL drop of Matrigel containing organoids (Matrigel dome) was placed in each well of a tissue cultured-treated 24-well plate, cross-linked at 37°C for 20 min, and overlaid with Advanced DMEM/F-12 supplemented with B27, N2, Hepes, penicillin/streptomycin, L-glutamine (all Thermofisher), EGF (50 ng/mL; R&D systems), Noggin (100 ng/mL; R&D systems) and R-Spondin (500 ng/mL; R&D systems). For apical-out organoids, hindgut spheroids were collected and placed in suspension culture in non-tissue culture-treated 6-well plates. The plates were coated with 1% Pluronic solution in PBS (Sigma-Aldrich) for 2 h at 37°C, to avoid cell-surface adherence. The same medium as for apical-in organoids was used, but in this case, it was supplemented with 2% Matrigel. The suspension cultures of apical-out organoids were performed either in a normoxic (21% O2) or in a hypoxic (5% O2) incubator (PHCbi) and they are referred to as suspension or suspension hypoxia, respectively.
Bacteria culture
Lactobacillus casei and Bifidobacterium longum (kindly provided by John Pender’s lab at Maastricht University) were cultivated in de Man, Rogosa and Sharpe (MRS) broth (Thermofisher) at 37°C in an anaerobic chamber. To evaluate bacterial growth in organoid media, L. casei and B. longum were anaerobically grown in MRS broth overnight at 37°C. The following day, the concentration of bacteria was established by optical density measured at a wavelength of 600 nm (OD600). Bacteria were diluted to 103/mL in organoid medium and incubated for 0, 6, 12, and 24 h at 5% O2, 5% CO2 at 37°C. Next, 0.1 and 1 mL of the culture were used to make pour plates using MRS agar (Sigma-Aldrich). Colonies were counted after 48 h of anaerobic incubation at 37°C.
Co-culture of organoids with bacteria
Following differentiation, apical-out intestinal organoids were placed in 35 mm petri dishes (pre-coated with a 1% Pluronic solution). The concentration of bacteria was established by OD600. 107 or 108 bacteria were added to the same 35 mm petri dish. The organoid-bacteria systems were co-cultured for 12 h at 37°C in hypoxic conditions (5% O2). Following that, organoids were washed with PBS and collected for downstream experiments.
Epithelial barrier integrity
To test the epithelial barrier integrity, the permeability of the fluorescence marker 4 kDa Fluorescein isothiocyanate (FITC)-labeled dextran (Sigma-Aldrich) was evaluated. Intact organoids were collected and incubated in a solution containing 2 mg/mL 4 kDa FITC-dextran for 30 min at room temperature (RT). To disrupt the barrier integrity, organoids were treated with 2 mM ethylenediamine tetraacetic acid (EDTA; VWR) in Hanks’ balanced salt solution (w/o calcium and magnesium; Thermofisher) on ice for 15 min. Afterward, they were resuspended in the same FITC-dextran solution as intact organoids. Organoids were then mounted and immediately imaged using a confocal laser scanning microscope (Leica TCS SP8).
Fatty acid absorption assay
Initially, apical-in organoids were incubated with 5 mM EDTA in PBS for 1 h on a shaking platform at 4°C, in order to remove the surrounding Matrigel. Both apical-in and apical-out organoids were then washed with DMEM without phenol red and treated with a solution containing 5 μM fluorescent fatty acid analog C1-BODIPY-C12 (Thermofisher) and 5 μM fatty-acid-free BSA (Sigma-Aldrich) for 30 min at 37°C. Next, the organoids were fixed in 4% paraformaldehyde (VWR) in PBS for 30 min and stained for actin (phalloidin) and cell nuclei (4′,6-diamidino-2-phenylindole; DAPI). Finally, organoids were imaged with a confocal laser scanning microscope (Leica TCS SP8). The intracellular fluorescent signal from C1-BODIPY-C12 was quantified in single confocal z-scans using QuPath 0.3.2.
RNA isolation and quantitative Real-Time PCR (qPCR)
The total RNA was isolated from the organoids with the RNeasy Mini Kit (Qiagen) according to manufacturer’s guidelines. The cDNA was synthesized from RNA using the iScript cDNA Synthesis Kit (Bio-Rad). All samples were analyzed on a CFX96 real-time PCR detection system (Bio-Rad) using the iQ SYBR Green Supermix (Bio-Rad). All gene expression levels were normalized using the hypoxanthine phosphoribosyltransferase (HPRT) housekeeping gene. Data analysis followed the 2−ΔΔCt method. The results are representative of three independent experiments. The primer sequences are listed in the Supplemental Material.
Scanning Electron Microscopy (SEM)
Organoids were chemically fixed with 1.5% glutaraldehyde (Merck) in 0.067 M cacodylate (Acros Organics) buffered to pH 7.4 and 1% sucrose (Merck) for 3 h at RT. Subsequently, they were washed with 0.1 M cacodylate buffer and postfixed with a mixture of 1% osmium tetroxide (Agar Scientific) and 1.5% potassium ferricyanide (Merck) in the same buffer, for 1 h in the dark at 4°C. After washing with Milli-Q water, the organoids were dehydrated in a graded series of ethanol (Merck) (70, 90, up to 100%) at RT and dried with hexamethyldisilazane (HMDS) (>99.9%, Sigma-Aldrich). Finally, the samples were mounted onto SEM stubs, coated with a thin layer of gold by a sputter coater SC7620 (Quorum Technologies) and examined with an electron microscope (Jeol JSM-IT200).
Transmission Electron Microscopy (TEM)
Organoids were chemically fixed with 1.5% glutaraldehyde in 0.067 M cacodylate buffered to pH 7.4 and 1% sucrose for 3 h at RT. Following that, they were washed with 0.1 M cacodylate buffer and postfixed with a mixture of 1% osmium tetroxide and 1.5% potassium ferricyanide in the same buffer, for 1 h in the dark at 4°C. After washing with Milli-Q water, the organoids were dehydrated in a graded series of ethanol (70, 90, up to 100%) at RT. Next, organoids were infiltrated with Epon, embedded in the same resin and polymerized for 48 h at 60°C. Using a diamond knife (DiATOME), ultrathin sections of 60 nm were cut on a Leica UC7 ultramicrotome. The sections were transferred to 50 Mesh copper grids and covered with a formvar and carbon film. They were then imaged on a Tecnai T12 Electron Microscope equipped with an Eagle 4 k × 4 k CCD camera (Thermofisher) and a Veleta 2 k × 2 k CCD camera (Olympus Soft Imaging).
Immunofluorescence and confocal microscopy
Organoids were fixed with 4% paraformaldehyde in PBS for 30 min and washed three times with PBS. Permeabilization was performed with 0.5% Triton X-100 (Merck) in PBS for 30 min and blocking with 5% donkey serum (VWR) in permeabilization solution for 1 h, all at RT. Primary antibodies were incubated overnight at 4°C and the following day secondary antibodies were added and incubated for 2 h at RT. Nuclei were counterstained with DAPI and actin with phalloidin. A full list of antibodies is provided in the Supplemental Material. All samples were imaged with a confocal laser scanning microscope (Leica TCS SP8) and the images were processed with ImageJ.
Oxygen measurements
Self-adhesive sensor dots (PreSens Precision Sensing GmbH) were autoclaved (121°C, 15 min) and batch calibrated using a two-point calibration in oxygen-free water and air-saturated water, according to the manufacturer’s guidelines. The oxygen-free standard was made by dissolving Na2SO3 (1 g) and Co(NO3)2 standard solution (50 µL) (ρ(Co) = 1000 mg/L; in nitric acid 0.5 mol/L) in water (100 mL). Air-saturated water was obtained by blowing air into a stirred water-filled beaker for 20 min under agitation. We observed no significant changes in the signal acquisition of non- vs autoclaved sensor dots, suggesting that sterilization did not affect the sensing capability. Next, sensor dots were glued onto 24-well plate wells pre-coated with 1% Pluronic solution in PBS (Sigma-Aldrich) for 2 h at 37°C, and seeded with organoids embedded in a drop of Matrigel or suspended in cell culture medium. Plates were incubated at 37°C for 7 days at normoxic (21% O2) and hypoxic (5% O2) conditions. Cell culture medium was refreshed after 4 days of culture. The concentration of dissolved oxygen was measured every 5 min from the bottom side of the 24-well plate by using a fluorescence transmitter (Oxy-SMA, PreSens Precision Sensing GmbH) connected to polymeric optic fibers and processed by using the PreSens Measurement Studio 2 software.
Statistical analysis
Statistical analyses were performed in GraphPad Prism 9 software. Student’s two-tailed t-test with Welch’s correction and one- or two-way ANOVA were used to determine statistical significance. Significant differences were defined as p < 0.05. p Values of statistical significance are represented as ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05. Error bars in figures indicate standard error of the mean (S.E.M.).
Results
Differentiation of apical-out organoids in hypoxia
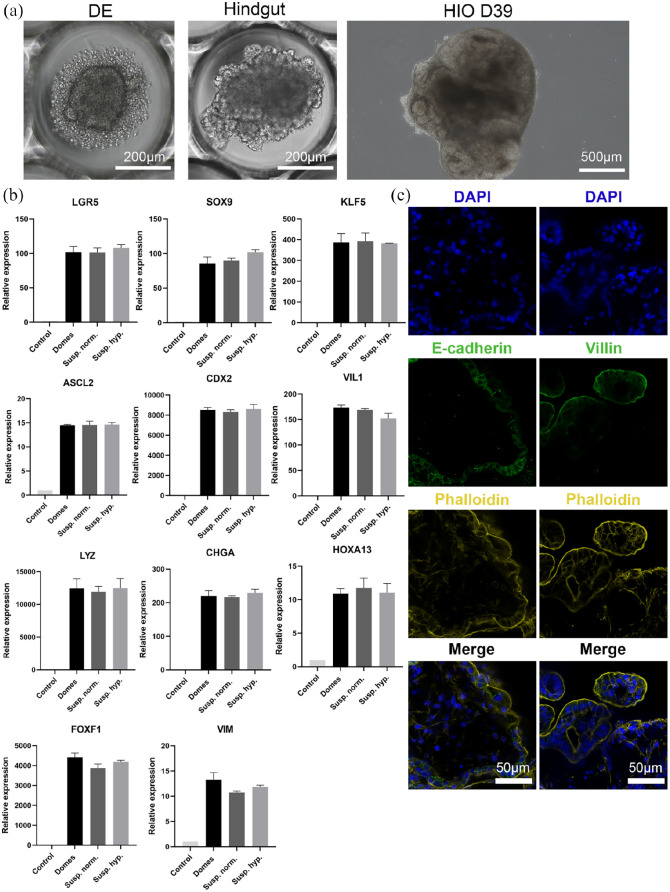
Recently, human intestinal organoid models with reversed polarity have been described in order to facilitate the access to the apical surface of the organoids.28,29 Based on our existing reversed polarity intestinal organoid model,29 here, we developed apical-out intestinal organoids in hypoxic conditions (5% O2), aiming to create a more physiologically relevant in vitro model for host-microbiome and host-pathogen interaction studies. Briefly, human embryonic stem cells (H9 cells) were aggregated to create homogeneous EBs using polymer film-based microwell arrays (Figure 1(a), Supplemental Figure 1). In the next days, EBs were differentiated stepwise, first toward definitive endoderm and then toward hindgut. Ultimately, hindgut spheroids were removed from the microwells and placed in a suspension culture system to differentiate them further toward intestinal organoids. This system provides a scalable platform to produce up to 7000 intestinal organoids with reversed polarity, from a single 24-well plate. Scaled-up production of apical-out organoids can be particularly useful for high-throughput downstream applications, such as drug screenings.
Figure 1.
Apical-out small intestinal organoid differentiation in hypoxic conditions. (a) Bright-field images indicating each step of the directed differentiation of ESCs toward intestinal organoids (Definitive endoderm → Hindgut → Human Intestinal Organoids). (b) qRT-PCR analysis demonstrated the expression levels of proliferation genes (LGR5, SOX9, KLF5, and ASCL2), intestinal differentiation genes (CDX2, VIL1, LYZ, CHGA, and HOXA13) and mesenchymal genes (FOXF1 and VIM) in Matrigel-embedded (domes), suspension normoxia, and suspension hypoxia organoids. Untreated H9 cells were used as controls. Statistical analysis showed no significant difference between the three organoid models at any of the time-points. Error bars indicate mean ± S.E.M. (n = 3). (c) Confocal microscopy demonstrated the expression of the basolateral marker E-cadherin (green, left column) in the inner part of the organoids, whereas the apical markers phalloidin (yellow, both columns) and Villin (green, right column) in the outer part of the organoids, thus indicating a successful reversal of epithelial polarity.
For the proper function of the intestine, maintenance of structural organization and presence of diverse epithelial cell lineages are pivotal. Previous reports have shown that pluripotent stem cell-derived organoids recapitulate closely the cellular composition of the in vivo intestine.29,40 To assess the differentiation capacity and maturation level of apical-out organoids in hypoxic conditions, we performed gene expression analysis for both proliferation and differentiation of intestinal cell lineages after 30 days in culture and compared them with apical-in organoids grown embedded in Matrigel domes and with apical-out organoids grown in normoxic conditions (21% O2) (Figure 1(b)). Specifically, we evaluated the expression of the leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5), the sex determining region Y-box 9 (SOX9), the Krueppel-like factor 5 (KLF5) and the Achaete scute-like 2 (ASCL2), all indicators of proliferation. For differentiation, the expression of the intestinal differentiation marker Caudal Type Homeobox 2 (CDX2), the enterocyte brush border marker Villin 1 (VIL1), the Paneth cell marker Lysozyme (LYZ), and the enteroendocrine cell marker Chromogranin A (CHGA) were examined. Presence of mesenchyme was identified by the expression of the distal hindgut mesoderm marker Homeobox A13 (HOXA13) and the mesenchymal markers Forkhead Box F1 (FOXF1) and Vimentin (VIM). The expression levels of all these markers were similar in all three systems (Matrigel domes, suspension, and suspension hypoxia) and no statistically significant differences were identified. We also performed immunofluorescence stainings to visualize the expression of the proliferation marker Ki67 and phalloidin, the intestinal differentiation marker CDX2, the goblet cell marker Mucin 2 (MUC2) and the enteroendocrine marker Synaptophysin (Supplemental Figure 2). Collectively, these results indicate that apical-out intestinal organoids can be differentiated efficiently in a hypoxic environment.
Next, we examined the structural organization of these organoids. Confocal microscopy demonstrated the expression of the basolateral surface marker E-cadherin in the inner part of the apical-out organoids. In contrast, the expression of phalloidin, which visualizes F-actin, and Villin, which visualizes the apical brush border of the enterocytes, were identified in the outer part of the organoids (Figure 1(c)). Transmission electron microscopy (TEM) verified the reversal of epithelial polarity. Microvilli were found on the exterior surface of the organoids, facing the surrounding culture medium (Supplemental Figure 3). Overall, these data confirm that organoids cultured in suspension in hypoxic conditions, have a reversed epithelial polarity, similar to organoids grown in normoxic conditions.
HIF-1α-induced effects on the organoids
Intestine is exposed to a unique oxygen gradient from the highly vascularized subepithelial mucosa toward the hypoxic lumen.41 The hypoxia-inducible factors (HIFs) are the main regulators of oxygen homeostasis, mediating both oxygen delivery and adaptation to oxygen deprivation.42 HIFs are heterodimers consisting of an oxygen sensitive α-subunit and a constitutively expressed β-subunit. Under normoxic conditions, the HIF-α subunits are hydroxylated but under hypoxic conditions, this function is inhibited, thus leading to stabilization of HIF-α subunits that heterodimerize with the HIF-β subunits.32 The HIF-1α isoform contributes to the proper function of the intestinal barrier and the maintenance of mucosal homeostasis.43 In this study, we aimed to evaluate the response of apical-out organoids upon exposure to hypoxia, by investigating the expression of HIF-1α and its downstream targets.
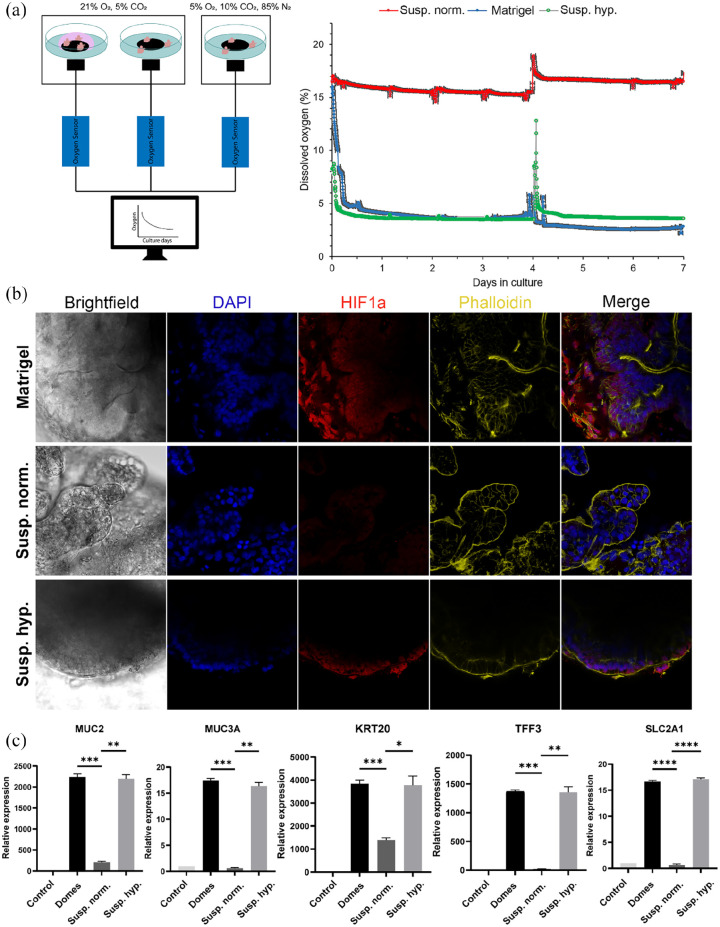
Initially, we assessed the oxygen levels in each different culture system (Matrigel-embedded, suspension and suspension hypoxia) (Figure 2(a)). To do that, we measured the concentration of dissolved oxygen using an oxygen optic sensor over a period of 7 days. On the one hand, organoids cultured in suspension in a hypoxic incubator (5% O2) had similar oxygen levels as organoids embedded in Matrigel and cultured in a normoxic incubator (⁓3–5% O2). On the other hand, the oxygen levels in organoids cultured in suspension in a normoxic incubator were much higher (15–18% O2). These results are consistent with previous studies showing that inside a Matrigel dome, the oxygen levels range between 2.8% and 9.7%44 and in suspension cultures, the oxygen concentration in cell culture medium is about 18% for a standard normoxic incubator and about 2% for a 2% O2 incubator.33
Figure 2.
HIF-1α-induced effects on apical-out small intestinal organoids. (a) Sensor measurements of oxygen levels in organoids cultured in Matrigel domes, suspension normoxia, and suspension hypoxia over a period of 7 days. For organoids grown in suspension normoxia, the dissolved oxygen concentration was about 15%–18%, whereas in Matrigel-embedded and suspension hypoxia cultures, the dissolved oxygen concentration was about 3%–5%. The peak at day 4 corresponds to medium refreshment. Error bars indicate mean ± S.E.M. (n = 3). (b) Immunofluorescence stainings indicated the expression of HIF-1α in low oxygen conditions (Matrigel-embedded and hypoxia suspension). No expression was identified in organoids grown in suspension normoxia. (c) Comparison of gene expression levels of HIF-1α targets, including MUC2, MUC3A, KRT20, TFF3, and SLC2A1 between the three culture conditions. All these genes were significantly upregulated in low oxygen conditions. Error bars indicate mean ± S.E.M. (n = 3).
After the estimation of oxygen levels in the different organoid culture systems, we aimed to evaluate the protein expression of HIF-1α (Figure 2(b)). Immunofluorescence staining indicated increased HIF-1α expression in organoids grown embedded in Matrigel and in hypoxia suspension. HIF-1α expression was detected in both the nucleus and the cytoplasm of the intestinal organoids’ cells. However, no expression was identified in suspension organoids cultured in a normoxic incubator, since the high levels of oxygen do not allow for HIF-1α stabilization. Following the confirmation of HIF-1α expression, we evaluated the expression of certain HIF-1α target genes (Figure 2(c)). A number of mucosal barrier formation-related genes are critically regulated by HIF-1α, including the Mucins 2 and 3A (MUC2 and MUC3A) and the intestinal trefoil factor 3 (TFF3).45 We indeed found significant upregulation of these genes in both Matrigel-embedded and suspension hypoxia organoids. Higher expression levels of keratin 20 (KRT20), a marker of mature enterocytes and goblet cells, were detected in hypoxia, which is in accordance with previously reported data.46 Finally, the glucose transporter solute carrier family 2 member 1 (SLC2A1, also known as GLUT1) is also responsive to hypoxia.47 This was confirmed in our organoids grown in low oxygen conditions (Matrigel-embedded and suspension hypoxia). Collectively, these results show that the apical surface of suspension hypoxia organoids is exposed and responds to the hypoxic environment, similar to the in vivo situation. Upon exposure to the hypoxic environment, HIF-1α was activated and the expression of its downstream targets was upregulated, too.
Barrier integrity in hypoxic apical-out organoids
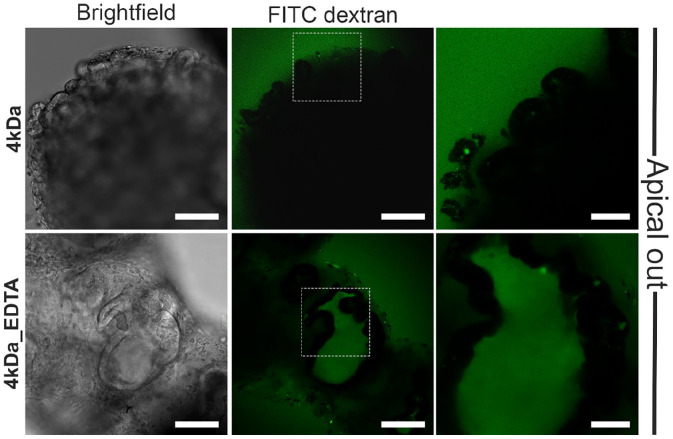
One of the fundamental functions of the intestinal epithelium is to act as a physical and biochemical barrier between the luminal contents and the underlying tissue. Disruption of the intestinal barrier has been associated with various diseases, such as inflammatory bowel disease and irritable bowel syndrome. To evaluate the epithelial barrier function in our hypoxic apical-out organoids, we performed a fluoresceinyl isothiocyanate (FITC)-dextran diffusion assay. This is a common way to evaluate barrier function both in vivo,48,49 in vitro.50,51 Apical-out organoids were incubated in a 4 kDa FITC-dextran (FITC-D4) solution for 30 min. Subsequently, we observed its diffusion into the organoid lumen using a confocal microscope (Figure 3). This experiment showed that the apical-out organoids cultured in hypoxic conditions excluded the FITC-D4, thus indicating intact epithelial barrier integrity. As a positive control, we treated organoids with EDTA, a chelating agent known to disrupt tight junctions and compromise barrier integrity.52 Treatment of apical-out organoids with 2 mM EDTA for 15 min led to disruption of barrier integrity, as suggested by the diffusion of FITC-D4 in the intercellular spaces of the organoids. Collectively, these results support that apical-out small intestinal organoids grown in hypoxic conditions demonstrate intact epithelial barrier function.
Figure 3.
Epithelial barrier integrity in hypoxia apical-out organoids. Confocal microscopy demonstrated that no diffusion of the 4 kDa FITC-dextran solution in untreated organoids (top row) occurred, thus indicating strong barrier integrity. In contrast, treatment of organoids with 2 mM EDTA (bottom row) disrupted the junctions and the dextran diffused into the intercellular space. Scale bars: 100 μm (left and middle) and 50 μm (right).
Polarized nutrient absorption
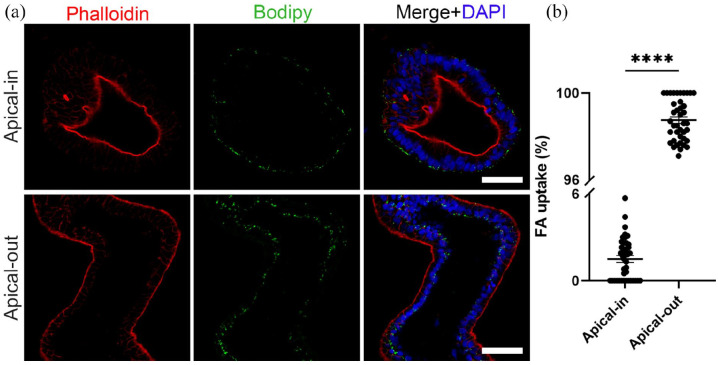
The formation of a strong epithelial barrier is key to another major function of the intestinal epithelium, the controlled nutrient absorption. This function is mediated by certain transport proteins that are located in the apical and/or basal membrane domains.53 Fatty acids enter into the apical membranes of the enterocytes through the following transport proteins: cluster of differentiation 36 (CD36, also known as fatty acid translocase), plasma membrane-associated fatty acid-binding protein (FABPpm) and/or fatty acid transport proteins 1–6 (FATP1–6).54 Once inside the enterocytes, fatty acids are transported toward the endoplasmic reticulum where they contribute to the synthesis of phospholipids, triacylglycerols, and cholesterol esters. These lipids are assembled into chylomicrons or stored in the cytosol as lipid droplets.55 To assess the fatty acid uptake in apical-out organoids grown in hypoxic conditions, we used the fluorescent fatty acid analog C1-BODIPY-C12 and compared its uptake to the uptake in apical-in organoids. Both apical-in and apical-out organoids were incubated with a solution containing the BODIPY dye for 30 min. Afterward, organoids were fixed and stained with phalloidin (indicating F- actin) and DAPI (indicating nuclei; Figure 4). Visualization with a confocal microscope and subsequent quantification demonstrated strong fluorescent signal only in apical-out organoids, thus showing that these organoids can successfully absorb the fatty acid analog from the surrounding medium (>96% uptake). In apical-in organoids, the fluorescent signal was weak, thus indicating that there was no uptake of fatty acids in these organoids. Overall, these results support the presence of active fatty acid transport proteins directly accessible in the outer apical surface of apical-out organoids grown in hypoxia.
Figure 4.
Polarized nutrient uptake: (a) representative images of apical-in and apical-out organoids incubated with the fluorescent fatty acid (FA) analog C1-BODIPY-C12 (green). Only apical-out organoids took up the BODIPY. DAPI (blue) marked the nuclei and phalloidin (red) the apical side of the epithelium. Scale bars: 50 μm and (b) quantification of the FA uptake in apical-in and apical-out organoids. Error bars indicate mean ± S.E.M. (n = 4).
Co-culture of organoids with Lactobacillus casei
Lactobacilli are among the dominant bacteria in the gut and can be found in several dietary sources (i.e. kefir, yoghurt, sourdough bread etc.). They provide numerous benefits to the host, such as the enhancement of gut barrier integrity, the strengthening of tight junctions, and the regulation of mucin expression and immune responses.56 L. casei is a facultative anaerobic strain that is proven very promising for the prevention of intestinal inflammation and the protection of the mucosal barrier.57 Here, we aimed to evaluate the effects of L. casei cells and metabolites on small intestinal organoids with apical-out orientation in hypoxic conditions. To this end, we either co-cultured organoids with 107 or 108 bacteria cells or we added 10 μg/mL bacteria lysates to the organoid medium for 12 h. Higher amounts of bacteria (>108) have proven to be toxic and led to severe cell death in the organoids.
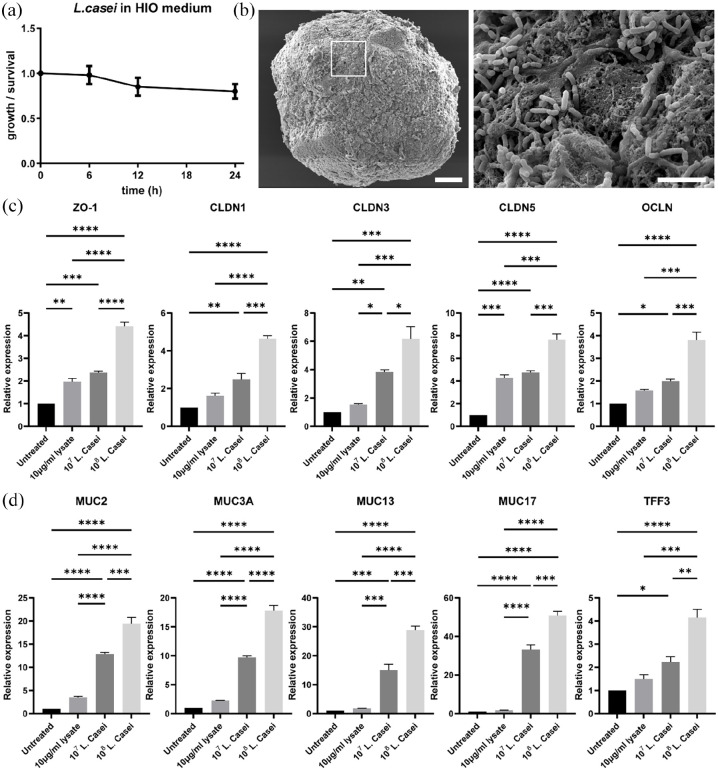
Prior to the co-culture experiments, we cultured the bacteria in organoid medium in hypoxia, to evaluate their growth over time (Figure 5(a)). Even though bacteria did not seem to replicate during the 24 h incubation period, more than 80% of the bacteria survived. Since L. casei is an anaerobic strain, we assume that a possible reason for the small amount of bacterial cells’ death are the oxygen levels differences that arise during the handling (e.g. measuring concentration, transfer in the organoid medium). Alternatively, another reason could be the reduced fitness of bacteria in the organic medium of the organoids. Then, we aimed to identify whether the L. casei cells colonize the apical epithelial surface of the small intestinal organoids. Hence, we performed SEM imaging and observed that L. casei cells attached on the apical surface of the organoids (Figure 5(b)), which is in accordance with previous in vivo and in vitro studies.58
Figure 5.
Co-culture of organoids with L. casei: (a) anaerobic culture of L. casei cells in human intestinal organoid (HIO) medium. Error bars indicate mean ± S.D. (n = 4). (b) SEM microscopy showed colonization of L. casei bacteria on the apical surface of the organoids. White box represents the area magnified in the corresponding image on the right. Scale bars: 100 μm and 5 μm. (c and d) qRT-PCR analysis demonstrated the expression levels of the junction markers: ZO-1, OCLN, CLDN1, CLDN3, and CLDN5 (c) and the mucins markers: MUC2, MUC3A, MUC13, MUC17, and TFF3 (d) in organoids co-cultured with L. casei-derived lysates and two concentrations of L. casei cells (107 and 108). Error bars indicate mean ± S.E.M. (n = 3).
To evaluate the effects of L. casei on the barrier integrity of apical-out organoids, we performed gene expression analysis for the junction markers zonula occludens 1 (ZO-1), claudin-1, 3 and 5 (CLDN1, CLDN3, CLDN5), and occludin (OCLN) (Figure 5(c)). ZO-1 and OCLN mediate the transport of large molecules up to 6 nm via the Leak Pathway, whereas CLDN1, CLDN3, and CLDN5 regulate the transport of smaller ions and solutes (up to 0.8 nm) via the Pore Pathway.59,60 All these markers were found upregulated when L. casei cells or lysates were added to the culture. This response was “dose-dependent,” meaning that increasing amounts of bacteria led to increasing gene expression of junction markers. Probiotic bacteria strains have also been found to affect mucin expression, thus regulating the properties of the mucus layer and indirectly the intestinal immune system.57 To examine whether this is occurring in apical-out intestinal organoids, we conducted quantitative real-time PCR for major secreted and membrane-bound mucins and mucin-related genes. Our analysis included MUC2, MUC3A, MUC13, MUC17, and TFF3. Similar to the junction markers, we found the expression of these genes significantly upregulated and positively correlated with the increasing numbers of microorganisms added. In summary, these results indicate that the presence of L. casei cells or lysates enhances the barrier formation and mucus production in apical-out organoids, when compared to untreated organoids. Noteworthy, it is important to highlight that viable bacterial cells have a much stronger effect on the organoids when compared to the lysates. To achieve sufficient quantities of protein content in lysates (10 μg/mL), approximately 3.4 × 108 bacteria were required. This is more than three times higher than the amount of bacteria co-cultured with the organoids and yet the effects were less robust. Thus, this indicates the importance of direct organoid-bacteria contact. Overall, apical-out small intestinal organoids grown in hypoxia can be used to explore further the mechanisms underlying the probiotic effects of L. casei on the intestinal epithelium.
Co-culture of organoids with Bifidobacterium longum
Together with lactobacilli, bifidobacteria are among the first colonizers of the neonatal gut and rank among the 10 most dominant bacteria. Bifidobacteria are probiotics and they can be found in many food types, including yogurt, kefir, seaweed, and whole grains. They are known to prevent the invasion of pathogens by modulating the pro-inflammatory responses,37 promote intestinal barrier integrity,61 and improve the mucosal barrier function.62 B. longum is a nonpathogenic, microaerotolerant anaerobic strain that is considered to be one of the earliest colonizers of the infants’ gastrointestinal tract. In this study, we investigated the effects of B. longum cells and metabolites on apical-out intestinal organoids in anaerobic conditions. Similar to L. casei, we either performed co-culture of organoids with 107 or 108 bacteria cells or we added 10 μg/mL bacteria lysates to the organoid medium for 12 h. Higher numbers of B. longum bacteria were cytotoxic for the organoids.
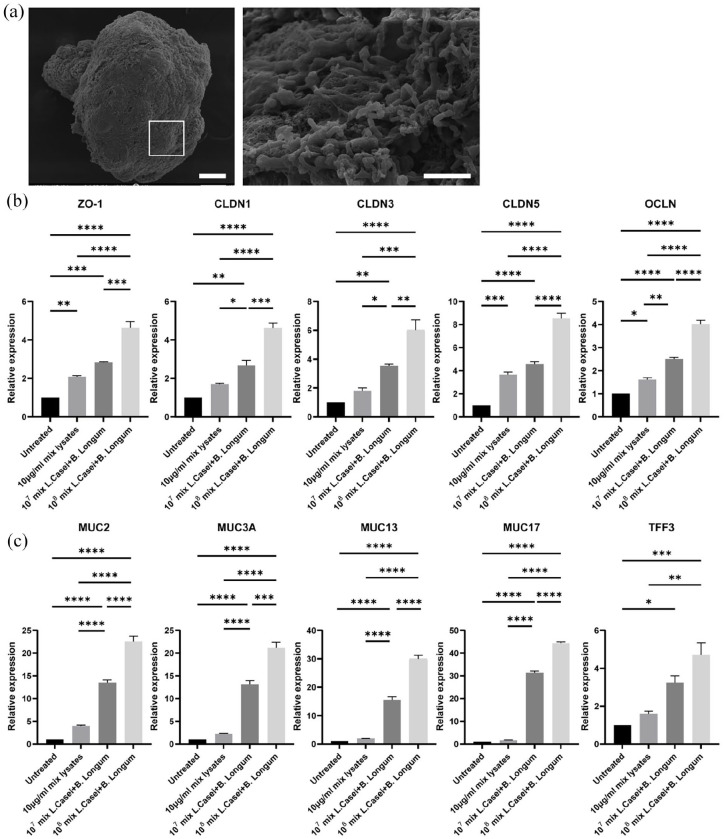
Preceding the co-culture of bacteria with the organoids, we also evaluated the growth of B. longum in organoid medium in hypoxia (Figure 6(a)). We observed that up to 12 h later, the bacteria moderately replicated in organoid medium, whereas after 24 h they stopped proliferating, but they maintained high viability levels (99%). According to previous studies, B. longum adherence to the gastrointestinal tract is important to colonize the gut and exert their probiotic effects.63,64 To explore whether B. longum attached to the surface of apical-out organoids we performed SEM analysis (Figure 6(b)). The results indicated that B. longum adhered to the apical surface of the organoids, thus showing successful colonization.
Figure 6.
Co-culture of organoids with B. longum. (a) Anaerobic culture of B. longum cells in intestinal organoid medium. Error bars indicate mean ±S.D. (n = 4). (b) SEM microscopy indicated colonization of B. longum bacteria on the apical surface of the organoids. White box represents the area magnified in the corresponding image on the right. Scale bars: 100and 5 μm (c and d) qRT-PCR analysis demonstrated the expression levels of the junction markers ZO-1, OCLN, CLDN1, CLDN3, and CLDN5 (c) and the mucins markers MUC2, MUC3A, MUC13, MUC17, and TFF3 (d) in organoids co-cultured with B. longum-derived lysates and two concentrations of B. longum cells (107 and 108). Error bars indicate mean ± S.E.M. (n = 3).
We also aimed to evaluate the probiotic effects of B. longum on the apical-out small intestinal organoids. Similar to L. casei, we initially assessed the effect on barrier integrity. We conducted quantitative analysis of gene expression for the junction markers ZO-1, CLDN1, CLDN3, CLDN5, and OCLN (Figure 6(c)). In comparison to the untreated organoids, they were all significantly upregulated when bacterial cells or lysates were added to the culture. Also in the case of B. longum, there was a positive correlation between the number of bacteria and the gene expression levels of the junction markers. Next, to evaluate whether the presence of B. longum has an impact on the mucous modulation, we analyzed the gene expression of MUC2, MUC3A, MUC13, MUC17, and TFF3 (Figure 6(d)). A dose-dependent trend toward increased expression of these markers was observed upon addition of bacterial cells or lysates. Together these results indicate that the B. longum bacteria can successfully colonize the apical-out organoids and establish probiotic effects in vitro. Also in the case of B. longum, the direct contact of organoids with bacterial cells, led to more robust effects. Collectively, small intestinal organoids with reversed polarity, grown in hypoxia are a suitable in vitro model to unravel unknown mechanisms underlying the probiotic effects of B. longum on the intestinal epithelium.
Triple co-culture of organoids, L. casei and B. longum
Intestinal microbiota is a complex, dynamic population of microorganisms that consists of thousands of different species. Alterations in the microbiota has been associated with chronic immune disorders, such as inflammatory bowel disease, obesity, and diabetes. Lactobacilli and bifidobacteria are among the dominant species and the most widely used probiotic bacteria in food supplements.37 Probiotic supplements usually consist of a combination of bacteria, and they are beneficial for the host immune health. In this study, apart from studying the effects of single bacteria strains, we aimed to evaluate whether we can recapitulate the probiotic effects of a combination of L. casei and B. longum on apical-out small intestinal organoids. This is a step further toward mimicking closer the in vivo situation, where thousands of different bacteria co-reside in the gastrointestinal tract.
Apical-out small intestinal organoids were either co-cultured with 107 or 108 bacteria cells or incubated with 10 μg/mL bacteria lysates for 12 h in anaerobic conditions. The ratio between the amounts of bacteria cells or lysates added, was 50:50 (L. casei : B. longum). Also in the case of multiple bacteria strains, we observed bacterial cell adherence on the apical surface of the organoids, using electron microscopy (Figure 7(a)). Similar to the effects of single bacteria strains, we identified significantly elevated gene expression of the junction-related markers ZO-1, CLDN1, CLDN3, CLDN5, and OCLN (Figure 7(b)) and the mucin-related markers MUC2, MUC3A, MUC13, MUC17, and TFF3 (Figure 7(c)). These results indicate that the two bacteria strains can successfully interact with and colonize the apical surface of human small intestinal organoids and improve barrier formation and mucin regulation.
Figure 7.
Triple co-culture of organoids with L. casei and B. longum. (a) SEM microscopy indicated colonization of L. casei and B. longum bacteria on the apical surface of the organoids. The white box in the left image represents the area magnified in the corresponding image on the right. Scale bars: 100and 5 μm. (b and c) qRT-PCR analysis demonstrated the expression levels of the junction markers ZO-1, OCLN, CLDN1, CLDN3, and CLDN5 (b) and the mucins markers MUC2, MUC3A, MUC13, MUC17, and TFF3 (c) in organoids co-cultured with a 50:50 mix of L.casei- and B.longum-derived lysates (final concentration 10 μg/ml) and a 50:50 mix of L.casei and B. longum cells (final concentrations: 107 and 108). Error bars indicate mean ± S.E.M. (n = 3).
Discussion
In the past few years, the role of the gut microbiome in health and disease has attracted a lot of interest. However, this remains a rather unexplored field with about 71% of the species lacking a culture representative.65 Hence, there is an urgent need for physiologically relevant and robust in vitro models to shed light on the complex gut-microbiome interactions. In this study, we developed for the first time, an apical-out small intestinal organoid model in hypoxia. These organoids contain all the major intestinal cell types and mimic structural and functional properties of the in vivo intestine. The directly accessible apical membrane in the outer surface of the organoids facilitates, amongst others, nutrient, drug or other compound screenings in high-throughput, since thousands of organoids with reversed polarity can be derived from a single 24-well plate. Here, we focused on the study of host-microbiome interactions. A unique advantage of this reversed polarity intestinal organoid model is that the apical surface is exposed to a hypoxic environment, thus recapitulating closer the in vivo situation. These culture conditions enable the studies of the interactions with anaerobic microorganisms, which constitute the vast majority of the gut microbiota species.
Lately, intestinal organoids with apical-out orientation have been described using human,28 chicken,30 and porcine31 adult stem cells. However, in none of these models, the apical surface was exposed to a hypoxic environment. Thus, co-cultures with anaerobes is not optimal, since obligate anaerobes will not be able to survive and facultative anaerobes show differences in their growth in the presence of higher or “normal” (normoxic) oxygen concentrations.66 For the study of anaerobic strains, usually microinjections are performed into the hypoxic lumen of apical-in organoids.13,67 Alternatively but no longer in a 3D organoid context, bacteria have been co-cultured with 2D monolayers of intestinal epithelial cells in cell culture devices (inserts or microfluidic systems), which were designed to control oxygen concentrations and gradients.68,69 However, microinjections are tedious, and monolayers do not recapitulate the 3D architecture of the in vivo tissues. Our group has previously established an apical-out small intestinal organoid model using pluripotent stem cells in normoxia.29 Here, we followed the same stepwise differentiation protocol but in hypoxic conditions. The organoids can be differentiated with the same high efficiency as in normoxia and show a reversed epithelial polar organization. Adding on to the presence of the major small intestinal cell types and proper structural organization, we demonstrated that our hypoxia-tolerant, apical-out intestinal organoids recapitulate functional characteristics of the intestine. Specifically, in the in vivo intestine, the adaptation to hypoxia is mainly regulated by hypoxia-inducible factors (HIFs). After verifying the presence of reduced oxygen concentration in our culture system, we identified increased protein expression of the HIF-1α isoform in the hypoxia organoids. Gene expression analysis showed that HIF-1α targets are significantly upregulated as well. Therefore, apical-out small intestinal organoids actively respond and adapt to low oxygen conditions, via similar mechanisms as in vivo.32 This is particularly important since deficiencies of HIF-1α have been associated with pathological conditions, including inflammatory bowel disease and colorectal cancer.32,70 Similar to other apical-out organoid models, we demonstrated that these hypoxia-tolerant organoids form a tight epithelial barrier, which is one of the main functions of the intestine.71 Additionally, successful apical-specific nutrient uptake was verified by the absorbance of a fluorescent fatty acid analog. In summary, these results indicate that we have established an effective and robust protocol to reverse epithelial polarity in hypoxia. This method can be successfully adapted to various culture conditions and constitutes a valuable in vitro tool for the study of nutrient uptake, drug absorption, and host-microbiome interactions. The versatility of this system is particularly useful for the study of the complex microbiome, where different microorganisms require different culture conditions.
The human gut microbiota is composed of about 1014 microorganisms (mostly anaerobic) and is crucial for the nutrition and health status of the organism.37,72 Probiotics are living microorganisms, which are known to provide health benefits for the host. These benefits include the improvement of gut barrier formation, maintenance of mucosal homeostasis, and immunomodulation.37 In this study, we co-cultured for the first time apical-out organoids with the anaerobic strains L. casei and B. longum. We identified that these probiotic bacteria can successfully colonize the apical surface of the organoids and have beneficial effects on them. We also highlighted the importance of direct contact of organoids with the bacterial cells. Collectively, these results show that this novel hypoxia-tolerant organoid model can be a valuable tool to explore the mechanisms underlying the probiotic effects of these microorganisms in greater depth. This would be of great interest for the production of more efficient probiotic supplements, the demand of which has immensely increased in the past decades.73 Furthermore, since the probiotic benefits have mainly been investigated in pathogenic situations,73 this model can be useful to determine their importance for healthy individuals as well. Additionally, these organoids can be used to study the effects of other known bacterial strains or even aid the discovery of unknown ones through the co-culture with microbiota, derived straight from human stool specimens. For such studies, it would be interesting in the future to grow these apical-out organoids in even lower oxygen levels, since there are microorganisms that require <0.1% oxygen to survive. Further research and identification of bacterial strains will benefit the investigation of their synergistic effect on the host as well.
Gut microbiota consists of approximately 300–500 bacterial species, which comprise a complex ecosystem.74 Hence, apart from the study of single microorganisms, it is crucial to study how multiple species interact when they are cultured together and what their combined effects on the host are. Here, we performed a triple co-culture system with the hypoxia apical-out organoids and the probiotic strains L. casei and B. longum, and identified efficient adherence on the apical surface and active probiotic effects on the organoids. The interactions of different Lactobacillus and Bifidobacterium strains—the predominant species of gastrointestinal microbiota—have been studied before but using a colorectal adenocarcinoma cell line (Caco-2),37 which is less physiologically relevant than organoids. Various probiotic supplements include combinations of different bacterial strains, thus a high-throughput 3D in vitro model that facilitates the study of effects of multiple bacterial strains on a host can be a particularly useful tool for identifying new beneficial combinations of bacteria and testing new food supplements. Furthermore, in this study, we used a 50:50 ratio of the two bacteria strains, but it would be interesting in a larger combinatorial screen to systematically test different ratios of various bacteria strains. Finally, future steps could include the exposure of these apical-out organoids co-cultured with probiotics to different pathogenic bacteria to further assess the functionality of the mucosal barrier integrity.
To conclude, we have developed a novel, scalable hypoxia-tolerant, apical-out small intestinal organoid model. These organoids contain the major intestinal cell lineages and recapitulate structural and functional characteristics of the in vivo tissue. Specifically, they have distinct apical and basolateral surfaces, they form a strong barrier and perform polarized nutrient uptake. The directly accessible apical surface facilitates also the investigation of host-microbiome interactions, since microorganisms can simply be added to the culture medium. The hypoxic environment allows for the first time, the study of anaerobes using organoids with reversed polarity. Overall, this system has great potential to simplify and advance not only research related to host-microbiome and host-pathogen interactions, but also pharmaceutical and nutritional studies.
Supplemental Material
Supplemental material, sj-docx-1-tej-10.1177_20417314221149208 for Hypoxia-tolerant apical-out intestinal organoids to model host-microbiome interactions by Panagiota Kakni, Barry Jutten, Daniel Teixeira Oliveira Carvalho, John Penders, Roman Truckenmüller, Pamela Habibovic and Stefan Giselbrecht in Journal of Tissue Engineering
Footnotes
Author contributions: P.K. conceived the project and designed experiments, performed cell and organoid culture and characterization, fabrication of the microwells, immunohistochemistry, confocal microscopy, quantitative PCR experiments, sensor measurements, uptake assays, barrier integrity assays, organoid-bacteria co-cultures, SEM and TEM microscopy, data analysis, and prepared and edited the manuscript. B.J performed the bacteria culture and contributed to the co-culture with the organoids. D.T.O.C performed sensor experiments and data analysis. J.P., R.T. and P.H. contributed to scientific discussions and revised the paper. S.G. conceived the project and designed experiments, oversaw all the experiments, the interpretation of data and the paper preparation.
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: R.T and S.G are founders and shareholders of 300MICRONS GmbH.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Dutch Province of Limburg (program “Limburg INvesteert in haar Kenniseconomie/LINK”; SAS-2014-00837 and SAS-2018-02477) supported financially this research. The authors gratefully acknowledge the Gravitation Program “Materials Driven Regeneration,” funded by the Netherlands Organization for Scientific Research (024.003.013). Additionally, the authors would like to thank the Microscopy CORE Lab of Maastricht University for the support in the conduction of the SEM experiments.
ORCID iD: Panagiota Kakni  https://orcid.org/0000-0001-9435-8633
https://orcid.org/0000-0001-9435-8633
Supplemental material: Supplemental material for this article is available online.
References
- 1. Merker SR, Weitz J, Stange DE. Gastrointestinal organoids: How they gut it out. Dev Biol 2016; 420: 239–250. [DOI] [PubMed] [Google Scholar]
- 2. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 2014; 14(3): 141–153. [DOI] [PubMed] [Google Scholar]
- 3. Schneeberger K, Roth S, Nieuwenhuis EES, et al. Intestinal epithelial cell polarity defects in disease: lessons from microvillus inclusion disease. Dis Model Mech 2018; 11: dmm031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cani PD. Human gut microbiome: hopes, threats and promises. Gut 2018; 67: 1716–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jandhyala SM, Talukdar R, Subramanyam C, et al. Role of the normal gut microbiota. World J Gastroenterol 2015; 21: 8787–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kho ZY, Lal SK. The human gut microbiome – a potential controller of wellness and disease. Front Microbiol 2018; 9: 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh A, Poling HM, Spence JR, et al. Gastrointestinal organoids: a next-generation tool for modeling human development. Am J Physiol Liver Physiol 2020; 319: G375–G381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kakni P, Truckenmüller R, Habibović P, et al. Challenges to, and prospects for, reverse engineering the gastrointestinal tract using organoids. Trends Biotechnol 2022; 40: 932–944. [DOI] [PubMed] [Google Scholar]
- 9. Bozzetti V, Senger S. Organoid technologies for the study of intestinal microbiota–host interactions. Trends Mol Med 2022; 28: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Min S, Kim S, Cho SW. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp Mol Med 2020; 52: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hou Q, Ye L, Liu H, et al. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ 2018; 25(9): 1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shaffiey SA, Jia H, Keane T, et al. Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen Med 2015; 11: 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Son YS, Ki SJ, Thanavel R, et al. Maturation of human intestinal organoids in vitro facilitates colonization by commensal lactobacilli by reinforcing the mucus layer. FASEB J 2020; 34: 9899–9910. [DOI] [PubMed] [Google Scholar]
- 14. Hill DR, Huang S, Nagy MS, et al. Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. eLife 2017; 6: e29132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heo I, Dutta D, Schaefer DA, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 2018; 3(7): 814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Forbester JL, Goulding D, Vallier L, et al. Interaction of Salmonella enterica serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun 2015; 83: 2926–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilson SS, Tocchi A, Holly MK, et al. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol 2015; 8(2): 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang YG, Wu S, Xia Y, et al. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep 2014; 2: e12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karve SS, Pradhan S, Ward DV, et al. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS One 2017; 12: e0178966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. In J, Foulke-Abel J, Zachos NC, et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol 2016; 2: 48–62.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. VanDussen KL, Marinshaw JM, Shaikh N, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015; 64: 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, et al. Mutational signature in colorectal cancer caused by genotoxic pks+E. Coli. Nat 2020; 580: 269–273. 5807802 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leslie JL, Huang S, Opp JS, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun 2015; 83: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engevik MA, Yacyshyn MB, Engevik KA, et al. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 2015; 308: G510–G524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Engevik MA, Engevik KA, Yacyshyn MB, et al. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol 2015; 308: G497–G509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Engevik MA, Aihara E, Montrose MH, et al. Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth. Am J Physiol Gastrointest Liver Physiol 2013; 305: G697–G711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Co JY, Margalef-Català M, Monack DM, et al. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat Protoc 2021; 16: 5171–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Co JY, Margalef-Català M, Li X, et al. Controlling epithelial polarity: A human enteroid model for host-pathogen interactions. Cell Rep 2019; 26: 2509–2520.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kakni P, López-Iglesias C, Truckenmüller R, et al. Reversing epithelial polarity in pluripotent stem cell-derived intestinal organoids. Front Bioeng Biotechnol 2022; 10: 879024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nash TJ, Morris KM, Mabbott NA, et al. Inside-out chicken enteroids with leukocyte component as a model to study host–pathogen interactions. Commun Biol 2021; 4(1): 41–15. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Y, Yang N, Chen J, et al. Next-generation porcine intestinal organoids: an Apical-Out organoid model for swine enteric virus infection and Immune Response Investigations. J Virol 2020; 94: e01006-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singhal R, Shah YM. Oxygen battle in the gut: hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J Biol Chem 2020; 295: 10493–10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Newby D, Marks L, Lyall F. Dissolved oxygen concentration in culture medium: assumptions and pitfalls. Placenta 2005; 26: 353–357. [DOI] [PubMed] [Google Scholar]
- 34. Konjar Š, Pavšič M, Veldhoen M. Regulation of oxygen homeostasis at the intestinal epithelial barrier site. Int J Mol Sci 2021; 22: 9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rolfe RD, Hentges DJ, Campbell BJ, et al. Factors related to the oxygen tolerance of anaerobic bacteria. Appl Environ Microbiol 1978; 36: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hentges DJ. Anaerobes: general characteristics. In:Baron S. (ed.) Medical microbiology 4th ed. Springer, Galveston (TX): University of Texas Medical Branch at Galveston, 1996. https://www.ncbi.nlm.nih.gov/books/NBK7638/ (accessed 4 July 2022). [PubMed] [Google Scholar]
- 37. Candela M, Perna F, Carnevali P, et al. Interaction of probiotic Lactobacillus and bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol 2008; 125: 286–292. [DOI] [PubMed] [Google Scholar]
- 38. Giselbrecht S, Gietzelt T, Gottwald E, et al. 3D tissue culture substrates produced by microthermoforming of pre-processed polymer films. Biomed Microdevices 2006; 8: 191–199. [DOI] [PubMed] [Google Scholar]
- 39. Kakni P, Hueber R, Knoops K, et al. Intestinal organoid culture in polymer film-based Microwell arrays. Adv Biosyst 2020; 4: e2000126. [DOI] [PubMed] [Google Scholar]
- 40. Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011; 470: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Van Welden S, Selfridge AC, Hindryckx P. Intestinal hypoxia and hypoxia-induced signalling as therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol 2017; 14: 596–611. [DOI] [PubMed] [Google Scholar]
- 42. Zheng L, Kelly CJ, Colgan SP. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol 2015; 309: C350–C360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun L, Li T, Tang H, et al. Intestinal epithelial cells-derived hypoxia-inducible factor-1α is essential for the homeostasis of intestinal intraepithelial lymphocytes. Front Immunol 2019; 10: 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Okkelman IA, Foley T, Papkovsky DB, et al. Live cell imaging of mouse intestinal organoids reveals heterogeneity in their oxygenation. Biomaterials 2017; 146: 86–96. [DOI] [PubMed] [Google Scholar]
- 45. Kumar T, Pandey R, Chauhan NS. Hypoxia inducible factor-1α: the curator of Gut Homeostasis. Front Cell Infect Microbiol 2020; 10: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu X, Wu R, Shehadeh LA, et al. Severe hypoxia exerts parallel and cell-specific regulation of gene expression and alternative splicing in human mesenchymal stem cells. BMC Genomics 2014; 15: 303. DOI: 10.1186/1471-2164-15-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karhausen J, Furuta GT, Tomaszewski JE, et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Investig 2004; 114: 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Woting A, Blaut M. Small intestinal permeability and Gut-Transit time determined with low and high molecular weight fluorescein isothiocyanate-dextrans in C3H Mice. Nutrients 2018; 10: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baxter MFA, Merino-Guzman R, Latorre JD, et al. Optimizing fluorescein isothiocyanate dextran measurement as a biomarker in a 24-h feed restriction model to induce gut permeability in broiler chickens. Front Vet Sci 2017; 4: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kowapradit J, Opanasopit P, Ngawhirunpat T, et al. In vitro permeability enhancement in intestinal epithelial cells (Caco-2) monolayer of water soluble quaternary ammonium chitosan derivatives. AAPS PharmSciTech 2010; 11: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu P, Elamin E, Elizalde M, et al. Modulation of intestinal epithelial permeability by plasma from patients with Crohn’s disease in a three-dimensional cell culture model. Sci Rep 2019; 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zheng Y, Zuo Z, Chow AH. Lack of effect of beta-cyclodextrin and its water-soluble derivatives on in vitro drug transport across rat intestinal epithelium. Int J Pharm 2006; 309: 123–128. [DOI] [PubMed] [Google Scholar]
- 53. Kiela PR, Ghishan FK. Physiology of intestinal absorption and secretion. Best Pract Res Clin Gastroenterol 2016; 30: 145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang TY, Liu M, Portincasa P, et al. New insights into the molecular mechanism of intestinal fatty acid absorption. Eur J Clin Invest 2013; 43: 1203–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hussain MM. Intestinal lipid absorption and lipoprotein formation. Curr Opin Lipidol 2014; 25: 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Camilleri M. Human intestinal barrier: Effects of stressors, diet, prebiotics, and probiotics. Clin Transl Gastroenterol 2021; 12: e00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. La Fata G, Weber P, Mohajeri MH. Probiotics and the gut immune system: Indirect Regulation. Probiotics Antimicrob Proteins 2018; 10: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vélez MP, De Keersmaecker SC, Vanderleyden J. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol Lett 2007; 276: 140–148. [DOI] [PubMed] [Google Scholar]
- 59. Pearce SC, Al-Jawadi A, Kishida K, et al. Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol 2018; 16(1): 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Monaco A, Ovryn B, Axis J, et al. The epithelial cell leak pathway. Int J Mol Sci 2021; 22: 7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hsieh CY, Osaka T, Moriyama E, et al. Strengthening of the intestinal epithelial tight junction by bifidobacterium bifidum. Physiol Rep 2015; 3: e12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yao S, Zhao Z, Wang W, et al. Bifidobacterium longum: protection against inflammatory bowel disease. J Immunol Res 2021; 2021: 8030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xiong Y, Zhai Z, Lei Y, et al. A novel major pilin subunit protein FimM is involved in adhesion of Bifidobacterium longum BBMN68 to intestinal epithelial cells. Front Microbiol 2020; 11: 590435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Westermann C, Gleinser M, Corr SC, et al. A critical evaluation of bifidobacterial adhesion to the host tissue. Front Microbiol 2016; 7: 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Almeida A, Nayfach S, Boland M, et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol 2021; 39: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. André AC, Debande L, Marteyn BS. The selective advantage of facultative anaerobes relies on their unique ability to cope with changing oxygen levels during infection. Cell Microbiol 2021; 23: e13338. [DOI] [PubMed] [Google Scholar]
- 67. Williamson IA, Arnold JW, Samsa LA, et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and Luminal Physiology. Cell Mol Gastroenterol Hepatol 2018; 6: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng 2019; 3: 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim R, Attayek PJ, Wang Y, et al. An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 2019; 12: 015006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rohwer N, Jumpertz S, Erdem M, et al. Non-canonical HIF-1 stabilization contributes to intestinal tumorigenesis. Oncogene 2019; 38: 5670–5685. [DOI] [PubMed] [Google Scholar]
- 71. Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med 2018; 50: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science 2012; 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 73. Khalesi S, Bellissimo N, Vandelanotte C, et al. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr 2019; 73: 24–37. [DOI] [PubMed] [Google Scholar]
- 74. Quigley EMM, Eamonn D, Quigley MM. Gut bacteria in health and disease. Gastroenterol Hepatol (N Y) 2013; 9: 560. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tej-10.1177_20417314221149208 for Hypoxia-tolerant apical-out intestinal organoids to model host-microbiome interactions by Panagiota Kakni, Barry Jutten, Daniel Teixeira Oliveira Carvalho, John Penders, Roman Truckenmüller, Pamela Habibovic and Stefan Giselbrecht in Journal of Tissue Engineering