Abstract
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). We evaluated the anti-SARS-CoV-2 antibody levels, anti-spike (S)-immunoglobulin G (IgG) and anti-nucleocapsid (N)-IgG, and the neutralization activity of IgG antibody in COVID‑19‑convalescent plasma against variants of SARS-CoV-2, alpha, beta, gamma, delta, kappa, omicron and R.1 strains. The study included 30 patients with clinically diagnosed COVID-19. The anti-S-IgG and anti-N-IgG levels ranged from 30.0 to 555.1 and from 10.1 to 752.6, respectively. The neutralization activity (50% inhibition concentration: IC50) for the wild-type Wuhan strain ranged from < 6.3 to 81.5 µg/ml. IgG antibodies were > 100 µg/ml in 18 of 30 (60%) subjects infected with the beta variant. The IC50 values for wild-type and beta variants correlated inversely with anti-S-IgG levels (p < 0.05), but no such correlation was noted with anti-N-IgG. IgG antibodies prevented infectivity and cytopathic effects of six different variants of concern in the cell-based assays of wild-type, alpha, gamma, delta, kappa and R.1 strains, but not that of the beta and omicron strains. IgG is considered the main neutralizing activity in the blood, although other factors may be important in other body tissues.
Subject terms: SARS-CoV-2, Viral infection
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), which emerged in China at the end of 20191–5 and spread rapidly worldwide. Genetic variants of SARS-CoV-2 have emerged and are currently circulating around the world. They define three classes of SARS-CoV-2 variants6: the B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma), B.1.617.2 (delta) and B.1.1.529 (omicron) variants, which are classified as variants of concern (VOC), while B.1.617.1 (kappa) and R.1 variants are classified as variants of interest (VOI). Variant mutations in these viruses are associated with changes in the activity of receptor binding and reduced neutralization by antibodies7–9.
Patients infected with COVID-19 produce various antibodies, including immunoglobulin M (IgM), immunoglobulin G (IgG) and immunoglobulin A (IgA)10–12. Especially, IgG antibodies against the spike (S) protein containing the anti-receptor binding domain (RBD) and nucleocapsid (N) protein of SARS-CoV-2 prevent the acquisition of viral infection13. The amounts of anti-S-IgG and anti-N-IgG antibodies produced after natural SARS-CoV-2 infection in unvaccinated individuals are more than 10 times those in negative samples upon admission and more than 100 times those at convalescence14.
The purpose of this study was to determine the neutralization activities of IgG antibodies from COVID‑19‑convalescent plasma against variants of SARS-CoV-2, alpha, beta, gamma, delta, kappa, omicron and R.1 strains (Fig. 1).
Figure 1.
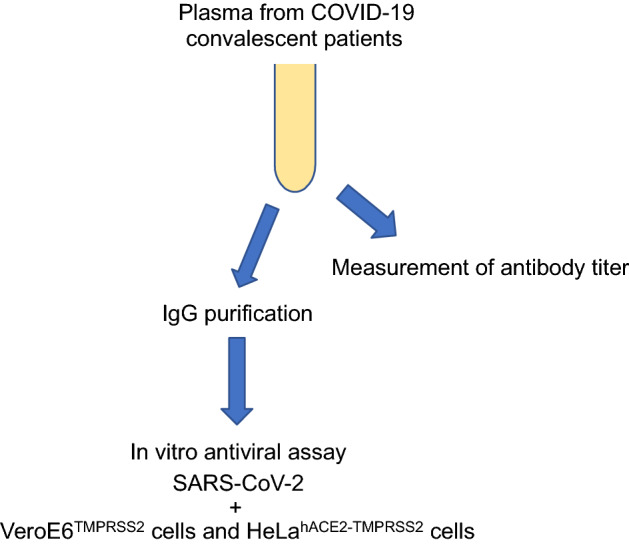
Schematic diagram of the IgG antibody neutralization assay of COVID-19 convalescent plasma.
Results
Anti-SARS-CoV-2 antibody titers
Figure 2 shows the levels of anti-S-IgG and anti-N-IgG measured in the 30 study patients. The anti-S-IgG and -N-IgG levels ranged from 30.0 to 555.1, and from 10.1 to 752.6, respectively.
Figure 2.
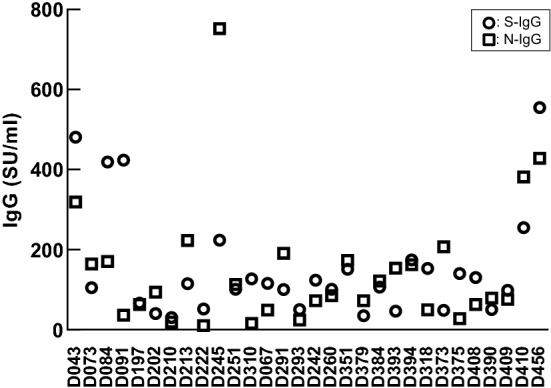
Anti-SARS-CoV-2 antibody titers in individual subjects.
Antiviral activity of IgG antibody in convalescent plasma samples
We evaluated the antiviral activity of IgG antibody in convalescent plasma against the wild-type (WT) and variants, alpha, beta, gamma, delta, kappa and R.1 strains. Table 1 shows the IC50 of IgG antibodies against WT and variant strains in VeroE6TMPRSS2 cells. The IC50 varied from < 6.3 to 81.5 µg/ml for the WT, alpha, gamma, delta, kappa and R.1 variants. On the other hand, the IC50 of IgG antibodies against the beta variant was > 100 µg/ml in 18 of 30 (60%) subjects. Figure 3 shows the mean IC50 of all the antibodies. The mean ± SD IC50 values for the alpha (43.68 ± 25.36 µg/ml in QHN001, 41.54 ± 26.52 µg/ml in QK002), gamma (32.86 ± 27.14 µg/ml), delta (44.14 ± 22.52 µg/ml) and R.1 (24.18 ± 13.62 µg/ml) variants were similar to that of WT (36.25 ± 22.72 µg/ml), but the values for the beta (82.86 ± 26.68 µg/ml) and kappa (63.19 ± 24.82 µg/ml) variants in VeroE6TMPRSS2 cells were clearly higher than WT. The mean IC50 of beta was significantly higher than those of WT, alpha, gamma, delta, and R.1 (p < 0.0001, Kruskal–Wallis test). The post-hoc Dunn’s multiple comparisons test showed statistically significant difference in mean IC50 between beta and WT (p < 0.0001), alpha (p < 0.0001), gamma (p < 0.0001), delta (p < 0.01), and R.1 (p < 0.0001), and also in the mean IC50 of kappa and WT (p < 0.01), gamma (p < 0.001), and R.1 (p < 0.0001).
Table 1.
Convalescent plasma antiviral activity of IgGs in VeroE6TMPRSS2.
| IgG | 05-2N | QHN001 | QK002 | TY8-612 | TY7-501 | K1734 | K5356 | 76,107 |
|---|---|---|---|---|---|---|---|---|
| B | B.1.1.7 | B.1.1.7 | B.1.351 | P.1 | B.1.617.2 | B.1.617.1 | R.1 | |
| WT | Alpha | Alpha | Beta | Gamma | Delta | Kappa | – | |
| D043 | 11.9 | 20.3 | 18.1 | 30.7 | 10.7 | 39.7 | 31.0 | 7.0 |
| D073 | 63.9 | 53.2 | 78.9 | > 100 | 58.3 | > 100 | > 100 | 42.2 |
| D084 | 28.8 | 29.4 | 23.7 | 73.6 | 14.6 | 59.3 | 74.0 | 14.3 |
| D091 | 54.5 | > 100 | > 100 | > 100 | 39.0 | > 100 | > 100 | 51.8 |
| D197 | 16.2 | 34.6 | 27.9 | > 100 | 64.1 | 41.2 | 71.1 | 39.1 |
| D202 | 74.7 | > 100 | 66.8 | > 100 | > 100 | 47.8 | 89.8 | 53.9 |
| D210 | 61.3 | 65.5 | 49.3 | > 100 | 54.9 | 55.8 | 93.6 | 51.6 |
| D213 | 29.5 | 26.5 | 20.2 | > 100 | 49.9 | 35.9 | 51.5 | 23.2 |
| D222 | 69.5 | 54.9 | 26.3 | > 100 | 22.0 | > 100 | > 100 | 29.5 |
| D245 | 22.9 | 18.5 | 10.6 | > 100 | 8.0 | 42.4 | 60.2 | 28.1 |
| D251 | 30.3 | 26.2 | 16.1 | > 100 | 6.9 | 29.1 | 52.6 | 23.0 |
| D310 | 28.4 | 17.2 | 15.0 | 59.2 | 9.3 | 36.7 | 61.7 | 28.9 |
| D067 | 81.5 | 64.2 | > 100 | > 100 | 85.2 | 42.0 | 47.2 | 25.6 |
| D291 | 36.0 | 28.6 | 48.7 | > 100 | 59.8 | 42.8 | 40.0 | 19.3 |
| D293 | 63.8 | 40.2 | 70.9 | > 100 | 87.8 | 59.7 | > 100 | 29.8 |
| D242 | 44.7 | 64.4 | 61.3 | > 100 | 21.7 | 58.6 | 56.1 | 24.6 |
| D260 | 19.2 | 13.6 | 7.5 | 18.6 | < 6.3 | 52.2 | 55.4 | < 6.3 |
| D351 | 40.2 | 32.2 | 22.1 | > 100 | 17.0 | 49.7 | 84.7 | 22.0 |
| D379 | 78.3 | > 100 | 62.5 | 67.8 | 53.0 | 29.2 | 34.2 | 14.4 |
| D384 | 20.0 | 36.3 | 27.4 | 85.9 | 12.7 | 30.3 | 47.8 | 20.3 |
| D393 | 43.6 | 64.1 | 54.9 | > 100 | 23.6 | 26.3 | 57.9 | 17.3 |
| D394 | 46.7 | 47.2 | 63.7 | > 100 | 46.3 | 42.6 | > 100 | 41.2 |
| D318 | 10.4 | 27.7 | 28.0 | 75.9 | 9.5 | 26.1 | 40.6 | 12.0 |
| D373 | 19.9 | 51.9 | 66.0 | > 100 | 20.0 | 18.0 | 34.5 | 15.5 |
| D375 | 17.1 | 33.2 | 39.0 | 78.5 | 18.1 | 25.3 | > 100 | 21.3 |
| D408 | 19.0 | 58.6 | 56.7 | 90.8 | 22.1 | 26.8 | 38.5 | 13.9 |
| D390 | 27.6 | 53.2 | 41.6 | > 100 | 41.0 | 27.9 | 42.9 | 25.8 |
| D409 | 7.6 | 13.6 | 10.1 | 17.1 | < 6.3 | 42.7 | 63.7 | 8.1 |
| D410 | 13.6 | 22.1 | 16.0 | 52.1 | 10.2 | 20.3 | 36.5 | 9.1 |
| D456 | < 6.3 | 13.0 | 16.9 | 35.7 | 7.4 | 15.9 | 30.3 | < 6.3 |
| mAb1414 | 0.72 | > 10 | NT | > 10 | > 10 | > 10 | > 10 | 5.12 |
| mAb2414 | 5.78 | > 10 | NT | > 10 | 3.46 | > 10 | > 10 | > 10 |
| mAb40591 | 3.27 | 5.46 | NT | > 10 | 0.88 | 2.62 | > 10 | 1.35 |
| pAbA19215 | < 0.63 | < 0.63 | NT | > 10 | > 10 | < 0.63 | 5.56 | < 0.63 |
Data are IC50 (µg/ml) values.
Figure 3.
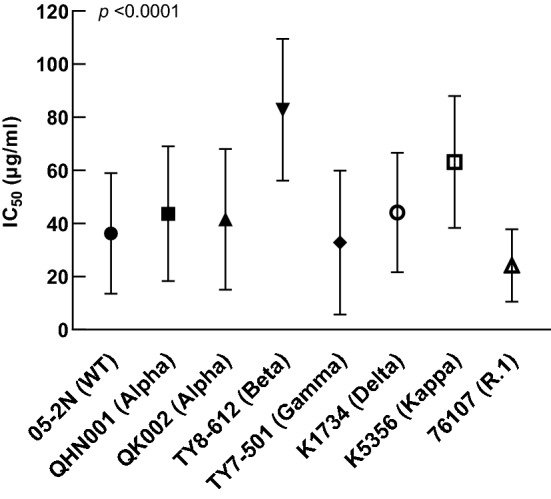
Antiviral activity of IgG antibodies in convalescent plasma against wild-type and each variant. Data are mean ± standard deviation.
We also evaluated the activities of anti-SARS-CoV-2 antibodies. All the four tested antibodies showed potent activities against the WT strain, however, the activities were markedly lower in most variant strains, especially mAb1414 and 2414 nullified activity, in almost all tested variants (Table 1). Table 2 shows the IC50 of IgG antibodies15 against WT and variants strains in HeLahACE2-TMPRSS2 cells. Note the higher IC50 values for the beta and omicron variants relative to the WT.
Table 2.
Convalescent plasma antiviral activity of IgGs in HeLahACE2-TMPRSS2.
| Cell | IgG | 05-2N | TY8-612 | K1734 | 929-1N |
|---|---|---|---|---|---|
| B | B.1.351 | B.1.617.2 | B.1.1.529 | ||
| WT | Beta | Delta | Omicron | ||
| VeroE6 TMPRSS2 | D043 | 11.9 | 30.7 | 39.7 | n.d. |
| D073 | 63.9 | > 100 | > 100 | n.d. | |
| D084 | 28.8 | 73.6 | 59.3 | n.d. | |
| D091 | 54.5 | > 100 | > 100 | n.d. | |
| HeLa hACE2-TMPRSS2 | D043 | 7.9 | 17.5 | 9.4 | 17.9 |
| D073 | 40.7 | > 100 | 53.7 | > 100 | |
| D084 | 8.9 | 65.3 | 22.8 | > 100 | |
| D091 | 30.0 | 93.1 | 72.4 | 59.0 |
Data are IC50 (µg/ml) values.
Correlation between neutralizing activity against WT/beta variant and antibody titers
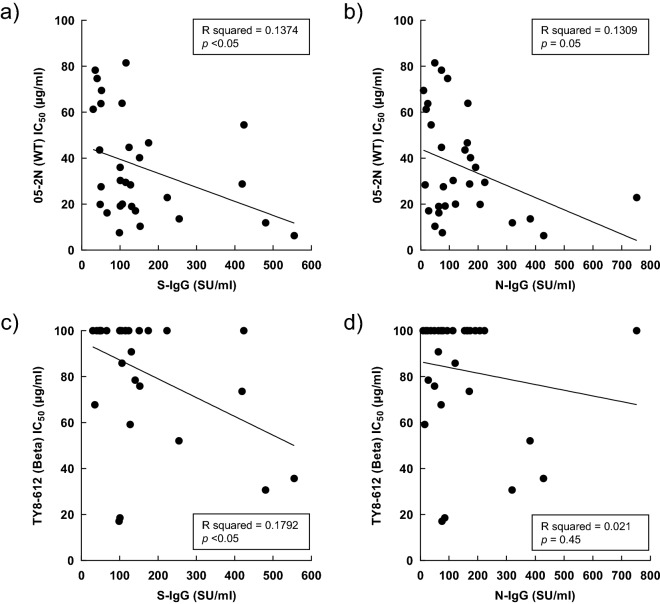
Finally, we compared the correlation between neutralizing activities against WT, beta variant and antibody titers. The IC50 values in WT and beta correlated significantly with anti-S-IgG levels (p < 0.05), but not with anti-N-IgG levels (Fig. 4a–d).
Figure 4.
Correlation between neutralizing activity against antibody titers and wild-type and beta variant. (a) Wild-type vs. anti-S-IgG levels. (b) Beta variant vs. anti-S-IgG levels. (c) Wild-type vs. anti-N-IgG levels. (d) Beta variant vs. anti-N-IgG levels.
Discussion
Genetic variants of SARS-CoV-2, alpha, beta, gamma, delta, kappa, omicron and R.1 strains, have infected millions of people around the world6. We evaluated the antiviral activities of IgG antibodies against those variants obtained from convalescent plasma samples. The IgG antibodies prevented the infectivity and cytopathic effects of three different VOCs and two VOIs in the cell-based assays using various infectious variants, WT, alpha, gamma, delta, kappa, and R.1 strains, but not those of the beta and omicron strains. These results are somewhat similar to those reported on the neutralizing activities found in BNT162b2-vaccinated individuals, which demonstrated potent activities against the alpha, delta, and kappa variants in serum samples from responders, compared with relatively moderate activity against the beta strain16,17. As described in the Methods section, the tested plasma samples were obtained from patients who were infected with COVID-19 between June 2020 and March 2021, and thus, it is considered that most IgGs were from convalescent plasmas of non-VOCs (Nextstrain clade 20B). Therefore, it is reasonable that the results of vaccines (prepared based on the sequence of WT) and that of the convalescent plasma samples tended to be the same7,16–18. We evaluated the correlation between the levels of the antibodies and neutralization activities of IgG antibodies. The higher levels of anti-S-IgG tended to suppress acquisition of viral infection. Then, the anti-S-IgG levels correlated inversely with the IC50 values for the WT and beta variant. In other words, anti-S-IgG antibody prevented the infectivity and cytopathic effects in SARS-CoV-2 infection. These results indicate that anti-S-IgG antibody acts directly against the RBD of SARS-CoV-2, preventing viral entry into the cells. Similar results were reported on the correlation between anti-S-IgG levels and neutralizing activity of COVID‑19‑convalescent plasma19,20. In contrast, none of the anti-N-IgG levels correlated inversely with the IC50 values for the WT and beta variant. These results suggest that anti-N-IgG antibody is unlikely to prevent SARS-CoV-2 infection in cells. Nevertheless, one previous study reported that anti-N-IgG antibody correlated with the neutralizing efficacy of a SARS-CoV-2 pseudovirus in all randomly selected COVID‑19‑convalescent plasma units21. Further analysis of the correlation between anti-N-IgG antibody and neutralizing activity is warranted.
Our study has certain limitations. First, we did not determine the variants of SARS-CoV-2 from COVID‑19‑convalescent patients. However, WT and the initial alpha strains were consistent with the variants that had spread in Japan during the study period22. Second, the small sample size is a limitation of this study as it reduces the generalizability of the findings to a larger population. Further research with a larger sample size is needed to confirm these results. In this study, the IgG was purified and used to evaluate the activity of COVID‑19‑convalescent plasma, and the results correlated with S-IgG in the blood. IgG is considered to be the main neutralizing activity in the blood, although other factors may be important in neutralizing the activity in the lungs, mucous membranes, and other tissues.
Materials and methods
Patients
The study subjects were 30 patients (22 men and 8 women, median age 54; range: 25–69 years) diagnosed clinically with COVID-19 for the first time after visiting the National Center for Global Health and Medicine (NCGM), Tokyo, Japan, between June 2020 and March 2021. Twenty five of the 30 (83.3%) patients developed co-existing severe pneumonia, and of these, 5 required positive pressure ventilation, and 3 of the latter also required treatment with extracorporeal membrane oxygenation. Plasma test samples were obtained between 33 and 316 days (median 96 days) after the onset of clinical features of COVID-19 infection. The Human Ethics Committee of the NCGM approved the study (#NCGM-G-003472–02) and each patient provided a written informed consent. The study also conformed to the principles of the Declaration of Helsinki.
Measurement of anti-SARS-CoV-2 antibody titers
Samples from each participant were analyzed for the levels of two anti-SARS-CoV-2 antibodies (anti-S-IgG and anti-N-IgG) using the chemiluminescence enzyme immunoassay (CLEIA) platforms (HISCL) purchased from Sysmex Co. (Kobe, Japan) as reported previously14.
Cells, viruses, antibodies and isolation of IgG fractions from COVID‑19‑convalescent patients
Vero-E6TMPRSS2 cells23 and HeLahACE2-TMPRSS2 cells24 were obtained from Japanese Collection of Research Bioresources (JCRB) Cell Bank (Osaka, Japan). Each cell type was maintained in D-MEM supplemented with 10% FCS, 100 μg/ml of penicillin, 100 μg/ml of streptomycin, and 1 mg/mL of G418. PANGO lineage B, wild type (WT) Wuhan strain [SARS-CoV-2 NCGM-05-2N (SARS-CoV-205-2N)] and B.1.1.529 (omicron) variants [hCoV-19/Japan/IC-2279/2021 SARS-CoV-2 NCGM-929-1N (SARS-CoV-2929-1N)] were isolated from nasopharyngeal swabs of a patient with COVID-19, who was admitted to the NCGM7,13,16,17. Seven clinically isolated SARS-CoV-2 mutant strains were used in the present study: two B.1.1.7 (alpha) variants [hCoV-19/Japan/QHN001/2020 (SARS-CoV-2QHN001, GISAID accession ID; EPI_ISL_804007) and hCoV-19/Japan/QK002/2020 (SARS-CoV-2QK002, GISAID Accession ID; EPI_ISL_768526)] and a B.1.351 (beta) variant [hCoV-19/Japan/TY8-612-P0/2021 (SARS-CoV-2TY8-612, GISAID accession ID; EPI_ISL_1123289)] and a P.1 (gamma) variant [hCoV-19/Japan/TY7-501/2021 (SARS-CoV-2TY7-501, GISAID Accession ID; EPI_ISL_833366)] were obtained from the National Institute of Infectious Diseases, Tokyo. The B.1.617.2 (delta) variant [hCoV-19/Japan/TKYK01734/2021 (SARS-CoV-21734, GISAID Accession ID; EPI_ISL_2080609)], B.1.617.1 (kappa) variant [TKYTK5356_2021 (SARS-CoV-25356, DDBJ Accession ID; LC633761)] and R.1 variant [hCoV-19/Japan/TKY76107/2021 (SARS-CoV-276107, GISAID Accession ID; EPI_ISL_1041946)] were provided by Tokyo Metropolitan Institute of Public Health, Tokyo. Each variant was confirmed to contain each variant of concern-specific amino acid substitutions before the assays conducted in the present study (vide infra). The mAb1414, mAb2414 and mAb40591, anti-SARS-CoV-2 monoclonal antibodies, were purchased from Active Motif (Carlsbad, CA) and Sino Biological (Beijing, China), respectively. The pAbA19215, anti-SARS-CoV-2 polyclonal antibody was purchased from ABclonal (Woburn, MA). Plasma or serum samples were collected from patients, and IgG fractions were purified using a spin column-based antibody purification kit (Cosmo Bio, Tokyo) according to the instructions provided by the manufacturer. Briefly, serum or plasma was collected, heat-inactivated for 30 min at 56 °C, and spin columns were centrifuged at 3500 rpm for 5 min. The IgG fractions in the supernatants were eluted and collected.
Antiviral assays
The neutralizing activities of IgG fractions from COVID‑19‑convalescent plasma were determined by quantifying the IgG antibody suppression of the cytopathic effect (CPE) of each SARS-CoV-2 strain in VeroE6TMPRSS2 cells and HeLahACE2-TMPRSS2 cells, using the procedures described previously7,13,16,17. Briefly, each of the purified IgG fraction was two-fold serially diluted in the culture medium. The diluted IgG fractions were incubated with 100 50% tissue culture infectious dose (TCID50) of the viruses at 37 °C for 20 min (final IgG dilution range: 6.3–100 µg/ml), after which the IgG-virus mixtures were inoculated into VeroE6TMPRSS2 cells and/or HeLahACE2-TMPRSS2 cells (1.0 × 104/well) in 96-well plates. The SARS-CoV-2 strains used in this assay were as follows: wild type strain, SARS-CoV-205-2N (PANGO lineage B), two alpha variants (SARS-CoV-2QHN001 and SARS-CoV-2QK002), beta variant SARS-CoV-2TY8-612, gamma variant SARS-CoV-2TY7-501, delta variant SARS-CoV-21734, kappa variant SARS-CoV-25356, omicron variant SARS-CoV-2929-1N and R.1 variant SARS-CoV-276107. After 3-day culture of the cells, the level of cytopathic effect (CPE) observed in SARS-CoV-2-exposed cells was determined using the WST-8 assay, employing Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). The IgG antibody dilution that yielded 50% inhibition of CPE was defined as the 50% Inhibition Concentration (IC50). Each of the purified IgG fractions was tested in duplicate.
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Differences between groups were analyzed for statistical significance using Kruskal–Wallis test. When the latter test was significant, post-hoc Dunn’s multiple comparisons test was applied. Correlations between two assays were analyzed for statistical significance using nonparametric Spearman test. A p value < 0.05 denoted the presence of statistically significant difference. All statistical analyses were performed using the GraphPad Prism software version 8 (GraphPad Software, San Diego, CA).
Institutional review board statement
The Ethics Committee at the NCGM approved the present study (#NCGM-G-003472-02). Each patient provided written informed consent. The study also conformed to the Declaration of Helsinki principles.
Acknowledgements
The authors thank Drs. Kenji Sadamasu, Mami Nagashima, Hiroyuki Asakura, and Mr. Isao Yoshida for providing two SARS-CoV-2 variants. We also thank Drs. Nobuyo Higashi-Kuwata and Shinichiro Hattori, as well as Ms. Mariko Kato for the expert help in parts of the study experiments.
Author contributions
Conceptualization, K.T., K.M. and H.M.; Methodology, K.T. and K.M.; Software, K.T. and K.M.; Validation, K.T. and K.M.; Formal Analysis, K.T. and K.M.; Investigation, K.T., K.M., K.M., Y.T. and T.H.; Resources, N.K., S.K. and N.O.; Data Curation, K.T., K.M. and Y.T.; Writing—Original Draft Preparation, K.T. and K.M.; Writing—Review & Editing, K.M. and H.M.; Supervision, H.G., N.O., S.O. and H.M.; Project Administration, H.M.; Funding Acquisition, K.M., Y.T. and H.M.
Funding
This research was supported in part by a grant from the Japan Agency for Medical Research and Development to K. Maeda (grant #JP20fk0108260, #20fk0108502) and to H. Mitsuya (grant #20fk0108502), and in part by a grant for MHLW Research on Emerging and Re-emerging Infectious Diseases and Immunization Program to K. Maeda (grant #JPMH20HA1006) from the Ministry of Health, Labor and Welfare, and in part by a grant for COVID-19 to H. Mitsuya (grant #19A3001), K. Maeda (grant #20A2003D) and Y. Takamatsu (grant #21A1007) from the Intramural Research Program of National Center for Global Health and Medicine, and in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health to H. Mitsuya.
Data availability
The datasets generated during and/or analyzed during the study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kiyoto Tsuchiya, Email: ktsuchiy@acc.ncgm.go.jp.
Hiroaki Mitsuya, Email: hmitsuya@hosp.ncgm.go.jp.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsuya H, Kokudo N. Sustaining containment of COVID-19: Global sharing for pandemic response. Glob. Health Med. 2020;2:53–55. doi: 10.35772/ghm.2020.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (Accessed January 2023).
- 7.Maeda K, Amano M, Uemura Y, Tsuchiya K, Matsushima T, Noda K, Shimizu Y, Fujiwara A, Takamatsu Y, Ichikawa Y, et al. Correlates of neutralizing/SARS-CoV-2-S1-binding antibody response with adverse effects and immune kinetics in BNT162b2-vaccinated individuals. Sci. Rep. 2021;11:22848. doi: 10.1038/s41598-021-01930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, De la Cruz E, Jorge A, et al. Neutralizing antibodies against the SARS-CoV-2 delta and omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022;28:481–485. doi: 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernández-Luis P, Aguilar R, Pelegrin-Pérez J, Ruiz-Olalla G, García-Basteiro AL, Tortajada M, Moncunill G, Dobaño C, Angulo A, Engel P. Decreased and heterogeneous neutralizing antibody responses against RBD of SARS-CoV-2 variants after mRNA vaccination. Front. Immunol. 2022;13:816389. doi: 10.3389/fimmu.2022.816389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, Yu X, Gao C, Zhang L, Zhai H, Hu Y, Liu E, Wang Q, Gao Y, Wei D, et al. Characterization of antibody responses to SARS-CoV-2 in convalescent COVID-19 patients. J. Med. Virol. 2021;93:2227–2233. doi: 10.1002/jmv.26646. [DOI] [PubMed] [Google Scholar]
- 11.Glück V, Grobecker S, Tydykov L, Salzberger B, Glück T, Weidlich T, Bertok M, Gottwald C, Wenzel JJ, Gessner A, et al. SARS-CoV-2-directed antibodies persist for more than six months in a cohort with mild to moderate COVID-19. Infection. 2021;49:739–746. doi: 10.1007/s15010-021-01598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takamatsu Y, Omata K, Shimizu Y, Kinoshita-Iwamoto N, Terada M, Suzuki T, Morioka S, Uemura Y, Ohmagari N, Maeda K, et al. SARS-CoV-2-neutralizing humoral IgA response occurs earlier but is modest and diminishes faster than IgG response. Microbiol. Spectr. 2022;21:e0271622. doi: 10.1128/spectrum.02716-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda K, Higashi-Kuwata N, Kinoshita N, Kutsuna S, Tsuchiya K, Hattori SI, Matsuda K, Takamatsu Y, Gatanaga H, Oka S, et al. Neutralization of SARS-CoV-2 with IgG from COVID-19-convalescent plasma. Sci. Rep. 2021;11:5563. doi: 10.1038/s41598-021-84733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noda K, Matsuda K, Yagishita S, Maeda K, Akiyama Y, Terada-Hirashima J, Matsushita H, Iwata S, Yamashita K, Atarashi Y, et al. A novel highly quantitative and reproducible assay for the detection of anti-SARS-CoV-2 IgG and IgM antibodies. Sci. Rep. 2021;11:5198. doi: 10.1038/s41598-021-84387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takamatsu Y, Imai M, Maeda K, Nakajima N, Higashi-Kuwata N, Iwatsuki-Horimoto K, Ito M, Kiso M, Maemura T, Takeda Y, et al. Highly neutralizing COVID-19 convalescent plasmas potently block SARS-CoV-2 replication and pneumonia in Syrian hamsters. J. Virol. 2022;96:e0155121. doi: 10.1128/jvi.01551-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amano M, Maeda K, Tsuchiya K, Shimada S, Mitsuya H. Third-dose BNT162b2 vaccination elicits markedly high-level SARS-CoV-2-neutralizing antibodies in vaccinees who poorly responded to second dose in Japan. J. Infect. Dis. 2022;226:2038–2039. doi: 10.1093/infdis/jiac209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amano M, Otsu S, Maeda K, Uemura Y, Shimizu Y, Matsuoka M, Shimada S, Mitsuya H. Neutralization activity of sera/IgG preparations from fully BNT162b2 vaccinated individuals against SARS-CoV-2 alpha, beta, gamma, delta and kappa variants. Sci. Rep. 2022;12:13524. doi: 10.1038/s41598-022-17071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh EE, Frenck RW, Jr, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santiago L, Uranga-Murillo I, Arias M, González-Ramírez AM, Macías-León J, Moreo E, Redrado S, García-García A, Taleb V, Lira-Navarrete E, et al. Determination of the concentration of IgG against the spike receptor-binding domain that predicts the viral neutralizing activity of convalescent plasma and serum against SARS-CoV-2. Biology. 2021;10:208. doi: 10.3390/biology10030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolscheid-Pommerich R, Bartok E, Renn M, Kümmerer BM, Schulte B, Schmithausen RM, Stoffel-Wagner B, Streeck H, Saschenbrecker S, Steinhagen K, et al. Correlation between a quantitative anti-SARS-CoV-2 IgG ELISA and neutralization activity. J. Med. Virol. 2022;94:388–392. doi: 10.1002/jmv.27287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedenberg AT, Pan CH, Diehl WE, Romeiser JL, Hwang GR, Leiton CV, Muecksch F, Shroyer KR, Bennett-Guerrero E. Neutralizing activity to SARS-CoV-2 of convalescent and control plasma used in a randomized controlled trial. Transfusion. 2021;61:1363–1369. doi: 10.1111/trf.16283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Outbreak.info. A standardized, open-source database of COVID-19 resources and epidemiology data. https://outbreak.info/ (Accessed January 2023).
- 23.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the study are available from the corresponding author on reasonable request.