Abstract
Ring-shaped hexameric helicases are essential motor proteins that separate duplex nucleic acid strands for DNA replication, recombination, and transcriptional regulation. Two evolutionarily distinct lineages of these enzymes, predicated on RecA and AAA+ ATPase folds, have been identified and characterized to date. Hexameric helicases couple NTP hydrolysis with conformational changes that move nucleic acid substrates through a central pore. How hexameric helicases productively engage client DNA or RNA segments and use successive rounds of NTPase activity to power translocation and unwinding have been longstanding questions in the field. Recent structural and biophysical findings are beginning to reveal commonalities in NTP hydrolysis and substrate translocation by diverse hexameric helicase families. Here, we review these molecular mechanisms and highlight aspects of their function that are yet to be understood.
Keywords: helicase, RecA, AAA+, ATPase, DNA replication
Introduction – Hexameric Helicase Phylogeny and Organization
Helicases are specialized enzymes that consume nucleoside triphosphates (NTPs) to separate nucleic acid strands. This activity can unwind DNA, RNA, and DNA-RNA duplexes and displace nucleic acid-binding proteins to facilitate essential cellular processes such as replication and transcription, translation, and splicing. Additionally, helicases are involved in unwinding higher-order nucleic acid structures such as G-quadruplexes, Holliday junctions, and triplexes (Guo et al., 2015; Jain et al., 2010; Mendoza et al., 2016; Tsaneva et al., 1993). Many helicases function within large protein complexes, and even serve as organizing hubs for such assemblies. Given their critical role in the cell, helicase dysfunction has been linked to molecular problems such as DNA damage and genomic instability, as well as human diseases such as premature aging, cancer predisposition, mitochrondriopathies, and immune disorders (Ellis et al., 1995; Goffart et al., 2009; Levitus et al., 2005; Levran et al., 2005; Litman et al., 2005; Weber et al., 1990; Weeda et al., 1990; Yu et al., 1996). As a consequence, helicases have been the focus of numerous studies aimed at understanding their striking biophysical behavior and at developing their utility as therapeutic targets.
All helicases belong to the P-loop NTPase superclass, a large group of nucleotide-binding proteins encompassing 75 different types of enzyme activities (Leipe et al., 2003; Longo et al., 2020). P-loop NTPases are distinguished by a characteristic αβα sandwich fold that possesses two conserved amino-acid sequence elements, termed the Walker A (WA, GXXGXGK(T/S)) and Walker B (WB, D(D/E)XX) motifs, which coordinate a bound Mg2+ ion and an associated NTP (usually ATP or GTP) (Fig. 1) (Abrahams et al., 1994; Story and Steitz, 1992; Story et al., 1992; Walker et al., 1982). P-loop NTPases can be divided into families based on the conservation of key catalytic resides and the insertion of auxiliary secondary structural elements. Helicases belong to a particular group known as the Additional Strand Catalytic Glutamate (ASCE) family (Iyer et al., 2004; Leipe et al., 2003). ASCE proteins are distinguished from other, more distantly related P-loop NTPases by a namesake conserved glutamate that is located within or adjacent to the WB motif (Fig. 1) (Subramanya et al., 1996; Thomsen and Berger, 2008). The ASCE group can be further divided into several well-known classes of enzymes, such as the ATP-binding cassette (ABC), Signal Transduction ATPases with Numerous Domains (STAND), ATPases Associated with diverse cellular Activities (AAA+), and RecA subfamilies (Iyer et al., 2004; Leipe et al., 2002, 2003, 2004).
Fig. 1: Hexameric helicase superfamilies and folds.
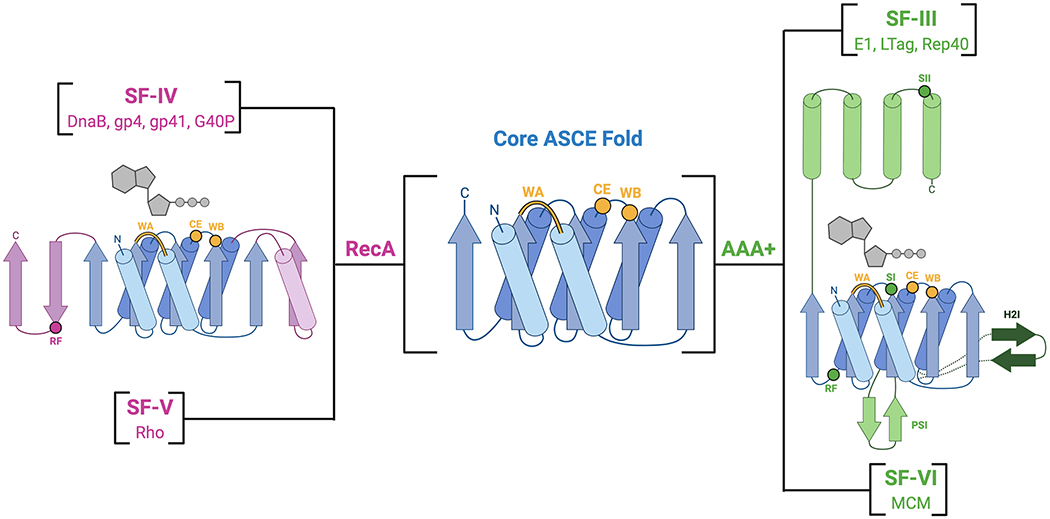
The core ASCE αβα binding fold (blue) has diverged into the RecA (pink) and AAA+ (green) families that make up all hexameric helicases. Select structural insertions and locations or specific motifs are indicated (yellow for ASCE, pink or green for RecA and AAA+, respectively). Dark green dotted lines and arrows show the helix two insert (H2I) that is specific to SF-VI and not found in SF-III helicases.
Based on their protein fold, oligomeric state, and translocation polarity, helicases have been classified into six superfamilies (SFs), referred to as SF-I to SF-VI (Gorbalenya and Koonin, 1993; Singleton et al., 2007). SF-I and -II helicases consist of a tandem, linked pair of RecA ATPase folds and contain many helicases of biomedical relevance such as XPD and SARS-CoV2 Nsp13, whereas SF-III to -VI enzymes are ring-shaped hexamers (Gorbalenya and Koonin, 1993; Ivanov et al., 2004; Lehmann, 2001; Singleton et al., 2007; Subramanya et al., 1996). SF-IV and SF-V proteins comprise the hexameric RecA helicases (Figs. 1, 2A and B), and exhibit a processive, 5’-3’ directionality (or ‘polarity’) of translocation along client nucleic acid substrates (LeBowitz and McMacken, 1986; Matson et al., 1983; Venkatesan et al., 1982). SF-IV contains many classically studied replicative helicases, such as bacterial DnaB, phage T7 gp4 helicase-primase, phage T4 gp41, phage SPP1 G40P, and the mitochondrial protein TWINKLE. The sole member of SF-V is a helicase known as Rho, a bacterial transcription termination factor that translocates along nascent RNA transcripts to unwind RNA-DNA hybrids and assist with RNA polymerase recycling and the control of gene expression (Brennan et al., 1987; Richardson, 2002). The AAA+ helicases, which processively translocate in the 3’-5’ direction, belong to either SF-III or SF-VI (Figs. 1, 2C and D) (Goetz et al., 1988; Seo et al., 1993). SF-III contains helicases from eukaryotic viruses, such as papillomavirus E1, simian virus 40 large tumor antigen (SV40 LTag), and adeno-associated virus Rep40. SF-VI is composed of the eukaryotic and archaeal replicative Mini-Chromosome Maintenance (MCM) helicases. In eukaryotes, the MCM helicase is made up of six paralogous subunits (Mcm2 through Mcm7), forming a heterohexameric helicase (Bochman and Schwacha, 2008; Kearsey and Labib, 1998; Labib and Diffley, 2001). MCMs are activated by the binding of Cdc45 and the GINS (Go-Ichi-Ni-San) complex to form a Cdc45-MCM-GINS (CMG) assembly that drives replication fork progression (Gambus et al., 2006; Ilves et al., 2010; Moyer et al., 2006).
Fig. 2: Overview of RecA and AAA+ hexameric helicase structures.
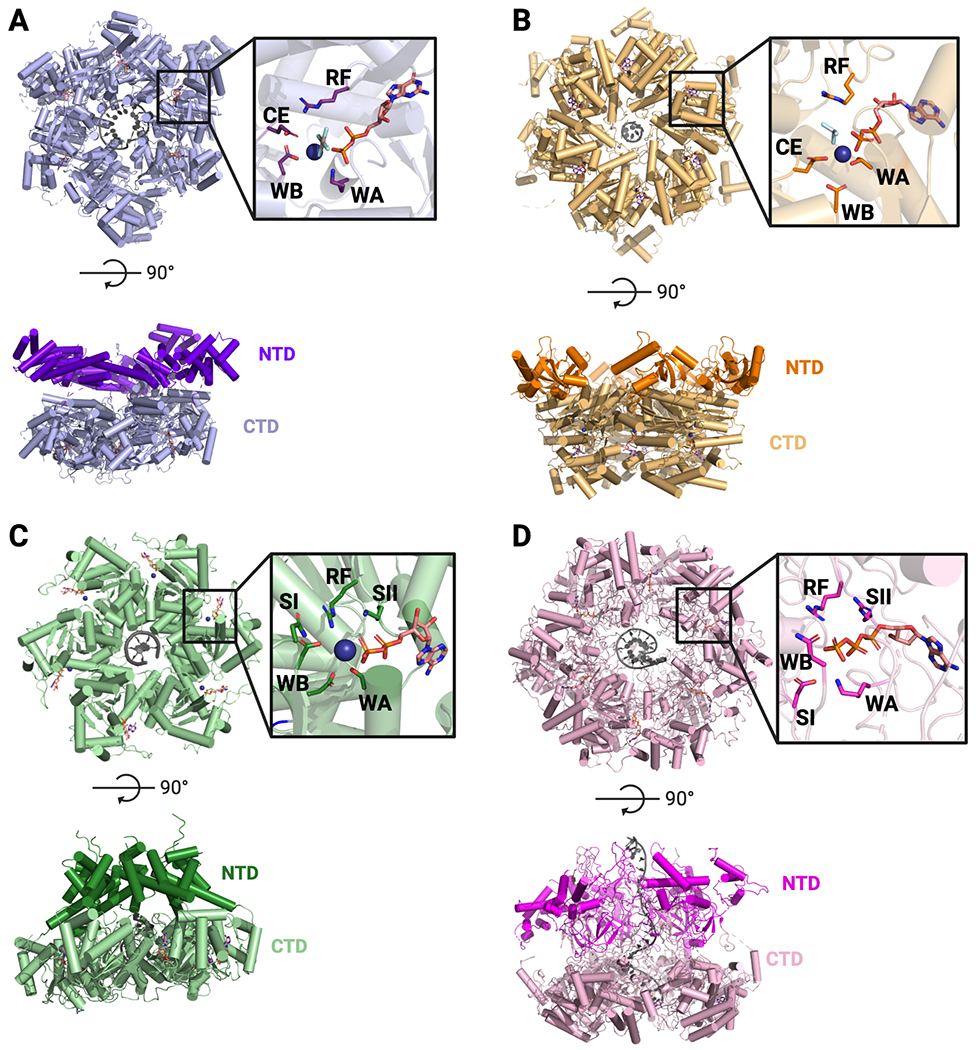
A. SF-IV – G. stearothermophilus DnaB bound to ssDNA and GDP•AlF4 (PDB 4ESV). B. SF-V – E. coli Rho bound to ADP•BeF3 and RNA (PDB 3ICE). C. SF-III – papillomavirus E1 bound to ADP and ssDNA (PDB 2GXA). D. SF-VI – D. melanogaster Mcm2-7 bound to ADP, ATP and ssDNA (PDB 6RAZ). For all – Top: Top-down view of hexameric rings and active sites (inset). Bottom: Side view showing N-and C-terminal domains. RF = Arginine Finger, WA = Walker A, WB = Walker B, CE = Catalytic Glutamate, SI = Sensor I, SII = Sensor II.
All hexameric helicases have a similar, though not necessarily evolutionarily conserved, arrangement for coordinating NTPs and nucleic acid substrates (Figs. 2, 3). The hexameric ring creates six subunit-subunit interfaces, each of which houses a bipartite ATPase center. These active sites are constructed by the WA and WB regions from one protomer and a catalytically essential basic amino acid, called the Arginine Finger (RF), in deference to G-protein nomenclature, from the neighboring subunit (Fig. 2) (Ahmadian et al., 1997; Scheffzek et al., 1996; Walker et al., 1982). Hexameric helicases also possess a set of pore loops that line their central channel to support nucleic acid binding and translocation (Fig. 3). The pore loops frequently contain arginine and lysine residues that interact non-specifically with the bound DNA or RNA substrate though the phosphodiester backbone (Enemark and Joshua-Tor, 2006; Fletcher et al., 2003; Itsathitphaisarn et al., 2012; McGeoch et al., 2005; Petojevic et al., 2015; Thomsen and Berger, 2009) The ATPase motor domains of hexameric helicases are frequently appended with different types of N- and/or C-terminal domains (e.g., winged-helix domains or oligonucleotide/oligosaccharide binding folds) that serve to augment helicase function (Brewster et al., 2008; Dolan et al., 1990; Dombroski and Platt, 1988; Fletcher et al., 2003; Nakayama et al., 1984)
Fig. 3: Nucleic acid binding and ATPase status of hexameric helicases.
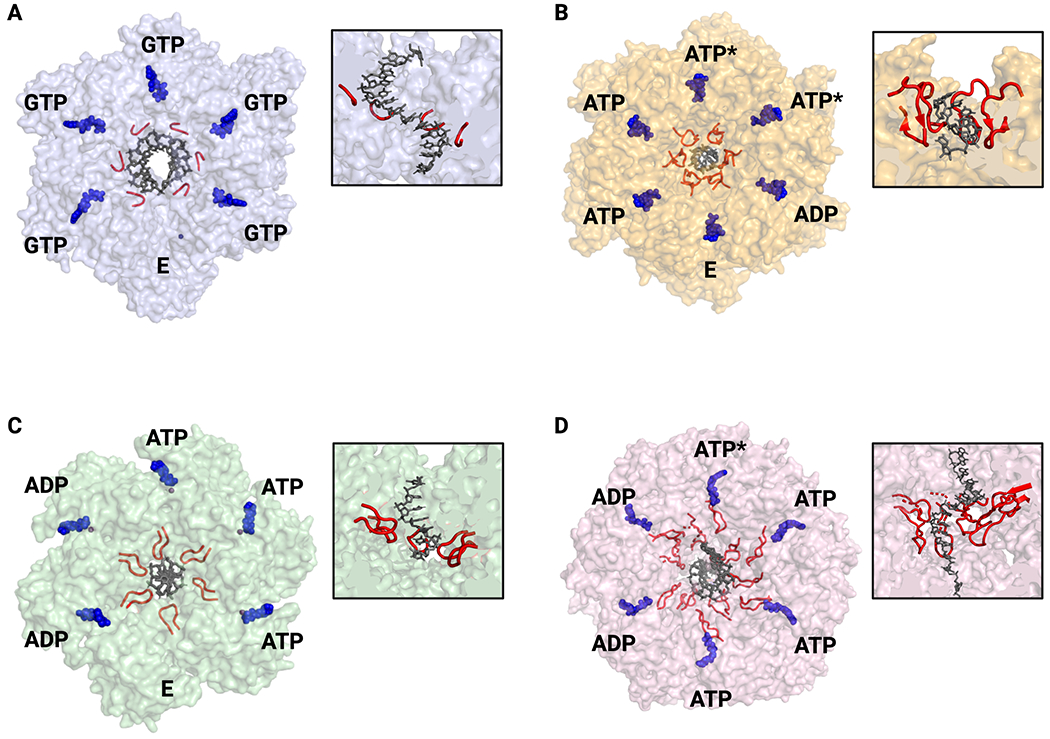
A. SF-IV – G. stearothermophilus DnaB. All GTP-bound active sites are in the same conformation and thus ATPase status around the ring cannot be assigned (PDB 4ESV). B. SF-V – E. coli Rho with ATPase state assigned by active site conformation and nucleotide status (PDB 3ICE). C. SF-III – papillomavirus E1 with ATPase state assigned by active site conformation (PDB 2GXA). D. SF-VI – D. melanogaster Mcm2-7 with ATPase state assigned by cryo-EM density and active site conformation (PDB 6RAZ). For all – nucleic acid substrates are shown in blue and pore loops in red. Nucleotide states: ATP – substrate bound; ADP – product bound; E – empty/exchangeable, ATP* - hydrolysis competent. All views are looking down from the 3’-5’ perspective. Insets show cutaway of helicase structure to highlight pore loops interactions with nucleic acid substrates.
Although hexameric RecA and AAA+ helicases are both ASCE ATPases, the two groups of enzymes differ in their relative placement of certain key catalytic residues. In RecA helicases, the RF lies on family specific structural insertion of two β strands at one edge of the ASCE domain, whereas in AAA+ helicases, this residue sits on a different, α-helical element (Fig. 1) (Lenzen et al., 1998; Singleton et al., 2000). AAA+ proteins also contain a Sensor I (SI) residue that resides on β4 of the central fold, as well as a Sensor II (SII) residue that is positioned on an α-helix that is part of a small helical subdomain that sits atop the principle ASCE fold (Fig. 1) (Guenther et al., 1997; Hanson and Whiteheart, 2005). The SI and SII motifs were originally identified as part of a large hydrogen bonding network that senses and responds to the presence of the γ phosphate on ATP, similar to the “Switch II” region of Ras (Brunger et al., 1990; Pai et al., 1990). The SI residue is generally polar and functions with the catalytic glutamate (CE) in the WB motif to coordinate a nucleophilic water molecule for hydrolysis (Hanson and Whiteheart, 2005). The SII residue is generally an arginine and helps to coordinate bound nucleotide and control conformational changes associated with hydrolysis (Hanson and Whiteheart, 2005). Hexameric RecA and AAA+ helicases also differ in the orientation of their core ASCE folds, which sit perpendicular to each other relative to their active sites (Wang, 2004). As a consequence, the nucleic acid-binding loops of the two protein families extend from different secondary structural positions with respect to the site of NTP turnover. This subunit rotation not only suggests that RecA and AAA+ ATPases evolved the ability to hexamerize independently of one another, but also changes the orientation of conserved catalytic motifs relative to the direction of force generation. Whether these structural differences contribute to some of the known mechanistic differences between the two helicase families is unknown.
Since the initial discovery that helicases could form hexamers, there has been significant interest in understanding the mechanism of these enzymes. A number of biochemical studies have shown that NTP binding and hydrolysis around the ring is highly cooperative (Crampton et al., 2004; Notarnicola and Richardson, 1993; Patel et al., 1994). These observations have led to questions regarding how NTP hydrolysis physically drives translocation and duplex unwinding, and what controls both helicase step size (the number of bases moved per ATP binding per hydrolysis event) and velocity.
NTP Hydrolysis in Hexameric Helicases
In all hexameric helicases, nucleotide binding and hydrolysis are coordinated to both local and global conformational changes. Historically, three types of models for hydrolytic cycling have been proposed for the operation of these enzymes (Lyubimov et al., 2011; Singleton et al., 2007). In one, known as the concerted model (Fig. 4A), all six subunits bind, hydrolyze, and release ATP in synchrony with each other. In a second, termed a ‘stochastic’ model (Fig. 4B), there appears to be no firm requirement for subunits to hydrolyze ATP in any particular order. The concerted model was derived from structural studies of AAA+ proteins such as SV40 LTag and HsIU (a protein unfoldase), which captured the ATPase rings in six-fold symmetric nucleotide-bound or apo states (Gai et al., 2004; Wang et al., 2001). Interestingly, these states were imaged in the absence of a client DNA or peptide; more recent structural findings (described below) indicate that these symmetrized forms likely reflect non-translocative pre- or post-substrate engagement states. By comparison, the ‘stochastic’ model was largely developed from biochemical studies of covalently linked protomers of ClpX, another AAA+ protein unfoldase, bearing distinct combinations of inactive ATPase centers (Martin et al., 2005). This effort showed that ClpX is highly tolerant to different permutations of WA, WB, and RF substitutions around the hexameric ring, demonstrating that there is a high degree of plasticity in the system.
Fig. 4: Models for NTP hydrolysis.
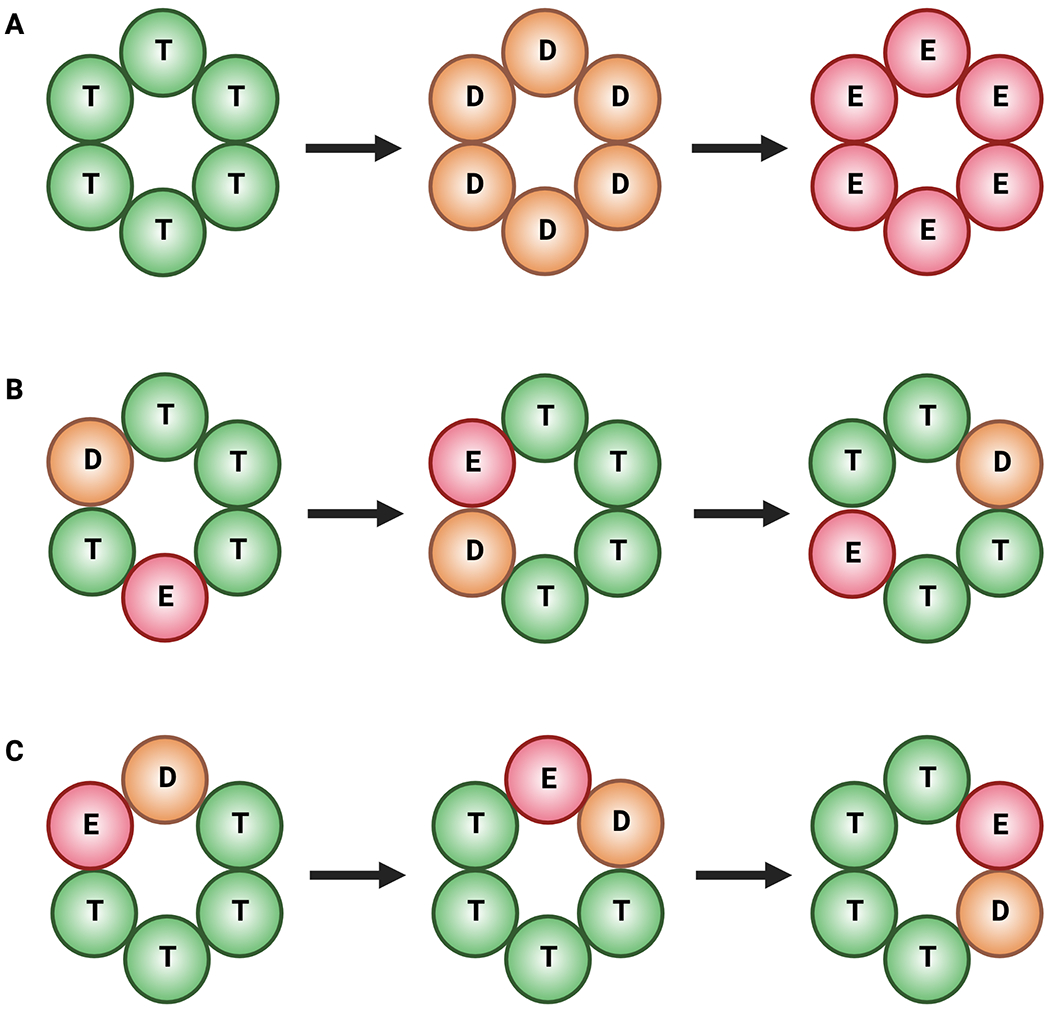
ATPase states: T (green) – ATP; D (orange) – ADP + Pi; E (red) – empty. A. Concerted hydrolysis. All sites fire in synchrony. B. Stochastic hydrolysis. Sites fire at random. C. Rotary hydrolysis. Sites fire in a sequential order around the ring.
The concerted and stochastic models differ from a third scheme, termed ‘rotary’ hydrolysis (Fig. 4C), in which nucleotide turnover by each active site proceeds in a sequential clockwise or counterclockwise order around the hexameric ring. An early inspiration for the rotary mechanism came from structural and single-molecule studies of the F1-ATPase, a heterohexameric RecA-family motor protein that is part of ATP synthase and generates ATP from the protonmotive force across the inner membrane of mitochondria (reviewed in (Walker, 2013)). A structure of the F1-ATPase showed that the nucleotide status and conformation of its three catalytic pockets (present in the β subunits, as the α subunits bind ATP noncatalytically) alternated about the ring between substrate (AMPPNP)-bound, product (ADP)-bound, or empty states (Abrahams et al., 1994). This arrangement suggested that hydrolysis would progress as a sequential wave around the ring in conjunction with the rotation of a central α-helical element inside the pore of the motor (known as the γ subunit, or ‘stalk’). Rotation was confirmed by direct visualization of single F1-ATPase motors in which a fluorescently labeled actin filament was attached to the γ subunit by streptavidin-biotin conjugation (Noji et al., 1997). Upon the addition of ATP, the labeled actin rotated counterclockwise and three molecules of ATP were hydrolyzed per full rotation, as predicted by the crystal structure (Abrahams et al., 1994; Adachi et al., 2000; Noji et al., 1997).
As structural, biochemical, and single-molecule studies of hexameric helicases have progressed, several lines of evidence have converged in support of RecA family enzymes operating by a tightly coupled rotary mechanism. A structure of the isolated helicase domain of T7 gp4 bound to four molecules of ADPNP•Mg2+ provided the first structure that was suggestive of rotary firing in a SF-IV helicase (Singleton et al., 2000). More recent structural studies of gp4 and DnaB bound to substrate single-stranded DNA (ssDNA) have provided snapshots of hexamers with a single empty active site (Fig. 3A), a feature that rules out a concerted hydrolysis mechanism but that is consistent with a rotary firing scheme (Gao et al., 2019; Itsathitphaisarn et al., 2012). Biochemical studies involving heterooligomers of gp4 that disrupt NTP binding, hydrolysis, DNA binding, or intersubunit communication have shown that even a single defective subunit results in a clear loss of positive cooperativity, ruling out a purely stochastic firing model as well (Crampton et al., 2004; Notarnicola and Richardson, 1993; Patel et al., 1994). Single-molecule studies of G40P have shown that the binding of even a single ATPγS molecule can stall the helicase, also at odds with the behavior expected for a stochastic or concerted mechanism (Crampton et al., 2006; Schlierf et al., 2019). Structures of RNA-bound states of the SF-V helicase Rho, which shares ~50% sequence similarity to the F1-ATPase, align with its SF-IV cousins, revealing hexamers that contain a mix of empty, ATP-, and ADP-like states that progress in sequence around the ring (Fig. 3B) (Thomsen and Berger, 2009; Thomsen et al., 2016). Pre-steady state kinetic studies of Rho function have supported the observed proportionality of these states, in particular noting that there is a one to two-subunit lag between nucleotide hydrolysis and Pi release that is seen structurally; this lag has been reported for T7 gp4 as well (Adelman et al., 2006; Liao et al., 2005; Thomsen et al., 2016).
Like their RecA counterparts, there is significant evidence that AAA+ hexameric helicases operate by a rotary hydrolysis mechanism. The first crystal structure of a hexameric AAA+ helicase bound to ssDNA, the papillomavirus E1 protein (SF-III) displayed a series of ATPase states that progress from apo to ATP- and then ADP-like configurations in sequential order (Fig. 3C) (Enemark and Joshua-Tor, 2006). A subsequent single-molecule study of E1 showed that the inclusion of ADP led to a significant decrease in unwinding efficiency by increasing the number of paused and back-tracked molecules, suggesting that it cannot overcome a catalytically stalled subunit; this observation has been taken to argue against the helicase using a random or stochastic ATPase firing order (Lee et al., 2014), a mechanism that has been proposed to explain the tolerance of the AAA+ peptide translocase, ClpX, to misfiring subunits (Martin et al., 2005). As with SF-III proteins, structural studies of ssDNA-bound SF-VI MCM helicases have captured active site occupancies that also have supported a rotary mechanism (Fig. 3D) (Eickhoff et al., 2019). However, biochemical studies of MCMs in which nucleotide binding or hydrolysis has been disrupted in specific subunits in the heterohexamer have also shown that some active sites are more tolerant to disruption than others (Bochman et al., 2008; Eickhoff et al., 2019; Ilves et al., 2010). This latter observation suggests that, similar to ClpX, some AAA+ helicases retain sufficient plasticity to overcome an inactivated subunit even if they normally function by a rotary mechanism (Martin et al., 2005). Why and how some hexameric AAA+ complexes may be more tolerant to misfiring than RecA hexamers is unknown. Interestingly, when viewed down the pore in the direction of translocation (toward the 3’ nucleic acid end for SF-IV/V helicases and toward the 5’ end for SF-III/VI helicases) the order of apo, ATP-, and ADP-bound states around the ring is inverted between RecA and AAA+ enzymes (Figs. 3B vs. C–D) (Thomsen and Berger, 2009). This difference in pattern suggests that the two classes of helicases move in opposing directions along a DNA or RNA chain by having an active site firing sequence that progresses either counterclockwise (RecA) or clockwise (AAA+).
Translocation by Hexameric Helicases
Processive helicases use ATP turnover to power their translocation along substrate DNA. Several models have been proposed to account for this movement. In one, known as the inchworm model, each round of catalysis causes one portion of the helicase to step forward into a single-stranded/double-stranded nucleic acid junction (the ‘fork’) and away from a rearward segment of the helicase, after which the forward portion freezes and the back element steps forward. It is now believed that non-hexameric SF-I and -II helicases move in this manner (reviewed in (Gao and Yang, 2020) and (Singleton et al., 2007)). An important feature of the inchworm model is that the net movement of the nucleic acid substrate through the helicase is a sliding motion that results from the coordinated hand-off of each base from the front end of the motor to the back. A levering movement analogous to what might be expected for an inchworm mechanism, called the ‘pumpjack’ (Fig. 5A), has been proposed for the replicative CMG helicase on the basis of different conformational states seen for the assembly in absence of DNA (Yuan et al., 2016). In these structures, the C-terminal AAA+ motor domains were observed to move up and down, pulling the GINS- and Cdc45-bound N-terminal domain along with them in a ratcheting manner.
Fig. 5: Models for translocation.
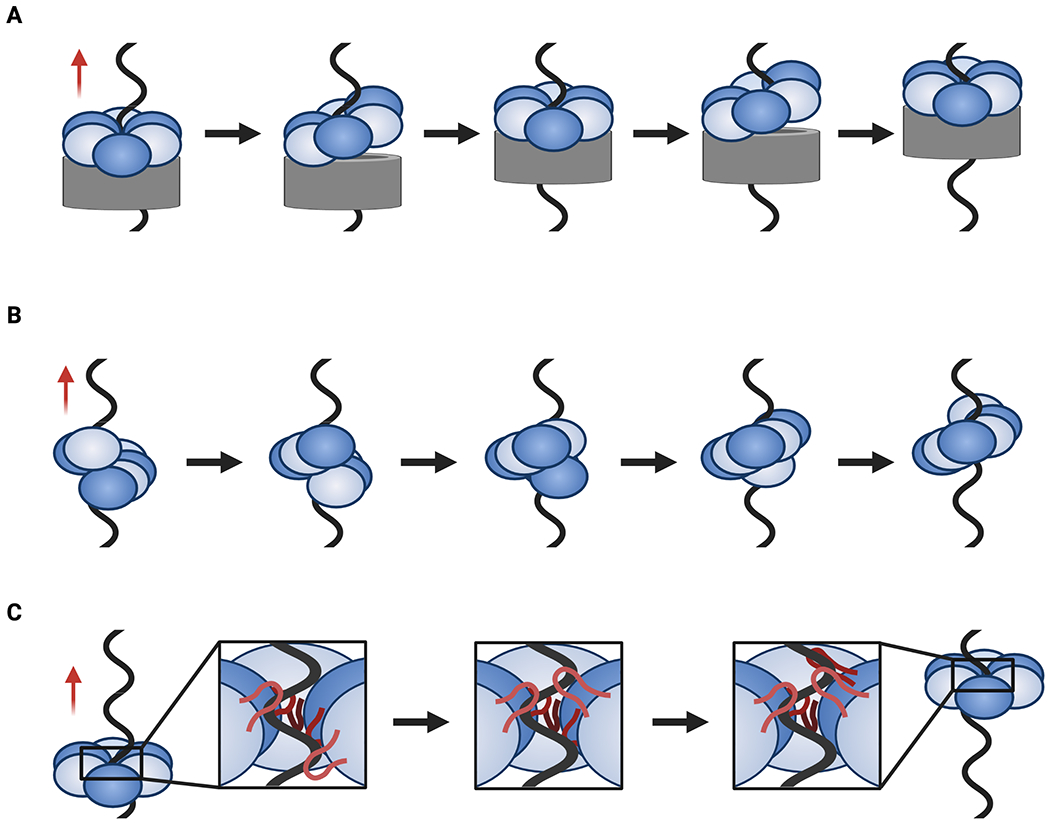
A. Pumpjack translocation where a stable N-terminal region (gray) encircles ssDNA while the C-terminal motor regions moves up and down as a levering unit. B. Hand-over-hand translocation in which rounds of ATP binding, hydrolysis, and product release coincide with an entire subunit moving from one end of the helicase spiral to the other (bottom-to-top movement as depicted here). C. Concerted escort translocation in which DNA binding loops (red, inset) form a spiral around DNA and rounds of ATP binding, hydrolysis, and product release are coupled to upward and downward loop movements (bottom-to-top movement as depicted here). Red arrows indicate the direction of translocation.
Two other translocation schemes proposed for hexameric helicases have been called the hand-over-hand and coordinated escort models, both of which are inherently rotary (Fig. 5B, C) (Enemark and Joshua-Tor, 2006; Itsathitphaisarn et al., 2012). In these schemes, cycles of ATP binding, hydrolysis, and product release are tied to conformational changes and alterations in DNA binding affinity within the hexameric ring that sequentially advance each subunit into the fork. In the hand-over-hand model, this movement reflects a large translational motion of an entire ATPase domain or helicase subunit (Fig. 5B). For the coordinated escort model, this action is performed primarily by the DNA-binding pore loops and some smaller, rocking motions in their associated ATPase domains (Fig. 5C). A feature shared by both models is that there is no sliding or hand-off of DNA from one subunit to the next. Instead, each subunit of the helicase appears to grab either one or two nucleobases in the substrate (depending on the superfamily) and then carry that cargo through helicase pore until it is released. In maintaining subunit/nucleic-acid contacts as the substrate passes through the pore, these two approaches contrast with an alternative, “revolution” model proposed for the translocation of dsDNA by the ring-shaped phi29 packaging protein, which has suggested that duplex DNA is actively passed from one subunit to the next in an ATP-coupled manner to facilitate movement (Schwartz et al., 2013).
SF-IV RecA helicases such as DnaB and gp4 appear to operate by a hand-over-hand mechanism that drives translocation in a 5’-3’ direction. When bound to substrate DNA, both enzymes take the form of a short spiral in which the ATPase domains of the top- and bottom-most protomers, sometimes referred to as the ‘seam’ subunits, are separated by a narrow gap (Gao et al., 2019; Itsathitphaisarn et al., 2012). This gap is spanned by a linker element that extends from the 5’-proximal subunit to dock against the 3’-proximal subunit, topologically sealing the helicase ring around the DNA. The 3’ seam subunit is not bound to ATP or substrate DNA. A recent structure of gp4 with ssDNA shows that there is a network of residues that connect the pore loops to a nucleophilic water in the active site, establishing an allosteric relay between DNA binding and NTP hydrolysis (Gao et al., 2019). Interestingly, tightening of the gp4 active sites along the spiral seems to be matched by a tightening of the helicase’s interactions with DNA, a relationship also noted in biochemical studies of gp4 and a structure of ssDNA-bound DnaB (Gao et al., 2019; Hingorani and Patel, 1993; Itsathitphaisarn et al., 2012). This feature of SF-IV helicases suggests that product release causes the 5’ (fork-distal) subunit of the seam to dissociate from DNA and to bind ATP when it reassociates with DNA at the 3’ (fork-proximal) portion of the spiral.
Similar to SF-IV RecA helicases, structural studies have indicated that SF-VI MCM helicases also form a spiral when bound to ssDNA and operate by a hand-over-hand mechanism to translocate in a 3’-5’ direction (Eickhoff et al., 2019; Meagher et al., 2019). A recent cryo-EM study of ssDNA-bound CMG imaged four different states of the ATPase, revealing that the motor domains of ssDNA-bound MCM helicases form a spiral in which different subunits (either Mcm2, 3, or 4) occupy the 5’ (fork-proximal) position at the seam (Eickhoff et al., 2019). In one of these models, the two 3’-proximal subunits (Mcm5 and 3) are tightly associated with each other through ATP binding, but whereas Mcm3 is seen to contact DNA, Mcm5 does not. This observation suggests that Mcm5 and Mcm3 may be able to translocate together as a unit, potentially illustrating why ATP hydrolysis can be required at some MCM subunit interfaces but dispensable at others (Bochman et al., 2008; Eickhoff et al., 2019; Ilves et al., 2010; Schwacha and Bell, 2001). Variability in the number of subunits that translocate from the bottom to the top of the ATPase spiral or that are needed to productively associate with nucleotide could provide a means for CMG to accommodate lesions or small disruptions in the tracking strand during DNA translocation and unwinding. Interestingly, studies using methylphosphonate chemistry have shown that CMG contacts only one strand during translocation over dsDNA, similar to the duplex DNA packaging motor of phase phi29 (Aathavan et al., 2009; Langston and O’Donnell, 2019).
It has been suggested that a hand-over-hand mechanism is universal to AAA+ translocases, with structures of peptide-associated unfoldases and translocases providing support for such claims (Puchades et al., 2020). However, the E1 helicase does not form a lock-washer structure nor is it completely planar (Enemark and Joshua-Tor, 2006). Instead, the pore loops of its six subunits form a spiral that tracks along the phosphodiester backbone of a tightly coiled ssDNA substrate bound in the central pore of the enzyme. Although there is some rotational, out-of-plane displacement between each pair of E1 subunits, the pore loops also appear to lever up and down depending on the nucleotide status of their associated protomers. As a consequence, translocation by E1 appears to rely on a coordinated escort mechanism in which a loop dives toward the 5’ DNA end, engages a base, and then moves upward, remaining associated with that base until it is released at the end of the ATPase cycle for that subunit. As with SF-IV and -VI helicases, DNA engagement occurs concomitantly with ATP binding and DNA release with product release. Interestingly, the Rho transcription termination factor seems to work by a mix of hand-over-hand and coordinated escort. Like E1, there is no opening or seam in the Rho ring, but five of its ATPase subunits form a pronounced out-of-plane spiral and, like SF-IV and -IV enzymes, the sixth appears capable of moving parallel to the central axis of the helicase as it transits from the 5’- to the 3’-proximal position in the ring (Thomsen and Berger, 2009). At the same time, the RNA-binding pore loops in Rho undergo a levering motion that is linked to status of their associated ATPase centers (Thomsen et al., 2016). The hybrid nature of Rho is intriguing, as it suggests that the coordinated escort and hand-over-hand mechanisms are two extremes of a continuum of local and global molecular movements that couple a rotary ATPase cycle to nucleic acid translocation.
Unwinding and Step Size
Historically, two energetic models have been proposed for how helicases use nucleotide turnover to promote duplex unwinding (Fig. 6) (Lohman and Bjornson, 1996). In one, called the active or power-stroke model, ATP hydrolysis is tied to a structural change in the helicase that performs mechanical work to physically separate base pairs in a nucleic acid duplex. This action in principle would allow a translocating enzyme to plow through several base pairs per ATP turnover event; since the free energy from ATP hydrolysis under standard conditions is ~ −7 kcal/mol and melting of a single base pair costs ~1.6 kcal/mol, a power-stroke enzyme operating at 100% efficiency could conceivably melt up to 4 or 5 base pairs per ATP consumed (Fig. 6A) (Von Hippel and Delagoutte, 2001). In the second model, termed a passive or Brownian ratchet mechanism, the substrate duplex transiently opens or frays due to thermal fluctuations, exposing a single-stranded nucleic acid segment that can then be captured by the helicase in a conformational step that is linked to the NTPase cycle (Fig. 6B). The energetic cost associated with spontaneous base pair melting limits helicase movement to a maximum step size of a few bases per NTP turnover. Quantitative models that account for the degree of passiveness or activeness in hexameric helicases have suggested that there may be a spectrum of activities that range from highly active to highly passive. The ratio of helicase velocity during translocation on ssDNA or ssRNA strands as compared to the rate of duplex unwinding as a function of varying force and GC content has been proposed as a means of mapping helicase activity on this spectrum (Manosas et al., 2010).
Fig. 6: Power stroke vs. Brownian ratchet unwinding mechanisms.
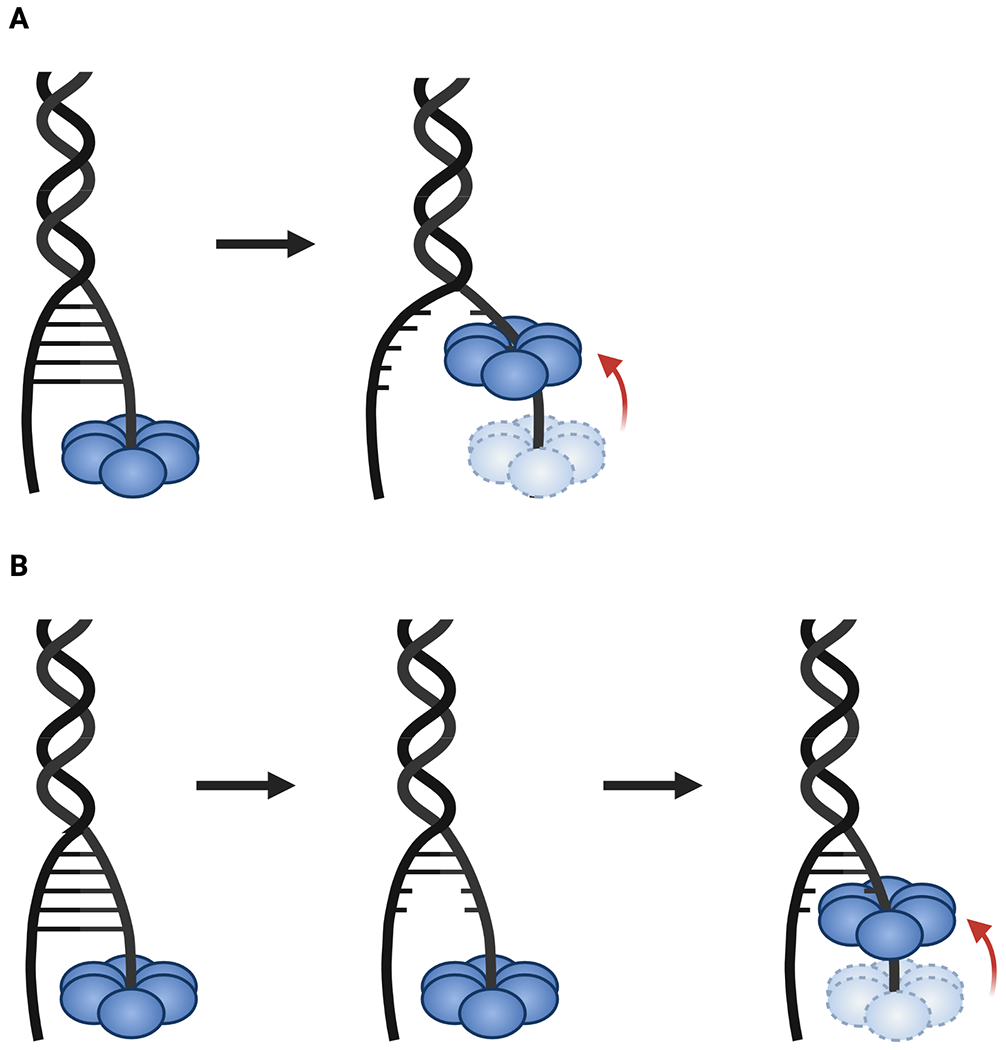
A. In the power stroke model, ATP hydrolysis by the helicase is tied to conformational changes that actively push it forward into a duplex, forcibly splitting paired bases to unwind the substrate. B. Passive unwinding involves local fraying of base pairs due to thermal or torsional fluctuations that expose ssDNA, which the helicase then captures through an ATP-dependent movement.
To date, studies have shown that DNA unwinding by SF-IV helicases is passive (Johnson et al., 2007; Lionnet et al., 2007; Manosas et al., 2010). Enzymes such as DnaB or gp4 often display dsDNA unwinding rates that are significantly slower than ssDNA translocation rates and that vary significantly with GC content (Jeong et al., 2004; Johnson et al., 2007; Kim et al., 2002, 1996; Ribeck et al., 2010). While these helicases tend to unwind DNA at a constant rate, they are prone to pausing, especially when encountering duplexes of increased GC content (Johnson et al., 2007; Lionnet et al., 2007; Ribeck et al., 2010; Schlierf et al., 2019; Syed et al., 2014). Interestingly, many SF-IV helicases show increased ATPase and unwinding activity when bound to a partner polymerase (Bird et al., 2000; Kim et al., 1996; Pandey and Patel, 2014; Stano et al., 2005). For example, the gp4 helicase shows a marked decrease in pausing and slipping when engaged to an actively synthesizing T7 DNA polymerase (even up to 50% GC content), indicating that enzymatic coupling between nucleotide addition (by the polymerase) and hydrolysis (by the helicase) increases unwinding efficiency (Pandey and Patel, 2014). The T7 DNA polymerase contains low level strand displacement capabilities, and it has been suggested that the positioning of the leading strand DNA polymerase at the fork with the helicase allows for synergistic unwinding at the forefront of the replisome in bacteria and phages (Gao et al., 2019; Pandey and Patel, 2014; Stano et al., 2005). Bacterial primase, known as DnaG, also stimulates the ATPase and unwinding activities of DnaB however, this effect is seen with the helicase binding domain of DnaG alone, suggesting that this stimulatory affect is allosteric, rather than coupled to RNA polymerization by the primase.
Although there has been debate about the step size of RecA-family hexameric helicases, most of the available data points to a small value that might be expected for a passive mechanism of unwinding. For example, the structure of Rho bound to ssRNA shows that the pore loops bind to the phosphodiester backbone in single nucleotide increments, suggesting a step size of one (Thomsen and Berger, 2009). With respect to phage SF-IV helicases, there have been variable reports for either one or two nucleotide steps based on single molecule (G40P) or structural studies (gp4), respectively (Gao et al., 2019; Schlierf et al., 2019). The available data indicate that in discussing step size by a hexameric helicases it is important to distinguish between physical and kinetic step sizes. Physical step size is the number of base pairs traveled in a conformational cycle of a given helicase, whereas the kinetic step size refers to the number of base pairs traveled between successive rate-limiting steps (Patel and Donmez, 2006). In considering RecA-family hexameric helicases, structural data indicate that a single enzyme would take either a 6-nucleotide (SF-V) or 12-nucleotide (SF-IV) step between the point where it releases and rebinds its nucleic acid substrate, while the helicase ring as a whole would advance by steps of only 1 or 2 nucleotides into the fork to promote unwinding. Interestingly, the pitch of DNA bound to DnaB and gp4 is similar to that seen for FtsK, suggesting that the core stepping mechanism may be conserved between SF-IV helicases and RecA-type dsDNA translocases (Gao et al., 2019; Itsathitphaisarn et al., 2012; Jean et al., 2020). Consistent with this idea, the pores of SF-IV hexamers are sufficiently large to accommodate a paired duplex and members of this helicase family are known to possess the ability to traverse dsDNA segments without melting the complementary paired strands (Itsathitphaisarn et al., 2012; Kaplan, 2000). It is only when SF-IV helicases are loaded onto the 5’ssDNA-end a fork bearing a sufficiently large unpaired 3’ tail that they unwind DNA by sterically excluding the non-translocated strand from the helicase pore (Kaplan, 2000). Interestingly, biochemical studies have indicated that the excluded strand can interact with the exterior surface of both DnaB and gp4 (Carney et al., 2017; Galletto et al., 2004; Hingorani and Patel, 1993); however, biochemical and structural studies of the T7 replisome have also indicated that the leading strand polymerase can engage the excluded strand right at the junction where ssDNA enters the helicase pore (Gao et al., 2019; Nandakumar et al., 2015), sequestering DNA in a manner that would prevent wrapping. It has been suggested that the wrapping of the excluded strand may serve as a regulatory mechanism to slow helicase advancement when the motor is uncoupled from the polymerase (Perera et al., 2019b). Additional work is needed to investigate this issue further.
A picture for the unwinding mechanism of AAA+ helicases derives from both structural and single-molecule studies. Structural efforts have suggested a subunit step size of 6 nucleotides for SF-III helicases and 12 nucleotides for SF-VI enzymes, with corresponding hexamer steps of 1 and 2 nucleotide nucleotides, respectively (Eickhoff et al., 2019; Enemark and Joshua-Tor, 2006; Meagher et al., 2019). As with their RecA counterparts, the small hexamer steps are consistent with a more passive mechanism of unwinding. Analyses of CMG have revealed non-uniform DNA unwinding rates, with periods of pausing and backwards translocation that result in DNA reannealing akin to a biased random walk (Burnham et al., 2019). Retrograde helicase movement was observed during long stretches of time and on extended DNAs, suggesting that the behavior is not due to unproductive ATP hydrolysis events. These results further indicate that hexameric AAA+ helicases unwind DNA in a relatively passive manner. Interestingly, cryo-EM structures have shown that dsDNA can enter far into the N-terminal region of CMG, and that loops in this region seem to divert the displaced strand to a possible exit tunnel between Mcm3 and Mcm5 (Yuan et al., 2020). As with SF-IV helicases, it has been suggested that the exterior surface of MCM helicases may associate with the excluded strand, an interaction that has been proposed to prevent backsliding (Costa et al., 2008; Graham et al., 2011, 2016, 2018; Rothenberg et al., 2007). However, structural studies of eukaryotic replisome components suggest that the Pol α–primase complex sits close to the N-terminal (fork-proximal) tier of CMG through a interaction with the Ctf4/AND-1 scaffolding protein (Georgescu et al., 2017; Simon et al., 2014; Sun et al., 2015b; Yuan et al., 2019; Zhu et al., 2007). This placement would position Pol α–primase close to the point where the excluded strand is ejected from CMG during DNA unwinding (a polymerase/helicase juxtaposition somewhat analogous to that seen for T7 gp4), thus preventing wrapping from occurring. Future work will be needed to resolve the fate of the displaced strand and the role of any interactions it might have with the MCM exterior.
Hexameric Helicase Loading
The ring-like structure of hexameric helicases creates a topological challenge for the binding of nucleic acid substrates. For replicative hexameric helicases (SF-III, -IV, and -VI), replication origins are embedded in the middle of circular or very long linear DNAs; for SF-V enzymes, target RNA segments are flanked by large ribonucleoprotein complexes, such as RNA polymerase and the ribosome. Both situations prevent threading of the helicase ring onto a free DNA or RNA end, and instead necessitate a discrete loading strategy. Interestingly, these strategies vary widely between and even within hexameric helicase superfamilies.
One means used by nature to grapple with helicase loading is to assemble the enzyme at its target locus from monomeric subunits (Fig. 7A). SF-III helicases such as LTag and E1 appear to follow this approach, although they can proceed through somewhat distinct assembly processes (Fouts et al., 1999; Mastrangelo et al., 1989). Dedicated origin binding domains on the N-termini of LTag and E1 initially form doubly dimeric or trimeric intermediates, respectively, which initially bind to double-stranded origins (Meinke et al., 2007; Schuck and Stenlund, 2005). In LTag, this “double dimer” then recruits 8 more protomers in an ATP-dependent fashion to form a head-to-head double hexamer (reviewed in (Hickman and Dyda, 2005)). How this complex transitions to encircling ssDNA and separates into two active hexameric helicases is not yet understood. The N-terminus of E1 also binds to double-stranded origins but higher-order oligomerization is initially blocked by a molecular chaperone – the E2 protein – which binds to the AAA+ domains of E1 (Abbate et al., 2004). ATP binding by E1 is thought to promote E2 dissociation and aid E1 oligomerization, forming a head-to-head “double trimer” intermediate and can hydrolyze ATP to locally untwist and melt the origin (Schuck and Stenlund, 2011). How this intermediate leads to further assembly of E1 double hexamers or active E1 hexamers is unknown, although it has been suggested that this transition may be regulated by a C-terminal tail present in the papillomavirus E1 enzyme (Schuck and Stenlund, 2015).
Fig. 7: Strategies for loading hexameric helicases.
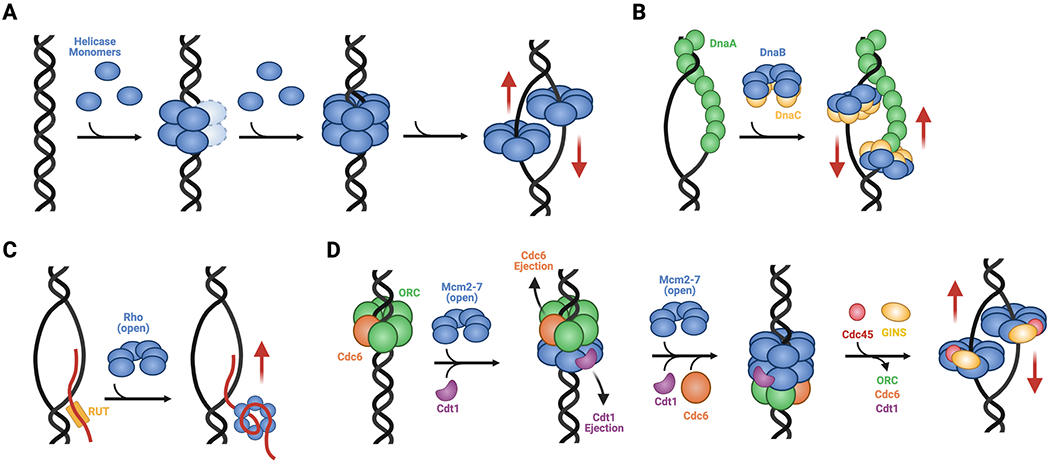
A. Ring assembly. SF-III helicases such as LTag and E1 are assembled at viral replication origins, proceeding through head-to-head intermediate complexes into single hexamers that encircle single DNA strands. B. Chaperoned rings opening. The SF-IV DnaB helicase from E. coli forms pre-assembled hexamers that are physically opened by DnaC and loaded onto a ssDNA bubble formed by the replication initiator, DnaA. C. Self-regulated ring closure. The SF-V helicase Rho forms a pre-opened hexamer that is recruited to target RNAs by Rho utilization (rut) sequences; once bound, the threading of ssRNA into the helicase channel promotes ring closure. D. Chaperoned ring closure. SF-VI helicases like MCMs form pre-opened hexameric rings that are loaded onto duplex replication origins by ORC, Cdc6, and Cdt1. This process establishes a closed-ring, head-to-head double hexamer intermediate that, when activated by Cdc45 and GINS to form the CMG complex, melts the origin and isomerizes into single hexamers that encircle ssDNA. Red arrows indicate direction of post-loading translocation. Helicase bypass following loading occurs for SF-III, -IV, and -VI enzymes.
The loading mechanisms of RecA family SF-IV hexameric helicases are highly varied. In contrast to SF-III enzymes, SF-IV helicases are loaded onto single-stranded nucleic acids. However, in at least one instance, as exemplified by B. subtilis DnaC (a DnaB-family helicase), loading has been reported to use a ring-assembly strategy (Velten et al., 2003). In this particular system, an ATP-dependent factor known as DnaI (an AAA+ ATPase) and two ATP-independent factors, DnaB (so-named for historical reasons and not to be confused with DnaB helicases) and DnaD, assist with the formation of helicase hexamers around DNA (Soultanas, 2002; Velten et al., 2003); the single-stranded substrate is generated in bacteria by a replication initiation protein known as DnaA (Bramhill and Kornberg, 1988). The mechanism of loading for B. subtilis DnaC contrasts with that of E. coli DnaB, which pre-assembles into closed-ring hexamers that require a factor known as DnaC (also an AAA+ ATPase and an ortholog of B. subtilis DnaI) for loading (Fig. 7B) (Kobori and Kornberg, 1982). Structural studies have shown that E. coli DnaC physically opens DnaB rings as a means to allow ssDNA to enter the helicase pore, and that once DNA binds, the loader-bound helicase isomerizes into a conformation similar to that seen in translocation-competent states of the enzyme (Arias-Palomo et al., 2013, 2019; Itsathitphaisarn et al., 2012). During the initiation of DNA replication, two DnaB6C6 complexes are loaded per origin, with the DnaB hexamers facing each other (Fang et al., 1999). In E. coli, the loaded DnaB molecules do not appear to form any specific interactions with each other (Fang et al., 1999), although a double-hexamer intermediate has been observed for one bacterial species of DnaB (Helicobacter pylori) in the absence of DNA (Stelter et al., 2012). For replication to proceed, the two DnaB helicases must translocate past each other, the mechanism of which is not well-understood. Interestingly, a majority of bacterial species lack DnaC/DnaI loader proteins, and instead use an ATP-independent chaperone called DnaC/I antecedent (DciA) (Brézellec et al., 2016). How DciA facilitates helicase loading has yet to be established. The SF-IV helicases of certain phages, such as T4 gp41 and SPP1 G40P, are thought utilize ATP-independent loading factors that may work by a ring-opening strategy (e.g., gp59 in T4 and G39P in SPP1) (Bailey et al., 2003; Mueser et al., 2000). However, at least one phage enzyme (T7 gp4 helicase-primase) is thought to be able to pre-assemble into a hexameric or heptameric oligomer that is capable of self-loading onto DNA, and it has been proposed that the ssDNA binding site on the primase domain facilitates ring opening (Ahnert, 2000). Some phages also use dedicated proteins to hijack and load host DnaB proteins onto viral replication origins; the P protein of phage lambda, which cracks open E. coli DnaB hexamers in a manner similar to that of DnaC but without a need for ATP is one such example (Chase et al., 2018). Moreover, at least one DnaB homolog (from H. pylori) has been shown to be capable of bypassing a DnaC-dependent loading mechanism in vivo, operating through an as yet unknown mechanism (Soni et al., 2003, 2005).
Two groups of helicases, SF-V and SF-VI, have been demonstrated to form self-regulated pre-opened hexameric rings prior to loading (Bochman and Schwacha, 2010; Costa et al., 2011; Lyubimov et al., 2012; Skordalakes and Berger, 2003; Yu et al., 2000). The promiscuous or inappropriate binding of substrate nucleic acid segments is controlled by different means in these systems. Open Rho rings can be recruited to and loaded on client RNAs either by the association of the helicase’s N-terminal RNA binding domains with cytosine-rich ‘rut’ (rho utilization) sequences (Fig. 7C), or by interacting with partner proteins such as RNA polymerase and NusG (Allison et al., 1998; Martinez et al., 1996a, 1996b; Said et al., 2021). By comparison, MCM hexamers rely on an elaborate loading mechanism that couples helicase deposition and activation with discrete cell cycle stages. In eukaryotes, a heterohexameric AAA+ complex known as the origin recognition complex (ORC), works in conjunction with Cdc6 (another AAA+ ATPase) and Cdt1 to recruit MCM rings to replication origins and align them around duplex DNA (Fig. 7D) (On et al., 2018; Parker et al., 2017). Like SF-III helicases, MCM rings are initially loaded as head-to-head double hexamers (Abid Ali et al., 2017; Noguchi et al., 2017); interestingly, the N-terminal domains that support inter-hexamer interactions are evolutionarily unrelated between SF-III and SF-VI helicases, indicating that the double-hexamer loading strategy arose independently during evolution (Fletcher et al., 2003). MCM loading occurs in the early G1 phase of the cell cycle, and the double hexamers persist until S phase, at which point they are activated by kinase-dependent phosphorylation and the binding of the Cdc45 and GINS accessory factors (Chang et al., 2015; Diffley et al., 1994; Labib, 2010; Remus and Diffley, 2009). These events promote DNA melting and isomerization of the MCM double hexamer into translocation competent single hexamers around ssDNA (Douglas et al., 2018). How the MCM transitions between double and single hexamer states and from encircling dsDNA to ssDNA is not yet understood at a structural level. Interestingly, the head-to-head nature of the loaded MCM DH indicates that matured CMG assemblies must bypass each other during the start of translocation and replication, similar to the action of their SF-III helicase counterparts and bacterial DnaB enzymes. The fact that all cellular replicative helicases, as well as those of some viruses, manifest this bypass behavior suggests that there has been some evolutionary pressure to adopt such a mechanism, such as ensuring the complete and efficient replication of origin regions (Fang et al., 1999).
Roadblock and Damage Bypass
During strand synthesis, replicative hexameric helicases must contend with roadblocks in the form of both chemical modifications and stalled or covalently attached proteins. Several hexameric helicases (e.g., DnaB, MCM) have been shown to be capable of actively translocating over small lesions, such as cyclobutene pyrimidine dimers and alkyl phosphotriesters, with varying effects on polymerase stalling (Byun et al., 2005; Suhasini et al., 2012; Sun et al., 2015a; Taylor and Yeeles, 2018). In T7 gp4 and CMG, this tolerance is diminished by bulky lesions such as interstrand cross-links or DNA-protein adducts, which have been shown to block helicase progression (Brown and Romano, 1989; Duxin et al., 2014; Fu et al., 2011). Interestingly, CMG has been shown to be able to bypass DNA-protein crosslinks, even when the lesions are present on the translocating strand (Sparks et al., 2019). This bypass activity is often aided by monomeric SF-I helicases and translocases that either help remove the block or promote CMG migration over the barrier (Huang et al., 2013; Schauer et al., 2020; Sparks et al., 2019; Yardimci et al., 2012). Surprisingly, SV40 LTag is able to actively navigate past large covalent protein-DNA blocks on the translocation strand without removing the adduct, suggesting that the ring briefly opens to continue unwinding past the block (Yardimci et al., 2012). How lesions affect hexameric helicase unwinding energetics and how auxiliary partners modulate the function of these enzymes to promote ring integrity and lesion bypass remains a frontier area of inquiry.
Concluding remarks
Although the two predominant types of hexameric helicases, RecA and AAA+, appear to have evolved independently of one another, members of these classes nonetheless display some organizational and functional similarities (Table 1). All hexameric helicases form asymmetric rings when bound to a nucleic acid substrate in which bipartite NTPase centers reside at subunit interfaces. Hexameric helicases also appear to use a sequential, rotary NTP hydrolysis mechanism to drive substrate translocation; however, the allosteric couplings that communicate nucleotide status between active sites and coordinate NTP turnover with DNA or RNA movement is poorly understood and likely differs between the two enzyme families. A deeper understanding of this allosteric relay is needed to help explain why some hexameric helicases possess sufficient plasticity to tolerate inactive subunits. It also remains to be determined whether NTPase activity is relatively monotonic during helicase operation, or whether it occurs in bursts, as has been described for the phi29 dsDNA translocase (Moffitt et al., 2009).
Table 1. Summary of mechanisms utilized by hexameric helicase superfamilies.
Distinct evolutionary lineages of hexameric helicases have evolved surprisingly similar mechanisms of NTP hydrolysis and duplex unwinding. Modes of helicase translocation exist on a spectrum from hand-over-hand, with a full separation of subunits, to coordinated escort with a closed, planar, ring. Ring-shaped helicases also employ a wide variety of loading strategies, although many rely on dedicated partner proteins.
| Superfamily | Hydrolysis | Translocation | Unwinding | Loading |
|---|---|---|---|---|
| III | Rotary | Coordinated escort | Passive | Monomer assembly on dsDNA |
| IV | Rotary | Hand-over-hand | Passive | Partner protein mediated loading on ssDNA |
| V | Rotary | Hybrid hand-over-hand and coordinated escort | Passive | Self- or partner protein-mediated loading on ssRNA |
| VI | Rotary | Hand-over-hand | Passive | Partner-protein mediated loading on dsDNA |
Interestingly, hexameric helicases thus far appear to operate by a relatively passive translocation mechanism. This stands in contrast to some monomeric SF-I and -II helicases, which have been suggested to be capable of exerting a true power stroke (Manosas et al., 2010). A common theme seen with in vitro measurements of hexameric helicase unwinding rates is that these speeds rarely meet the rates required for in vivo function (Burnham et al., 2019; Jeong et al., 2004; Kim et al., 1996). Additionally, many hexameric helicases are fast at translocating along single nucleic acid strands, but slow considerably when tasked with duplex unwinding (Jeong et al., 2004; Kim et al., 2002; Manosas et al., 2010). The binding of partner proteins has been observed to dramatically increase the ATPase and DNA unwinding rates of many different hexameric helicases, particularly those involved with replication (Bird et al., 2000; Kim et al., 1996; Pandey and Patel, 2014; Stano et al., 2005). The mechanisms responsible for these effects remain a fascinating area of study.
Many hexameric helicases also depend on specialized nucleic acid recruitment and loading approaches that help regulate access to nucleic acid substrates. Although these strategies vary, it is intriguing that many bacterial species and all eukaryotes use a homologous suite of AAA+ proteins – DnaA and DnaC/DnaI in bacteria, ORC and Cdc6 in eukaryotes – to coordinate the deposition of replicative hexameric helicases onto chromosomal origins. This group of proteins belongs to a phylogenetically linked clade of ‘initiator’ factors within the AAA+ superfamily, even though the DnaB and MCM helicases they act on are evolutionarily distinct. How replication initiation factors evolved their highly distinct helicase recruitment and loading mechanisms is not known, nor is it fully understood how, following loading, replicative helicases switch from a translocation-restricted state to a processive, DNA unwinding configuration. Future studies will be needed to more fully resolve these questions.
The realization that evolutionarily distinct lineages of helicases have converged on similar – or at least analogous – operational mechanisms is intriguing. Some of this congruence, such as the apparent ubiquity of a rotary NTP hydrolysis/translocation mechanism, may reflect some type of selective advantage, perhaps involving chemomechanical coupling efficiency and/or enzyme processivity. Similarly, the common reliance on partner proteins for controlling helicase loading, activation, and the bypass of roadblocks undoubtedly stems from a need for additional levels of helicase regulation in cells. Future work in these areas will be necessary to derive new fundamental insights into hexameric helicase function and evolution.
Acknowledgements
We thank all members of the Berger lab for their helpful discussion in the preparation of this review.
Declarations of Interest
The authors report no declarations of interest. This work was supported by the NIGMS (R37-GM071747 to JMB and T32-GM008403 to AJF) and NSF (DGE-1746891 to AJF).
References
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, and Bustamante C (2009). Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 461, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbate EA, Berger JM, and Botchan MR (2004). The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 18, 1981–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abid Ali F, Douglas ME, Locke J, Pye VE, Nans A, Diffley JFX, and Costa A (2017). Cryo-EM structure of a licensed DNA replication origin. Nat. Commun 8, 2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams JP, Leslie AGW, Lutter R, and Walker JE (1994). Structure at 2.8 Â resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628. [DOI] [PubMed] [Google Scholar]
- Adachi K, Yasuda R, Noji H, Itoh H, Harada Y, Yoshida M, and Kinosita K (2000). Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl. Acad. Sci. U. S. A 97, 7243–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman JL, Jeong Y-J, Liao J-C, Patel G, Kim D-E, Oster G, and Patel SS (2006). Mechanochemistry of Transcription Termination Factor Rho. Mol. Cell 22, 611–621. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, and Wittinghofer A (1997). Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol 4, 686–689. [DOI] [PubMed] [Google Scholar]
- Ahnert P (2000). A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 19, 3418–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison TJ, Wood TC, Briercheck DM, Rastinejad F, Richardson JP, and Rule GS (1998). Crystal structure of the RNA-binding domain from transcription termination factor rho. Nat. Struct. Biol 5, 352–356. [DOI] [PubMed] [Google Scholar]
- Arias-Palomo E, O’Shea VL, Hood IV, and Berger JM (2013). The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 153, 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Palomo E, Puri N, Murray VLO, Yan Q, and Berger JM (2019). Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA. Mol. Cell 74, 173–184.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Sedelnikova SE, Mesa P, Ayora S, Waltho JP, Ashcroft AE, Baron AJ, Alonso JC, and Rafferty JB (2003). Structural analysis of Bacillus subtilis SPP1 phage helicase loader protein G39P. J. Biol. Chem 278, 15304–15312. [DOI] [PubMed] [Google Scholar]
- Bird LE, Pan H, Soultanas P, and Wigley DB (2000). Mapping Protein-Protein Interactions within a Stable Complex of DNA Primase and DnaB Helicase from Bacillus stearothermophilus †. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, and Schwacha A (2008). The Mcm2-7 Complex Has In Vitro Helicase Activity. Mol. Cell 31, 287–293. [DOI] [PubMed] [Google Scholar]
- Bochman ML, and Schwacha A (2010). The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 ‘gate.’ Nucleic Acids Res. 38, 6078–6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Bell SP, and Schwacha A (2008). Subunit Organization of Mcm2-7 and the Unequal Role of Active Sites in ATP Hydrolysis and Viability. Mol. Cell. Biol 28, 5865–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D, and Kornberg A (1988). Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52, 743–755. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Dombroski AJ, and Platt T (1987). Transcription termination factor rho is an RNA-DNA helicase. Cell 48, 945–952. [DOI] [PubMed] [Google Scholar]
- Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, and Chen XS (2008). Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci 105, 20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brézellec P, Vallet-Gely I, Possoz C, Quevillon-Cheruel S, and Ferat JL (2016). DciA is an ancestral replicative helicase operator essential for bacterial replication initiation. Nat. Commun 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WC, and Romano LJ (1989). Benzo[a]pyrene-DNA adducts inhibit translocation by the gene 4 protein of bacteriophage T7. J. Biol. Chem 264, 6748–6754. [PubMed] [Google Scholar]
- Brunger AT, Milburn MV, Tong L, DeVos AM, Jancarik J, Yamaizumi Z, Nishimura S, Ohtsuka E, and Kim SH (1990). Crystal structure of an active form of RAS protein, a complex of a GTP analog and the HRAS p21 catalytic domain. Proc. Natl. Acad. Sci 87, 4849–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham DR, Kose HB, Hoyle RB, and Yardimci H (2019). The mechanism of DNA unwinding by the eukaryotic replicative helicase. Nat. Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee M, Walter JC, and Cimprich KA (2005). Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney SM, Gomathinayagam S, Leuba SH, and Trakselis MA (2017). Bacterial DnaB helicase interacts with the excluded strand to regulate unwinding. J. Biol. Chem 292, 19001–19012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Riera A, Evrin C, Sun J, Li H, Speck C, and Weinreich M (2015). Cdc6 ATPase activity disengages Cdc6 from the pre-replicative complex to promote DNA replication. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase J, Catalano A, Noble AJ, Eng ET, Olinares PD, Molloy K, Pakotiprapha D, Samuels M, Chait B, des Georges A, et al. (2018). Mechanisms of opening and closing of the bacterial replicative helicase. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, van Duinen G, Medagli B, Chong J, Sakakibara N, Kelman Z, Nair SK, Patwardhan A, and Onesti S (2008). Cryo-electron microscopy reveals a novel DNA-binding site on the MCM helicase. EMBO J. 27, 2250–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, and Berger JM (2011). The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol 18, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton DJ, Guo S, Johnson DE, and Richardson CC (2004). The arginine finger of bacteriophage T7 gene 4 helicase: Role in energy coupling. Proc. Natl. Acad. Sci 101, 4373–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton DJ, Mukherjee S, and Richardson CC (2006). DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell 21, 165–174. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, and Rowley A (1994). Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Dolan JW, Marshall NF, and Richardson JP (1990). Transcription termination factor rho has three distinct structural domains. J. Biol. Chem 265, 5747–5754. [PubMed] [Google Scholar]
- Dombroski AJ, and Platt T (1988). Structure of rho factor: an RNA-binding domain and a separate region with strong similarity to proven ATP-binding domains. Proc. Natl. Acad. Sci 85, 2538–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Ali FA, Costa A, and Diffley JFX (2018). The mechanism of eukaryotic CMG helicase activation. Nature 555, 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dewar JM, Yardimci H, and Walter JC (2014). Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159, 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff P, Kose HB, Martino F, Petojevic T, Abid Ali F, Locke J, Tamberg N, Nans A, Berger JM, Botchan MR, et al. (2019). Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome. Cell Rep. 28, 2673–2688.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye T-Z, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, and German’ J (1995). The Bloom’s Syndrome Gene Product Is Homologous to RecQ Helicases. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, and Joshua-Tor L (2006). Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275. [DOI] [PubMed] [Google Scholar]
- Fang L, Davey MJ, and O’Donnell M (1999). Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell 4, 541–553. [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, and Chen XS (2003). The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Mol. Biol 10, 160–167. [DOI] [PubMed] [Google Scholar]
- Fouts ET, Yu X, Egelman EH, and Botchan MR (1999). Biochemical and Electron Microscopic Image Analysis of the Hexameric E1 Helicase. J. Biol. Chem 274, 4447–4458. [DOI] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Schärer OD, and Walter JC (2011). Selective Bypass of a Lagging Strand Roadblock by the Eukaryotic Replicative DNA Helicase. Cell 146, 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai D, Zhao R, Li D, Finkielstein CV, and Chen XS (2004). Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60. [DOI] [PubMed] [Google Scholar]
- Galletto R, Jezewska MJ, and Bujalowski W (2004). Unzipping Mechanism of the Double-stranded DNA Unwinding by a Hexameric Helicase: The Effect of the 3′ Arm and the Stability of the dsDNA on the Unwinding Activity of the Escherichia coli DnaB Helicase. J. Mol. Biol 343, 101–114. [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, and Labib K (2006). GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol 8, 358–366. [DOI] [PubMed] [Google Scholar]
- Gao Y, and Yang W (2020). Different mechanisms for translocation by monomeric and hexameric helicases. Curr. Opin. Struct. Biol 61, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Cui Y, Fox T, Lin S, Wang H, De Val N, Zhou ZH, and Yang W (2019). Structures and operating principles of the replisome. Science (80-. ). 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, Yurieva O, Li H, and O’Donnell ME (2017). Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci 114, E697–E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz GS, Dean FB, Hurwitz J, and Matson SW (1988). The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem 263, 383–392. [PubMed] [Google Scholar]
- Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, and Spelbrink JN (2009). Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet 18, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, and Koonin EV (1993). Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol 3, 419–429. [Google Scholar]
- Graham BW, Schauer GD, Leuba SH, and Trakselis MA (2011). Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res. 39, 6585–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BW, Tao Y, Dodge KL, Thaxton CT, Olaso D, Young NL, Marshall AG, and Trakselis MA (2016). DNA Interactions Probed by Hydrogen-Deuterium Exchange (HDX) Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Confirm External Binding Sites on the Minichromosomal Maintenance (MCM) Helicase. J. Biol. Chem 291, 12467–12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BW, Bougoulias ME, Dodge KL, Thaxton CT, Olaso D, Tao Y, Young NL, Marshall AG, and Trakselis MA (2018). Control of Hexamerization, Assembly, and Excluded Strand Specificity for the Sulfolobus solfataricus MCM Helicase. Biochemistry 57, 5672–5682. [DOI] [PubMed] [Google Scholar]
- Guenther B, Onrust R, Sali A, O’Donnell M, and Kuriyan J (1997). Crystal Structure of the δ′ Subunit of the Clamp-Loader Complex of E. coli DNA Polymerase III. Cell 91, 335–345. [DOI] [PubMed] [Google Scholar]
- Guo M, Hundseth K, Ding H, Vidhyasagar V, Inoue A, Nguyen CH, Zain R, Lee JS, and Wu Y (2015). A distinct triplex DNA unwinding activity of ChlR1 helicase. J. Biol. Chem 290, 5174–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, and Whiteheart SW (2005). AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol 6, 519–529. [DOI] [PubMed] [Google Scholar]
- Hickman AB, and Dyda F (2005). Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol 15, 77–85. [DOI] [PubMed] [Google Scholar]
- Hingorani MM, and Patel SS (1993). Interactions of Bacteriophage T7 DNA Primase/Helicase Protein with Single-Stranded and Double-Stranded DNAs. Biochemistry 32, 12478–12487. [DOI] [PubMed] [Google Scholar]
- Von Hippel PH, and Delagoutte E (2001). A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell 104, 177–190. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, and Seidman MM (2013). The DNA Translocase FANCM/MHF Promotes Replication Traverse of DNA Interstrand Crosslinks. Mol. Cell 52, 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, and Botchan MR (2010). Activation of the MCM2-7 Helicase by Association with Cdc45 and GINS Proteins. Mol. Cell 37, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, and Steitz TA (2012). The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, and Ziebuhr J (2004). Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol 78, 5619–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, and Aravind L (2004). Evolutionary history and higher order classification of AAA+ ATPases. In Journal of Structural Biology, (Academic Press; ), pp. 11–31. [DOI] [PubMed] [Google Scholar]
- Jain A, Bacolla A, Chakraborty P, Grosse F, and Vasquez KM (2010). Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry 49, 6992–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean NL, Rutherford TJ, and Löwe J (2020). FtsK in motion reveals its mechanism for double-stranded DNA translocation. Proc. Natl. Acad. Sci. U. S. A 117, 14202–14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Levin MK, and Patel SS (2004). The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl. Acad. Sci. U. S. A 101, 7264–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Bai L, Smith BY, Patel SS, and Wang MD (2007). Single-Molecule Studies Reveal Dynamics of DNA Unwinding by the Ring-Shaped T7 Helicase. Cell 129, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DL (2000). The 3’-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol 301, 285–299. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, and Labib K (1998). MCM proteins: evolution, properties, and role in DNA replication. Biochim. Biophys. Acta - Gene Struct. Expr 1398, 113–136. [DOI] [PubMed] [Google Scholar]
- Kim DE, Narayan M, and Patel SS (2002). T7 DNA helicase: A molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol 321, 807–819. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, Mchenry CS, and Marians KJ (1996). Coupling of a Replicative Polymerase and Helicase: A-DnaB Interaction Mediates Rapid Replication Fork Movement template simultaneously unwinding the duplex DNA ahead of the fork and manufacturing the primers for. [DOI] [PubMed] [Google Scholar]
- Kobori JA, and Kornberg A (1982). The Escherichia coli dnaC Gene Product 111. PROPERTIES OF THE dnaB-dnaC PROTEIN COMPLEX*. [PubMed] [Google Scholar]
- Labib K (2010). How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24, 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, and Diffley JF. (2001). Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev 11, 64–70. [DOI] [PubMed] [Google Scholar]
- Langston LD, and O’Donnell ME (2019). An explanation for origin unwinding in eukaryotes. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBowitz JH, and McMacken R (1986). The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem 261, 4738–4748. [PubMed] [Google Scholar]
- Lee SJ, Syed S, Enemark EJ, Schuck S, Stenlund A, Ha T, and Joshua-Tor L (2014). Dynamic look at DNA unwinding by a replicative helicase. Proc. Natl. Acad. Sci. U. S. A 111, E827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR (2001). The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 15, 15–23. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, and Aravind L (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol 317, 41–72. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, and Aravind L (2003). Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol 333, 781–815. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, and Aravind L (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: Multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol 343, 1–28. [DOI] [PubMed] [Google Scholar]
- Lenzen CU, Steinmann D, Whiteheart SW, and Weis WI (1998). Crystal Structure of the Hexamerization Domain of N-ethylmaleimide–Sensitive Fusion Protein. Cell 94, 525–536. [DOI] [PubMed] [Google Scholar]
- Levitus M, Waisfisz Q, Godthelp BC, Vries Y. de, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, et al. (2005). The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet 37, 934–935. [DOI] [PubMed] [Google Scholar]
- Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, et al. (2005). The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet 37, 931–933. [DOI] [PubMed] [Google Scholar]
- Liao JC, Jeong YJ, Kim DE, Patel SS, Oster G, and Kim YJ (2005). Mechanochemistry of T7 DNA helicase. J. Mol. Biol 350, 452–475. [DOI] [PubMed] [Google Scholar]
- Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, and Croquette V (2007). Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. U. S. A 104, 19790–19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, and Cantor SB (2005). BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8, 255–265. [DOI] [PubMed] [Google Scholar]
- Lohman TM, and Bjornson KP (1996). Mechanisms of helicase-catalyzed DNA unwinding. [DOI] [PubMed] [Google Scholar]
- Longo LM, Jabłońska J, Vyas P, Kanade M, Kolodny R, Ben-Tal N, and Tawfik DS (2020). On the emergence of P-Loop NTPase and Rossmann enzymes from a Beta-Alpha-Beta ancestral fragment. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AY, Strycharska M, and Berger JM (2011). The nuts and bolts of ring-translocase structure and mechanism. Curr. Opin. Struct. Biol 21, 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AY, Costa A, Bleichert F, Botchan MR, and Berger JM (2012). ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc. Natl. Acad. Sci 109, 11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Xi XG, Bensimon D, and Croquette V (2010). Active and passive mechanisms of helicases. Nucleic Acids Res. 38, 5518–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, and Sauer RT (2005). Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature 437, 1115–1120. [DOI] [PubMed] [Google Scholar]
- Martinez A, Opperman T, and Richardson JP (1996a). Mutational analysis and secondary structure model of the RNP1-like sequence motif of transcription termination factor Rho. J. Mol. Biol 257, 895–908. [DOI] [PubMed] [Google Scholar]
- Martinez A, Burns CM, and Richardson JP (1996b). Residues in the RNP1-like sequence motif of Rho protein are involved in RNA-binding affinity and discrimination. J. Mol. Biol 257, 909–918. [DOI] [PubMed] [Google Scholar]
- Mastrangelo IA, Hough PVC, Wall JS, Dodson M, Dean FB, and Hurwrtz J (1989). ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338, 658–662. [DOI] [PubMed] [Google Scholar]
- Matson SW, Tabor S, and Richardson CC (1983). The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem 258, 14017–14024. [PubMed] [Google Scholar]
- McGeoch AT, Trakselis MA, Laskey RA, and Bell SD (2005). Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol 12, 756–762. [DOI] [PubMed] [Google Scholar]
- Meagher M, Epling LB, and Enemark EJ (2019). DNA translocation mechanism of the MCM complex and implications for replication initiation. Nat. Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke G, Phelan P, Moine S, Bochkareva E, Bochkarev A, Bullock PA, and Bohm A (2007). The Crystal Structure of the SV40 T-Antigen Origin Binding Domain in Complex with DNA. PLoS Biol. 5, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza O, Bourdoncle A, Boulé JB, Brosh RM, and Mergny JL (2016). G-quadruplexes and helicases. Nucleic Acids Res. 44, 1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, and Bustamante C (2009). Intersubunit coordination in a homomeric ring ATPase. Nature 457, 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, and Botchan MR (2006). Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U. S. A 103, 10236–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueser TC, Jones CE, Nossal NG, and Hyde CC (2000). Bacteriophage T4 gene 59 helicase assembly protein binds replication fork DNA. The 1.45 Å resolution crystal structure reveals a novel α-helical two-domain fold. J. Mol. Biol 296, 597–612. [DOI] [PubMed] [Google Scholar]
- Nakayama N, Arai N, Kaziro Y, and Arai K (1984). Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J. Biol. Chem 259, 88–96. [PubMed] [Google Scholar]
- Nandakumar D, Pandey M, and Patel SS (2015). Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi Y, Yuan Z, Bai L, Schneider S, Zhao G, Stillman B, Speck C, and Li H (2017). Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc. Natl. Acad. Sci 114, E9529–E9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noji H, Yasuda R, Yoshida M, and Kinosita K (1997). Direct observation of the rotation of F1-ATPase. Nature 386, 299–302. [DOI] [PubMed] [Google Scholar]
- Notarnicola SM, and Richardson CC (1993). The nucleotide binding site of the helicase/primase of bacteriophage T7. Interaction of mutant and wild-type proteins. J. Biol. Chem 268, 27198–27207. [PubMed] [Google Scholar]
- On KF, Jaremko M, Stillman B, and Joshua-Tor L (2018). A structural view of the initiators for chromosome replication. Curr. Opin. Struct. Biol 53, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, and Wittinghofer A (1990). Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9, 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M, and Patel SS (2014). Helicase and Polymerase Move Together Close to the Fork Junction and Copy DNA in One-Nucleotide Steps. Cell Rep. 6, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Botchan MR, and Berger JM (2017). Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit. Rev. Biochem. Mol. Biol 52, 107–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, and Donmez I (2006). Mechanisms of helicases. J. Biol. Chem 281, 18265–18268. [DOI] [PubMed] [Google Scholar]
- Patel SS, Hingorani MM, and Ng WM (1994). The K318A Mutant of Bacteriophage T7 DNA Primase-Helicase Protein Is Deficient in Helicase But Not Primase Activity and Inhibits Primase-Helicase Protein Wild-Type Activities by Heterooligomer Formation. Biochemistry 33, 7857–7868. [DOI] [PubMed] [Google Scholar]
- Perera HM, Behrmann MS, Hoang JM, Griffin WC, and Trakselis MA (2019a). Contacts and context that regulate DNA helicase unwinding and replisome progression. pp. 183–223. [DOI] [PubMed] [Google Scholar]
- Perera HM, Behrmann MS, Hoang JM, Griffin WC, and Trakselis MA (2019b). Contacts and context that regulate DNA helicase unwinding and replisome progression. pp. 183–223. [DOI] [PubMed] [Google Scholar]
- Petojevic T, Pesavento JJ, Costa A, Liang J, Wang Z, Berger JM, and Botchan MR (2015). Cdc45 (cell division cycle protein 45) guards the gate of the eukaryote replisome helicase stabilizing leading strand engagement. Proc. Natl. Acad. Sci. U. S. A 112, E249–E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchades C, Sandate CR, and Lander GC (2020). The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol. Cell Biol 21, 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, and Diffley JF (2009). Eukaryotic DNA replication control: Lock and load, then fire. Curr. Opin. Cell Biol 21, 771–777. [DOI] [PubMed] [Google Scholar]
- Ribeck N, Kaplan DL, Bruck I, and Saleh OA (2010). DnaB helicase activity is modulated by DNA geometry and force. Biophys. J 99, 2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP (2002). Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta - Gene Struct. Expr 1577, 251–260. [DOI] [PubMed] [Google Scholar]
- Rothenberg E, Trakselis MA, Bell SD, and Ha T (2007). MCM Forked Substrate Specificity Involves Dynamic Interaction with the 5′-Tail. J. Biol. Chem 282, 34229–34234. [DOI] [PubMed] [Google Scholar]
- Said N, Hilal T, Sunday ND, Khatri A, Bürger J, Mielke T, Belogurov GA, Loll B, Sen R, Artsimovitch I, et al. (2021). Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. Science (80-. ). 371, eabd1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer GD, Spenkelink LM, Lewis JS, Yurieva O, Mueller SH, van Oijen AM, and O’Donnell ME (2020). Replisome bypass of a protein-based R-loop block by Pif1. Proc. Natl. Acad. Sci 117, 30354–30361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Lautwein A, Kabsch W, Reza Ahmadian M, and Wittinghofer A (1996). Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature 384, 591–596. [DOI] [PubMed] [Google Scholar]
- Schlierf M, Wang G, Chen XS, and Ha T (2019). Hexameric helicase G40P unwinds DNA in single base pair steps. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, and Stenlund A (2005). Assembly of a double hexameric helicase. Mol. Cell 20, 377–389. [DOI] [PubMed] [Google Scholar]
- Schuck S, and Stenlund A (2011). Mechanistic Analysis of Local Ori Melting and Helicase Assembly by the Papillomavirus E1 Protein. Mol. Cell 43, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, and Stenlund A (2015). A Conserved Regulatory Module at the C Terminus of the Papillomavirus E1 Helicase Domain Controls E1 Helicase Assembly. J. Virol 89, 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, and Bell SP (2001). Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell 8, 1093–1104. [DOI] [PubMed] [Google Scholar]
- Schwartz C, De Donatis GM, Zhang H, Fang H, and Guo P (2013). Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology 443, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YS, Muller F, Lusky M, and Hurwitz J (1993). Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci 90, 702–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinković D, et al. (2014). A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, and Wigley DB (2000). Crystal structure of T7 gene 4 ring indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, and Wigley DB (2007). Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem 76, 23–50. [DOI] [PubMed] [Google Scholar]
- Skordalakes E, and Berger JM (2003). Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell 114, 135–146. [DOI] [PubMed] [Google Scholar]
- Soni RK, Mehra P, Choudhury NR, Mukhopadhyay G, and Dhar SK (2003). Functional characterization of Helicobacter pylori DnaB helicase. Nucleic Acids Res. 31, 6828–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni RK, Mehra P, Mukhopadhyay G, and Kumar Dhar S (2005). Helicobacter pylori DnaB helicase can bypass Escherichia coli DnaC function in vivo. Biochem. J 389, 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P (2002). A functional interaction between the putative primosomal protein Dnal and the main replicative DNA helicase DnaB in Baccillus. Nucleic Acids Res. 30, 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, Chistol G, Gao AO, Räschle M, Larsen NB, Mann M, Duxin JP, and Walter JC (2019). The CMG Helicase Bypasses DNA-Protein Cross-Links to Facilitate Their Repair. Cell 176, 167–181.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, and Patel SS (2005). DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 435, 370–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter M, Gutsche I, Kapp U, Bazin A, Bajic G, Goret G, Jamin M, Timmins J, and Terradot L (2012). Architecture of a Dodecameric Bacterial Replicative Helicase. Structure 20, 554–564. [DOI] [PubMed] [Google Scholar]
- Story RM, and Steitz TA (1992). Structure of the reca protein-adp complex. Nature 355, 374–376. [DOI] [PubMed] [Google Scholar]
- Story RM, Weber IT, and Steitz TA (1992). The structure of the e. Coli recA protein monomer and polymer. Nature 355, 318–325. [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Bird LE, Brannigan JA, and Wigley DB (1996). Crystal structure of a DExx box DNA helicase. Nature 384, 379–383. [DOI] [PubMed] [Google Scholar]
- Suhasini AN, Sommers JA, Yu S, Wu Y, Xu T, Kelman Z, Kaplan DL, and Brosh RM (2012). DNA repair and replication fork helicases are differentially affected by alkyl phosphotriester lesion. J. Biol. Chem 287, 19188–19198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Pandey M, Inman JT, Yang Y, Kashlev M, Patel SS, and Wang MD (2015a). T7 replisome directly overcomes DNA damage. Nat. Commun 6, 10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, and O’Donnell ME (2015b). The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol 22, 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed S, Pandey M, Patel SS, and Ha T (2014). Single-Molecule Fluorescence Reveals the Unwinding Stepping Mechanism of Replicative Helicase. Cell Rep. 6, 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MRG, and Yeeles JTP (2018). The Initial Response of a Eukaryotic Replisome to DNA Damage. Mol. Cell 70, 1067–1080.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, and Berger JM (2008). Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases: MicroReview. Mol. Microbiol 69, 1071–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, and Berger JM (2009). Running in Reverse: The Structural Basis for Translocation Polarity in Hexameric Helicases. Cell 139, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, Lawson MR, Witkowsky LB, Qu S, and Berger JM (2016). Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor. Proc. Natl. Acad. Sci. U. S. A 113, E7691–E7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaneva IR, Müller B, and West SC (1993). RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc. Natl. Acad. Sci. U. S. A 90, 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, and Polard P (2003). A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell 11, 1009–1020. [DOI] [PubMed] [Google Scholar]
- Venkatesan M, Silver LL, and Nossal NG (1982). Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J. Biol. Chem 257, 12426–12434. [PubMed] [Google Scholar]
- Walker JE (2013). The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans 41, 1–16. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, and Gay NJ (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J (2004). Nucleotide-dependent domain motions within rings of the RecA/AAA+ superfamily. J. Struct. Biol 148, 259–267. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, and Chung CH (2001). Nucleotide-dependent conformational changes in a protease-associated ATPase HslU. Structure 9, 1107–1116. [DOI] [PubMed] [Google Scholar]
- Weber CA, Salazar EP, Stewart SA, and Thompson LH (1990). ERCC2: cDNAs cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3. EMBO J. 9, 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G, van Ham RCA, Vermeulen W, Bootsma D, van der Eb AJ, and Hoeijmakers JHJ (1990). A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne’s syndrome. Cell 62, 777–791. [DOI] [PubMed] [Google Scholar]
- Yardimci H, Wang X, Loveland AB, Tappin I, Rudner DZ, Hurwitz J, van Oijen AM, and Walter JC (2012). Bypass of a protein barrier by a replicative DNA helicase. Nature 492, 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. (1996). Positional cloning of the Werner’s syndrome gene. Science (80-. ). 272, 258–262. [DOI] [PubMed] [Google Scholar]
- Yu X, Horiguchi T, Shigesada K, and Egelman EH (2000). Three-dimensional reconstruction of transcription termination factor rho: Orientation of the N-terminal domain and visualization of an RNA-binding site. J. Mol. Biol 299, 1279–1287. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Bai L, Sun J, Georgescu R, Liu J, O’Donnell ME, and Li H (2016). Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol 23, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Georgescu R, Santos R de LA, Zhang D, Bai L, Yao NY, Zhao G, O’Donnell ME, and Li H (2019). Ctf4 organizes sister replisomes and Pol α into a replication factory. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Georgescu R, Bai L, Zhang D, Li H, and O’Donnell ME (2020). DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat. Commun 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, and Dutta A (2007). Mcm10 and And-1/CTF4 recruit DNA polymerase to chromatin for initiation of DNA replication. Genes & Dev. 21, 2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]


