Abstract
Objective:
To determine the efficacy of an orally delivered phosphate-rich polymer, Pi-PEG, to prevent surgical site infection (SSI) in a mouse model of spontaneous wound infection involving gut-derived pathogens.
Background:
Evidence suggests that pathogens originating from the gut microbiota can cause postoperative infection via a process by which they silently travel inside an immune cell and contaminate a remote operative site (Trojan Horse Hypothesis). Here, we hypothesize that Pi-PEG can prevent SSIs in a novel model of postoperative SSIs in mice.
Methods:
Mice were fed either a standard chow diet (high fiber/low fat, SD) or a western-type diet (low fiber/high fat, WD), and exposed to antibiotics (oral clindamycin/intraperitoneal cefoxitin). Groups of mice had Pi-PEG added to their drinking water and SSI incidence was determined. Gross clinical infections wound cultures and amplicon sequence variant analysis of the intestinal contents and wound were assessed to determine the incidence and source of the developing SSI.
Results:
In this model, consumption of a WD and exposure to antibiotics promoted the growth of SSI pathogens in the gut and their subsequent presence in the wound. Mice subjected to this model drinking water spiked with Pi-PEG were protected against SSIs via mechanisms involving modulation of the gut-wound microbiome.
Conclusions:
A nonantibiotic phosphate-rich polymer, Pi-PEG, added to the drinking water of mice prevents SSIs and may represent a more sustainable approach in lieu of the current trend of greater sterility and the use of more powerful and broader antibiotic coverage.
Keywords: microbiome, surgical site infection, gut, nonantibiotic
Despite mandated bundles of care to reduce surgical site infections (SSIs) including antibiotics, skin cleansing, rescrubbing before closure, glove changing, and use of wound protectors, SSIs continue to rise with as many as 50% demonstrating resistance to the antibiotics chosen for prophylaxis.1,2 Although it is generally presumed that most SSIs arise from some type of intraoperative contamination event,3 under currently mandated protocols for asepsis in elective surgery, precisely where the pathogens that cause SSI to originate from and precisely how they cause infection remains unclear.2
Given the lack of concrete data to define the precise etiopathogenesis of SSIs, our laboratory introduced the idea that SSIs may also develop postoperatively from pathogens that originate from among the gut microbiota via a Trojan Horse–type mechanism.4 In this scenario, pathogenic species hidden within the gut microbiome (ie, intestinal crypts)5 can be taken up by immune cells where they then silently travel in the bloodstream and home to a remote injury site to release their infectious payload. Under such circumstances, blood cultures that only detect the presence of viable (ie, cultivatable) extracellular microbes will remain negative. Yet along this pathogenic continuum, multiple contingencies must first be fulfilled in order for a clinical SSI to develop. First, pathogenic species must predominate among the gut microbiota; second, when immune cells engulf a gut pathogen, a state of mutual “tolerance” must develop such that neither the immune cell eliminates the pathogen nor the pathogen causes apoptosis of the immune cell; finally, the pathogen carrying immune cell must successfully “home” to a tissue injury site where the appropriate environmental “cues’ exist that allow for their growth and invasion. Previously we created a model to recapitulate all these events such that the full complement of considered contingencies were met to result in a clinical SSI by tracking the fate of green fluorescent protein-labeled methicillin-resistant Staphylococcus aureus following its stable colonization via oral gavage.4
The aims of the present study were to test the efficacy of a nonantibiotic orally administered phosphate-rich copolymer, Pi-PEG, to prevent SSIs in a mouse model of spontaneous wound infection arising from among the gut microbiota. We developed a novel, de novo synthesized Pi-PEG (ie, ABA-PEG20k-Pi20) that is a high–molecular-weight polyethylene glycol (PEG) A-B-A structure covalently bonded to Pi to durably embed Pi into the host-pathogen environment of the gut.6,7 The PEG moiety itself functions as a barrier-enhancing agent that can physically shield bacteria away from the epithelial surface.8–10 The Pi content, when covalently bonded onto the PEG carrier, can durably embed Pi into the host-pathogen interaction and can suppress bacterial virulence directly by affecting phosphosensory and phosphoregulatory pathways11 that connect to quorum sensing signaling.12 Pi-PEG does not affect bacterial growth thus allowing the normal microbiota to proliferate and competitively exclude and suppress low abundance yet potentially pathogenic species. In the present study, we sought to test Pi-PEG in a model of SSI development in which mice consume a western diet (WD) and are exposed to antibiotics commonly used clinically (cefoxitin, clindamycin). Therefore, aim of this study was to demonstrate the efficacy of oral administration of Pi-PEG as an additive to the drinking water of mice to prevent SSIs in a novel model of SSI pathogenesis.
METHODS
Mouse Model of Spontaneous Gut Microbiota–derived SSI
To disrupt the gut microbiome as might develop in humans at high risk for a postoperative SSI (obesity, hyperglycemia, recent exposure to antibiotics, major tissue injury, presence of prosthetic/foreign material), we used an original model created in our laboratory in which mice are fed a low-fiber/high-fat diet and treated with oral and parenteral antibiotics.13 Briefly, C57BL/6 male mice aged 10 weeks (Charles River Laboratories) were used in all experiments and maintained in accordance with the guidelines at the University of Chicago (Protocol 72090). Mice were co-housed under standardized laboratory conditions in a temperature-controlled room (22–24°C) with a 12-hour light/dark diurnal cycle. Mice were allowed food and water ad libitum.
The experimental design is outlined in Figure 1. Mice were maintained for 6 weeks on either high fat/low fiber WD (Bioserve S3282, sterile pellets) or a standard chow diet (SD). After 6 weeks, both groups were randomly assigned to receive antibiotics (WD+Abx; SD+Abx) versus no antibiotics (WD; SD). Mice receiving antibiotics were treated with subcutaneous cefoxitin (40 mg/kg) and oral clindamycin (100 mg/kg) once daily for 2 days before surgery and for 2 days after surgery.
Figure 1. Mouse model of SSI. Experimental design.
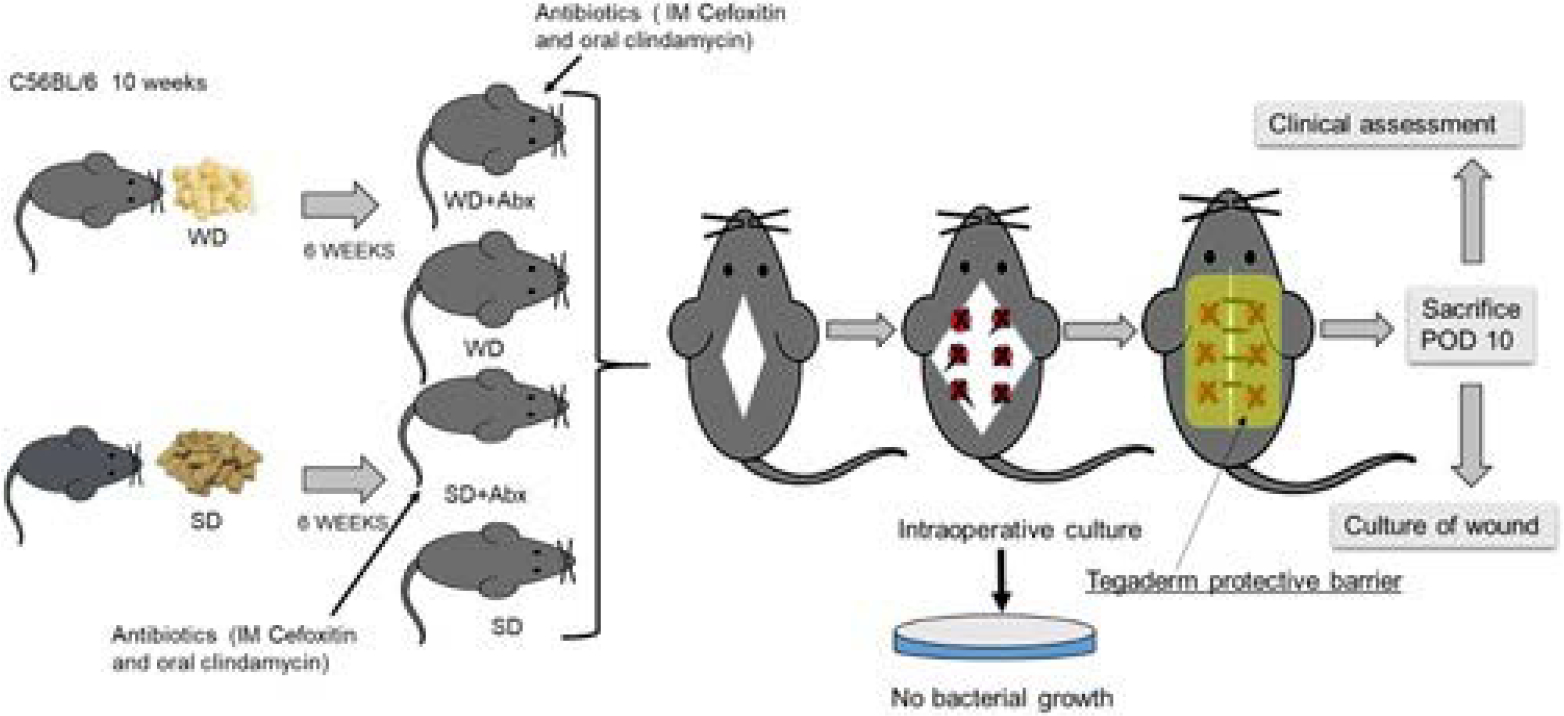
C56BL/6, 10 weeks mice were randomly assigned to WD (n=44) and SD (n=37) for 6 weeks. After 6 weeks, mice were randomly divided into 4 groups: SD (n=18), SD+Abx (n=19), WD (n=29), and WD+Abx (n=15). A muscle wound injury was then performed in the dorsum of mice by exposing the underlying muscle, cauterizing six 1 mm spots along the exposed bed, and then placing a silk suture in each spot. On POD10, all mice were sacrificed and the clinical wound score was determined. Finally, wound swabs were performed for culture. IM indicates intramuscular.
To test the efficacy of Pi-PEG to prevent infection in this model, selected reiterative experiments were performed in which 1% Pi-PEG was added to the drinking 10 days before wound creation. WD+Abx groups of mice were randomly assigned to plain water and water supplemented with 1% Pi-PEG.
On the day of surgery, all mice were anesthetized with intraperitoneal ketamine (100 mg/kg) and xylazine (10 mg/kg). A wide area of hair over the surgical site (dorsum) was removed. Next, the skin was decontaminated with 10% w/w Providone-Iodine Prep Pad (NDC 10819-3883-1) and 70% Isopropyl Alcohol (BD). A midline back skin incision was made, the skin was undermined on both sides to expose the paraspinal muscles bilaterally. At 3 small locations (2–3 mm) on each side of the spine, the muscle was injured with the bovie cautery following which a suture of 5-O silk was applied. Finally, just before the wound, a culture swab was taken and streaked onto Tryptic soy agar. The skin was then closed with 6-O Nylon. A Tegaderm (3M) dressing film (2.75 × 2.35 inches) was applied to cover and protect the incision site until sacrifice. All animals were resuscitated with 1 mL 0.9% normal saline subcutaneously. The antibiotics were continued until postoperative day (POD) 2. All mice recovered from the surgery completely and appeared healthy up to the day of sacrifice on POD10. At sacrifice, the back wound was opened and a clinical wound healing score (CWHS) (0–6) was calculated for each mouse using the following parameters: discharge: no discharge=0, serous discharge=1, purulent discharge=2; wound edge appearance: healed=0, partially dehiscense=1, full dehiscense=2 and inflammation/abscess: no inflammation=0, mild inflammation=1, severe inflammation with abscess=2 (Fig. 2A). After calculation of the CWHS, a wound swab was taken and cultured on Tryptic Soy Broth agar. Finally, a microbial score was calculated for each mouse on a scale of 0 to 2 as follows: no growth=0, monomicrobial growth=1, polymicrobial growth=2 (Fig. 2B). A combined “clinical-microbial wound infection score” (CMWIS) (total 0–8) was then calculated for each mouse by combining the CWHS and microbial scores. Last, at sacrifice, the wound muscle and cecal content were collected and analyzed for microbiota genetic composition via 16S rRNA sequencing. Mice were then euthanized as per the on-file Protocol 72090.
Figure 2.
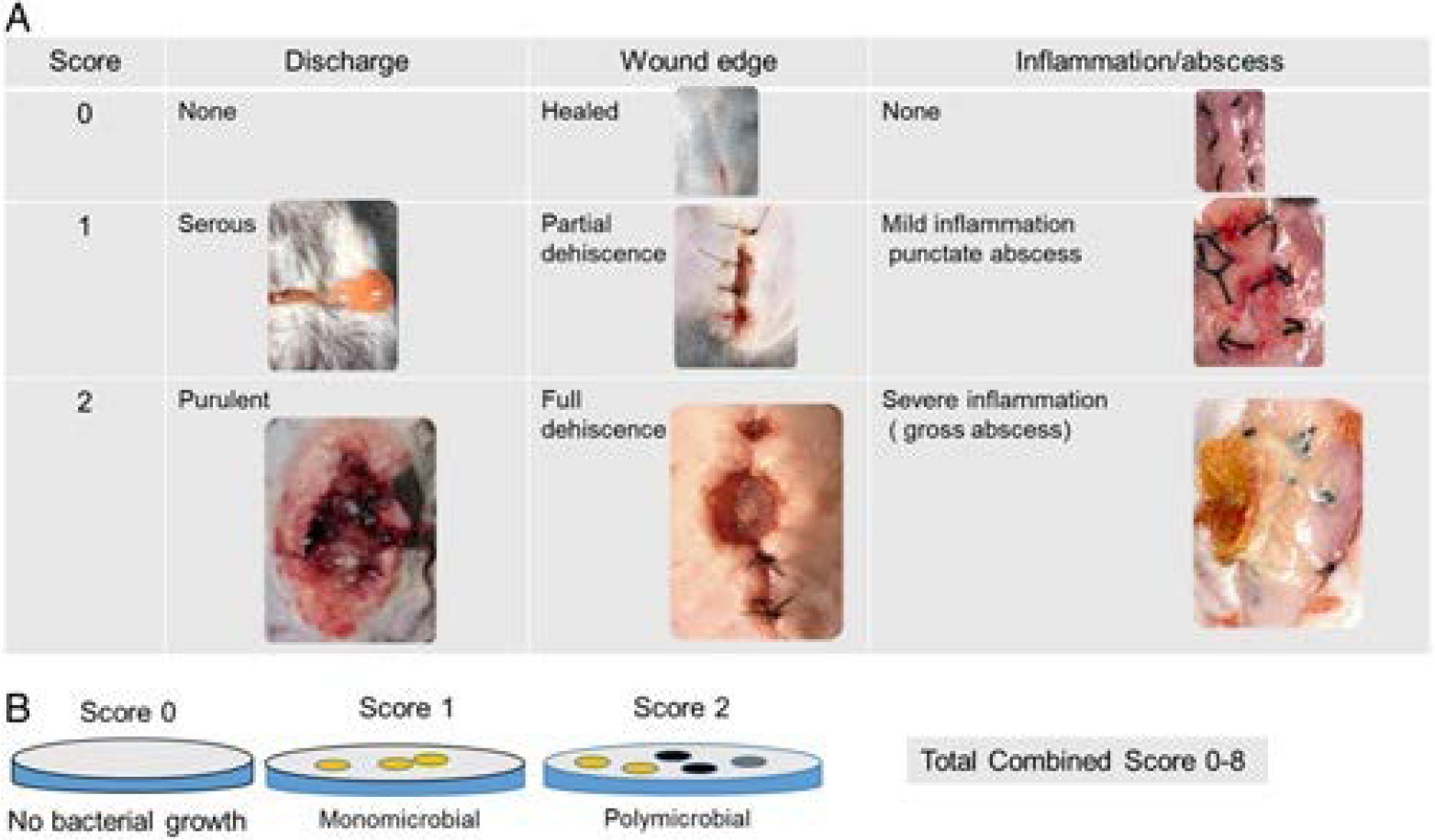
Clinicomicrobial wound score. A, Clinical wound scoring (total 0–6) includes the following descriptions: discharge estimation: 0—discharge none; 1—serous discharge, 2—purulent discharge; wound edge appearance: 0—healed; 1—partial dehiscence; 2—full dehiscence; inflammation: 0—no inflammation/abscess, 1—mild inflammation, and 2—severe inflammation/gross abscess. B, Microbial wound scoring (total 0–2) includes: 0—no culturable bacteria; 1—monomicrobial growth, and 2—polymicrobial growth. CMWIS=Clinical wound score+microbial wound score.
Preparation of Pi-PEG Solution
Phosphorylated PEG compound with a defined ABA (hydrophilic/hydrophobic/hydrophilic structures), ABA-PEG20k-Pi20 (herein named as Pi-PEG), was synthesized as previously described.6 Pi-PEG was dissolved in tap water to a final concentration 1%. Mice were allowed at libitum access to either normal drinking water or drinking water spiked with 1% Pi-PEG. The final solution is clear, odorless, and tasteless.
Microbiota Analysis of the Wound and Blood
Wound swabs and blood were cultured on Tryptic soy agar. After overnight incubation in 37°C, plates were inspected for the growth of organisms. Bacterial indentification was performed on Vitek MS (MALDI-TOF) (bioMérieux Inc., Durham, NC). Susceptibility testing of bacteria was performed by the Vitek 2 XL system (bioMérieux Inc.) using the AST-GN75 card.
Isolation of DNA and 16S rRNA Sequencing
Tissue samples were homogenized and DNA was extracted using the QIAmp DNA kit (Qiagen). For cecal contents, DNA extraction was done using QIAmp PowerFecal proDNA kit (Qiagen). For library preparation, DNA was amplified using the barcoded 12-bp Golay primer set designed for the Earth Microbiome Project (EMP).14 A mitochondrial PNA blocker (PNA Bio) was used to prevent amplification of the murine mitochondrial region. For each sample, PCR was performed using the EMP primers, mPNA, AccuStart II PCR ToughMix, and the extracted DNA (Quntabio). The results of the quantification were then used to normalize the amount of DNA for sequencing, and to ensure each amplicon was represented evenly during sequencing. The volumes were then sequentially consolidated into a single tube using an OpenTrons liquid-handler running a custom Python script. Finally, an aliquot of the final pool was taken, and the DNA was purified using the Agencourt AMPure XP PCR* purification system (Beckman-Coulter). The samples were run on an Illumina MiSeq at UIUC (150 bp ×2).
16S rRNA Sequence Data Analysis
For 16S rRNA analysis, the paired-end reads were joined using the join_paired_ends.py script followed by quality filtering and demultiplexing using the split_libraries_fastq.py script in QIIME.15 The final set of demultiplexed sequences were then selected for amplicon sequence variant (ASV) using the DeBlur pipeline.16 To improve downstream network analysis, ASVs present in ≤10 samples were removed. The data were rarefied to a depth of 10,000 reads per sample. Alpha and beta diversity were analyzed using the Phyloseq and MicrobiomeSeq package in the statistical software R. For alpha diversity—the Shannon index was used and for beta diversity—NMDS plots that were generated based on a weighted UniFrac dissimilarity matrix. Assessment of the statistical significance of alpha and beta diversity trends was performed using permutational multivariate analysis of variance. To determine significantly different ASVs between groups of interest, the analyses of composition of microbiome pipeline was used at a P value (false discovery rate) cutoff of <0.05.
Statistical Analysis
Data from the study was reported as mean±SD; statistical significance was determined by analysis of variance or t test, where appropriate. P value <0.05 was considered statistically significant unless otherwise stated. Statistical analysis for the composite wound healing score was performed on the mean CMWIS/group by analysis of variance.
RESULTS
Mice Consuming a WD Develop Impaired Wound Healing
Mice consuming a WD and exposed to antibiotics demonstrated significantly impaired wound healing and SSI compared with mice consuming a SD. In the group of mice (WD+Abx), the highest CWHS was observed. Representative images of CWHS are displayed in Figure 3A with corresponding scores in Figure 3B. The CWHS positively correlated with the level of incidence for positive wound cultures (Fig. 3C). The combined score expressed as an infection/healing wound score (CMWIS) is presented in (Fig. 3D). Among all across all treatment groups, blood cultures were negative (data not shown).
Figure 3.
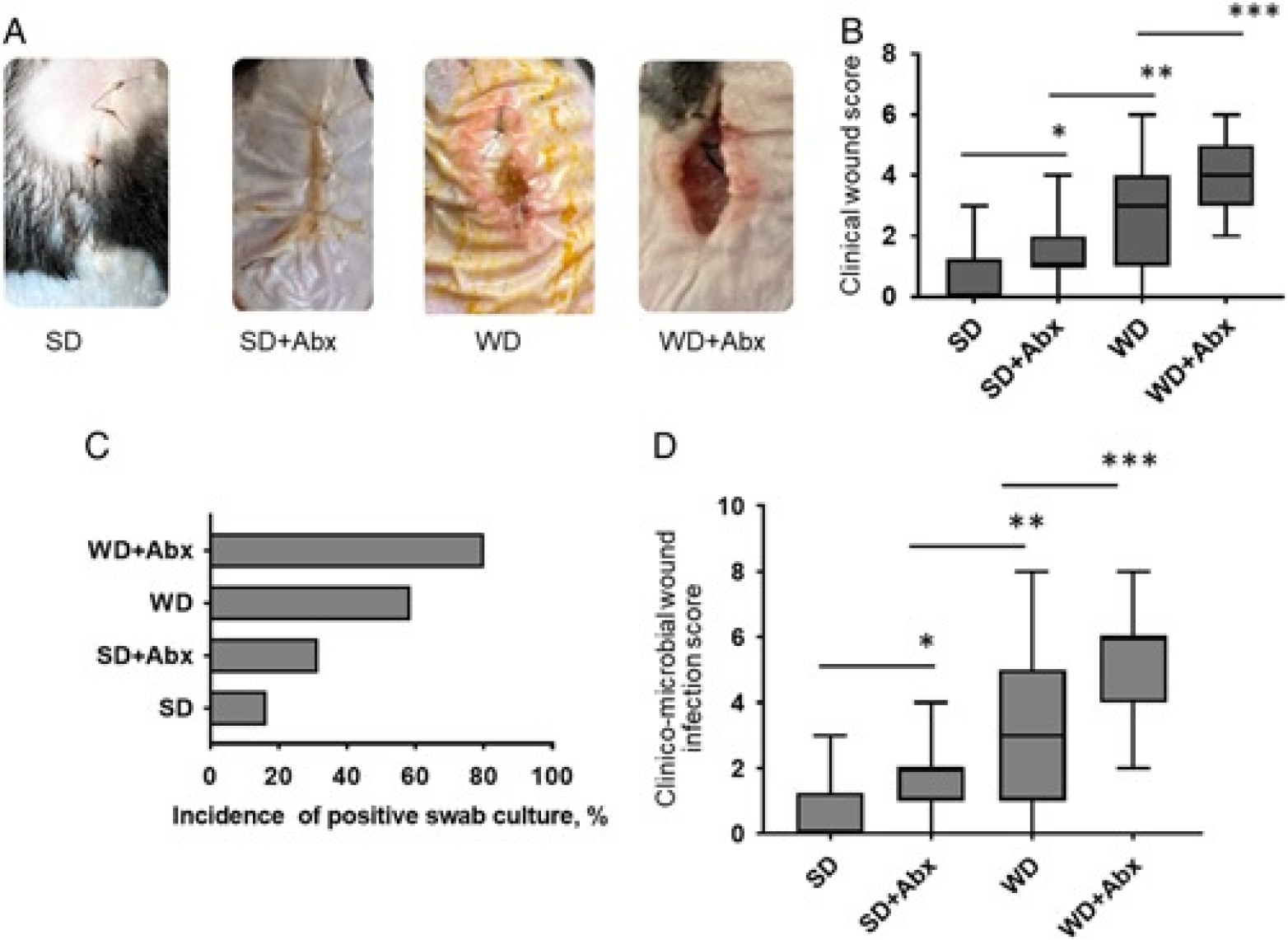
Wound healing/infection results. A, Representative images of wounds among treatment groups. B, Clinical wound scores. *P=0.0096, **P=0.0147, ***P=0.0174, Mann-Whitney test for paired columns. P<0.0001 (1-way analysis of variance). C, Percentage of the incidence of positive wound (via swab) cultures. D, Clinicomicrobial wound infection scores. *P=0.0237, **P=0.0148, ***P=0.016, Mann-Whitney test for paired columns. P<0.0001 (1-way analysis of variance). SD: n=18 mice, SD+Abx: n=19, WD: n=29, WD+Abx: n=15.
WD and Antibiotics Treatment Predisposed SSI With Polymicrobial Organisms
Cultured wound swabs performed at a sacrifice when the surgical site was opened on POD10, demonstrated polymicrobial growth in 73% of mice in the WD+Abx group, 21% of mice in the WD treated group, 10% in the SD+Abx group, and 0% in the SD group (Fig. 4A). Predominant species were identified as methicillin-sensitive Staphylococcus aureus (MSSA) in SD-fed and WD-fed mice. In WD-fed mice, in addition to MSSA, Pseudomonas aeruginosa, Enterobacter cloacae complex, Enterococcus faecalis, Klebisella oxytoca and Escherichia coli were identified. MSSA was no longer detected after antibiotic treatment independent of diet, whereas E. cloacae complex become the predominant pathogen. In SD+Abx treated mice, E. faecalis and Enterococcus gallinarum were observed, while in WD+Abx treated mice, E. faecalis, Proteus mirabilis, E. coli, K. oxytoca, and E. gallinarum were identified (Fig. 4B).
Figure 4.
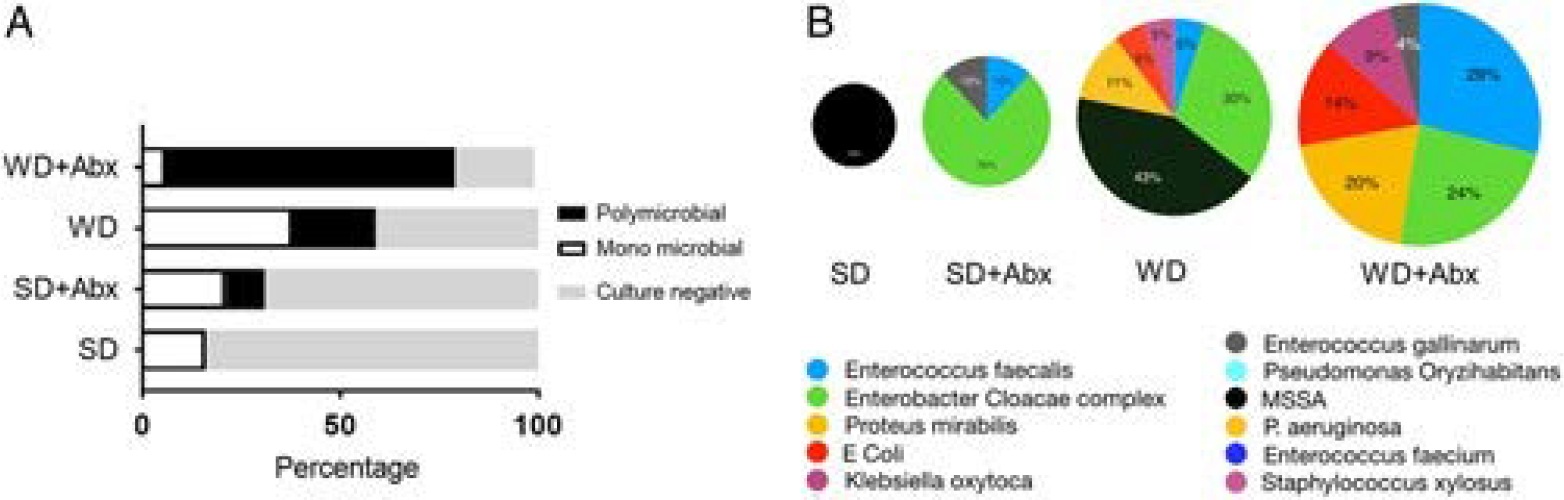
Wound culture analyses. A, Relative percentages of polymicrobial and monomicrobial wound infections between treatment groups. B, Species identification of cultured bacteria from wound swabs.
Postoperative Contamination of the Operative Incision With SSI Bacteria in This Model Results in a Low Negligible Wound Infection Rate
To further prove that the wound infections in this model are NOT due to postoperative contamination of the closed operative wound site, reiterative studies were performed in chow-fed mice undergoing the back injury model (back muscle exposure, with 6 cautery injury site plus suture placement). Once the operative site was closed, wounds were swabbed daily for 3 days with a 200 μL of a cocktail of 3 most common isolates causing SSIs in this model: E. faecalis, E. cloacae complex, and Proteus mirabilis at 107 cfu/mL in 10 mice. Results indicate that 80% of mice in this group did not develop clinical infections when sacrificed on POD10, whereas 2 mice developed infections with a relatively low (compared with WD+abx treated mice—see Figs 3D,5B) clinical microbial wound infection score (Supplemental Fig. S1).
Figure 5.
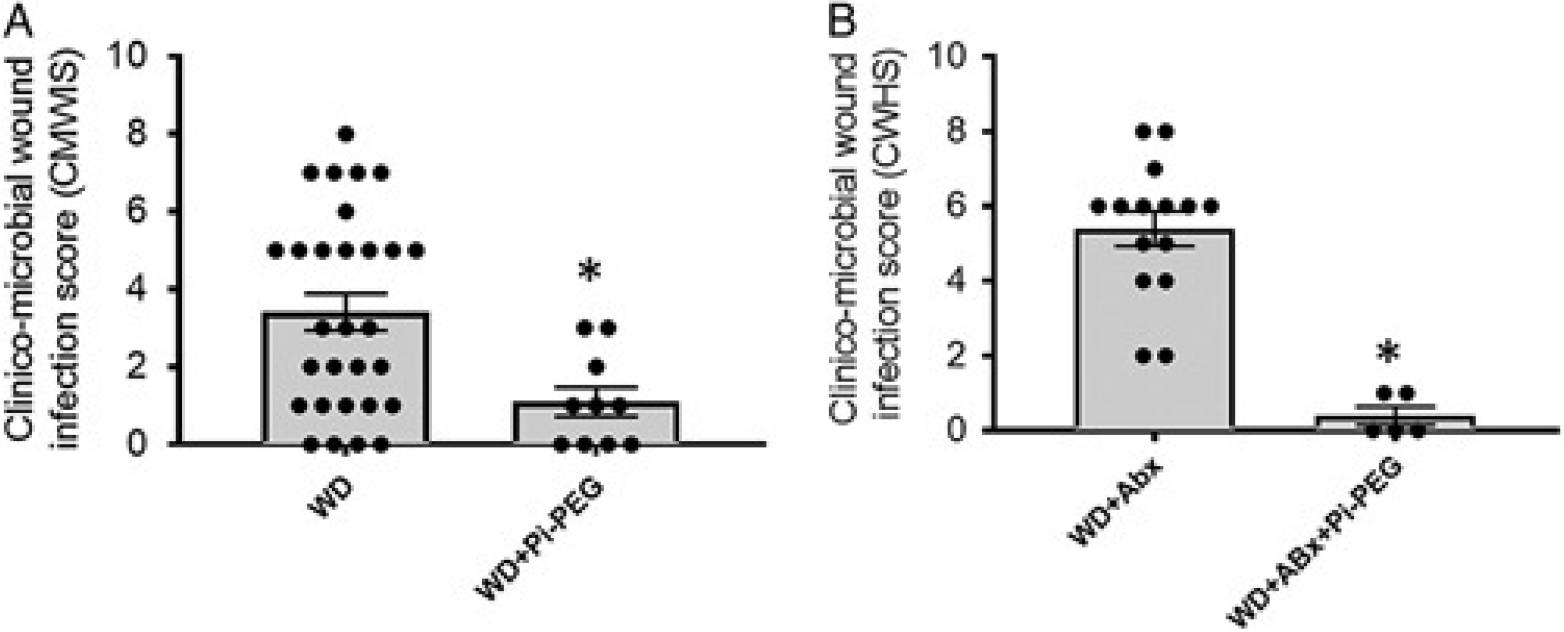
Mice drinking water supplemented with 1% Pi-PEG do not develop surgical wound healing/infection complications. CMWIS is significantly decreased by oral 1% Pi-PEG in mice fed WD (n=29, WD and n=10, WD+Pi-PEG, P=0.010) (A) and mice fed WD and treated with antibiotics (n=15, WD+Abx and n=5, WD+Abx+Pi-PEG, *P<0.0001) (B).
Pi-PEG Improved Wound Healing in Both WD and WD+Abx Treatment Groups
Drinking water spiked with 1% Pi-PEG was well tolerated in all mice; they appeared normal, consumed water at volumes equal to their comparator groups, displayed normal growth, and did not develop diarrhea. Wound healing was significantly improved in mice drinking 1% Pi-PEG (Fig. 5A,B). The composite CMWIS score was 3.4±0.6 (WD, n=29) versus 1.19±0.37 (WD+Pi-PEG, n=10), P=0.009 (Fig. 5A) and 5.4±1.2 (WD+Abx, n=15) versus 0.48±0.21 (WD+Abx+Pi-PEG, n=5), P=0.001 (Fig. 5B). None of the wound swabs in the mice drinking water spiked with Pi-PEG were culture positive.
ASV Analysis Indicates That the Strains From the Gut and Wound Share Genetic Homology and Oral Pi-PEG Is Preventative
To determine if bacteria at the operated surgical site at the taxonomic strain level, shared identity with those present in the gut, ASV analysis was performed.17 In mice fed SD, ASVs in the wound corresponded to E. coli, Staphylococcus, and Bacteroides at an abundance of 0.1–0.4 (Fig. 6A). In WD-fed mice ASVs (ie, of similar identity in the wound and gut) consisted of Proteus and Staphylococcus seen at much higher abundances than WD-fed mice, reaching a relative abundance up to 0.98 (Fig. 6B). After WD-fed mice were treated with antibiotics, the wounds were dominated by Enterobacter (Fig. 6C) sharing identity to ASVs present in the gut. In mice drinking Pi-PEG treated with WD+Abx, the previously seen Proteobacteria genera Proteus and Enterobacter ASVs were no longer present (Fig. 6D) in the wounds.
Figure 6.
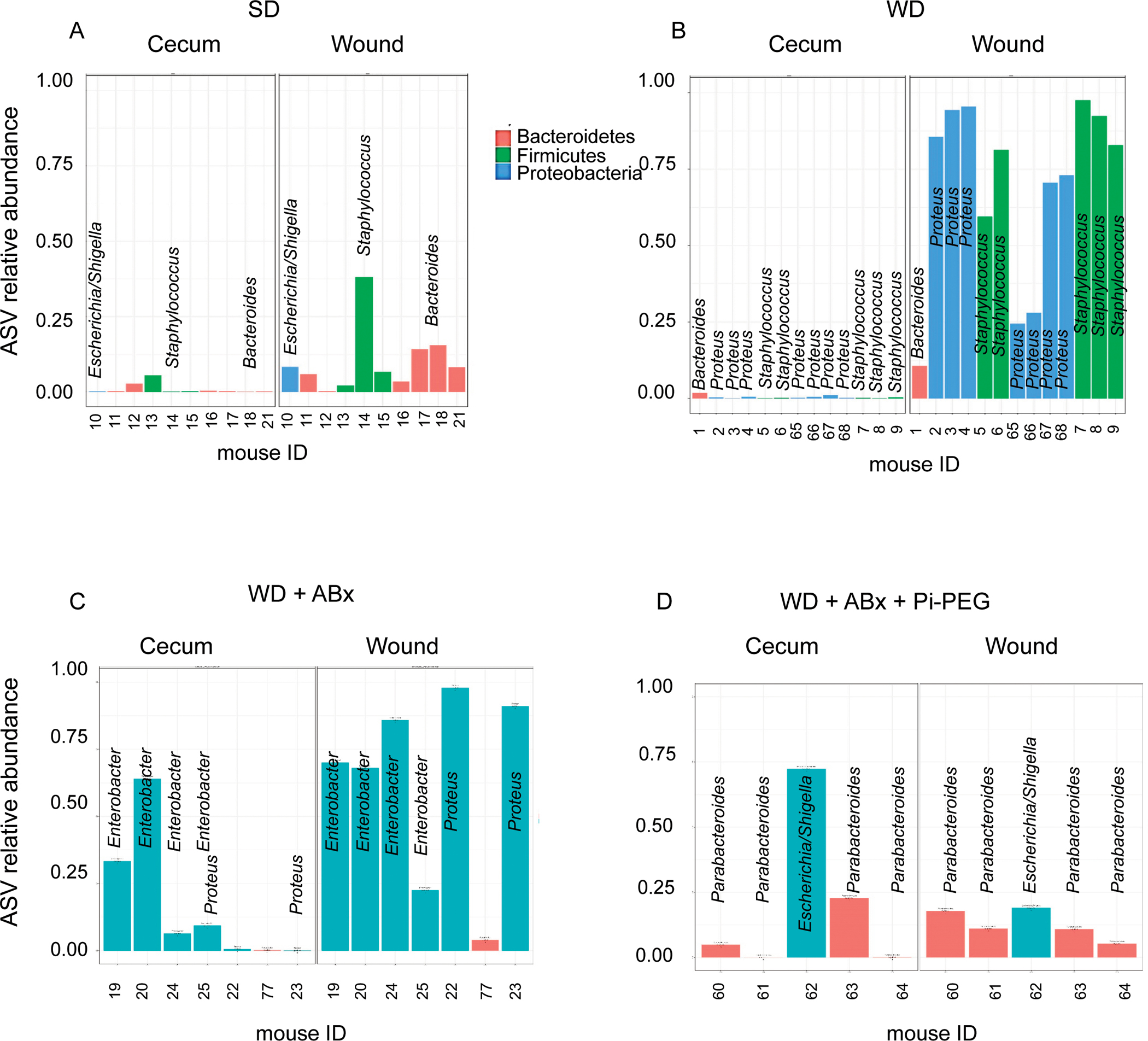
16S rRNA ASV analysis of gut and wound microbiota. A, ASVs common to the gut and wound microbiota in SD mice. B, ASVs common to gut and wound in WD mice. C, ASVs common to gut and wound in WD+Abx mice. D, ASVs common to gut and wound in WD+Abx mice drinking 1% Pi-PEG.
Pi-PEG Significantly Minimizes the Abundance of Proteobacteria Species in the Wound
To determine if the loss of these bacterial species following Pi-PEG was the result of a reduction in the abundance of Proteobacteria in the gut, we compared the operational taxonomic unit relative abundance in the cecum and wound microbiota in mice fed WD without a surgical wound, in mice fed WD with a surgical wound, in mice treated with WD+Abx with a surgical wound, and in mice treated with WD+Abx+Pi-PEG with a surgical wound. In mice fed a WD and treated with antibiotics, Pi-PEG does not reduce the abundance of Proteobacteria in the gut (Fig. 7A), however, it does appear to reduce the abundance of Proteobacteria in the wound (Fig. 7B). These results suggest that the creation of surgical wound (ie, the wound back model herein described) has a minimal impact on the gut microbiota (WD alone vs WD+wound,Fig.7A); however, certain gut species (mainly Proteus) have the ability to reach remote surgical wounds (Fig. 7A,B). Antibiotic treatment appears to further exacerbate the presence of these bacterial species as Proteobacteria were observed to dominate both in the gut and wound. Finally, Pi-PEG does not impact the composition of the gut microbiota, but significantly minimizes the abundance of Proteobacteria species in the wound suggesting that Pi-PEG may function to prevent remote SSIs by somehow preventing the method by which gut bacteria travel to the wound site (ie, by preventing their uptake in immune cells, their ability to survive the journey, their successful implantation into the remote wound site). Further molecular-based studies are underway to determine the precise mechanism (s) of action of this dual-component agent.
Figure 7.
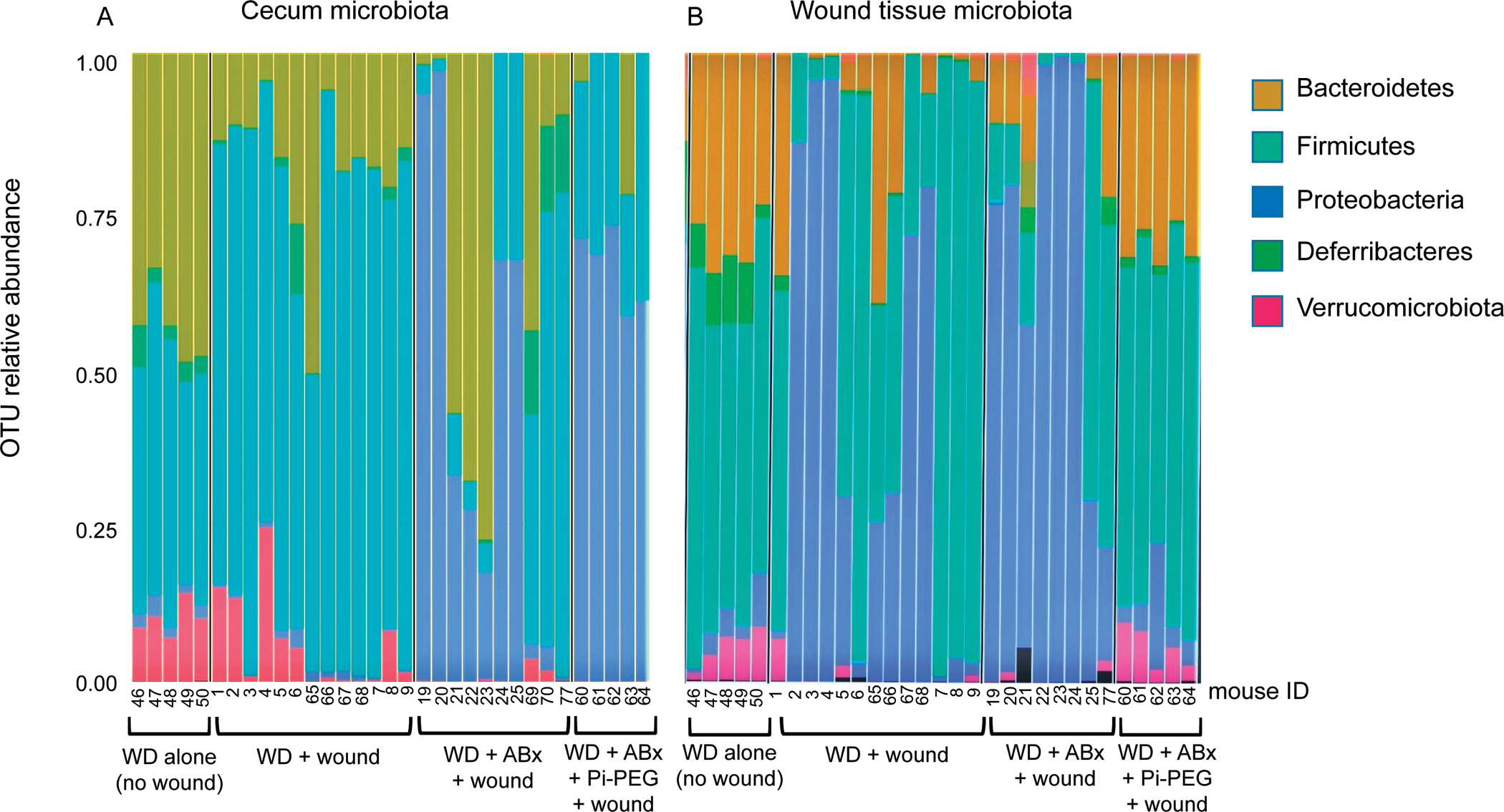
16S rRNA operational taxonomic unit (OTU) analysis of gut and wound microbiota. Composition of microbiota at the phylum level in the gut (A) and wound (B) of mice on a WD diet exposed to different treatments.
DISCUSSION
In this study, we demonstrate that Pi-PEG provides a gut-centered treatment for reducing SSIs arising at wound sites remote from the gut contents. Previous work from our laboratory demonstrates that Pi-PEG durably embeds phosphate along the entire gut acting as a surrogate mucin-like agent in which microbial growth is not affected, bacterial virulence is suppressed via quorum sensing inhibition and there is the physical distancing of the luminal microbes aways from the epithelial surface.18 In this manner, Pi-PEG can enhance the barrier function of the intestinal epithelial surface and shield against invading microbes. As a result, orally delivered Pi-PEG has the potential to prevent gut pathogens from migrating to remote operative sites perhaps via the Trojan Horse Hypothesis of SSI, although this latter mechanism remains to be proven in the current model. We have previously shown that the ABA structure polyethelene glycol (a core of Pi-PEG) can durably adhere to both bacterial and host cells8,9 and thus may prevent phagocytosis of bacteria by neutrophils/immune cells. Alternatively, delivery of phosphate bound to the ABA PEG core to the intestinal tract may attenuate the virulence of pathogens19 and thus impair their ability to degrade the PEG via “pegylases.”20
The rationale for Pi-PEG as an agent to prevent SSIs arising from pathogens among the gut microbiota is based on multiple previous reports from our laboratory indicating that bacteria have evolved highly conserved phosphosensory and phosphoregulatory mechanisms to “sense” the local concentration of phosphate and respond accordingly.11 Via the well-described pstS-PhoB system of phosphate acquisition and regulation, we observed that when local phosphate concentrations are abundant, pathogenic bacteria become insensate to incoming host “cues” that activate their virulence circuits.19,21 When phosphate levels are abundant, bacteria do not express pstS-PhoB, however when local phosphate is depleted, virulence expression is activated (ie, invasion, immune subversion) in order for bacteria to acquire phosphate from intracellular stores (ie, ATP, DNA). Work from our laboratory has elucidated critical connections between pstS and quorum sensing circuits that allow for this response.11 Recognition of this response in pathogenic bacteria is critical given that surgical injury alone causes the release of phosphatonins which increase the renal excretion of phosphate leading to intestinal mucosal phosphate depletion—a finding that occurs in the absence of any change in the serum phosphate concentration.21 As a result, we developed Pi-PEG recognizing that neither oral nor intravenous inorganic phosphate administration would be able to prevent mucosal phosphate depletion or replete it.
Important to the overall goals of this study to prove the efficacy of the Pi-PEG compound was the development of a clinically relevant animal model in which an SSI developed remote from direct contamination by gut contents yet still involved them. Although controlling all the variables that are possible contaminants in this model was challenging, based on a series of previous studies,13,22,23 we reasoned that most patients in whom a clinical SSI develops are consuming a WD leading to overweight and are invariably exposed to antibiotics as part of routine asepsis practice. While it could be argued that diet quality and type and duration of antibiotics could have affected the SSI incidence in this model, we were careful to make sure that the SSIs that developed in this model were meaningful to the clinical experience of surgeons. This required combining both visual inspections of the wound and culture results to calculate a blended scoring system (ie, CMWIS).
Across all wounds in an operative site (skin, wound, organ space, etc.), the precise origin of the pathogen that eventually is causative to the clinical SSI remains unknown. While intraoperative contamination at the time of surgery is invariably assumed to be the major source of a clinical SSI, and certainly can occur, in the current era of sterilization and antibiotic use, the precise source and mechanism by which pathogens contaminate a wound to the point that clinical infection occurs, remains unknown. The molecular-level evidence to fully understand this process would involve tracking the vectorial movement of pathogens from their site of origin, defining their survival against a competent immune systems, and determining how they overcome the natural resistance of host tissues to which they home. This clearly would require exhaustive genetic-level tracking (ie, whole-genome sequencing) at the taxonomic strain level, of all potential pathogens present at all potential origin sites (gums, nose, gut, skin, operative equipment/environment, personnel, etc.). Although the present study provided some evidence in this regard, clearly much more work is needed to prove that SSIs develop from pathogens that originate from among the gut microbiota following surgery. In addition, more detailed mechanistic studies are needed to understand the precise mechanism(s) of action by which orally administered Pi-PEG functions to prevent SSIs both experimentally and clinically.
A major limitation of this study is that the Trojan Horse mechanism as the main pathway by which gut pathogens cause clinically meaningful SSI in this model was not fully confirmed. For example, in the present study we did not culture circulating immune cells as we had done previously; nor did we track gavaged green fluorescent protein-labeled bacteria and determine that those microbes present in the gut were the same bacteria that traveled to the surgically created wound via the bloodstream.4 Another limitation of the present study is that although the Pi-PEG in this model was highly efficacious in preventing an SSI, its precise mechanism of action and the relative contribution of the Pi versus the PEG remain unknown. In addition, variability in the number of animals assigned to each group was a function of performing reiterative experiments to achieve statistical significance while at the same time limiting the overall use of animals. This could potentially introduce error. Last, without time-stamped comprehensive daily tracking of all potential sources of the SSI pathogens in this model (mouth, gut, skin, urine, etc.), it is not possible to conclude that, despite genetic similarities of between the gut and wound infecting pathogens, that they necessarily originated from the gut. Further work is underway to more precisely define the microbial dynamics in this model and the precise mechanism of action of Pi-PEG.
We did not treat chow-fed mice with Pi-PEG as the incidence of SSIs in this group was extremely low and would have required large numbers of mice to achieve statistical significance. Similarly, based on the 16s rRNA data, there does appear to be a protective effect of Pi-PEG on the antibiotic effect in terms of the gut microbiota (Fig. 7).
In summary, results from the present study provide foundational knowledge upon which to base a strategy to prevent SSIs using an orally administered Pi-PEG. This approach complements rather than supplants all ongoing efforts to prevent SSIs. Further work, including clinical trials, will be needed to confirm the efficacy and safety of this approach.
Supplementary Material
Supplemental figure S1. Clinical microbial wound infection score
REFERENCES
- 1.Teillant A, Gandra S, Barter D, et al. Potential burden of antibiotic resistance on surgery and cancer chemotherapy antibiotic prophylaxis in the USA: a literature review and modelling study. Lancet Infect Dis. 2015;15:1429–1437. [DOI] [PubMed] [Google Scholar]
- 2.Alverdy JC, Hyman N, Gilbert J. Re-examining causes of surgical site infections following elective surgery in the era of asepsis. Lancet Infect Dis. 2020;20:e38–e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wills BW, Smith WR, Arguello AM, et al. Association of surgical jacket and bouffant use with surgical site infection risk. JAMA Surg. 2020;155:323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krezalek MA, Hyoju S, Zaborin A, et al. Can methicillin-resistant Staphylococcus aureus silently travel from the gut to the wound and cause postoperative infection? Modeling the “Trojan Horse Hypothesis”. Ann Surg. 2018;267:749–758. [DOI] [PubMed] [Google Scholar]
- 5.Zaborin A, Penalver Bernabe B, Keskey R, et al. Spatial compartmentalization of the microbiome between the lumen and crypts is lost in the murine cecum following the process of surgery, including overnight fasting and exposure to antibiotics. mSystems. 2020;5:e00377–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao J, Zaborin A, Poroyko V, et al. De novo synthesis of phosphorylated triblock copolymers with pathogen virulence-suppressing properties that prevent infection-related mortality. ACS Biomater Sci Eng. 2017;3:2076–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Mao J, Nagao M, et al. Structure and dynamics of lipid membranes interacting with antivirulence end-phosphorylated polyethylene glycol block copolymers. Soft Matter. 2020;16:983–989. [DOI] [PubMed] [Google Scholar]
- 8.Wu L, Zaborina O, Zaborin A, et al. High-molecular-weight polyethylene glycol prevents lethal sepsis due to intestinal Pseudomonas aeruginosa. Gastroenterology. 2004;126:488–498. [DOI] [PubMed] [Google Scholar]
- 9.Valuckaite V, Seal J, Zaborina O, et al. High molecular weight polyethylene glycol (PEG 15–20) maintains mucosal microbial barrier function during intestinal graft preservation. J Surg Res. 2013;183:869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edelstein A, Fink D, Musch M, et al. Protective effects of nonionic triblock copolymers on bile acid-mediated epithelial barrier disruption. Shock. 2011;36:451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaborin A, Romanowski K, Gerdes S, et al. Red death in caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA. 2009;106:6327–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaborin A, Defazio JR, Kade M, et al. Phosphate-containing polyethylene glycol polymers prevent lethal sepsis by multidrug-resistant pathogens. Antimicrob Agents Chemother. 2014;58:966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyoju SK, Zaborin A, Keskey R, et al. Mice fed an obesogenic western diet, administered antibiotics, and subjected to a sterile surgical procedure develop lethal septicemia with multidrug-resistant pathobionts. mBio. 2019;10:e00903–e00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir A, McDonald D, Navas-Molina JA, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valuckaite V, Zaborina O, Long J, et al. Oral PEG 15–20 protects the intestine against radiation: role of lipid rafts. Am J Physiol Gastrointest Liver Physiol. 2009;297:G1041–G1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaborina O, Holbrook C, Chen Y, et al. Structure-function aspects of PstS in multi-drug-resistant Pseudomonas aeruginosa. PLoS Pathog. 2008;4:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins LD, Cook KA, Cain RB. Microbial degradation of polyethylene glycols. J Appl Bacteriol. 1979;47:75–85. [DOI] [PubMed] [Google Scholar]
- 21.Long J, Zaborina O, Holbrook C, et al. Depletion of intestinal phosphate after operative injury activates the virulence of P aeruginosa causing lethal gut-derived sepsis. Surgery. 2008;144:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaines S, van Praagh JB, Williamson AJ, et al. Western diet promotes intestinal colonization by collagenolytic microbes and promotes tumor formation after colorectal surgery. Gastroenterology. 2020;158:958.e2–970.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Berg FF, van Dalen D, Hyoju SK, et al. Western-type diet influences mortality from necrotising pancreatitis and demonstrates a central role for butyrate. Gut. 2021;70:915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure S1. Clinical microbial wound infection score


