Abstract
Recombinant adenovirus serotype 5 (Ad5) vector has been widely applied in vaccine development targeting infectious diseases, such as Ebola virus disease and coronavirus disease 2019 (COVID-19). However, the high prevalence of preexisting anti-vector immunity compromises the immunogenicity of Ad5-based vaccines. Thus, there is a substantial unmet need to minimize preexisting immunity while improving the insert-induced immunity of Ad5 vectors. Herein, we address this need by utilizing biocompatible nanoparticles to modulate Ad5–host interactions. We show that positively charged human serum albumin nanoparticles ((+)HSAnp), which are capable of forming a complex with Ad5, significantly increase the transgene expression of Ad5 in both coxsackievirus-adenovirus receptor-positive and -negative cells. Furthermore, in charge- and dose-dependent manners, Ad5/(+)HSAnp complexes achieve robust (up to 227-fold higher) and long-term (up to 60 days) transgene expression in the lungs of mice following intranasal instillation. Importantly, in the presence of preexisting anti-Ad5 immunity, complexed Ad5-based Ebola and COVID-19 vaccines significantly enhance antigen-specific humoral response and mucosal immunity. These findings suggest that viral aggregation and charge modification could be leveraged to engineer enhanced viral vectors for vaccines and gene therapies.
Keywords: Adenovirus serotype 5, Vaccine, Preexisting immunity, Nanoparticles, Transgene expression
1. Introduction
Recombinant viral vectors, including adenoviruses, vesicular stomatitis virus, measles virus, and modified vaccinia Ankara, have been used extensively for vaccine development targeting infectious diseases [1], [2]. In the case of coronavirus disease 2019 (COVID-19), as of 9 September 2022, 27 of the 172 vaccines that entered clinical trials were based on viral vectors. The outstanding biological features of recombinant adenovirus vectors include good safety profiles, broad tissue tropism, and immunogenicity to induce robust adaptive immune responses. These features provide a universal platform for vaccine design. A replication-defective human adenovirus serotype 5 (Ad5)-based COVID-19 vaccine (Ad5-nCoV) expressing the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is safe and immunogenic in healthy adults, conferring protective efficacy against symptomatic COVID-19 cases [3], [4], [5]. The World Health Organization has issued an Emergency Use Listing for Ad5-nCoV (Convidecia), expanding access to COVID-19 vaccination in lower income countries. Ad5-based vaccines also elicit protective immunity against other pathogens, such as Ebola virus [6], [7], [8], [9], Zika virus [10], [11], and Mycobacterium tuberculosis [12], [13].
By expressing antigens of interest in host cells, Ad5-based vaccines can induce robust humoral and T-cell immunity. However, Ad5 frequently infects humans and generates preexisting immunity (PEI) in populations worldwide. This immunity can compromise the immune response of Ad5-based vaccines and remains a major obstacle in achieving optimal vaccine efficacy [4], [5], [14]. Depending on the geographic location, the global seroprevalence of Ad5 can be as high as 60%–98% [15], [16]. Several strategies have been explored to circumvent PEI against adenoviruses. In one strategy, low-seroprevalence adenoviruses are used as alternative vaccine vectors [13], [17], [18]. However, vaccines based on rare serotypes of adenovirus have exhibited lower immunogenicity than Ad5-based vaccines [19]. In the second strategy, heterologous prime-boost vaccinations are used, which are more immunogenic than homologous regimens [14], [20]. Determining the suitable prime-boost combination is time consuming, precluding the rapid development of vaccines, which is desirable under emergency circumstances. In the third strategy, chemical conjugation or encapsulation is used to shield adenovirus from preexisting neutralizing antibodies [21], [22], [23]. However, engineering adenovirus capsids while maintaining the biological functions in vivo is challenging.
Epidemiological studies have shown that exposure to inhaled airborne particles increases the incidence of viral respiratory infections [24], [25]. In vivo studies have also shown that different types of nanoparticles, including carbon, silver, and titanium dioxide, affect host susceptibility to viral infections [26], [27], [28], [29]. These studies indicate that nanoparticles with desirable properties have the potential to be used to improve the immune response of viral vectored vaccines delivered by the respiratory route. We envision that, through surface engineering, positively charged nanoparticles can interact with negatively charged Ad5 and increase Ad5–cell interaction by decreasing the electrostatic repulsion between Ad5 and the cell membrane. This would result in increased transgene expression and enhanced immune response to overcome PEI.
Herein, we explore this possibility using positively charged human serum albumin nanoparticles, designated as (+)HSAnp. Albumin has been demonstrated to be a universal protein scaffold for biomolecule delivery because of its natural biological properties and excellent safety profile, as exemplified by the approval of Abraxane [30]. In this study, human serum albumin (HSA) was used as the protein core. HSA was encapsulated in a polymeric shell via in situ polymerization [31], conjugated with an acryloyl group, and further polymerized with monomers and crosslinkers. The surface charge of (+)HSAnp was modulated by adjusting the relative amounts of the positively charged and neutral monomers.
2. Materials and methods
2.1. Cells lines and Ad5 vaccines
A549 cells, 3T3 cells, Hela cells, and angiotensin-converting enzyme 2 (ACE2)-expressing cell line (ACE2-293T cells) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific), penicillin (100 units·mL−1), and streptomycin (100 μg·mL−1). Human myeloid leukemia monocytes (THP-1) were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS, 2 mmol·L–1 L-glutamine (Thermo Fisher Scientific), penicillin (100 units·mL−1), and streptomycin (100 μg·mL−1).
The E1/E3-deleted replication-defective Ad5-based Ebola vaccine (Ad5-EBOV; expressing Ebola virus glycoprotein (GP)) and Ad5-nCoV (expressing SARS-CoV-2 S protein) vaccines were prepared as described previously [8], [32]. Ad5-luc (expressing luciferase), Ad5-green fluorescent protein (GFP) (expressing GFP), and Ad5 (empty vector) were purchased from Hanbio (China). Virus particle (VP) concentrations of Ad5 were measured using ultraviolet–visible (UV–Vis) spectrophotometry following treatment with 0.1% sodium dodecyl sulfate (SDS). One unit of optical density at 260 nm (1 cm pathlength) corresponds to 1.1 × 1012 VP·mL−1. The concentration of infectious units (IFU) was measured with a commercial kit (AdenoX Rapid Titer Kit; TaKaRa, Japan).
2.2. Preparation of bone marrow-derived dendritic cell (BMDC) and bone marrow-derived macrophage (BMM)
Primary mouse BMDCs and BMMs were isolated and differentiated as described previously. In brief, bone marrow cells isolated from the femurs and tibias of BALB/c mice were cultured in R10 containing 20 ng·mL−1 granulocyte macrophage colony stimulating factor (GM-CSF) and interleukin-4 (IL-4). On day 3 and day 5, complete and half of the culture medium were replaced, respectively. On day 7, non-adherent and loosely adherent cells were harvested as BMDCs, and adherent cells were harvested as BMMs.
2.3. Preparation of (+)HSAnp
(+)HSAnp were prepared by in situ polymerization as described previously [31], [33]. In brief, HSA (Sigma-Aldrich, USA) was conjugated with acryloyl groups through a reaction with N-acryloxy succinimide (Sigma-Aldrich) in molar ratios of 1:20 at room temperature (RT) for 1 h. Modified HSA was then encapsulated in a polymer shell formed by radical polymerization using acrylamide, N-(3-aminopropyl) methacrylamide (Apm), bis-methylacrylamide, and ammonium persulfate/tetramethylethylenediamine at RT for 1 h. The zeta potential of (+)HSAnp was adjusted by the adding an appropriate amount of Apm. The molar ratios of HSA, acrylamide (the neutral monomer), Apm (the positively charged monomer) and bis-methylacrylamide (the crosslinker) in (+)HSAnp-1, (+)HSAnp-2, (+)HSAnp-3, and (+)HSAnp-4 were 1:3000:100:300, 1:3000:200:300, 1:3000:300:300, and 1:3000:400:300, respectively. The positively charged (+)HSAnp was purified by means of ion-exchange chromatography. The protein concentrations of (+)HSAnp were determined by means of bicinchoninic acid assay (BCA). The purity of (+)HSAnp was analyzed via high-performance liquid chromatography (HPLC) with a G3000SWXL column (Tosoh, Japan). Dynamic light scattering (DLS) was used to measure the size and zeta potential of HSA and (+)HSAnp.
2.4. Preparation and characterization of Ad5/(+)HSAnp
The Ad5/(+)HSAnp complex was formed by mixing Ad5 and (+)HSAnp in 10 mmol·L–1 phosphate buffer (PB; pH 7.0) for 10 min at RT. To characterize the Ad5/(+)HSAnp, 3 × 1010 viral particles of Ad5-nCoV were incubated with 33 ng, 330 ng, 3.3 μg, or 33 μg of (+)HSAnp (1:1.0 × 10, 1: 1.0 × 102, 1: 1.0 × 103, or 1: 1.0 × 104 molar ratio), respectively, in 1 mL of buffer. Size and zeta potential measurements of Ad5 and Ad5/(+)HSAnp were performed on the Zetasizer Nano instrument (Malvern, UK).
2.5. Negative-stain electron microscopy
Samples of (+)HSAnp, Ad5-EBOV, or Ad5-EBOV/(+)HSAnp were loaded onto carbon-coated copper grids and incubated for 1 min. The grid was then stained with 2% (w/v) uranyl acetate. Micrographs were acquired using a transmission electron microscope (JEM-1200EX; JEOL, Japan).
2.6. In vitro transgene expression
A549 cells, 3T3 cells, Hela cells, THP-1, BMDC, and BMM were seeded into 96-well cell culture plates (Corning, USA) at a density of 1 × 104 cells·well−1. After overnight incubation, the cells were transfected with Ad5-luc or Ad5-GFP (1 × 105 IFU·well−1), either alone or complexed with (+)HSAnp (100 ng·well−1). The cells were incubated for 24 h, followed by transgene expression analysis. The luciferase expression of Ad5-luc was measured using a luciferase assay kit (Promega, USA). The GFP expression of Ad5-luc was measured by means of fluorescence imaging or flow cytometry (FCM).
2.7. Cell viability test
Cells were seeded into 96-well plates (1 × 104 cells·well−1) 1 day before the addition of (+)HSAnp (0–10 μg·mL−1) or Ad5-luc (1 × 105 IFU·well−1) complexed with (+)HSAnp (1 μg·mL−1). After 24 h of incubation, the cell viability was measured with a cell-counting kit-8 (CCK-8; Solarbio, China).
2.8. Cellular uptake of (+)HSAnp
The (+)HSAnp was labeled with fluorescein isothiocyanate (FITC; Sigma-Aldrich). A549, 3T3, and Hela cells were plated in 96-well plates at a density of 1 × 104 cells·well−1 and incubated overnight. After 24 h of incubation with FITC-labeled (+)HSAnp (100 ng·well−1), the cells were stained with Hoechst 33258 (Invitrogen, USA). Fluorescence images were captured using a cell imaging multi-mode reader (Cytation 5; BioTek, USA), and the cellular FITC intensity was determined by means of quantitative analysis according to the instrument’s instructions.
2.9. Quantitative real-time polymerase chain reaction (qRT-PCR)
Hela and 3T3 cells were plated in six-well plates at a density of 3 × 105 cells·well−1 and incubated overnight. Cells were infected with Ad5-luc (3 × 106 IFU·well−1) either alone or complexed with (+)HSAnp-4 (1 μg·mL−1). After 2 h of incubation with Ad5-luc/(+)HSAnp-4, the cells were rinsed with phosphate-buffered saline (PBS) and harvested for genomic DNA isolation (Qiagen, Germany). TaqMan-based qRT-PCR assays were performed according to the manufacturer’s instructions (Thermo Fisher Scientific). The primers and probes used in the qRT-PCR reactions were Ad5_forward (TTGCGTCGGTGTTGGAGA), Ad5_reverse (AGGCCAAGATCGTGAAGAACC), and Ad5_probe (carboxyfluorescein (FAM)-CTGCACCACATTTCGGCCCCAC-carboxytetramethylrhodamine (TAMRA)). Serially diluted and purified Ad5-luc was used for the standards in an absolute quantitation qRT-PCR assay. The cellular uptakes of Ad5-luc were normalized to the genomic DNA amounts of cells.
2.10. RNA interference
Double-stranded small interfering RNAs (siRNAs) with two nucleotide overhangs targeted against human coxsackievirus-adenovirus receptor (CAR) and a negative control siRNA were used in this study, as described previously [34]. The siRNAs targeting CAR gene (CXADR; siCXADR), including siCXADR-1 (sense sequence: 5′-GCUACAUCGGCAGUAAUCAdTdT-3′; antisense sequence: 5′-UGAUUACUGCCGAUGUAGCdTdT-3′) and siCXADR-2 (sense sequence: 5′-GGUGGAUCAAGUGAUUAUUdTdT-3′; antisense sequence: 5′-AAUAAUCACUUGAUCCACCdTdT-3′), and the negative control siRNA (siCon) (sense sequence: 5′-UUCUCCGAACGUGUCACGUdTdT-3′; antisense sequence: 5′-ACGUGACACGUUCGGAGAAdTdT-3′) were purchased from Sangon Biotech (China). A549 cells were seeded into 24-well plates at a density of 1 × 105 cells·well−1. After overnight incubation, the cells were transfected with siRNAs and Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. After 24 h, CXADR knockdown was analyzed by means of fluorescence imaging and FCM.
2.11. Animal experiments
Female BALB/c mice (5–6 weeks old) were obtained from Beijing Vital River Laboratory Animal Technologies Co., Ltd. (China), and were cared for at the Animal Center of the Beijing Institute of Biotechnology under guidelines approved by the Institutional Animal Care and Use Committee (ethics numbers IACUC-DWZX-2017–020 and IACUC-DWZX-2020–059).
2.12. PEI model
BALB/c mice were intranasally inoculated with a single dose of an empty Ad5 vector (1 × 107 IFU·20 μL−1). After 4 weeks, serum samples were collected and evaluated via enzyme-linked immunosorbent assay (ELISA) to determine whether high-level antibodies against the Ad5 vector were present.
2.13. In vivo bioluminescence imaging (BLI)
BALB/c mice (n = 5) were intranasally administered 20 μL samples containing 5 × 106 IFU Ad5-luc alone or Ad5-luc complexed with 0.25, 0.50, 1.00, 2.00, or 5.00 μg (+)HSAnps (1:3.0 × 103, 1:6.0 × 103, 1:1.2 × 104, 1:2.4 × 104, or 1:5.0 × 104 molar ratio) in the presence or absence of PEI, respectively. Before in vivo BLI (IVIS Spectrum; PerkinElmer, USA), the mice were given an intraperitoneal injection of luciferin (Promega, USA) at a dose of 150 mg·kg−1 and were anesthetized through the inhalation of isoflurane. For kinetic analysis, the mice were examined with BLI at 1, 5, 10, 30, 60, and 120 days post administration. To determine the biodistribution, Ad5-luc/(+)HSAnp-4-inoculated mice were injected with luciferin, sacrificed to harvest organs (lung, heart, liver, kidney, and spleen), and examined with BLI at 10 days post administration. The bioluminescent signal was measured in relative light units (RLU) within the region of interest (ROI).
2.14. Immunization
For Ad5-EBOV, BALB/c mice (n = 10) were immunized intranasally with 5 × 106 IFU of Ad5-EBOV or Ad5-EBOV complexed with 1 μg (+)HSAnp-4 (1:1.2 × 104 molar ratio). Serum samples were collected at 2, 4, 6, 10, and 14 weeks post vaccination and assayed for Ad5-specific and GP-specific immunoglobulin G (IgG) titers. Mice were sacrificed at week 14 post vaccination, followed by an assay of GP-specific antibodies in bronchoalveolar lavage fluid (BALF) and the detection of splenic T-cell response.
For Ad5-nCoV, BALB/c mice (n = 6) were immunized intranasally with 5 × 106 IFU of Ad5-nCoV or Ad5-nCoV complexed with 1 μg of (+)HSAnp-4 (1:1.2 × 104 molar ratio). Serum samples were collected at weeks 2, 4, 8 and 12 post vaccination and assessed for S-specific IgG levels and inhibition of interaction between the SARS-CoV-2 receptor binding domain (RBD) and human ACE2 (hACE2). Neutralized antibodies against SARS-CoV-2 pseudotyped viruses were evaluated at week 4 post vaccination. Mice were sacrificed at week 12 post vaccination, followed by an assay of S-specific antibodies in BALF and the detection of splenic T-cell response.
2.15. Enzyme-linked immunosorbent assay
For antigen-specific antibody assays, 96-well plates were coated with 1 μg·mL−1 recombinant EBOLA GP lacking the transmembrane domain (GP ΔTM) (IBT Services, USA), 2 μg·mL−1 purified Ad5 hexon, or 1 μg·mL−1 recombinant SARS-CoV-2 S protein (Sino Biological, China) overnight at 4 °C. The plates were then washed with PBS containing 0.1% Tween 20 (PBST) and blocked with PBS containing 5% non-fat milk for 1 h at 37 °C. Serially diluted immune serum or BALF samples were added to the wells and incubated for 1 h at 37 °C. After washing with PBST, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or IgA (1:10 000 dilution; Abcam, UK) was added and incubated for 1 h at 37 °C. The plates were washed with PBST before the addition of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution (Solarbio), and reactions were stopped by adding stop solution (Solarbio). Absorbances at 450 nm (A450) were measured using a microplate reader (SPECTRA MAX 190; Molecular Device, USA). Antibody titers were determined as the highest reciprocal dilution that reached A450 ≥ 2.1-fold over negative background values. For the SARS-CoV-2 S-specific IgG ELISA, a neutralizing monoclonal antibody (40591-MM43; Sino Biological, China) was used to develop a standard curve for the determination of IgG concentration in serum or BALF samples.
2.16. Enzyme-linked immunospot assay
The antigen-specific cellular immune response induced by Ad5-EBOV was evaluated using an interferon (IFN)-γ enzyme-linked immunospot (ELISpot) kit (MabTech, Sweden). Cells from splenocytes were seeded into precoated plates at a density of 2 × 105 well−1 and incubated with peptide pool spanning EBOLA GP (2 μg·mL−1 of each peptide) for 16 h at 37 °C and 5% CO2. The plates were washed with PBST before the addition of the biotin-conjugated antibody against mouse IFN-γ. After 1 h of incubation at RT, the plates were washed, and streptavidin-conjugated HRP was added to the wells. 3-Amino-9-ethylcarbazole solution (BD Biosciences, USA) was used as the substrate, and the air-dried plates were measured in an ELISpot reader (SinSage Technology, China).
2.17. RBD–hACE2 blocking assay
Antibodies blocking the interaction between RBD and hACE2 were detected with a magnetic chemiluminescent immunoassay kit (Realmind Biotech, China). Serum samples (1:20 dilution) were assayed, and PBS was used as a control. Inhibition rate was defined as (1 – RLUsample/RLUPBS) × 100%.
2.18. Pseudovirus neutralization assay
The SARS-CoV-2 pseudotyped virus bearing the S protein from wild-type (WT), delta (B.1.617.2) variant, or omicron variant (BA.1.1.529) was prepared as described previously [32]. Serially diluted heat-inactivated serum samples were mixed with the titrated pseudovirus for 1 h at 37 °C and were then added to ACE2-293 T cells seeded in 96-well plates. After 48 h, the cells were lysed, and luminescence was measured. Median effective concentration (EC50) neutralization titers were calculated as the sample dilution at which luminescence readings were equal to 50% of virus-only control wells.
2.19. Biocompatibility test
BALB/c mice (n = 5) were immunized intranasally with a single dose of (+)HSAnp-4 (1 μg·mouse–1), Ad5-nCoV (5 × 106 IFU·mouse–1), Ad5-nCoV/(+)HSAnp-4 (Ad5-nCoV, 5 × 106 IFU·mouse–1; (+)HSAnp-4, 1 μg·mouse–1), or PBS as a control. One day and 7 days later, the whole blood and lungs were collected from the immunized mice for blood test and histology analysis. The serum levels of creatinine (CR), blood urea nitrogen (BUN), and uric acid (UA) were measured for renal function assessment. The serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) were evaluated for liver function analysis. Red blood cell (RBC) count, hemoglobin (HGB), and platelet (PLT) count were quantified for hematology assessment.
2.20. Statistical analysis
Data are presented as means ± standard error of the mean (SEM). Statistical differences were determined with GraphPad Prism 9 using a two-tailed unpaired t-test, Pearson correlation test, one-way analysis of variance (ANOVA) with Dunn’s multiple comparison test, or two-way ANOVA with Šidák’s multiple comparison test. A P value less than 0.05 was considered to be significant.
3. Results and discussion
3.1. Characterization of the Ad5/(+)HSAnp complex
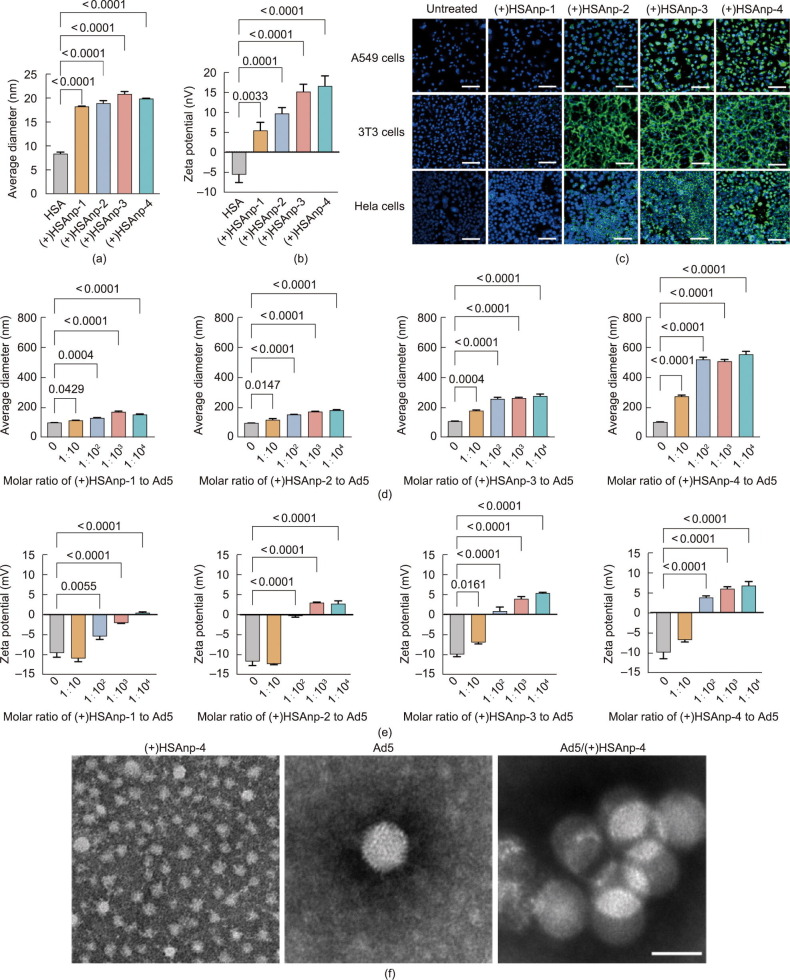
In situ polymerization provides an appealing platform for synthesizing functional biocompatible nanoparticles by generating polymers on protein surfaces [31]. As previously described [33], protein nanoparticles with similar sizes but varying surface potentials were prepared by adjusting the molar ratio of the polymerized charged monomer to the neutral monomer. In this study, the molar ratios of HSA, neutral monomer, and charged monomer in (+)HSAnp-1, (+)HSAnp-2, (+)HSAnp-3, and (+)HSAnp-4 were 1:3000:100, 1:3000:200, 1:3000:300, and 1:3000:400, respectively. Unmodified proteins were removed using ion exchange chromatography (Fig. S1 in Appendix A). Characterization of the purified (+)HSAnps was performed via DLS. The measured size of the (+)HSAnps was approximately 20 nm (Fig. 1 (a)). The zeta potential of (+)HSAnp-1, (+)HSAnp-2, (+)HSAnp-3, and (+)HSAnp-4 was +5.5, +9.8, +15.2, and +16.6 mV, respectively (Fig. 1(b)). The positively charged surfaces of (+)HSAnps enabled highly efficient intracellular delivery, which was charge dependent (Fig. 1(c) and Fig. S2 in Appendix A). The (+)HSAnps exhibited low cytotoxicity (Fig. S3 in Appendix A).
Fig. 1.
Characterization of the Ad5/(+)HSAnp complex. (a, b) DLS measurements of (a) average sizes and (b) zeta potentials of (+)HSAnp (n = 3). (c) Fluorescence microscopy images of cells treated with FITC labeled as (+)HSAnps for 24 h (scale bar:100 μm). (d, e) DLS measurements of (d) average sizes and (e) zeta potentials of Ad5 and the Ad5/(+)HSAnp complex (n = 3). (f) Representative transmission electron microscopy images of (+)HSAnp-4, Ad5, and Ad5/(+)HSAnp-4 (scale bar: 100 nm). Data are presented as means ± SEM. Statistical differences were determined using one-way ANOVA with Dunn’s multiple comparison test.
Adenoviruses are nonenveloped icosahedral viruses (90–100 nm) with negatively charged protein shells. At RT, positively charged (+)HSAnps formed complexes with Ad5 in 10 mmol·L–1 PB (pH 7.0). DLS measurements showed that the Ad5/(+)HSAnp complexes significantly increased in size as the molar ratio or surface potential of (+)HSAnps increased (Fig. 1(d)). The diameter of Ad5/(+)HSAnp-4 reached 560 nm, while Ad5 had a diameter of approximately 105 nm. Moreover, the surface potential of Ad5/(+)HSAnps was modulated. The formation of positively charged Ad5/(+)HSAnps can be achieved by increasing the molar ratio and surface potential of (+)HSAnps (Fig. 1(e)). The zeta potential of Ad5/(+)HSAnp-4 (1:1.0 × 104 molar ratio) was approximately +7 mV, which was significantly higher than that of Ad5 (approximately –10 mV). These results indicate that (+)HSAnps induced viral aggregation of Ad5, which was confirmed by transmission electron microscopy (Fig. 1(f)).
3.2. Nanocomplexed Ad5 enhances transgene expressions in vitro
To investigate whether Ad5/(+)HSAnps could enhance the delivery of Ad5, we first evaluated the impact of (+)HSAnps on the transgene expression of Ad5 encoding firefly luciferase (Ad5-luc) in vitro. The cell experiments were performed at a multiplicity of infection (MOI) of 10. As shown in Fig. 2 (a), (+)HSAnps significantly increased the transgene expression of Ad5-luc in several cell types. The enhanced transgene expression correlated with the surface potential of (+)HSAnp. In 3T3 cells, (+)HSAnp-2, (+)HSAnp-3, and (+)HSAnp-4 enhanced expression by 5.4, 8.3, and 9.8 times (Fig. 2(a)), respectively. Similar results were observed when Ad5 was used to encode GFP (Ad5-GFP). FCM and fluorescence microscopy showed that Ad5-GFP/(+)HSAnp significantly increased cellular GFP fluorescence (Figs. 2(b)–(g)). Compared with Ad5-GFP, Ad5-GFP/(+)HSAnp-1, Ad5-GFP/(+)HSAnp-2, Ad5-GFP/(+)HSAnp-3, and Ad5-GFP/(+)HSAnp-4 significantly improved the percentage of GFP-positive HeLa cells from 58% to 64%, 82%, 84%, and 85%, respectively (Fig. S4 in Appendix A). We subsequently explored whether Ad5/(+)HSAnp could enhance transgene expression in antigen-presenting cells, which play critical roles in the innate and adaptive immune responses. As shown in Figs. 2(c), (d), (f), and (g) and Fig. S4, transgene expression of Ad5-GFP/(+)HSAnp-4 was significantly higher than that of Ad5-GFP in BMDCs and BMMs, respectively. Moreover, no detectable cytotoxicity of Ad5/(+)HSAnp was observed (Fig. S5 in Appendix A).
Fig. 2.
Ad5/(+)HSAnp enhance transgene expressions in vitro. (a) Transgene expressions of Ad5-luc in complex with or without (+)HSAnp in Hela cells, A549 cells, 3T3 cells, and THP-1 (n = 5). (b–d) FCM analysis of Ad5-GFP/(+)HSAnp enhancing transgene expression of GFP in (b) Hela cells, (c) BMDCs, and (d) BMMs (n = 3). (e–g) Fluorescence microscopy of Ad5-GFP/(+)HSAnp-4 enhancing transgene expression of GFP in (e) Hela cells, (f) BMDCs, and (g) BMMs (scale bar: 100 μm). Cell experiments were performed at a MOI of 10. Data are presented as means ± SEM. Statistical differences in parts (a–d) were determined using ANOVA with Dunn’s multiple comparison test. Luc intensity: luciferase bioluminescence intensity; MFI: mean fluorescence intensity.
3.3. Investigation of factors impacting enhanced transgene expression of Ad5/(+)HSAnp
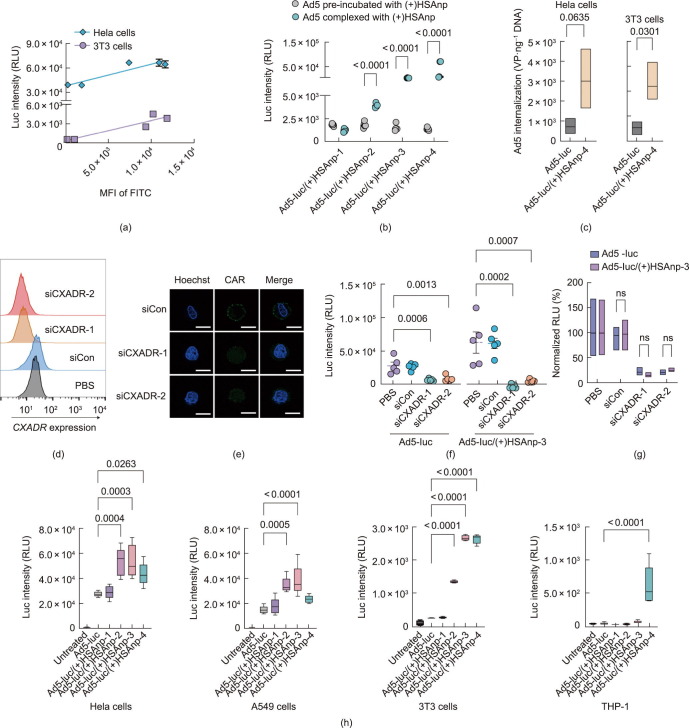
To investigate the cellular mechanism underlying the enhanced transgene expression of Ad5/(+)HSAnp, we first performed a correlation analysis and showed that the transgene expression of Ad5-luc/(+)HSAnp was positively correlated with the cellular internalization efficiency of FITC-labeled (+)HSAnp. The Pearson correlation coefficient in HeLa and 3T3 cells was 0.93 and 0.94, respectively (Fig. 3 (a)). Pre-incubation with (+)HSAnp did not mediate higher transgene expression of Ad5-luc in 3T3 cells (Fig. 3(b)), suggesting that a complex formation between Ad5 and (+)HSAnp is critical for the enhanced expression. We further explored whether the enhanced transgene expression of Ad5-luc/(+)HSAnp was associated with increased cellular internalization of Ad5. qRT-PCR was performed, with the cellular internalization of Ad5 normalized to the amount of cellular genomic DNA. As shown in Fig. 3(c), Ad5-luc/(+)HSAnp-4 increased the cell entry of Ad5 in Hela and 3T3 cells by 4.2 and 4.3 times that of Ad5, respectively.
Fig. 3.
Investigation of the cellular mechanism underlying the enhanced transgene expressions of Ad5/(+)HSAnp. (a) Correlation analysis of the transgene expression of Ad5-luc and the cellular internalization of (+)HSAnp-4 in Hela and 3T3 cells (n = 5). (b) Transgene expression of Ad5-luc pre-incubated or complexed with (+)HSAnp in 3T3 cells (n = 4). (c) qRT-PCR analysis of the cellular internalizations of Ad5 and Ad5/(+)HSAnp-4 in Hela and 3T3 cells (n = 3). (d) FCM and (e) fluorescence microscopy analysis of the knock-down of CXADR mediated by RNA interference in A549 cells (scale bar: 10 μm). (f) Absolute and (g) normalized transgene expression of Ad5-luc and Ad5-luc/(+)HSAnp-3 in CXADR-knockdown A549 cells (n = 5). (h) Transgene expression of Ad5-luc and Ad5-luc/(+)HSAnp co-incubated with Ad5-neutralizing serum in Hela cells, A549 cells, 3T3 cells, and THP-1 (n = 5). Ad5-luc and Ad5-luc/(+)HSAnp were mixed with anti-Ad5 serum (diluted at 1:4000) for 1 h at RT before transfection into cells. The serum was obtained from BALB/c mice intranasally immunized with an empty Ad5 vector at 4 weeks post administration. Cell experiments were performed at an MOI of 10. Data are presented as means ± SEM. Two-way ANOVA with Šidák’s multiple comparison test was used to determine the significance in parts (b) and (g). Two-tailed unpaired t-test was used to determine the significance within each cell line in (c). One-way ANOVA with Dunn’s multiple comparison test was used to determine the significance of the indicated comparisons within each group in (f) and (h). ns: no significance.
CAR is the primary receptor for adenovirus [35]. The capsid of Ad5 mainly consists of hexons and fibers. Fibers bind to CAR and mediate the attachment of Ad5 to host cells. Hexon is the most abundant capsid protein of Ad5 and is the main target of Ad5-neutralizing antibodies (Nabs) [36]. In contrast to the HeLa and A549 cells, the 3T3 cells and THP-1 do not express CAR [37], [38], [39]. These findings were consistent with the observations that the transgene expression of Ad5-luc in 3T3 cells and THP-1 was much lower than that in HeLa and A549 cells (Fig. 2(a)).
BMDCs and BMMs have been defined as CAR-deficient cells [40], [41], [42]. To examine whether (+)HSAnp facilitates the cell entry of Ad5 by employing other receptors in CAR-positive cells, we knocked down the CXADR through RNA interference [34] in A549 cells (Figs. 3(d) and (e)). Compared with PBS-treated cells, CXADR knockdown resulted in significantly reduced transgene expression of both Ad5-luc and Ad5-luc/(+)HSAnp (Figs. 3(f) and (g)). This finding suggests that CAR is still the primary receptor for Ad5-luc/(+)HSAnp in CAR-positive cells. As Ad5-luc/(+)HSAnp significantly enhanced cellular internalization and transgene expression of Ad5 in 3T3 cells (Figs. 2(a) and 3(c)), it is likely that a CAR-independent pathway is involved in the cellular entry of Ad5/(+)HSAnp into CAR-negative cells, although this speculation requires further investigation. Notably, we demonstrated that Ad5/(+)HSAnp inhibited the neutralizing activity of Ad5 neutralizing antibodies. In the presence of Ad5 Nabs, Ad5-luc/(+)HSAnp significantly increased transgene expression in both CAR-positive cells (Hela and A549 cells) and CAR-negative cells (3T3 cells and THP-1) (Fig. 3(h)).
3.4. Ad5/(+)HSAnp enhances transgene expression in vivo
To examine whether the in vitro findings also apply in vivo, BLI was performed to measure the transgene expression of Ad5-luc/(+)HSAnp in BALB/c mice. Following nasal instillation, Ad5-luc mainly expressed luciferase at the nasal site, whereas Ad5-luc/(+)HSAnp-3 and Ad5-luc/(+)HSAnp-4 robustly expressed luciferase in the upper chest (Fig. 4 (a)). A biodistribution analysis revealed that Ad5-luc/(+)HSAnp-4 cells mainly expressed luciferase in the lungs, rather than the heart, liver, kidney, or spleen (Figs. 4(b) and (c)). A quantitative analysis demonstrated that the in vivo enhancement of transgene expression was charge- and dose-dependent, with Ad5-luc/(+)HSAnp-4 identified as the top-performing complex (Figs. 4(d) and (e), and Figs. S6 and S7 in Appendix A). Compared with Ad5-luc, Ad5-luc/(+)HSAnp-4 induced up to 227-fold higher luciferase expression in the lungs 10 days following intranasal administration (Fig. 4(d)).
Fig. 4.
Transgene expression of Ad5/(+)HSAnp in vivo. (a) In vivo BLI of BALB/c mice at 1 day and 10 days post intranasal administration with Ad5-luc or Ad5-luc/(+)HSAnp (n = 5). (b, c) Biodistribution analysis of Ad5-luc/(+)HSAnp at 10 days post intranasal administration (n = 5). (d) Charge-dependence of (+)HSAnp in enhancing the transgene expression of Ad5-luc/(+)HSAnp in mice lungs at 10 days post intranasal administration (n = 5). (e) Dose-dependence of (+)HSAnp-4 in enhancing the transgene expression of Ad5-luc/(+)HSAnp-4 in mice lungs at 10 days post intranasal administration (n = 5). (f) Time-course analysis of the transgene expression of Ad5-luc/(+)HSAnp in mice lungs in the absence of PEI (n = 5). (g) Quantitative transgene expressions of Ad5-luc administrated 2 h before, 2 h after, or complexed with (+)HSAnp-4 (n = 4) at 10 days post intranasal administration. (h) Time-course analysis of the transgene expression of Ad5-luc/(+)HSAnp in mice lungs in the presence of PEI (n = 5). (i) Representative image of BALB/c mice at 5 days post intranasal administration in the presence of PEI (n = 5). Data are presented as means ± SEM. The bioluminescent signals in parts (b), (d), and (e) were measured after lung tissue harvest and are shown as RLU (photons·s−1). The bioluminescent signals in parts (f), (g), and (h) were measured in vivo and are shown as RLU (photons s−1·cm·–2·sr–1). The statistical differences in parts (d), (e), and (g) were determined using ANOVA with Dunn’s multiple comparison test. Statistical differences in (f) and (h) were determined using a two-way ANOVA with Šidák’s multiple comparison test.
We further investigated the in vivo kinetics of the transgene expression of Ad5-luc/(+)HSAnp. Time-course measurements detected transgene expression of Ad5-luc/(+)HSAnp-4 as early as 1 day post administration. Expression peaked on day 10 and remained higher than that of Ad5-luc until 60 days following administration (Fig. 4(f)). In addition, the in vivo transgene expressions of Ad5-luc administrated 2 h before, 2 h after, or complexed with (+)HSAnp-4 were compared at 10 days following intranasal administration. As shown in Fig. 4(g) and Fig. S8 in Appendix A, separate administration of Ad5-luc and (+)HSAnp-4 did not enhance transgene expression, which is consistent with the in vitro data (Fig. 3(b)).
In vivo enhancement of the transgene expression of nanocomplexed Ad5-luc was further evaluated in a PEI model established by the intranasal administration of 1 × 107 IFU of an empty Ad5 vector 4 weeks before intranasal instillation with Ad5-luc or Ad5-luc/(+)HSAnp. As shown in Figs. 4(h) and (i), Ad5-luc/(+)HSAnp-4 mediated 2.1- and 2.4-fold higher transgene expression in mouse lungs than Ad5-luc at 1 and 5 days, respectively, after intranasal administration.
3.5. Nanocomplexed Ad5-based Ebola and COVID-19 vaccines overcome PEI
The magnitude and persistence of antigen expression have a significant influence on adenoviral-vectored vaccine potency [43]. We investigated whether enhanced antigen expression mediated by Ad5/(+)HSAnp could induce a stronger immune response to Ad5-based vaccines in the absence and presence of PEI. We first explored this possibility using an Ad5-EBOV [8], [9] encoding an envelope GP. Untreated mice (naïve) and mice with high PEI were immunized intranasally with a single dose of 5 × 106 IFU of Ad5-EBOV or Ad5-EBOV complexed with 1 μg of (+)HSAnp-4 (1:1.2 × 104 molar ratio). The presence of PEI was confirmed by the detection of high serum IgG titers to Ad5 after the intranasal administration of 1 × 107 IFU of an empty Ad5 vector (Fig. 5 (a)). As expected, owing to PEI, PEI/Ad5-EBOV generated lower GP-specific serum IgG titers than naïve/Ad5-EBOV. In the absence of PEI—probably due to saturated immunogenicity—Ad5-EBOV/(+)HSAnp-4 did not induce a significantly higher antibody response than the response produced by Ad5-EBOV (Fig. 5(b)). However, in the presence of PEI, Ad5-EBOV/(+)HSAnp-4 induced 8.2-, 6.0-, 7.5-, 4.8-, and 5.5-fold higher GP-specific serum IgG titers than Ad5-EBOV at 2, 4, 6, 10, and 14 weeks following administration, respectively (Fig. 5(b)). To assess mucosal immunity, BALF was collected at week 14 following vaccination. As shown in Figs. 5(c) and (d), in the presence of PEI, Ad5-EBOV/(+)HSAnp-4 induced 9.6- and 6.9-fold higher GP-specific IgG and IgA titers in BALF than Ad5-EBOV, respectively. The cellular immune responses of intranasal delivery of Ad5-EBOV and Ad5-EBOV/(+)HSAnp-4 were tested using an IFN-γ ELISpot assay. However, in both the absence (naïve) and presence of PEI, no significant differences were observed between Ad5-EBOV and Ad5-EBOV/(+)HASap-4 groups (Fig. S9 in Appendix A).
Fig. 5.
Complexed Ad5-based Ebola and COVID-19 vaccines overcome PEI. (a–d) BALB/c mice (n = 10) were immunized intranasally with a single dose of 5 × 106 IFU of Ad5-EBOV or Ad5-EBOV complexed with 1 μg (+)HSAnp-4 (1:1.2 × 104 molar ratio) in the absence (naïve) or presence of PEI. (a) Ad5-specific and (b) GP-specific serum IgG titers were measured at specific time points post vaccination. BALF was collected at week 14 post vaccination and assessed for GP-specific (c) IgG and (d) IgA titers. (e–m) In the presence of PEI, BALB/c mice were immunized intranasally with a single dose of 5 × 106 IFU of Ad5-nCoV or Ad5-nCoV complexed with 1 μg (+)HSAnp-4 (1:1.2 × 104 molar ratio). (e) Ad5-specific serum IgG titers were measured before vaccination. (f) SARS-CoV-2 S-specific IgG serum concentrations were assessed at 2, 4, 8, and 12 weeks post vaccination. BALF was collected at week 12 post vaccination and assessed for (g) S-specific IgG and (h) IgA titers. (i) A biochemical assay was used to measure the serum inhibition of RBD-hACE2 interactions. (j–o) Serum neutralizing antibody titers against (j, m) SARS-CoV-2 WT, (k, n) delta variant, and (l, o) omicron variant were measured by means of pseudovirus neutralization assays at 4 weeks post vaccination. Data are presented as means ± SEM. Statistical differences in parts (b), (f), and (i) were determined using a two-way ANOVA with Šidák’s multiple comparison test. Statistical differences in (c) and (d) were determined using ANOVA with Dunn’s multiple comparison test. Statistical differences in parts (g), (h), (m), (n), and (o) were determined using a two-tailed unpaired t-test.
We further explored the potential of (+)HSAnp to enhance the immunogenicity of Ad5-nCoV [3], [4], [5]. BALB/c mice with PEI (Fig. 5(e)) were intranasally administered a single dose (5 × 106 IFU) of Ad5-nCoV or Ad5-nCoV complexed with 1 μg (+)HSAnp-4 (1:1.2 × 104 molar ratio). Antibody responses were assessed at 2, 4, 8, and 12 weeks following vaccination. Ad5-nCoV/(+)HSAnp-4 induced a robust immune response. The S-specific IgG serum concentration (μg·mL−1) reached 93.9 at 4 weeks and 99.0 at 8 weeks, representing a 2.3- and 1.9-fold increase, respectively, compared with Ad5-nCoV (Fig. 5(f)). Ad5-nCoV/(+)HSAnp-4 also induced higher specific IgA titers in BALF (Figs. 5(g) and (h)). The results of a biochemical assay based on blocking the interaction between RBD and hACE2 demonstrated that Ad5-nCoV/(+)HSAnp-4 induced higher levels of neutralizing antibodies than Ad5-nCoV at 2 and 4 weeks following vaccination (Fig. 5(i)). Moreover, in a pseudovirus neutralization assay measuring neutralizing antibodies against SARS-CoV-2 WT (Fig. 5(j)), delta variant (Fig. 5(k)), and omicron variant (Fig. 5(l)), the serum neutralization titers in the Ad5-nCoV/(+)HSAnp-4-vaccinated group were 2.2-, 2.8-, and 3.3-fold higher, respectively, than those in the Ad5-nCoV-vaccinated group at 4 weeks following vaccination (Figs. 5(m)–(o)).
3.6. In vivo biocompatibility of Ad5/(+)HSAnp nanocomplexes
Finally, we assessed whether the intranasal delivery of Ad5/(+)HSAnp-4 was well tolerated in mice. Blood tests were performed to assess renal function, liver function, and hematological profiles, including measurements of CR, BUN, UA, AST, ALT, and ALP, as well as RBC, HGB, and PLT. As shown in Fig. 6 (a), there were no significant differences in blood test results between the Ad5-nCoV/(+)HSAnp-4 and Ad5-nCoV vaccinated groups at 1 day and 7 days post-immunization. Furthermore, compared with that of PBS-treated mice, the histological analysis of the lungs of Ad5-nCoV/(+)HSAnp-4-vaccinated mice showed no abnormalities (Fig. 6(b)).
Fig. 6.
Evaluation of the safety profile of Ad5/(+)HSAnp. (a) BALB/c mice (n = 5) were intranasally administrated with a single dose of PBS, 1 μg of (+)HSAnp-4, 5 × 106 IFU of Ad5-nCoV, or 5 × 106 IFU of Ad5-nCoV complexed with 1 μg of (+)HSAnp-4 (1:1.2 × 104 molar ratio). (a) Blood tests and (b) histological analysis of lungs performed at 1 day and 7 days post administration (scale bar: 1000 μm). Data are presented as means ± SEM. Statistical differences were determined using ANOVA with Dunn’s multiple comparison test.
4. Discussion
In this work, by using (+)HSAnp, we demonstrated that biocompatible nanoparticles with desirable properties can complex with Ad5 and modulate Ad5–host interactions. Ad5/(+)HSAnp achieved robust lung-targeted transgene expression via the intranasal route, further enhancing the immunogenicity of Ad5-EBOV and Ad5-nCoV in the presence of PEI. Rather than relying on the common method of seeking a rare serotype of adenovirus to bypass preexisting anti-vector antibodies, our work attempts to address the problem of PEI by exploiting nanomaterial strategies. Nanotechnology approaches benefits vaccine development in many ways, including efficient antigen delivery, highly multivalent display, and strong stimulation of the immune system [44], [45]. For example, lipid nanoparticles have been developed as key components of messenger RNA (mRNA) vaccines against SARS-CoV-2 [46].
Viruses are naturally proteinaceous nanoparticles that package nucleic acid genomes. Through long-term evolution, viruses have evolved the ability to efficiently infect host cells by optimizing their surface properties and morphologies. In the present study, we demonstrate that the biological functions of virus-based vectors can be further improved via complexation with functional nanoparticles. (+)HSAnp modulates the surface properties of Ad5 and induces viral aggregation, which likely enhances antiviral resistance in body fluids. Previous studies have demonstrated that the aggregation of Ad2 in source water enhanced resistance to chlorine compared with monodispersed viruses [47]. In the present study, Ad5/(+)HSAnp enhanced the cellular uptake efficiency of Ad5 in both CAR-positive and -negative cells. This expanded receptor tropism facilitated increased transgene expression by Ad5/(+)HSAnp in vivo. A self-assembled Ad5/(+)HSAnp complex can easily be prepared without the need for sophisticated chemical conjugation or encapsulation. As (+)HSAnp interacts with Ad5 via electrostatic attraction rather than specific interactions, we assume that this strategy has the potential to be used for other nanoparticles and viral vectors. In contrast to previous studies using polymers or liposomes coated with adenovirus [23], [48], [49], [50], [51], we achieved the surface remodeling of Ad5 by modulating the aggregation state and surface potential. These findings provide an alternative avenue for addressing PEI; namely, the use of biocompatible nanoparticles to enhance the bioactivity of viral vectors. Cationic polymers and liposomes are small molecules that adhere to the surface of Ad5 through electrostatic interactions. Charge shielding is the mechanism underlying the enhanced transduction of adenoviruses. In contrast, the size of (+)HSAnp (approximately 20 nm) provides a larger surface to interact with Ad5 and induce viral aggregation. This offers an extra mechanism to enhance the cellular entry of Ad5. Our results are consistent with those of previous studies on retroviruses, in which larger molecular-weight polymers showed higher viral transfection efficiency due to the induction of viral aggregation [52].
Adenoviruses are nonenveloped viruses that can contact cells only through interactions between the viral capsid proteins and cellular receptors. By inhibiting capsid-receptor binding, preexisting anti-Ad5 antibodies dampen the cell entry of Ad5 vectors and compromise the immunogenicity of Ad5-based vaccines. At both the cellular and animal levels, we demonstrated that the Ad5/(+)HSAnp complex enhanced the transgene expression of Ad5 in the presence of Ad5-neutralizing antibodies. As (+)HSAnps induced viral aggregation and charge modification of Ad5, Ad5/(+)HSAnp could overcome PEI in two potential ways. First, viral aggregation has been regarded as the cause of the unneutralized fraction of the infectious virus [53]. It is likely that the aggregation results in steric hindrance of antibodies binding to the capsid proteins of viruses. Moreover, even if the surface of viral aggregates is recognized by antibodies, the aggregates may undergo morphological changes upon contact with cells, allowing unneutralized fractions to infect the cells. Second, the positively charged Ad5/(+)HSAnp complex can settle and attach to negatively charged cell membranes through electrostatic interactions, expanding the ways in which Ad5 interacts with cells, rather than relying solely on interactions between the viral capsid and CAR. Therefore, anti-Ad5 antibodies cannot completely neutralize the cell entry of Ad5/(+)HSAnp. However, the observation that Ad5/(+)HSAnp-4 did not significantly enhance the antigen-specific cellular immunity requires further investigation.
Inhalation is a desirable route for vaccine and drug delivery, because it is noninvasive and efficient for the prevention and treatment of respiratory diseases. Our recent study demonstrated that aerosolized Ad5-nCoV generated by a nebulizer was well tolerated and immunogenic in adults [54]. In this study, we show that the intranasal administration of Ad5-nCoV/(+)HSAnp enhanced the immune response in a PEI model. These results agree with previous findings that nanoparticle carriers can facilitate biomacromolecules in overcoming biological barriers following respiratory delivery [55], [56]. Surface charge is a crucial parameter for the cellular uptake and biodistribution of nanoparticle carriers [57]. Both in vitro and in vivo, the enhanced transgene expression of complexed Ad5 was dependent on the cationic charge of (+)HSAnp. These observations are consistent with recent discoveries that cationic formulations are essential in the design of lung-targeted carriers for mRNA delivery [58].
In summary, the data presented here demonstrate that viral aggregation and charge modification induced by biocompatible nanoparticles can enhance the transgene expression of viral vectors. Nanocomplexed Ad5 robustly expressed genes of interest in mouse lungs and overcame PEI by intranasal instillation. This study offers new insights into viral vector-based vaccines and gene therapies for future translational studies.
Acknowledgments
This work was supported in part by the grant from National Natural Science Foundation of China (82171818, 81703048, 82041019, and 82101919), the grant from Defense Industrial Technology Development Program of China (JCKY2020802B001) and Beijing Municipal Science and Technology Commission (Z201100005420024).
Authors’ contribution
Wei Chen, Junjie Xu, and Lihua Hou supervised and conceptualized the study. Yilong Yang, Peng Lv, and Xiaofan Zhao performed the preparation and characterization of nanocomplexed Ad5. Yilong Yang, Fangze Shao, and Ruihua Li performed the in vitro transgene expression of nanocomplexed Ad5. Shipo Wu, Yudong Wang, and Jun Zhang performed the immunological assays. Yilong Yang and Fangze Shao performed the in vivo bioluminescence imaging and biocompatibility assays. Yilong Yang, Shipo Wu, Xiaopeng Zhang, and Jianmin Li analyzed the data. Yilong Yang and Shipo Wu wrote the paper with assistance from all coauthors.
Compliance with ethics guidelines
Beijing Institute of Biotechnology has submitted the provisional patent application related to use of (+)HSAnp to improve immunogenicity of Ad5- based vaccines. Yilong Yang, Shipo Wu, Yudong Wang, Fangze Shao, Peng Lv, Ruihua Li, Xiaofan Zhao, Jun Zhang, Xiaopeng Zhang, Jianmin Li, Lihua Hou, Junjie Xu, and Wei Chen declare that they have no conflicts of interest or financial conflicts to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2022.12.007.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Excler J.L., Saville M., Berkley S., Kim J.H. Vaccine development for emerging infectious diseases. Nat Med. 2021;27(4):591–600. doi: 10.1038/s41591-021-01301-0. [DOI] [PubMed] [Google Scholar]
- 2.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halperin S.A., Ye L., MacKinnon-Cameron D., Smith B., Cahn P.E., Ruiz-Palacios G.M., et al. Final efficacy analysis, interim safety analysis, and immunogenicity of a single dose of recombinant novel coronavirus vaccine (adenovirus type 5 vector) in adults 18 years and older: an international, multicentre, randomised, double-blinded, placebo-controlled phase 3 trial. Lancet. 2022;399(10321):237–248. doi: 10.1016/S0140-6736(21)02753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396(10249):479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu F.C., Li Y.H., Guan X.H., Hou L.H., Wang W.J., Li J.X., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395(10240):1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grobusch M.P., Goorhuis A. Safety and immunogenicity of a recombinant adenovirus vector-based Ebola vaccine. Lancet. 2017;389(10069):578–580. doi: 10.1016/S0140-6736(16)32619-8. [DOI] [PubMed] [Google Scholar]
- 7.Ledgerwood J.E., DeZure A.D., Stanley D.A., Coates E.E., Novik L., Enama M.E., et al. Chimpanzee adenovirus vector Ebola vaccine. N Engl J Med. 2017;376(10):928–938. doi: 10.1056/NEJMoa1410863. [DOI] [PubMed] [Google Scholar]
- 8.Zhu F.C., Hou L.H., Li J.X., Wu S.P., Liu P., Zhang G.R., et al. Safety and immunogenicity of a novel recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in China: preliminary report of a randomised, double-blind, placebo-controlled, phase 1 trial. Lancet. 2015;385(9984):2272–2279. doi: 10.1016/S0140-6736(15)60553-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhu F.C., Wurie A.H., Hou L.H., Liang Q., Li Y.H., Russell J.B., et al. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: a single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;389(10069):621–628. doi: 10.1016/S0140-6736(16)32617-4. [DOI] [PubMed] [Google Scholar]
- 10.Guo Q., Chan J.F., Poon V.K., Wu S., Chan C.C., Hou L., et al. Immunization with a novel human type 5 adenovirus-vectored vaccine expressing the premembrane and envelope proteins of Zika virus provides consistent and sterilizing protection in multiple immunocompetent and immunocompromised animal models. J Infect Dis. 2018;218(3):365–377. doi: 10.1093/infdis/jiy187. [DOI] [PubMed] [Google Scholar]
- 11.Bullard B.L., Corder B.N., Gordon D.N., Pierson T.C., Weaver E.A. Characterization of a species E adenovirus vector as a Zika virus vaccine. Sci Rep. 2020;10(1):3613. doi: 10.1038/s41598-020-60238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smaill F., Jeyanathan M., Smieja M., Medina M.F., Thanthrige-Don N., Zganiacz A., et al. A human type 5 adenovirus-based tuberculosis vaccine induces robust T cell responses in humans despite preexisting anti-adenovirus immunity. Sci Transl Med. 2013;5(205):205ra134 doi: 10.1126/scitranslmed.3006843. [DOI] [PubMed] [Google Scholar]
- 13.Van Zyl-Smit R.N., Esmail A., Bateman M.E., Dawson R., Goldin J., van Rikxoort E., et al. Safety and immunogenicity of adenovirus 35 tuberculosis vaccine candidate in adults with active or previous tuberculosis. A randomized trial. Am J Respir Crit Care Med. 2017;195(9):1171–1180. doi: 10.1164/rccm.201603-0654OC. [DOI] [PubMed] [Google Scholar]
- 14.Fausther-Bovendo H., Kobinger G.P. Pre-existing immunity against Ad vectors: humoral, cellular, and innate response, what's important? Hum Vaccin Immunother. 2014;10(10):2875–2884. doi: 10.4161/hv.29594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mennechet F.J.D., Paris O., Ouoba A.R., Salazar Arenas S., Sirima S.B., Takoudjou Dzomo G.R., et al. A review of 65 years of human adenovirus seroprevalence. Expert Rev Vaccines. 2019;18(6):597–613. doi: 10.1080/14760584.2019.1588113. [DOI] [PubMed] [Google Scholar]
- 16.Yu B., Zhou Y., Wu H., Wang Z., Zhan Y., Feng X., et al. Seroprevalence of neutralizing antibodies to human adenovirus type 5 in healthy adults in China. J Med Virol. 2012;84(9):1408–1414. doi: 10.1002/jmv.23325. [DOI] [PubMed] [Google Scholar]
- 17.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against COVID-19. N Engl J Med. 2021;384(23):2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbink P., Lemckert A.A., Ewald B.A., Lynch D.M., Denholtz M., Smits S., et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol. 2007;81(9):4654–4663. doi: 10.1128/JVI.02696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol. 2009;21(3):346–351. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh R., Al-Jamal K.T., Lacerda L., Kostarelos K. Nanoengineering artificial lipid envelopes around adenovirus by self-assembly. ACS Nano. 2008;2(5):1040–1050. doi: 10.1021/nn8000565. [DOI] [PubMed] [Google Scholar]
- 22.O'Riordan C.R., Lachapelle A., Delgado C., Parkes V., Wadsworth S.C., Smith A.E., et al. PEGylation of adenovirus with retention of infectivity and protection from neutralizing antibody in vitro and in vivo. Hum Gene Ther. 1999;10(8):1349–1358. doi: 10.1089/10430349950018021. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Q., Han J., Zhao D., Gong T., Zhang Z., Sun X. Protection of adenovirus from neutralizing antibody by cationic PEG derivative ionically linked to adenovirus. Int J Nanomedicine. 2012;7:985–997. doi: 10.2147/IJN.S27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domingo J.L., Rovira J. Effects of air pollutants on the transmission and severity of respiratory viral infections. Environ Res. 2020;187 doi: 10.1016/j.envres.2020.109650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazzolla Gatti R., Velichevskaya A., Tateo A., Amoroso N., Monaco A. Machine learning reveals that prolonged exposure to air pollution is associated with SARS-CoV-2 mortality and infectivity in Italy. Environ Pollut. 2020;267 doi: 10.1016/j.envpol.2020.115471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sattler C., Moritz F., Chen S., Steer B., Kutschke D., Irmler M., et al. Nanoparticle exposure reactivates latent herpesvirus and restores a signature of acute infection. Part Fibre Toxicol. 2017;14(1):2. doi: 10.1186/s12989-016-0181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H., Zheng X., Nicholas J., Humes S.T., Loeb J.C., Robinson S.E., et al. Single-walled carbon nanotubes modulate pulmonary immune responses and increase pandemic influenza a virus titers in mice. Virol J. 2017;14(1):242. doi: 10.1186/s12985-017-0909-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borrego B., Lorenzo G., Mota-Morales J.D., Almanza-Reyes H., Mateos F., López-Gil E., et al. Potential application of silver nanoparticles to control the infectivity of Rift Valley fever virus in vitro and in vivo. Nanomedicine. 2016;12(5):1185–1192. doi: 10.1016/j.nano.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 29.Smallcombe C.C., Harford T.J., Linfield D.T., Lechuga S., Bokun V., Piedimonte G., et al. Titanium dioxide nanoparticles exaggerate respiratory syncytial virus-induced airway epithelial barrier dysfunction. Am J Physiol Lung Cell Mol Physiol. 2020;319(3):L481–L496. doi: 10.1152/ajplung.00104.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paik P.K., James L.P., Riely G.J., Azzoli C.G., Miller V.A., Ng K.K., et al. A phase 2 study of weekly albumin-bound paclitaxel (Abraxane®) given as a two-hour infusion. Cancer Chemother Pharmacol. 2011;68(5):1331–1337. doi: 10.1007/s00280-011-1621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan M., Du J., Gu Z., Liang M., Hu Y., Zhang W., et al. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat Nanotechnol. 2010;5(1):48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- 32.Wu S., Zhong G., Zhang J., Shuai L., Zhang Z., Wen Z., et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat Commun. 2020;11(1):4081. doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu D., Yang Y., Xu P., Xu D., Liu Y., Castillo R., et al. Real-time quantification of cell internalization kinetics by functionalized boluminescent nanoprobes. Adv Mater. 2019;31(39):e1902469. doi: 10.1002/adma.201902469. [DOI] [PubMed] [Google Scholar]
- 34.Werk D., Schubert S., Lindig V., Grunert H.P., Zeichhardt H., Erdmann V.A., et al. Developing an effective RNA interference strategy against a plus-strand RNA virus: silencing of coxsackievirus B3 and its cognate coxsackievirus-adenovirus receptor. Biol Chem. 2005;386(9):857–863. doi: 10.1515/BC.2005.100. [DOI] [PubMed] [Google Scholar]
- 35.Roelvink P.W., Lizonova A., Lee J.G., Li Y., Bergelson J.M., Finberg R.W., et al. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72(10):7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumida S.M., Truitt D.M., Lemckert A.A., Vogels R., Custers J.H., Addo M.M., et al. Neutralizing antibodies to adenovirus serotype 5 vaccine vectors are directed primarily against the adenovirus hexon protein. J Immunol. 2005;174(11):7179–7185. doi: 10.4049/jimmunol.174.11.7179. [DOI] [PubMed] [Google Scholar]
- 37.Nilchian A., Plant E., Parniewska M.M., Santiago A., Rossignoli A., Skogsberg J., et al. Induction of the coxsackievirus and adenovirus receptor in macrophages during the formation of atherosclerotic plaques. J Infect Dis. 2020;222(12):2041–2051. doi: 10.1093/infdis/jiaa418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomko R.P., Xu R., Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94(7):3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma P., Kolawole A.O., Wiltshire S.M., Frondorf K., Excoffon K. Accessibility of the coxsackievirus and adenovirus receptor and its importance in adenovirus gene transduction efficiency. J Gen Virol. 2012;93(Pt 1):155–158. doi: 10.1099/vir.0.036269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okada N., Tsukada Y., Nakagawa S., Mizuguchi H., Mori K., Saito T., et al. Efficient gene delivery into dendritic cells by fiber-mutant adenovirus vectors. Biochem Biophys Res Commun. 2001;282(1):173–179. doi: 10.1006/bbrc.2001.4527. [DOI] [PubMed] [Google Scholar]
- 41.Kaner R.J., Worgall S., Leopold P.L., Stolze E., Milano E., Hidaka C., et al. Modification of the genetic program of human alveolar macrophages by adenovirus vectors in vitro is feasible but inefficient, limited in part by the low level of expression of the coxsackie/adenovirus receptor. Am J Respir Cell Mol Biol. 1999;20(3):361–370. doi: 10.1165/ajrcmb.20.3.3398. [DOI] [PubMed] [Google Scholar]
- 42.Worgall S., Worgall T.S., Kostarelos K., Singh R., Leopold P.L., Hackett N.R., et al. Free cholesterol enhances adenoviral vector gene transfer and expression in CAR-deficient cells. Mol Ther. 2000;1(1):39–48. doi: 10.1006/mthe.1999.0013. [DOI] [PubMed] [Google Scholar]
- 43.Coughlan L. Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Front Immunol. 2020;11:909. doi: 10.3389/fimmu.2020.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fries C.N., Curvino E.J., Chen J.L., Permar S.R., Fouda G.G., Collier J.H. Advances in nanomaterial vaccine strategies to address infectious diseases impacting global health. Nat Nanotechnol. 2021;16(4):1–14. doi: 10.1038/s41565-020-0739-9. [DOI] [PubMed] [Google Scholar]
- 45.Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat Nanotechnol. 2020;15(8):646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 46.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6(12):1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kahler A.M., Cromeans T.L., Metcalfe M.G., Humphrey C.D., Hill V.R. Aggregation of adenovirus 2 in source water and impacts on disinfection by chlorine. Food Environ Virol. 2016;8(2):148–155. doi: 10.1007/s12560-016-9232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendez N., Herrera V., Zhang L., Hedjran F., Feuer R., Blair S.L., et al. Encapsulation of adenovirus serotype 5 in anionic lecithin liposomes using a bead-based immunoprecipitation technique enhances transfection efficiency. Biomaterials. 2014;35(35):9554–9561. doi: 10.1016/j.biomaterials.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wortmann A., Vöhringer S., Engler T., Corjon S., Schirmbeck R., Reimann J., et al. Fully detargeted polyethylene glycol-coated adenovirus vectors are potent genetic vaccines and escape from pre-existing anti-adenovirus antibodies. Mol Ther. 2008;16(1):154–162. doi: 10.1038/sj.mt.6300306. [DOI] [PubMed] [Google Scholar]
- 50.Wagner J., Li L., Simon J., Krutzke L., Landfester K., Mailänder V., et al. Amphiphilic polyphenylene dendron conjugates for surface remodeling of adenovirus 5. Angew Chem Int Ed Engl. 2020;59(14):5712–5720. doi: 10.1002/anie.201913708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji Z., Xie Z., Zhang Z., Gong T., Sun X. Engineering intravaginal vaccines to overcome mucosal and epithelial barriers. Biomaterials. 2017;128:8–18. doi: 10.1016/j.biomaterials.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Davis H.E., Rosinski M., Morgan J.R., Yarmush M.L. Charged polymers modulate retrovirus transduction via membrane charge neutralization and virus aggregation. Biophys J. 2004;86(2):1234–1242. doi: 10.1016/S0006-3495(04)74197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallis C., Melnick J.L. Virus aggregation as the cause of the non-neutralizable persistent fraction. J Virol. 1967;1(3):478–488. doi: 10.1128/jvi.1.3.478-488.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu S., Huang J., Zhang Z., Wu J., Zhang J., Hu H., et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect Dis. 2021;21(12):1654–1664. doi: 10.1016/S1473-3099(21)00396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel A.K., Kaczmarek J.C., Bose S., Kauffman K.J., Mir F., Heartlein M.W., et al. Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv Mater. 2019;31(8):e1805116. doi: 10.1002/adma.201805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mastorakos P., da Silva A.L., Chisholm J., Song E., Choi W.K., Boyle M.P., et al. Highly compacted biodegradable DNA nanoparticles capable of overcoming the mucus barrier for inhaled lung gene therapy. Proc Natl Acad Sci USA. 2015;112(28):8720–8725. doi: 10.1073/pnas.1502281112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fromen C.A., Rahhal T.B., Robbins G.R., Kai M.P., Shen T.W., Luft J.C., et al. Nanoparticle surface charge impacts distribution, uptake and lymph node trafficking by pulmonary antigen-presenting cells. Nanomedicine. 2016;12(3):677–687. doi: 10.1016/j.nano.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat Nanotechnol. 2020;15(4):313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.