Abstract
Background
The coronavirus disease 2019 (COVID-19) and community-acquired pneumonia (CAP) present a high degree of similarity in chest computed tomography (CT) images. Therefore, a procedure for accurately and automatically distinguishing between them is crucial.
Methods
A deep learning method for distinguishing COVID-19 from CAP is developed using maximum intensity projection (MIP) images from CT scans. LinkNet is employed for lung segmentation of chest CT images. MIP images are produced by superposing the maximum gray of intrapulmonary CT values. The MIP images are input into a capsule network for patient-level pred iction and diagnosis of COVID-19. The network is trained using 333 CT scans (168 COVID-19/165 CAP) and validated on three external datasets containing 3581 CT scans (2110 COVID-19/1471 CAP).
Results
LinkNet achieves the highest Dice coefficient of 0.983 for lung segmentation. For the classification of COVID-19 and CAP, the capsule network with the DenseNet-121 feature extractor outperforms ResNet-50 and Inception-V3, achieving an accuracy of 0.970 on the training dataset. Without MIP or the capsule network, the accuracy decreases to 0.857 and 0.818, respectively. Accuracy scores of 0.961, 0.997, and 0.949 are achieved on the external validation datasets. The proposed method has higher or comparable sensitivity compared with ten state-of-the-art methods.
Conclusions
The proposed method illustrates the feasibility of applying MIP images from CT scans to distinguish COVID-19 from CAP using capsule networks. MIP images provide conspicuous benefits when exploiting deep learning to detect COVID-19 lesions from CT scans and the capsule network improves COVID-19 diagnosis.
Keywords: Maximum intensity projection, Capsule network, COVID-19, Community-acquired pneumonia, Computed tomography
1. Introduction
Coronavirus disease 2019 (COVID-19) was first observed in humans in late 2019 and has since spread across the globe [1,2]. The real-time polymerase chain reaction (RT-PCR) test is the gold standard for diagnosing COVID-19 through the detection of the novel coronavirus nucleic acid [3]. The PCR results of patients affected by detoxification concentration always show a false negative, with multiple nucleic-acid tests required to confirm the diagnosis [4,5]. Computed tomography (CT) images are used to detect and diagnose chest lesions, such as ground-glass opacities and crazy-paving [[6], [7], [8]]. However, in CT images, community-acquired pneumonia (CAP) and COVID-19 appear similar, and it is challenging to distinguish them, even for experienced radiologists.
Deep learning methods, especially convolutional neural networks (CNNs), have significant feature representation abilities and provide effective tools in diagnosing COVID-19 and distinguishing COVID-19 from CAP through computer-aided systems via chest X-rays and CT images. Rahaman et al. [1] investigated 15 different state-of-the-art pre-trained CNN models, including VGG, ResNet, InceptionNets, DenseNets, MobileNet, and Xception, for recognizing COVID-19 from chest X-rays. Moreover, a combination of DarkNet and AlexNet [9] and a pretrained DenseNet-121 model [10] have been used to diagnose COVID-19 patients using chest X-rays. In contrast to chest X-rays, CT images allow more details to be obtained with no overlapping tissues and provide an effective tool for distinguishing COVID-19 from CAP. Three-dimensional (3D) volume analysis and two-dimensional (2D) slice analysis have been combined to provide CT images for the classification of COVID-19 and CAP [11]. Qi et al. [12] proposed a multiple-instance learning method to distinguish COVID-19 from CAP, while Qi and his colleagues [13] developed a fully automatic deep-learning pipeline that can accurately distinguish COVID-19 from CAP using CT images by mimicking the diagnostic process of radiologists. On the basis of the above methods, more robust and advanced deep-learning models should be developed to improve the diagnosis of COVID-19.
Although deep learning using CT images and chest X-rays achieves high-accuracy diagnosis and classification of COVID-19 from CAP, few studies take the routine workflow of radiologists into account. A more effective and explainable procedure is required given the high degree of similarity between chest CT images of COVID-19 and CAP. Initially, radiologists in clinical practice quickly scan maximum intensity projection (MIP) images to identify lesion candidates (high-CT-value region) for additional testing on particular slices. Inspired by this procedure, we propose a novel CNN-based method that uses MIP images from CT scans and a capsule network. The purpose of this study is to explore whether MIP images generated from CT scans could help capsule networks to improve performance in terms of distinguishing COVID-19 from CAP and enhance the efficiency of COVID-19 diagnosis.
Histological studies have demonstrated that the pulmonary vasculature is affected by pneumonia, and that a high prevalence of thromboembolic disease is a hallmark of severe COVID-19 infection [14]. Existing studies have focused on the lung parenchymal involvement of CT imaging features, with few studies exploring the role of the pulmonary vascular system in COVID-19 [15]. A previous study indicates that the pulmonary vascular system in COVID-19 is redistributed from smaller to larger vessels [16].
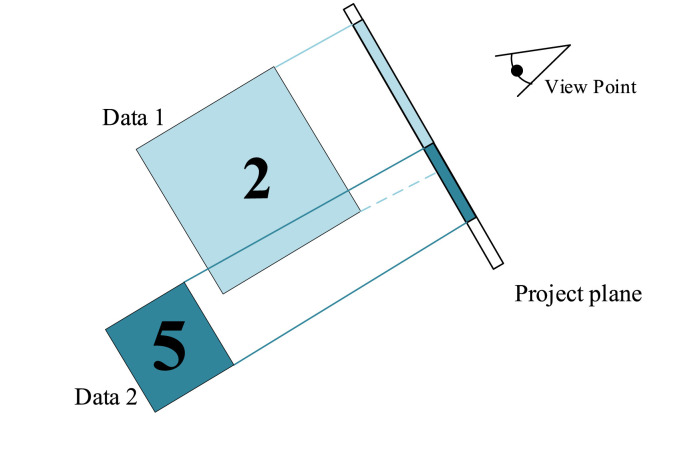
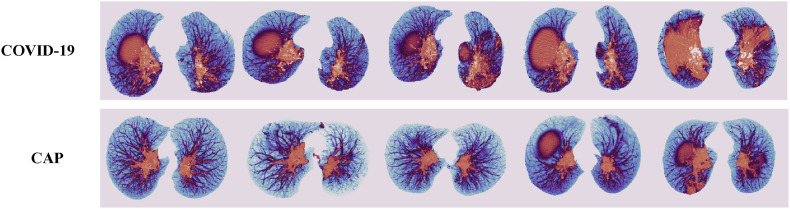
MIP [17] is a widely used postprocessing technique for CT images, such as in the detection of lung nodules, for which it enhances the visualization [18]. An MIP image is a two-dimensional representation acquired using the fluoroscopic method, which means that it is obtained by calculating the maximum density of pixels that pass over each ray of the object. In CT imaging, when X-rays pass through a section of raw tissue, the pixel value with the highest density along this camera direction is retained and projected onto a two-dimensional plane to obtain a reconstructed MIP image, as shown in Fig. 1 . Therefore, the MIP technique provides an excellent response for representing the vascular morphology. MIP images can show even small density changes, accurately reflect the condition of blood vessels, and can distinguish the calcification of vessel walls. In our method, MIP is used to identify changes in the vascular morphology and determine the presence of lesions in CT images. Some examples of CT scans in COVID-19 and CAP patients using the MIP method are shown in Fig. 2 .
Fig. 1.
Schematic overview of MIP rendering. The maximum intensities along rays originating in the viewpoint are projected.
Fig. 2.
Some examples of MIP images of COVID-19 and CAP.
In this paper, we describe a DenseNet-121-based capsule network for distinguishing COVID-19 from CAP using MIP images generated from CT scans. Excellent performance is achieved on multi-center datasets. The main contributions of this work are as follows. (1) We annotate lung masks of COVID-19 cases and train LinkNet to segment these masks. (2) MIP images are produced by superposing the maximum gray of intrapulmonary CT values to represent the morphological changes in vessels and lesions of COVID-19. (3) We propose an MIP-based framework for distinguishing COVID-19 from CAP. (4) We demonstrate the feasibility of combining the clinical screening method and capsule network to improve the performance of distinguishing COVID-19 from CAP and enhance the diagnosis of COVID-19.
The remainder of this paper is organized as follows. Section 2 introduces multiple CT image datasets, and gives an overview of the proposed method, lung segmentation, MIP image acquisition, and the capsule network. An ablation study and details of the training and evaluation of models are also described in Section 2. Section 3 presents the ablation experiment results, discusses the performance of the proposed method on multiple datasets, and compares the proposed method with state-of-the-art COVID-19 classification methods. Section 4 discusses the challenges of lung segmentation in COVID-19 cases, and outlines the advantages of the proposed method, its limitations, and future work. Finally, Section 5 summarizes the proposed model and suggests ideas for future research.
2. Related work
2.1. Deep learning in COVID-19 diagnosis
With the rise of deep learning methods in medical image processing, techniques have also been developed for COVID-19, normal, and CAP cases using X-rays and CT images [19,20]. Nwosu and his colleagues [21] proposed a two-channel residual neural network with a semi-supervised learning strategy to classify normal, pneumonia, and COVID-19 images via chest X-rays. Waheed et al. [22] developed a variant generative adversarial network (GAN), CovidGAN, to generate synthetic X-ray images for the classification of normal and COVID-19 cases. Ouyang et al. [23] integrated online attention with 3D ResNet-34 in developing a dual-sampling attention network for distinguishing COVID-19 from CAP. Nagi and his colleagues [20] utilized a custom deep learning model, namely a modified version of MobileNet-v2, and an extended Xception model for the classification of COVID-19, lung opacity, and normal X-ray images. The above methods employed conventional X-ray images, which do not provide a significant amount of detail in the lungs.
CT images provide a more detailed view of the lungs, soft tissue, and blood vessels [24]. Mohammed et al. [25] proposed an integrated method for selecting the optimal deep learning model based on a novel crow swarm optimization algorithm for COVID-19 diagnosis using CT images. Saeed et al. [26] proposed a method based on complex fuzzy hyper-soft sets, which is a formulation of complex fuzzy (CF) and hyper-soft sets for the classification of COVID-19 and non-COVID-19. Mahmoudi et al. [27] proposed a CNN-based method for detecting and quantifying COVID-19 using CT images, while another study utilized a modified U-Net for COVID-19 lung infection segmentation [28]. Ibrahim et al. [29] investigated hybrid deep learning methods that can quickly and accurately identify COVID-19 from non-COVID-19 using lung CT images. They developed a diagnosis system starting with the segmentation of lung CT scan images and ending with disease prediction, giving a reliable COVID-19 prediction method.
2.2. Capsule network in medical image classification
Capsule networks provide a novel approach to creating synthetic neurons. Several studies have demonstrated how widely capsule networks are used in several sectors [30], including image classification. Capsule networks have achieved state-of-the-art performance on datasets such as CIFAR-10 [31,32], Fashion MNIST [33], MNIST [34], and SVHN [35]. Different feature extractors can be employed with capsule networks, including DenseNet [36], ResNet [37], Res2Net and SE-Block [38], and ResNet-v2 [39]. Recently, capsule networks have been exploited for COVID-19 diagnosis [40,41]. Gupta et al. [42] proposed the COVID-WideNet for detecting COVID-19 from non-COVID-19 cases based on a capsule network with two convolutional layers and three capsule layers with less-trainable parameters. Li and his colleagues [43] proposed a novel capsule network with a non-iterative and parameterized multi-head attention routing algorithm to replace the traditional iterative dynamic routing process. This method extracts more generalized representation features from X-ray images, thus improving the classification of COVID-19, pneumonia, and normal cases.
3. Materials and methods
3.1. Dataset
Data were acquired from different hospitals and publicly available datasets. Table 1 summarizes these datasets. The details are as follows.
-
●
The lab dataset contains 168 CT scans from 56 patients with COVID-19 and 165 scans from 100 patients with CAP. These images were taken between December 2019 and March 2020 at the General Hospital of the Yangtze River Shipping and Affiliated Hospital of Guizhou Medical University. RT-PCR tests were used to diagnose the COVID-19 patients.
-
●
The China Consortium of Chest CT Image Investigation (CC–CCII) dataset consists of 2716 CT scans (or patients), in which 1245 CT scans (or patients) were diagnosed with COVID-19 and 1471 CT scans (or patients) were diagnosed with CAP [44].
-
●
The Cancer Imaging Archive (TCIA) dataset compromises 629 CT scans from 538 patients with COVID-19 [45].
-
●
The Dongguan dataset consists of 236 CT scans from 158 patients with COVID-19. The dataset was obtained from Wanjiang People's Hospital. The patients were scanned by GE Medical Systems CT and Philips HOST-100196 CT.
Table 1.
Summary of CT scans of COVID-19 and CAP in the datasets.
| Dataset | Category |
Total | |
|---|---|---|---|
| COVID-19 | CAP | ||
| Lab dataset | 161/35 | 165/31 | 326 |
| CC-CCII | 1245 | 1471 | 2716 |
| TCIA | 629 | – | 629 |
| Dongguan dataset | 236 | – | 236 |
3.2. Overview of the study procedure
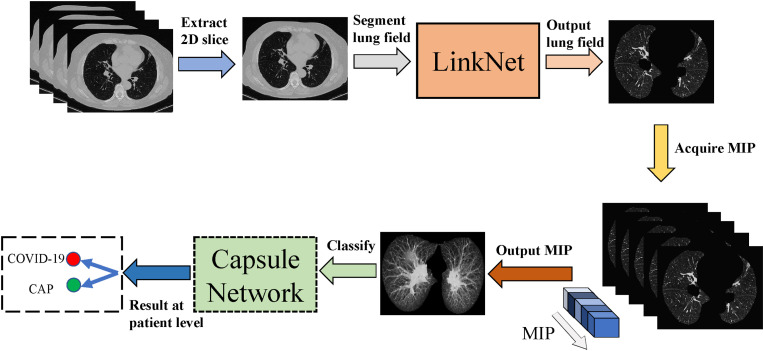
Fig. 3 represents the overall workflow of the proposed method for distinguishing COVID-19 from CAP using MIP images generated by CT scans and a capsule network. First, 2D slices are extracted from CT scans for segmenting the lung mask. Segmentation methods including LinkNet, U-Net, Recurrent Residual CNN-based U-Net (R2U-Net), Attention U-Net, U-Net++, and CE-Net are applied to the 2D CT images. Second, MIP images are produced from the superposition of maximum gray in intrapulmonary CT values. Finally, the MIP images are input into the capsule network for patient-level prediction of the final COVID-19 diagnosis. The feature extractors of the capsule network consist of ResNet-50, Inception-V3, and DenseNet-121. Details of each step of the proposed method are described in the following sections.
Fig. 3.
Workflow of the proposed method for distinguishing COVID-19 and CAP.
3.3. Lung segmentation
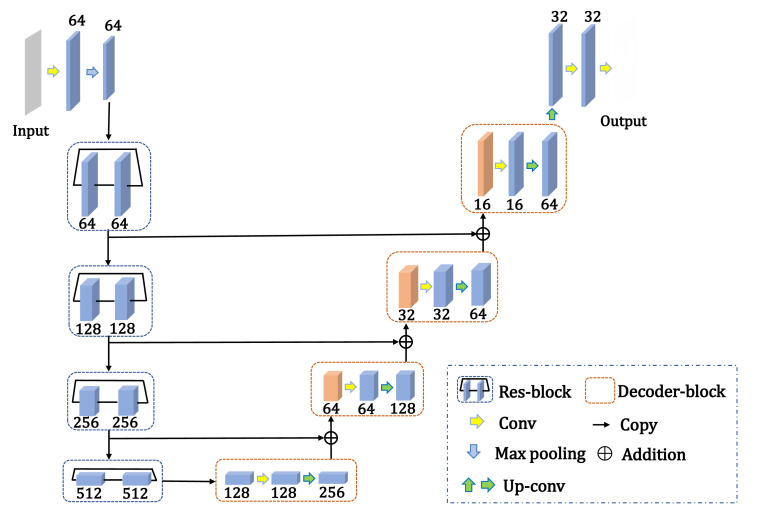
As the CT scans were acquired from different CT scanners and hospitals, a fixed window (window level = −300 HU, window width = 1400 HU) was set and the CT images were normalized to the range [0, 1]. The lung masks for 161 CT scans of COVID-19 were annotated in a semi-automatic way. First, the Pulmonary Toolkit (https://github.com/tomdoel/pulmonarytoolkit) was employed to initially segment the mask of the lung field. The mask was then manually modified by radiologists. The preprocessed CT images and annotations of the lung mask were fed into the segmentation model for training and evaluation. LinkNet [46] was used to segment the lung parenchyma. The network architecture is depicted in Fig. 4 . Similar to U-Net [47], LinkNet is composed of an encoder and a decoder, which are interconnected by an addition operation. In the encoder part, LinkNet uses ResNet-34 [48] pretrained on the ImageNet dataset [49], with the fully connected layer and the global average pooling layer removed. The decoder has five blocks, each consisting of a 2 × 2 up-sampling layer followed by two sets of layers, each containing a convolution layer, batch normalization layer, and rectified linear unit (ReLU) activation layer. In the first four blocks of the decoder, feature maps are generated from the corresponding part in the encoder and added to the feature map after up-sampling. Finally, a 3 × 3 convolution layer followed by sigmoid activation is applied to output the binary masks of the lung field.
Fig. 4.
Architecture of LinkNet for lung segmentation.
The binary cross-entropy (BCE) was applied as the loss function in the segmentation network. This function is expressed as follows:
where represents the prediction of the network and represents the ground-truth (annotated lung mask).
The morphological filling method, namely the “Find Contours” function in the OpenCV library, was applied to enhance the performance of lung mask segmentation. Five existing segmentation networks, i.e., U-Net [47], R2U-Net [50], Attention U-Net [51], U-Net++ [52,53], and CE-Net, were compared with our lung segmentation model.
3.4. Acquisition of MIP images
In clinical practice, radiologists usually examine axial sections of patients’ CT images to differentiate between COVID-19 lesions and CAP. Moreover, MIP images are routinely used by radiologists to improve the detection of COVID-19. In our method, MIP images are acquired through the superposition of maximum CT values at each coordinate from a stack of consecutive slices.
3.5. MIP-based capsule network for prediction of COVID-19 and CAP
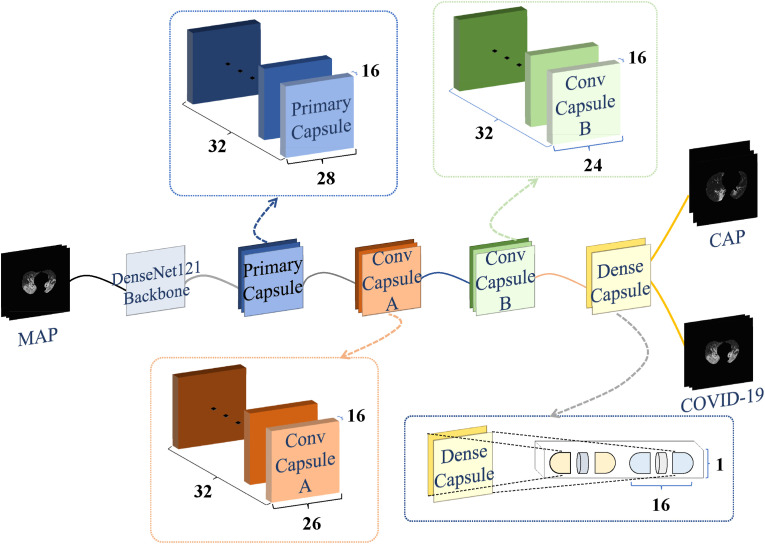
As shown in Fig. 5 , using an MIP image as input, the capsule network was trained for the patient-level prediction of COVID-19 and CAP. The capsule network consists of a feature extraction module (DenseNet-121 backbone) and a capsule module including a primary capsule, convolutional capsules A and B, and a dense capsule.
Fig. 5.
Architecture of the capsule network for the prediction of COVID-19 and CAP.
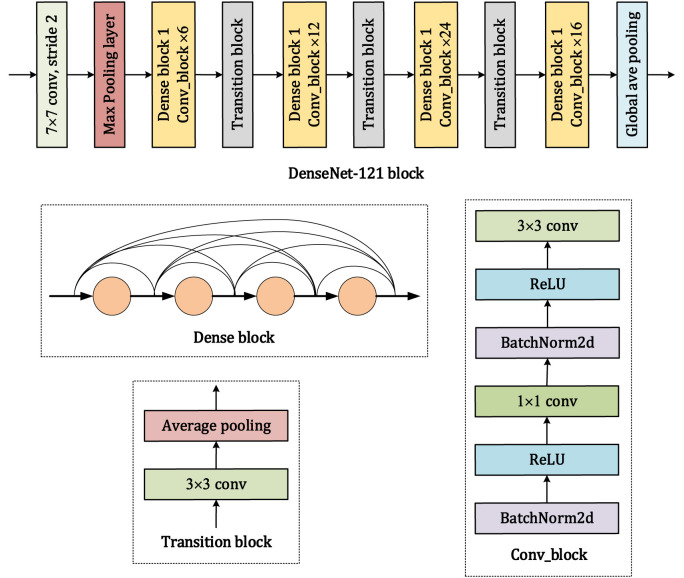
The input image measures 512 × 512 pixels. The first three stages of the pretrained DenseNet-121 [48] are used as a feature extraction block. As shown in Fig. 6 , the feature extraction block is comprised of a convolutional layer with 7 × 7 filters, a max pooling layer, four dense blocks, three transition layers, and a global average pooling layer. The dense block consists of different numbers of conv_block units, which perform batch normalization, ReLU activation, and convolution with 1 × 1 and 3 × 3 filters. A transition layer is placed between adjacent dense blocks. Finally, 1024 features are output from the DenseNet-121 block and transmitted to the primary capsule layer.
Fig. 6.
Architecture of DenseNet-121 in the capsule network.
Each capsule module consists of a primary capsule layer and two convolution capsule layers. The primary capsule layer is preceded by a convolutional layer (512 kernels of size 1 × 1), which processes the output features of DenseNet-121. Dynamic routing is followed by the primary capsule layer, which is used to reshape the output of the former convolutional block.
The probability of two categories is obtained by the dense capsule layer. The norm of the two capsules (i.e., the output of the capsule network) is input into a SoftMax operation to produce the final prediction.
A spread loss function reduces the sensitivity of training to the model initialization and super-parameters. The following spread loss function was used to train the network:
where and represent the activation values of the target and the i-th position from the target, respectively.
3.6. Ablation experiments
Three comparative experiments were conducted. The first attempted to determine whether the segmentation of the lung field improves the performance of distinguishing COVID-19 from CAP. For this, the MIP images generated by the original CT images without lung segmentation and the intrapulmonary CT images were input to the capsule network for prediction.
The second experiment attempted to determine whether the MIP images are useful. Following our previous study [13], all intrapulmonary slices were directly fed into the capsule network and the slice-level predictions were output. Majority voting was then utilized to produce the final patient predictions.
The third experiment examined whether the capsule network affects the classification performance of COVID-19 and CAP. In this experiment, the capsule modules were replaced by the vanilla DenseNet-121 blocks.
3.7. Experimental setup
During the experiments on the lung segmentation network, we marked the lung fields on 161 CT scans, including 10,280 image slices of COVID-19. The dataset was divided into training, validation, and testing sets at a ratio of 7:1:2. To train the capsule network on our lab dataset, 333 MIP images generated from 333 CT scans were divided into training, validation, and testing sets at a ratio of 8:1:1. The CC-CCII, TCIA, and Dongguan datasets were used as external independent datasets for testing. Data augmentation was implemented in the training stage via scaling and random rotation in the horizontal and vertical directions. Early stopping was adopted to alleviate the problem of overfitting when the validation accuracy did not increase over five epochs.
For the segmentation task, the batch size was set to 8, the initial learning rate was 1 × 10−4, the Adam optimizer was used, and the number of epochs was fixed to 50. For the classification task, the batch size was set to 16, the number of epochs was 25, the initial learning rate was 1 × 10−4, the Adam optimizer was used, and the number of dynamic routing iterations of the capsule network was set to 3. The experiments were implemented using PyTorch as a deep learning framework. The models were trained on a workstation with an Intel Core I7-9700 3.00 GHz CPU and four NVIDIA GeForce RTX 2080 Ti GPUs.
3.8. Evaluation metrics
The intersection over union (IoU) and Dice coefficient were used to evaluate the lung segmentation performance. The accuracy, precision, sensitivity, specificity, and area under the curve (AUC) were used to evaluate the classification models.
For the IoU and Dice metrics, TP is the number of true positives and FN is the number of false negatives. In the classification metrics, TP indicates the number of COVID-19 patients correctly classified as COVID-19 patients by the proposed model, FP (false positive) denotes the number of CAP patients falsely classified as COVID-19 patients, FN denotes the number of COVID-19 patients falsely classified as CAP patients, and TN (true negative) indicates the number of CAP patients correctly classified as CAP patients.
4. Results
4.1. Lung segmentation
The lung segmentation performance using the six different models is summarized in Table 2 . LinkNet achieves the best performance, with IoU and Dice coefficient scores of 0.967 and 0.983. This confirms that LinkNet is beneficial for lung segmentation. Conversely, R2U-Net gives the minimum IoU (0.928) and Dice coefficient (0.962).
Table 2.
Performance of the six lung segmentation networks.
| Model | IoU | Dice |
|---|---|---|
| U-Net [54] | 0.962 | 0.980 |
| LinkNet | 0.967 | 0.983 |
| R2U-Net [50] | 0.928 | 0.962 |
| Attention U-Net | 0.951 | 0.974 |
| U-Net++ [52,53] | 0.936 | 0.966 |
| CE-Net | 0.964 | 0.981 |
* Bold font indicates the network with the best performance.
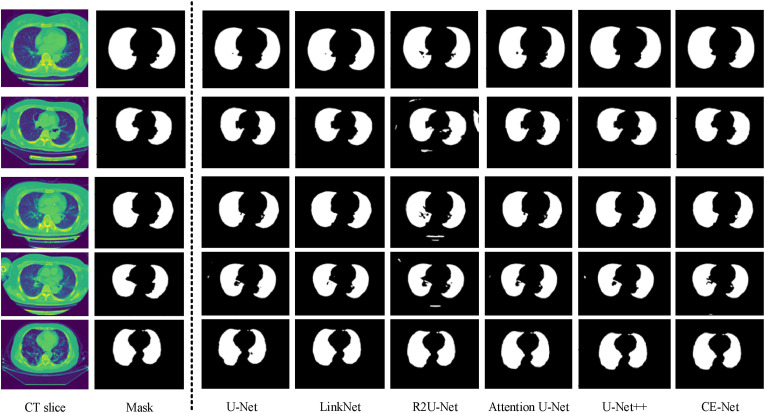
Some examples of the lung segmentation results extracted from COVID-19 CT images are depicted in Fig. 7 . Under-segmented regions can be observed in the results given by U-Net, R2U-Net, Attention U-Net, U-Net++, and CE-Net. Overall, parts of the CT scanner bed are incorrectly detected and segmented as lung field regions.
Fig. 7.
Examples of lung segmentation using different networks.
4.2. Prediction of capsule network using three different backbones on lab dataset
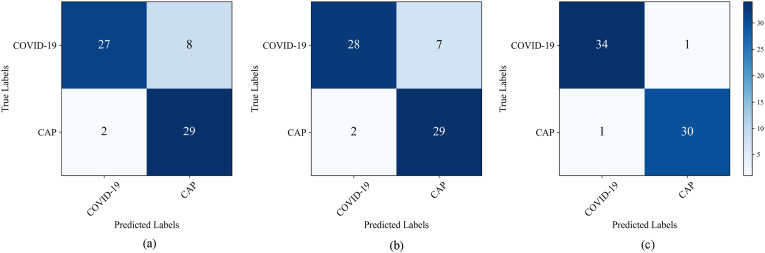
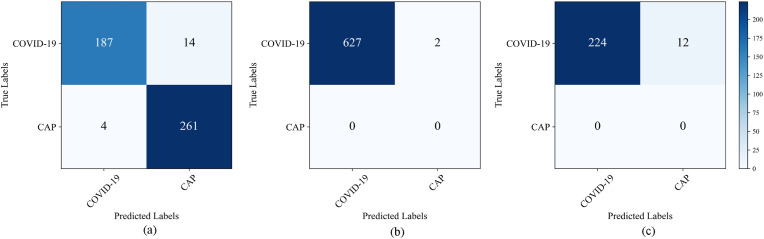
Fig. 8 shows the confusion matrix of the capsule network on the lab dataset with different feature extraction modules (ResNet-50, Inception-V3, and DenseNet-121). This shows which CNN feature extractor achieves the best classification accuracy. The testing dataset contains 66 MIP images, 35 for COVID-19 and 31 for CAP. The ResNet-50, Inception-V3, and DenseNet-121 modules correctly identified 27, 28, and 34 COVID-19 patients and 29, 29, and 30 CAP patients, respectively.
Fig. 8.
Confusion matrix for the classification of COVID-19 and CAP using three different feature extraction modules. (a) ResNet-50; (b) Inception-V3; (c) DenseNet-121.
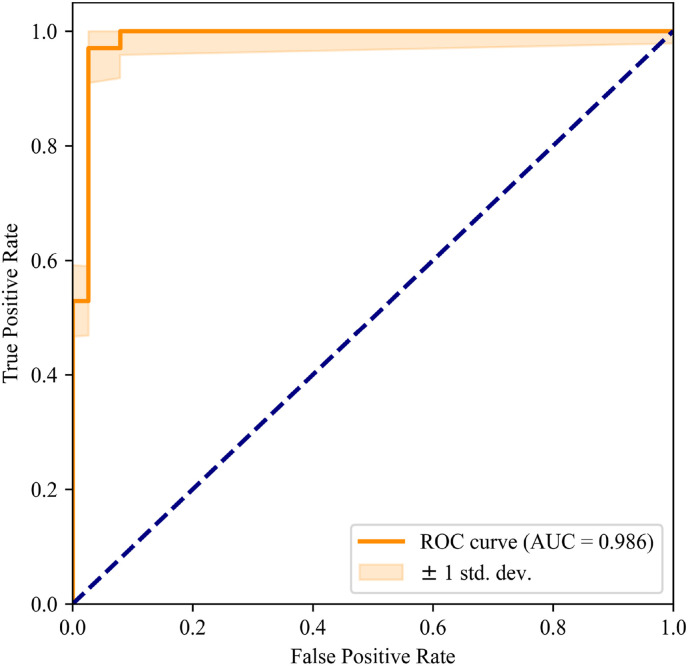
The number of parameters, accuracy, precision, sensitivity, and specificity scores using the ResNet-50, Inception-V3, and DenseNet-121 modules are reported in Table 3 . The capsule network using DenseNet-121 achieves the best performance in classifying COVID-19 and CAP with the fewest training parameters (accuracy of 97.0%, precision of 0.971, sensitivity of 0.971, specificity of 0.968, and AUC of 0.986; see Fig. 9 ). In second place, Inception-V3 uses the most training parameters to achieve 86.4% accuracy and 0.883 AUC. The ResNet-50 backbone gives the minimum accuracy (84.9%), precision (0.931), sensitivity (0.771), and specificity (0.935). We can see that the capsule network with the DenseNet-121 backbone produces the best overall performance on the COVID-19 and CAP categories.
Table 3.
Performance comparison of different feature extraction modules using capsule network.
| Model | Params. (M) | Accuracy | Precision | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|
| ResNet-50 | 9.63 | 0.849 | 0.931 | 0.771 | 0.935 | 0.910 |
| Inception-V3 | 9.92 | 0.864 | 0.933 | 0.800 | 0.935 | 0.883 |
| DenseNet-121 | 8.04 | 0.970 | 0.971 | 0.971 | 0.968 | 0.986 |
* Bold font indicates the best value among the three models.
Fig. 9.
ROC curve of the capsule network with DenseNet-121.
4.3. Predictions for other datasets
Fig. 10 and Table 4 summarize the performance of the capsule network on the CC-CCII, TCIA, and Dongguan datasets. The experiments confirm the capsule network is beneficial and robust for COVID-19 diagnosis. The CC-CCII dataset consists of COVID-19 and CAP cases. As shown in Fig. 9(a), for the COVID-19 category, 187 of 201 items are correctly predicted, while for the CAP category, the proposed method correctly classifies 261 of 265 items. Moreover, the accuracy, precision, sensitivity, specificity, and AUC in terms of distinguishing the two categories are 0.961, 0.979, 0.930, 0.985, and 0.971, respectively. The TCIA and Dongguan datasets only consist of COVID-19 patients. As shown in Fig. 10(b) and (c), the number of correctly predicted COVID-19 cases is 627 out of 629 and 224 out of 236 cases. The accuracy of COVID-19 diagnosis with the TCIA and Dongguan datasets is 99.7% and 94.9%, respectively. In this case, we conclude that the proposed method is beneficial and robust for the diagnosis of COVID-19 on multiple datasets.
Fig. 10.
Confusion matrix on other datasets. (a) CC-CCII dataset; (b) TCIA dataset; (c) Dongguan dataset.
Table 4.
Performance comparison on different datasets.
| Dataset | Accuracy | Precision | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|
| CC-CCII | 0.961 | 0.979 | 0.930 | 0.985 | 0.971 |
| TCIA | 0.997 | – | – | – | – |
| Dongguan | 0.949 | – | – | – | – |
4.4. Results of the ablation experiment
Ablation experiments were conducted to analyze the proposed method's key components. As shown in Table 5 , the main components are the lung segmentation, MIP image generation, and capsule network. Without applying lung segmentation before the acquisition of MIP images, the accuracy decreases to 63.5%. In this situation, the MIP images are directly generated by the CT scan and may include redundant information, such as the patient table and bone information. If we use the raw CT images rather than the MIP images (i.e., the patient-level prediction is produced by majority voting on the predictions of the capsule network via each slice of the CT images), the accuracy is only 85.7%, approximately 11.3% lower than when using the MIP images. Removing the capsule network and only using the DenseNet-121 network for classification reduces the accuracy to 81.8%. Thus, we conclude that lung segmentation, MIP images, and the capsule network are all beneficial to distinguishing COVID-19 from CAP using CT images. The three components of the proposed method help boost the classification performance.
Table 5.
Performance comparison with different pretraining blocks.
| Ablation experiment | Accuracy |
|---|---|
| Lung segmentation (−) | 63.5% |
| MIP (−) | 85.7% |
| Capsule networks (−) | 81.8% |
| Our method | 97.0% |
4.5. Comparison of our method with current state-of-the-art techniques
We investigated other state-of-the-art methods for classifying COVID-19 and compared them with our proposed method. The results in Table 6 show that our proposed method achieves an accuracy of 0.970, outperforming all state-of-the-art methods except for our previous approach. The sensitivity of our method is higher than or comparable to that of the pipeline mimicking radiologist [13], a combination of CNN and SVM [55], multi-instance learning and a long short-term memory (LSTM) network [56], weakly supervised multi-scale learning [4], [12], a 2D CNN [57], a semi-supervised learning strategy with multi-view fusion [58], the BigBiGAN framework [59], the pretrained EfficientNet-b7 [60], and 3D ResNet-34 with attention modules [23].
Table 6.
Performance of our method against state-of-the-art methods.
| Ref | Dataset | Method | Performance |
|||
|---|---|---|---|---|---|---|
| Acc. | Sen. | Spe. | AUC | |||
| Our proposed method | 156 patients (56 COVID-19 and 100 CAP) |
|
0.970 | 0.971 | 0.968 | 0.986 |
| HU et al., 2022 [4] | 450 patient scans (150 of COVID-19, CAP and NP) |
|
0.891 | 0.870 | 0.862 | 0.906 |
| Qi et al., 2022 [13] | 157 patients (57 COVID-19 and 100 CAP) |
|
0.971 | 0.959 | 0.981 | 0.992 |
| Ibrahim et al., 2022 [29] | 2984 patients (COVID-19: 1396; non-COVID-19: 1588) |
|
0.95 | 0.95 | 0.96 | |
| Erdal et al., 2022 [55] | 2496 CT scans (1428 COVID‐19 and 1068 CAP) |
|
0.932 | 0.858 | 0.993 | 0.984 |
| Xu et al., 2022 [56] | 515 patients (204 COVID-19 and 311 CAP) |
|
– | 0.862 | 0.980 | 0.956 |
| Li et al., 2022 [61] | 4352 CT scans (1292 COVID-19, 1735 CAP, and 1325 non-pneumonia) |
|
– | 0.885 | 0.940 | 0.955 |
| Zhu et al., 2022 [58] | 2522 patients (1495 COVID-19 and 1027 CAP) |
|
0.920 | 0.931 | 0.904 | 0.963 |
| Qi et al., 2021 [12] | 241 patients (COVID-19: 141; CAP: 100) |
|
0.959 | 0.972 | 0.941 | 0.955 |
| Javaheri et al., 2021 [57] | 335 CT scans (111 COVID-19, 115 CAP, 109 Normal) |
|
0.933 | 0.909 | 1.00 | 0.940 |
| Basset et al., 2021 [60] | 305 CT scans (169 COVID-19; 60 CA; 76 Normal) |
|
0.968 | – | – | 0.988 |
| Ouyang et al., 2020 [23] | - 3645 CT images (COVID-19: 2565; CAP: 1080) |
|
0.875 | 0.869 | 0.901 | 0.944 |
| Song et al., 2020 [59] | 201 CT images (COVID-19: 98; non-COVID-19: 103) |
|
– | 0.920 | 0.910 | 0.972 |
4.6. Interpretation of our method using t-SNE
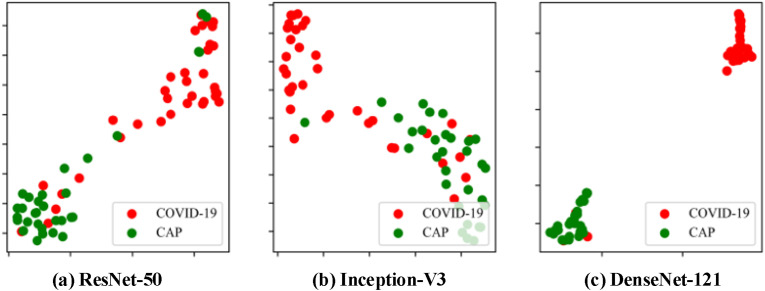
As described in Fig. 11 , the full complexity of distinguishing COVID-19 from CAP can be illustrated by visualizing the parameter space of patients with COVID-19 and CAP from our lab testing data using the t-distributed stochastic neighbor embedding (t-SNE) method [62]. Although COVID-19 and CAP produce highly similar CT images, the predictions of our proposed capsule network using three different backbones have a significant non-overlap. The medoid of the COVID-19 group lies farther away from that of the CAP group in Fig. 10(c) than in Fig. 10(a) and (b). As expected, this demonstrates that our proposed method with the DenseNet-121 backbone has an extremely high discriminative power between COVID-19 and CAP.
Fig. 11.
Visualization of COVID-19/CAP parameter space with the t-SNE method using a capsule network with different backbones. Each dot represents a patient and its color represents the group. Red dots represent patients with COVID-19 and green dots represent patients with CAP. (a) ResNet-50; (b) Inception-V3; (c) DenseNet-121.
5. Discussion
5.1. Challenges in lung segmentation and distinguishing COVID-19 from CAP using CT images
Lung segmentation is a crucial preprocessing step for the classification network. It reduces the impact of tissues outside the lung field and enables the capsule network to focus on the lesions within the lung field. Moreover, the intrapulmonary parenchyma is the prerequisite for the generation of MIP images. However, segmenting the lung field in COVID-19 and CAP patients is challenging due to the impact of lesions in CT images. Six off-the-shelf CNN models (U-Net, LinkNet, R2U-Net, Attention U-Net, U-Net++, and CE-Net) were employed for lung segmentation. LinkNet outperformed the other five networks, achieving a Dice coefficient of 0.983 and an IoU of 0.967. These scores are greater than or comparable to previous results obtained using DenseNet-161 U-Net [63], Lung Seg-Net [64], and three-stage segmentation [65].
CT is one of the most widely used imaging methods in clinical practice [[66], [67], [68], [69]] and plays an important role in the diagnosis of CAP and epidemiological studies [70]. Ground-glass opacities, consolidation, and peripheral and bilateral involvement have been observed in CT images of COVID-19 [71]. However, in CT images, COVID-19 exhibits many similarities to CAP, i.e., high similarity to pneumonia of different types (especially in the early stage) and large variations in different stages of the same type [72,73]. Hence, it is vital to develop an automatic deep learning-based diagnosis algorithm specific to COVID-19. In our previous studies [12,13], the automatic pipeline mimicking radiologist and multiple-instance learning (MIL) methods were developed for distinguishing COVID-19 from CAP. These methods achieved accuracy scores of 97% and 95% and AUC scores of 0.992 and 0.955, respectively. Our proposed method outperformed the MIL method and produced comparable results to the automatic pipeline [13]. This is also evident from our t-SNE analysis and visualization of the distribution of COVID-19 and CAP cases in Fig. 10, which illustrate the complexity of the parameter space.
5.2. Advantages of MIP images
In this work, the MIP method was developed to consider the routine workflow of radiologists. In clinical practice, radiologists quickly scan the MIP images to determine the pneumonia candidates for additional investigations on specific slices. Postprocessing through MIP [17] transfers 3D voxels to the plane of projection at their highest intensity. As this improves the visibility of nodules in comparison to the presence of bronchi and vasculature, it is frequently utilized for the identification of lung nodules [74]. Inspired by this initial procedure, we proposed an MIP-based capsule network approach. The results presented herein demonstrate that the combination of MIP and CNN improves the accuracy and efficiency of COVID-19 classification.
5.3. Limitations and further studies
The proposed approach still has certain limitations. First, the size of the dataset is small, and so a more diverse population from more centers is required to verify the performance of the proposed method. Second, gathering more subjects exhibiting other pneumonia subtypes, lung diseases, and even healthy individuals would help improve the diagnosis ability. Third, building a larger dataset with linked CT and clinical data, especially data on underlying disorders, would allow for further research of the diagnosis system and the development of further functionality, including assessments of the severity of the disease. Fourth, the current method only focuses on the analysis of the lung parenchyma in CT images. Some segmentation methods of lung airways and vessels have been established using deep learning-based methods. The analysis of airways and vessels in COVID-19 patients using CT images could be considered. Finally, more advanced methods, such as GANs [75] and ensemble learning [76], could improve the diagnosis performance of COVID-19.
6. Conclusion
This work proposed a novel method of distinguishing COVID-19 from CAP using MIP images from CT scans and a capsule network. The performance of the method demonstrates the significance and effectiveness of MIP images for COVID-19 detection in CT scans. This study has demonstrated that MIP images provide conspicuous benefits when exploiting CNNs to detect COVID-19 lesions.
To the best of our knowledge, there is a high degree of similarity between COVID-19 and CAP in chest CT images. This increases the need for a diagnosis system that distinguishes COVID-19 from CAP in CT images. The proposed method combines the advantages of MIP images and capsule networks to address this issue. This system could reduce radiologists’ workloads by significantly decreasing the number of scans that need to be manually evaluated. This work provides a new direction for the usage of CT scans in COVID-19 diagnosis.
The main limitation of the present study is the small set of training data. Although data were obtained from multiple centers, the overall size of the data is relatively small. Gathering more subjects exhibiting other pneumonia subtypes would help improve the range of diagnosis. Moreover, assessments of the severity of COVID-19 should be included in further research on the diagnosis system. The proposed diagnosis system can be extended to other diseases and other modalities, such as the transfer learning of the approach for lung cancer classification.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was waived because this was a prospective study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China [grant numbers 82072008, 82270044]; the Fundamental Research Funds for the Central Universities [grant numbers N2119010, N2124006-3]; and the Foshan Special Project of Emergency Science and Technology Response to COVID-19 [grant number 2020001000376].
References
- 1.Rahaman M.M., Li C., Yao Y., Kulwa F., Rahman M.A., Wang Q., Qi S., Kong F., Zhu X., Zhao X. Identification of COVID-19 samples from chest X-Ray images using deep learning: a comparison of transfer learning approaches. J. X Ray Sci. Technol. 2020;28(5):821–839. doi: 10.3233/XST-200715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., Wang M., Fang E.F., Menpes-Smith W., Xia J. Weakly supervised deep learning for covid-19 infection detection and classification from CT images. IEEE Access. 2020;8:118869–118883. [Google Scholar]
- 5.Farooq M., Hafeez A. 2020. Covid-resnet: A Deep Learning Framework for Screening of Covid19 from Radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 6.Li J., Zhao G., Tao Y., Zhai P., Chen H., He H., Cai T. Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19. Pattern Recogn. 2021;114 doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng H., Xiong R., He R., Lin W., Hao B., Zhang L., Lu Z., Shen X., Fan T., Jiang W. CT imaging and clinical course of asymptomatic cases with COVID-19 pneumonia at admission in Wuhan, China. J. Infect. 2020;81(1):e33–e39. doi: 10.1016/j.jinf.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ardakani A.A., Kwee R.M., Mirza-Aghazadeh-Attari M., Castro H.M., Kuzan T.Y., Altintoprak K.M., Besutti G., Monelli F., Faeghi F., Acharya U.R. A practical artificial intelligence system to diagnose COVID-19 using computed tomography: a multinational external validation study. Pattern Recogn. Lett. 2021;152:42–49. doi: 10.1016/j.patrec.2021.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulati A. MedRxiv; 2020. LungAI: A Deep Learning Convolutional Neural Network for Automated Detection of COVID-19 from Posteroanterior Chest X-Rays. [Google Scholar]
- 10.Haritha D., Pranathi M.K., Reethika M. 2020 5th International Conference on Computing, Communication and Security (ICCCS) IEEE; 2020. COVID detection from chest X-rays with DeepLearning: CheXNet; pp. 1–5. [Google Scholar]
- 11.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid AI Development Cycle for the Coronavirus (Covid-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring Using Deep Learning CT Image Analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
- 12.Qi S., Xu C., Li C., Tian B., Xia S., Ren J., Yang L., Wang H., Yu H. DR-MIL: deep represented multiple instance learning distinguishes COVID-19 from community-acquired pneumonia in CT images. Comput. Methods Progr. Biomed. 2021;211 doi: 10.1016/j.cmpb.2021.106406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Q., Qi S., Wu Y., Li C., Tian B., Xia S., Ren J., Yang L., Wang H., Yu H. Fully automatic pipeline of convolutional neural networks and capsule networks to distinguish COVID-19 from community-acquired pneumonia via CT images. Comput. Biol. Med. 2022;141 doi: 10.1016/j.compbiomed.2021.105182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotzinger D.C., Qanadli S.D. Should vascular abnormalities Be integrated into the chest CT imaging signature of coronavirus disease 2019? Chest. 2021;159(5):2107–2108. doi: 10.1016/j.chest.2020.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poletti J., Bach M., Yang S., Sexauer R., Stieltjes B., Rotzinger D.C., Bremerich J., Sauter A.W., Weikert T. Automated lung vessel segmentation reveals blood vessel volume redistribution in viral pneumonia. Eur. J. Radiol. 2022;150 doi: 10.1016/j.ejrad.2022.110259. [DOI] [PubMed] [Google Scholar]
- 16.Lins M., Vandevenne J., Thillai M., Lavon B.R., Lanclus M., Bonte S., Godon R., Kendall I., De Backer J., De Backer W. Assessment of small pulmonary blood vessels in COVID-19 patients using HRCT. Acad. Radiol. 2020;27(10):1449–1455. doi: 10.1016/j.acra.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prokop M., Shin H.O., Schanz A., Schaefer-Prokop C.M. Use of maximum intensity projections in CT angiography: a basic review. Radiographics. 1997;17(2):433–451. doi: 10.1148/radiographics.17.2.9084083. [DOI] [PubMed] [Google Scholar]
- 18.Zheng S., Guo J., Cui X., Veldhuis R.N., Oudkerk M., Van Ooijen P.M. Automatic pulmonary nodule detection in CT scans using convolutional neural networks based on maximum intensity projection. IEEE Trans. Med. Imag. 2019;39(3):797–805. doi: 10.1109/TMI.2019.2935553. [DOI] [PubMed] [Google Scholar]
- 19.Abdulkareem K.H., Mostafa S.A., Al-Qudsy Z.N., Mohammed M.A., Al-Waisy A.S., Kadry S., Lee J., Nam Y. Automated system for identifying COVID-19 infections in computed tomography images using deep learning models. J. Healthc. Eng. 2022;2022 doi: 10.1155/2022/5329014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Nagi A.T., Awan M.J., Mohammed M.A., Mahmoud A., Majumdar A., Thinnukool O. Performance Analysis for COVID-19 Diagnosis using custom and state-of-the-art deep learning models. Appl. Sci. 2022;12(13):6364. [Google Scholar]
- 21.Nwosu L., Li X., Qian L., Kim S., Dong X. 2021. Semi-supervised Learning for COVID-19 Image Classification via ResNet. arXiv preprint arXiv:2103.06140. [Google Scholar]
- 22.Waheed A., Goyal M., Gupta D., et al. Covidgan: data augmentation using auxiliary classifier gan for improved covid-19 detection[J] IEEE Access. 2021;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imag. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 24.Ye Z., Zhang Y., Wang Y., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur. Radiol. 2020;30(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammed M.A., Al-Khateeb B., Yousif M., Mostafa S.A., Kadry S., Abdulkareem K.H., Garcia-Zapirain B. Novel crow swarm optimization algorithm and selection approach for optimal deep learning COVID-19 diagnostic model. Comput. Intell. Neurosci. 2022;2022 doi: 10.1155/2022/1307944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeed M., Ahsan M., Saeed M.H., Rahman A.U., Mehmood A., Mohammed M.A., Jaber M.M., Damaševičius R. An optimized decision support model for COVID-19 diagnostics based on complex fuzzy hypersoft mapping. Mathematics. 2022;10(14):2472. [Google Scholar]
- 27.Mahmoudi R., Benameur N., Mabrouk R., Mohammed M.A., Garcia-Zapirain B., Bedoui M.H. A deep learning-based diagnosis system for COVID-19 detection and pneumonia screening using CT imaging. Appl. Sci. 2022;12(10):4825. [Google Scholar]
- 28.Shamim S., Awan M.J., Mohd Zain A., Naseem U., Mohammed M.A., Garcia-Zapirain B. Automatic COVID-19 lung infection segmentation through modified unet model. J. Healthc. Eng. 2022;2022 doi: 10.1155/2022/6566982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Ibrahim D.A., Zebari D.A., Mohammed H.J., Mohammed M.A. Effective hybrid deep learning model for COVID‐19 patterns identification using CT images. Expet Syst. 2022 doi: 10.1111/exsy.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawan S., Rajan J. Capsule Networks for Image Classification: A Review. Neurocomputing. 2022;509:102–120. [Google Scholar]
- 31.Fuchs A., Pernkopf F. 2020. Wasserstein Routed Capsule Networks. arXiv preprint arXiv:2007.11465. [Google Scholar]
- 32.Zhao L., Wang X., Huang L. 2020. An Efficient Agreement Mechanism in CapsNets by Pairwise Product. arXiv preprint arXiv:2004.00272. [Google Scholar]
- 33.Gu J. Interpretable graph capsule networks for object recognition. Proc. AAAI Conf. Artif. Intell. 2021;35(2):1469–1477. [Google Scholar]
- 34.Lenssen J.E., Fey M., Libuschewski P. Group equivariant capsule networks. Adv. Neural Inf. Process. Syst. 2018;31 [Google Scholar]
- 35.Sabour S., Frosst N., Hinton G.E. Dynamic routing between capsules. Adv. Neural Inf. Process. Syst. 2017;30 [Google Scholar]
- 36.Phaye S.S.R., Sikka A., Dhall A., Bathula D.R. Asian Conference on Computer Vision. Springer; 2018. Multi-level dense capsule networks; pp. 577–592. [Google Scholar]
- 37.Jia B., Huang Q., DE-CapsNet A diverse enhanced capsule network with disperse dynamic routing. Appl. Sci. 2020;10(3):884. [Google Scholar]
- 38.Ren H., Su J., Lu H. 2019. Evaluating Generalization Ability of Convolutional Neural Networks and Capsule Networks for Image Classification via Top-2 Classification. arXiv preprint arXiv:1901.10112. [Google Scholar]
- 39.Yang S., Lee F., Miao R., Cai J., Chen L., Yao W., Kotani K., Chen Q., Rs-CapsNet An advanced capsule network. IEEE Access. 2020;8:85007–85018. [Google Scholar]
- 40.Saif A., Imtiaz T., Rifat S., Shahnaz C., Zhu W.-P., Ahmad M.O. CapsCovNet: a modified capsule network to diagnose Covid-19 from multimodal medical imaging. IEEE Trans. Artif. Intell. 2021;2(6):608–617. doi: 10.1109/TAI.2021.3104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan H., Xu X., Zheng T., Li Z., Zhao M., Cui X. DenseCapsNet: detection of COVID-19 from X-ray images using a capsule neural network. Comput. Biol. Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta P., Siddiqui M.K., Huang X., Morales-Menendez R., Pawar H., Terashima-Marin H., Wajid M.S. COVID-WideNet—a capsule network for COVID-19 detection. Appl. Soft Comput. 2022;122 doi: 10.1016/j.asoc.2022.108780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F., Lu X., Yuan J. MHA-CoroCapsule: multi-head attention routing-based capsule network for COVID-19 chest X-ray image classification. IEEE Trans. Med. Imag. 2021;41(5):1208–1218. doi: 10.1109/TMI.2021.3134270. [DOI] [PubMed] [Google Scholar]
- 44.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark K., Vendt B., Smith K., Freymann J., Kirby J., Koppel P., Moore S., Phillips S., Maffitt D., Pringle M. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imag. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaurasia A., Culurciello E. 2017 IEEE Visual Communications and Image Processing (VCIP) IEEE; 2017. Linknet: exploiting encoder representations for efficient semantic segmentation; pp. 1–4. [Google Scholar]
- 47.Ronneberger O., Fischer P., Brox T., U-net . International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 48.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 49.Fei-Fei L., Deng J., Li K. ImageNet: constructing a large-scale image database. J. Vis. 2009;9(8) 1037-1037. [Google Scholar]
- 50.Alom M.Z., Yakopcic C., Hasan M., et al. Recurrent residual U-Net for medical image segmentation[J] J. Med. Imaging. 2019;6(1) doi: 10.1117/1.JMI.6.1.014006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oktay O., Schlemper J., Folgoc L.L., Lee M., Heinrich M., Misawa K., Mori K., McDonagh S., Hammerla N.Y., Kainz B. 2018. Attention U-Net: Learning where to Look for the Pancreas. arXiv preprint arXiv:1804.03999. [Google Scholar]
- 52.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. Unet++: a nested u-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Unet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imag. 2019;39(6):1856–1867. doi: 10.1109/TMI.2019.2959609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaziya H., Shyamala K., Zaheer R. 2018 International Conference on Communication and Signal Processing (ICCSP) IEEE; 2018. Automatic lung segmentation on thoracic CT scans using U-Net convolutional network. 0643-0647. [Google Scholar]
- 55.İn E., Geçkil A.A., Kavuran G., Şahin M., Berber N.K., Kuluöztürk M. Using artificial intelligence to improve the diagnostic efficiency of pulmonologists in differentiating COVID‐19 pneumonia from community‐acquired pneumonia. J. Med. Virol. 2022;94(8):3698–3705. doi: 10.1002/jmv.27777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu F., Lou K., Chen C., Chen Q., Wang D., Wu J., Zhu W., Tan W., Zhou Y., Liu Y. An original deep learning model using limited data for COVID‐19 discrimination: a multicenter study. Med. Phys. 2022;49(6):3874–3885. doi: 10.1002/mp.15549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Javaheri T., Homayounfar M., Amoozgar Z., Reiazi R., Homayounieh F., Abbas E., Laali A., Radmard A.R., Gharib M.H., Mousavi S.A.J. CovidCTNet: an open-source deep learning approach to diagnose covid-19 using small cohort of CT images. NPJ Dig. Med. 2021;4(1):29. doi: 10.1038/s41746-021-00399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Q., Zhou Y., Yao Y., Sun L., Shi F., Shao W., Zhang D., Shen D. Semi-supervised multi-view fusion for identifying CAP and COVID-19 with unlabeled CT images. IEEE Trans. Emerg. Topics Comput. Intell. 2022:1–13. [Google Scholar]
- 59.Song J., Wang H., Liu Y., Wu W., Dai G., Wu Z., Zhu P., Zhang W., Yeom K.W., Deng K. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur. J. Nucl. Med. Mol. Imag. 2020;47(11):2516–2524. doi: 10.1007/s00259-020-04929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdel-Basset M., Hawash H., Moustafa N., Elkomy O.M. nnTwo-stage deep learning framework for discrimination between COVID-19 and community-acquired pneumonia from chest CT scans. Pattern Recogn. Lett. 2021;152:311–319. doi: 10.1016/j.patrec.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van der Maaten L., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;986:2579–2605. [Google Scholar]
- 63.Qiblawey Y., Tahir A., Chowdhury M.E., Khandakar A., Kiranyaz S., Rahman T., Ibtehaz N., Mahmud S., Maadeed S.A., Musharavati F. Detection and severity classification of COVID-19 in CT images using deep learning. Diagnostics. 2021;11(5):893. doi: 10.3390/diagnostics11050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pawar S.P., Talbar S.N. LungSeg-Net: lung field segmentation using generative adversarial network. Biomed. Signal Process Control. 2021;64 [Google Scholar]
- 65.Osadebey M., Andersen H.K., Waaler D., Fossaa K., Martinsen A.C., Pedersen M. Three-stage segmentation of lung region from CT images using deep neural networks. BMC Med. Imag. 2021;21(1):112. doi: 10.1186/s12880-021-00640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie H., Shan H., Cong W., Liu C., Zhang X., Liu S., Ning R., Wang G. Deep efficient end-to-end reconstruction (DEER) network for few-view breast CT image reconstruction. IEEE Access. 2020;8:196633–196646. doi: 10.1109/access.2020.3033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brenner D.J., Hall E.J. Computed tomography—an increasing source of radiation exposure. N. Engl. J. Med. 2007;357(22):2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 68.Subhalakshmi R., Balamurugan S.A.A., Sasikala S. Deep learning based fusion model for COVID-19 diagnosis and classification using computed tomography images. Concurr. Eng. Res. Appl. 2022;30(1):116–127. doi: 10.1177/1063293X211021435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhuyan H.K., Chakraborty C., Shelke Y., Pani S.K. COVID‐19 diagnosis system by deep learning approaches. Expet Syst. 2021 doi: 10.1111/exsy.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim Y., Lee M.-J., Kim M.-J. European Congress of Radiology-ECR; 2015. Comparison of Radiation Dose from X-Ray, CT, and PET/CT in Paediatric Patients with Neuroblastoma Using a Dose Monitoring Program. 2015. [Google Scholar]
- 71.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fung T.S., Liu D.X. Similarities and dissimilarities of COVID-19 and other coronavirus diseases. Annu. Rev. Microbiol. 2021;75(1):1–29. doi: 10.1146/annurev-micro-110520-023212. [DOI] [PubMed] [Google Scholar]
- 73.Abdel-Basset M., Hawash H., Moustafa N., Elkomy O.M. Two-stage deep learning framework for discrimination between COVID-19 and community-acquired pneumonia from chest CT scans. Pattern Recogn. Lett. 2021;152:311–319. doi: 10.1016/j.patrec.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park E.-A., Goo J.M., Lee J.W., Kang C.H., Lee H.J., Lee C.H., Park C.M., Lee H.Y., Im J.-G. Efficacy of computer-aided detection system and thin-slab maximum intensity projection technique in the detection of pulmonary nodules in patients with resected metastases. Invest. Radiol. 2009;44(2):105–113. doi: 10.1097/RLI.0b013e318190fcfc. [DOI] [PubMed] [Google Scholar]
- 75.Monkam P., Qi S., Ma H., Gao W., Yao Y., Qian W. Detection and classification of pulmonary nodules using convolutional neural networks: a survey. IEEE Access. 2019;7:78075–78091. [Google Scholar]
- 76.Zhang B., Qi S., Monkam P., Li C., Yang F., Yao Y.-D., Qian W. Ensemble learners of multiple deep CNNs for pulmonary nodules classification using CT images. IEEE Access. 2019;7:110358–110371. [Google Scholar]